Abstract
Plastid and mitochondrial RNAs in vascular plants are subjected to cytidine‐to‐uridine editing. The model plant species Arabidopsis thaliana (Arabidopsis) has two nuclear‐encoded plastid‐targeted organelle RNA recognition motif (ORRM) proteins: ORRM1 and ORRM6. In the orrm1 mutant, 21 plastid RNA editing sites were affected but none are essential to photosynthesis. In the orrm6 mutants, two plastid RNA editing sites were affected: psbF‐C77 and accD‐C794. Because psbF encodes the β subunit of cytochrome b 559 in photosystem II, which is essential to photosynthesis, the orrm6 mutants were much smaller than the wild type. In addition, the orrm6 mutants had pale green leaves and reduced photosynthetic efficiency. To investigate the functional relationship between ORRM1 and ORRM6, we generated orrm1 orrm6 double homozygous mutants. Morphological and physiological analyses showed that the orrm1 orrm6 double mutants had a smaller plant size, reduced chlorophyll contents, and decreased photosynthetic efficiency, similar to the orrm6 single mutants. Although the orrm1 orrm6 double mutants adopted the phenotype of the orrm6 single mutants, the total number of plastid RNA editing sites affected in the orrm1 orrm6 double mutants was the sum of the sites affected in the orrm1 and orrm6 single mutants. These data suggest that ORRM1 and ORRM6 are in charge of distinct sets of plastid RNA editing sites and that simultaneous mutations in ORRM1 and ORRM6 genes do not cause additional reduction in editing extent at other plastid RNA editing sites.
Keywords: multiple organellar RNA editing factors, organelle RNA recognition motif proteins, organelle zinc finger proteins, pentatricopeptide repeat proteins, plastid RNA editing, RNA editing interacting proteins
1. INTRODUCTION
RNA editing is a post‐transcriptional process through which discrete changes are introduced to RNA sequences. In plants, RNA editing is restricted to plastids and mitochondria and is viewed as a correction mechanism to compensate for mutations in the haploid organelle genomes (Lu, 2018; Shi, Hanson, & Bentolila, 2017a; Sun, Bentolila, & Hanson, 2016; Takenaka, Zehrmann, Verbitskiy, Hartel, & Brennicke, 2013). A common type of plant organelle (plastid and mitochondrion) RNA editing is cytidine‐to‐uridine (C‐to‐U) deamination. Different plant species may have different plastid and mitochondrial C‐to‐U RNA editing sites (Bentolila, Oh, Hanson, & Bukowski, 2013; Chateigner‐Boutin & Small, 2007; Ruwe, Castandet, Schmitz‐Linneweber, & Stern, 2013). In mammalian species, C‐to‐U RNA editing is catalyzed by the APOBEC‐type cytidine deaminase (Xu & Messing, 2006). However, in plant plastids and mitochondria, C‐to‐U RNA editing is carried out by the RNA editing complex, which contains at least four types of editing factors: pentatricopeptide repeat (PPR) and PPR‐related proteins, RNA editing interacting proteins/multiple organellar RNA editing factors (RIPs/MORFs), organelle zinc finger (OZ) proteins, and organelle RNA recognition motif (ORRM) proteins (Lu, 2018; Shi, Hanson, et al., 2017; Sun et al., 2016; Takenaka et al., 2013).
PPR proteins are ubiquitously present in eukaryotes (Barkan & Small, 2014; Fujii & Small, 2011). Although most eukaryotes contain less than a dozen PPR proteins, land plants contain > 400 PPR proteins and all of them are predicted or have been shown to be localized to the plastids or mitochondria (Lurin et al., 2004). PPR‐E‐DYW‐type PPRs, which contain a PPR domain, an E (extension) domain, and a DYW domain, and PPR‐E‐type PPRs, which contain a PPR domain and an E domain, were found to be involved in C‐to‐U RNA editing in land plant plastids and mitochondria (Chateigner‐Boutin et al., 2013; Lu, 2018; Okuda et al., 2009; Okuda, Myouga, Motohashi, Shinozaki, & Shikanai, 2007; Sun et al., 2016). DYW1, a PPR‐related protein, was also found to participate in this process (Boussardon et al., 2012, 2014). The PPR domain has the ability to bind to RNAs in a sequence‐specific manner (Okuda, Nakamura, Sugita, Shimizu, & Shikanai, 2006; Okuda & Shikanai, 2012; Schallenberg‐Rüdinger, Kindgren, Zehrmann, Small, & Knoop, 2013; Tasaki, Hattori, & Sugita, 2010; Williams‐Carrier, Kroeger, & Barkan, 2008). The DYW domain has a [HXE(X)nCXXC] motif, which is homologous to the signature zinc finger motif in classic cytidine deaminases (Boussardon et al., 2014; Faivre‐Nitschke, Grienenberger, & Gualberto, 1999). Therefore, PPR‐ and PPR‐related proteins are prime candidates for C‐to‐U deamination in land plant organelles (Lu, 2018). In line with the sequence‐specific binding between a PPR protein and the corresponding RNA target, PPR proteins are rarely found to interact with each other. However, a number of PPR proteins have been found to interact with other RNA editing factors, including RIPs/MORFs (Bentolila et al., 2012; Takenaka et al., 2012; Wagoner, Sun, Lin, & Hanson, 2015; Zhang et al., 2014), ORRM1 (Sun et al., 2013), and OZ1 (Sun et al., 2015).
RIP/MORF proteins contain conserved RIP/MORF domains and are only present in the plastids and mitochondria of flowering plants (Lu, 2018; Takenaka et al., 2013). The Arabidopsis thaliana (Arabidopsis) nuclear genome encodes nine functional RIPs/MORFs (Bentolila et al., 2012; Takenaka et al., 2012). Among them, RIP2/MORF2 and RIP9/MORF9 are plastid‐targeted, RIP1/MORF8 is dually targeted to plastids and mitochondria, and the other six are mitochondrion‐targeted (Bentolila et al., 2012; Takenaka et al., 2012). RIPs/MORFs do not contain any RNA‐binding domains; however, RIPs/MORFs may form homodimers or heterodimers (Takenaka et al., 2012; Zehrmann et al., 2015). Furthermore, RIPs/MORFs were found to interact with other types of RNA editing factors, including PPRs (Bentolila et al., 2012; Takenaka et al., 2012; Zhang et al., 2014), OZ1 (Sun et al., 2015), and ORRMs (Hackett et al., 2017; Shi, Germain, Hanson, & Bentolila, 2016b; Shi, Hanson, & Bentolila, 2015). Therefore, it was proposed that RIPs/MORFs act as a scaffold, bridging different components of the RNA editing complex together (Lu, 2018).
OZ proteins contain Ran‐binding‐protein2 (RanBP2, CXXCX10CXXC)‐type zinc finger domains and are found in many land plants (Sun et al., 2015). RanBP2‐type zinc fingers are capable of binding to RNAs in a sequence‐specific manner (Nguyen et al., 2011). The Arabidopsis nuclear genome encodes four OZ proteins: OZ1, OZ2, OZ3, and OZ4, which contain two, two, three, and four RanBP2‐type zinc fingers, respectively (Sun et al., 2015). OZ3 is predicted to be targeted to the mitochondrion; OZ1, OZ2, and OZ4 are predicted to be plastid‐targeted. The plastid localization of OZ1 has been experimentally confirmed (Sun et al., 2015). OZ1 has been found to interact with itself and other plastid RNA editing factors, such as RIP1/MORF8, ORRM1, and ORRM6 (Hackett et al., 2017; Sun et al., 2015).
