Abstract
Amyotrophic lateral sclerosis (ALS) is a debilitating and fatal disorder that can be caused by mutations in the superoxide dismutase 1 (SOD1) gene. Although ALS is currently incurable, CRISPR base editors hold the potential to treat the disease through their ability to create nonsense mutations that can permanently disable the expression of the mutant SOD1 gene. However, the restrictive carrying capacity of adeno-associated virus (AAV) vectors has limited their therapeutic application. In this study, we establish an intein-mediated trans-splicing system that enables in vivo delivery of cytidine base editors (CBEs) consisting of the widely used Cas9 protein from Streptococcus pyogenes. We show that intrathecal injection of dual AAV particles encoding a split-intein CBE engineered to trans-splice and introduce a nonsense-coding substitution into a mutant SOD1 gene prolonged survival and markedly slowed the progression of disease in the G93A-SOD1 mouse model of ALS. Adult animals treated by this split-intein CRISPR base editor had a reduced rate of muscle atrophy, decreased muscle denervation, improved neuromuscular function, and up to 40% fewer SOD1 immunoreactive inclusions at end-stage mice compared to control mice. This work expands the capabilities of single-base editors and demonstrates their potential for gene therapy.
Keywords: CRISPR-Cas9, base editor, gene editing, split intein, AAV, ALS, SOD1, gene therapy, neurodegeneration
Lim et al. establish a trans-splicing system to deliver CRISPR base editors in vivo that enables treatment of a mouse model of ALS. They show that base editing can increase survival and slow the progression of disease.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder characterized by the loss of motor neurons in the spinal cord and brain that invariably results in progressive muscle atrophy, paralysis, and ultimately death.1 There is no cure for ALS and current therapies provide only modest benefits to patients,2,3 underscoring the need for strategies to treat this destructive disorder.
While most cases of ALS are sporadic, dominant mutations in the Cu-Zn superoxide dismutase 1 (SOD1) gene are responsible for ∼20% of inherited or familial forms of the disease.4 To date, more than 100 mutations in the SOD1 gene have been implicated in ALS, and although the role of mutant SOD1 in the pathogenesis of the disorder remains incompletely understood, it is thought that mutant SOD1 expression in both motor neurons5 and non-neuronal cells5, 6, 7, 8, 9, 10, 11, 12 is a driving factor behind the development of the disease and its progression. Accordingly, antisense oligonucleotides (ASOs) and RNA interference (RNAi) have been used to knock down the expression of mutant SOD1 and improve therapeutic outcomes in animal models of the disorder.13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 However, owing to their transient life cycle, ASOs may require a lifetime of administrations to the cerebrospinal fluid (CSF) to sustain their therapeutic effect, which could pose a physical and financial burden to patients. Furthermore, neither ASOs nor RNAi has the capacity to permanently correct the underlying root genetic cause of a disorder such as SOD1-linked ALS, which could ultimately limit their therapeutic effectiveness.
Genome-editing technologies, including RNA-guided CRISPR-Cas9 nucleases,25, 26, 27, 28 offer an alternative approach for treating ALS by providing a means to permanently disable the function of the mutant SOD1 gene. For instance, we previously demonstrated that a CRISPR-Cas9 system could be delivered to the spinal cord of a mouse model of ALS to reduce the expression of the mutant SOD1 gene via genome editing and delay the onset of the disease.29 However, potential clinical applications of Cas9 nucleases are limited in part by their reliance on DNA breaks, which could lead to deleterious large deletions and chromosomal rearrangements,30 and non-homologous end joining, a stochastic DNA repair pathway that could give rise to undesirable and potentially mutagenic outcomes.31
CRISPR single-base editors, which can be used to introduce targeted C>T or A>G base alterations in the absence of a double-strand DNA break,32 have emerged as an alternative to CRISPR-Cas9 nucleases that could overcome some of these limitations. To date, the most widely used base editors consist of fusions of a Cas9 nickase with a rat APOBEC1 cytidine deaminase domain, which can induce deamination of a target cytosine to facilitate its conversion to a thymine33 or a TadA adenine deaminase that can convert a target adenosine to a guanosine.34 Unlike CRISPR-Cas9 and other gene-editing modalities that rely on homology-directed repair to facilitate single-base substitutions, cytidine and adenine base editors employ base excision and DNA mismatch repair, two DNA repair pathways that are active in both dividing and non-dividing cells and thus could be used to introduce single-base edits in post-mitotic and slowly dividing cells,35,36 such as those within the nervous system. Given these advantages, as well as their ability to introduce single-base alterations that can silence gene expression,37,38 we reasoned that CRISPR base editors could be harnessed to permanently disrupt the expression of the mutant SOD1 gene in vivo and treat ALS.
In this study, we establish an intein-based trans-splicing system that enables the in vivo delivery of cytidine base editors (CBEs) consisting of the Streptococcus pyogenes Cas9 (SpCas9) protein. We show that this split-intein CBE can lower mutant SOD1 in vivo and provide therapeutic benefit to a mouse model of SOD1-linked ALS. This work thus expands the capabilities of CRISPR base editors and illustrates their potential for treating a neurodegenerative disorder.
Results
Disruption of SOD1 Gene Expression by a CBE
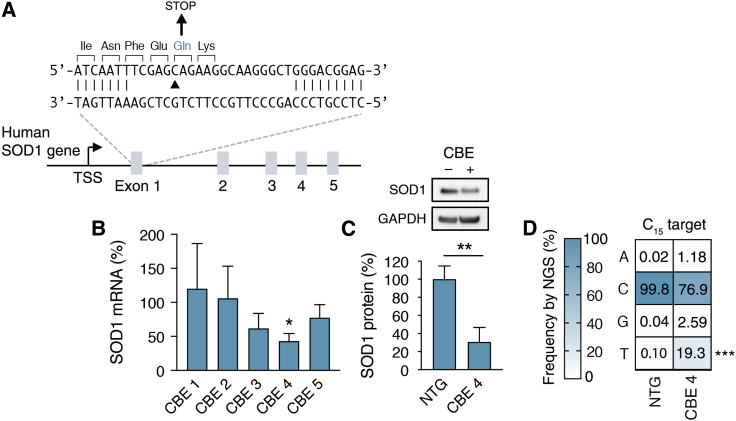
In order to determine whether base editing could be used to reduce the expression of the SOD1 gene, we searched the human SOD1 coding sequence for CBE target sites containing CAA, CAG, and CGA codons located within the canonical CBE editing window that could be converted to nonsense mutations following targeted C>T base substitutions.37,38 We identified two codons that met this criterion and designed five single guide RNAs (sgRNAs) targeting these two sites (Figure S1), each for a third-generation CBE consisting of the rat APOBEC1 catalytic domain and the uracil DNA glycosylase inhibitor protein fused to either the D10A SpCas9 nickase variant33 or the Cas9 D10A nickase variant from Staphylococcus aureus39 (SaCas9, specifically the KKH variant of SaCas940). These sgRNAs do not overlap with any common SOD1 mutation and thus target both wild-type and mutant SOD1, which could enable treatment of the disorder in a mutation-independent manner. Notably, no adverse effects were reported in ALS patients treated with an ASO targeting both SOD1 mRNAs,41 supporting the feasibility of a non-allele-specific strategy for treating SOD1-ALS.
We first tested whether the designed base editors could lower SOD1 in human embryonic kidney (HEK) 293T cells. According to qRT-PCR, the most efficient CBE targeted exon 1 of the SOD1 gene using the SpCas9 protein and reduced SOD1 mRNA by ∼50% compared to a non-targeted base editor (p < 0.05) (Figures 1A and 1B). Western blot corroborated these findings and revealed an ∼65% decrease in SOD1 protein at the same time point (p < 0.01) (Figure 1C). To confirm base editing, we deep sequenced the SOD1 gene from transfected HEK293T cells. We found that by 6 days post-transfection ∼19% of the analyzed reads contained the target C>T substitution (p < 0.001) (Figure 1D), as well as a C>T edit at a bystander cytosine that resulted in only a silent mutation (Figure S2). Deep sequencing further revealed that non-T product formation was less than 3.5% at the target cytosine (that is, cytosine 15) and less than 6.9% at the bystander cytosine (that is, cytosine 19) for this CBE (Figure S2).
