Abstract
Aneuploidy (i.e., abnormal chromosome number) is the leading cause of miscarriage and congenital defects in humans. Moreover, aneuploidy is ubiquitous in cancer. The deleterious phenotypes associated with aneuploidy are likely a result of the imbalance in the levels of gene products derived from the additional chromosome(s). Here, we summarize the current knowledge on how the presence of extra chromosomes impacts gene expression. We describe studies that have found a strict correlation between gene dosage and transcript levels as wells as studies that have found a less stringent correlation, hinting at the possible existence of dosage compensation mechanisms. We conclude by peering into the epigenetic changes found in aneuploid cells and outlining current knowledge gaps and potential areas of future investigation.
Keywords: : aneuploidy, chromosome, dosage compensation, gene expression
Aneuploidy: why do we care?
Aneuploidy is the condition of a cell or organism possessing an abnormal chromosome number (see Table 1 for a glossary of terms used in this section and throughout the article). In humans, aneuploidy is recognized as the leading cause of miscarriage and, in the rare cases in which it is compatible with live birth (e.g., trisomy 21 or mosaic aneuploidies), it results in severe congenital defects [1]. Aneuploidy is also a hallmark of cancer, and cancer cells frequently display complex karyotypes with chromosome numbers well above the normal, diploid number [2,3]. It is worth noting that monosomies (with the exception of the X chromosome) are never observed in humans and also very rarely observed in miscarriages [4], presumably because this condition is not compatible with cell survival or implantation of the fertilized egg. Similarly, hyperdiploidy (higher than normal chromosome number) is more frequently observed in cancer cells than hypodiploidy (lower than normal chromosome number) [2,3]. Thus, in most cases in which aneuploidy occurs, cells will possess a number of chromosomes that is above the norm. This invokes the question as to whether dosage compensation mechanisms (i.e., mechanisms by which the expression of certain genes is modulated to compensate for differences in gene dosage) come into play when extra chromosomes are present due to aneuploidy.
Table 1. . Glossary of important terms used in this article.
| Term | Definition/description |
|---|---|
| Diploid | Possessing two sets of chromosomes |
| Haploid | Possessing one set of chromosomes |
| Euploid | Possessing a correct number of chromosomes |
| Aneuploid | Possessing an incorrect number of chromosomes |
| Hyperdiploid | Possessing a number of chromosomes higher than normal |
| Hypodiploid | Possessing a number of chromosomes lower than normal |
| Trisomy | Three copies of a given chromosome in a diploid background |
| Monosomy | One copy of a given chromosome in a diploid background |
| Disomy | Two copies of a chromosome in a haploid background |
| Karyotype | Characteristic chromosome complement of a eukaryotic organism |
Experimental models of aneuploidy: correlation between gene/chromosome copy number & gene expression levels
The path that leads from gene to final product can be subject to regulation at many levels, but in this review we will mainly focus on transcriptional regulation of gene expression. Numerous studies over the past decade have used experimental models to investigate the effects of aneuploidy on gene expression. One such model is the budding yeast, where haploid strains carrying single or double disomies can be generated by various methods [5,6]. In general, these budding yeast models have shown that the RNA corresponding to the genes on the aneuploid chromosome of disomic budding yeast strains is proportionally increased compared with haploid strains (Figure 1A) [5,6]. These findings are consistent with a previous study showing that gene expression profiling successfully identified aneuploid budding yeast strains [7], indicating that there is good correlation between gene/chromosome copy number and gene expression levels. Gene expression analysis of trisomic mouse embryonic fibroblasts [8] is consistent with the findings in aneuploid budding yeast. Indeed, the expression of genes on the trisomic chromosome was found to be about 50% higher than the expression of genes from the same chromosome in diploid cells [8]. Finally, a study performed in human cells carrying defined aneuploidies found that transcript levels generally scale up with chromosome number [9], which is consistent with the observation that there is a global upregulation of genes on chromosome 21 in Down syndrome fetuses [10]. In addition, many studies have reported a direct correlation between copy number variations and changes in gene expression in cancer cells [11–17].
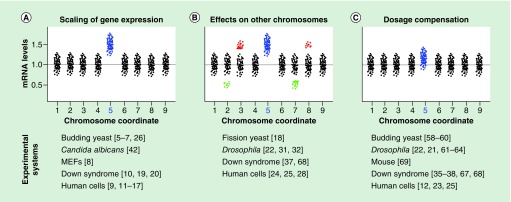
Figure 1. . Possible patterns of aneuploidy-dependent changes in gene expression.