ORRM proteins are a subfamily of organelle‐localized RNA recognition motif (RRM) proteins (Lu, 2018; Shi, Hanson, et al., 2017). RRM proteins are capable of binding to RNAs and are present in viruses, bacteria, and eukaryotes (Maris, Dominguez, & Allain, 2005). The Arabidopsis nuclear genome encodes six ORRM proteins: ORRM1 and ORRM6 are plastid‐targeted; ORRM2, ORRM3, ORRM4, and ORRM5 are mitochondrion‐targeted (Lu, 2018; Shi, Hanson, et al., 2017). ORRM1 contains an ORRM domain at the C‐terminus and two truncated RIP/MORF domains at the N‐terminus; ORRM2 and ORRM6 contain an ORRM domain at the C‐terminus; ORRM3, ORRM4, and ORRM5 contain an N‐terminal ORRM domain and a C‐terminal glycine‐rich domain (Hackett et al., 2017; Shi, Bentolila, & Hanson, 2016a; Shi, Castandet, Germain, Hanson, & Bentolila, 2017b; Shi, Germain, et al., 2016; Shi et al., 2015; Sun et al., 2013).
Recombinant ORRM1 protein was found to bind preferentially to ORRM1‐dependent RNA editing sites in vitro, via the ORRM domain (Sun et al., 2013). The duplicated RIP/MORF moiety of ORRM1 was found to be required for the interaction between ORRM1 and selective plastid‐targeted PPR proteins, such as CHLORORESPIRATORY REDUCTION28 (CRR28) and ORGANELLE TRANSCRIPT PROCESSING82 (OTP82) (Sun et al., 2013). ORRM1 was also found to interact with other plastid‐targeted editing factors, including RIP1/MORF8, RIP2/MORF2, and OZ1 (Sun et al., 2015). The loss‐of‐function orrm1‐1 Arabidopsis mutant displayed near‐complete loss of editing at 12 plastid RNA editing sites and substantial reduction in editing extent at nine plastid RNA editing sites (Sun et al., 2013). Although a large number of plastid RNA editing sites were affected, the orrm1‐1 mutant did not display any phenotypic defect under standard growth conditions (Sun et al., 2013).
Unlike the orrm1‐1 mutant, the loss‐of‐function orrm6‐1 and orrm6‐2 Arabidopsis mutants showed near‐complete loss of editing at psbF‐C77 and substantial reduction in editing extent at accD‐C794 (Hackett & Lu, 2017; Hackett et al., 2017). psbF encodes the β subunit of cytochrome b 559, an essential component of photosystem II (PSII). Consequently, the orrm6‐1 and orrm6‐2 mutants displayed reduced PSII photochemical efficiency, small and pale green leaves, and stunted growth (Hackett & Lu, 2017; Hackett et al., 2017). Consistent with the plastid RNA editing pattern in the orrm6‐1 and orrm6‐2 mutants, recombinant ORRM6 protein was found to bind preferentially to synthetic RNAs flanking 40 nucleotides upstream and 19 nucleotides downstream of psbF‐C77 and accD‐C794 (Hackett et al., 2017). Furthermore, ORRM6 was found to interact with itself and other plastid RNA editing factors, including RIP1/MORF8, RIP2/MORF2, RIP9/MORF9, and OZ1 (Hackett et al., 2017).
As mentioned above, the orrm1‐1 single mutant showed substantial reduction in editing extent at nine plastid RNA editing sites (Sun et al., 2013). This begs the question whether other plastid‐targeted ORRM or ORRM‐like protein(s), such as ORRM6, is responsible for residual editing at these nine plastid RNA editing sites in the orrm1‐1 single mutant. The orrm6 single mutants displayed substantial reduction in editing extent at accD‐C794 (Hackett & Lu, 2017; Hackett et al., 2017). This raises the question whether other plastid‐targeted ORRM or ORRM‐like protein(s), such as ORRM1, is responsible for residual editing at accD‐C794 in the orrm6 single mutants. Furthermore, recombinant ORRM6 protein showed some binding activity toward the synthetic RNA flanking psbE‐C214, a plastid RNA editing site not affected by the loss‐of‐function mutations in the ORRM6 gene (Hackett et al., 2017). This made us consider whether ORRM6 could function at additional plastid RNA editing sites that are not identified by loss‐of‐function mutations in the ORRM6 gene. To investigate the functional relationship between the two plastid‐targeted ORRM proteins and explore the possible existence of ORRM‐like proteins in the Arabidopsis plastid, we generated orrm1 orrm6 double homozygous Arabidopsis mutants, examined their plastid RNA editing pattern, perform a series of morphological and physiological analyses, and compared them with the wild type and the single mutants. The results showed that ORRM1 and ORRM6 are in charge of distinct sets of plastid RNA editing sites.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
Arabidopsis thaliana (Arabidopsis) T‐DNA insertion lines orrm1‐1 (SALK_072648), orrm6‐1 (SAIL_763_A05), and orrm6‐2 (WiscDsLox485‐488P23) in the Columbia ecotype were obtained from the Arabidopsis Biological Resource Center (Sessions et al., 2002; Woody, Austin‐Phillips, Amasino, & Krysan, 2007). Homozygosity of the orrm1‐1, orrm6‐1, and orrm6‐2 single mutants and the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants was confirmed by PCR, using the Phire Plant Direct PCR kit (Thermo Scientific) and genotyping primers listed in Table S1. Plants were grown in a growth chamber (Percival Scientific) on a 12‐hr light/12‐hr dark photoperiod. The light intensity was 150 μmol photons m−2 s−1, the temperature was 22°C, and the relative humidity was 50%. Unless otherwise stated, plants used for photographing, pigment extraction and measurements, chlorophyll fluorescence, as well as leaf total RNA extraction and subsequent RT‐PCR and Sanger sequencing, were four weeks old.
2.2. Measurements of pigment contents
Total chlorophyll and carotenoid were extracted from rosette leaves with 80% acetone in 2.5 mM HEPES‐KOH, pH7.5, and the amounts (mg) of chlorophyll a and b and carotenoid per gram of fresh tissues were measured on a BioMate 3S UV‐Visible spectrophotometer (Thermo Scientific) at four wavelengths: 470, 646, 654, and 663 nm (Wellburn, 1994).
2.3. Chlorophyll fluorescence measurements
Chlorophyll fluorescence parameter F v /F m (maximum photochemical efficiency of PSII) was determined on dark‐adapted plants at room temperature with the MAXI version of the IMAGING‐PAM M‐Series chlorophyll fluorescence system (Heinz Walz GmbH), as described previously (Lu, Hall, & Last, 2011). Fv/Fm is calculated as follows: Fv/Fm = (F m − F o)/F m, where F v, F m, and F o are variable, maximal, and minimal fluorescence of dark‐adapted leaves, respectively (Nath, O'Donnell, & Lu, 2017; Nath, Wessendorf, & Lu, 2016).
2.4. Analysis of plastid RNA editing by Sanger sequencing
Total RNA was extracted from Arabidopsis rosette leaves using the RNeasy plant mini kit (QIAGEN), digested with the RNase‐free DNase I (QIAGEN), and reverse‐transcribed with random primers (Promega) and Moloney murine leukemia virus reverse transcriptase (Promega) to generate the mRNA:cDNA hybrids, as described previously (Clark & Lu, 2015). The transcript regions encompassing the Arabidopsis plastid RNA editing sites were amplified using Phusion High‐Fidelity DNA Polymerase (New England Biolabs) and PCR amplification/Sanger sequencing primers listed in Table S1. The resulting PCR products were sequenced at the Michigan State University Genomics Facility, using the Sanger method and the PCR amplification/Sanger sequencing primers listed in Table S1.