Figure 1.
CBE-Mediated Base Editing Reduces SOD1 Gene Expression
(A) Schematic of the human SOD1 gene and the CBE 4 target site. Arrowhead indicates the target cytosine within the Gln codon. Unpaired bases in the target site indicate the protospacer. TSS, transcriptional start site. (B) Human SOD1 mRNA in HEK293T cells by quantitative qRT-PCR 12 days after transfection with CBEs. Data are normalized to a non-targeted CBE (n = 3). (C) (Top) Western blot of human SOD1 protein in cell lysate from HEK293T cells 12 days after transfection with CBE 4. (Bottom) Quantitation of western blot. SOD1 protein in each lane was normalized to GAPDH protein. Values were normalized to a non-targeted CBE (n = 3). (D) Plot showing the mean frequency change of the target cytosine in the SOD1 gene by deep sequencing in HEK293T cells 6 days after transfection with CBE 4 or a non-targeted control (NTG), n = 3. Error bars indicate SD. *p < 0.05, **p < 0.01, ***p < 0.001; (B and C) two-tailed unpaired t test; (D) one-tailed unpaired t test.
To evaluate CBE specificity, we identified 10 candidate off-target sites in the human genome that differed from the target site in SOD1 by up to four base mismatches and contained at least one cytosine within the CBE editing window (Figure S3A). Deep sequencing showed no appreciable increase in C>T base mutations within or near the editing window in 9 of the 10 off-target sites compared to control cells, with C>T editing (∼4%) detected only at a non-protein coding sequence that differed from the SOD1 target by three bases near the 5′ end of the protospacer (Figure S3B). Taken together, these results demonstrate that CRISPR base editors can be used to create an early stop codon in the SOD1 gene to lower its expression.
Establishment of a Split-Intein CBE Architecture that Enables Efficient Single-Base Editing
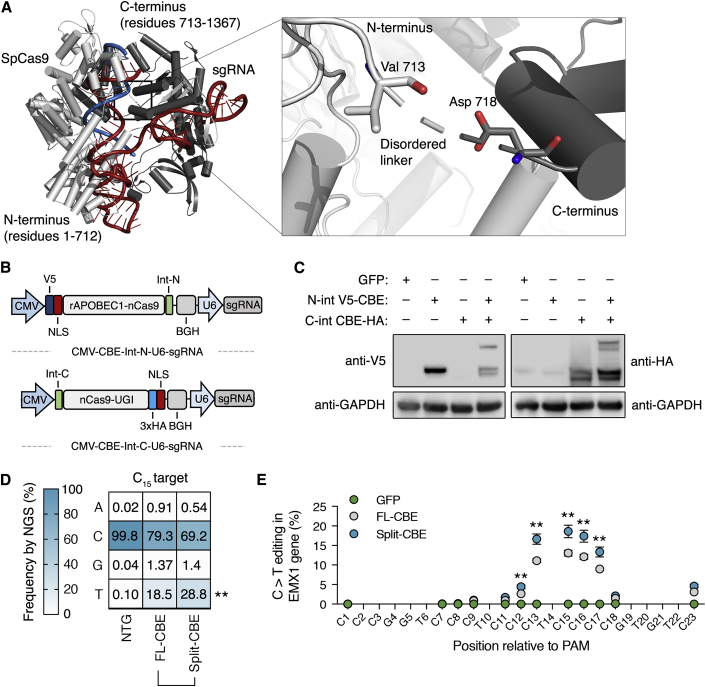
We next sought to determine whether base editors could reduce SOD1 in an animal model following in vivo delivery using an adeno-associated virus (AAV) vector. However, while AAVs are promising therapeutic gene delivery vehicles,42 particularly for many neurological disorders,43, 44, 45 they possess a limited carrying capacity that restricts their ability to deliver a full-length base editor.46 To overcome this limitation, we created a split-intein SpCas9-based CBE compatible with dual AAV particle delivery. Specifically, we fused N- and C-intein fragments from the DnaB protein of Rhodothermus marinus47 to a CBE that we split into two halves at Val 957, a residue located within a disordered linker connecting the α-helical recognition (REC) lobe and the nuclease (NUC) lobe of the SpCas9 protein (corresponding to residues 712–717)48 (Figures 2A and 2B). Following co-delivery, these two intein-containing halves are expected to participate in a protein trans-splicing reaction that will reassemble the full-length CBE. Importantly, unlike a split-intein-based approach that was used to deliver a SaCas9-based CBE,49 our strategy supports the delivery of base editors consisting of the SpCas9 protein, the most widely used Cas9 variant to date.
Figure 2.
Split-Intein CBEs Can Reconstitute and Edit the Human Genome
(A) Crystal structure of the SpCas9 protein complexed with sgRNA (red) and target DNA (blue). Inset shows the disordered linker that connects the REC lobe and the NUC lobe. Light gray indicates N-terminal SpCas9 domain; dark gray indicates C-terminal SpCas9 domain (PDB: 4OO8).48 (B) Schematic of the engineered split-intein CBE two-plasmid system. Abbreviations are as follows: CMV, cytomegalovirus promoter; NLS, nuclear localization signal; Int-N, N-terminal intein domain; Int-C, C-terminal intein domain: V5, V5 epitope tag; 3× HA, three tandem repeats of the human influenza hemagglutinin (HA) epitope tag. (C) Western blot of HEK293T cells transfected with the N-terminal split-intein CBE domain, the C-terminal, or both. The western blots were probed with anti-HA or anti-V5 antibodies to detect reconstitution of the full-length base editors. (D) Plot showing the mean frequency change of the target cytosine in the SOD1 gene by deep sequencing in HEK293T cells 6 days after transfection with the full-length CBE, the split-intein CBE, or a non-targeted control (NTG), n = 3. (E) Mean C>T conversion frequency of editing-window cytosines in the EMX1 gene by deep sequencing in HEK293T cells 6 days after transfection with full-length CBE or split-intein CBE (n = 3). Error bars indicate SD. **p < 0.01; (D and E) two-tailed unpaired t test.
According to western blot, co-expression of the split-intein SpCas9-CBE in HEK293T cells resulted in intein-mediated protein trans-splicing and reconstitution of the full-length base editor at efficiencies near 30% (Figure 2C). Moreover, we measured significantly increased editing using the split-intein CBE compared to the full-length base editor, as deep sequencing showed that ∼29% of the analyzed reads from HEK293T cells transfected with the spit-intein CBE had the target C>T edit in the SOD1 gene, whereas ∼19% of the analyzed reads from cells transfected with the full-length CBE had the target modification (p = 0.001) (Figure 2D). Furthermore, compared to the full-length enzyme, deep sequencing revealed no decrease in product purity at the target cytosine following editing by the split-intein CBE (p > 0.1 for each outcome) (Figures 2D and S4) and a small increase in C>T editing at only 1 of the 10 previously identified off-target sites in HEK293T cells (Figure S3B). These results thus indicate that split-intein-mediated protein reconstitution did not dramatically decrease CBE specificity.
To determine the generality of this architecture, we tested the ability of the split-intein SpCas9-CBE to induce exon skipping by mutating conserved splice acceptor sites in PIK3CA exon 5, RELA exon 7, and JAG1 exon 9 in HEK293T cells.50 RT-PCR showed that the split-intein CBE induced exon skipping ∼2- to 6-fold more efficiently than did the native CBE at each target (Figure S5). We next compared the editing window of the split-intein CBE to the full-length single-base editor. Using deep sequencing to measure C>T edits within a cytosine-rich target sequence in the EMX1 gene, we found that the split-intein CBE retained its canonical editing window and, compared to the full-length enzyme, more efficiently edited each cytosine within this window (p < 0.01 for each position) (Figure 2E). With the exception of a 0.014%–0.05% increase of an A>G substitution at cytosine 17, we saw no difference in the product purity for any edited cytosine using the split-intein CBE versus the full-length editor at the EMX1 target (p > 0.1 for these outcomes) (Figure S6). Altogether, these results demonstrate that split-intein CBEs can reconstitute and efficiently edit the human genome, including the human SOD1 gene. These results also show that split-intein CBEs have editing characteristics similar to the parental single-base editor.