The diagrams show a number of different scenarios that have been reported for how aneuploidy impacts gene expression. The examples illustrated here depict such scenarios in hypothetical diploid cells with a haploid chromosome number of 9 and carrying an extra copy of chromosome 5 (highlighted in blue). (A) Many studies have found that acquisition of an extra chromosome results in a proportional increase in the expression of genes carried on that chromosome. This scaling of gene expression may affect all the genes (as in the example shown here) or only a subset of genes on the aneuploid chromosome. (B) Other studies have reported that, in addition to an increase in the expression of genes carried by the extra chromosome, aneuploidy can also perturb the expression levels of genes on other chromosomes. Both overexpression (red) and underexpression (green) of genes on other chromosomes have been reported. (C) A number of studies have also reported some kind of dosage compensation, by which the genes on the aneuploid chromosome are not expressed at the expected levels based on gene dosage. It should be noted that dosage compensation in response to aneuploidy has been reported to occur for variable numbers of genes (from a few genes to the majority of genes on the aneuploid chromosome), depending on context. Moreover, the degree of compensation may vary. Finally, dosage compensation may occur simultaneously with effects on other chromosomes. The bottom half of the figure reports lists of organisms in which each of the different scenarios have been observed (often in combination with other effects) and corresponding key references. The references listed for ‘Down syndrome’ include studies in both human and mouse models.
However, many other studies, using a variety of experimental models, have reported that certain genes on the aneuploid chromosomes display expression levels similar to those observed in the euploid condition ([18–23], see next section for a detailed discussion) and that aneuploidy can also influence the expression of genes on other chromosomes [24–28], as already suggested by studies in aneuploid plants nearly 100 years ago [29]. The effect of aneuploidy on the expression of genes on other chromosomes (Figure 1B) may reflect the aneuploidy-induced altered expression of genes whose products can control gene expression broadly [30]. For instance, the aneuploidy-linked genes could be transcriptional regulators or genes responsible for controlling chromatin state (e.g., DNA methylases or histone acetylases/deacetylases). Evidence in support of this has emerged from studies in Drosophila melanogaster that reported a phenomenon, named the ‘inverse effect’, in which aneuploidy for a certain chromosome results in reduced expression of genes on other chromosomes [21,31,32], although in some of these studies expression of only a limited number of genes from the nonaneuploid chromosomes was analyzed and the analysis was not performed at the transcript level. Further insight on the effects of aneuploidy on chromosomes other than the aneuploid one comes from studies in mouse models of Down syndrome, in which the mice carry one of two possible segmental aneuploidies that are syntenic with human chromosome 21. These models have revealed that trisomy 21 (cause of Down syndrome) results, as expected, in overexpression (compared with the level in the euploid state) of the genes on the aneuploid chromosome [19,20]. However, the overexpressed genes include transcription factors and methylation pathway genes, which in turn can result in altered expression of or epigenetic changes in downstream target genes on various chromosomes [19,33–39]. Other studies have highlighted the upregulation of certain pathways, regardless of the aneuploidy. Specifically, upregulation of genes that are part of stress response pathways, including proteotoxic stress, were reported for a number of experimental models of aneuploidy [8,40,41]. However, this may be linked to a physiological response to unbalanced gene dosage.
Additional insight into the link between aneuploidy and gene expression can be gathered from experimental setups in which aneuploidy emerges as a result of experimentally imposed selective pressure. In these contexts, aneuploidy emerges because of the need to increase the levels of certain gene products that will confer resistance/promote survival under the selective conditions in question. For instance, aneuploidy was found to be a frequent adaptation of Candida albicans to the antifungal drug fluconazole [42,43]. The aneuploidy appeared to confer resistance by promoting the overexpression of the fluconazole target and of a transcriptional regulator of efflux pump genes [44]. Other studies have also reported the emergence of aneuploidy in yeast strains in response to a number of genetic or environmental perturbations [26,45–48]. Some examples of similar phenomena have been reported in human cells. For instance, human colonic epithelial cells cultured in low serum conditions acquired an extra copy of chromosome 7 and this adaptation was attributed to the overexpression of the EGF receptor, whose gene maps on chromosome 7 [49]. Trisomy 7 was also reported to emerge in human neural progenitor cells upon EGF withdrawal [50]. If aneuploidy-dependent adaptation is driven by the overexpression of a single or a few genes, then one would expect the expression of the remaining genes on that chromosome to be downregulated in order to mitigate the imbalance that would result from overexpressing the hundreds of genes carried by the aneuploid chromosome. Although this question has not been directly investigated, some information is available. For instance, transcriptomic analysis indicated that in Candida albicans with extra copies of chromosome 5L, most genes on the extra chromosome 5L segment were expressed at the levels expected based on copy number [42]. Similarly, yeast strains that acquired aneuploidy as an adaptation to various genetic perturbations displayed a proportional increase in expression for genes along the entire aneuploid chromosome, although a few genes were found to deviate from the average overexpression levels [26,46]. Thus, it appears that, when aneuploidy emerges as a result of experimentally imposed selection, the expression of both beneficial/driver genes and passenger genes is upregulated and no evidence of downregulation of passenger genes has been reported so far.
