SUMMARY
Sporadic gliomas in companion dogs provide a window on the interaction between tumorigenic mechanisms and host environment. We compared the molecular profiles of canine gliomas with those of human pediatric and adult gliomas to characterize evolutionarily conserved mammalian mutational processes in gliomagenesis. Employing whole-genome, exome, transcriptome, and methylation sequencing of 83 canine gliomas, we found alterations shared between canine and human gliomas such as the receptor tyrosine kinases, TP53 and cell-cycle pathways, and IDH1 R132. Canine gliomas showed high similarity with human pediatric gliomas per robust aneuploidy, mutational rates, relative timing of mutations, and DNA-methylation patterns. Our cross-species comparative genomic analysis provides unique insights into glioma etiology and the chronology of glioma-causing somatic alterations.
In Brief
Amin et al. characterize the molecular landscape of canine gliomas and compare it with that of human pediatric and adult gliomas, revealing high similarity between human pediatric and canine gliomas. The cross-species analysis identifies conserved glioma drivers and aneuploidy as a hallmark of high-grade disease.
Graphical Abstract

INTRODUCTION
The natural history of cancer is marked by temporal acquisition of diverse genetic and epigenetic aberrations. The inevitable intratumoral and interpatient heterogeneity among evolving cancer cells poses a major obstacle in our understanding of cancer evolution and designing effective treatment strategies (Alizadeh et al., 2015). Recent developments in high-throughput lineage tracing, organoid cultures, and patient-derived xenografts have provided better resolution of heterogeneity and driver events. Nonetheless, in the absence of natural host response, preclinical in vitro and rodent models are unable to fully recapitulate a spontaneously evolving tumor’s life history. This limitation challenges the accuracy of predicting therapeutic responses in these preclinical models, especially response to immunotherapies (Buque and Galluzzi, 2018).
Somatic evolution of cancers may follow convergent patterns across mammalian species by selecting cells that carry beneficial mutations in highly conserved regions, i.e., genes and their regulatory non-coding regions enabling one or more cancer hallmarks (Hanahan and Weinberg, 2011). Unlike induced cancer models, comparative genomics of spontaneous tumors across species provides a unique advantage to identify defects in such shared, evolutionarily constrained regions (Lindblad-Toh et al., 2011) and to evaluate the importance of host context in the tumor’s evolution. In addition to their natural tumorigenesis, spontaneous cancers in dogs are marked by the presence of a fully functional tumor microenvironment (Khanna et al., 2006; LeBlanc et al., 2016). Cancer cells are subject to clonal selection and drift, and the resulting tumor is molded by selection pressure from the tissue context (DeGregori, 2017; Fortunato et al., 2017). This Darwinian adaptation may select for somatic alterations in evolutionarily conserved regions in both dogs and humans that are relevant to tumorigenesis.
Sporadic gliomas occur in companion dogs at frequencies similar to those in humans (Snyder et al., 2006; Song et al., 2013). Genomic characterization of canine glioma has a distinct merit, in that dogs are diagnosed in the adult stage of life but with an age distribution that is comparable with human pediatric disease. This seeming conundrum in fact creates an opportunity to compare somatic drivers and their relative timing in canine glioma with those in human glioma. Studies involving comparative genomics of spontaneous canine cancers have already enabled identification of breed-specific, disease-risk loci under strong evolutionary constraints and with known roles in human cancer, e.g., germline FGF4 retrogene expression in chondrodysplasia (Parker et al., 2009), somatic BRAF V600E mutation in canine invasive transitional cell carcinoma of the bladder (Decker et al., 2015b), recurrent somatic SETD2 mutations in canine osteosarcoma (Sakthikumar et al., 2018), and TP53 pathway alterations in canine melanoma (Hendricks et al., 2018; Wong et al., 2019). Earlier studies in canine gliomas have characterized somatic copy-number alterations syntenic with those in human adult gliomas (Dickinson et al., 2016) and have identified genetic susceptibility factors near genes such as CAMKK2, P2RX7, and DVL2 (Mansour et al., 2018; Truve et al., 2016).
Here, we have performed comparative genomic, transcriptomic, and epigenetic profiling across three population structures, canine glioma, human adult glioma, and human pediatric glioma, to study somatic evolutionary traits of glioma across two species and in different age groups. We leveraged genomic profiles to infer molecular life history in order to understand cross-species convergent evolution of glioma (Aktipis et al., 2013; Stearns, 1992).
RESULTS
Human Glioma Driver Events Are Frequently Found in Canine Disease
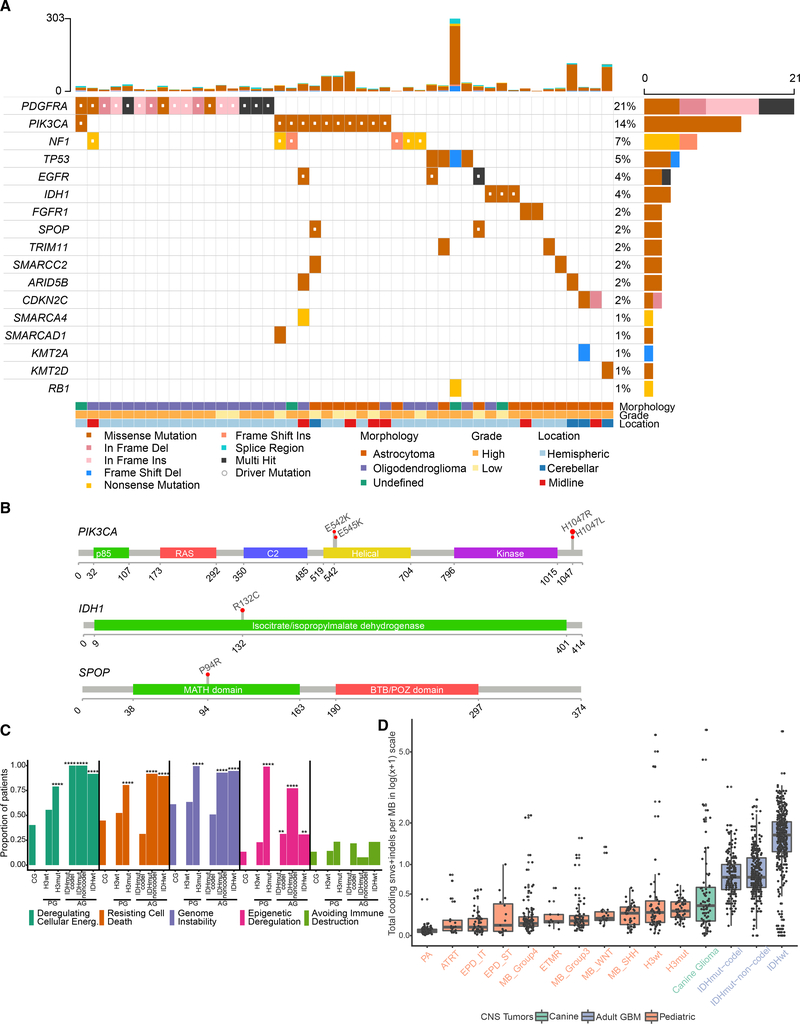
We performed whole-genome, exome, transcriptome, and methylation sequencing (373 libraries) on canine gliomas (n = 83) and germline (n = 67) samples from 83 dogs (NCBI SRA accession: PRJNA579792), with all samples obtained via necropsy. Using the recently updated criteria for diagnostic histopathological classification (Koehler et al., 2018), 46 cases were classified as oligodendroglioma, 31 cases as astrocytoma, and 6 cases as undefined glioma (Table S1). We defined a common set of 81 cases for which whole-genome and exome data were available with minimum of 30× coverage in exome regions (Table S1 and Figure S1A; STAR Methods). From mutation calls derived from all 81 cases, we detected somatic mutational driver events using dNdS (Martincorena et al., 2017), MuSiC2 (Dees et al., 2012), and a semi-supervised comparison with known cancer drivers in human adult and human pediatric cancers (Bailey et al., 2018; Gröbner et al., 2018; Mansour et al., 2018) (Figure 1A and Table S2; STAR Methods). We detected mutations in genes associated with human pediatric (Mackay et al., 2017) and adult glioma (Brennan et al., 2013; Ceccarelli et al., 2016) such as the TP53, PDGFRA, PIK3CA, and EGFR (Figure S1B), as well as recurrent hotspot and mutually exclusive mutations with high oncogenic impact according to the Catalog of Somatic Mutations in Cancer (COSMIC) database (Tate et al., 2019) in PIK3CA H1047R/L (n = 8), PDGFRA K385I/M (n = 6), IDH1 R132C (n = 3), and SPOP P94R (n = 1; 1 shared with PIK3CA H1047R) (Figure 1B and Table S3). These mutations were also identified as being under positive selection or as significantly mutated genes using the dNdS (Martincorena et al., 2017) approach (Table S2) and thus indicating driver mutations of canine gliomas. Mutations affecting the IDH1 R132 codon are a defining characteristic of low-grade adult gliomas (Cancer Genome Atlas Research Network et al., 2015) and were detected infrequently in pediatric and canine gliomas (n = 3/81). Overall, 36/81 (44%) of canine gliomas carried at least one significantly mutated gene. This proportion was comparable with published findings in human pediatric gliomas (114/217, 52%, chi-square p value 0.54) (Gröbner et al., 2018) but contrasted with the frequency at which adult gliomas contain at least one significantly mutated gene alteration (753/812, 93%, chi-square p value 0.0004). To demonstrate similarity between canine gliomas and human gliomas, we summarized levels of somatic coding mutations, high-level copy amplifications and deep deletions in gene sets reflecting previously reported cancer hallmarks (Table S4). We tallied weighted pathway contributions per cohort (canine, adult, pediatric) by the number of coding mutations within each cohort and genes per pathway. Adult glioma is commonly separated into subtypes on the basis of IDH mutation as well as chromosome arm 1p and 19q deletion, resulting in three subtypes: (1) IDH wild type; (2) IDH mutant with codeletion (IDHmut-codel); and (3) IDH mutant without codeletion (IDHmut-noncodel) (Louis et al., 2016). Pediatric high-grade gliomas are separated based on histone H3 mutation status into two subtypes: histone H3 gene mutant (H3 mutant) versus wild type (H3 wild type) (Louis et al., 2016). We did not include low-grade pediatric gliomas in our comparison due to the paucity of somatic alterations in these glioma types (Jones et al., 2013; Pollack et al., 2019; Zhang et al., 2013). We found that canine gliomas were most similar to pediatric H3 wild-type gliomas at the pathway alteration level, i.e., comparable hallmark enrichment with no significant difference between groups. Pediatric H3 mutant, adult IDH wild type, and IDHmut-noncodel gliomas showed increased frequency of gene mutations in cancer hallmarks such as deregulating cellular energetics, genomic instability, and resisting cell death (Figures 1C and S1C). Among all 11 cancer hallmarks tested, “avoiding immune destruction” scored low across both canine and human gliomas (Table S4), potentially owing to the immune-cold nature of gliomas (Boussiotis and Charest, 2018; Brown et al., 2018).