Split-Intein CBEs Provide Therapeutic Benefit to a Mouse Model of ALS
We next evaluated whether the split-intein CBE could lower SOD1 and slow disease progression in the G93A-SOD1 mouse model of ALS. The G93A-SOD1 strain that we used in this study carries ∼25 copies of a ubiquitously expressed human SOD1G93A transgene51 and develops an especially aggressive neurodegenerative disorder that involves the accumulation of inclusions that are immunoreactive by SOD1 antibodies, in addition to progressive muscle atrophy, motor neuron loss, and the eventual denervation of neuromuscular junctions.51,52 This mouse model shows the first signs of disease typically around 94 days of age and succumbs to the disorder on average by 126 days of age.
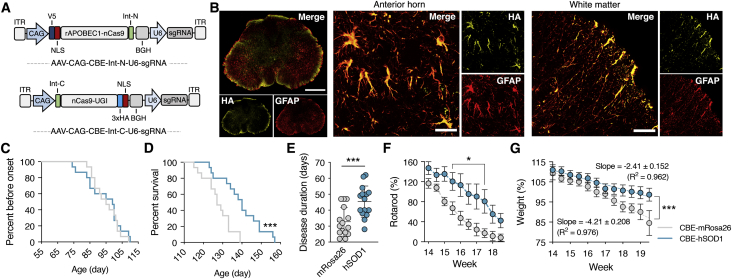
Astrocytes have been reported to be a determinant of disease progression in SOD1-linked ALS,6, 7, 8, 9, 10 as disrupting mutant SOD1 expression in these cells using Cre-mediated gene excision can sharply slow late-stage disease progression in a model of the disorder.9 We thus packaged the N- and C-terminal split-intein CBEs into AAV9, a naturally occurring AAV serotype that can efficiently transduce spinal cord astrocytes following intrathecal delivery,22,53,54 and injected 56- to 60-day-old G93A-SOD1 mice with 8 × 1010 particles each of an N- and C-terminal AAV9 vector encoding either the sgRNA targeting the human SOD1 gene (AAV9-CBE-human SOD1 [hSOD1]) or the mouse Rosa26 genomic locus (AAV9-CBE-Rosa26) into the lumbar CSF (Figure 3A). We note that no editing was expected in the mouse SOD1 gene due to a base mismatch in the protospacer adjacent motif (PAM) of the native mouse target sequence that would prevent Cas9 from binding.
Figure 3.
In Vivo Base Editing Improves Therapeutic Outcomes in the G93A-SOD1 Mouse Model of ALS
(A) Schematic of AAV vectors encoding the split-intein CBE. Abbreviations are as follows: ITR, inverted terminal repeat; CAG, cytomegalovirus early enhancer/chicken β-actin promoter; NLS, nuclear localization signal; Int-N, N-terminal intein domain; Int-C, C-terminal intein domain, V5, V5 epitope tag; 3× HA, three tandem repeats of the human influenza hemagglutinin (HA) epitope tag. (B) Representative immunofluorescence staining of the lumbar spinal cord 4 weeks after G93A-SOD1 mice were injected with 8 × 1010 particles of each AAV encoding the N- and C-terminal split-intein CBE. Scale bars, 500 μm, left; 30 μm, anterior horn; 50 μm, white matter. (C) Percent before disease onset, (D) percent survival, (E) disease duration, (F) rotarod, and (G) weight of G93A-SOD1 mice injected with 8 × 1010 particles of dual AAV encoding the hSOD1- or mRosa26-targeting N- and C-terminal split-intein CBE. (C–E and G) hSOD1 (n = 15) and mRosa26 (n = 15); (F) hSOD1 (n = 10) and mRosa26 (n = 10). (F and G) Mean rotarod and weights for each mouse were normalized to day 63 values for the same mice. (G) Linear regression analysis was used to calculate the rate of weight loss. Values indicate means and error bars indicate (E) SD or (F and G) SEM. *p < 0.05, **p < 0.01, ***p < 0.001; (C and D) log-rank Mantel-Cox test; (E) one-tailed unpaired t test; (F) two-way ANOVA followed by a Bonferroni post hoc test.
Immunofluorescence analysis revealed widespread CBE expression throughout the spinal cord by 4 weeks post-injection (Figure 3B). Specifically, we determined that ∼80% of the cells in the anterior horn expressing the reactive astrocyte marker glial fibrillary acidic protein (GFAP) were positive for the C-terminal CBE via its hemagglutinin (HA) epitope tag (Figure S7) and that ∼70% of the analyzed dual GFAP+ HA+ cells were immunoreactive for the V5 epitope encoded by the N-terminal CBE (Figure S8), indicating dual AAV transduction. Flow cytometry analysis of dissociated tissue sections further indicated that ∼9.5% of the analyzed spinal cord cells were HA+ (Figure S9).
Similar to a previous report showing that AAV9 injection to the CSF of the lumbar spine of G93A-SOD1 mice favors the transduction of astrocytes,22 we observed limited CBE expression in NeuN+ and Tuj1+ neurons, as well as Iba1+ microglia (Figure S10). In particular, we estimated that less than ∼10% of choline acetyltransferase (ChAT)-positive motor neurons expressed the C-terminal CBE. Notably, we observed no difference in the percentage of transduced GFAP+ cells between animals injected with AAV9-CBE-hSOD1 or AAV9-CBE-mRosa26 (Figure S7).
Consistent with the immunofluorescence findings showing minimal transduction of ChAT+ motor neurons and prior work demonstrating that mutant SOD1 expression in these cells is a driver behind onset and early disease progression,5 we observed no difference in disease onset, which we defined as the point when animals reached peak weight, in mice injected with AAV9-CBE-hSOD1 (93.5 ± 11.0 days) or AAV9-CBE-mRosa26 (94.9 ± 7.9 days; p > 0.7) (Figure 3C). However, compared to control animals, mice injected with AAV9-CBE-hSOD1 had an ∼11% increase in mean survival (hSOD1, 139.4 ± 10.8 days; mRosa26, 127 ± 7.2 days; p < 0.001) (Figure 3D) and an ∼39% increase in disease duration, which we calculated as the length of time between onset and end stage for each mouse (hSOD1, 45.4 ± 9.7 days; mRosa26, 32.7 ± 9.0 days; p < 0.001) (Figure 3E). Additionally, while we observed no difference in early disease for each cohort (a parameter previously defined9 as the time from onset to 10% weight loss; p > 0.1) (Figure S11A), we found that, compared to control animals, G93A-SOD1 mice injected with AAV9-CBE-hSOD1 had an ∼85% increase in the duration of late disease (from 10% weight loss to end stage; hSOD1, 16.6 ± 9.5 days; mRosa26, 8.9 ± 6.3 days; p = 0.01) (Figure S11B). Importantly, we observed that base editor-treated male and female G93A-SOD1 mice showed a similar increase in disease duration compared to control animals (Figure S12), indicating that the observed slowing in disease progression was not the result of sex differences.
Compared to control animals, G93A-SOD1 mice treated by AAV9-CBE-hSOD1 had improved motor function by rotarod (p = 0.01) (Figure 3F) and improved hindlimb grip strength after week 17, which marked the beginning of late disease for both treated and untreated animals (p < 0.05) (Figure S13). Linear regression analysis further showed that G93A-SOD1 mice injected with AAV9-CBE-hSOD1 lost weight ∼43% slower during the period lasting from onset (week 14) to the point at which all untreated animals succumbed to disease (hSOD1, −2.41 ± 0.152 weight % per measurement; mRosa26, −4.22 ± 0.208 weight % per measurement; p < 0.0001) (Figure 3G). Moreover, we found that from the onset of late-stage disease (week 17 for both groups) to end stage, mice treated by base editing lost weight ∼75% slower than did control animals (hSOD1, −1.203 ± 0.0.09 weight % per measurement; mRosa26: −5.14 ± 0.604 weight % per measurement; p < 0.001; Figure S14). Altogether, these results demonstrate that base editing can slow the rate with which neuromuscular function and weight decline in G93A-SOD1 mice.