In summary, using experimental models of aneuploidy, a number of studies have shown that generally gene expression scales with gene/chromosome copy number (Figure 1A). Similar observations were reported for yeast strains that acquired aneuploidy as a result of experimentally imposed stresses or genetic manipulations. However, many other studies have revealed that the correlation between gene/chromosome copy number and gene expression is not always so direct and various degrees of variations in gene expression have been reported. This will be discussed in the next section.
Aneuploidy & dosage compensation: when gene expression does not scale with chromosome copy number
We will start this section by describing dosage compensation, a mechanism by which the expression of certain genes is modulated to compensate for differences in gene dosage. The classical example of dosage compensation is that observed in organisms with XY sex determination systems. In such organisms, females carry twice as many X chromosomes, and hence twice as many X-linked genes, as males. Dosage compensation mechanisms establish a balance between males and females in the expression of X-linked genes [51–53]. This is thought to be necessary because the X chromosome in such organisms is typically a rather large chromosome, with a relatively large number of genes, not all of which are responsible for sex determination [51,52]. Conversely, the Y chromosome is typically small, largely heterochromatic, and carries genes that are mainly involved in determination of primary and secondary sex characters [51,52]. Three major mechanisms of sex chromosome dosage compensation have been studied in detail and these include: inactivation of one of the X chromosomes in female embryos, which is the mechanism found in mammals [54]; doubling of transcription of genes on the single male X chromosome, which is the mechanism acting in D. melanogaster [55]; halving the expression of genes on the two X chromosomes in hermaphrodites, which is the mechanism observed in Caenorhabditis elegans [53,56], where individuals exist as either hermaphrodites (XX) or males (X0). The remainder of this section will discuss whether similar dosage compensation mechanisms can come into play to reduce the expression of genes on extra chromosomes found in aneuploid cells/organisms.
Like sex chromosome systems, aneuploidy creates a situation in which the genes carried by certain chromosomes are present in higher than normal doses. In the studies described in the previous section, copy number and gene expression levels were reported to be generally well correlated; nevertheless, some genes appeared to be compensated (i.e., the amount of transcripts was lower than what would be expected based on copy number) [18–20,23]. In fact, in certain experimental models, this dosage compensation phenomenon appears to be broad and affect a large fraction of the aneuploid gene content (Figure 1C). For instance, despite some controversy still exists [57], recent studies have highlighted that partial dosage compensation may occur in wild aneuploid yeast strains. In one study, 11–36% of genes on the extra chromosome were found to exhibit lower than expected levels of expression [58,59], whereas another study reported that only approximately 25% of genes with increased copy number showed a corresponding increase in mRNA levels [60]. Moreover, analysis of certain widely used D. melanogaster cell lines (e.g., S2 and Kc167) that display high levels of aneuploidy revealed that some degree of dosage compensation can be observed at both the transcriptional and the translational levels [61,62]. Dosage compensation was also observed in Drosophila strains carrying specific partial or whole-chromosome aneuploidies [21,22,63]. For instance, the levels of X-linked transcripts in triple X Drosophila metafemales were found to be similar to the levels in normal XX females [21]. Similarly, strains carrying partial or whole-chromosomes aneuploidies for autosomes displayed buffering of gene expression so that the transcript levels deviated from those expected based on gene dosage and were closer to transcript levels in wild-type animals [63]. For aneuploidies affecting the 4th chromosome, gene expression buffering was shown to depend on a chromatin-binding protein known as Painting of Fourth or POF [63,64], whereas in other cases this dosage compensation is believed to result from overexpression of negative regulators on the aneuploid chromosome [65,66]. Similar dosage compensation/buffering effects have been observed in human and mouse models of aneuploidy, as summarized below.
In mammals, trisomy 21 in human and mouse models of Down syndrome have been particularly valuable in understanding dosage compensation in aneuploid cells. For example, 73% of the genes on chromosome 21 produced transcripts at lower than the expected 1.5-fold level in lymphoblastoid cells from Down syndrome patients, while 22% escaped dosage compensation and their corresponding mRNAs appeared at the expected 1.5-fold increase [67]. Similarly, 46% of the genes displayed transcript levels at lower than the expected 1.5-fold increase, while 25% were increased to the expected 1.5-fold level in mouse Down syndrome cell models, in which mouse embryonic stem cells with an additional human chromosome 21 were differentiated into neuronal cells [68]. In the partial trisomy mouse model Ts65Dn, which carries the orthologs of approximately 50% of the genes on human chromosome 21, 55% of the genes were found to be expressed at levels significantly lower than 1.5-fold, whereas 37% of the genes were expressed at the expected level of 1.5-fold, although the percentages and gene identities in each category varied between different tissues [19,20].