Figure 1. Comparative Somatic Landscape of Canine and Human Gliomas.
(A) Somatic variants in canine gliomas. Top bar plot shows patient-wise frequency of somatic variants (n = 46 of 81 canine patients) and right-side bar plot shows gene-wise frequency of somatic variant types. Bottom annotations show relevant patient-specific annotations.
(B) Gene lollipop plots showing recurrent hotspot mutations for three genes: PIK3CA, IDH1, and SPOP. All hotspot mutations are ortholog to validated COSMIC mutations in human cancers.
(C) Hallmark enrichment of somatic cancer drivers (mutations and copy-number alterations) across canine glioma (CG) and WHO molecular subtypes of human adult (IDH wild-type, IDHmut-codel, IDHmut-noncodel) and pediatric (H3-mutant and H3 wild-type) high-grade glioma. y axis represents proportion of patients in the respective cohort harboring mutations in selected five hallmarks. Two-sided Fisher’s exact test was used for comparison of proportions between cohorts. p values less than the threshold (p < 0.05) are shown (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
(D) Somatic mutation rate across canine and human brain tumors: Box plot showing somatic mutation rates as coding mutations per megabase in log1p or log(x+1) scale. x axis shows 11 types of pediatric brain tumors (Gröbner et al., 2018), canine glioma (n = 81), adult pediatric high-grade gliomas separated by H3 mutant and H3 wild type, and adult gliomas separated by IDH mutation and 1p/19q codeletion status (far right). Each box spans the first and third quartiles with the median in the center. The lower and upper whiskers extend up to 1.5 times interquartile range, and values outside whiskers are outliers. PA, pilocytic astrocytoma; ATRT, atypical teratoid rhabdoid tumor; EPD_ST, ependymoma supratentorial; ETMR, embryonal tumors with multilayered rosettes; MB, medulloblastoma. Tumors are sorted in ascending order by increasing mutation rate.
We compared mutation burden between canine and a variety of human pediatric and adult cohorts using coding mutation rates from 4,761 human patients (Bailey et al., 2018; Ceccarelli et al., 2016; Gröbner et al., 2018; Mansour et al., 2018) (STAR Methods). The somatic mutation rate of canine glioma (0.34 coding mutations per megabase; 95% confidence interval [CI]: 0.15–0.6) was similar to that of human pediatric gliomas (Figures 1D and S1D). High-grade canine gliomas (n = 63/81) had mutation rates comparable with those of pediatric H3-mutant and H3 wild-type subtypes (0.34, 0.27, 0.25 coding mutations per megabase, respectively; Wilcoxon p value 0.18 and 0.1; Figure S1E), but significantly lower than in human adult IDH-mutant and IDH wild-type gliomas (0.77 and 1.67 coding mutations per megabase, respectively; Wilcoxon p values of 8 × 10−9 or less). Low mutation burden has been linked to fewer mutations in cancer-driving genes (Martincorena et al., 2017) and may explain the relative paucity of significantly mutated genes observed in canine gliomas, including weaker positive selection (q > 0.1) for known and mutated cancer genes (n = 50; Figure S1F). These results demonstrate that the landscape of somatic single-nucleotide variants is similar to that of human glioma, and suggests that canine glioma aligns more closely with human pediatric glioma than with adult disease.
Aneuploidy Is a Major Driver of Canine and Pediatric High-Grade Glioma
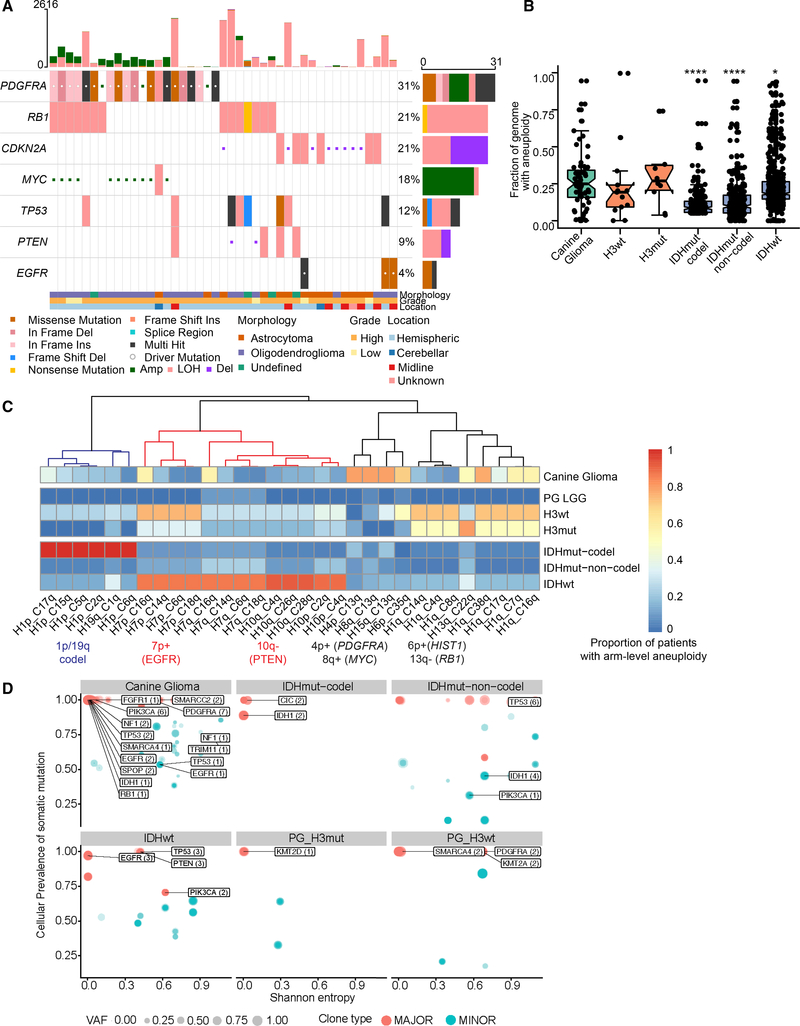
We compared the DNA copy-number landscape of glioma across species with a focus on the >50% of canine gliomas (45/81) without evidence of significantly mutated genes. No focal copy-number amplifications were detected among canine gliomas. Human glioma tumor-suppressor gene CDKN2A/B was homozygously deleted in 8/67 (12%, all astrocytomas), and PTEN in 2/67 (3%) of canine glioma genomes (Figures 2A and S2A). Together, 67/81 (83%) patients with canine glioma contained somatic mutations and/or focal copy alterations in known human glioma drivers (Figure S1E). Contrasting with the limited presence of focal DNA copy-number alterations was the high frequency of arm-level copy gains (canine chromosomes 7q, 13q, 16q, 20q, 34q, 35q, and 38q) and arm-level losses (canine chromosomes 1q, 5q, 12q, 22q, and 26q) (Figure S2B). The most frequent arm-level alteration comprised the shared syntenic regions of glioma drivers PDGFRA, KIT, and MYC (Figure S2C) and typically resulted in more than four copies of these genes (canine 13q+; 11/67 cases, 16%). Other common arm-level alterations included PIK3CA (canine 34q+) and the HIST1 cluster (canine 35q+) as well as hemizygous loss of heterozygosity of tumor-suppressor genes TP53, RB1, and PTEN (Figure 2A).
Figure 2. Aneuploidy Is a Major Driver of High-Grade Gliomas.
(A) Focal somatic copy alterations in canine gliomas (n = 43 of 67 canine patients). Squared symbol in cell suggests either amplification (>4 copies) or deep deletion (2 copy loss) based on GISTIC2 gene-level calls (STAR Methods). Top bar plot shows patient-wise frequency of somatic variants and copy-number alterations, and right-side bar plot shows driver-wise frequency of somatic variant types, including copy-number alterations. Bottom annotations show relevant patient-specific annotations.