CRISPR Base Editing Can Reduce Mutant SOD1 Reactive Inclusions in the Spinal Cord and Protect Motor Neurons and Neuromuscular Junctions
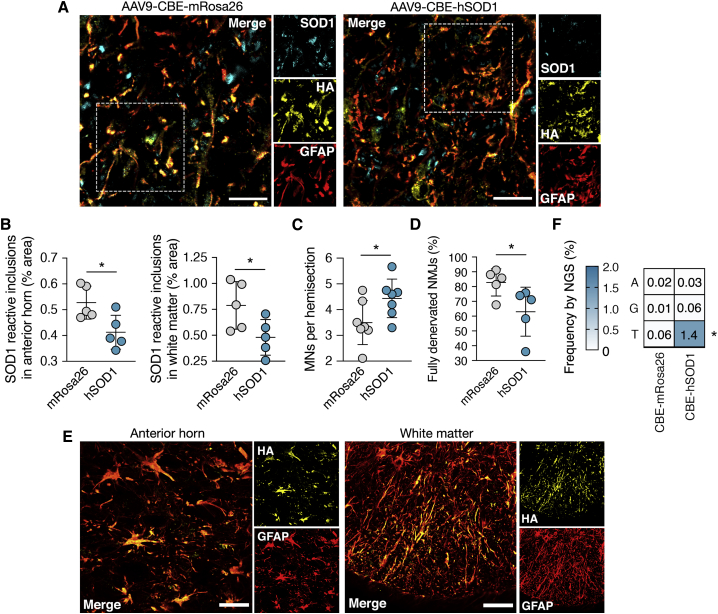
Disrupting mutant SOD1 expression in astrocytes using Cre-mediated gene excision has been reported to reduce the accumulation of SOD1 immunoreactive inclusions in the gray and white matter of the spinal cord.55 In order to determine whether base editing decreased mutant SOD1 in vivo, we used immunostaining to measure the total area occupied by inclusions that are immunoreactive for a SOD1 antibody in spinal cord sections from end-stage animals. Compared to control mice, we found that animals treated by AAV9-CBE-hSOD1 had ∼40% fewer SOD1 reactive inclusions in the ventral white matter (p < 0.05) and ∼28% fewer inclusions in the anterior gray column (p < 0.05) (Figures 4A and 4B), indicating that base editing reduced mutant SOD1 protein. Immunofluorescence analysis of the anterior horn further revealed that, compared to controls, GFAP+ cells from treated mice were markedly smaller and less abundant at end stage (Figure S15), suggesting that base editing decreased astrogliosis and hypertrophy. However, no difference in activated microglia was observed in treated versus control animals (Figure S16).
Figure 4.
Split-Intein CBE-Mediated Base Editing Reduces Mutant SOD1 Reactive Inclusions in the Spinal Cord and Protects Motor Neurons and Neuromuscular Junctions
(A) Representative immunofluorescence staining of end-stage spinal cord sections from G93A-SOD1 mice injected with 8 × 1010 particles each of dual AAV encoding the hSOD1- or mRosa26-targeting N- and C-terminal split-intein CBEs. Scale bars, 15 μm. (B) Quantitation of area occupied by SOD1 reactive inclusions in the anterior horn (left) and white matter (right) of spinal cord sections from end-stage treated (hSOD1) or untreated (mRosa26) G93A-SOD1 mice (n = 5). (C and D) Mean number of (C) ChAT+ motor neurons (MNs) per lumbar spinal cord hemisection and (D) fully denervated neuromuscular junctions (NMJs) from tibialis anterior muscle sections from end-stage treated or untreated G93A-SOD1 mice (C, n = 7; D, n = 7). (E) Representative immunofluorescence staining of spinal cord sections from end-stage G93A-SOD1 animals showing expression of the split-intein CBE. Scale bars, 30 μm, anterior horn; 50 μm, white matter. (F) Plot showing the mean frequency change of the target cytosine in the SOD1 gene in the cervical spinal cord from G93A-SOD1 mice. hSOD1 (n = 4); mRosa26 (n = 5). Values indicate means and error bars indicate SD. *p < 0.05; (B–D and F) one-tailed unpaired t test.
Compared to control mice, we found that animals treated by base editing had ∼27% more ChAT+ motor neurons in the lumbar spinal cord (p < 0.05) (Figures 4C and S17) and ∼23% fewer fully denervated neuromuscular junctions (p < 0.05) (Figures 4D and S18), the latter of which we quantified as the percentage of α-bungarotoxin-positive acetylcholine receptor (AChR) clusters that did not overlap with any synaptophysin-positive nerve terminals in tibialis anterior (TA) muscle sections. These results thus indicate that base editing improved motor neuron survival and partially preserved nerve terminals in G93A-SOD1 mice.
We also examined CBE expression in spinal cord sections from end-stage animals. Similar to the immunofluorescence findings from 4 weeks post-injection, we found that ∼75% of GFAP+ cells in the anterior horn of end-stage sections were HA+ (Figures 4E and S19), indicating that base editor expression was likely maintained throughout the disease. This finding is consistent with prior work demonstrating that spinal cord astrocytes in SOD1-ALS mice transduced by an AAV9 vector can survive for the duration of the disorder and continuously express transgenes encoded by the AAV episome.16
Finally, we evaluated base editing in vivo. Deep sequencing of the human SOD1G93A transgene amplified from dissected whole spinal cord tissue from end-stage animals, which included a mix of transduced and non-transduced cells, showed that on average ∼1.2% of the analyzed reads from mice injected with AAV9-CBE-hSOD1 had the target C>T edit versus ∼0.06% in control mice (p < 0.05) (Figure 4F). Given that we estimated that only ∼6.5% of spinal cord cells were transduced by both AAV vectors, these results indicate that editing likely occurred at an efficient rate within the cells that expressed the full-length CBE even in the context of the high transgene copy number background of the G93A-SOD1 mouse. To evaluate off-target editing in vivo, we deep sequenced eight candidate off-target sites in the mouse genome that differed by the on-target site by up to three base mismatches. No increase in C>T editing was seen at any of these sites in treated versus control animals (p > 0.1) (Figure S20).
In conclusion, we have developed a trans-splicing system that enables in vivo delivery by AAV of single-base editors consisting of the SpCas9 protein. Our results demonstrate that a split-intein CBE can be used to lower mutant SOD1 in the spinal cord and improve therapeutic outcomes in a mouse model of SOD1-linked ALS.
Discussion
CRISPR base editors can be used to introduce targeted C>T or A>G alterations in DNA in the absence of mutagenic double-strand DNA breaks33,34 and hold promise for treating genetic disorders, including SOD1-linked ALS, a fatal neurodegenerative disease characterized by motor neuron loss and progressive muscle atrophy that results from the toxic gain of function of mutant SOD1. In this study, we demonstrate that intrathecal delivery of dual AAV vector particles encoding a split-intein CBE engineered to trans-splice and induce a nonsense mutation into a hSOD1G93A transgene improved survival and slowed disease progression (particularly late disease) in the G93A-SOD1 mouse model of ALS. Base editing reduced the accumulation of SOD1 immunoreactive inclusions in certain areas of the spinal cord by up to 40% and decreased the rate of muscle atrophy and muscle denervation. This work thus establishes that CRISPR base editors can be used to treat a neurodegenerative disorder.
ALS is an adult-onset disease that most often affects individuals between 40 and 60 years of age.1 Given that many persons are diagnosed with ALS following the onset of initial symptoms and that mutant SOD1 expression in astrocytes is an established driver of disease progression,6, 7, 8, 9, 10 we employed a targeting strategy that would enable editing within these cells in order to slow the disease. Accordingly, we used AAV9, a naturally occurring AAV serotype recently approved by the US Food and Drug Administration (FDA) to treat spinal muscular atrophy43 and capable of efficiently transducing spinal cord astrocytes following an intrathecal injection,22,53,54 to deliver the split-intein CBE in vivo. In support of the rationale for this approach, a previous study reported that the selective excision of the mutant SOD1 gene in GFAP+ cells resulted in an ∼15% increase in survival and an ∼144% increase in the duration of the late-stage phase of disease compared to control mice.9 Although our studies were performed in a different model of SOD1-ALS, we observed a similar trend: in vivo base editing in G93A-SOD1 mice increased survival by ∼11% and increased the duration of late disease by ∼85%. Thus, this proof-of-concept study demonstrates the potential of CRISPR base editing technology to treat SOD1-ALS.