Studies from other trisomy models both in human and mouse generally support the notion that gene expression does not correlate strictly with gene copy number in aneuploid cells. In trisomy 13 human fetal cells, the average levels of transcripts from the trisomic chromosome were only increased by approximately 10% compared with levels in euploid cells [25]. In trisomy 8 acute myeloid leukemia cells, the transcript levels for genes on chromosome 8 were found to be only increased by approximately 32% [12]. Furthermore, in a mouse model with segmental trisomy 17, the transcript levels for genes in the trisomic region were increased in the brain by only approximately 20% on average, although some genes showed the expected 50% increase [23]. Interestingly, a more recent study using the same mouse segmental trisomy 17 model found that transcript levels for the aneuploid genes were approximately 60% higher than normal in the liver, reflecting the change in gene dosage [69]. However, in early pachytene spermatocytes, the increase in transcript levels was only approximately 18%, suggesting the possible presence of dosage compensation mechanisms in germ cells [69]. Overall, these data indicate that whether dosage compensation occurs or not in response to aneuploidy may depend on many factors, including the identity of the genes/chromosome, the species in which aneuploidy occurs, or even the specific tissue or cellular microenvironment.
Little is known about whether dosage compensation can occur at the translational level, partly due to the technical limitations of current proteomic tools in accurately detecting 1.5-fold changes. Nevertheless, recent studies demonstrated that the abundance of certain proteins in human aneuploid cells can be reduced toward diploid levels even when the levels of transcripts are largely proportional to the chromosome copy number [9,70]. It is believed that the compensation at the protein level may derive from increased protein degradation, rather than reduced protein synthesis [70].
Overall, there have been many reports suggesting that dosage compensation can occur in aneuploid cells (Figure 1C). However, many open questions still remain regarding the mechanisms that may ameliorate imbalanced gene expression and restore cellular homeostasis in aneuploid cells: is there a cellular sensor that detects extra chromosomal material? If so, what is it? How does the transcriptional/translational machinery handle the extra chromosomal material? Do changes in chromosome number activate chromosome-specific chromatin remodeling (that could, in turn, control gene expression)? To start addressing this last question, we will next focus on the epigenetic changes associated with aneuploidy.
The consequences of aneuploidy on the epigenetic signature
Epigenetic regulation occurs mainly through DNA methylation and histone modifications of various types (i.e., methylation, acetylation, phosphorylation, ubiquitination, sumoylation, ADP ribosylation and deamination among others) [71,72]. Changes in chromatin structure by epigenetic processes alter the accessibility of the transcription machinery, and ultimately result in changes in gene expression patterns [71]. Methylated cytosine, or 5-methylcytocine, is often found in clusters of CpG dinucleotides, known as CpG islands. CpG islands are frequently located within, or in close proximity of gene promoters [73], and gene expression becomes silenced when CpG islands are methylated. In contrast to this single type of DNA epigenetic mark, histone modifications occur in a variety of types and locations within histone molecules. The best-studied modifications are those that are on the histone N-terminal ‘tail’ regions, which are accessible on the nucleosome surface [71]. Each type of histone modification plays a distinct role in the regulation of gene expression, and can be found in defined genomic regions. For example, H3K4me3 and H3K36me3 are both found on transcriptionally active genes. H3K4me3 marks are enriched in the promoter region, whereas H3K36me3 marks are enriched in the gene body [71,74]. Conversely, H3K9me and H3K27me marks are both found on transcriptionally inactive genes, but H3K9me is enriched in the promoter region while H3K27me is enriched around the transcriptional start site [71,74]. The sum of all epigenetic modifications determines the local chromatin structure and hence the downstream gene expression patterns. The epigenetic marks can be stably maintained or dynamically regulated in a context-specific manner and three types of enzymes control the epigenetic signature in space and time. These enzymes are generally referred to as writers, which establish the marks, erasers, which remove the marks, and readers, which interpret the marks.
Currently, very little is known about if/how aneuploidy alters epigenetic marks, and hence downstream gene expression. To investigate this problem, a number of studies have taken advantage of constitutional aneuploidies and/or mouse models, but the analysis has exclusively focused on DNA methylation status and not on histone modifications. Several recent studies in this area of investigation revealed that trisomy 21 results in DNA hypermethylation in the promoter regions of genes across all autosomes [34–37,75,76]. None of these studies reported chromosome-specific changes in DNA methylation, with one exception, where promoter hypermethylation was reported to be enriched on chromosome 21 [36]. Locations of DNA hypermethylation appear to be cell-type or developmental-stage specific, although there appears to be no consensus among different studies. One possible mechanism by which trisomy 21 may promote genome-wide DNA hypermethylation is by acting in trans on other chromosomes [37]. This idea is consistent with the fact that a number of genes on chromosome 21 are involved in DNA methylation pathways [19,39]. Moreover, the DNA demethylases TET2 and TET3 were reported to be downregulated in Down syndrome placenta and fetal skin fibroblasts [35,75], indicating that DNA hypermethylation may appear during early development and be maintained into adulthood. Another study exploited a case of constitutional trisomy 8 mosaicism and compared trisomic and euploid fibroblasts from the same individual [77]. The study reported that cells with trisomy 8 displayed DNA hypomethylation in gene-poor regions of chromosome 8, but no changes in DNA methylation of gene promoter regions on chromosome 8 [77]. Moreover, trisomy 8 did not appear to affect the levels of DNA methylation on other chromosomes [77]. An older study examined DNA methylation status in SW48 colon cancer cells, which are trisomic for chromosome 7, 14, and a subregion of 10q [78]. The study found that SW48 cells exhibited DNA hypomethylation almost exclusively in gene-poor regions of the genome compared with euploid fibroblasts [78], although fibroblasts may not be the best control, given that DNA methylation profiles may vary in different cell types.