2.5. Accession numbers
Sequence data of related genes/proteins can be found in the GenBank/EMBL databases under the following accession numbers: ORRM1, At3g20930; ORRM6, At1g73530.
3. RESULTS
3.1. Identification of orrm1 orrm6 double homozygous mutants
To explore the functional relationship between ORRM1 and ORRM6, we created orrm1 orrm6 double homozygous Arabidopsis mutants. The orrm1‐1 (Sun et al., 2013) and orrm6‐1 and orrm6‐2 (Hackett & Lu, 2017; Hackett et al., 2017) homozygous mutants were crossed, and the resulting F2 populations were screened for orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double homozygous mutants. Genotyping was performed by amplifying DNA directly from two‐week‐old plants, using the Phire Plant Direct PCR kit (Thermo Scientific). The orrm1‐1, orrm6‐1, and orrm6‐2 alleles were genotyped with primer combinations SALK_072648LP + SALK_072648RP and SALK_072648RP + LBa1, SAIL_763_A05LP + SAIL_763_A05RP and SAIL_763_A05RP + LB3, and WiscDsLox485‐488P23LP + WiscDsLox485‐488P23RP and WiscDsLox485‐488P23LP + p745, respectively (Table S1). Self‐fertilized F3 seeds harvested from double homozygous F2 plants were used to grow plants for downstream analyses.
3.2. The orrm1 orrm6 double mutants adopted the phenotype of the orrm6 single mutants
As reported in a previous study (Sun et al., 2013), the orrm1‐1 mutant did not show any phenotypic defect, presumably because none of the plastid RNA transcripts affected in the orrm1‐1 mutant is essential. The orrm1‐1 mutant was actually slightly bigger than the Columbia wild type. However, it is not clear whether loss‐of‐function mutation in the ORRM1 gene causes changes in pigment contents and photosynthetic efficiency. The orrm6‐1 and orrm6‐2 mutants were substantially smaller than the wild type, and they displayed reduced PSII photochemical efficiency, small and pale green leaves, and stunted growth (Hackett et al., 2017), presumably because psbF, one of the two plastid RNA transcripts affected in the orrm6 mutants, encodes an essential PSII subunit. To examine whether loss‐of‐function mutation in the ORRM1 gene causes changes in fresh weights, leaf numbers, pigment contents, and photosynthetic efficiency and whether simultaneous loss‐of‐function mutations in ORRM1 and ORRM6 genes result in additive effects, we compared phenotypes, measured fresh weights of the above‐ground portion of the plants, counted rosette leaf numbers, determined pigment contents, and measured photosynthetic parameters in four‐week‐old wild type, orrm1 and orrm6 single and double mutants.
The orrm1‐1 single mutant was indeed larger than the Columbia wild type (Figure 1a). The phenotype of the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐1 double mutants largely resembled the orrm6‐1 and orrm6‐2 single mutants: They were much smaller than the wild type, with small and pale green leaves and retarded growth (Figure 1a). The orrm1‐1 single mutant had a significantly heavier fresh weight (Figure 1b) and a significantly larger rosette leaf number (Figure 1c) than the wild type grown at the same time under the same conditions. This suggests that the orrm1‐1 single mutant is truly bigger and possibly more advanced in its development than the wild type. The orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants had statistically similar fresh weights (Figure 1b) and statistically similar leaf numbers (Figure 1c) as the orrm6‐1 and orrm6‐2 single mutants. This suggests that the orrm1 orrm6 double mutants are indeed phenotypically similar to the orrm6 single mutants.
Figure 1.
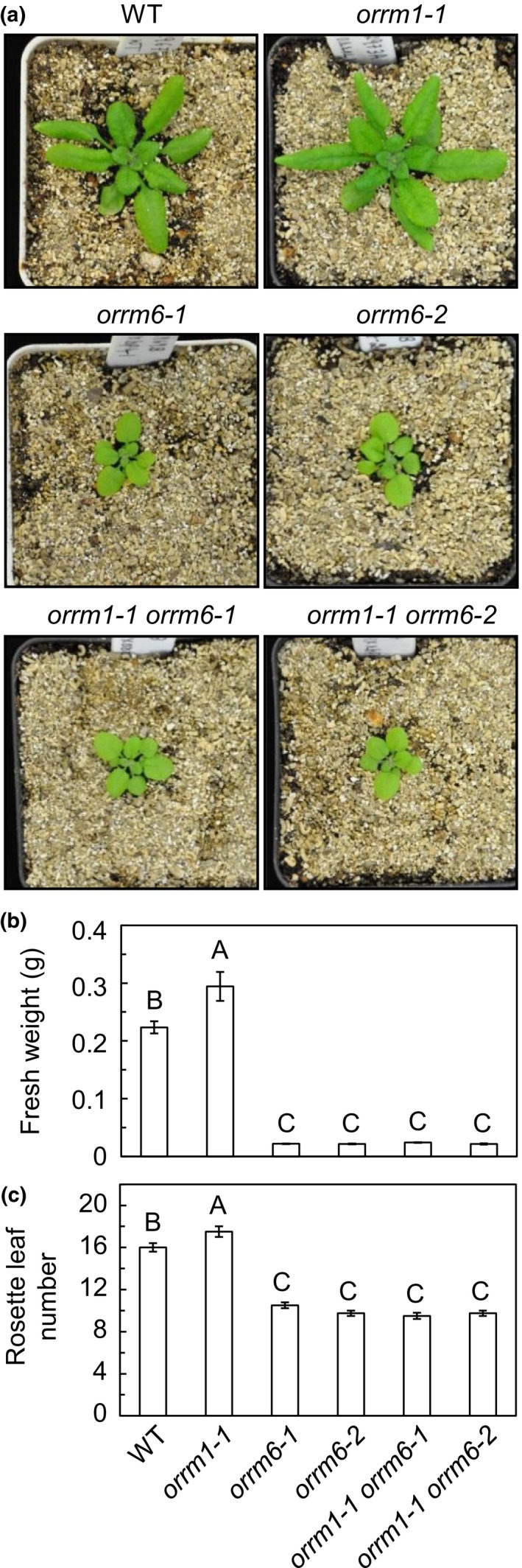
Morphology of 4‐week‐old wild type, orrm1 and orrm6 single mutants, and orrm1 orrm6 double mutants. (a) Images of 4‐week‐old plants. (b) Fresh weights of 4‐week‐old plants. Data are presented as means ± SE (n = 6). (c) Rosette leaf numbers of four‐week‐old plants. Data are presented as means ± SE (n = 4). Values not connected by the same uppercase letters are significantly different (Student's t test, p < .05). Plants used for photographing, fresh weights, leaf number counting, pigment extraction, chlorophyll fluorescence analysis, and RNA extraction were grown on a 12‐hr light/12‐hr dark photoperiod with an irradiance of 150 μmol photons m−2 s−1 during the light period
The contents of chlorophyll a, chlorophyll b, and total chlorophyll in the orrm1‐1 mutant were 12%, 20%, and 14% higher than those in the wild type, respectively (Figure 2a–c). Due to the differential increase in the contents of chlorophyll a and b, the chlorophyll a/b ratio in the orrm1‐1 mutant was slightly lower than that in the wild type (Figure 2d). The contents of chlorophyll a, chlorophyll b, and total chlorophyll in the orrm6‐1 and orrm6‐2 mutants were approximately 21%, 6%, and 18% lower than those in the wild type, respectively (Figure 2a–c). Due to the differential decrease in the contents of chlorophyll a and b, the chlorophyll a/b ratio in the orrm6‐1 and orrm6‐2 mutants was 14% lower than that in the wild type (Figure 2d). The chlorophyll a, chlorophyll b, and total chlorophyll contents and the chlorophyll a/b ratio in the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants resembled the orrm6‐1 and orrm6‐2 single mutants: They were significantly lower than that in the wild type but statistically similar to those in the orrm6 single mutants (Figure 2a–d). These results are consistent with the pale green pigmentation observed in the orrm6 single mutants and the orrm1 orrm6 double mutants (Figure 1a).