Given that the restrictive carrying capacity of AAV prevents the packaging of a full-length CRISPR base editor, we employed intein-mediated protein trans-splicing to reconstitute a SpCas9-based CBE following dual AAV particle delivery. Specifically, we inserted inteins from the DnaB protein of R. marinus into a disordered linker (residues 712–717) of the SpCas9 protein that connects its two distinct lobes, the REC lobe and the NUC lobe.48 Interestingly, despite a ∼30% reconstitution efficiency, we observed increased C>T editing using the split-intein CBE compared to the full-length single-base editor. While the exact reason for the increase in editing remains unknown, we suspect that the peptide sequence generated by the intein-mediated protein trans-splicing reaction may stabilize the disordered linker in the SpCas9 protein, which could increase its ability to bind or nick DNA. In addition to enhanced C>T editing, we observed no appreciable increase in off-target editing using the split-intein CBE and found that it possessed a nearly identical editing window to the parental editor, indicating that inserting the DnaB-based intein moiety into SpCas9 did not negatively impact DNA targeting or editing specificity. Thus, our results suggest that the split-intein SpCas9-based architecture described herein is broadly applicable and could be used to deliver other recently emerged Cas9-based technologies that exceed the carrying capacity of a single AAV vector particle.56 Of note, in addition to intein-mediated protein trans-splicing, which has been used to deliver an SaCas9-CBE to the liver49 and an SpCas9-ABE to cells in culture,57 viral vector trans-splicing, an approach that relies on intermolecular recombination or ligation of dual AAV vector genomes, has also been used to deliver an SpCas9-ABE to the muscle.58
Although they offer considerable advantages over CRISPR-Cas9 nucleases, recent reports have suggested that CBEs can generate off-target single-nucleotide variations in DNA59,60 and RNA.61,62 To this end, rat APOBEC1 catalytic domains with reduced RNA editing activity have been developed that could help alleviate these concerns.61 In particular, these recently described domains could be incorporated into the split-intein CBE architecture that we describe herein to further refine its specificity. Additionally, some of these same engineered domains were found to possess more restrictive editing widows than the native APOBEC1 domain, which in the future could help to reduce or eliminate the bystander editing that we and others have observed.
Strategies for reducing mutant SOD1 protein hold promise for treating SOD1-linked ALS. However, to date, more than 100 different mutations in the SOD1 gene have been linked to the disease. Although CRISPR base editors hold the potential to seamlessly correct certain disease-causing mutations, the genetic heterogeneity of SOD1-ALS poses a serious technical and economic challenge that could prevent treatment of the disease in a mutation-specific manner. To this end, we note that a non-allele-specific strategy could be used to treat SOD1-ALS, as a phase I clinical trial for an ASO designed to target both wild-type and mutant SOD1 mRNA is underway, underscoring the potential of such an approach. However, a gene knockout-and-replace therapy63,64 could be employed in place of a non-allele-specific strategy to overcome any toxicity that may result from disrupting the wild-type SOD1 gene within the spinal cord.
To analyze in vivo base editing, we deep sequenced genomic DNA harvested from dissected bulk spinal cord tissue, which we estimated consisted of ∼7% dual-transduced cells. This estimate was based on the fact that: (1) flow cytometry indicated that ∼9.5% of dissociated spinal cord cells were positive for the HA tag encoded by the C-terminal CBE, and (2) immunofluorescence staining of spinal cord sections indicated that ∼70% of dual GFAP+ and HA+ cells were also positive for the V5 epitope tag encoded by the N-terminal CBE. From the deep sequencing analysis of the bulk tissue, we found that ∼1.2% of all analyzed SOD1 reads contained the target C>T edit, suggesting an “effective editing rate” of ∼20% in the dual-transduced cells. Although purifying adult spinal cord astrocytes from whole tissue remains a challenging task, future studies involving the use of transgenic animals expressing a fluorescent reporter gene in the target cell type could enable the isolation of enough material by fluorescence-activated cell sorting (FACS) to analyze base editing outcomes in a highly enriched population of cells, which in turn could support a more detailed analysis of off-target editing in DNA and RNA. Additionally, while we observed no signs of discomfort or toxicity in animals treated by the CBE, further studies will also be necessary to fully characterize their safety and tolerability.
Animals treated by base editing had ∼40% fewer SOD1 reactive inclusions in the ventral white matter and ∼28% fewer inclusions in the anterior gray column. Although the exact identity of all the cells with decreased inclusions was not established, it is possible that disrupting mutant SOD1 expression in astrocytes, the primary cell type transduced by AAV9 in this study, led to a decrease in inclusions in neighboring cells. This observation is consistent with previous studies that demonstrated that disrupting mutant SOD1 in astrocytes alone using Cre-mediated gene excision reduced the accumulation of SOD1 inclusions in the spinal cord by 60%–90% in the G85R-SOD1 mouse model of ALS.55 Given the potential implications of this finding in the treatment of ALS, additional studies are needed to further explore this possibility.
Efficient gene delivery to the cells involved in SOD1-linked ALS is crucial to the success of a CRISPR-based treatment for the disorder, particularly for strategies that rely on dual AAV transduction. In the present study, we used AAV9 to deliver the split-intein CBE largely to spinal cord astrocytes, which are thought to contribute to motor neuron loss and influence the later stage of the disease.6, 7, 8, 9, 10 However, many other cell types influence SOD1-ALS, including motor neurons, which can determine disease onset and early disease progression,5 as well as microglia5,11 and certain oligodendrocyte progenitor cells.12 Thus, optimizing gene delivery65 to these and other cell populations involved in disease onset and progression will be important in advancing a gene therapy for the disorder to the clinic. Such vector optimization could also be used to boost dual transduction efficiency. Similarly, directed evolution or other protein engineering strategies could be employed to optimize the compatibility of the DnaB inteins with the CBE to improve its reconstitution efficiency and in vivo editing capabilities.
In conclusion, we have demonstrated that base editing can be used to treat a mouse model of ALS. Our split-intein approach for delivering SpCas9-CBEs through dual AAV vector particles could enable gene therapies for other neurological and neurodegenerative disorders.
Materials and Methods
Plasmid Construction
The plasmid encoding BE3 (pCMV-BE3) was a gift from David R. Liu (Addgene, 73021). For experiments performed in HEK293T cells, DNAs encoding (1) the N-split-intein CBE (that is, rAPOBEC1 with residues 2–712 of SpCas9 and the N-intein fragment from the DnaB protein of R. marinus) and (2) the C-split-intein CBE (that is, the C-intein fragment from the DnaB protein of R. marinus with residues 713–1371 of SpCas9, the uracil glycosylase inhibitor, and an SV40 nuclear localization signal sequence) were custom synthesized as gBlock gene fragments (Integrated DNA Technologies [IDT]) and cloned into pCMV-BE3 between the NotI and BspEI restriction sites for the C-intein and between the EcoRV and PmeI restriction sites for the N-intein by Gibson assembly using a reaction mix prepared by our laboratory, as described.66
The pAAV plasmids pAAV-CAG-N-Int-CBE-U6-sgRNA and pAAV-CAG-C-Int-CBE-U6-sgRNA were generated by cloning gBlock gene fragments encoding the DNA sequence above with a CAG promoter to drive expression of the split-intein CBE between the XbaI and NotI restriction sites of pX602 (a gift from Feng Zhang; Addgene, 107055) by Gibson assembly. The amino acid sequence of each protein is provided in Figure S21.
