Abstract
The N-degron pathway is an emerging target for anti-tumor therapies, because of its capacity to positively regulate many hallmarks of cancer, including angiogenesis, cell proliferation, motility, and survival. Thus, inhibition of the N-degron pathway offers the potential to be a highly effective anti-cancer treatment. With the use of a small interfering RNA (siRNA)-mediated approach for selective downregulation of the four Arg/N-degron-dependent ubiquitin ligases, UBR1, UBR2, UBR4, and UBR5, we demonstrated decreased cell migration and proliferation and increased spontaneous apoptosis in cancer cells. Chronic treatment with lipid nanoparticles (LNPs) loaded with siRNA in mice efficiently downregulates the expression of UBR-ubiquitin ligases in the liver without any significant toxic effects but engages the immune system and causes inflammation. However, when used in a lower dose, in combination with a chemotherapeutic drug, downregulation of the Arg/N-degron pathway E3 ligases successfully reduced tumor load by decreasing proliferation and increasing apoptosis in a mouse model of hepatocellular carcinoma, while avoiding the inflammatory response. Our study demonstrates that UBR-ubiquitin ligases of the Arg/N-degron pathway are promising targets for the development of improved therapies for many cancer types.
Keywords: N-degron pathway, UBR-ubiquitin ligases, hepatocellular carcinoma, chemotherapy, apoptosis, UBR1, UBR2, UBR4, UBR5
Graphical Abstract
With the use of a small interfering RNA (siRNA)-mediated approach for selective downregulation of the Arg/N-degron pathway in a murine hepatocellular carcinoma model, Leboeuf et al. demonstrated potentiated action of chemotherapy. Since components of this pathway are ubiquitously expressed, targeting the N-degron pathway has the potential to increase effectiveness of therapy in many cancer types.
Introduction
Many significant advances in cancer therapy have emerged in the last few years, offering promising solutions for previously untreatable diseases. However, some cancers, such as hepatocellular carcinoma (HCC), remain refractory to current therapies. The high functional capacity of the liver and the absence of pathological symptoms complicate early diagnosis of HCC,1 resulting in a high occurrence of inoperable patients. Additionally, many patients develop resistance to sorafenib, and only recently, multiple kinase-inhibitor regorafenib and two different checkpoint inhibitors were approved for patients who progress after sorafenib.2 Despite this, the median survival following diagnostic for inoperable patients is 6 to 8 months,1 highlighting the need to develop novel medications and approaches. Recent developments in RNA therapy resulted in efficient long-term drugs for hereditary and metabolic liver diseases that outperform common small molecules and antibodies.3 As naked RNA is quickly excreted by kidneys and has poor delivery efficiency, carriers are needed to deliver the small interfering RNA (siRNA) to the cells of interest. However, development of these delivery systems remains the main challenge today.4 Among several systems for siRNA delivery in vivo, lipid nanoparticles (LNPs) are robust, efficient, and clinically validated and provide efficient delivery to the liver.5,6 During circulation in the bloodstream, LNPs are covered by a protein corona, including apolipoproteins that drive specific receptor-mediated delivery of LNPs to hepatocytes.7 Enormous efforts in optimizing siRNA-containing lipid nanoparticles for hepatic-specific silencing8 resulted in the development of a Food and Drug Administration (FDA)-approved, siRNA-LNP-based drug targeting the TTR gene for the treatment of hereditary transthyretin amyloidosis.9 This technology can be applied to downregulate other vulnerable molecular pathways and targets directly in diseased cells.
In recent years, the Arg/N-degron pathway emerged as a proteolytic system relevant to perturbations that underlie cancer.10, 11, 12 Through regulated degradation of specific proteins, the Arg/N-degron pathway (previously known as the Arg/N-end rule pathway) mediates a strikingly large number of biological functions.13,14 Recent studies revealed a major anti-apoptotic function of the Arg/N-degron pathway through its ability to selectively degrade specific proapoptotic protein fragments, including Asp-BRCA1, Arg-BIMEL, Cys-TRAF1, and Cys-RIPK1.15,16 Apoptosis is critical for removing unneeded or diseased cells; therefore, blocking apoptosis is vital to cancer cell survival. Conversely, the promotion of cell death is a key aspect of most cancer therapies. The anti-apoptotic activity of the Arg/N-degron pathway suggests that its upregulation may suppress apoptosis and thus promote malignant phenotypes. Indeed, recent studies have demonstrated that UBR5 (EDD), one of the key ubiquitin ligases of this pathway, is often amplified in liver, lung, and ovarian carcinomas, supporting a model wherein UBR5 could be targeted to decrease cancer cell survival.17 Another ubiquitin ligase of the Arg/N-degron pathway, UBR2, has been shown to be upregulated in tumors in response to cachectic stimuli, including proinflammatory cytokines.18 Finally, loss-of-function genetic studies in model organisms indicate that this pathway positively regulates vascular development and cell motility.19, 20, 21 These and other data argue that the Arg/N-degron pathway plays an important role in the positive regulation of cancer cell proliferation, motility, and survival. Therefore, inhibition of this pathway offers the potential to be a highly effective anti-tumor treatment.
The recognition components of the N-degron pathway are specific E3 ubiquitin ligases (N-recognins) that can target destabilizing N-terminal amino acids termed N-degrons.14,22,23 Both human and mouse have four known N-recognins in the Arg branch of this pathway, UBR1, UBR2, UBR4, and UBR5, that are functionally redundant24, 25, 26, 27 and essential for early embryonic development.21,28, 29, 30 These proteins are structurally dissimilar and belong to different families of ubiquitin ligases, which makes the development of a universal, small-molecule inhibitor extremely challenging, if possible.31, 32, 33, 34, 35 Several attempts were made to regulate the N-degron pathway by small molecules. Di-peptides as well as heterovalent ligands were developed to mimic N-degrons and competitively bind N-recognins, preventing binding to their natural substrates.32,33,36 Other compounds, such as tannic acid and merbromin, were found to inhibit ATE1, a critical enzyme of the Arg branch of this pathway, acting before the E3 ligases, specifically and effectively disturbing ATE1 activities in the cell.37 siRNAs were also used to downregulate individual N-recognins of the Arg/N-degron pathway in vitro, illustrating their distinctive roles in the degradation of particular targets.38, 39, 40 However, these studies were confined to in vitro settings, resulting in transient or partial inhibition of the N-degron pathway.
Here, we investigate the UBR-ubiquitin ligases of the Arg/N-degron pathway as new targets for cancer therapy using clinically proven RNA interference (RNAi) technology.41 Downregulation of the Arg/N-degron pathway both in vitro and in vivo induces a profound effect on proliferation, migration, and apoptosis of cancer cells. Prolonged downregulation of the E3 ligases of the Arg/N-degron pathway is well tolerated by normal tissue at a variety of doses. Moreover, we show that downregulation of the UBR-ubiquitin ligases, in combination with apoptosis-inducing drugs, decreases progression of liver cancer in the mouse model.
Results
Knockdown of UBR-Ubiquitin Ligases In Vitro
In order to achieve targeted silencing of the mouse Ubr1, Ubr2, Ubr4, and Ubr5 mRNA, the best-scored 10 siRNA against each UBR-ubiquitin ligase was screened in Hepa 1–6 cells (Figure S1A). siRNAs were selected to avoid off-target activity based on several known criteria.42, 43, 44 The two most highly potent and effective siRNA probes for each UBR-ubiquitin ligase were chosen and used in further studies (Figure S1B). The low half-maximal inhibitory concentration (IC50) allows for using low concentrations of siRNA, which additionally decreases the possibility of off-target effects. A concentration of 0.25 nM of each siRNA was found to be sufficient to reach 70%–80% downregulation of the protein after 72 h of exposure (Figures 1A and 1B) in Hepa 1–6 and AML-12 cells.