(B) Comparative aneuploidy score: box plots showing fraction of genome with aneuploidy (y axis) for canine gliomas (n = 67), H3-mutant (n = 10), and H3 wild-type (n = 13) pediatric high-grade gliomas, and human adult glioma (n = 969), separated by IDH mutation and 1p/19q codeletion status. Each box spans the first and third quartiles with the median in the center. The lower and upper whiskers extend up to 1.58 times interquartile rage divided by square root of samples per box plot (displayed as dots; STAR Methods), and values outside whiskers are outliers. p values were calculated using two-sided Wilcoxon rank-sum non-parametric test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
(C) Aneuploidy metrics across shared syntenic regions of canine and human genome: Heatmap showing comparative aneuploidy across three cohorts. Each column shows the proportion of patients with the most variable arm-level aneuploidy (present or absent) for a given shared syntenic region. x-axis label shows syntenic chromosome arms for human (H) and canine (C) genome. Each row represents canine glioma and molecular subgroups of human high-grade pediatric and adult glioma as detailed in (B), plus pediatric low-grade gliomas (PG_LGG). Colored dendrogram branches (blue, red, and black) represent three aneuploidy clusters described in the main text. Corresponding glioma driver alterations are highlighted below syntenic chromosome arms.
(D) Scatterplot showing distribution of somatic glioma driver genes with respect to their cellular prevalence (cancer cell fraction) and intratumoral heterogeneity (Shannon entropy) across canine and molecular subtypes of human pediatric and adult gliomas. Each circle represents a clonal cluster assignment per tumor sample. Size of the circle represents a major (1 clone) versus minor subclones (ranging from 2 to 4). Labeled genes represent glioma drivers shown in Figure 1A. Darker to lighter blue scale for circle and driver genes it may contain (arrows) represents the increase in intratumoral heterogeneity as measured by Shannon entropy.
We quantified the prevalence of aneuploidy across the canine, human pediatric, and adult glioma populations (Taylor et al., 2018). For copy-number estimation, matched tumor-normal whole-genome sequencing profiles from canine (n = 67) and pediatric gliomas (n = 50) (Mansour et al., 2018), and Affymetrix SNP6 profiles for adult gliomas (n = 969) (Ceccarelli et al., 2016) were analyzed (STAR Methods). We calculated aneuploidy as the proportion of the copy-number segmented genome that was non-diploid (STAR Methods). Canine glioma independent of tumor grade had a median of 25% genome aneuploidy, which was significantly higher than that in adult IDH-mutant tumors (8%–9% of genome) and marginally higher than in adult IDH wild-type glioma (18% of genome) (Figure 2B). In contrast, pediatric H3 wild type (19% of genome) and H3 mutant (26% of genome) showed rates of aneuploidy comparable with that of canine glioma. We then searched for aneuploidy within syntenic regions, which may be subject to selection pressure during gliomagenesis. We mapped canine chromosome arms to their human counterparts and used unsupervised hierarchical clustering of the most variable syntenic aneuploid regions to identify regions of shared aneuploidy (Figure 2C). The analysis revealed three aneuploidy clusters. The first cluster (blue dendrogram) consisted of human 1p/19q codeletions seen commonly in adult IDH-mutant gliomas but observed in 20%–36% of canine (across four canine chromosomes of cases) and 25% of H3 wild-type and H3-mutant human pediatric gliomas. The second cluster (red dendrogram) consisted of arm-level aneuploidy of human 7p (EGFR) and 10q (PTEN) arms characteristic of human adult IDH wild-type (86% and 92% of patients, respectively) and pediatric H3-mutant and H3 wild-type gliomas (33% and 75% of patients) for which 5% and 14% of canine gliomas showed arm-level aneuploidy in the EGFR and PTEN regions, respectively. None of three IDH1 mutant canine gliomas shared these syntenic aberrations, suggesting a mutually exclusive pattern as observed in human gliomas. The third cluster (black dendrogram) consisted of human 4p/8q and syntenic canine 13q arm, which contains the genes PDGFRA and MYC, amplified in 78% of canine gliomas. The ACVR1 and the HIST1 genes are frequently mutated in pediatric high-grade gliomas and in particular H3.1K27M diffuse intrinsic pontine glioma (Mackay et al., 2017). We observed loss of the syntenic human 2q/canine 36q region (containing ACVR1) within 37%, 28%, and 17% of canine, pediatric H3 wild-type, and H3-mutant gliomas, respectively. In contrast, this alteration was not observed in human pediatric or adult IDH-mutant glioma and was present in 6% of IDH wild-type adult gliomas. Similarly, human chromosome arm 6p/canine chromosome arm 35q, containing the HIST1 gene cluster, was frequently amplified in canine gliomas (70%) and pediatric H3 wild-type (50%) and H3-mutant gliomas (13%) but not in pediatric low-grade or adult gliomas (<5%).
We measured intratumoral heterogeneity using the Shannon Diversity Index per each patient tumor sample across canine gliomas and different molecular subtypes of human gliomas. Shannon entropy value correlated with the proportion of variants per subclone and the total number of subclones in a tumor sample, i.e., values near zero indicated lower intratumoral diversity (homogeneity or a dominant clone), while values closer to 1 or higher were associated with increased diversity and tumors consisting of more than one subclone (Wolf et al., 2019). We found that the canine gliomas showed a relatively bimodal distribution, with 25% of canine gliomas (15/60; 7 cases had no resolved clonal structure for intratumoral heterogeneity analysis) being very heterogeneous (Shannon Diversity Index ≥ 0.45—third quartile) while remaining cases showed patterns suggesting clonal dominance (median Shannon Diversity Index 0.03, 95% CI: 0.02–0.07). Intratumoral heterogeneity of canine gliomas was comparable with adult IDH wild type or IDHmut-noncodels (unpaired two-tailed Wilcoxon test p value >0.18). In contrast, canine gliomas had significantly higher heterogeneity over H3 wild-type (p value 0.002) and H3-mutant (p value 0.007) pediatric gliomas (Figure S2D).
To better understand the potential functional versus non-functional nature of intratumor heterogeneity (Jamal-Hanjani et al., 2017), we asked whether frequent driver mutations found in canine gliomas (Figure 1A) are part of major (dominant) versus minor clone, and how these driver events compare with measured heterogeneity across molecular subtypes in human pediatric and adult gliomas (Williams et al., 2016). We observed that among less heterogeneous tumor samples (Shannon entropy near 0), shared driver events across canine and adult gliomas are part of major clones, including PIK3CA mutations in canine gliomas, IDH1/2 mutations in IDH-mutant adult gliomas, and EGFR somatic mutations in adult IDH wild-type GBM (Figure 2D). Among tumor samples with increased heterogeneity, we found mutations in PDGFRA in canine gliomas (n = 7/60) and H3 wild-type pediatric gliomas (n = 2/14), whereas mutations in TP53 (n = 9/20) and PTEN (3/20) were seen among IDH wild-type and IDHmut-noncodel patients. We did not observe significant enrichment of driver events within minor clone(s).
Collectively, the observed high degree of aneuploidy and clonal nature of somatic drivers in canine glioma may suggest progressive genomic instability. Comparing the aneuploidy score among canine gliomas with high versus low coding mutational rate showed significant increases (Figure S2E; Wilcoxon p value 0.006) in aneuploidy among patients with a high mutational rate, suggesting that an underlying mutational process promotes genomic instability during gliomagenesis.
