Abstract
MicroRNAs that are overexpressed in cystic fibrosis (CF) bronchial epithelial cells (BEC) negatively regulate CFTR and nullify the beneficial effects of CFTR modulators. We hypothesized that it is possible to reverse microRNA-mediated inhibition of CFTR using CFTR-specific target site blockers (TSBs) and to develop a drug-device combination inhalation therapy for CF. Lead microRNA expression was quantified in a series of human CF and non-CF samples and in vitro models. A panel of CFTR 3′ untranslated region (UTR)-specific locked nucleic acid antisense oligonucleotide TSBs was assessed for their ability to increase CFTR expression. Their effects on CFTR activity alone or in combination with CFTR modulators were measured in CF BEC models. TSB encapsulation in poly-lactic-co-glycolic acid (PLGA) nanoparticles was assessed as a proof of principle of delivery into CF BECs. TSBs targeting the CFTR 3′ UTR 298–305:miR-145-5p or 166–173:miR-223-3p sites increased CFTR expression and anion channel activity and enhanced the effects of ivacaftor/lumacaftor or ivacaftor/tezacaftor in CF BECs. Biocompatible PLGA-TSB nanoparticles promoted CFTR expression in primary BECs and retained desirable biophysical characteristics following nebulization. Alone or in combination with CFTR modulators, aerosolized CFTR-targeting TSBs encapsulated in PLGA nanoparticles could represent a promising drug-device combination therapy for the treatment for CFTR dysfunction in the lung.
Keywords: microRNA, CFTR, target site blocker, nebulised PLGA nanoparticles, RNA sequencing, iPSC-derived CF and CFTR gene-corrected bronchosperes, ALI culture, CFTR modulators, Primary bronchial epithelial cells, High content screening
Precise targeting of specific miR-145-5p or miR-223-3p binding sites with target site blockers encapsulated in biocompatible nanoparticles restores CFTR activity and enhances CFTR modulator action in p.Phe508del/p.Phe508del CF bronchial epithelial cells.
Introduction
Cystic fibrosis (CF) is caused by mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene.1,2 CFTR encodes a channel that mediates chloride and other anion transport across the apical membrane of epithelial cells. Mutations are classified in classes I to VI according to the mechanisms by which they affect CFTR protein synthesis, folding, trafficking, or function, with p.Phe508del being the most common CF-causing mutation worldwide.3 The p.Phe508del mutation encodes a misfolded CFTR protein that is retained within the endoplasmic reticulum and degraded; only a small proportion reaches the apical membrane of the cell. In order to develop CF, an individual must carry two mutant CFTR alleles. The p.Phe508del mutation may be carried on both alleles, i.e., homozygous for p.Phe508del, or on one allele in conjunction with another disease-causing CFTR mutation, i.e., heterozygous for p.Phe508del. Although considered a multi-organ disorder, CF morbidity and mortality are primarily associated with chronic lung disease, where absent or dysfunctional CFTR leads to depletion of the airway surface liquid and mucus dehydration, resulting in impaired mucociliary clearance, chronic infection, and inflammation.
We and others have shown that CFTR is post-transcriptionally regulated by microRNAs (miRNAs) (reviewed in Glasgow et al.4); the lead miRNAs that have been experimentally validated as direct regulators of wild-type and/or p.Phe508del human CFTR are miR-101-3p,5, 6, 7 miR-145-5p,7, 8, 9 miR-223-3p,9 miR-494-3p,5,8, 9, 10 and miR-509-3p.10,11 Two studies have demonstrated that antagomir- or peptide nucleic acid (PNA)-based inhibitors of miR-145-5p can enhance CFTR expression and/or function in CF bronchial epithelial cells (BECs),12,13 and another demonstrated PNA-mediated inhibition of miR-509-3p.14 However, these strategies, while effective in vitro, could have undesired off-target effects on other miR-145-5p or miR-509-3p-regulated genes, as well as causing co-inhibition of other miRNAs.15
Target site blockers (TSBs) are locked nucleic acid antisense oligonucleotides that specifically compete with miRNAs for the binding to individual miRNA recognition elements (MREs) of a target mRNA, hence preventing them from gaining access to those sites. A TSB targeting the miR-9 MRE in Anoctamin 1 (ANO1/TMEM16A) increases ANO1-mediated chloride efflux and mucociliary clearance in CF models.16 Here, we designed a set of CFTR-specific TSBs and assessed their activity as effective blockers of miRNA-mediated inhibition of CFTR. We quantified their effects on CFTR expression and activity in a variety of in vitro and ex vivo human CF BEC models, alone or in combination with two currently available CFTR therapies, ivacaftor/lumacaftor and ivacaftor/tezacaftor.
Ivacaftor is a CFTR potentiator that increases the open channel probability of CFTR.17 Lumacaftor is a CFTR corrector that improves intracellular trafficking of misfolded CFTR to the apical surface of the cells.18 Together, they are authorized for the treatment of p.Phe508del/p.Phe508del in CF children >2 years. Tezacaftor, another CFTR corrector, when used in combination with ivacaftor in p.Phe508del homozygotes or heterozygotes with a residual function CFTR allele, can deliver similar clinical benefits.19,20 The proinflammatory CF lung milieu can increase expression of miRNAs that regulate CFTR9,10 and, as has been shown for miR-145-5p, can nullify correction by ivacaftor/lumacaftor.12 Whether TSBs that target specific miRNA binding sites within the CFTR 3′ UTR can enhance ivacaftor/lumacaftor or ivacaftor/tezacaftor correction of CFTR has not been tested.
An ideal therapeutic to treat the pulmonary manifestations of CF would be delivered locally to the lung. Therefore, we encapsulated TSBs within poly-lactic-co-glycolic acid (PLGA) nanoparticles and assessed their toxicity, function, and delivery to CF BECs. Importantly, the PLGA-TSB nanoparticles were also coupled with a nebulizer that can generate stable and respirable aerosols, capable of targeted delivery throughout the lung,21,22 to demonstrate how such a drug-device combination might be translated to the clinic.
Results
All studies were performed using non-CF or p.Phe508del/p.Phe508del cells unless otherwise stated.
Lead miRNA Expression in CF BEC Lines, Primary Cells, and Bronchial Brushings
Endogenous levels of the five lead miRNAs previously reported as CFTR direct regulators (i.e., miR-101-3p,5, 6, 7 miR-145-5p,7, 8, 9 miR-223-3p,9 miR-494-3p,5,8, 9, 10 and miR-509-3p10,11) were measured in a range of in vitro, ex vivo, and clinical samples.