Figure 2.
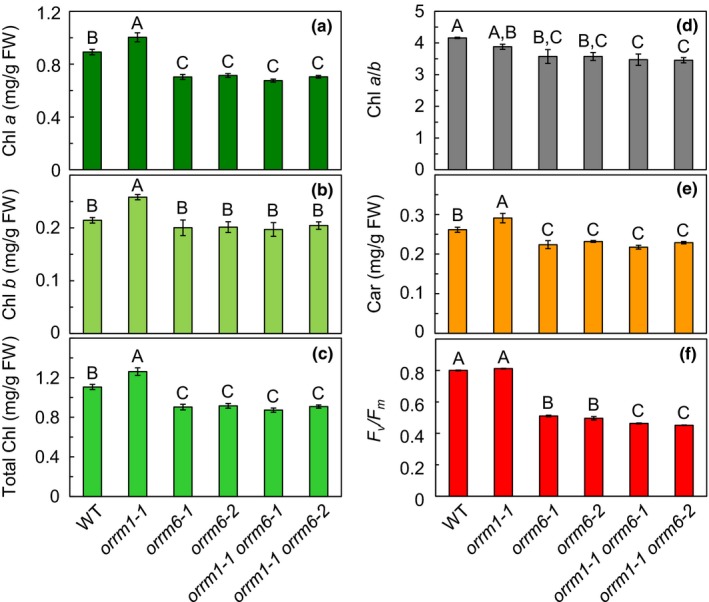
Pigment contents and chlorophyll fluorescence of 4‐week‐old plants. (a–e) Chlorophyll a (a), chlorophyll b (b), total chlorophyll (c), chlorophyll a/b ratio (d), carotenoid (e), and Fv/Fm (f) of 4‐week‐old plants. Chlorophyll and carotenoid were extracted and determined as described by Wellburn (1994). Measurements of chlorophyll fluorescence parameters were performed with the IMAGING‐PAM M‐Series chlorophyll fluorescence system (Heinz Waltz) on dark‐adapted plants. Data are presented as means ± SE (n = 5 for pigment contents and n = 4 for chlorophyll fluorescence parameters). Values not connected by the same uppercase letters are significantly different (Student's t test, p < .05). Plants used for pigment extraction and fluorescence analysis were grown on a 12‐hr light/12‐hr dark photoperiod with an irradiance of 150 μmol photons/m−2 s−1 during the light period. Chl, chlorophyll. Car, carotenoid. FW, fresh weight. WT, wild type
The carotenoid level in the orrm1‐1 mutant was 11% higher than that in the wild type; the carotenoid level in the orrm6‐1 and orrm6‐2 mutants was 14% and 11% lower than that in the wild type (Figure 2e). The level of carotenoid in the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants resembled the orrm6‐1 and orrm6‐2 single mutants: It was significantly lower than that in the wild type but statistically similar to that in the orrm6 single mutants (Figure 2e).
To assess whether the orrm1 and orrm6 single and double mutants have defects in PSII, we determined F v /F m, an indicator of the maximum photochemical efficiency of PSII (Baker, Harbinson, & Kramer, 2007). F v /F m in the orrm1‐1 mutant was statistically similar to that in the wild type; however, F v /F m in the orrm6‐1 and orrm6‐2 mutants was 36%–38% lower than that in the wild type (Figure 2f). F v /F m in the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants was 42%–44% lower than that in the wild type and 9% lower than that in the corresponding orrm6 single mutants (Figure 2e). The substantial decreases in F v /F m in the orrm6 single mutants and the orrm1 orrm6 double mutants are consistent with significant reductions of editing extent at the psbF‐C77 site in these mutants (Hackett & Lu, 2017; Hackett et al., 2017).
Taken together, the orrm1 orrm6 double mutants had similar levels of chlorophyll a, chlorophyll b, total chlorophyll, chlorophyll a/b ratio, carotenoid, and F v /F m as the orrm6 single mutants, suggesting that the orrm1 orrm6 double mutants adopted the phenotype of the orrm6 single mutants.
3.3. The total number of plastid RNA editing sites affected in the double mutants was the sum of sites affected in the single mutants
We examined the editing patterns of 34 validated plastid RNA editing sites (Table S2) (Chateigner‐Boutin & Small, 2007) with high‐fidelity PCR amplification and Sanger sequencing, using primers designed previously (Table S1) (Cai et al., 2009; Hackett et al., 2017). The orrm1‐1 single mutant was previously found to show near‐complete loss of editing at 12 plastid RNA editing sites: accD‐C1568, matK‐C640, ndhB‐C467, ndhB‐C586, ndhB‐C836, ndhB‐C872, ndhD‐C674, ndhD‐C878, ndhD‐C887, ndhG‐C50, rpoB‐C2432, and rps12‐intron (Sun et al., 2013). In this study, these 12 plastid RNA editing sites displayed similar loss of editing in the orrm1‐1 single mutant and the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants but were unchanged in the orrm6‐1 and orrm6‐2 single mutants (Figure 3). In addition to these 12 sites, the orrm1‐1 single mutant showed reduction in editing extent at nine plastid RNA editing sites: clpP‐C559, ndhB‐C746, ndhB‐C830, ndhB‐C1255, ndhD‐C2, rpoA‐C200, rpoB‐C338, rpoB‐C551, and rps14‐C149 (Sun et al., 2013). In this study, these nine plastid RNA editing sites displayed similar reduction in editing extent in the orrm1‐1 single mutant and the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants but was unchanged in the orrm6‐1 and orrm6‐2 single mutants (Figure 3).
Figure 3.