DNA Damage-Related Mutational Processes Shape Somatic Driver Landscape and Maintain Genomic Instability
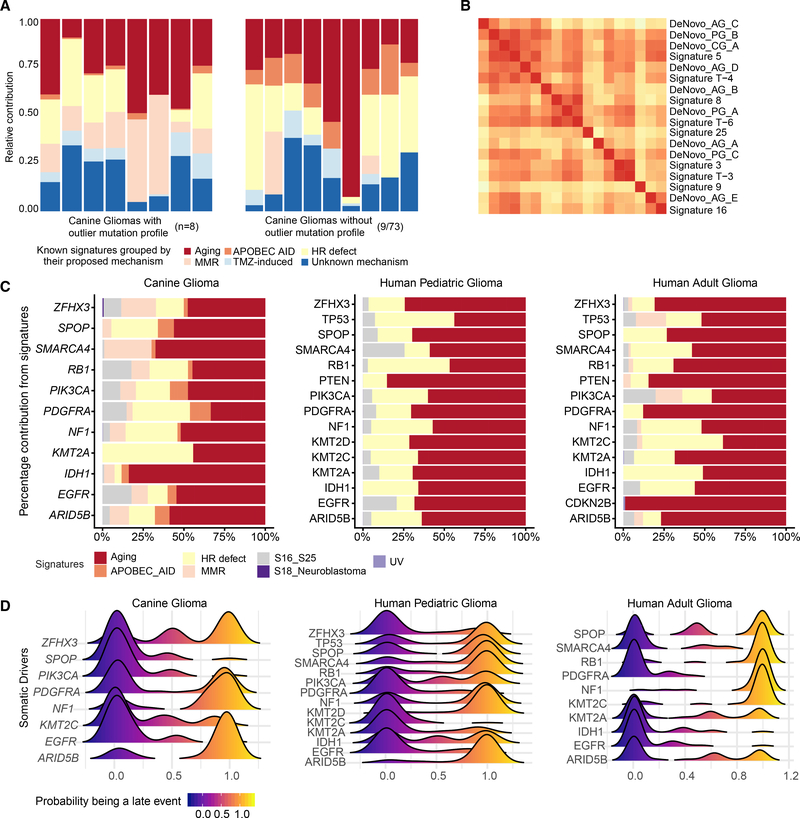
We leveraged known mutational signatures from adult (COSMIC v2, 1 to 30) and pediatric cancers (T1 to T12) to estimate and compare underlying mutational processes across canine and human gliomas (Alexandrov et al., 2013; Gröbner et al., 2018; Mansour et al., 2018) (Table S5). The most enriched signatures across all canine gliomas (Figure S3A) were associated with aging (COSMIC signature 1, pediatric signature T1), mismatch repair deficiency (COSMIC signature 15), APOBEC-AID (COSMIC signature 2, 9), homologous repair defect signatures (COSMIC signature 8, pediatric signature T3), and signatures with unknown relevance (COSMIC signature 12, pediatric signature T10 and T11). Among the nine canine gliomas with the highest mutation rates (median coding mutation rate of 0.55 per megabase) (Figure 3A), there was significant (Wilcoxon p value 0.025) enrichment of two additional mismatch repair signatures (pediatric signature T9 or COSMIC signature 6, 15) (Figure S3B). A frameshift indel in mismatch repair gene MSH6 was detected in one case with an outlier mutation frequency (coding mutation rate of 5.04 per MB) (Figure S3C). Among the remaining cases (median coding mutation rate of 0.25 per MB), homologous repair defect or “BRCAness” signatures (COSMIC signature 3 or pediatric signature T3, COSMIC signature 8 or pediatric signature T6) were the second most prominent signatures after clock-like signatures (COSMIC signature 1, 5). Homologous repair defect signatures have been reported to be enriched in pediatric high-grade gliomas with higher genomic instability (Gröbner et al., 2018). The known human signatures were validated by clustering de novo constructed signatures for all three cohorts (canine, human adult, and pediatric gliomas). Independent of cohort type, we observed significant cosine similarity (>0.8; Figures 3B and S3D) of de novo signatures with known homologous repair defect mutational processes (including COSMIC signature 3/pediatric signature T3, COSMIC signature 8/pediatric signature T6), further implying a role for these mutational processes in cross-species gliomagenesis.
Figure 3. Molecular Life History Analysis Using Mutational Signatures and Timing Analysis.
(A) Deconvolution of known human mutational signatures on canine glioma somatic variant data. Stacked bar plots show relative contribution of known human mutational signatures in individual canine patients. Signature contributions were aggregated based on their grouping into proposed mechanism. Only signatures with a relative contribution of more than a third quartile per sample are shown in the plot. Plot on the left side shows eight cases with highest mutational frequency (based on outlier mutational profile, STAR Methods) and plot on the right side shows nine representative cases with median signature contribution within interquartile range. Signatures with no proposed mechanism are grouped into the unknown category. APOBEC AID, activation-induced cytidine deaminases; HR defect, homologous repair defect; MMR, mismatch repair; TMZ-induced, alkylating agent temozolomide-associated signature.
(B) Hierarchical clustering of cosine similarities between known human mutational signatures and de novo signatures constructed using available whole-genome data from canine (CG), pediatric (PG), and adult (AG) data. Higher cosine similarity (red color) indicates higher resemblance of de novo signature to known mutational signature. Only one of three cluster groups are shown here; the complete clustering is shown in Figure S3D.
(C) Horizontal stacked bar plots represent percentage contribution of signature groups (x axis) for somatic driver mutations (y axis) found in canine and human gliomas. Each of seven signature groups represents a combination of one of more known human signatures. S16_S25 and S18_Neuroblastoma: signatures were previously described by Gröbner et al. (2018).
(D) Molecular timing of somatic drivers across canine and human gliomas: Stacked density plots, one per each of three cohorts, shows probability (x axis) of a driver event (y axis) being a late event in tumor evolution and value of <0.5 being an earlier event. Density plots for each driver event were calculated based on pairwise winning probability (where win is defined as an early event) as used in sports statistics (Bradley-Terry model). Winning probabilities were subtracted from 1 to display early events on the left side of the plot.
Next, we determined the relative contribution of mutational processes (with deconvoluted human signatures as a proxy) in generating mutations within significantly mutated genes, thus to identify the dominant mutational process(es) active during tumor evolution (Figure 3C). Although clock-like processes (COSMIC signature 1, 5) largely contributed to an age-related increase in mutations, including in driver genes, we found that homologous repair defect signatures (COSMIC signature 3, 8) contributed (26%, 21/81 cases) to driver mutations across all three cohorts, emphasizing that homologous repair defect can not only serve as a potential source for driver mutations but also fuel progressive genomic instability along with observed high aneuploidy (Blank et al., 2015; Targa and Rancati, 2018) in high-grade gliomas across all three cohorts.
Comparative Molecular Timing Analysis Highlights Context-Specific Early and Late Drivers of Gliomagenesis
We inferred the sequential order of somatic alterations during gliomagenesis by estimating clonality of glioma driver events (Figure 3D) (Jolly and Van Loo, 2018; Shinde et al., 2018). In brief, significantly mutated genes were timed as occurring early (clonal) to late (subclonal) during tumor evolution based on their cancer cell fraction after accounting for tumor purity, ploidy, and copy-number status (STAR Methods). We observed clonal PDGFRA and EGFR mutations as the only shared and early event across all three cohorts. Subsequent whole chromosome 13 amplification bearing the PDGFRA mutant allele marked the emergence of the most recent common ancestor in six canine gliomas (Figure S3E), which grew to be a dominant clone at the time of diagnosis. IDH1 mutation marks an initiating event in IDH-mutant human gliomas (Barthel et al., 2018). Correspondingly, IDH1 mutations were ubiquitously timed as an initiating event (cancer cell fraction [CCF] > 0.9) in three canine and three human adult IDH1 mutant cases, and as an early event in one case of pediatric glioma (CCF = 0.83). We observed NF1 frameshift mutations mostly as a late event across all cohorts, whereas PIK3CA mutations appeared as an early event for canine and human pediatric gliomas. Although the relatively uniform timing patterns of these known glioma drivers suggest convergent evolution in varied contexts, i.e., presence of hotspot mutations in shared drivers (PDGFRA, PIK3CA) during clonal evolution of glioma across two species and different age groups, we also observed an oscillating pattern of timing and consequent underlying natural selection for a set of epigenetic drivers in the lysine methyltransferase (MLL) family (Rao and Dou, 2015). MLL3 (KMT2C) gene mutations were clonal events in canine and pediatric gliomas but subclonal in adult gliomas, whereas ARID5B mutations showed the inverse pattern (Figure 3D). MLL family genes include some of the most commonly mutated genes in pediatric cancers, including gliomas (Huether et al., 2014; Sturm et al., 2014), but not in adult gliomas (Bailey et al., 2018).
Canine Gliomas Are Classified as Pediatric Glioma by DNA Methylation
We hypothesized that epigenetic deregulation in canine gliomas may carry a tumor-specific methylation pattern reflecting underlying tumor pathology, as has been observed across human brain tumors (Capper et al., 2018). We leveraged reduced representation bisulfite sequencing of canine gliomas to generate genome-wide DNA-methylation profiles to classify canine gliomas according to a classification model widely used for human brain tumors (Capper et al., 2018). As the human brain tumor classifier was developed using the Illumina human 450k array platform, we developed a logistic regression model to enable classification of the sequencing-based canine DNA-methylation profiles. We found that the model classified 35/45 (78%) of canine samples as pediatric glioma (Figure 4). Six of 45 (13%) samples were classified as IDH wild-type adult glioma, and 4/45 (9%) samples were classified as IDH-mutant adult glioma. Of the three samples carrying an IDH1 R132 mutation, one was classified as IDH-mutant adult glioma, with a classification probability of 99%, while a second IDH-mutant sample had a relatively high classification probability for IDH-mutant adult glioma (40%), in parallel with a 57% pediatric glioma classification probability. The third sample had a low classification probability for IDH-mutant adult glioma (13%) and was classified as pediatric glioma with an 84% probability. Although the majority of canine samples were classified as pediatric glioma, the age of diagnosis of the patients in our canine cohort exceeded the age of sexual maturity in canines, which is reached between 10 months and 2 years of age (Thompson et al., 2017). The distribution of age of diagnosis of canine tumors classified as pediatric suggests that classification was a function of methylation profile similarity rather than chronological age. Adult human high-grade glioma tends to be restricted to the cerebral hemispheres, whereas pediatric high-grade gliomas occur throughout the central nervous system with about half of pediatric high-grade gliomas occurring in midline locations (Mackay et al., 2017). Of ten midline canine tumors (six cerebellar, four midline), eight were classified by DNA methylation as pediatric glioma and two were labeled as adult IDH mutant (Figure S4A).
Figure 4. Classification of Canine Gliomas Using Human Brain Tumor Methylation Classifier.