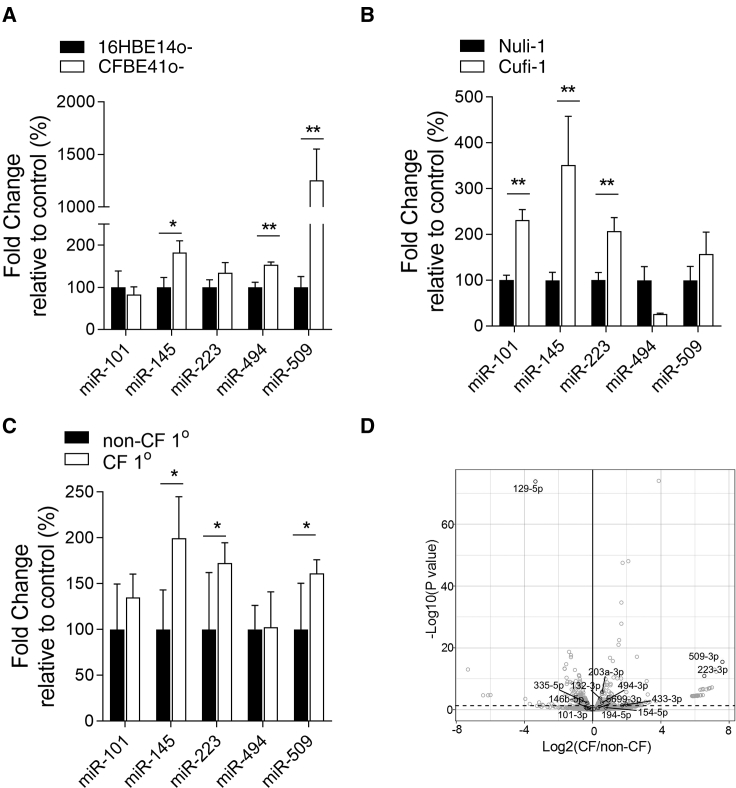
When comparing CF versus non-CF BECs, all miRNAs except miR-101-3p were increased, some significantly, in CFBE41o− versus 16HBE14o− (Figure 1A), and all except miR-494-3p were increased in Cufi-1 versus Nuli-1 and also in adult primary CF versus non-CF cells as measured by miRNA TaqMan assay (Figures 1B and 1C). miR-223-3p and miR-509-3p were significantly increased in ALI (air liquid interface) culture of CFBE41o− cells stably transfected with Phe508del versus wild-type CFTR as measured by RNA sequencing (Figure 1D).
Figure 1.
miRNA Levels in CF and Non-CF Cell Lines and Primary Bronchial Epithelial Cells
Relative expression levels, shown as fold change relative to control (%) of miRNAs were determined by qRT-PCR using individual TaqMan assays and normalized to U6snRNA in (A) CFBE41o− versus 16HBE14o−, (B) Cufi-1 versus Nuli-1, or (C) adult CF versus non-CF primary cells (n = 6 in triplicate for each cell line). Data are presented as mean ± SEM and were compared by one-way ANOVA (Dunnett’s multiple comparisons test *p < 0.05,**p < 0.01). (D) Volcano plot generated from miRNA-seq showing differential expression of miRNAs in CFBE41o− ALI-cultured cells stably transfected with Phe508del (CF) compared to WT (non-CF) CFTR. miRNAs conserved in mammals (black, TargetScanHuman release 7.2); non-conserved miRNAs (gray). p < 0.05 corresponding to −Log10 > 1.3. n = 6/group from different cultures. Dashed line indicates p = 0.05.
Lead miRNA levels were also measured by miRNA profiling in n = 6 each CF and non-CF BECs from children, where miR-145-5p and miR-223-3p were increased in CF (7.4-fold, p = 0.028, and 4.1-fold, p = 0.04, respectively), miR-101-3p showed no difference, and miR-494-3p/miR-509-3p were not available on the platform/not detectable (GEO: GSE128861). Overall, the lead miRNAs are detectable at variably increased levels in all CF BECs analyzed by different methods. As no single miRNA can be identified as being unique, an inhibition strategy to individually target each of the lead miRNAs was undertaken.
TSB4 and TSB6 Effectively Block miRNA-Mediated Repression of a CFTR 3′ UTR Luciferase Reporter Gene and Increase Endogenous CFTR Protein Levels in CF BECs
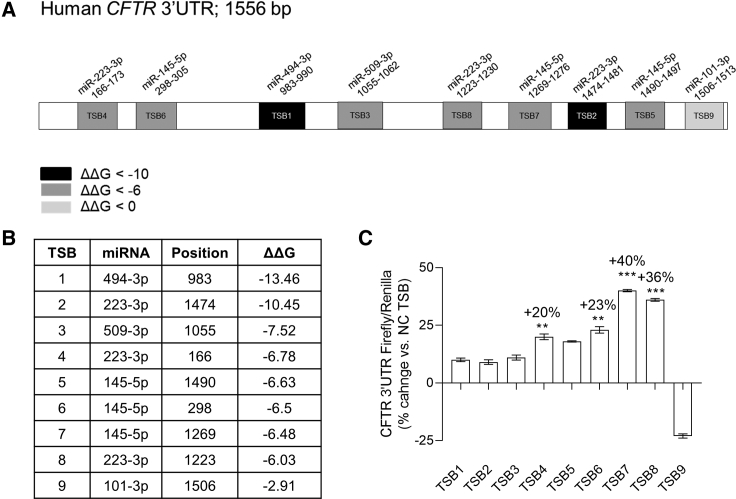
In silico predictions using PITA identified all potential MREs for the lead miRNAs in the CFTR 3′ UTR: miR-101-3p × 5 sites, miR-145-5p × 8 sites, miR-223-3p × 9 sites, miR-494-3p × 6 sites, and miR-509-3p × 3 sites. From this list, nine sites were selected against which TSBs were designed with an arbitrary ΔΔG cutoff of from −13.46 to −6.03 and the miR-101-3p site at position 1506 (with a ΔΔG of −2.91) (Figures 2A and 2B; Table S1). The efficacy of the TSBs at increasing CFTR 3′ UTR luciferase expression in CFBE41o− cells was screened. Luciferase was significantly increased after transfection with TSB4, TSB6, TSB7, and TSB8 versus NC (non-targeting control) TSB (Figure 2C).
Figure 2.
CFTR-Specific TSB Design, Screening, and Selection
(A) Visual map of the in silico predictions of miRNA-responsive elements (MREs) of the five lead miRNAs (i.e., miR-101-3p, miR-145-5p, miR-223-3p, miR-494-3p, and miR-509-3p) within the CFTR 3′ UTR. (B) Table showing the prediction of the target sites of the lead miRNAs in the CFTR 3′ UTR according to the PITA algorithm. ΔΔG is ΔG(duplex) − ΔG(open), where ΔG(duplex) is the hybridization energy of the miRNA to the binding site and ΔG(open) is the energy required to open the local RNA secondary structure around the binding site. The more negative the ΔΔG, the stronger the expected binding of a miRNA to that site. (C) CFTR 3′ UTR luciferase activity reported as % change in relative light units (RLU) in CFBE41o− cells (n = 3 in triplicate). Data are presented as mean ± SEM and were compared by one-way ANOVA (Dunnett’s multiple comparisons test **p < 0.01, ***p < 0.001). Samples transfected with non-targeting control (NC) TSB were used as reference and set at 0%.