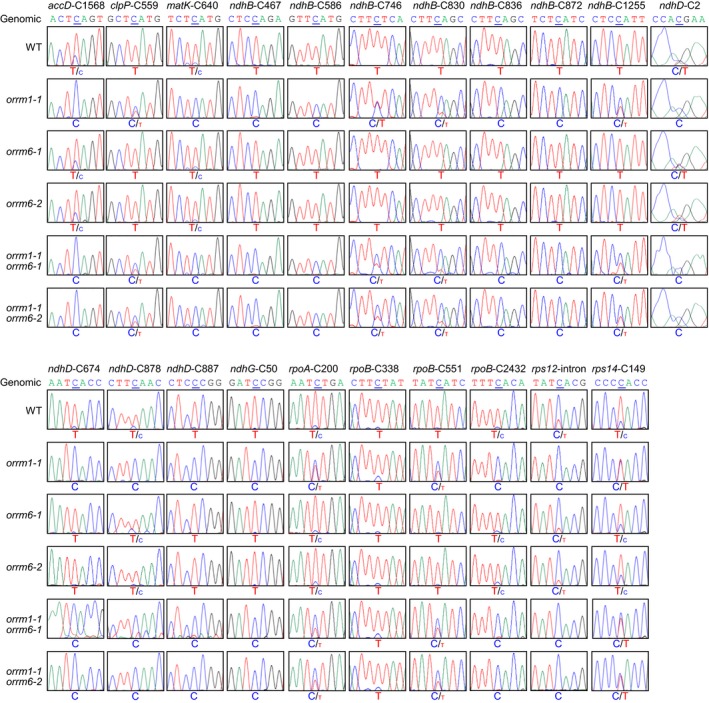
Sanger sequencing of 21 plastid RNA editing sites affected by the loss‐of‐function mutation in ORRM1. RT‐PCR products surrounding the editing sites were directly sequenced. The seven‐nucleotide sequences encompassing the cytidine target (underlined) were shown. The corresponding genomic sequences of these sites were displayed as controls
The orrm6‐1 and orrm6‐2 single mutants were previously found to show substantial reduction in editing extent at accD‐C794 and near‐complete loss of editing at psbF‐C77 (Hackett et al., 2017). In this study, these two plastid RNA editing sites displayed similar reduction in editing extent in the orrm6‐1 and orrm6‐2 single mutants and the orrm1‐1 orrm6‐1 and orrm6‐1 orrm6‐2 double mutants but were unchanged in the orrm1‐1 single mutant (Figure 4).
Figure 4.
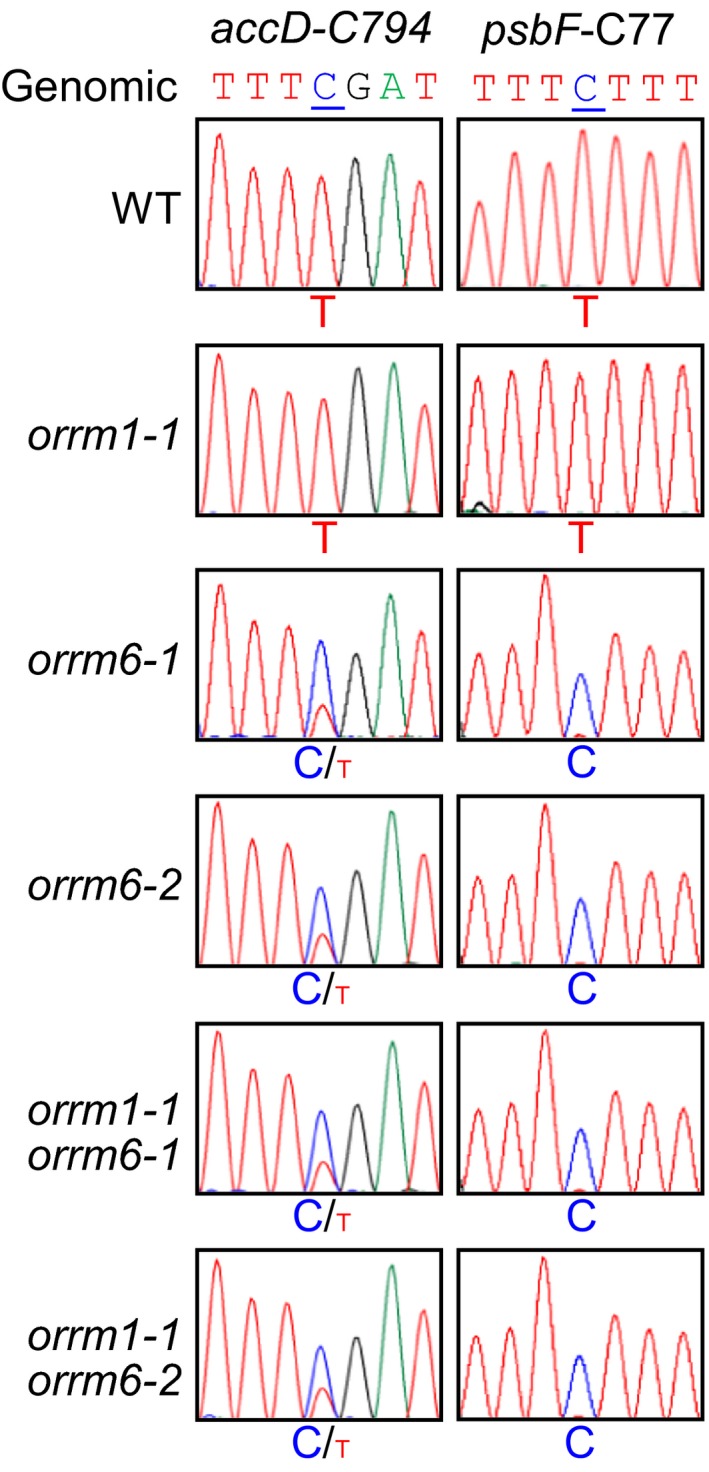
Sanger sequencing of two plastid RNA editing sites affected by loss‐of‐function mutations in ORRM6. RT‐PCR products surrounding the editing sites were directly sequenced. The seven‐nucleotide sequences encompassing the cytidine target (underlined) were shown. The corresponding genomic sequences of these sites were displayed as controls
Among the 34 validated plastid RNA editing sites, 11 sites were not affected in either orrm1 or orrm6 mutants: atpF‐C92, ndhB‐C149, ndhB‐C1481, ndhD‐C383, ndhF‐C290, petL‐C5, psbE‐C214, psbZ‐C50, rpl23‐C89, rpoC1‐C488, and rps14‐C80 (Figure 5) (Hackett & Lu, 2017; Hackett et al., 2017; Sun et al., 2013). These 11 sites were not affected in the orrm1‐1 orrm6‐1 and orrm1‐1 orrm6‐2 double mutants (Figure 5). Taken together, the total number of plastid RNA editing sites affected in the orrm1 orrm6 double mutants was the sum of sites affected in the orrm1 and orrm6 single mutants. This suggests that simultaneous mutations in the two plastid‐targeted ORRM proteins do not cause additional loss or reduction in editing extent at other plastid RNA editing sites.
Figure 5.