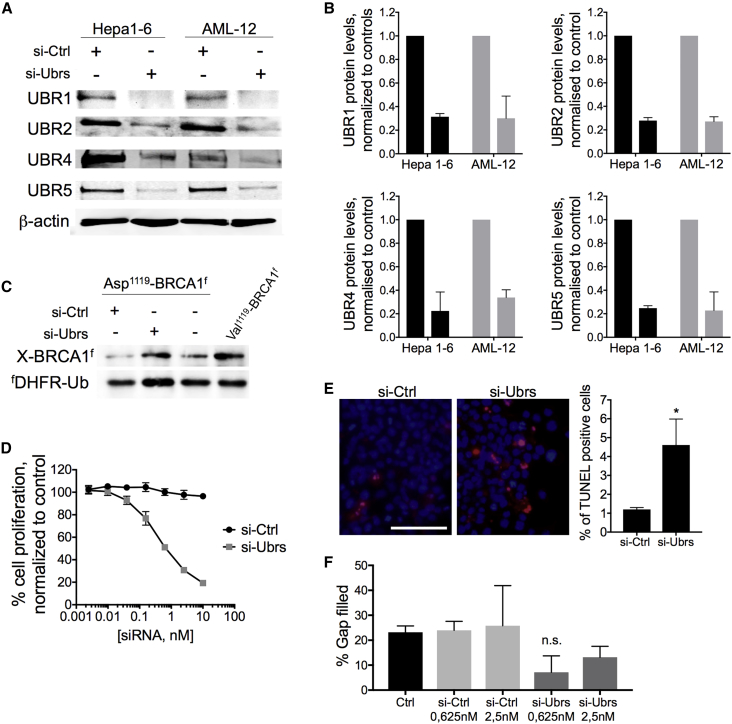
Figure 1.
Effect of the Downregulation of Ubr-Ubiquitin Ligases of the Arg/N-Degron Pathway In Vitro
(A) Western blot analysis of UBR1, UBR2, UBR4, and UBR5 proteins in Hepa 1–6 and AML-12 cells after 72 h of exposure to 1 nM siRNA. (B) Quantification of protein Ubr expression in Hepa 1–6 and AML-12 cells after 72 h of exposure to 1 nM siRNA. (C) Asp1119-BRCA1f (produced from fDHFR-UbR48-Asp1119-BRCA1f) and Val1119-BRCA1f were expressed in Hepa 1–6 cells transfected with siRNA. Protein extracts were analyzed by SDS-PAGE and western blot against FLAG. (D) Analysis of cell proliferation by the neutral red assay on Hepa 1–6 cells after 72 h of exposure to siRNA. (E) Analysis of cell death TUNEL assay in Hepa 1–6 cells after 72 h of exposure to 10 nM siRNA. Red nuclei represent TUNEL-positive cells. Scale bar, 100 μm. (F) Migration assay performed on Hepa 1–6 cells after 72 h of exposure to siRNA against Ubrs. Results show mean ± SD. p values were determined by a Student’s test (*p = 0.01).
Next, the Asp1119-BRCA1 degradation reporter was used to prove that the observed downregulation of Arg/N-degron ubiquitin ligases is sufficient to decrease the functional activity of the pathway.15 This reporter system is comprised of a reference FLAG-tagged derivative of the mouse dihydrofolate reductase (fDHFR-UbR48) coupled to the FLAG-tagged known target of the Arg/N-degron pathway. Cotranslational cleavage of the Ub fusion by deubiquitylases produces, at the initial equimolar ratio, both the test protein with a desired N-terminal residue and the reference protein fragment. In normal conditions, the proapoptotic fragment of the BRCA-1 protein (Asp-BRCA1) is degraded by the Arg/N-degron pathway and should therefore accumulate if the pathway is impaired. Indeed, less degradation of the proapoptotic fragment of BRCA1 was observed when Hepa 1–6 cells were treated with siRNA against Ubrs than in the controls (Figure 1C), indicating that the functional activity of the N-degron pathway is inhibited in our system.
The Arg/N-degron pathway is known to positively regulate cell proliferation and migration.19,21 Neutral red and scratch assays were used to confirm that downregulation of UBR1, UBR2, UBR4, and UBR5 affected these biological processes in Hepa 1–6 cells after 72 h of exposure to siRNA. Indeed, both cell proliferation and migration were decreased after transfection with siUbrs compared to si control (siCtrl) (Figures 1D–1F; Figure S2A). On the other hand, the Arg/N-degron pathway is responsible for the degradation of proapoptotic fragments.15 Therefore, downregulation of the UBR-ubiquitin ligases should translate into an increase of spontaneous apoptosis in siRNA-transfected cells. Spontaneous apoptosis was increased by 3%–8% compared to controls, as demonstrated by the TUNEL assay (Figure 1E) and Annexin V staining (Figure S3). We observed no positive 7-aminoactinomycin D (7AAD) cells, indicating that only the apoptotic cell death pathway was increased in siRNA-Ubr-treated cells. The in vitro results were also validated using a second set of highly potent Ubr siRNAs (Figure S2), confirming the on-target effect of siRNA-mediated downregulation on cell proliferation, migration, and apoptosis.
UBR-Ubiquitin Ligase Knockdown in Healthy Mouse Liver
The selected siRNAs against UBR1, UBR2, UBR4, and UBR5 were individually formulated into C12-200 lipid nanoparticles, previously validated in mice and nonhuman primates,45 and evaluated 72 h post-treatment for activity in vivo at a dose of 0.25 mg/kg each (Figure S1C). As formulated siRNA in LNPs enables multiple gene silencing in vivo, we made LNP bearing either one or four siRNA duplexes targeting UBR-ubiquitin ligases. Due to their size (80–90 nm) and almost neutral charge, C12-200 siRNA-LNPs easily pass through the fenestrae of the endothelium layer, separating hepatocytes from blood, and are further internalized by hepatocytes via macropinocytosis.45 The biological distribution of C12-200 siRNA LNP after intravenous (i.v.) injection in mice has been thoroughly assessed in previous studies, confirming RNAi-mediated silencing specifically in the liver.46 The most efficient LNP-siRNAs (si1-6, si2-9, si4-7, and si5-4; Table S1) were selected to perform dose-response, target-recovery, and tissue-biodistribution experiments (Figures S1D–S1F), as well as all further experiments in this study. Single administration of LNP-siUbr at doses ranging from 0.125–0.5 mg/kg resulted in profound knockdown of Ubr mRNA levels (80%–85%) in the liver of naive animals, 72 h postinjection (Figure S1D). None of the doses tested resulted in death of the animals, indicating that we did not reach the maximum tolerated dose. No significant target downregulation was detected in the spleen, kidney, lungs, and bone marrow. However, 50%–60% knockdown of Ubrs was observed in the visceral adipose tissue (Figure S1F), possibly due to delivery to peripheral macrophages.47 Maximal mRNA and protein downregulation in the liver occurred 3 days after injection, followed by a slow recovery (Figure S1E), and silencing to more than 60% lasted at least 10 days, which allows a convenient once-per-week regimen for multiple dosing.
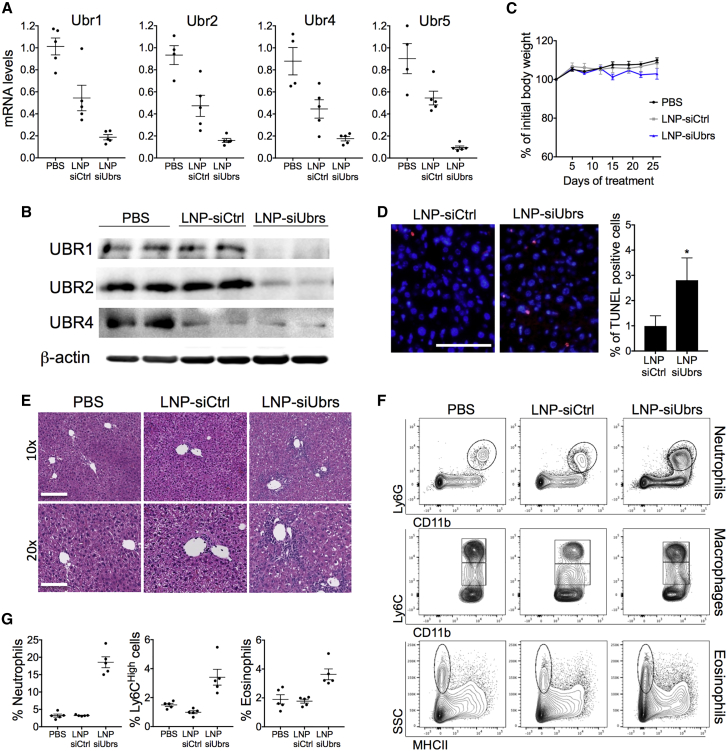
Chronic treatment with LNPs revealed that long-term downregulation of UBR-ubiquitin ligases can be achieved in the liver of healthy mice without inducing significant toxicity to this organ (Figure 2). C57BL/6 females received biweekly injections of LNPs loaded with siRNA (1.4 mg/kg total) for 4 weeks. Although the levels of ALT, AST, and ALP were increased in the mice receiving LNP-siUbrs (Table S3), no significant weight change or differences in the behavior of the mice were observed between the groups (Figure 2C), indicating that long-term administration of LNP-siUbrs was well tolerated. 75%–90% downregulation of Ubr1, Ubr2, Ubr4, and Ubr5 mRNA in the liver was achieved, compared to the PBS control group; however, a decrease of the mRNA levels of all Ubrs was also seen in the control siRNA group. Nonetheless, a drop in Ubr mRNA levels was not observed in other long-term treatments with LNPs loaded with the siRNA against luciferase at a less or equal dose (see Figure S5), and no phenotypical differences were observed between the healthy animals and the LNP-siCtrl-treated animals. Treatment with the control LNP slightly decreased the level of UBR4 protein, which could be the result of accelerated lysosomal degradation of UBR4.48 Indeed, multiple injections with LNPs can lead to significant lipid accumulation in liver cells and increased autophagy, which in turn, initiates lysosome degradation.49 Finally, chronic downregulation of UBR-ubiquitin ligases led to a slight but significant increase of apoptotic cells and enlarged intercellular space in the liver (Figures 2D and 2E), which confirms that the function of the Arg/N-degron pathway is diminished in this organ.