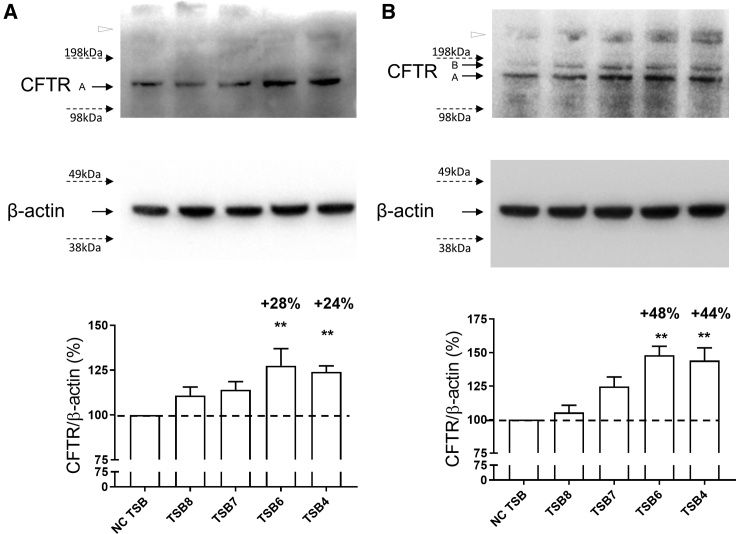
The levels of endogenous CFTR protein following TSB4/6/7/8 transfection were quantified in CF cell lines. TSB7 and TSB8 transfection upregulated CFTR protein; however, only TSB4 and TSB6 significantly increased unglycosylated band A and/or core glycosylated band B CFTR protein levels in CFBE41o− (p = 0.0077 for TSB4 and p = 0.0041 for TSB6; Figures 3A and S1A) and Cufi-1 (p = 0.0026 for TSB4 and p = 0.0014 for TSB6; Figures 3B and S1A) cells compared to NC TSB. These cells are Phe508del and therefore do not express a fully glycosylated mature CFTR band C of 170–220 kDa,23 which would be an indicator of CFTR folding and maturation, although there is a faint unidentified high-molecular-weight band (>198 kDa, open triangle in Figures 3A and 3B) that may reflect this.
Figure 3.
Effect of CFTR-Specific TSBs on CFTR Protein Expression
Western blot of endogenous CFTR protein following TSB transfection (100 nM) in (A) CFBE41o− (n = 5) and (B) Cufi-1 (n = 3) CF bronchial epithelial cells. Quantification after internal normalization with β-actin is displayed together with a representative image. In both CF cell lines, CFTR migrated with a band at ∼130 kDa that corresponds to the immature (i.e., unglycosylated band A) form and was used for quantification. In Cufi-1, CFTR band B (core glycosylated band B) was also visible in some experiments. Data are presented as mean ± SEM and were compared by one-way ANOVA (Dunnett’s multiple comparisons test), **p < 0.01. Samples transfected with non-targeting control (NC) TSB are reported as reference and set at 100%.
Subsequent studies focused on TSB4 and TSB6. To confirm that these TSBs were specific for CFTR, site-specific mutagenesis of the miR-223-3p MRE at position 166–173, and the miR-145-5p MRE at 298–305 was performed. No significant increase in luciferase by TSB4 or TSB6 was observed from plasmids carrying mutated MREs (Figure S2). Sequence alignment of human and murine CFTR mRNA transcripts indicates that MREs for TSB4 and TSB6 are not conserved in mice (data not shown).
TSB4 and TSB6 Enhance CFTR Channel Function in CF BECs
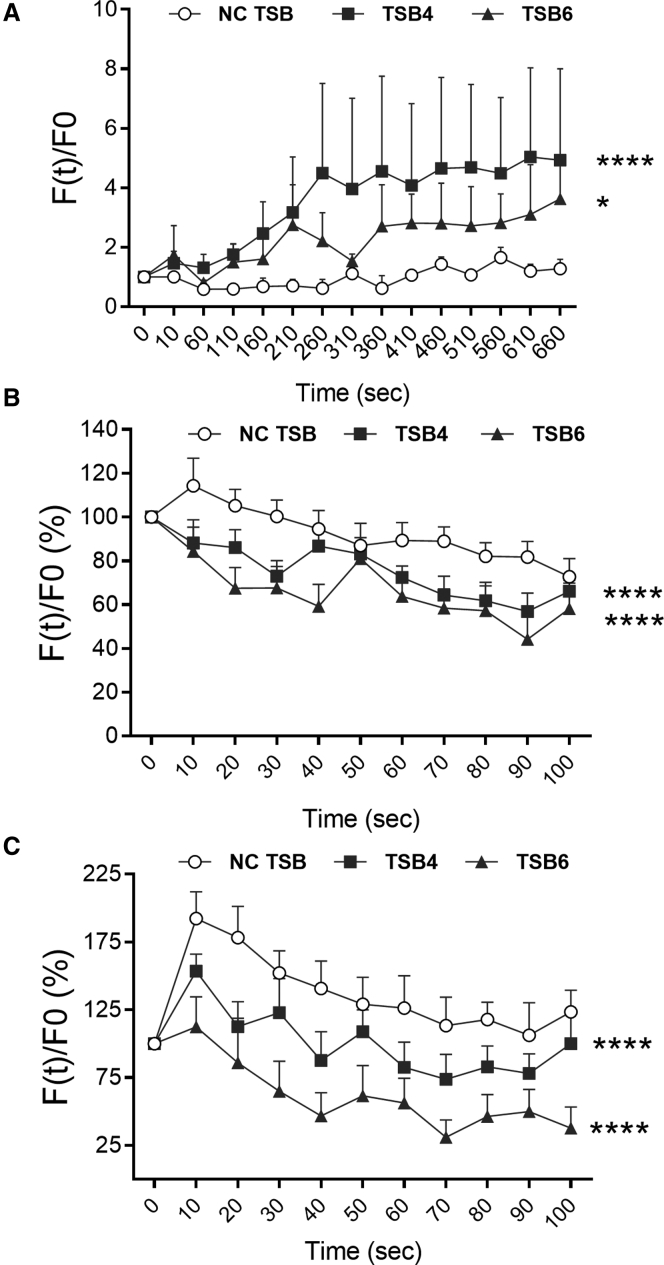
We assessed the effect of TSB4 and TSB6 on CFTR function using two established methods. The chloride-sensitive dye methoxyquinolinium bromide (MQAE) is quenched by intracellular Cl− ions ([Cl−]i), therefore increasing fluorescence corresponds to decreasing [Cl−]i and improvement of CFTR ion channel activity. After transfection with TSB4 or TSB6 in CFBE41o− cells, fluorescence significantly increased over time (2.7-fold, p = 0.0001, and 1.4-fold, p = 0.048, compared to non-targeting control [NC] TSB) (Figure 4A).
Figure 4.
CFTR Functional Assays in CF Bronchial Epithelial Cells
CFTR activity 48 h post-transfection with a non-targeting control (NC) TSB, TSB4, or TSB6 (100 nM) was assessed with the chloride-sensitive dye MQAE in (A) CFBE41o− cells and the halide-sensitive YFP in (B) CFBE41o− and (C) Cufi-1 cells. All experiments were performed after stimulation of CFTR with 20 μM of forskolin (FSK) (n > 3 per cell line, in triplicate). Data are presented as background-corrected fluorescence values normalized for the initial fluorescence (i.e., baseline fluorescence for MAQE assay and fluorescence measured before addition of I− for YFP assay) over time and compared by two-way ANOVA (Dunnett’s multiple comparisons test *p < 0.05, ****p < 0.0001).
These results were validated with the yellow fluorescent protein (YFP)-based CFTR functional assay. YFP fluorescence is quenched by intracellular I−; therefore, decreased fluorescence after applying iodide corresponds to higher influx of I− and hence to enhanced CFTR anion channel activity. After co-transfection of the YFP plasmid with TSB4 or TSB6, significant decreases in fluorescence (p = 0.0001 for both) were evident both in CFBE41o− (−18% and −31% when compared to NC TSB) and Cufi-1 (−35% and −70%) cells over time (Figures 4B and 4C). Taken together, these data indicate that the inhibition of CFTR-specific miRNAs can improve CFTR anion channel activity.