Heatmap displaying results of L2-regularized, logistic regression classification of canine methylation profiles (n = 45). Each column of the heatmap represents a sample, and each row in the top panel is the probability that that sample falls under a given subtype classification. The classification with the highest probability in a given sample has a symbol with symbol color, size, and shape denoting sample histology, tumor grade, and anatomical location, respectively. Panels below the probability heatmap show the tumor purity, somatic mutation rate, and age for the samples. The horizontal line on the age subpanel denotes the age of maturity for canines (2 years). See also Figure S4.
DNA-methylation profiles have been used to estimate molecular age (Pai et al., 2011). We used this approach to compare the level of age acceleration in canine and human glioma. No significant difference was observed in inferred DNA-methylation age between canine tumors classified as adult glioma versus those classified as pediatric among tumors with a classification probability greater than 50% (5.945 versus 5.958, p value 0.9125), consistent with the lack of correlation observed between canine methylation classification and chronological age. The normalized mean age acceleration was significantly higher for human pediatric glioma samples (2.5) than either human adult glioma (0.8) or canine glioma samples (−0.18) (Figure S4B). Unlike human samples, the DNA-methylation-inferred age did not correlate with chronological age for canine samples (Pearson correlation coefficient 0.21), which may reflect limitations in the aging clock model derived for canids, rather than biological differences in canine tumor methylation. The DNA-methylation profile of canine glioma further corroborates the evidence that glioma in dogs is generally more similar to human pediatric glioma than human adult glioma.
Immune Microenvironment
As spontaneous tumors arising in immune-competent hosts, canine gliomas represent an excellent resource through which to improve our understanding of how the immune system responds to and affects brain tumor development. To obtain a baseline understanding of how the canine glioma (n = 11) immune microenvironment compares with that of adult (n = 11) and pediatric human gliomas (n = 5), we used immunohistochemistry to profile the frequency of the various immune populations including monocytes (CD14), T cells (CD3), B cells (CD79A), macrophage/microglia (IBA1), and M2 polarized innate immune cells (CD163) using antibodies that had been validated for cross-species staining. Tissue segmentation was performed so that cell quantification was analyzed in a total of 33,029 fields within the gliomas. Notably, there are many key shared immunological features between the human and canine gliomas such as the relative scarcity of both CD3+ T cell and CD79A+ B cells and a marked predominance of IBA1+ macrophage/microglia and CD163+ M2 polarized innate immune cells, especially in high-grade and pediatric gliomas as previously described (Wei et al., 2019), indicating that dogs with spontaneously arising gliomas may be valid models for the testing of immune therapeutics (Figures 5A and 5B). Our immunohistochemistry results converged with the relative immune cell fractions derived from RNA-sequencing data by using the leukocyte gene signaturebased CIBERSORT deconvolution method (Newman et al., 2015), which we applied on gene expression profiles from human adult (n = 703), pediatric (n = 92), and canine glioma (n = 40) (Figure S5). The relative immune cell fractions found in each glioma type were well correlated with one another, with the low-grade pediatric glioma exhibiting the lowest correlation with high-grade canine glioma (Rho = 0.83).
Figure 5. Immunohistochemistry of Canine and Human Gliomas.
(A) Representative hematoxylin & eosin and immunohistochemistry staining of human adult (n = 11), canine (n = 11), and human pediatric gliomas (n = 5) using antibodies against T cells (CD3), macrophage/microglia (IBA1), M2 polarized innate immune cells (CD163), monocytes (CD14), and B cells (CD79A). Scale bars, 50 μm.
(B) Violin plots represent the density of percentage positivity by field (y axis) for each of five antibodies described in (A). The points are the mean value of percentage positivity per patient within each of three cohorts, i.e., human adult (n = 11), canine (n = 11), and human pediatric gliomas (n = 5). Patients were grouped into high- versus low-grade gliomas in the absence of available molecular subtype data.
See also Figure S5.
DISCUSSION
Comparative genomic oncology is a robust approach for identifying evolutionarily conserved drivers and for studying the natural history of spontaneous tumors in an immune-competent host, e.g., in domestic dogs (Decker et al., 2015a; Frampton et al., 2018; Tollis et al., 2017). Our cross-species analysis using comprehensive molecular profiling of sporadic gliomas highlights two key findings. First, convergent evolution of gliomas is observed across canine, human pediatric, and human adult gliomas, with shared molecular traits such as shared hotspot and mutually exclusive mutations in PDGFRA and PIK3CA, and in genes associated with the p53 and cell-cycle pathways, among others. This is further supported by aneuploidy being prevalent among canine and human pediatric high-grade gliomas, which are potentially under selection pressure within shared syntenic regions of the genome. Also, DNA damagerelated mutational processes such as homologous recombination defects constitute a major source for progressive genomic instability, and generate somatic variations upon which natural selection acts to produce shared molecular and histopathological features of glioma. Second, the molecular landscape of canine gliomas resembles that of human pediatric gliomas based on the observed pattern of somatic alterations among non-shared drivers and DNA-methylation patterns. We did not observe canine counterparts of rare human glioma variants such as pleomorphic xanthoastrocytoma, giant cell glioblastoma, or pilocytic astrocytoma, by histopathology or by association in somatic drivers. To make a definitive claim that the canine gliomas are similar to one of the major categories of either adult or pediatric molecularly defined gliomas, additional characterization studies are needed that compare canine and human glioma in terms of cellular states (Neftel et al., 2019) as defined by single-cell transcriptomics.
Convergent evolution can reflect a footprint of adaptation to similar selective pressures (Fortunato et al., 2017). While such convergence is well appreciated in human cancers, and in particular treatment-resistant cancers (Venkatesan et al., 2017), our observation of such molecular and phenotypic convergence across two species provides a strong indicator of variations under selective pressures exerted by the tissue or ecological context (DeGregori, 2017; Schneider et al., 2017). We note that convergent evolution should not discount a possibility of drivers unique to canine gliomas, especially within the context of germline variants (Mansour et al., 2018; Truve et al., 2016) and non-coding regulatory regions (Lindblad-Toh et al., 2011; Villar et al., 2015). Characterization of such species-specific drivers can be of much value to identify evolutionary linchpins, which if abrogated can drive oncogenesis with similar histopathological and clinical traits. Further studies are needed to help understand how the time point at which tissue samples used in our comparative analysis were obtained, necropsy for canine samples, and diagnosis for human samples affects our results.
The molecular life history of a tumor is marked by multiple, often successive aberrations in genes (Armitage and Doll, 1954; Nowell, 1976). Accordingly, cancer is largely a disease of old age except in cases with early exposures to mutagens, e.g., germline or acquired defects in one or more hallmarks of cancer (Hanahan and Weinberg, 2011). The median age of occurrence for canine gliomas in our cohort was 9 years, i.e., dogs in their adult stage of life. However, we demonstrate that canine gliomas have a significantly lower somatic mutation rate and, consequently, a lower number of significantly mutated genes than adult human gliomas. The mutation burden of canine glioma is also less than what has been reported for other canine cancers, although a direct comparison would require additional standardization of sequencing and data-preprocessing methods (Hendricks et al., 2018; Lorch et al., 2019; Sakthikumar et al., 2018).
Canine gliomas harbor significantly higher aneuploidy than adult human high-grade gliomas, and are more similar to human pediatric gliomas (Gröbner et al., 2018; Mackay et al., 2017). We find additional support for aneuploidy as a major driver in canine and pediatric H3-mutant and H3 wild-type high-grade gliomas with the observation of aneuploidy in regions of shared synteny containing the HIST1 and PDGFRA genes, known pediatric glioma drivers (Gröbner et al., 2018; Mackay et al., 2017), and in noting shared homologous repair defects as a mutational process that could drive genomic instability (Blank et al., 2015; Targa and Rancati, 2018). Recent efforts to engineer aneuploidy have provided better understanding of the functional role of aneuploidy and how it can be targeted in cancer (Bakhoum and Cantley, 2018; Taylor et al., 2018). Canine high-grade gliomas carrying aneuploidy, especially among syntenic regions carrying the HIST1 and ACVR1 genes, can be utilized as a preclinical model for such functional screening as well as to validate recent studies showing its role in immune evasion (Bakhoum et al., 2018; Davoli et al., 2017).
Tissue context and tumor microenvironment are critical factors for tumorigenesis (Haigis et al., 2019; Wang et al., 2017), and current models are unable to accurately represent the development of spontaneous tumors (Buque and Galluzzi, 2018). This renders preclinical evaluation ineffective and increases costs of clinical trials and results in minimal yields for patients. Preclinical trials of dog glioma patients enable identification of evolutionarily constrained and potentially targetable drivers, but simultaneously benefit dogs with glioma by offering treatment options that otherwise are prohibitive due to associated healthcare costs (LeBlanc et al., 2016). Future efforts leveraging results from the comparative genomics of glioma to study immune-mediated host responses can shed light on the complex interplay between the tumor and host immune response and also aid in optimizing ongoing parallel canine clinical trials (Addissie and Klingemann, 2018) in order to improve an otherwise limited response to immunotherapies in canine and human gliomas. Our findings of canine gliomas with low tumor mutational burden but with a clonal nature of somatic drivers would be relevant in development of a preclinical model to dissect the interplay between mutation burden and immune escape during tumor evolution due to loss of clonal neoantigens (McGranahan and Swanton, 2019; Wolf et al., 2019). With respect to the immune microenvironment, differences in immune cell gene expression patterns between species could confound immune cell comparisons by under- or overestimating the presence of specific immune cell types. Despite these potential differences, comparative transcriptomic analyses of mouse and human immune cells have shown that the cells in each species exhibit a high degree of global conservation with one another, and signatures derived from murine immune cells have provided accurate immune infiltration estimates in human cancer types (Shay et al., 2013; Varn et al., 2017). Thus, the estimates in this study provide a baseline for how the relative fractions of major immune cells compare among adult, pediatric, and canine gliomas. We would have liked to have done further immune characterization including expression profiling of immune checkpoint response markers such as PD-1+ tumor-infiltrating lymphocytes and PD-L1 immune and tumor expression, but these antibodies have not been validated in canines. Moving forward, signatures derived from canine immune cells will be of value in examining the presence of more specific immune cell types.