Figure 2.
Chronic Downregulation of Ubr-Ubiquitin Ligases in the Liver Induces Inflammation
Mice were treated with LNP-siRNA for 4 weeks, 2 times/week. The combined dose of LNP-siUbr siRNA was 0.3 mg/kg for Ubr1, -2, and -5 and 0.5 mg/kg for Ubr4, for a total of 1.4 mg/kg. Mice treated with LNP-siCtrl (luciferase) received 1.4 mg/kg. (A) mRNA and (B) protein levels of Ubr1, Ubr2, Ubr4, and Ubr5 in the liver of LNP-treated mice. Protein levels of UBR5 are undetectable in the liver. (C) Weight curve of animals treated with LNPs, measured before each injection and averaged for the group. Measures were compared as a percentage of change from day 0. (D) Analysis of cell death by the TUNEL assay in livers of LNP-treated mice. Red nuclei represent TUNEL-positive cells. Scale bar, 100 μm. p values were determined by a Mann-Whitney test (*p < 0.05). (E) H&E stain of mice treated with LNPs or vehicle. Scale bars, 200 μm (10 times) and 100 μm (20 times). (F) Representative flow cytometry analysis of neutrophil, macrophage, and eosinophil populations in the liver of mice treated with LNPs. (G) Percent of neutrophil, Ly6Chigh macrophage, or eosinophil populations in the livers of LNP or vehicle-treated mice, gated on CD45+ cells. Results show mean ± SEM; n = 5 mice per group.
One noticeable result is the infiltration of mononuclear cells in the livers of mice treated with LNPs containing siRNA against UBRs (Figure 2E). The infiltrating cells were identified by flow cytometry as neutrophils, Ly6Chigh macrophages, and eosinophils (Figures 2F and 2G), indicating the presence of chronic inflammation in the liver. These cell populations were also increased in the spleen of the same animals (Figures S4B and S4C); however, this seems to be a consequence of the inflammation present in the liver rather than the influence of the Arg/N-degron pathway in the spleen, since no downregulation of Ubr mRNA was observed in the spleen (Figure S4A).
Knockdown of the N-degron Pathway in a Spontaneous Model of HCC
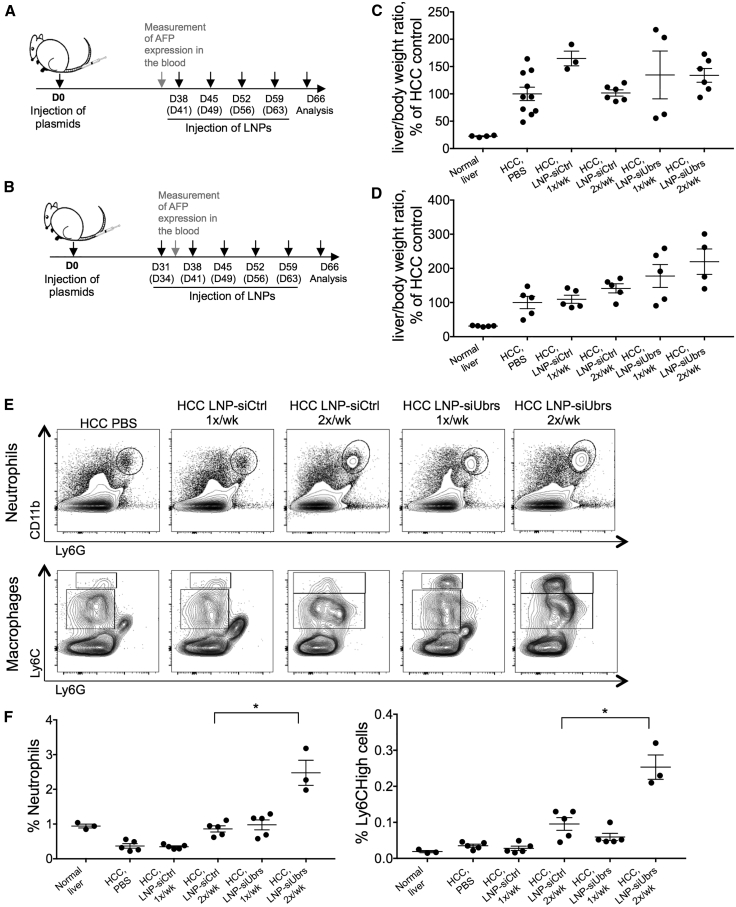
To evaluate the effects of downregulation of the Arg/N-degron pathway in the context of liver cancer, we used a spontaneous model of HCC developed by Tward et al.50 Briefly, plasmids containing human MET and ΔN90-β-catenin and the transposase Sleeping Beauty are delivered to hepatocytes by hydrodynamic injection, and the oncogenes are stably integrated into the genome by the Sleeping Beauty transposase. By the 5th week after injection, α-fetoprotein becomes detectable in the serum of tumor-bearing mice, and by the end of the 10th week, the liver is greatly enlarged up to a liver-to-body mass ratio of 15% to 40% versus 4%–5% in nontumor control animals. We previously demonstrated efficient delivery of siRNA to the tumor nodules that develop in the plasmid-injected mice.46 As UBR5 and UBR2 have been reported to be overexpressed in some types of cancer,17,18 we assessed the level of mRNA expression of all four UBR-ubiquitin ligases and found that none was upregulated in our specific HCC model (Figure S5). The impact of downregulating the Arg/N-degron pathway on tumor load was evaluated by injecting lipid nanoparticles containing siRNA against Ubrs or luciferase at a total dose of 1.4 mg/kg into tumor-bearing mice for 4 or 5 weeks, once or twice per week (Figures 3A and 3B and Figure S5). In all regimens tested, downregulation of the targeted UBR-ubiquitin ligases with a high LNP dose led to an increase in liver/body weight ratio and tumor load (Figures 3C and 3D). Increased populations of neutrophils and Ly6Chigh cells were also observed in the spleens of HCC animals treated with LNP-siUbrs twice a week (Figures 3E and 3F), demonstrating increased inflammation in these animals, a well-known driver of HCC development and progression. As a consequence, we re-evaluated the regimen of LNP injections and chose a lower dose and frequency for the ensuing experiments to mitigate the observed inflammation.
Figure 3.
Downregulation of UBR-Ubiquitin Ligases in HCC Aggravates Tumor Load and Inflammation
Schematic representation of the experiment: timeline of tumor induction (injection of oncogene-encoding plasmids) and repeated injections of LNP-formulated siRNA. LNPs were injected for 4 weeks (A) or 5 weeks (B), once or twice a week. Tissues were collected for analysis on day 66 after tumor induction. (C and D) Liver/body mass ratio analysis of mice bearing HCC, treated for 4 weeks (C) or 5 weeks (D) with LNPs. Ratios were calculated to the ratios of the HCC control group. (E) Representative flow cytometry analysis of neutrophil and Ly6Chigh populations in the spleen of HCC mice treated with LNPs. (F) Percent of neutrophil and Ly6Chigh macrophage populations in the spleen of HCC mice treated with LNP, gated on CD45+ cells. Results show mean ± SEM; n = 3–10 mice per group. p values were determined by a Mann-Whitney test (*p < 0.05).