We also tested CFTR activity in a 3D organoid model of lung-specific CFTR function based on forskolin-induced swelling (FIS) of bronchospheres generated from p.Phe508del/p.Phe508del and syngeneic gene-corrected (p.Phe508del/wild-type) induced pluripotent stem cells (iPSCs). miR-145-5p and miR-223-3p were expressed in the CF organoids and increased in the gene-corrected bronchospheres (Figures S3A and S3B). Although robust FIS was observed in p.Phe508del/p.Phe508del bronchospheres in response to CFTR modulators (data not shown), there was no immediately observable response to either TSB4 or TSB6, likely due to low transfection efficiency in whole organoids.
TSB4 and TSB6 Enhance CFTR Modulator Effects on CFTR Activity in CF BECs
miR-145-5p levels are elevated in CF bronchoalveolar lavage fluid (BALF)-derived exosomes,12 and Pseudomonas-conditioned medium or interleukin-1β (IL-1β) can increase expression of miR-145 and/or miR-223 in CFBE41o− cells,9 thereby likely decreasing CFTR expression in the inflamed CF lung milieu. Figure S4 shows that CF BALF induces a similar effect by significantly increasing miR-145-5p and miR-223-3p expression in CFBE41o− and Cufi-1 cells. Thus, adjunct therapeutic targeting of the miR-145-5p and miR-223-3p binding sites in the CFTR 3′ UTR with existing CF modulators could hold merit.
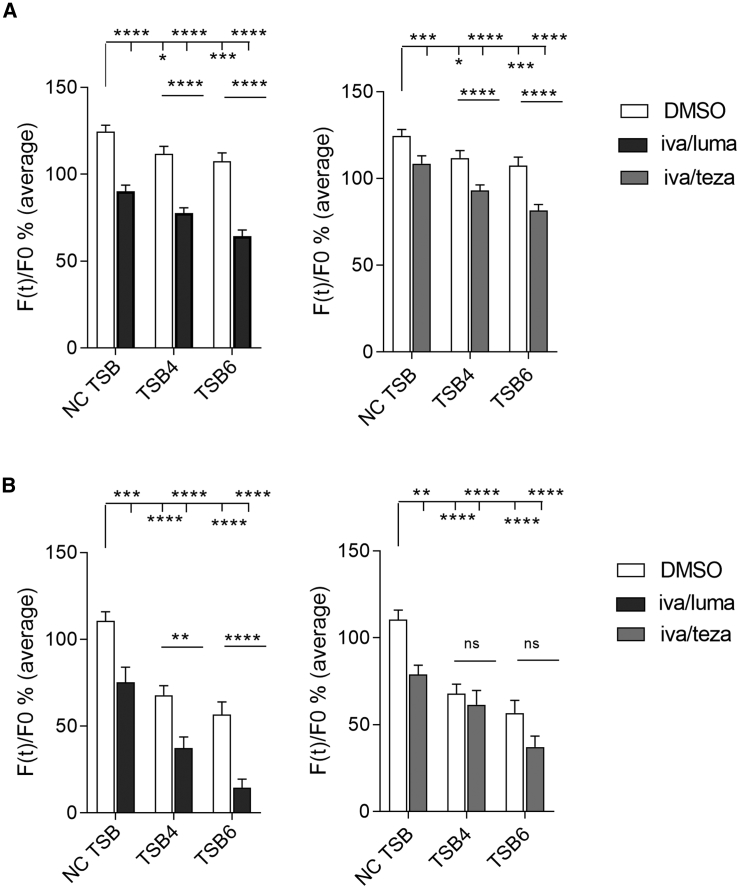
CF BECs were transfected with TSBs and treated with ivacaftor/lumacaftor or ivacaftor/tezacaftor or DMSO control, and YFP fluorescence was assessed. Transfection with TSB4 or TSB6 enhanced the CFTR modulator effects in both CFBE41o− and Cufi-1 cells (Figure 5). Ivacaftor/lumacaftor or ivacaftor/tezacaftor had no effect on miR-145-5p or miR-223-3p levels in CFBE41o− or Cufi-1 cells (Figure S5).
Figure 5.
YFP Assay in CF Bronchial Epithelial Cells Transfected with TSBs in Combination with CFTR Modulators
(A) CFBE41o− and (B) Cufi-1 cells were transfected with a non-targeting control (NC) TSB, TSB4, or TSB6 (100 nM) then immediately treated with DMSO or ivacaftor/lumacaftor (black) or ivacaftor/tezacaftor (gray) for 48 h. All experiments were performed after stimulation of CFTR with 20 μM of forskolin (FSK), (n > 3 per cell line, in triplicate). Data are presented as average of the background-corrected fluorescence values normalized for the initial fluorescence (i.e., fluorescence measured before addition of I−) and compared by two-way ANOVA (Dunnett’s multiple comparisons test *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Biocompatible PLGA Nanoparticles Encapsulating TSB4 and TSB6 Increase CFTR in Primary CF BECs
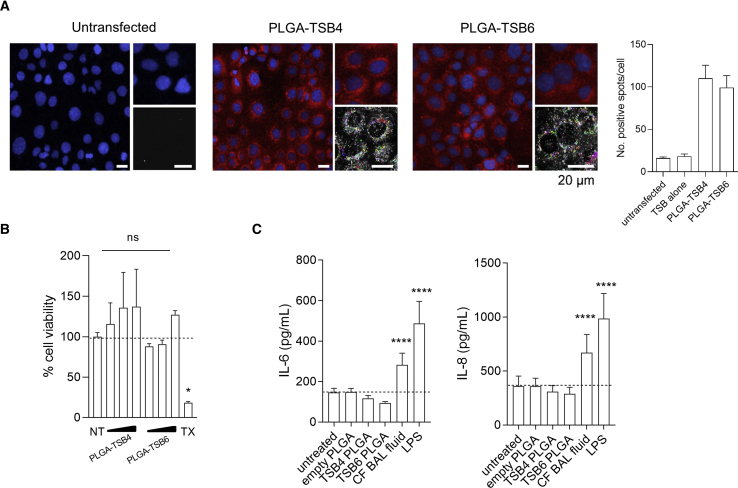
PLGA nanoparticles encapsulating TSB4 and TSB6 were formulated. High content screening (HCS) confocal images demonstrate cellular uptake and a primarily cytosolic distribution of rhodamine-conjugated TSB4- or TSB6-PLGA nanoparticles by CFBE41o− cells (Figure 6A). Both particles show similar uptake with an average number of spots/cell of 110.2 ± 15.3 for TSB4 and 99.3 ± 14.2 for TSB6. By comparison, untransfected cells have 16.1 ± 1.4 spots/cell. The PLGA-TSB nanoparticles were non-toxic to CFBE41o− cells and did not induce proinflammatory IL-6 or IL-8 responses (Figures 6B and 6C).
Figure 6.
Delivery and Toxicity of PLGA-TSBs in CFBE41o− Cells
(A) Representative images (scale, 20 μm) and quantitation of high content screening of rhodamine-conjugated PLGA-TSB4 or PLGA-TSB6 nanoparticles (red) (containing the equivalent to 100 nM of each TSB) into CFBE41o− cells (n = 12–24 wells each). Cell nuclei are stained blue. An example of the nanoparticles being detected and segmented within the population of cells using “find spots” is shown in the bottom right of each image. Effect of PLGA-TSBs on (B) viability at 48 h (TX, Triton X-100 positive control), and (C) IL-6 and IL-8 cytokine release from CFBE41o− cells after 24 h, n = 3 (CF BALF 1% and LPS 10 μg/mL, positive controls). Data are presented as mean ± SEM, *p < 0.05 and ****p < 0.0001.