Oligonucleotides encoding sgRNA sequences were custom synthesized (IDT), phosphorylated by T4 polynucleotide kinase (New England Biolabs [NEB]) for 30 min at 37°C, and annealed by incubation at 95°C for 5 min followed by cooling on ice for 10 min. DNA duplexes were then ligated into the BbsI restriction sites of pSP-gRNA (Addgene, 47108)67 or the BsaI restriction sites of pAAV-CAG-N-Int-CBE-U6-sgRNA and pAAV-CAG-C-Int-CBE-U6-sgRNA using T4 ligase (NEB). Correct insertion of the sgRNA targeting sequence was verified by Sanger sequencing (Roy J. Carver Biotechnology Center, University of Illinois, Urbana, IL, USA).
Cell Culture and Transfection
HEK293T cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Life Technologies) and 1% (v/v) antibiotic-antimycotic (Anti-Anti; Life Technologies) in a humidified 5% CO2 atmosphere at 37°C. Transfections were performed in 24-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At the time of transfection, 1 μg of DNA was transfected per well. Transfection efficiency was routinely measured to be ∼80% by fluorescence microscopy according to EGFP expression.
qPCR
To analyze SOD1 expression, RNA was extracted using the RNeasy mini kit (QIAGEN). Then, 50 ng of total RNA was used for a reverse transcription reaction using a high-capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer’s instructions. 40 ng of cDNA template was then used per qPCR reaction in technical triplicates using the iTaq Universal SYBR Green Supermix (Bio-Rad). Results were normalized to GAPDH expression and the average fold change was calculated using the 2−ΔΔCT method.
To analyze exon skipping, RNA was harvested from cells using the RNeasy Plus mini kit (QIAGEN) and cDNA was synthesized using the qScript cDNA synthesis kit (Quanta Biosciences) using 0.5–1 μg of DNA with a cycling protocol recommended by the manufacturer. PCR was performed with 50 ng of cDNA in 25-μL reactions using the KAPA2G robust PCR kit (Kapa Biosystems, Roche) with cycling parameters recommended by the manufacturer. PCR products were visualized on a 2% agarose gel stained with ethidium bromide and visualized using a ChemiDoc-It2 (UVP).
Western Blot
Cells were lysed using NuPAGE LDS (lithium dodecyl sulfate) sample buffer (1×; Invitrogen) or by radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate, pH 8.0), and protein concentration was determined using the Pierce BCA (bicinchoninic acid) protein assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. 15 μg of protein was electrophoresed by SDS-PAGE and electrophoretically transferred onto a polyvinylidene fluoride (PVDF) or a nitrocellulose membrane in transfer buffer (20 mM Tris-HCl, 150 mM glycine, and 20% [v/v] methanol) for 1.5 h at 100 V. Membranes were blocked with 5% (v/v) blotting-grade blocker (Bio-Rad) or 5% (w/v) nonfat dry milk in Tris-buffered saline (TBS) (20 mM Tris-HCl, 150 mM NaCl, and 0.1%, pH 7.5) with 0.05% Tween 20 (TBS-T) for 1 h and then incubated with primary antibodies in blocking solution. The following primary antibodies were used: rabbit anti-hSOD1 (1:2,000; Cell Signaling Technology, 2770S), anti-HA (1:1,000; Cell Signaling Technology, 3724), anti-V5 (1:1,000; Cell Signaling Technology, 13202S), anti-GAPDH (1:1,000; Cell Signaling Technology, 2118S), or rabbit anti-β-actin (1:1,000; Cell Signaling Technology, 4970S). After the overnight incubation, membranes were washed three times with TBS-T and incubated with goat anti-rabbit horseradish peroxidase conjugate (1:4,000, Thermo Fisher Scientific, 65-6120) or goat anti-rabbit horseradish peroxidase conjugate (1:2,000; Cell Signaling Technology, 7074p2) in blocking solution for 1 h at room temperature. Membranes were washed three times with TBS-T and developed using a SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) or a Clarity western enhanced chemiluminescence (ECL) substrate (Bio-Rad) and visualized by automated chemiluminescence using the ChemiDoc XRS+ (Bio-Rad). Band intensity was quantitated using Image Lab software (Bio-Rad) or ImageJ and normalized to control protein in each lane.
AAV Vector Production
AAV vector was manufactured according to our protocols.68 HEK293T cells were seeded onto 15-cm plates at a density of 2.5 × 107 cells per plate in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) Anti-Anti. After 16 h, cells were transfected with 15 μg of pAAV-CAG-N-int-CBE-U6-sgRNA or pAAV-CAG-C-int-CBE-U6-sgRNA, 15 μg of pAAV9, and 15 μg of pHelper using 135 μL of polyethylamine (1 μg/μL). XmaI digest was used to confirm the integrity of the pAAV plasmids. Cells were harvested at 48 h after transfection by manual dissociation using a cell scraper and centrifuged at 4,000 × g for 5 min at room temperature. Cells were then resuspended in lysis buffer (50 mM Tris-HCl and 150 mM NaCl, pH 8.0) and freeze-thawed three times using liquid nitrogen and a 37°C water bath. Cell lysate was incubated with 10 U of Benzonase (Sigma-Aldrich) per 1 mL of cell lysate for 30 min at 37°C. We then centrifuged the lysate at 20,000 × g for 30 min at room temperature. The resulting supernatant was overlaid onto an iodixanol density gradient, and virus was isolated by ultracentrifugation. Following extraction, AAV was washed three times with 15 mL of PBS with 0.001% Tween 20 using an Ultra-15 centrifugal filter unit (Amicon) at 3,000 × g and concentrated to ∼100–150 μL. Virus was stored at 4°C and the genomic titer was determined by quantitative real-time PCR using SYBR Green (Sigma-Aldrich).
Injections
All animal procedures were approved by the Illinois Institutional Animal Care and Use Committee (IACUC) at the University of Illinois and conducted in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Eight-week-old G93A-SOD1 mice bred from male G93A-SOD1 mice (B6SJL-Tg(SOD1*G93A)1Gur/J; Jackson Laboratory, 002726) and female B6SJLF1/J mice (Jackson Laboratory, 100012) were injected with 8 × 1010 vector genomes of AAV9-CAG-N-Int-CBE-U6-sgRNA and AAV9-CAG-C-Int-CBE-U6-sgRNA in 10 μL of PBS with 0.001% Tween 20 into the mouse lumbar subarachnoid space between L5 and L6 vertebrate using a Hamilton syringe with a 22s gauge 2-inch needle. Animals were genotyped for the presence of the hSOD1G93A transgene by PCR using genomic DNA purified from an ear clip using the primers hSOD1 forward (5′-CATCAGCCCTAATCCATCTGA-3′) and hSOD1 reverse (5′-CGCGACTAACAATCAAAGTGA-3′). Treatment and control groups were sex balanced and litter matched.
Behavior
All measurements were performed by a blinded investigator. Starting 1 week after injections, mice were weighed biweekly and monitored for changes in physical appearance. Disease onset and late disease were retrospectively defined as the days at which animals reached peak weight and lost 10% of their peak weight, respectively. Motor coordination was measured biweekly using a Rotamex-5 rotarod (Columbus Instruments). Animals were placed onto an apparatus programmed to accelerate from 4 to 40 rpm in 180 s, and the latency to fall was recorded. Each session comprised three trials. Hindlimb strength was measured biweekly using a grip strength meter (Harvard Apparatus). Mice were scruffed to firmly latch onto a pull bar. Mice were then gently pulled in the opposite direction and the maximum force exerted prior to the release of the bar was recorded for each animal. Each session comprised three measurements. All data were normalized to the starting mouse weight at day 63 and the starting rotarod time and hindlimb grip strength at day 63. End stage was determined as the point when the animals could no longer turn themselves over within 10 s of being placed on their back, lost more than 20% of their peak weight, or had complete paralysis. Mice were provided with wet mashed food in their cages at the first sign of hindlimb paralysis and were monitored daily thereafter.