Knockdown of the Arg/N-degron Pathway Potentiates the Action of Apoptosis-Inducing Drugs
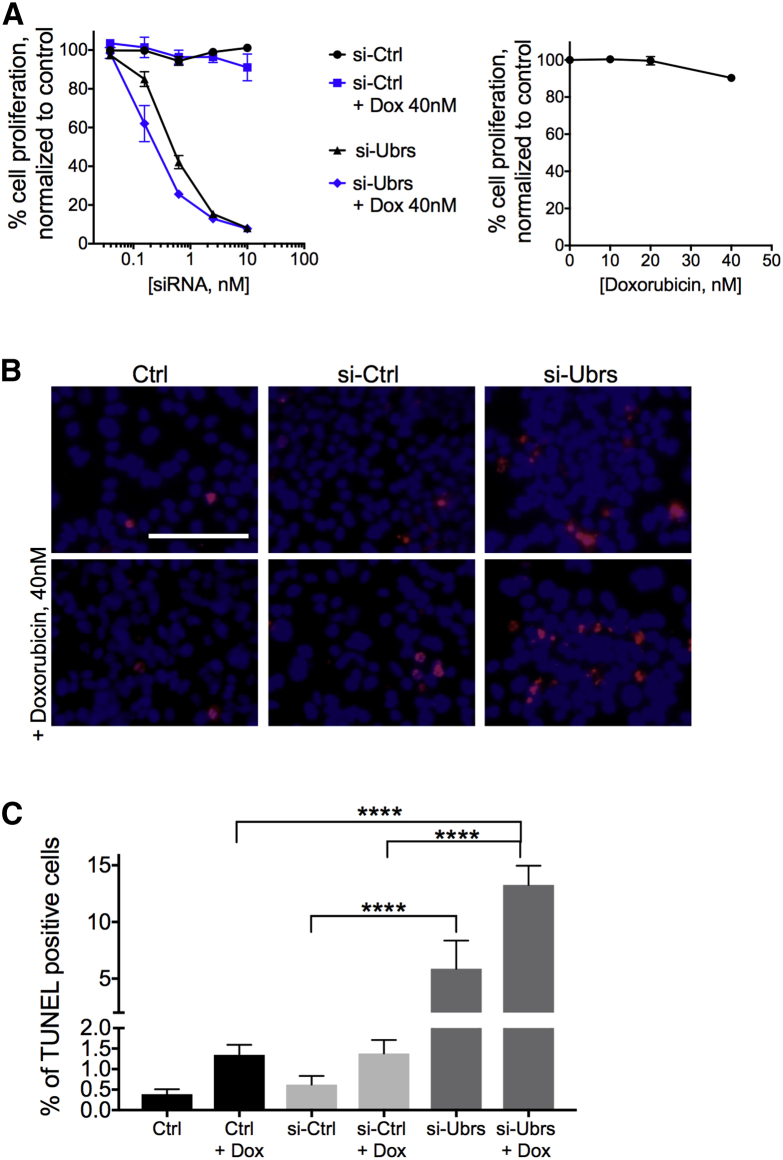
In order to capitalize on the effects of the Arg/N-degron pathway downregulation on proliferation and apoptosis in cancer cells while avoiding increased inflammation, the following strategy was adopted: a combinatorial treatment with apoptosis-inducing drugs combined with a reduced dose of siRNA against UBR1, UBR2, UBR4, and UBR5. This approach was first tested in vitro to assess the impact of downregulating UBRs in the presence of staurosporine or doxorubicin on proliferation and apoptosis. Hepa 1–6 cells were transfected with siRNA for 72 h before the addition of low doses of doxorubicin (Figure 4) or staurosporine (Figure S6) for an additional 48 h or 24 h, respectively. Proliferation was inhibited at lower doses of siRNA against UBRs when doxorubicin is added to the cells (Figure 4A). Indeed, at 0.156 nM of Ubr-siRNA, when proliferation of the cells is inhibited by 15%–25%, addition of doxorubicin at doses that do not affect proliferation (right graph) accentuates this phenotype by another 15%–20%. As for apoptosis, when cells are treated with siRNA against UBR-ubiquitin ligases alone, we see an increase of 4%–5% of apoptotic cells compared to controls. However, when the siRNA are used in combination with apoptosis-inducing drugs, apoptosis increases by 11.5%–13.5% compared to controls (Figures 4B and 4C). Therefore, the combinatorial approach decreases proliferation and increases apoptosis at lower doses of siRNA and apoptosis-inducing drugs that would otherwise be inefficient alone, demonstrating that both treatments act in synergy to kill cancer cells.
Figure 4.
Knockdown of UBR-Ubiquitin Ligases of the Arg/N-degron Pathway Potentiates the Action of Apoptosis-Inducing Drugs In Vitro
(A) Proliferation of Hepa 1–6 cells after exposure to siRNA and doxorubicin (left) or doxorubicin alone (right). (B) Analysis of cell death by TUNEL in Hepa 1–6 cells after 72 h of exposure to 1 nM siRNA against Ubrs and an additional 48 h to doxorubicin. Red nuclei represent TUNEL-positive cells. Scale bar, 100 μm. (C) Quantification of the results in (B). Results show mean ± SD. p values were determined by a one-way ANOVA (****p < 0.0001).
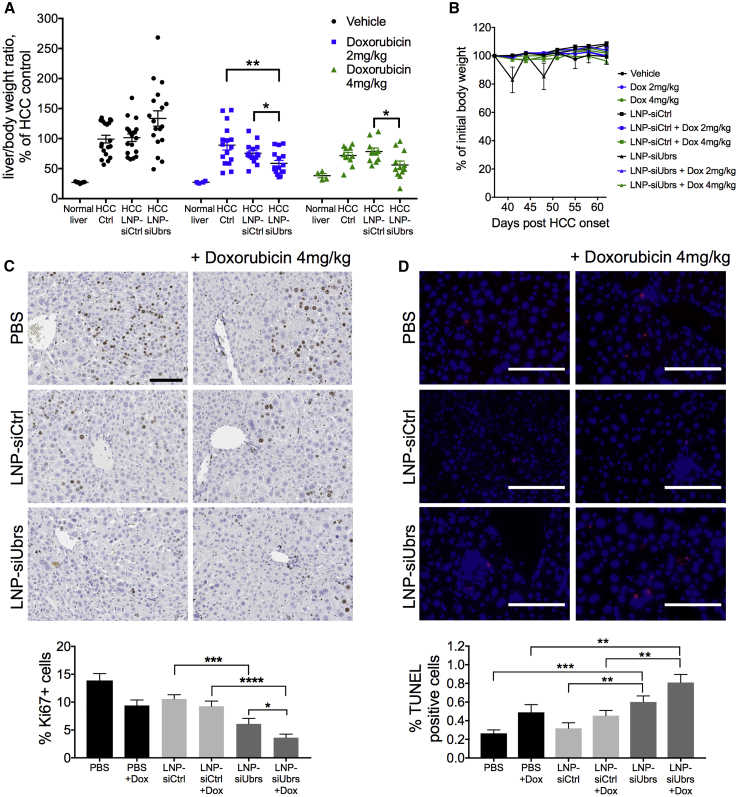
Finally, the combinatorial approach was tested in the spontaneous mouse model of HCC. Mice with HCC were alternatively injected with LNP-siRNA at 1 mg/kg total and/or doxorubicin (2 mg/kg or 4 mg/kg) every 3–4 days for a total of 4 weeks, starting with LNP-siRNA on day 37 after induction of the cancer (Figure S7A). We found that prolonged downregulation of the Arg/N-degron pathway, combined with the use of a chemotherapeutic agent, led to inhibition of HCC progression. Relative liver weights were decreased by an average of 30% compared to controls (Figure 5A), and tumor load was significantly lower than that observed with doxorubicin alone and that of the control LNP-siRNA with doxorubicin. Morphology of the HCC livers treated with LNP-siUbrs and doxorubicin resembled normal liver tissue, contrarily to all other groups, indicating less proliferation and less damage to the tissue (Figure S7B). The reduced dose of LNPs used in this case allowed avoidance of an increase of inflammatory immune cells (Figure S7C). Additionally, doxorubicin does not have an immediate effect on inflammation nor does it affect the viability of immune cells, as demonstrated by the equal or increased population of neutrophils, macrophages, or dendritic cells in the spleens of mice treated with LNPs and doxorubicin (Figure S7C). To demonstrate the effect of the combinatorial treatment on tumor cell proliferation and survival, we stained fixed liver sections for Ki67, a marker of cellular proliferation (Figure 5C). In the animals treated with LNP-siUbrs, Ki67-positive cells were decreased by an average of 6% compared to controls, and the addition of doxorubicin increases the difference to 9% (p = 0.0001, Mann-Whitney test). Finally, doxorubicin treatment resulted in a significant increase of TUNEL-positive staining in the liver of HCC-bearing mice treated with LNP-siUbrs. (Figure 5D). Together, these data suggest that the combined effects of the downregulation of the Arg/N-degron pathway, with the use of chemotherapy on the progression of hepatocellular carcinoma, are not mediated by modulation of the inflammatory response but rather by an inhibitory effect on proliferation and an increase of apoptosis.