Ideally, such a therapeutic would be aerosolized to the lung. Previously, we described the biophysical characteristics of these PLGA-TSB nanoparticles.24 Here, their aerosol characteristics were assessed using the Aerogen Solo nebulizer. They showed no significant differences in their droplet sizes (Table S2). The volume median diameter was approximately 4.4 μM for both and so can be considered a highly respirable aerosol. Nor was there a difference of note in the aerosol output rates (0.51 and 0.48 mL/min). Given the standard 2.5- or 3-mL dose volume for inhaled medications, this equates to a delivery time of approximately 5 to 6 min for both formulations.
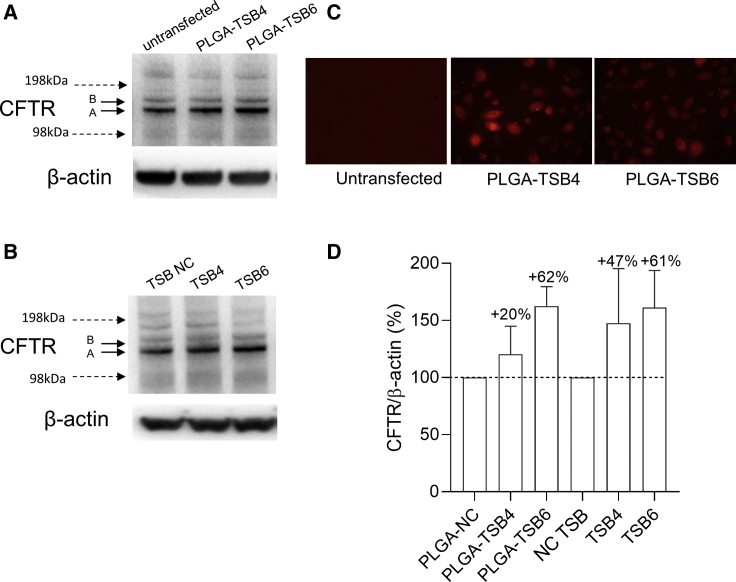
Finally, a proof-of-principle study demonstrated that PLGA-TSB4 and PLGA-TSB6 could increase CFTR in primary CF BECs (20% and 62%, respectively) and that fluorescent versions of the PLGA-TSB nanoparticles transfect well into the same cells (Figures 7A, 7C, 7D, and S1B). Unencapsulated TSB4 and TSB6 also increased CFTR protein expression in primary CF BECs (47% and 61%, respectively) (Figures 7B, 7D, and S1B).
Figure 7.
Effect of TSBs or PLGA-TSBs on CFTR Protein Expression in Primary CF Cells
Western blot of CFTR and β-actin protein 72 h following (A) PLGA-TSB (containing the equivalent of 100 nM of each TSB) or (B) TSB (100 nM) transfection into primary CF bronchial epithelial cells. (C) Representative epifluorescence microscopy images of transfection of rhodamine-conjugated PLGA-TSB4 or -TSB6 into primary CF bronchial epithelial cells at 24 h after transfection. (D) Quantification of CFTR after internal normalization with β-actin (n = 2). Control samples are reported as reference and set at 100%.
Discussion
These studies show that CFTR-specific TSBs can modulate Phe508del CFTR in CF BECs to enhance its expression and anion permeability. miR-145-5p and miR-223-3p showed enhanced expression in Phe508 homozygous CF BEC lines and BECs from children and adults with CF. These miRNAs were also expressed in CF and CFTR gene-corrected iPSC-derived CF lung organoids. TSBs precisely targeting specific individual binding sites of these miRNAs in the CFTR 3′ UTR were effective at enhancing CFTR protein levels in two Phe508del homozygous BEC lines. Delivery of the TSBs encapsulated in PLGA nanoparticles also increased CFTR expression in primary CF BECs, providing a proof of principle for aerosol delivery of TSBs as an inhalation therapy to the CF lung. We concentrated our studies on human CF models because murine CFTR is not regulated by the same miRNAs as human CFTR.
Previously, TSBs targeting unspecified miR-101 and miR-145 sites in the CFTR 3′ UTR demonstrated increased CFTR expression and function in CF nasal epithelial cells.7 However, there are five and eight sites each for miR-101-3p and miR-145-5p in the CFTR 3′ UTR. Therefore, in this work we took into account not only these 13 binding sites but all other predicted binding sites for the lead miRNAs that have been demonstrated to regulate CFTR. In total, 31 individual MREs were considered, and TSBs were designed to the nine sites predicted to be most functional. Ultimately, independently inhibiting two specific MREs was found to be most effective: the miR-145-5p site located at 298–305 and miR-223-3p at 166–173. There was no additive effect of combining TSB4 and TSB6 (data not shown), possibly due to steric hindrance between the closely located MREs. Although TSB1 and TSB2 did increase the luciferase signal, the effects did not reach significance, notwithstanding their MREs' negative ΔΔG values. Indeed, we expected these target MREs to be strongly inhibited by TSBs 1 and 2, since miR-494 and miR-223, respectively, were predicted to bind to CFTR 3′ UTR with the highest strength. Possible reasons for this may be (1) that the site targeted by TSB1 is a false positive, i.e., although PITA predicts the MRE at position 983–990 in the CFTR 3′ UTR as a potential miR-494-3p binding site, it has not been experimentally validated, (2) that TSB2 may be unable to block a strong miRNA-MRE interaction, i.e., although the MRE located at 1474–1481 is an experimentally validated miR-223 site,9 for some unknown reason TSB2 was incapable of inhibiting the binding of the miRNA, or (3) the secondary structure of the chimeric construct (firefly luciferase + CFTR 3′ UTR) might not be suitable for the binding of those TSBs.
The TSBs work by blocking the binding of miR-145-5p and miR-223-3p to specific sites in the CFTR 3′ UTR and specifically target CFTR expression at the post-transcriptional level rather than via transcriptional upregulation of CFTR.7 CFTR protein levels were increased by the TSBs in two Phe508del homozygous BEC lines. Patient idiosyncrasy, site of lung tissue of origin, or cell culture conditions that ultimately affect CFTR glycosylation may be responsible for the observed difference in glycosylation pattern between the two cell lines; however, we do not know the precise reason. The increased anion flux in CF BECs observed following individual TSB treatment most likely arises from an increased number of functional Phe508del CFTR channels in the plasma membrane, rather than changes in channel selectivity or gating.
PLGA is a synthetic polymer approved by the US FDA and European Medicine Agency in various drug-delivery systems in humans. It is widely used due to its inherent biocompatibility, low toxicity and immunogenicity, and biodegradability.24 In vivo, PLGA is removed by the citric acid cycle. PLGA drug carriers can achieve sustained cytoplasmic delivery via rapid escape from endolysosomes. Previously, we reported a comprehensive biophysical investigation of the PLGA-TSB4 and PLGA-TSB6 nanoparticles24 (therein termed LNA1 and 2, respectively). Size distribution, polydispersity index, and zeta potential revealed the nanoparticle diameters were ∼200 nm, and transmission electronic microscopy confirmed they had a monodisperse particle size distribution. The surface charge was neutral to slightly negative, and a high amount of each TSB was encapsulated by the PLGA. When nebulized, there were no differences in particle size versus prior to nebulization. The work presented here builds on that study. The data demonstrate functional properties of the PLGA-TSB nanoparticles in primary CF BECs, as well as desirable volumetric median diameter and fine particle fraction values that are indicative of highly respirable aerosols with nebulizer output rates that would ensure reasonable treatment times. This is a key consideration in the CF patient population where the daily burden of aerosol and other therapies is already high.