In summary, our study shows that the comparative molecular life history of gliomas details conserved drivers of glioma at both the genetic and epigenetic levels, with aneuploidy as a major hallmark of high-grade disease. Our results effectively position preclinical models of spontaneous canine glioma for use in understanding glioma drivers, and evaluating therapies targeting aneuploidy as well as immunotherapies, with relevance to all human gliomas and pediatric disease in particular.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for data resources should be directed to and will be fulfilled by the Lead Contact, Roel Verhaak (roel. verhaak@jax.org). This study did not generate new unique reagents.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Canine Patients and Tissue Samples
Tissue samples from canine patients with gliomas were acquired with material transfer agreements from Auburn University College of Veterinary Medicine, Colorado State University, Texas A&M College of Veterinary Medicine & Biomedical Sciences, UC Davis School of Veterinary Medicine and Virginia-MD College of Veterinary Medicine. Tissue samples from resected tumor (n=83) and matched normal tissue (n=67 or paired cases) were collected at the surgical treatment or immediately following euthanasia. There were also four additional dog patients where we had adequate DNA and RNA for methylation (n=48) and RNA-seq (n=40) profiling but unable to do WGS/Exome sequencing because of failed library preparation (Table S1). Matched normal tissue were from post-necropsy sample of contra-lateral healthy brain tissue (n=38), white blood cells (n=13), and remaining 17 samples from other tissues. Samples were archived in snap-frozen (n=37/67 paired cases; n=8/16 tumor-only cases) and Formalin-Fixed Paraffin-Embedded (FFPE, n=30/67 paired cases; n=8/16 tumor-only cases) state. Samples were then shipped to sequencing core facilities for sample preparation, quality control and sequencing (see STAR Methods Details below).
METHOD DETAILS
Published Data Sources
For comparison to human glioma, we downloaded both - raw sequencing data and processed tables for human pediatric and adult gliomas with appropriate controlled-data access agreements where needed. We used published mutation rates (Figure 1D) and mutational signatures (Figure 3) from pan-cancer datasets from adults (n=3,281) and pediatric (n=961) cohorts (Alexandrov et al., 2013; Bailey et al., 2018; Gröbner et al., 2018). For aneuploidy and molecular life history analysis (details below), we downloaded raw sequencing data and analyzed whole genomes from 53 pediatric gliomas (Ma et al., 2018; St. Jude Cloud Pediatric Cancer Genome Project, https://pecan.stjude.cloud), SNP6 data from adult gliomas – IDHwt (n=517), IDHmut-codel (n=171), and IDHmut-noncodel (n=281) cases (Ceccarelli et al., 2016), as well as whole genomes from 23 adult GBMs (Brennan et al., 2013). For coding mutation rate calculation, we used a subset of TCGA glioma set where exome/whole genome based variant calls were available: IDHwt (n=371), IDHmut-non-codel (n=268), and IDHmut-codel (n=169).
Sample Preparation
DNA/RNA extraction - Genomic DNA and total RNA of fresh frozen tissue and FFPE tissue from paraffin scrolls were were extracted simultaneously using AllPrep DNA/RNA Mini Kit (Qiagen) and AllPrep DNA/RNA FFPE Kit (Qiagen) according to the manufacturer’s instructions, respectively. Additional DNase treatment was performed on-column for RNA purification. WGS sample preparation - 200–400ng of DNA was sheared to 400bp using a LE220 focused-ultrasonicator (Covaris) and size selected using Ampure XP beads (Beckman Coulter). The fragments were treated with end-repair, A-tailing, and ligation of Illumina compatible adapters (Integrated DNA Technologies) using the KAPA Hyper Prep Kit (Illumina) (KAPA Biosystems/ Roche). For FFPE samples, 5 to 10 cycles of PCR amplification were performed. Quantification of libraries were performed using real-time qPCR (Thermo Fisher). Libraries were sequenced paired end reads of 151bp on Illumina Hiseq X-Ten (Novogene). WES sample preparation - Sample were prepared as described above in the WGS sample preparation, targeting 200bp with PCR amplification. Target capture was performed using SeqCap EZ Canine Exome Custom Design (canine 140702_canFam3_exomeplus_BB_EZ_HX1 probe set) (Broeckx et al., 2015) (Roche Nimblegen). Briefly, WGS libraries were hybridized with capture probes using Nimblegen SepCap EZ Kit (Roche Nimblegen) according to manufacturer’s instruction. Captured fragments were PCR amplified and purified using Ampure XP beads.
Quantification of libraries were performed using real-time qPCR (Thermo Fisher). Libraries were sequenced paired end of 76bp on Hiseq4000 (Illumina). RNA-seq sample preparation - RNA-seq libraries were prepared with KAPA Stranded mRNA-Seq kit (Kapa Biosystem/ Roche) according to manufacturer’s instruction. First, poly A RNA was isolated from 300ng total RNA using oligo-dT magnetic beads. Purified RNA was then fragmented at 85°C for 6 mins, targeting fragments range 250–300bp. Fragmented RNA is reverse-transcribed with an incubation of 25°C for 10mins, 42°C for 15 mins and an inactivation step at 70°C for 15mins. This was followed by second strand synthesis at 16°C, 60 mins. Double stranded cDNA (dscDNA) fragments were purified using Ampure XP beads (Beckman). The dscDNA were then A-tailed, and ligated with illumina compatible adaptors (IDT). Adaptor-ligated DNA was purified using Ampure XP beads. This is followed by 10 cycles of PCR amplification. The final library was cleaned up using AMpure XP beads. Quantification of libraries were performed using real-time qPCR (Thermo Fisher). Sequencing was performed on Hiseq4000 (Illumina) generating paired end reads of 75bp. Reduced Representation Bisulfite Sequencing (RRBS) sample preparation - Library preparation for RRBS was performed using Premium RRBS Kit (Diagenode) according to manufacturer’s instructions. Briefly, 100ng of DNA was used for each sample, which was enzymatically digested, end-repaired and ligated with an adaptor. Subsequently, 8 samples with different adaptors were pooled together and subjected to bisulfite treatment. After purification steps following bisulfite conversion, the pooled DNA was amplified with 9–14 cycles of PCR and then cleaned up with Ampure XP beads. Quantification of libraries were performed using real-time qPCR (Thermo Fisher). Libraries were sequenced single end 101bp on Hiseq2500 (Illumina).
Sequencing Alignments, QC, and Fingerprinting
DNA alignments - DNA alignments for whole genome (WGS) and exome sequencing was done using bwa-mem (version 0.7.15-r1140) (Fleshner and Chernett, 1997) with -M -t 12 argument and against CanFam3.1 reference genome from UCSC, https://genome.ucsc.edu/cgi-bin/hgGateway?db=canFam3 (md5: 112bc809596d22c896d7e9bcbe68ede6). For each sample, fastq files were aligned per read group and then merged using Picard tools (v2.18.0, http://broadinstitute.github.io/picard) SortSam command to make an interim bam file. Final, analysis-ready bam file per sample – tumor and normal bam, if available – was created by series of steps following best practices guidelines from GATK4 (version 4.0.8.1) (DePristo et al., 2011), namely MarkDuplicates, Indel Realignment, and Base Quality Score Recalibration (BQSR). Alignment QC metrics were calculated using GATK4 DepthOfCoverage (for WGS) and CollectHSMetrics (for exome) as well as Qualimap (version 2.2.1) (Okonechnikov et al., 2016) bamqc for merged bam files. Coverage statistics were also based on regions of interest (ROIs) which consisted of exonic region-level annotations for biotypes: protein-coding gene, microrna, lincrna, and pseudogene from Ensembl gene annotations for canine genome (v91 and higher). We flagged samples as failed QC if merged bam file has a genome-wide coverage of < 30% or > 75% of ROIs have 30% or lesser coverage. Accordingly, three samples (of three cases) failed QC step and they were removed from all analyses. Note that 77 cases in patient tissues and samples section represent all cases which passed QC at WGS, exome, RNA-seq, and methylation level data preprocessing. RNA alignments - Raw fastq files from paired-end RNA-seq assay for 41 tumor samples and 3 matched normal tissue samples were first preprocessed through fastp (version 0.19.5) (Chen et al., 2018) to perform read-based quality pruning, including adapter trimming. Resulting fastq files were then used as input for kallisto quant (version 0.45.0) – a pseudoalignment based method to quantify RNA abundance at transcript-level in transcripts per million (TPM) counts format. We then used sleuth R package (version 0.30.0) (Pimentel et al., 2017) to output model-based, gene-level normalized TPM matrix which was also corrected for potential batch effects due to RNA-seq data derived from two sequencing core facilities and tissue archival (snap-frozen vs FFPE). Detailed workflow, including command-line parameters for model fitting are in extended methods section. RRBS alignments - Raw fastq files from RRBS assay for 45 tumor samples were processed through FastQC (version 0.11.7, https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and Trim Galore (version 0.5.0, https://github.com/FelixKrueger/TrimGalore) for quality control, filtering low quality base calls, and adapter trimming. Trimmed reads were then mapped to a bisulfite converted reference genome (canFam3.1, obtained from Ensembl release 85) using the Bismark Bisulfite Mapper (v0.19.1) with the Bowtie2 short read aligner (v2.2.3) (Krueger and Andrews, 2011), allowing for one non-bisulfite mismatch per read. Cytosine methylation calls were made for the mapped reads using the Bismark methylation extractor (version 0.19). The resulting methylation values were obtained as β-values, calculated as the ratio of methylated to total reads at a given CpG site. DNA fingerprinting – DNA fingerprinting for each of WGS and exome tumor-normal and tumor-only bam files was done using NGSCheckMate tool (version 1.0.0) (Lee et al., 2017). Germline snps in protein-coding regions was used as a variant reference panel to allow simultaneous fingerprinting of WGS and exome libraries. NGSCheckMate does sample pairing QC based on shared germline variants found in samples (tumor and normal tissue from the same patient) and also model difference between samples (or libraries) based on sequencing depth-dependent variation in allelic fraction of reference variants. Fingerprint results for WGS and exome samples from 77 canine glioma did not show mixture of tumor-normal or cross-patient sample contamination (See Figure S1F).