The studies described above also underscored a potential role of a new type of DNA methylation, 5-hydroxymethylcytosine (5 hmC), and found that the overall levels of 5 hmC were decreased in both trisomy 21 and trisomy 8 cells [37,77]. Although it is still a matter of debate whether 5 hmC plays specific functions or is a passive intermediate, 5 hmC is more commonly found in gene bodies of active genes, in contrast to 5-methylcytocine that is typically detected in CpG islands [79]. This led to the hypothesis that 5 hmC may regulate gene expression by modulating chromatin accessibility for the transcriptional machinery [79–81]. Therefore, the effect of aneuploidy on gene expression may also depend on how the specific aneuploidy impacts the levels of 5 hmC.
Finally, certain epigenetic changes have been extensively studied in cancer cells, many of which are aneuploid. A detailed review of this topic goes beyond the scope of this article, but we provide here a brief summary of a few key findings. Epigenetic hallmarks of cancer cells include DNA hypomethylation in genomic regions that contain repetitive sequences and are relatively gene poor (reviewed in [82–84]) and de novo DNA hypermethylation in selected CpG islands (reviewed in [82–84]). Histone modification patterns in cancer cells are less studied, and no cancer-specific changes in histone modifications have been reported. Nonetheless, alterations in histone modifications can be observed both at the global level across the genome and at specific gene loci in a cell type- or cancer type-specific manner (reviewed in [85,86]). Unfortunately, the karyotypes of cancer cells are very complex, and this makes it challenging to decipher whether the observed changes in epigenetic marks are due to the abnormal chromosome number, or other cellular abnormalities that arise during the acquisition of the transformed phenotype. Nevertheless, the hypomethylation of gene-poor genomic regions is observed both in cancer cells and in cells with simpler aneuploidies, such as fibroblasts with trisomy 8 [77] and SW48 cells [78], raising the possibility that this type of epigenetic signature may be driven by aneuploidy.
Overall, an extra chromosome appears to lead to a common epigenetic signature, characterized by global DNA hypomethylation of gene-poor regions, DNA hypermethylation of promoter regions and a general depletion of 5 hmC, with some exceptions [34–37,75–78].
Conclusion
Studies on the effects of aneuploidy on gene expression have not always produced consistent results. Some of these discrepancies may be due to differences in experimental models, data collection and/or data analysis [65]. However, many studies performed over the years in different experimental models of aneuploidy have led to the conclusion that gene expression generally scales with gene/chromosome copy number (Figure 1A & B). At the same time, many studies have also reported substantial dosage compensation that will result in gene expression levels that are below those expected based on gene/chromosome copy number (Figure 1C). Whether or not dosage compensation is observed may depend on the identity of the aneuploid chromosome or on the context (i.e., tissue or developmental stage) [19,20,67–69]. Alternatively, in some situations dosage compensation may require time to be established. This idea could explain the observation that populations of yeast cells carrying the same aneuploidy display great phenotypic heterogeneity, but in strains harboring ‘chronic’ aneuploidies this heterogeneity is lower [87]. It is possible that, in some contexts, newly formed aneuploid cells may require time to adjust to the presence of extra chromosomal material and activate mechanisms that reduce the negative effects of aneuploidy. The time required for this ‘adjustment’ may vary depending on the organism, the conditions in which the organism grows and/or the tissue (for multicellular organisms) in which the aneuploidy occurs.
Future perspective
Although many studies have reported dosage compensation in the context of aneuploidy, the underlying mechanisms remain unknown. One possible mechanism by which dosage compensation may occur in the context of aneuploidy is via epigenetic regulation of gene expression. Interestingly, a DNA methylation signature has been recurrently observed in aneuploid cells. However, DNA methylation status is only a small part of the overall epigenetic signature. Indeed, histone post-translational modifications are known to heavily impact gene expression, but so far not much is known about histone modifications in aneuploid cells. Systematic and detailed histone modification maps in aneuploid cells will be needed to better understand the epigenetic consequences of aneuploidy. Furthermore, it is currently unclear whether the epigenetic changes observed in aneuploid cells are a direct consequence of gaining chromosomes.
Another area of future interest will be the effect of aneuploidy on nuclear architecture and higher order chromatin arrangements [88,89]. So far, very little is known in this area, although a recent study found that the nuclei of aneuploid cells tend to be larger and the positioning of the aneuploid chromosome within the nucleus tends to be slightly more variable as compared with diploid cells [90].