Here, we also provide the first report of a synergistic effect of specific miRNA binding site inhibition in combination with CFTR correctors, to enhance anion flux through Phe508del-CFTR. CF BECs display increased functionally active Phe508del-CFTR when treated with a combination of TSBs and CFTR modulators. These findings indicate a therapeutic value in selectively inhibiting the activity of specific miRNAs in CF BECs to potentiate the effectiveness of CFTR modulators. This is important because not only are miRNAs that regulate CFTR increased basally in CF BECs, but the proinflammatory CF lung milieu increases expression of CFTR-specific miRNAs, as we have shown here and elsewhere.9 This has also been demonstrated for miR-145-5p in CF BALF exosomes, where it was first reported that increased miR-145-5p nullifies CFTR correction by ivacaftor/lumacaftor.12
Traditional CF therapies target the loss of function of CFTR. A phase II clinical trial with 116 CF patients has shown potential therapeutic efficacy when administering a CFTR-encoding plasmid contained in cationic liposomes via nebulization.25 Moreover, gene-based preclinical studies are ongoing to test novel viral26,27 and non-viral28, 29, 30, 31 formulations to overcome the delivery issues due to the natural barriers that the human body, and in particular the CF lung, offer toward exogenous nucleic acids. As an alternative to gene therapy, FDA-approved pharmaceutical modulators can restore function to mutant CFTR; however, optimal therapy and effective personalized medicine may require combinations of CFTR correctors with adjunct therapies. Here, we found a potentiating effect of two TSBs on CFTR modulators that enhance Phe508del-CFTR function. CF BECs transfected with TSBs potentiated the anion permeability effects of ivacaftor/lumacaftor or ivacaftor/tezacaftor on Phe508del-CFTR. Enhanced anion flux through Phe508del-CFTR when the CFTR modulators were applied after TSB4 or TSB6 transfection indicate that miR-223-3p and miR-145-5p, respectively, exert an inhibitory effect on CFTR function. When this inhibition is relieved with TSB transfection, the efficacy of ivacaftor/lumacaftor on CFTR function was enhanced up to 5-fold. The ability of TSBs to enhance up to 3-fold the corrector effects of ivacaftor/tezacaftor on Phe508del-CFTR function further strengthens the relevance of inhibition of specific MREs as an adjunct therapy for treating CF lung disease. Importantly, the TSBs were designed to be sequence specific to CFTR; therefore, off-target effects are predicted to be very limited, and their encapsulation into FDA-approved PLGA nanoparticles provides a biocompatible and safe delivery tool. Moreover, because TSBs function in a CFTR mutation-independent way they have the potential to be used as adjunct therapies for individuals with all classes of CFTR mutations.
There are limitations to this study. It was not possible to perform all assays in all of the models used, and functional analyses other than CFTR were beyond the scope of the current work. The lack of an observable effect of the TSBs in bronchospheres due to poor transfection was disappointing and is a challenge that faces the entire field in the development of anti-sense oligonucleotide delivery using in vitro organoid models of disease.32, 33, 34 To our knowledge, there is no description of locked nucleic acid delivery to organoids in the literature, and new methods will need to be developed to study their effects. Mucus-penetrating studies were also beyond the scope of this work; however, such studies will be important in the next steps of assessing this drug-device combination.35
Overall, the findings suggest that alone or in combination with CFTR modulators, aerosolized CFTR-targeting TSBs encapsulated in PLGA nanoparticles could represent a promising therapeutic strategy for the treatment for CFTR dysfunction in CF bronchial epithelium. In conclusion, the precise and selective inhibition of specific miRNAs binding to the CFTR 3′ UTR in CF BECs may offer an alternative or adjunct therapy to improve airway CFTR function in individuals homozygous for the p.Phe508del CFTR mutation.
Materials and Methods
Additional details are provided in the Supplemental Materials and Methods.
Cells, Patient Samples, and Treatments
Non-CF (16HBE14o−, Nuli-1) and CF (CFBE41o−, Cufi-1) human BECs, CFBE41o− stably transduced with wild-type (WT)-CFTR or Phe508del-CFTR, primary human BECs isolated from the proximal airways of six non-CF (five males, mean age 41.6 ± 8.0 years) and six CF adults (two males, mean age 29.3 ± 1.1) or purchased commercially, BECs isolated from six non-CF and six CF children (all male, mean age CF, 3 ± 2.5 years; non-CF, 3.3 ± 1.5 years), and CF and syngeneic gene-corrected (p.Phe508del/wild-type) bronchospheres generated from induced pluripotent stem cells were studied. All CF cells/samples were p.Phe508del/p.Phe508del. Where indicated, cells were treated with CFTR modulators (5 μM each) alone or in combination with TSBs or 1% CF bronchoalveolar lavage fluid (BALF). Ethics committee approval and informed consent were granted for all clinical samples.
miRNA Quantification
A TaqMan microRNA reverse transcription kit and predesigned TaqMan microRNA assays (Thermo Fisher Scientific, Waltham, MA, USA) were used for individual miRNA quantification on the LC480 LightCycler (Roche, Basel, Switzerland). Profiling of CF and non-CF pediatric samples was performed with TaqMan OpenArray human microRNA panels on the QuantStudio 12K flex system (Thermo Fisher Scientific, Waltham, MA, USA); GEO: GSE128861. Following library generation, RNA sequencing of CFBE41o− stably transduced with WT-CFTR or Phe508del-CFTR was performed on the Illumina NextSeq 500 (GEO: GSE128912).36, 37, 38
TSBs
TSBs were designed (Exiqon, now QIAGEN, Hilden, Germany) to compete with miR-101-3p, miR-145-5p, miR-223-3p, miR-494-3p, and miR-509-3p binding to specific sites in the CFTR 3′ UTR (Table S1).
Luciferase Assay
CFBE41o− cells were co-transfected with TSBs and wild-type39 or mutant CFTR 3′ UTR luciferase reporter plasmids. Luciferase activity was assessed by dual-luciferase reporter assay (Promega, Madison, WI, USA).
CFTR Assays
CFTR and β-actin were visualized in total protein extracts by western blot. CFTR activity in cell lines was measured using N-(ethoxycarbonylmethyl)-6- MQAE9 and YFP-H148Q/I152L.40,41 FIS was measured in CF bronchospheres.42
PLGA Nanoparticles
PLGA-TSB nanoparticles were formulated,24 and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and IL-6 and IL-8 ELISA were performed as before.43 Nanoparticles were analyzed by HCS.44 Following nebulization (Aerogen Solo), aerosol characteristics were measured using laser diffraction.45
Statistical Analysis
Analyses were performed using GraphPad PRISM 7.0. Results are expressed as mean ± SEM and compared as indicated. Differences were considered statistically significant when p ≤ 0.05.
Accession Numbers
Profiling data from n = 6 pediatric CF and non-CF BECs have been deposited with the Gene Expression Omnibus repository under accession number GEO: GSE128861. RNA sequencing data from ALI culture of CFBE41o− cells stably transfected with Phe508del versus wild-type CFTR have been deposited with the Gene Expression Omnibus repository under accession number GEO: GSE128912.