Somatic Variant Calling
Somatic variant calls were called on the merged whole genome and exome bam files using three callers: GATK4 (version 4.0.8.1) (McKenna et al., 2010) Mutect2 (Cibulskis et al., 2013), VarScan2 (version 2.4.2), and LoFreq (version 2.1.3.1) (Wilm et al., 2012). Matching and fingerprint validated WGS and exome files per sample were merged using Picard tools (v2.18.0, http://broadinstitute.github.io/picard), MergeSamFiles command. Three somatic callers were then run in either paired tumor – matched normal (n=57) or tumor-only (n=20) mode. Mutect2 was first run in panel-of-normals (PON) mode using 57 matched normal samples. Resulting PON file was used for calling somatic variant calls using Mutect2 in both, paired and tumor-only mode along with options: –germline-resources 58indiv.unifiedgenotyper.recalibrated_95.5_filtered.pass_snp.fill_tags.vcf.gz –af-of-alleles-not-in-resource 0.008621. Tumor-only Mutect2 mode was run using default arguments and paired Mutect2 calls had following arguments: –initial-tumor-lod 2.0 –normal-lod 2.2 –tumor-lod-to-emit 3.0 –pcr-indel-model CONSERVATIVE. Throughout the process of using GATK4 based tools, including Mutect2, we followed best practices guidelines (DePristo et al., 2011) where practical for canine genome, e.g., in contrast to human genome, population level resources are limited for canine genome. VarScan2 paired mode was run with a command: somatic and arguments: –min-coverage 8 –min-coverage-normal 6 –min-coverage-tumor 8 –min-reads2 2 –min-avg-qual 15 –min-var-freq 0.08 –min-freq-for-hom 0.75 –tumor-purity 1.0 –strand-filter 1 –somatic-p-value 0.05 –output-vcf 1. VarScan2 tumor-only mode was run using command: mpileup2cns and arguments: –min-coverage 8 –min-reads2 2 –min-avg-qual 15 –min-var-freq 0.08 –min-freq-for-hom 0.75 –strand-filter 1 –p-value 0.05 –variants –output-vcf 1. LoFreq paired mode was run using command: somatic and arguments: –threads 4 –call-indels –min-cov 7 –verbose and tumor-only mode was run using command: call and arguments: –call-indels –sig 0.05 –min-cov 7 –verbose -s. Resulting raw somatic calls - single nucleotide variants (SNV) and small insertions and/or deletions (Indels) - from three callers were then subject to filtering based on caller-specific filters and hard filters. Briefly, Mutect2 calls were subject to extensive filtering based on germline risk, artifacts arising due to sequencing platforms, tissue archival (FFPE), repeat regions, etc. See extended methods and https://software.broadinstitute.org/gatk/documentation/article?id=11136). VarScan2 somatic filters were applied as per developer’s guidelines (Koboldt et al., 2013). Hard filters were based upon filtering out variants present in dbSNP and PONs created via GATK4 Mutect2. Filtered somatic calls from three callers (in VCF version 4.2 format) were then subject to consensus somatic calls using SomaticSeq (version 3.1.0) (Fang et al., 2015) in majority voting mode with priority given to Mutect2 filtered (PASS) calls followed by consensus voting based on calls present in VarScan2 and LoFreq filtered calls. Resulting consensus VCF file for 77 cases were finally converted to Variant Effect Predictor (VEP version 91) (McLaren et al., 2016) annotated vcfs and Mutation Annotation Format (MAF, https://docs.gdc.cancer.gov/Data/File_Formats/MAF_Format) using vcf2maf utility (https://github.com/mskcc/vcf2maf). Annotated VCFs and MAFs were used for all of downstream analyses.
Significantly Mutated Genes (SMGs) Analysis
SMG analysis in canine gliomas (Figures 1A, 1C, and 2A) with paired tumor-normal samples (n=57) was performed using dNdS (Martincorena et al., 2017) and MuSiC2 (version 0.2) (Dees et al., 2012). We excluded tumor-only cases for being conservative in SMG analysis and minimize false-positives. Also, MuSiC2 required matched normal tissue required matched normal tissue for SMG analysis. Detailed parameters for SMG analysis are in extended methods. Detailed output from both methods are in Table S2.
Cancer Hallmark Analysis
Cancer hallmarks were defined according to published ten hallmarks (Hanahan and Weinberg, 2011) and one additional hallmark, i.e. epigenetic (Imielinski et al., 2012). A pool of 268 known glioma (Ceccarelli et al., 2016; Mackay et al., 2017) and pan-cancer driver genes (Gröbner et al., 2018; Bailey et al., 2018) were mapped to hallmarks following a previously published computer-assisted manual curation method (Table S4) (Iorio et al., 2018). Based on WHO molecular classification of brain tumors, somatic SNV and copy-number data from patients with human adult gliomas (AG) (Ceccarelli et al., 2016) were stratified into IDHwt (n=373), IDHmut-codel (n=169) and IDHmut-noncodel (n=268) subgroups while corresponding data from patients with human pediatric glioma (PG) (Mackay et al., 2017) were subgrouped based on mutations in Histone H3 gene, namely H3mut (n=200) and high-grade H3wt (n=126). For canine patients with glioma (CG), we used somatic mutations and copy-number calls from 67 cases with paired tumor-normal samples. For each of the six cohorts coding mutations were mapped to eleven hallmarks and coverage adjusted relative proportions of patients harboring an alteration in a given hallmark were calculated. For comparisons between cohorts a two-sided Fisher’s exact test was applied (Table S4).
Quantifying Somatic Mutation Rates
Somatic mutations (SNVs and Indels) rate was estimated within coding genes and adjusted based on relative per-base coverage with minimum coverage of 30X in coding regions (Figure 1D). Coding mutation rates for human pediatric (n=961) and adult cancers (n=3,800, includes 811 adult gliomas) were taken from published studies (Ceccarelli et al., 2016; Gröbner et al., 2018; Mansour et al., 2018).
Somatic Copy Number Segmentation
Somatic copy-numbers were called for paired tumor-normal cases (n=56) using HMMCopy tool (version 1.22.0) using author’s recommendations. In brief, GC counts and mappability files for CanFam3.1 genome were generated with 1000 bp window size. Read counts for each of tumor and normal bam files were generated using 1000 bp window size. Resulting count, mappability and count files were feed into HMMCopy algorithm (http://bioconductor.org/packages/release/bioc/html/HMMcopy.html) and segmentations were called using Viterbi algorithm. Segmented copy number calls were used to generate Integrated Genome Viewer (IGV) copy-number plots and GISTIC2 (version 2.0.22) based somatic copy number significance (Mermel et al., 2011), including calling gene-level deep deletions, loss-of-heterozygosity (LOH), and amplifications (Figure 2A) as well as inferring aneuploidy metrics (Figures 2B and 2C). Segmented copy number for pediatric gliomas (n=53) were called by using cloud-based TitanCNA workflow (https://dxapp.verhaaklab.com/dnanexus_ngsapp). Segmented copy number for adult gliomas were derived from SNP6 based platform from the TCGA Broad Firehose platform (version stddata__2016_01_28) with following download urls: http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/GBM/20160128/gdac.broadinstitute.org_GBM.Merge_snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.Level_3.2016012800.0.0.tar.gz and http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/LGG/20160128/gdac.broadinstitute.org_LGG.Merge_snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.Level_3.2016012800.0.0.tar.gz Only primary tumor cases from TCGA GBM (n=577) and TCGA LGG (n=513) cohort were used for downstream analyses, i.e., pathway analysis (Figure 1C) and aneuploidy metrics (Figures 2B–2D).