Understanding the impact of various epigenetic marks on both dosage compensation and nuclear organization in aneuploid cells will be the next big challenge. Indeed, epigenetic modifications regulate not only downstream gene expression, but also chromatin compression and compartmentalization, as well as nucleosome dynamics, nuclear architecture and 3D chromatin state [71,91–93]. Therefore, defining the relationship between aneuploidy and epigenetic regulation may be the key to understanding many unexplored or underexplored questions about the impact of aneuploidy on cell physiology.
Executive summary.
Aneuploidy: why do we care?
Aneuploidy is the leading cause of miscarriage in humans and, in cases in which it is compatible with live birth, it causes severe congenital defects.
Aneuploidy is a hallmark of cancer.
Cancer cells frequently possess complex karyotypes with chromosome numbers above the norm.
Experimental models of aneuploidy: correlation between chromosome copy number & gene expression levels
Generally, gene expression levels scale up with gene/chromosome copy number in many experimental models of aneuploidy.
There is strong evidence that in many cases, aneuploidy can also perturb the expression of genes on other chromosomes.
Aneuploidy & dosage compensation: when gene expression does not scale with chromosome copy number
Dosage compensation is a mechanism by which the expression of certain genes is modulated to compensate for differences in gene dosage.
Some degree of dosage compensation has been reported for many experimental models of aneuploidy, including yeast, mouse, human, and fly.
In some cases, dosage compensation is not complete. Specifically, the expression of genes on the aneuploid chromosome is not reduced all the way to the level seen in euploid cells, but it is reduced to levels substantially below the expected 50% increase.
The decreased (compared with the expected) expression levels may occur for most or only a subset of genes on the aneuploid chromosome.
The consequences of aneuploidy on the epigenetic signature
A number of studies found that trisomy 21 results in DNA hypermethylation in the promoter regions of genes across all chromosomes. This may be due to the trisomy 21-dependent overexpression of genes involved in DNA methylation.
Some studies have reported DNA hypomethylation of gene-poor regions of the genome in aneuploidy models.
Cancer cells, many of which are aneuploid, display DNA hypomethylation in regions of the genome that contain repetitive sequences and are gene poor and de novo DNA hypermethylation of selected CpG islands.
Acknowledgments
The authors acknowledge the members of the Cimini and Kojima Labs for useful discussion and feedback.
Footnotes
Financial & competing interest disclosure
Work in the Kojima Lab is supported by grant R01GM126223 from NIH (to S Kojima). Work in the Cimini Lab is supported by the ICTAS Center for Engineered Health at Virginia Tech, a Dean’s Discovery Fund grant from the Virginia Tech College of Science, and grant MCB-1517506 from the National Science Foundation (to D Cimini). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2(4), 280–291 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18(6), 658–667 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 1786(1), 32–40 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Hardy K, Hardy PJ. 1(st) trimester miscarriage: four decades of study. Transl. Pediatr. 4(2), 189–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres EM, Sokolsky T, Tucker CM. et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317(5840), 916–924 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Pavelka N, Rancati G, Zhu J. et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468(7321), 321–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes TR, Roberts CJ, Dai H. et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25(3), 333–337 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Williams BR, Prabhu VR, Hunter KE. et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322(5902), 703–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics 81(5), 457–467 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Phillips JL, Hayward SW, Wang Y. et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 61(22), 8143–8149 (2001). [PubMed] [Google Scholar]
- 12.Virtaneva K, Wright FA, Tanner SM. et al. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolated trisomy 8 and normal cytogenetics. Proc. Natl Acad. Sci. USA 98(3), 1124–1129 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollack JR, Sorlie T, Perou CM. et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl Acad. Sci. USA 99(20), 12963–12968 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsafrir D, Bacolod M, Selvanayagam Z. et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 66(4), 2129–2137 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Gao C, Furge K, Koeman J. et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc. Natl Acad. Sci. USA 104(21), 8995–9000 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Liu Y, Wang P. et al. Integrative analysis of DNA copy number and gene expression in metastatic oral squamous cell carcinoma identifies genes associated with poor survival. Mol. Cancer 9, 143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu TP, Lai LC, Tsai MH. et al. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS ONE 6(9), e24829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chikashige Y, Tsutsumi C, Okamasa K. et al. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct. Funct. 32(2), 149–161 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Kahlem P, Sultan M, Herwig R. et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 14(7), 1258–1267 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• In one of the mouse models of Down syndrome (Tn65Dn), the expression of genes on the extra chromosome was found to increase proportionally to gene copy number in nine tissues examined. Several genes escaped this rule by being expressed at either lower or higher than the expected 1.5-fold level, but the subset of genes displaying such deviant behavior varied by tissue type.