Author Contributions
C.D.S. performed study design, data collection, data analysis, data interpretation, figure design, and writing. E.F.F., I.O., N.M., and M.B.C. performed data collection, data analysis, data interpretation, and figure design. R.G. performed data collection, data analysis, data interpretation, figure design, and funding acquisition. S.V. performed data collection, data analysis, and data interpretation. A.G. performed data collection, data analysis, figure design, and funding acquisition. K.H. and F.H. performed data collection, data analysis, data interpretation, figure design, and writing. F.M. performed data analysis, data interpretation, and figure design. R.R., D.C.H., and J.C.S. performed data collection and data analysis. B.J.H. performed writing and funding acquisition. B.L. and S.A.C. performed data collection and funding acquisition. P.M. performed data collection, writing, and funding acquisition. R.M. and A.S.-U. performed data collection, data analysis, data interpretation, figure design, writing, and funding acquisition. C.M.G. performed conceptualization, study design, data analysis, data interpretation, figure design, writing, and funding acquisition.
Conflicts of Interest
The authors have no relevant conflicts of interest to declare other than the receipt of funding from the named agencies to carry out the current work.
Acknowledgments
Thank you to Andrew Berical and Rhiannon Werder in the Center for Regenerative Medicine, Boston University and Boston Medical Center and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, and Ruby Wang from Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA for technical assistance. Thank you to Dr. Nicoletta Pedemonte from the Gaslini Institute (Genova) who kindly donated the plasmid encoding YFP-H148Q/I152L protein. This work was supported by Cystic Fibrosis Foundation Therapeutics (GREENE15XXO to C.M.G.); the National Children's Research Centre (C/13/1 to C.M.G.), the NIH (R01HL144539 to A.S.-U.); the Cystic Fibrosis Foundation (SWIATE18G0 to A.S.-U.), Horizon2020 MSCA-IF (award 707771 GENDER-CF to A.G.), and the Irish Research Council (GOIPG/2015/2393 to R.G.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.02.001.
Supplemental Information
References
- 1.Rommens J.M., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 2.Riordan J.R., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 3.Bosch B., De Boeck K. Searching for a cure for cystic fibrosis. A 25-year quest in a nutshell. Eur. J. Pediatr. 2016;175:1–8. doi: 10.1007/s00431-015-2664-8. [DOI] [PubMed] [Google Scholar]
- 4.Glasgow A.M.A., De Santi C., Greene C.M. Non-coding RNA in cystic fibrosis. Biochem. Soc. Trans. 2018;46:619–630. doi: 10.1042/BST20170469. [DOI] [PubMed] [Google Scholar]
- 5.Megiorni F., Cialfi S., Dominici C., Quattrucci S., Pizzuti A. Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS ONE. 2011;6:e26601. doi: 10.1371/journal.pone.0026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viart V., Bergougnoux A., Bonini J., Varilh J., Chiron R., Tabary O., Molinari N., Claustres M., Taulan-Cadars M. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J. 2015;45:116–128. doi: 10.1183/09031936.00113214. [DOI] [PubMed] [Google Scholar]
- 8.Gillen A.E., Gosalia N., Leir S.H., Harris A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 2011;438:25–32. doi: 10.1042/BJ20110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oglesby I.K., Chotirmall S.H., McElvaney N.G., Greene C.M. Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in ΔF508 cystic fibrosis airway epithelium. J. Immunol. 2013;190:3354–3362. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- 10.Ramachandran S., Karp P.H., Osterhaus S.R., Jiang P., Wohlford-Lenane C., Lennox K.A., Jacobi A.M., Praekh K., Rose S.D., Behlke M.A. Post-transcriptional regulation of cystic fibrosis transmembrane conductance regulator expression and function by microRNAs. Am. J. Respir. Cell Mol. Biol. 2013;49:544–551. doi: 10.1165/rcmb.2012-0430OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amato F., Seia M., Giordano S., Elce A., Zarrilli F., Castaldo G., Tomaiuolo R. Gene mutation in microRNA target sites of CFTR gene: a novel pathogenetic mechanism in cystic fibrosis? PLoS ONE. 2013;8:e60448. doi: 10.1371/journal.pone.0060448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutful Kabir F., Ambalavanan N., Liu G., Li P., Solomon G.M., Lal C.V., Mazur M., Halloran B., Szul T., Gerthoffer W.T. MicroRNA-145 Antagonism Reverses TGF-β Inhibition of F508del CFTR Correction in Airway Epithelia. Am. J. Respir. Crit. Care Med. 2018;197:632–643. doi: 10.1164/rccm.201704-0732OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri E., Tamanini A., Jakova T., Gasparello J., Manicardi A., Corradini R., Sabbioni G., Finotti A., Borgatti M., Lampronti I. A Peptide Nucleic Acid against MicroRNA miR-145-5p Enhances the Expression of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) in Calu-3 Cells. Molecules. 2017;23:E71. doi: 10.3390/molecules23010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amato F., Tomaiuolo R., Nici F., Borbone N., Elce A., Catalanotti B., D’Errico S., Morgillo C.M., De Rosa G., Mayol L. Exploitation of a very small peptide nucleic acid as a new inhibitor of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. BioMed Res. Int. 2014;2014:610718. doi: 10.1155/2014/610718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finotti A., Gasparello J., Fabbri E., Tamanini A., Corradini R., Dechecchi M.C., Cabrini G., Gambari R. Enhancing the Expression of CFTR Using Antisense Molecules against MicroRNA miR-145-5p. Am. J. Respir. Crit. Care Med. 2019;199:1443–1444. doi: 10.1164/rccm.201901-0019LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonneville F., Ruffin M., Coraux C., Rousselet N., Le Rouzic P., Blouquit-Laye S., Corvol H., Tabary O. MicroRNA-9 downregulates the ANO1 chloride channel and contributes to cystic fibrosis lung pathology. Nat. Commun. 2017;8:710. doi: 10.1038/s41467-017-00813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Goor F., Hadida S., Grootenhuis P.D., Burton B., Cao D., Neuberger T., Turnbull A., Singh A., Joubran J., Hazlewood A. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Goor F., Hadida S., Grootenhuis P.D., Burton B., Stack J.H., Straley K.S., Decker C.J., Miller M., McCartney J., Olson E.R. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elborn J.S., Ramsey B.W., Boyle M.P., Konstan M.W., Huang X., Marigowda G., Waltz D., Wainwright C.E., VX-809 TRAFFIC and TRANSPORT study groups Efficacy and safety of lumacaftor/ivacaftor combination therapy in patients with cystic fibrosis homozygous for Phe508del CFTR by pulmonary function subgroup: a pooled analysis. Lancet Respir. Med. 2016;4:617–626. doi: 10.1016/S2213-2600(16)30121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowe S.M., Daines C., Ringshausen F.C., Kerem E., Wilson J., Tullis E., Nair N., Simard C., Han L., Ingenito E.P. Tezacaftor-Ivacaftor in Residual-Function Heterozygotes with Cystic Fibrosis. N. Engl. J. Med. 2017;377:2024–2035. doi: 10.1056/NEJMoa1709847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dugernier J., Hesse M., Vanbever R., Depoortere V., Roeseler J., Michotte J.B., Laterre P.F., Jamar F., Reychler G. SPECT-CT Comparison of Lung Deposition using a System combining a Vibrating-mesh Nebulizer with a Valved Holding Chamber and a Conventional Jet Nebulizer: a Randomized Cross-over Study. Pharm. Res. 2017;34:290–300. doi: 10.1007/s11095-016-2061-7. [DOI] [PubMed] [Google Scholar]