Figure 5.
Chemotherapy in HCC Is More Efficient Together with Downregulation of the Arg/N-degron Pathway
(A) Liver/body mass ratio analysis of mice bearing HCC treated for 4 weeks with LNPs at 1 mg/kg, with or without doxorubicin treatment; n = 10–18 mice per group. Results show mean ± SEM. (B) Weight curve of animals treated with LNPs, measured before each injection and averaged for the group. Measures were compared as a percentage of change from the first injection day. (C) Liver sections stained for the expression of Ki67 as a measure of cell proliferation; n = 4 mice per group. (D) Analysis of cell death by TUNEL assay in liver sections. Red nuclei represent TUNEL-positive cells. Scale bars, 100 μm; n = 2–3 mice per group. Results show mean ± SD. p values were determined by Mann-Whitney tests (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Discussion
Since its discovery, RNAi technology has been extensively used to interrogate molecular pathways in both normal physiology and disease and is now a cornerstone in the development of new treatments for cancer. Additionally, many studies show promising results using a combination of targeted gene downregulation with immune-checkpoint inhibitors51 or with small molecule anti-cancer drugs, particularly those that can be used as carriers for siRNA.52,53 With the use of a siRNA approach, we investigated how Ubr-ubiquitin ligases of the Arg/N-degron pathway are valuable targets for cancer therapy and how downregulation of this pathway could potentiate the action of chemotherapeutic drugs.
The N-degron pathway regulates several hallmarks of cancer, including sustained cell proliferation, activated migration, and resisting cell death.54 UBR1, UBR2, UBR4, and UBR5 ubiquitin ligases control the functionality of the Arg branch of the pathway and define its specificity, making them the prime targets for therapeutic intervention in this pathway. Our in vitro results show that downregulation of the UBR-ubiquitin ligases by siRNA in a hepatocellular carcinoma cell line leads to decreased proliferation and migration, as well as increased apoptosis of these cells, which is consistent with the known effects of the N-degron pathway. We also demonstrated the accumulation of a known substrate containing a N-degron, the proapoptotic fragment of BRCA1, proving that siRNA-mediated downregulation of the four ubiquitin ligases efficiently reduces the functional activity of the pathway. With the use of lipid nanoparticle delivery, we showed for the first time that sustained, simultaneous downregulation of the UBR ubiquitin ligases of the Arg/N-degron pathway can be achieved in mature adult tissue, without significant toxicity to the animal, allowing study of the role of this pathway in normal physiology, as well as in cancer development and progression. LNP-siRNA allowed prolonged downregulation of UBR1, UBR2, UBR4, and UBR5 in the mouse liver without excessive apoptosis of mature hepatocytes, indicating that slow, proliferating normal adult cells can endure a dysfunctional N-degron pathway for an extended period of time. These findings are corroborated by the Ate1−/flox mouse, where partial ablation of the pathway is well tolerated by mature, adult tissues with low proliferation rates, such as the kidney, brain, heart, and liver, suggesting an opportunity for pharmacological intervention in cancer, which has a much faster proliferation rate and should be more sensitive to the loss of the N-degron pathway, even partially.55,56 Although injections of LNP-siUbrs were well tolerated, the high dose of nanoparticles given to these mice caused infiltration of inflammatory cells, such as inflammatory monocytes (Ly6Chigh), neutrophils, and eosinophils, in the liver of LNP-siUbr-treated animals. Despite recent advances in designing more efficient lipids45,57 and less immunogenic particles and siRNA,58,59 the LNP-siRNA itself remains a light trigger for the immune system. However, this effect is clearly exacerbated when the Arg/N-degron pathway is impaired, as no significant infiltration of mononuclear cells was detected in the LNP-siCtrl-treated mice. We also showed increased liver size and tumor load in HCC animals treated with LNP-siUbrs compared to controls, and this was most likely due to the inflammation observed in the same animals. The Arg/N-degron pathway has been shown to regulate the activation of the NLRP1B inflammasome, which is required for the response against the anthrax lethal factor.60 Although LNPs and siRNA most likely trigger the immune system through a different mechanism, a similar interaction between the N-degron pathway and the inflammasome could be foreseen. Thus, there is a delicate balance between using enough siRNA to cause sufficient downregulation of the pathway to see an effect on its function without causing inflammation. An alternative strategy would be the use of GalNAc conjugates as carriers for the siRNA for liver-targeted delivery, as these molecules deliver specifically to hepatocytes while avoiding immune activation.61
A newer approach in the treatment of cancer is using a combination of chemotherapeutic drugs with specific pathway inhibitors. This allows the harnessing of the power of both strategies, while overcoming therapy resistance and avoiding major side-effects. In our case, use of siRNA against Ubrs to downregulate the Arg/N-degron pathway sensitizes cells to apoptosis and reduces proliferation, which in turn, enhances the effects of the chemotherapeutic drug. In vitro, lower doses of both the siRNA and doxorubicin are needed to more profoundly reduce proliferation and increase apoptosis than the single-agent controls. In the HCC model, downregulation of Ubr-ubiquitin ligases and doxorubicin synergizes in controlling tumor load by reducing proliferation and increasing apoptosis. A previous study, demonstrating a synergistic antitumor effect using a UBR1 inhibitor in combination with shikonin, a necropsis-inducing drug, in a colon cancer mouse model,62 supports our claim that downregulation of the Arg/N-degron pathway can boost chemotherapy efficacy in various cancers. However, in our liver cancer model, all four UBR-ubiquitin ligases of the Arg/N-degron should be downregulated in order to potentiate the effect of doxorubicin. Indeed, downregulation of either UBR1 or UBR5 in HCC mice led to decreased tumor load, but this decrease was not improved with the addition of doxorubicin (data not shown).
The combinatorial approach allows the use of a different regimen for the injection of LNP-siRNA: a lower dose injected weekly sufficiently downregulates the Arg/N-degron pathway while avoiding the inflammatory response seen with the higher dose given twice per week. Since the expression of Arg/N-degron pathway components is mostly confined to highly proliferating cells55 and known to be overexpressed in some types of cancer cells, even partial downregulation of protein levels of UBRs is enough to sensitize cells to the chemotherapy, enhancing the efficiency of the drug. The net effect of siRNA-mediated UBR1, UBR2, UBR4, and UBR5 inhibition, in combination with doxorubicin, is to significantly inhibit tumor proliferation and increase tumor cell apoptosis, resulting in lower tumor load in HCC mice.
Our study is a proof of concept and a demonstration that UBR1, UBR2, UBR4, and UBR5 are promising targets for the development of novel cancer therapies. Downregulation of the Arg/N-degron pathway potentiates the action of chemotherapy, improving the outcome of the treatment. As the N-degron pathway is involved in proliferation, migration, apoptosis, and DNA damage repair, any drug or treatment that interferes with these pathways could be boosted by prior or concomitant downregulation of the N-degron pathway. Since components of this pathway are ubiquitously expressed, targeting the N-degron pathway has the potential to increase effectiveness of therapy in many cancer types.