- 20.Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 14(7), 1268–1274 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Using the mouse model Tn65Dn and quantifying gene expression levels in six different tissues, this study revealed a complex regulation of gene expression that is not always proportional to gene copy number. About a third of the genes were expressed at the expected 1.5-fold level, whereas the remaining genes were expressed at levels that were higher or lower than expected.
- 21.Sun L, Johnson AF, Donohue RC, Li J, Cheng J, Birchler JA. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl Acad. Sci. USA 110(18), 7383–7388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L, Johnson AF, Li J, Lambdin AS, Cheng J, Birchler JA. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc. Natl Acad. Sci. USA 110(41), 16514–16519 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vacik T, Ort M, Gregorova S. et al. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc. Natl Acad. Sci. USA 102(12), 4500–4505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]; • In a mouse model of segmental aneuploidy, the expression of genes on the extra chromosome fragment was expressed at levels that were only 20% above the levels in the euploid state. This indicates partial dosage compensation.
- 24.Ben-David U, Arad G, Weissbein U. et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 5, 4825 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Fitzpatrick DR, Ramsay J, Mcgill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 11(26), 3249–3256 (2002). [DOI] [PubMed] [Google Scholar]; • Shows that in human trisomic fetal cells the genes on the trisomic chromosome displayed mild overexpression (dosage compensation) and the expression of many genes on other chromosomes was altered.
- 26.Rancati G, Pavelka N, Fleharty B. et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135(5), 879–893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raznahan A, Parikshak NN, Chandran V. et al. Sex-chromosome dosage effects on gene expression in humans. Proc. Natl Acad. Sci. USA 115(28), 7398–7403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Upender MB, Habermann JK, Mcshane LM. et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 64(19), 6941–6949 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blakeslee AF. Types of mutations and their possible significance in evolution. Am. Naturalist 55(638), 254–267 (1921). [Google Scholar]
- 30.Nicholson JM, Cimini D. How mitotic errors contribute to karyotypic diversity in cancer. Adv. Cancer Res. 112, 43–75 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Devlin RH, Holm DG, Grigliatti TA. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 118(1), 87–101 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birchler JA, Hiebert JC, Krietzman M. Gene expression in adult metafemales of Drosophila melanogaster. Genetics 122(4), 869–879 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolvetang EJ, Wilson TJ, Sanij E. et al. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum. Mol. Genet. 12(3), 247–255 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Kerkel K, Schupf N, Hatta K. et al. Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 6(11), e1001212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sailani MR, Santoni FA, Letourneau A. et al. DNA-methylation patterns in trisomy 21 using cells from monozygotic twins. PLoS ONE 10(8), e0135555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comparison of DNA methylation patterns between monozygotic twins discordant for trisomy 21 reveals that the level of DNA methylation was higher in individuals with trisomy 21 compared with euploid individuals. Consistently, individuals with trisomy 21 displayed overexpression of DNA methyltransferase (DNMT3B and DNMT3L) and downregulation of DNA demethylases (TET2 and TET3).
- 36.Bacalini MG, Gentilini D, Boattini A. et al. Identification of a DNA methylation signature in blood cells from persons with Down syndrome. Aging 7(2), 82–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendioroz M, Do C, Jiang X. et al. Trans effects of chromosome aneuploidies on DNA methylation patterns in human Down syndrome and mouse models. Genome Biol. 16, 263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letourneau A, Santoni FA, Bonilla X. et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508(7496), 345–350 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Do C, Xing Z, Yu YE, Tycko B. Trans-acting epigenetic effects of chromosomal aneuploidies: lessons from Down syndrome and mouse models. Epigenomics 9(2), 189–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc. Natl Acad. Sci. USA 109(31), 12644–12649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donnelly N, Storchova Z. Causes and consequences of protein folding stress in aneuploid cells. Cell Cycle 14(4), 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313(5785), 367–370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5(10), e1000705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68(3), 624–641 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Millet C, Ausiannikava D, Le Bihan T, Granneman S, Makovets S. Cell populations can use aneuploidy to survive telomerase insufficiency. Nat. Commun. 6, 8664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaya A, Gerashchenko MV, Seim I, Labarre J, Toledano MB, Gladyshev VN. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proc. Natl Acad. Sci. USA 112(34), 10685–10690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu G, Yong MY, Yurieva M. et al. Gene essentiality is a quantitative property linked to cellular evolvability. Cell 163(6), 1388–1399 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Morard M, Macias LG, Adam AC. et al. Aneuploidy and ethanol tolerance in Saccharomyces cerevisiae. Front. Genet. 10, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ly P, Eskiocak U, Kim SB. et al. Characterization of aneuploid populations with trisomy 7 and 20 derived from diploid human colonic epithelial cells. Neoplasia 13(4), 348–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sareen D, Mcmillan E, Ebert AD. et al. Chromosome 7 and 19 trisomy in cultured human neural progenitor cells. PLoS ONE 4(10), e7630 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6(2), 149–162 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell 124(5), 901–914 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Meyer BJ. X-Chromosome dosage compensation. WormBook (2005) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu. Rev. Genet. 31, 571–610 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Baker BS, Gorman M, Marin I. Dosage compensation in Drosophila. Annu. Rev. Genet. 28, 491–521 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Meyer BJ, Mcdonel P, Csankovszki G, Ralston E. Sex and X-chromosome-wide repression in Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 69, 71–79 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Torres EM, Springer M, Amon A. No current evidence for widespread dosage compensation in S. cerevisiae. Elife 5, e10996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hose J, Yong CM, Sardi M, Wang Z, Newton MA, Gasch AP. Dosage compensation can buffer copy-number variation in wild yeast. Elife 4, e05462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that aneuploidy is common in wild yeast isolates. In these isolates, many of the amplified genes are expressed at levels that are lower than would be expected based on copy number.