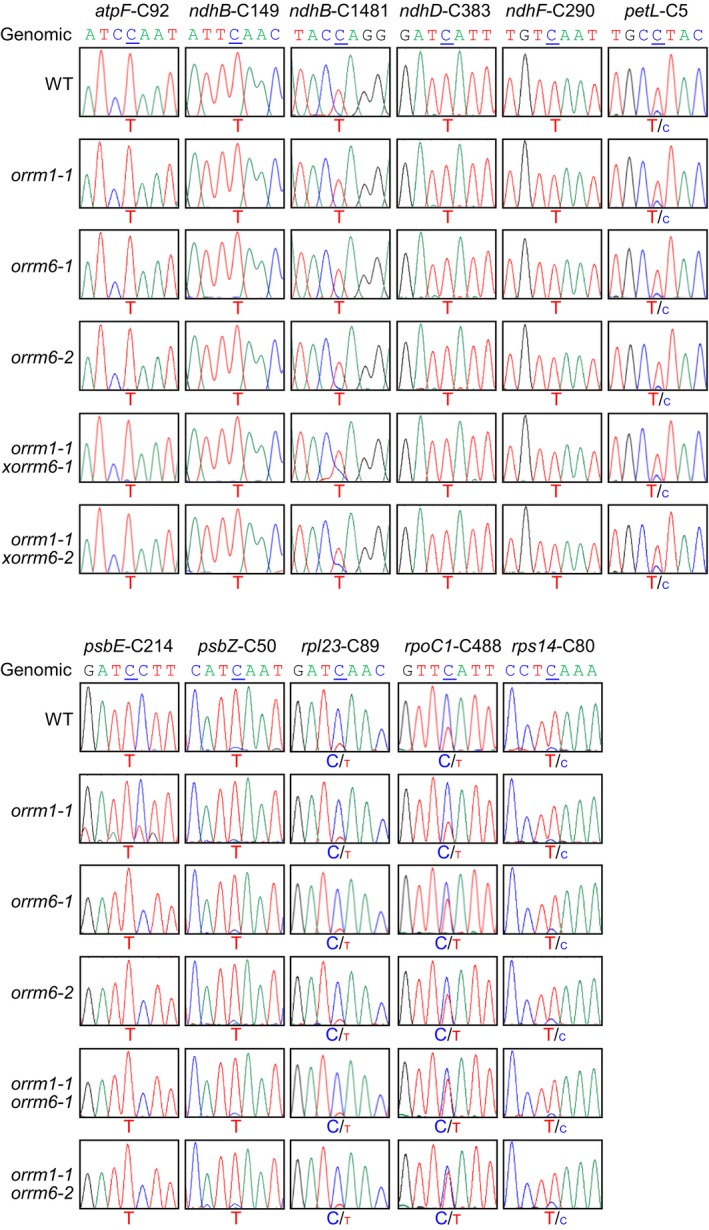
Sanger sequencing of 11 plastid RNA editing sites not affected in orrm1 or orrm6 mutants. RT‐PCR products surrounding the editing sites were directly sequenced. The seven‐nucleotide sequences encompassing the cytidine target (underlined) were shown. The corresponding genomic sequences of these sites were displayed as controls
4. DISCUSSION
The loss‐of‐function mutation in the ORRM1 and ORRM6 genes resulted in near‐complete loss or substantial reduction in editing at 21 and two plastid RNA editing sites, respectively (Hackett & Lu, 2017; Hackett et al., 2017; Sun et al., 2013). The 12 plastid RNA editing sites that showed near‐complete loss of editing in the orrm1 single mutant displayed similar loss of editing in the orrm1 orrm6 double mutants but were unchanged in the orrm6 single mutants (Figure 3). This suggests that ORRM1 is the sole ORRM protein at these 12 plastid RNA editing sites. The nine plastid RNA editing sites that showed substantial reduction in editing extent displayed similar reduction in editing extent in the orrm1 orrm6 double mutants but was unchanged in the orrm6 single mutants (Figure 3). This suggests that ORRM6 is not responsible for residual editing at these nine plastid RNA editing sites in the orrm1 single mutant. The psbF‐C77 RNA editing site that showed near‐complete loss of editing in the orrm6 single mutants displayed similar loss of editing in the orrm1 orrm6 double mutants but was unchanged in the orrm1 single mutant (Figure 4). This suggests that ORRM6 is the sole ORRM protein at psbF‐C77. The accD‐C794 RNA editing site that showed substantial reduction in editing extent in the orrm6 single mutants displayed similar reduction in editing extent in the orrm1 orrm6 double mutants but was unchanged in the orrm1 single mutant (Figure 4). This suggests that ORRM1 is not responsible for the residual editing at accD‐C794 in the in the orrm6 single mutants. The 11 plastid RNA editing sites that were not affected in either orrm1 or orrm6 mutants remained unchanged in the orrm1 orrm6 double mutants (Figure 5). This suggests that neither ORRM1 nor ORRM6 functions at these 11 plastid RNA editing sites. Taken together, the results in this study indicate that ORRM1 and ORRM6 are in charge of distinct sets of plastid RNA editing sites and that simultaneous mutations in ORRM1 and ORRM6 genes do not cause additional reduction in editing extent at other plastid RNA editing sites. This is consistent with the lack of physical interaction between ORRM1 and ORRM6 proteins in the reciprocal bimolecular fluorescence complementation assay (Hackett et al., 2017).
ORRM1 was found to interact directly with PPR proteins CRR28 and OTP82, via its duplicated RIP/MORF moiety (Figure 6a) (Sun et al., 2013). CRR28 is necessary for editing at ndhB‐467 and ndhD‐878 (Okuda et al., 2009), while OTP82 is required for editing at ndhG‐50 and ndhB‐836 (Okuda et al., 2010). These four plastid RNA editing sites were affected in the orrm1 single mutant and the orrm1 orrm6 double mutants (Figure 3). Interestingly, ORRM1 did not appear to interact directly with PPR protein ORGANELLE TRANSCRIPT PROCESSING81 (OTP81), which is required for efficient editing at accD‐C1568, matK‐C640, ndhB‐C872, rpoB‐C2432, and rps12‐intron (Wagoner et al., 2015). These five plastid RNA editing sites were affected in the orrm1 single mutant and the orrm1 orrm6 double mutants (Figure 3). These observations suggest that ORRM1 may interact with some PPR proteins directly via its own RIP/MORF domains, or indirectly associate with other PPR proteins.
Figure 6.
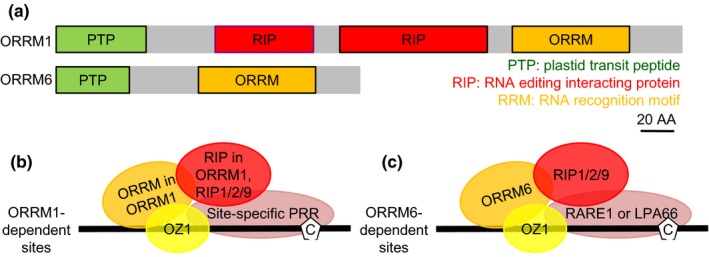
Domain composition of ORRM1 and ORRM6 and models for the editing complexes at the plastid RNA editing sites affected by mutations in ORRM1 and ORRM6. (a) Domain composition of full‐length ORRM1 and ORRM6 proteins. (b) Model for the editing complexes at the 21 ORRM1‐dependent plastid RNA editing sites: accD‐C1568, clpP‐C559, matK‐C640, ndhB‐C467, ndhB‐C586, ndhB‐C746, ndhB‐C830, ndhB‐C836, ndhB‐C872, ndhB‐C1255, ndhD‐C2, ndhD‐C674, ndhD‐C878, ndhD‐C887, ndhG‐C50, rpoA‐C200, rpoB‐C338, rpoB‐C551, rpoB‐C2432, rps12‐intron, and rps14‐C149. (c) Model for the editing complexes at the two ORRM6‐dependent plastid RNA editing sites: accD‐C794 and psbF‐C77. The PPR protein at accD‐C794 and psbF‐C77 is RARE1 and LPA66, respectively. For simplicity, only one name is shown for proteins with multiple names (e.g., “RIP1” for RIP1/MORF8). Black lines represent transcripts; the letter C in white pentagons represents the cytidine target
ORRM1 was found to interact with plastid‐targeted RIPs/MORFs (Sun et al., 2015). The 21 plastid RNA editing sites affected in the orrm1 single mutant and the orrm1 orrm6 double mutants (Figure 3) were also affected in the rip1, rip2, and/or rip9 mutants (Bentolila et al., 2012; Takenaka et al., 2012). RIP1/MORF8 was found to interact with the plastid‐targeted PPR protein OTP81 (Wagoner et al., 2015). RIP2/MORF2 and RIP9/MORF9 were found to interact with plastid‐targeted PPR proteins CRR28, DYW1, OTP81, and OTP82 (Wagoner et al., 2015; Zhang et al., 2014). CRR28 is required for editing at ndhB‐467 and ndhD‐878 (Okuda et al., 2009); DYW1 is essential for editing at ndhD‐C2 (Boussardon et al., 2012, 2014); OTP81 is necessary for efficient editing at accD‐C1568, matK‐C640, ndhB‐C872, rpoB‐C2432, and rps12‐intron (Wagoner et al., 2015); OTP82 is needed for editing at ndhG‐50 and ndhB‐836 (Okuda et al., 2010). These ten plastid RNA editing sites were substantially affected in the orrm1 single mutant and the orrm1 orrm6 double mutants (Figure 3).