Materials and Methods
Cell Culture and Transfections
Mouse Hepa 1–6 and AML-12 (ATCC, Manassas, VA) cell lines were grown in DMEM, supplemented with 10% fetal bovine serum (FBS) (Gibco, Waltham, MA) and 2 mM L-glutamine (Gibco) without antibiotics. Cells were split when they reached 80% confluency. Plasmids were transfected using Lipofectamine 2000 (Invitrogen, Waltham, MA), and siRNAs were transfected using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions.
siRNA Description and LNP Formulation
We designed chemically modified siRNAs targeting mouse Ubr1, Ubr2, Ubr4, or Ubr5 mRNA sequences (10 siRNAs per gene) (NCBI accession codes GenBank: NM_009461.2, GenBank: NM_001177374.1, GenBank: NM_001160319.1, and GenBank: NM_001081359.3). siRNAs were selected to avoid off-target activity based on several known criteria.42, 43, 44 Candidate 19-mer sequences were aligned against the Reference Sequence (RefSeq) mRNA database and estimated for their off-target binding capability. In particular, siRNAs were ranked based on the number/positions of the mismatches in the seed region, mismatches in the nonseed region, and mismatches in the cleavage-site position. The resulted sequences were further filtered to avoid known microRNA (miRNA) motifs and immune-stimulatory sequence motifs.44,63 Introduction of 2′-O-methyl (2′-OMe) pyrimidine nucleotides in siRNA further reduces immune response and off-target effects44,64 and together with the use of 3′-internucleotide phosphorothioates, increases stability against nucleases. Potency and efficacy of siRNA targeting Ubr1, Ubr2, Ubr4, and Ubr5 were studied by transfection in Hepa 1–6 cells, followed by quantitative PCR (qPCR) analysis after 24 h. siRNA with the lowest IC50 and highest downregulation of the target was selected for further studies (Table S1). The control siRNA targets the firefly luciferase gene (siCtrl).
siRNAs were formulated in lipid nanoparticles (LNPs), as previously described.46 Briefly, LNPs were formed by mixing solutions of siRNA in acidic water buffer and lipids in ethanol (3:1, v:v), using the microfluidic NanoAssemblr device (Precision NanoSystems), as described earlier.65 siRNAs (0.4 mg/mL for individual siRNA or 0.1 mg/mL of each for a mix of four ones) are first diluted in a 10-mM citrate buffer (pH 3.0), while lipids (C12-20045:1,2-distearoyl-sn-glycero-3-phosphocholine [DSPC] [850365; Avanti Polar Lipids]:cholesterol [С8667; Sigma-Aldrich]:PEG-lipid [880150; Avanti Polar Lipids]) are prepared at a 50:10:38.5:1.5-mol:mol ratio in ethanol (8.83 mg/mL total). siRNA and lipid solutions were mixed at a 10-mL/min rate, according to the manufacturer’s protocol. The final ratio of siRNA:C12-200 after mixing is 1:5, w:w. The LNP suspension was diluted in PBS buffer, dialyzed against 500 volumes of PBS at pH 7.4 in 20,000 molecular weight (MW) cutoff dialysis cassettes (66012; Thermo Fisher Scientific) overnight, and filtered through a polyethersulfone (PES) syringe filter (0.2 μm pores) (3915; Corning). Particle size and zeta potential measurements were performed using a Zetasizer Nano ZSP (Malvern Panalytical), according to the manufacturer’s protocol (Figure S8; Table S4). Zeta potential measurements were conducted under neutral pH conditions. Reported values are the average of 10–25 runs. siRNA entrapment efficiency was determined using the Quant-iT RiboGreen reagent (R11491; Thermo Fisher Scientific), as described earlier.66
cDNA Synthesis and qPCR
RNA was prepared by disrupting liver tissue in TRIzol (Invitrogen, Waltham, MA), using the MP FastPrep-24 instrument and Lysing Matrix D (MP Biomedicals, Solon, OH), followed by precipitation with isopropanol, according to the manufacturer’s instructions. cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA). Levels of mRNA were assessed by qPCR using SYBR Green (Thermo Fisher Scientific), using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA), in the QuantStudio 5 thermocycler (Applied Biosystems). The mRNA levels were normalized to the level of the mouse housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase [mGAPDH]) and to the average value of the control group. Specific primers are listed in Table S2.
Functional Assay of the N-degron Pathway
Hepa 1–6 cells were plated at 40% confluency in 24-well plates and transfected using Lipofectamine 2000 with plasmids encoding native (fDHFR-UbR48-Asp1119-BRCA1f) or stable (fDHFR-UbR48-Val1119-BRCA1f) versions of the flag-tagged proapoptotic BRCA1 fragment,15 with or without siRNA at 2.5 nM. After 72 h, cells were lysed with 1% SDS, 5 mM DTT, containing a complete protease-inhibitor mixture (Roche, Indianapolis, IN), followed by heating at 95°C for 10 min. Samples were diluted in lithium dodecyl sulfate (LDS) sample buffer, heated at 95°C for 10 min, and followed by SDS 10% PAGE. Membranes were incubated with anti-FLAG M2 antibody (Sigma, St. Louis, MO) and visualized using SuperSignal West Femto reagents (Thermo Scientific), according to the manufacturer’s instructions, on a Fusion Solo S (Vilber Lourmat, Marne-la-Vallée, France) imager.
Cell Proliferation, Migration, and Apoptosis Assays
Hepa 1–6 cells were plated at a density required to reach 80% confluency in 72 h or 96 h, transfected with siRNA against luciferase (control) or Ubrs for 72 h at concentrations mentioned in the text, and treated or not with doxorubicin (Millipore-Sigma) for 48 h or staurosporine (Millipore-Sigma) for 24 h. The neutral red assay (adapted from Fautz et al.67) was used to assess proliferation in Hepa 1–6 cells. After treatment, culture media were replaced by a solution of 50 μg/ml neutral red (Millipore-Sigma) in DMEM, supplemented with 10% FBS for 3 h at 37°C, 5% CO2. Cells were then washed once with PBS, and the dye was extracted from the cells by adding elution buffer (50% EtOH, 1% acetic acid). After 5 min of incubation, the elution buffer was transferred onto a new plate and absorbance read at 540 nm.
Migration was assessed using a scratch assay, where a monolayer of cells was scraped using a pipette tip. Photos were taken after the initial scratch and 24 h later. Migration was evaluated by calculating the distance between the edges of the scratch.
Apoptosis was measured using the fluorescein-based In Situ Cell Death Detection Kit TMR red (Roche). Treated cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100/0.25% Tween-20 for 7 min on ice, and assayed according to the manufacturer’s protocol. Cells were counterstained using Hoechst 33342 (Thermo Scientific) and analyzed by fluorescence microscopy using the EVOS FL imaging system (Thermo Fisher Scientific).
Animal Care and Treatments
All animal care and procedures were carried out according to the relevant National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee and the Office of Laboratory Animal Research at Massachusetts Institute of Technology (MA, US) and the Institute of Developmental Biology (Moscow, Russia), where the present study was performed. C57BL/6 or FVB/N mice of 6–8 weeks of age were purchased from Charles River or bred in-house. Mice were housed at 22°C using a 12-h-light to 12-h-dark cycle, fed ad libitum with regular rodent chow. Lipid nanoparticles and doxorubicin were diluted in sterile saline and injected via tail vein (i.v.) at doses and regimes specified in the text. Hepatocellular carcinoma was induced in FVB/N mice, as previously described,46 using plasmids encoding human ΔN90-β-catenin, human MET, and Sleeping Beauty transposase. α-Fetoprotein levels in the serum were tested by western blot at day 35 (anti-α-fetoprotein [AFP]; R&D Systems Minneapolis, MN). Animals were euthanized on days 66–68 post-tumor induction unless otherwise mentioned. Serum for analysis and liver samples for histology and immunohistochemistry were collected; the rest of the liver was snap frozen and ground. Aliquots of homogenized liver were used for mRNA and protein analysis.
Histological and Immunohistochemical Analysis
Liver samples were fixed in 4% buffered paraformaldehyde and embedded with paraffin using standard procedures. 5-μm-Thick sections were subjected to hematoxylin and eosin (H&E) staining or immunohistochemistry. H&E staining was performed using a ThermoShandon Gemini automated stainer (Eosin and Hematoxylin from Leica Biosystems, Buffalo Grove, IL). The envision system (Dako) was used for indirect peroxidase reaction using 3,3′-diaminobenzidine (DAB) to detect Ki67 (BioCare Medical, Pacheco, CA).
TUNEL staining was performed on liver sections treated with proteinase K using the In Situ Cell Death Detection Kit, TMR red. The sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and analyzed by fluorescence microscopy using the EVOS FL imaging system.