- 22.Altenburg A.F., van de Sandt C.E., Li B.W.S., MacLoughlin R.J., Fouchier R.A.M., van Amerongen G., Volz A., Hendriks R.W., de Swart R.L., Sutter G. Modified Vaccinia Virus Ankara Preferentially Targets Antigen Presenting Cells In Vitro, Ex Vivo and In Vivo. Sci. Rep. 2017;7:8580. doi: 10.1038/s41598-017-08719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Meegen M.A., Terheggen S.W., Koymans K.J., Vijftigschild L.A., Dekkers J.F., van der Ent C.K., Beekman J.M. CFTR-mutation specific applications of CFTR-directed monoclonal antibodies. J. Cyst. Fibros. 2013;12:487–496. doi: 10.1016/j.jcf.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Fernández Fernández E., Santos-Carballal B., de Santi C., Ramsey J.M., MacLoughlin R., Cryan S.A., Greene C.M. Biopolymer-Based Nanoparticles for Cystic Fibrosis Lung Gene Therapy Studies. Materials (Basel) 2018;11:E122. doi: 10.3390/ma11010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alton E.W.F.W., Armstrong D.K., Ashby D., Bayfield K.J., Bilton D., Bloomfield E.V., Boyd A.C., Brand J., Buchan R., Calcedo R., UK Cystic Fibrosis Gene Therapy Consortium Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015;3:684–691. doi: 10.1016/S2213-2600(15)00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alton E.W., Beekman J.M., Boyd A.C., Brand J., Carlon M.S., Connolly M.M., Chan M., Conlon S., Davidson H.E., Davies J.C. Preparation for a first-in-man lentivirus trial in patients with cystic fibrosis. Thorax. 2017;72:137–147. doi: 10.1136/thoraxjnl-2016-208406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooney A.L., Thornell I.M., Singh B.K., Shah V.S., Stoltz D.A., McCray P.B., Jr., Zabner J., Sinn P.L. A Novel AAV-mediated Gene Delivery System Corrects CFTR Function in Pigs. Am. J. Respir. Cell Mol. Biol. 2019;61:747–754. doi: 10.1165/rcmb.2019-0006OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vituret C., Gallay K., Confort M.P., Ftaich N., Matei C.I., Archer F., Ronfort C., Mornex J.F., Chanson M., Di Pietro A. Transfer of the Cystic Fibrosis Transmembrane Conductance Regulator to Human Cystic Fibrosis Cells Mediated by Extracellular Vesicles. Hum. Gene Ther. 2016;27:166–183. doi: 10.1089/hum.2015.144. [DOI] [PubMed] [Google Scholar]
- 29.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haque A.K.M.A., Dewerth A., Antony J.S., Riethmüller J., Schweizer G.R., Weinmann P., Latifi N., Yasar H., Pedemonte N., Sondo E. Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis. Sci. Rep. 2018;8:16776. doi: 10.1038/s41598-018-34960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki S., Sun R., Bui H.H., Crosby J.R., Monia B.P., Guo S. Steric Inhibition of 5′ UTR Regulatory Elements Results in Upregulation of Human CFTR. Mol. Ther. 2019;27:1749–1757. doi: 10.1016/j.ymthe.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhise N.S., Gray R.S., Sunshine J.C., Htet S., Ewald A.J., Green J.J. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31:8088–8096. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellor H.R., Davies L.A., Caspar H., Pringle C.R., Hyde S.C., Gill D.R., Callaghan R. Optimising non-viral gene delivery in a tumour spheroid model. J. Gene Med. 2006;8:1160–1170. doi: 10.1002/jgm.947. [DOI] [PubMed] [Google Scholar]
- 34.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacheco D.P., Butnarasu C.S., Briatico Vangosa F., Pastorino L., Visai L., Visentin S., Petrini P. Disassembling the complexity of mucus barriers to develop a fast screening tool for early drug discovery. J. Mater. Chem. B Mater. Biol. Med. 2019;7:4940–4952. doi: 10.1039/c9tb00957d. [DOI] [PubMed] [Google Scholar]
- 36.Swiatecka-Urban A., Brown A., Moreau-Marquis S., Renuka J., Coutermarsh B., Barnaby R., Karlson K.H., Flotte T.R., Fukuda M., Langford G.M., Stanton B.A. The short apical membrane half-life of rescued {Delta}F508-cystic fibrosis transmembrane conductance regulator (CFTR) results from accelerated endocytosis of {Delta}F508-CFTR in polarized human airway epithelial cells. J. Biol. Chem. 2005;280:36762–36772. doi: 10.1074/jbc.M508944200. [DOI] [PubMed] [Google Scholar]
- 37.Snodgrass S.M., Cihil K.M., Cornuet P.K., Myerburg M.M., Swiatecka-Urban A. Tgf-β1 inhibits Cftr biogenesis and prevents functional rescue of ΔF508-Cftr in primary differentiated human bronchial epithelial cells. PLoS ONE. 2013;8:e63167. doi: 10.1371/journal.pone.0063167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barberán-Soler S., Vo J.M., Hogans R.E., Dallas A., Johnston B.H., Kazakov S.A. Decreasing miRNA sequencing bias using a single adapter and circularization approach. Genome Biol. 2018;19:105. doi: 10.1186/s13059-018-1488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Santi C., Gadi S., Swiatecka-Urban A., Greene C.M. Identification of a novel functional miR-143-5p recognition element in the Cystic Fibrosis Transmembrane Conductance Regulator 3'UTR. AIMS Genet. 2018;5:53–62. doi: 10.3934/genet.2018.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galietta L.V., Jayaraman S., Verkman A.S. Cell-based assay for high-throughput quantitative screening of CFTR chloride transport agonists. Am. J. Physiol. Cell Physiol. 2001;281:C1734–C1742. doi: 10.1152/ajpcell.2001.281.5.C1734. [DOI] [PubMed] [Google Scholar]
- 41.Caputo A., Hinzpeter A., Caci E., Pedemonte N., Arous N., Di Duca M., Zegarra-Moran O., Fanen P., Galietta L.J. Mutation-specific potency and efficacy of cystic fibrosis transmembrane conductance regulator chloride channel potentiators. J. Pharmacol. Exp. Ther. 2009;330:783–791. doi: 10.1124/jpet.109.154146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vencken S., Foged C., Ramsey J.M., Sweeney L., Cryan S.A., MacLoughlin R.J., Greene C.M. Nebulised lipid-polymer hybrid nanoparticles for the delivery of a therapeutic anti-inflammatory microRNA to bronchial epithelial cells. ERJ Open Res. 2019;5:2. doi: 10.1183/23120541.00161-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh D.P., Murphy R.D., Panarella A., Raftery R.M., Cavanagh B., Simpson J.C., O’Brien F.J., Heise A., Cryan S.A. Bioinspired Star-Shaped Poly(l-lysine) Polypeptides: Efficient Polymeric Nanocarriers for the Delivery of DNA to Mesenchymal Stem Cells. Mol. Pharm. 2018;15:1878–1891. doi: 10.1021/acs.molpharmaceut.8b00044. [DOI] [PubMed] [Google Scholar]
- 45.Hibbitts A., O’Mahony A.M., Forde E., Nolan L., Ogier J., Desgranges S., Darcy R., MacLoughlin R., O’Driscoll C.M., Cryan S.A. Early-stage development of novel cyclodextrin-siRNA nanocomplexes allows for successful postnebulization transfection of bronchial epithelial cells. J. Aerosol Med. Pulm. Drug Deliv. 2014;27:466–477. doi: 10.1089/jamp.2013.1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.