Furthermore, ORRM1 was found to interact directly with OZ1 (Sun et al., 2015). The 21 plastid RNA editing sites affected in the orrm1 single mutant and the orrm1 orrm6 double mutants (Figure 3) were also affected in the oz1 mutant (Sun et al., 2015). Therefore, the editing complex at the 21 ORRM1‐dependent plastid RNA editing sites probably contains ORRM1, site‐specific PPR protein(s), RIPs/MORFs (RIP1/MORF8, RIP2/MORF2, and/or RIP9/MORF9), and OZ1 (Figure 6b).
Unlike ORRM1, ORRM6 does not contain any RIP/MORF domains (Figure 6a); therefore, ORRM6 is not expected to interact directly with PPR proteins. Indeed, ORRM6 failed to interact with plastid‐targeted PPR proteins LOW PSII ACCUMULATION66 (LPA66) and RARE1 (Hackett et al., 2017), which are required for editing at accD‐C794 and psbF‐C77, respectively (Cai et al., 2009; Robbins, Heller, & Hanson, 2009). These two plastid RNA editing sites were substantially affected in the orrm6 single mutant and the orrm1 orrm6 double mutants (Figure 4). ORRM6 was found to interact directly with RIP1/MORF8, RIP2/MORF2, RIP9/MORF9, and OZ1 (Hackett et al., 2017). The two ORRM6‐dependent plastid RNA editing sites (accD‐C794 and psbF‐C77) were also affected in rip1, rip2, rip9, and oz1 mutants (Sun et al., 2015; Takenaka et al., 2012). Therefore, the editing complex at the two ORRM6‐dependent plastid RNA editing sites probably contains ORRM6, site‐specific PPR protein (i.e., RARE1 for accD‐C794 and LPA66 for psbF‐C77), RIPs/MORFs (RIP1/MORF8, RIP2/MORF2, and/or RIP9/MORF9), and OZ1 (Figure 6c).
It remains unknown what plastid RRM‐containing protein(s) participate in RNA editing at the 11 plastid RNA editing sites that were not affected in the orrm1 and orrm6 single mutants (Figure 5). Two potential candidates are the 31 KD CHLOROPLAST PROTEIN A and B (i.e., CP31A and CP31B). CP31A and CP31B belong to a small group of chloroplast ribonucleoproteins that contain two RRM domains (Tillich et al., 2009). Among these 11 plastid RNA editing sites, five displayed reduced editing extent in cp31a cp31b single and/or double mutants: ndhB‐C1481, ndhF‐C290, petL‐C5, psbZ‐C50, and rpoC1‐C488 (Tillich et al., 2009). Therefore, CP31A and CP31B may serve as the RRM‐containing protein(s) at these five plastid RNA editing sites. However, the editing extent at the other six plastid RNA editing sites (atpF‐C92, ndhB‐C149, ndhD‐C383, psbE‐C214, rpl23‐C89, and rps14‐C80) was not affected by mutations in CP31A or CP31B. Further studies are needed to identify the plastid RRM‐containing protein(s) at these plastid RNA editing sites.
AUTHOR CONTRIBUTIONS
A.M.S., M.B.S., J.P.O., and Y.L. conducted experiments. A.M.S. analyzed data. Y.L. conceived the project, designed and supervised the experiments, and wrote the article.
Supporting information
Table S1
Table S2
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Amy T. Kobylarz for help with genotyping, Zoha Aqeel and Sanjna Chalasani for assistance with sequencing data analysis, Christopher D. Jackson for growth chamber management, and Maureen R. Hanson for comments on experimental design. This work was supported by the U.S. National Science Foundation (grant no. MCB‐1244008).
Searing AM, Satyanarayan MB, O′Donnell JP, Lu Y. Two organelle RNA recognition motif proteins affect distinct sets of RNA editing sites in the Arabidopsis thaliana plastid. Plant Direct. 2020;4:1–11. 10.1002/pld3.213
Funding information
This work was supported by the U.S. National Science Foundation (grant no. MCB‐1244008).
REFERENCES
- Baker, N. R. , Harbinson, J. , & Kramer, D. M. (2007). Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant, Cell and Environment, 30, 1107–1125. [DOI] [PubMed] [Google Scholar]
- Barkan, A. , & Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology, 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Bentolila, S. , Heller, W. P. , Sun, T. , Babina, A. M. , Friso, G. , van Wijk, K. J. , & Hanson, M. R. (2012). RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proceedings of the National Academy of Sciences, USA, 109, E1453–E1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila, S. , Oh, J. , Hanson, M. R. , & Bukowski, R. (2013). Comprehensive high‐resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genetics, 9, e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussardon, C. , Avon, A. , Kindgren, P. , Bond, C. S. , Challenor, M. , Lurin, C. , & Small, I. (2014). The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD‐1. New Phytologist, 203, 1090–1095. [DOI] [PubMed] [Google Scholar]
- Boussardon, C. , Salone, V. , Avon, A. , Berthome, R. , Hammani, K. , Okuda, K. , … Lurin, C. (2012). Two interacting proteins are necessary for the editing of the NdhD‐1 site in Arabidopsis plastids. The Plant Cell, 24, 3684–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, W. , Ji, D. , Peng, L. , Guo, J. , Ma, J. , Zou, M. , … Zhang, L. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiology, 150, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner‐Boutin, A.‐L. , Colas des Francs‐Small, C. , Fujii, S. , Okuda, K. , Tanz, S. K. , & Small, I. (2013). The E domains of pentatricopeptide repeat proteins from different organelles are not functionally equivalent for RNA editing. The Plant Journal, 74, 935–945. [DOI] [PubMed] [Google Scholar]
- Chateigner‐Boutin, A.‐L. , & Small, I. (2007). A rapid high‐throughput method for the detection and quantification of RNA editing based on high‐resolution melting of amplicons. Nucleic Acids Research, 35, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, T. J. , & Lu, Y. (2015). Analysis of loss‐of‐function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate‐derived amino acid contents. Plant Physiology, 168, 1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre‐Nitschke, S. E. , Grienenberger, J. M. , & Gualberto, J. M. (1999). A prokaryotic‐type cytidine deaminase from Arabidopsis thaliana: Gene expression and functional characterization. European Journal of Biochemistry, 263, 896–903. [DOI] [PubMed] [Google Scholar]
- Fujii, S. , & Small, I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytologist, 191, 37–47. [DOI] [PubMed] [Google Scholar]
- Hackett, J. B. , & Lu, Y. (2017). Whole‐transcriptome RNA‐seq, gene set enrichment pathway analysis, and exon coverage analysis of two plastid RNA editing mutants. Plant Signaling & Behavior, 12, e1312242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett, J. B. , Shi, X. , Kobylarz, A. T. , Lucas, M. K. , Wessendorf, R. L. , Hines, K. M. , … Lu, Y. (2017). An Organelle RNA Recognition Motif protein is required for photosystem II subunit psbF transcript editing. Plant Physiology, 173, 2278–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. (2018). RNA editing of plastid‐encoded genes. Photosynthetica, 56, 48–61. [Google Scholar]