Cell and Tissue Extracts and Western Blot
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and mouse liver samples were processed in lysis buffer (20 mM HEPES, 10% glycerol, 0.1 mM EDTA, 75 mM KCl, pH 7.9) containing complete protease inhibitor cocktail (Roche) using the MP FastPrep-24 instrument and lysing matrix D. The extracts were centrifuged at 12,000 g for 10 min at 4°C. Total protein concentrations in the supernatants were determined using the bicinchoninic assay (BCA) (Pierce, Waltham, MA), followed by the addition of a concentrated LDS sample buffer (to the final concentration of 1 × LDS sample buffer), heating at 95°C for 10 min, and SDS 5%–12% PAGE, with 50–100 μg total protein per lane. Fractionated proteins were analyzed by western blot using the following antibodies: anti-UBR1 (Abcam, Cambridge, MA, US), anti-UBR2 (Abcam), anti-UBR4 (Abcam), anti-EDD (Santa Cruz, Dallas, TX), anti-β-actin (Sigma). Western blots were visualized using the SuperSignal West Femto reagent, according to the manufacturer’s instructions.
Blood Biochemistry
Serum was collected by cardiac puncture, followed by centrifugation at 1700 g for 20 min. Biochemical analysis was performed by IDEXX Laboratories (Westbrook, Maine, US).
Flow Cytometry
Spleens were disrupted manually using the blunt end of a syringe plunger, whereas livers were incubated in Collagenase Type IV (0.5 mg/mL in Hank’s balanced salt solution [HBSS] with Ca2+, 5 mM HEPES, 0.5 mM EDTA) (Worthington, Lakewood, NJ) for 30 min before manual disruption and gradient separation using Optiprep (Cosmo Bio, Carlsbad, CA). Cells were resuspended at a density of 1 × 107 cells/mL in DMEM with 2% FBS and were incubated 15 min at room temperature with diluted monoclonal antibodies and then washed and resuspended in PBS with 2% FBS for immediate analysis. The following monoclonal antibodies were used from BioLegend (San Diego, CA): fluorescein isothiocyanate (FITC)-anti-CD11b (M1/70), phycoerythrin (PE)-anti-F4/80 (BM8), PerCPCy5.5-anti-CD11c (N418), PeCy7-anti-B220 (RA3-6B2), BV421-anti-Ly6G (1A8), BV510-anti-CD45 (30-F11), allophycocyanin (APC)-anti-I-A/I-E (M5/114.15.2), APCFire-anti-Ly6C (HK1.4). Annexin V/7AAD staining was performed according to the manufacturer (BioLegend). Data were acquired using a BD LSRFortessa, and analysis of flow cytometry data was performed using FlowJo software (BD Biosciences, San Jose, CA).
Statistical Analysis
Prism 7 (GraphPad Software, La Jolla, CA) was used for statistical analyses. Two-tailed, unpaired Mann-Whitney tests with 95% confidence bounds or one-way or two-way ANOVAs were used for statistical analysis unless otherwise indicated. A p value <0.05 was considered significant.
Author Contributions
D.L. performed the experiments, analyzed and interpreted the data, and wrote the manuscript. T.A. performed experiments. T.P. and L.R. formulated nanoparticles and performed experiments. D.G.A. provided conceptual advice. T.S.Z. and K.I.P. designed research, analyzed the data, and wrote the manuscript. All authors have read and approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The work was performed in the following cities: Moscow, Russia, and Cambridge, MA, US. The authors would like to thank Elena Smekalova and Mikhail Nesterchuk for advice and assistance in mouse experiments and Arsen Mikaelyan and Victor Kotelyanski for helpful comments on the project. We thank the Koch Institute Swanson Biotechnology Center histology core facility for technical support. This work was supported by the Next Generation Program: Skoltech-MIT joint projects and in part by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.01.021.
Supplemental Information
References
- 1.Bialecki E.S., Di Bisceglie A.M. Diagnosis of hepatocellular carcinoma. HPB. 2005;7:26–34. doi: 10.1080/13651820410024049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Personeni N., Pressiani T., Santoro A., Rimassa L. Regorafenib in hepatocellular carcinoma: latest evidence and clinical implications. Drugs Context. 2018;7:212533. doi: 10.7573/dic.212533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu B., Weng Y., Xia X.H., Liang X.J., Huang Y. Clinical advances of siRNA therapeutics. J. Gene Med. 2019;21:e3097. doi: 10.1002/jgm.3097. [DOI] [PubMed] [Google Scholar]
- 4.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials (Basel) 2017;7:77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zatsepin T.S., Kotelevtsev Y.V., Koteliansky V. Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int. J. Nanomedicine. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullis P.R., Hope M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D., Parayath N., Ganesh S., Wang W., Amiji M. The role of apolipoprotein- and vitronectin-enriched protein corona on lipid nanoparticles for in vivo targeted delivery and transfection of oligonucleotides in murine tumor models. Nanoscale. 2019;11:18806–18824. doi: 10.1039/c9nr05788a. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 9.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 10.Brower C.S., Piatkov K.I., Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol. Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shearer R.F., Iconomou M., Watts C.K., Saunders D.N. Functional Roles of the E3 Ubiquitin Ligase UBR5 in Cancer. Mol. Cancer Res. 2015;13:1523–1532. doi: 10.1158/1541-7786.MCR-15-0383. [DOI] [PubMed] [Google Scholar]
- 12.Mao J., Liang Z., Zhang B., Yang H., Li X., Fu H., Zhang X., Yan Y., Xu W., Qian H. UBR2 Enriched in p53 Deficient Mouse Bone Marrow Mesenchymal Stem Cell-Exosome Promoted Gastric Cancer Progression via Wnt/β-Catenin Pathway. Stem Cells. 2017;35:2267–2279. doi: 10.1002/stem.2702. [DOI] [PubMed] [Google Scholar]
- 13.Varshavsky A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA. 2019;116:358–366. doi: 10.1073/pnas.1816596116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piatkov K.I., Brower C.S., Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. USA. 2012;109:E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z., Payoe R., Fahlman R.P. The C-terminal proteolytic fragment of the breast cancer susceptibility type 1 protein (BRCA1) is degraded by the N-end rule pathway. J. Biol. Chem. 2012;287:7495–7502. doi: 10.1074/jbc.M111.301002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien P.M., Davies M.J., Scurry J.P., Smith A.N., Barton C.A., Henderson M.J., Saunders D.N., Gloss B.S., Patterson K.I., Clancy J.L. The E3 ubiquitin ligase EDD is an adverse prognostic factor for serous epithelial ovarian cancer and modulates cisplatin resistance in vitro. Br. J. Cancer. 2008;98:1085–1093. doi: 10.1038/sj.bjc.6604281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Lin R.K., Kwon Y.T., Li Y.P. Signaling mechanism of tumor cell-induced up-regulation of E3 ubiquitin ligase UBR2. FASEB J. 2013;27:2893–2901. doi: 10.1096/fj.12-222711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon Y.T., Kashina A.S., Davydov I.V., Hu R.-G., An J.Y., Seo J.W., Du F., Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 20.Kashina A. Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle. Anat. Rec. (Hoboken) 2014;297:1630–1636. doi: 10.1002/ar.22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An J.Y., Seo J.W., Tasaki T., Lee M.J., Varshavsky A., Kwon Y.T. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc. Natl. Acad. Sci. USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sriram S.M., Kwon Y.T. The molecular principles of N-end rule recognition. Nat. Struct. Mol. Biol. 2010;17:1164–1165. doi: 10.1038/nsmb1010-1164. [DOI] [PubMed] [Google Scholar]