Immunohistochemistry
Mice were transcardially perfused with PBS, and spinal cords and TA muscles were post-fixed overnight with 4% paraformaldehyde in PBS at 4°C. Spinal cords and muscles were cut into 40-μm coronal and sagittal sections, respectively, using a Leica CM3050 S cryostat. The sections were then transferred to a 48-well plate and stored in cryoprotectant at −20°C. Spinal cord and muscle sections were washed three times with PBS and incubated with blocking solution (PBS with 10% [v/v] donkey serum [Abcam] and 1% Triton X-100) for 2 h at room temperature and stained with primary antibodies in blocking solution for 72 h at 4°C. Spinal cord sections were then washed three times with PBS and incubated with secondary antibodies for 2 h at room temperature and washed three times with PBS. The same steps were carried out with the muscle sections, but they were instead incubated with Alexa Fluor 647-conjugated α-bungarotoxin with secondary antibodies. Stained sections were then mounted onto slides using VectaShield Hardset antifade mounting medium (Vector Laboratories) and visualized using a Leica TCS SP8 confocal microscope and Zeiss Observer Z1 microscope (Beckman Institute Imaging Technology Microscopy Suite, University of Illinois, Urbana, IL, USA). All image analyses were performed using ImageJ software.
The following primary antibodies were used: rabbit anti-hSOD1 (1:200; Cell Signaling Technology, 2770S), goat anti-ChAT (1:25; EMD Millipore, AB144P), goat anti-HA (1:250; GenScript, A00168), rabbit anti-HA (1:500; Cell Signaling Technology, 3724S), chicken anti-HA (1:500; Abcam, ab9111), rabbit anti-FLAG (1:500; Cell Signaling Technology, 14793S), rabbit anti-NeuN (1:500; Abcam, ab177487), rabbit anti-IbaI (1:500; Wako Pure Chemicals Industries, 019-19741), rat anti-Mac2 (1:500; Cedarlane, CL8942AP), mouse anti-β3-tubulin (1:1,000; Sigma-Aldrich, T8578), chicken anti-GFAP (1:1,000; Abcam, ab4674), and rabbit anti-synaptophysin 1 (1:500; Synaptic Systems, 101 002).
The following secondary antibodies were used: donkey anti-rabbit Cy3 (Jackson ImmunoResearch, 711-165-152), donkey anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch, 711-545-152), donkey anti-goat Cy3 (Jackson ImmunoResearch, 705-165-147), donkey anti-goat Alexa Fluor 488 (Thermo Fisher Scientific, A-11055), donkey anti-goat Alexa Fluor 647 (Jackson ImmunoResearch, 705-605-147), donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, 715-545-150), donkey anti-chicken Alexa Fluor 647 (Jackson ImmunoResearch, 703-605-155), donkey anti-chicken Cy3 (Jackson ImmunoResearch, 703-165-155), donkey anti-rat Cy3 (Jackson ImmunoResearch, 712-165-153), and donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, 715-545-150). Postsynaptic AChRs were detected using Alexa Fluor 647-conjugated α-bungarotoxin (1:500; Thermo Fisher Scientific, B35450).
To quantify the area occupied by SOD1 immunoreactive inclusions in spinal cord sections, an area within the ventral horn or the anterior white matter was highlighted in ImageJ and a pixel intensity threshold was applied to the region to quantify the area occupied by SOD1 immunoreactive inclusions. The total surface area occupied by the inclusions was normalized to the total area analyzed. Our analyses were performed exclusively in the anterior horn and the region of the white matter ventral to the anterior horn, and not throughout the entire coronal section. On average, 50 hemisections per mouse were analyzed. All measurements were performed by a blinded investigator.
Deep Sequencing
Amplicons for deep sequencing were generated by PCR using a KAPA2G robust PCR kit, as described above. Following validation of the quality of PCR products by gel electrophoresis, the PCR products were isolated using Agencourt AMPure XP PCR purification beads (Beckman Coulter). Indexed amplicon libraries for all samples were prepared using the KAPA HyperPrep kit (Kapa Biosystems, Roche) without shearing. Indexed HTS amplicons were then generated using a Nextera XT DNA library prep kit (Illumina) and quantitated by qPCR. Libraries were sequenced with a MiSeq nano flow cell for 251 cycles from each end of the fragment using a MiSeq reagent kit v2 (500 cycles). FASTQ files were created and demultiplexed using bcl2fastq v2.17.1.14 conversion software (Illumina). Deep sequencing was performed by the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois, Urbana, IL, USA.
DNA sequencing reads were demultiplexed by PCR primer sequences. Reads with Phred scores below 26 were then removed. Reads were then aligned to the human (GRCh38) or mouse (GRCm38) genome using Bowtie2 (with “-local” setting).69 SAMtools70 was used to sort and index aligned reads, and the Integrative Genomics Viewer (IGV)71 was used to for per-base variant calling using igvtools count function with window size set to 1, minimum mapping quality set to 30, and the “-bases” setting on.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8. SOD1 mRNA and protein were compared using an unpaired two-way t test. Disease onset, late disease onset, and survival Kaplan-Meier analyses were analyzed using the log-rank Mantel-Cox test. Rotarod and grip strength were compared using a two-way ANOVA followed by a Bonferroni post hoc test. Weight loss was analyzed using a linear regression analysis. SOD1 reactive inclusions, motor neuron survival, neuromuscular junction survival, and deep sequencing data were compared using a one-tailed unpaired t test.
Author Contributions
T.G. and P.P.-P. conceived of the study and designed experiments; A.K.B. designed sgRNAs and screened CBEs by qRT-PCR and western blot; M.G. and J.W. analyzed full-length CBEs by deep sequencing; M.G, J.W., and P.P.-P. conceived of and designed the split base editors; W.S.W. cloned the split base editors; W.S.W. and J.W. performed western blots to analyze split base editor reconstitution; M.G. and J.W. performed deep sequencing to quantify editing by the split base editors; M.G. performed studies to characterize the editing properties of the split base editors; M.G. performed exon skipping studies; C.K.W.L., A.K.B., J.E.P., and M.A.Z.C. packaged AAV vectors; C.K.W.L., A.K.B., and J.E.P. bred animals; A.K.B. and C.K.W.L. performed injections; C.K.W.L. and J.E.P. performed behavior experiments; C.K.W.L., J.E.P., and M.A.Z.C. performed immunohistochemistry; M.G. performed deep sequencing on spinal cord tissue; T.G. and C.K.W.L. prepared the figures and performed statistical analyses; and C.K.W.L., M.G, P.P.-P. and T.G wrote the manuscript.
Conflicts of Interest
J.W., M.G., W.W. and P.P.-P. are named inventors on patent applications related to split base editors. The remaining authors declare no competing interests.
Acknowledgments
We thank P. Jensen and S. Sirk for helpful discussion. C.K.W.L. was supported by a Roy J. Carver Fellowship in Engineering. M.G. was supported by a National Science Foundation Graduate Research Fellowship; M.A.Z.C. was supported by a University of Illinois Aspire Fellowship. This work was supported by the Muscular Dystrophy Association, United States (MDA602798 to T.G. and P.P.-P.), the Judith and Jean Pape Adams Foundation, United States (to T.G.), the American Heart Association, United States (17SDG33650087 to P.P.-P.), and by the National Institutes of Health, United States (R01GM127497 to P.P.-P.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.01.005.
Contributor Information
Pablo Perez-Pinera, Email: pablo@illinois.edu.
Thomas Gaj, Email: gaj@illinois.edu.