- 59.Gasch AP, Hose J, Newton MA, Sardi M, Yong M, Wang Z. Further support for aneuploidy tolerance in wild yeast and effects of dosage compensation on gene copy-number evolution. Elife 5, e14409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kvitek DJ, Will JL, Gasch AP. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4(10), e1000223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Malone JH, Powell SK. et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8(2), e1000320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z, Presgraves DC. Translational compensation of gene copy number alterations by aneuploidy in Drosophila melanogaster. Nucleic Acids Res. 45(6), 2986–2993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stenberg P, Lundberg LE, Johansson AM, Ryden P, Svensson MJ, Larsson J. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 5(5), e1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johansson AM, Stenberg P, Bernhardsson C, Larsson J. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26(9), 2307–2316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birchler JA. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma 123(5), 459–469 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl Acad. Sci. USA 109(37), 14746–14753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ait Yahya-Graison E, Aubert J, Dauphinot L. et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am. J. Hum. Genet. 81(3), 475–491 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang CC, Kazuki Y, Oshimura M, Ikeo K, Gojobori T. Gene dosage imbalance of human chromosome 21 in mouse embryonic stem cells differentiating to neurons. Gene 481(2), 93–101 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Jansa P, Homolka D, Blatny R, Mistrik M, Bartek J, Forejt J. Dosage compensation of an aneuploid genome in mouse spermatogenic cells. Biol. Reprod. 90(6), 124 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Borel C, Li L. et al. Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat. Commun. 8(1), 1212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32(1), 42–56 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol. 7(9), a025064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bird AP. CpG-rich islands and the function of DNA methylation. Nature 321(6067), 209–213 (1986). [DOI] [PubMed] [Google Scholar]
- 74.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 128(4), 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Jin S, Lee YK, Lim YC. et al. Global DNA hypermethylation in down syndrome placenta. PLoS Genet. 9(6), e1003515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Comparison of chorionic villus samples from subjects carrying a normal or Down syndrome fetus reveals DNA hypermethylation across all autosomes in Down syndrome samples. The authors propose that this may be partly due to downregulation of TET demethylases and REST/NRSF (a gene involved in transcriptional regulation).
- 76.Jones MJ, Farre P, Mcewen LM. et al. Distinct DNA methylation patterns of cognitive impairment and trisomy 21 in Down syndrome. BMC Med. Genomics 6, 58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davidsson J, Veerla S, Johansson B. Constitutional trisomy 8 mosaicism as a model for epigenetic studies of aneuploidy. Epigenetics Chromatin 6(1), 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber M, Davies JJ, Wittig D. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37(8), 853–862 (2005). [DOI] [PubMed] [Google Scholar]; • SW48 colon cancer cells, which are trisomic for chromosome 7, 14, and a subregion of 10q, exhibit DNA hypomethylation almost exclusively in gene-poor regions of the genome compared with euploid controls.
- 79.Shi DQ, Ali I, Tang J, Yang WC. New insights into 5 hmC DNA modification: generation, distribution and function. Front. Genet. 8, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin IH, Chen YF, Hsu MT. Correlated 5-Hydroxymethylcytosine (5 hmC) and gene expression profiles underpin gene and organ-specific epigenetic regulation in adult mouse brain and liver. PLoS ONE 12(1), e0170779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Wu X, Zhou Y. et al. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 46(6), 2883–2900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 21(35), 5400–5413 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8(9), a019505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 76(12), 3446–3450 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell 153(1), 38–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8(4), a019521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beach RR, Ricci-Tam C, Brennan CM. et al. Aneuploidy causes non-genetic individuality. Cell 169(2), 229–242 e221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gondor A, Ohlsson R. Chromosome crosstalk in three dimensions. Nature 461(7261), 212–217 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2(3), a003889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Braun R, Ronquist S, Wangsa D. et al. Single chromosome aneuploidy induces genome-wide perturbation of nuclear organization and gene expression. Neoplasia 21(4), 401–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Espada J, Esteller M. Epigenetic control of nuclear architecture. Cell. Mol. Life Sci. 64(4), 449–457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin 7(1), 25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu M, Ren B. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]