- 23.Tasaki T., Zakrzewska A., Dudgeon D.D., Jiang Y., Lazo J.S., Kwon Y.T. The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 2009;284:1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang C.S., Sukalo M., Batygin O., Addor M.C., Brunner H., Aytes A.P., Mayerle J., Song H.K., Varshavsky A., Zenker M. Ubiquitin ligases of the N-end rule pathway: assessment of mutations in UBR1 that cause the Johanson-Blizzard syndrome. PLoS One. 2011;6:e24925. doi: 10.1371/journal.pone.0024925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon Y.T., Xia Z., Davydov I.V., Lecker S.H., Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sultana R., Theodoraki M.A., Caplan A.J. Specificity in the actions of the UBR1 ubiquitin ligase in the degradation of nuclear receptors. FEBS Open Bio. 2013;3:394–397. doi: 10.1016/j.fob.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tasaki T., Mulder L.C.F., Iwamatsu A., Lee M.J., Davydov I.V., Varshavsky A., Muesing M., Kwon Y.T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon Y.T., Xia Z., An J.Y., Tasaki T., Davydov I.V., Seo J.W., Sheng J., Xie Y., Varshavsky A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders D.N., Hird S.L., Withington S.L., Dunwoodie S.L., Henderson M.J., Biben C., Sutherland R.L., Ormandy C.J., Watts C.K. Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol. Cell. Biol. 2004;24:7225–7234. doi: 10.1128/MCB.24.16.7225-7234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tasaki T., Kim S.T., Zakrzewska A., Lee B.E., Kang M.J., Yoo Y.D., Cha-Molstad H.J., Hwang J., Soung N.K., Sung K.S. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc. Natl. Acad. Sci. USA. 2013;110:3800–3805. doi: 10.1073/pnas.1217358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi W.S., Jeong B.C., Joo Y.J., Lee M.R., Kim J., Eck M.J., Song H.K. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura K. Inhibition of the Arg/N-end rule pathway-mediated proteolysis by dipeptide-mimetic molecules. Amino Acids. 2016;48:235–243. doi: 10.1007/s00726-015-2083-1. [DOI] [PubMed] [Google Scholar]
- 33.Lee M.J., Pal K., Tasaki T., Roy S., Jiang Y., An J.Y., Banerjee R., Kwon Y.T. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc. Natl. Acad. Sci. USA. 2008;105:100–105. doi: 10.1073/pnas.0708465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matta-Camacho E., Kozlov G., Li F.F., Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 2010;17:1182–1187. doi: 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- 35.Sriram S., Lee J.H., Mai B.K., Jiang Y., Kim Y., Yoo Y.D., Banerjee R., Lee S.H., Lee M.J. Development and characterization of monomeric N-end rule inhibitors through in vitro model substrates. J. Med. Chem. 2013;56:2540–2546. doi: 10.1021/jm400046q. [DOI] [PubMed] [Google Scholar]
- 36.Baker R.T., Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc. Natl. Acad. Sci. USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha S., Wang J., Buckley B., Wang Q., Lilly B., Chernov M., Kashina A. Small molecule inhibitors of arginyltransferase regulate arginylation-dependent protein degradation, cell motility, and angiogenesis. Biochem. Pharmacol. 2012;83:866–873. doi: 10.1016/j.bcp.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rageul J., Park J.J., Jo U., Weinheimer A.S., Vu T.T.M., Kim H. Conditional degradation of SDE2 by the Arg/N-End rule pathway regulates stress response at replication forks. Nucleic Acids Res. 2019;47:3996–4010. doi: 10.1093/nar/gkz054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cipolla L., Bertoletti F., Maffia A., Liang C.C., Lehmann A.R., Cohn M.A., Sabbioneda S. UBR5 interacts with the replication fork and protects DNA replication from DNA polymerase η toxicity. Nucleic Acids Res. 2019;47:11268–11283. doi: 10.1093/nar/gkz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S.T., Lee Y.J., Tasaki T., Hwang J., Kang M.J., Yi E.C., Kim B.Y., Kwon Y.T. The N-recognin UBR4 of the N-end rule pathway is required for neurogenesis and homeostasis of cell surface proteins. PLoS ONE. 2018;13:e0202260. doi: 10.1371/journal.pone.0202260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoy S.M. Patisiran: First Global Approval. Drugs. 2018;78:1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 43.Anderson E.M., Birmingham A., Baskerville S., Reynolds A., Maksimova E., Leake D., Fedorov Y., Karpilow J., Khvorova A. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei Y., Tuschl T. On the art of identifying effective and specific siRNAs. Nat. Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- 45.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., Qin J., Cantley W., Qin L.L., Racie T. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogorad R.L., Yin H., Zeigerer A., Nonaka H., Ruda V.M., Zerial M., Anderson D.G., Koteliansky V. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat. Commun. 2014;5:3869. doi: 10.1038/ncomms4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novobrantseva T.I., Borodovsky A., Wong J., Klebanov B., Zafari M., Yucius K., Querbes W., Ge P., Ruda V.M., Milstein S. Systemic RNAi-mediated Gene Silencing in Nonhuman Primate and Rodent Myeloid Cells. Mol. Ther. Nucleic Acids. 2012;1:e4. doi: 10.1038/mtna.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S.T., Tasaki T., Zakrzewska A., Yoo Y.D., Sa Sung K., Kim S.H., Cha-Molstad H., Hwang J., Kim K.A., Kim B.Y., Kwon Y.T. The N-end rule proteolytic system in autophagy. Autophagy. 2013;9:1100–1103. doi: 10.4161/auto.24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M., Zhang Y., Ren J. Autophagic Regulation of Lipid Homeostasis in Cardiometabolic Syndrome. Front. Cardiovasc. Med. 2018;5:38. doi: 10.3389/fcvm.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tward A.D., Jones K.D., Yant S., Cheung S.T., Fan S.T., Chen X., Kay M.A., Wang R., Bishop J.M. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc. Natl. Acad. Sci. USA. 2007;104:14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganesh S., Shui X., Craig K.P., Park J., Wang W., Brown B.D., Abrams M.T. RNAi-Mediated β-Catenin Inhibition Promotes T Cell Infiltration and Antitumor Activity in Combination with Immune Checkpoint Blockade. Mol. Ther. 2018;26:2567–2579. doi: 10.1016/j.ymthe.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saraswathy M., Gong S. Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Mater. Today. 2014;17:298–306. [Google Scholar]
- 53.Steinborn B., Truebenbach I., Morys S., Lächelt U., Wagner E., Zhang W. Epidermal growth factor receptor targeted methotrexate and small interfering RNA co-delivery. J. Gene Med. 2018;20:e3041. doi: 10.1002/jgm.3041. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 55.Brower C.S., Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS ONE. 2009;4:e7757. doi: 10.1371/journal.pone.0007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leu N.A., Kurosaka S., Kashina A. Conditional Tek promoter-driven deletion of arginyltransferase in the germ line causes defects in gametogenesis and early embryonic lethality in mice. PLoS ONE. 2009;4:e7734. doi: 10.1371/journal.pone.0007734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato Y., Matsui H., Yamamoto N., Sato R., Munakata T., Kohara M., Harashima H. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J. Control. Release. 2017;266:216–225. doi: 10.1016/j.jconrel.2017.09.044. [DOI] [PubMed] [Google Scholar]
- 59.Broering R., Real C.I., John M.J., Jahn-Hofmann K., Ickenstein L.M., Kleinehr K., Paul A., Gibbert K., Dittmer U., Gerken G., Schlaak J.F. Chemical modifications on siRNAs avoid Toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro. Int. Immunol. 2014;26:35–46. doi: 10.1093/intimm/dxt040. [DOI] [PubMed] [Google Scholar]
- 60.Chui A.J., Okondo M.C., Rao S.D., Gai K., Griswold A.R., Johnson D.C., Ball D.P., Taabazuing C.Y., Orth E.L., Vittimberga B.A., Bachovchin D.A. N-terminal degradation activates the NLRP1B inflammasome. Science. 2019;364:82–85. doi: 10.1126/science.aau1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y., Jo M., Schmidt J., Luo X., Prakash T.P., Zhou T., Klein S., Xiao X., Post N., Yin Z., MacLeod A.R. Enhanced Potency of GalNAc-Conjugated Antisense Oligonucleotides in Hepatocellular Cancer Models. Mol. Ther. 2019;27:1547–1557. doi: 10.1016/j.ymthe.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwalla P., Banerjee R. N-end rule pathway inhibition assists colon tumor regression via necroptosis. Mol. Ther. Oncolytics. 2016;3:16020. doi: 10.1038/mto.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frank-Kamenetsky M., Grefhorst A., Anderson N.N., Racie T.S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh C., Ou K., Belliveau N.M., Leaver T.J., Wild A.W., Huft J., Lin P.J., Chen S., Leung A.K., Lee J.B. Microfluidic-Based Manufacture of siRNA-Lipid Nanoparticles for Therapeutic Applications. Methods Mol. Biol. 2014;1141:109–120. doi: 10.1007/978-1-4939-0363-4_6. [DOI] [PubMed] [Google Scholar]
- 67.Fautz R., Husein B., Hechenberger C. Application of the neutral red assay (NR assay) to monolayer cultures of primary hepatocytes: rapid colorimetric viability determination for the unscheduled DNA synthesis test (UDS) Mutat. Res. 1991;253:173–179. doi: 10.1016/0165-1161(91)90130-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.