Supplemental Information
References
- 1.Rowland L.P., Shneider N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Bensimon G., Lacomblez L., Meininger V., ALS/Riluzole Study Group A controlled trial of riluzole in amyotrophic lateral sclerosis. N. Engl. J. Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 3.Abe K., Aoki M., Tsuji S., Itoyama Y., Sobue G., Togo M., Writing Group. Edaravone (MCI-186) ALS 19 Study Group Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16:505–512. doi: 10.1016/S1474-4422(17)30115-1. [DOI] [PubMed] [Google Scholar]
- 4.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Boillée S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 6.Haidet-Phillips A.M., Hester M.E., Miranda C.J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M.J., Foust K.D. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Song S., Miranda C.J., Braun L., Meyer K., Frakes A.E., Ferraiuolo L., Likhite S., Bevan A.K., Foust K.D., McConnell M.J. Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat. Med. 2016;22:397–403. doi: 10.1038/nm.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Kang S.H., Li Y., Fukaya M., Lorenzini I., Cleveland D.W., Ostrow L.W., Rothstein J.D., Bergles D.E. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat. Neurosci. 2013;16:571–579. doi: 10.1038/nn.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nizzardo M., Simone C., Rizzo F., Ulzi G., Ramirez A., Rizzuti M., Bordoni A., Bucchia M., Gatti S., Bresolin N. Morpholino-mediated SOD1 reduction ameliorates an amyotrophic lateral sclerosis disease phenotype. Sci. Rep. 2016;6:21301. doi: 10.1038/srep21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raoul C., Abbas-Terki T., Bensadoun J.C., Guillot S., Haase G., Szulc J., Henderson C.E., Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 16.Foust K.D., Salazar D.L., Likhite S., Ferraiuolo L., Ditsworth D., Ilieva H., Meyer K., Schmelzer L., Braun L., Cleveland D.W., Kaspar B.K. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 2013;21:2148–2159. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller T.M., Kaspar B.K., Kops G.J., Yamanaka K., Christian L.J., Gage F.H., Cleveland D.W. Virus-delivered small RNA silencing sustains strength in amyotrophic lateral sclerosis. Ann. Neurol. 2005;57:773–776. doi: 10.1002/ana.20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirren E., Aebischer J., Rochat C., Towne C., Schneider B.L., Aebischer P. SOD1 silencing in motoneurons or glia rescues neuromuscular function in ALS mice. Ann. Clin. Transl. Neurol. 2015;2:167–184. doi: 10.1002/acn3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borel F., Gernoux G., Cardozo B., Metterville J.P., Toro Cabrera G.C., Song L., Su Q., Gao G.P., Elmallah M.K., Brown R.H., Jr., Mueller C. Therapeutic rAAVrh10 mediated SOD1 silencing in adult SOD1G93A mice and nonhuman primates. Hum. Gene Ther. 2016;27:19–31. doi: 10.1089/hum.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoica L., Todeasa S.H., Cabrera G.T., Salameh J.S., ElMallah M.K., Mueller C., Brown R.H., Jr., Sena-Esteves M. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann. Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci. 2014;34:15587–15600. doi: 10.1523/JNEUROSCI.2037-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Yang B., Qiu L., Yang C., Kramer J., Su Q., Guo Y., Brown R.H., Jr., Gao G., Xu Z. Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum. Mol. Genet. 2014;23:668–681. doi: 10.1093/hmg/ddt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biferi M.G., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Bos C., Voit T., Ferry A., Barkats M. A new AAV10-U7-mediated gene therapy prolongs survival and restores function in an ALS mouse model. Mol. Ther. 2017;25:2038–2052. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iannitti T., Scarrott J.M., Likhite S., Coldicott I.R.P., Lewis K.E., Heath P.R., Higginbottom A., Myszczynska M.A., Milo M., Hautbergue G.M. Translating SOD1 gene silencing toward the clinic: a highly efficacious, off-target-free, and biomarker-supported strategy for fALS. Mol. Ther. Nucleic Acids. 2018;12:75–88. doi: 10.1016/j.omtn.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaj T., Ojala D.S., Ekman F.K., Byrne L.C., Limsirichai P., Schaffer D.V. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 2017;3 doi: 10.1126/sciadv.aar3952. eaar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mou H., Smith J.L., Peng L., Yin H., Moore J., Zhang X.O., Song C.Q., Sheel A., Wu Q., Ozata D.M. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017;18:108. doi: 10.1186/s13059-017-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyama T., Wilson D.M., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst.) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh W.H., Chiang H., Rees H.A., Edge A.S.B., Liu D.R. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 2018;9:2184. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuscu C., Parlak M., Tufan T., Yang J., Szlachta K., Wei X., Mammadov R., Adli M. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 38.Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., Ciccia A. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell. 2017;67:1068–1079.e4. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller T.M., Pestronk A., David W., Rothstein J., Simpson E., Appel S.H., Andres P.L., Mahoney K., Allred P., Alexander K. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.High K.A., Roncarolo M.G. Gene therapy. N. Engl. J. Med. 2019;381:455–464. doi: 10.1056/NEJMra1706910. [DOI] [PubMed] [Google Scholar]
- 43.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 44.Borel F., Gernoux G., Sun H., Stock R., Blackwood M., Brown R.H., Jr., Mueller C. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci. Transl. Med. 2018;10:eaau6414. doi: 10.1126/scitranslmed.aau6414. [DOI] [PubMed] [Google Scholar]
- 45.Hudry E., Vandenberghe L.H. Therapeutic AAV gene transfer to the nervous system: a clinical reality. Neuron. 2019;102:263. doi: 10.1016/j.neuron.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Gaj T., Epstein B.E., Schaffer D.V. Genome engineering using adeno-associated virus: basic and clinical research applications. Mol. Ther. 2016;24:458–464. doi: 10.1038/mt.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X.Q., Hu Z. A DnaB intein in Rhodothermus marinus: indication of recent intein homing across remotely related organisms. Proc. Natl. Acad. Sci. USA. 1997;94:7851–7856. doi: 10.1073/pnas.94.15.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 50.Gapinske M., Luu A., Winter J., Woods W.S., Kostan K.A., Shiva N., Song J.S., Perez-Pinera P. CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018;19:107. doi: 10.1186/s13059-018-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 52.Vinsant S., Mansfield C., Jimenez-Moreno R., Del Gaizo Moore V., Yoshikawa M., Hampton T.G., Prevette D., Caress J., Oppenheim R.W., Milligan C. Characterization of early pathogenesis in the SOD1G93A mouse model of ALS: part II, results and discussion. Brain Behav. 2013;3:431–457. doi: 10.1002/brb3.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bey K., Ciron C., Dubreil L., Deniaud J., Ledevin M., Cristini J., Blouin V., Aubourg P., Colle M.A. Efficient CNS targeting in adult mice by intrathecal infusion of single-stranded AAV9-GFP for gene therapy of neurological disorders. Gene Ther. 2017;24:325–332. doi: 10.1038/gt.2017.18. [DOI] [PubMed] [Google Scholar]
- 54.Gong Y., Berenson A., Laheji F., Gao G., Wang D., Ng C., Volak A., Kok R., Kreouzis V., Dijkstra I.M. Intrathecal adeno-associated viral vector-mediated gene delivery for adrenomyeloneuropathy. Hum. Gene Ther. 2019;30:544–555. doi: 10.1089/hum.2018.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Gutmann D.H., Roos R.P. Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum. Mol. Genet. 2011;20:286–293. doi: 10.1093/hmg/ddq463. [DOI] [PubMed] [Google Scholar]
- 56.Brooks A.K., Gaj T. Innovations in CRISPR technology. Curr. Opin. Biotechnol. 2018;52:95–101. doi: 10.1016/j.copbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Winter J., Luu A., Gapinske M., Manandhar S., Shirguppe S., Woods W.S., Song J.S., Perez-Pinera P. Targeted exon skipping with AAV-mediated split adenine base editors. Cell Discov. 2019;5:41. doi: 10.1038/s41421-019-0109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryu S.M., Koo T., Kim K., Lim K., Baek G., Kim S.T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 59.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 60.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 62.Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chadderton N., Millington-Ward S., Palfi A., O’Reilly M., Tuohy G., Humphries M.M., Li T., Humphries P., Kenna P.F., Farrar G.J. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol. Ther. 2009;17:593–599. doi: 10.1038/mt.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cideciyan A.V., Sudharsan R., Dufour V.L., Massengill M.T., Iwabe S., Swider M., Lisi B., Sumaroka A., Marinho L.F., Appelbaum T. Mutation-independent rhodopsin gene therapy by knockdown and replacement with a single AAV vector. Proc. Natl. Acad. Sci. USA. 2018;115:E8547–E8556. doi: 10.1073/pnas.1805055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaj T., Schaffer D.V. Adeno-associated virus-mediated delivery of CRISPR–Cas systems for genome engineering in mammalian cells. Cold Spring Harb. Protoc. 2016. 2016 doi: 10.1101/pdb.prot086868. prot086868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.