- Lu, Y. , Hall, D. A. , & Last, R. L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of Photosystem II in Arabidopsis thaliana . The Plant Cell, 23, 1861–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C. , Andres, C. , Aubourg, S. , Bellaoui, M. , Bitton, F. , Bruyere, C. , … Small, I. (2004). Genome‐wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell, 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris, C. , Dominguez, C. , & Allain, F. H. (2005). The RNA recognition motif, a plastic RNA‐binding platform to regulate post‐transcriptional gene expression. FEBS Journal, 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- Nath, K. , O'Donnell, J. P. , & Lu, Y. (2017). Chloroplastic iron‐sulfur scaffold protein NFU3 is essential to overall plant fitness. Plant Signaling & Behavior, 12, e1282023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, K. , Wessendorf, R. L. , & Lu, Y. (2016). A nitrogen‐fixing subunit essential for accumulating 4Fe‐4S‐containing Photosystem I core proteins. Plant Physiology, 172, 2459–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, C. D. , Mansfield, R. E. , Leung, W. , Vaz, P. M. , Loughlin, F. E. , Grant, R. P. , & Mackay, J. P. (2011). Characterization of a family of RanBP2‐type zinc fingers that can recognize single‐stranded RNA. Journal of Molecular Biology, 407, 273–283. [DOI] [PubMed] [Google Scholar]
- Okuda, K. , Chateigner‐Boutin, A. L. , Nakamura, T. , Delannoy, E. , Sugita, M. , Myouga, F. , … Shikanai, T. (2009). Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. The Plant Cell, 21, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, K. , Hammani, K. , Tanz, S. K. , Peng, L. , Fukao, Y. , Myouga, F. , … Shikanai, T. (2010). The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. The Plant Journal, 61, 339–349. [DOI] [PubMed] [Google Scholar]
- Okuda, K. , Myouga, F. , Motohashi, R. , Shinozaki, K. , & Shikanai, T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proceedings of the National Academy of Sciences, USA, 104, 8178–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, K. , Nakamura, T. , Sugita, M. , Shimizu, T. , & Shikanai, T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. Journal of Biological Chemistry, 281, 37661–37667. [DOI] [PubMed] [Google Scholar]
- Okuda, K. , & Shikanai, T. (2012). A pentatricopeptide repeat protein acts as a site‐specificity factor at multiple RNA editing sites with unrelated cis‐acting elements in plastids. Nucleic Acids Research, 40, 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, J. C. , Heller, W. P. , & Hanson, M. R. (2009). A comparative genomics approach identifies a PPR‐DYW protein that is essential for C‐to‐U editing of the Arabidopsis chloroplast accD transcript. RNA, 15, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe, H. , Castandet, B. , Schmitz‐Linneweber, C. , & Stern, D. B. (2013). Arabidopsis chloroplast quantitative editotype. FEBS Letters, 587, 1429–1433. [DOI] [PubMed] [Google Scholar]
- Schallenberg‐Rüdinger, M. , Kindgren, P. , Zehrmann, A. , Small, I. , & Knoop, V. (2013). A DYW‐protein knockout in Physcomitrella affects two closely spaced mitochondrial editing sites and causes a severe developmental phenotype. The Plant Journal, 76, 420–432. [DOI] [PubMed] [Google Scholar]
- Sessions, A. , Burke, E. , Presting, G. , Aux, G. , McElver, J. , Patton, D. , … Goff, S. A. (2002). A high‐throughput Arabidopsis reverse genetics system. The Plant Cell, 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Bentolila, S. , & Hanson, M. R. (2016a). Organelle RNA recognition motif‐containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signaling & Behavior, 11, e1167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Castandet, B. , Germain, A. , Hanson, M. R. , & Bentolila, S. (2017b). ORRM5, an RNA recognition motif‐containing protein, has a unique effect on mitochondrial RNA editing. Journal of Experimental Botany, 68, 2833–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Germain, A. , Hanson, M. R. , & Bentolila, S. (2016b). RNA recognition motif‐containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiology, 170, 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Hanson, M. R. , & Bentolila, S. (2015). Two RNA recognition motif‐containing proteins are plant mitochondrial editing factors. Nucleic Acids Research, 43, 3814–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Hanson, M. R. , & Bentolila, S. (2017a). Functional diversity of Arabidopsis organelle‐localized RNA‐recognition motif‐containing proteins. Wiley Interdisciplinary Reviews: RNA, 8(5), e1420. [DOI] [PubMed] [Google Scholar]
- Sun, T. , Bentolila, S. , & Hanson, M. R. (2016). The unexpected diversity of plant organelle RNA editosomes. Trends in Plant Science, 21, 962–973. [DOI] [PubMed] [Google Scholar]
- Sun, T. , Germain, A. , Giloteaux, L. , Hammani, K. , Barkan, A. , Hanson, M. R. , & Bentolila, S. (2013). An RNA recognition motif‐containing protein is required for plastid RNA editing in Arabidopsis and maize. Proceedings of the National Academy of Sciences, USA, 110, E1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. , Shi, X. , Friso, G. , Van Wijk, K. , Bentolila, S. , & Hanson, M. R. (2015). A zinc finger motif‐containing protein is essential for chloroplast RNA editing. PLoS Genetics, 11, e1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka, M. , Zehrmann, A. , Verbitskiy, D. , Hartel, B. , & Brennicke, A. (2013). RNA editing in plants and its evolution. Annual Review of Genetics, 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Takenaka, M. , Zehrmann, A. , Verbitskiy, D. , Kugelmann, M. , Härtel, B. , & Brennicke, A. (2012). Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proceedings of the National Academy of Sciences USA, 109, 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki, E. , Hattori, M. , & Sugita, M. (2010). The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. The Plant Journal, 62, 560–570. [DOI] [PubMed] [Google Scholar]
- Tillich, M. , Hardel, S. L. , Kupsch, C. , Armbruster, U. , Delannoy, E. , Gualberto, J. M. , … Schmitz‐Linneweber, C. (2009). Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proceedings of the National Academy of Sciences, USA, 106, 6002–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagoner, J. A. , Sun, T. , Lin, L. , & Hanson, M. R. (2015). Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat‐containing proteins are required for site‐specific chloroplast RNA editing. Journal of Biological Chemistry, 290, 2957–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn, A. R. (1994). The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology, 144, 307–313. [Google Scholar]
- Williams‐Carrier, R. , Kroeger, T. , & Barkan, A. (2008). Sequence‐specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA, 14, 1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody, S. T. , Austin‐Phillips, S. , Amasino, R. M. , & Krysan, P. J. (2007). The WiscDsLox T‐DNA collection: An Arabidopsis community resource generated by using an improved high‐throughput T‐DNA sequencing pipeline. Journal of Plant Research, 120, 157–165. [DOI] [PubMed] [Google Scholar]
- Xu, J.‐H. , & Messing, J. (2006). Maize haplotype with a helitron‐amplified cytidine deaminase gene copy. BMC Genetics, 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehrmann, A. , Härtel, B. , Glass, F. , Bayer‐Császár, E. , Obata, T. , Meyer, E. , … Takenaka, M. (2015). Selective homo‐ and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana . Journal of Biological Chemistry, 290, 6445–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Tang, W. , Hedtke, B. , Zhong, L. , Liu, L. , Peng, L. , … Lin, R. (2014). Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proceedings of the National Academy of Sciences, USA, 111, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Supplementary Material