Allele Specific Copy-Number Analysis
We derived allele-specific copy numbers and copy-number based clonality inference (including purity and ploidy estimates) using TitanCNA algorithm (version 1.19.1) (Ha et al., 2014). Snakemake (version 5.2.1) based workflow (Koster and Rahmann, 2018) was implemented using default arguments and genome-specific germline dbSNP resource (see under extended STAR Methods) (https://github.com/gavinha/TitanCNA/tree/master/scripts/snakemake) for WGS paired tumor-normal samples from 56 canine patients. For pediatric gliomas (n=53) and adult gbms with WGS data (n=23), allele-specific copy-number calls were used from TitanCNA workflow. Allele-specific copy-numbers were used for mutational signature and molecular timing analysis (Figure 3).
Aneuploidy Metrics
The simplest metric of aneuploidy was computed by taking the size of all non-neutral segments divided by the size of all segments. The resulting aneuploidy value indicates the proportion of the segmented genome that is non-diploid. In parallel, an arm-level aneuploidy score modeled after a previously described method was computed (Taylor et al., 2018). Briefly, adjacent segments with identical arm-level calls (−1, 0 or 1) were merged into a single segment with a single call. For each merged/reduced segment, the proportion of the chromosome arm it spans was calculated. Segments spanning greater than 80% of the arm length resulted in a call of either −1 (loss), 0 (neutral) or +1 (gain) to the entire arm, or NA if no contiguous segment spanned at least 80% of the arm’s length. For each sample the number of arms with a non-neutral event was finally counted. The resulting aneuploidy score is a positive integer with a minimum value of 0 (no chromosomal arm-level events detected) and a maximum value of 38 (total number of autosomal chromosome arms – given all of canine chromosomes are either acrocentric or telocentric).
Clustering Shared Syntenic Regions
Shared syntenic regions between CanFam3.1 and hg19 reference genome were downloaded from Ensembl BioMart (version 94) database using orthologous mapped Ensembl gene ids. Arm-level synteny was based on arm-level aneuploidy scores of shared syntenic regions in the respective, canine and human genomes. Hierarchical clustering of proportion of patients per molecular subtype having syntenic arm-level aneuploidy was then carried out for each of canine, human pediatric and adult cohort (Figure 2C).
Estimating Intra-tumoral Heterogeneity
We estimated patient-level ITH based on whole-genome derived subclonal structure and number of somatic variants in each of these subclones. Subclonal structure and cellular prevalence or cancer cell fraction of each tumor subclone (and underlying somatic variants) was derived using TITAN allele-specific copy number calls. Since accuracy of inferred subclonal structure depends largely on sequencing read depth and number of somatic variants per inferred subclone, we limited estimation of subclonal structure for maximum five subclones per patient given a minimum sequencing read depth of 30X for whole genome data we had across all three cohorts. Shannon entropy was then calculated using entropy function in the R package: entropy by taking number of somatic variants per subclone per patient as a vector. A resulting Shannon entropy value was used to plot figures along with cancer cell fraction and number of clones derived per patient. We acknowledge that our estimation of ITH and resolving subclonal structure can be improved with higher depth of sequencing (100X or more) to detect subclonal structures (number of clones) (Deshwar et al., 2015).
Mutational Signature Analysis
Mutational signature analysis was performed in two-parts. First, de-novo signatures (Figure 3B) were constructed for canine (n=81), human pediatric (n=53) and adult cohort (n=23) using somatic snvs from whole-genome data. Signatures were constructed using non-negative matrix factorization (nmf R package, version 0.21.0) with brunet approach and 100 iterations with expected range of signatures between 2 to 10. Optimal signatures were then selected using nmfEstimateRank function to match number of de-novo signatures (clusters) − 1 where inflection point for cophenetic correlation coefficient was observed. Accordingly, three de-novo signatures were found in canine and human pediatric gliomas while five in adult glioblastoma cohort. In the second part, known human mutational signatures from COSMIC (v2, n=30) and published pediatric cancer signature from two studies, T1 to T11 (Mansour et al., 2018) and P1 (Gröbner et al., 2018) were pooled together and used to deconvolute (MutationalPattern R package, version 1.6.2) mutational trinucleotide context (n=96) from somatic snvs in each of three cohorts. Somatic ultra-hypermutation cases from pediatric (n=3) and adult cohort (n=1) were excluded from signature analysis. Cosine similarities of known signatures with de-novo signatures was then calculated and clustered using hierarchical clustering (Figure 3B). Absolute and relative contribution of known signatures per sample was then quantified using fit_to_signatures function which finds the linear combination of signatures that closely resembles 96 context based mutational matrix by solving the nonnegative least-squares constraints problem. We then selected top contributing signatures per cohort based on signatures which contributed per sample to higher than 3rd quartile of median value of each signature’s contribution (rowMedian) per cohort (Figure S3A). Top contributing signatures were further calculated using outlier profling on canine patients showing highest mutational load (>3rd quartile of median coding mutation rate per megabase) and plotted in Figure 3A. Outlier sample detection was done using car::outlierTest function in R to label true outliers from entire cohort (2 cases) while correcting for confounding effects due to type of tissue archival (snap-frozen vs ffpe) and analysis type (tumor-matched normal versus tumor-only somatic variant calling). This was followed by second run of outlier by first excluding true outliers (2 cases), and then labelling outliers (six cases) based on chi-squared statistics using outliers::scores function in R. Signatures contributing to driver mutations (Figure 3C) were calculated based on first getting relative proportion of trinucleotide context per snv and then finding known signatures with maximum value for the same trinucleotide context. Known signatures were combined to a single group where they are shown in literature as potential underlying process, e.g., aging group is associated with COSMIC signature 1 and 5, and show significant cosine similarity (> 0.9) with pediatric signatures T1 and T4, respectively. Table S5 provides mapping between signature and known/proposed mechanisms, if any.
Molecular Timing Analysis and Natural History of Tumors
Probabilistic estimation of relative timing of driver mutations (among 79 observed somatic snvs in cancer driver genes) was based on existing methods (Gerstung et al., 2017; Jolly and Van Loo, 2018) with several steps carried out using Palimpsest R package (version 1.0.0; https://github.com/FunGeST/Palimpsest) (Shinde et al., 2018) and custom R scripts based on published approach (McGranahan et al., 2015): First step involved categorizing somatic drivers into clonal vs subclonal events using estimated cancer cell fraction (CCF) which is estimated fraction of cancer cells with a somatic snv. CCF per somatic snv was a product of variant allelic fraction (VAF) of a somatic snv, adjusted by local copy number of gene locus and whole tumor sample (ploidy) as well as purity estimate (tumor cell content) inferred from TitanCNA algorithm (Detailed under copy number estimation section above). A clonal (early) vs subclonal (late) mutation was then classified based on upper boundary of CCF was above 0.95 (clonal) or not (subclonal). Second, we timed copy number gain and copy-neutral LOH regions based on VAF of somatic snvs in these copy regions, i.e., early mutations prior to copy gain will have higher VAF relative to VAF of late mutations after copy gain. Third, we ordered mutations in four sequential categories: early clonal, early subclonal, late clonal, and late subclonal. We note here that early subclonal and late clonal categories are result of underlying parallel and/or convergent evolution of multiple clones (Venkatesan and Swanton, 2016) and/or a technical limitation (given ~60X depth of merged bam files and lack of spatial sequencing data) in resolving polyclonal structure of a tumor sample (Deshwar et al., 2015). We then tally frequency of each of these four categories per somatic driver mutation and get the average frequency of each category per driver mutation at cohort (canine, pediatric, adult) level. These average frequencies are converted to winning tables, similar to sports statistics where each driver mutation competes with remaining driver mutations with winning being an early somatic event based on order of events using clonality (Jolly and Van Loo, 2018) (step 3). Finally, a winning table is then passed to Bradley-Terry model (BradleyTerryScalable R package, version 0.1.0.9000) to estimate winning probability (driver event being an early event) based on a Bayesian maximum a posteriori probability (MAP) estimate. Resulting winning probability per driver mutation is subtracted from 1 to plot multiple density plots (ggridges R package, version: 0.5.1.9000) with X-axis now showing a probability of event being a late event (Figure 3D). We note that density plots are based on kernel density estimates and thus, may extend their tails (probability distribution) beyond 1 or less than zero (https://serialmentor.com/dataviz/histograms-density-plots.html).
Class Prediction Using Methylation Data
To compare the methylation patterns of human and canine glioma, the LIBLINEAR library was used to fit an L2-regularized logistic regression classifier. Model training and validation was performed on the human glioma samples and normal controls in the GSE109381 dataset (Capper et al., 2018), with the methylation status of CpGs located in regions of the human genome orthologous to canine CpG islands used to predict DNA methylation-based subtypes of glioma. The methylation categories designated as regression outcome variables were derived from the World Health Organization classification of gliomas: IDH-wild-type adult glioma, IDH-mutant, 1p/19q-intact adult glioma, IDH-mutant, 1p/19q-codeleted adult glioma, adult normal control, pediatric glioma, and pediatric normal control. After model fitting, the logistic regression classifier was applied to the canine samples, using the β-values of CpGs orthologous to the selected 11,495 Illumina 450K probes as input data. For classifier CpG sites in the canine samples with no methylation observations, β-values were predicted using the DNA module of the DeepCpG algorithm, a deep learning algorithm that predicts methylation state based on local DNA sequence context (Angermueller et al., 2017). The logistic regression classifier outputs the probability that a sample matches a given methylation category. Category probabilities were calculated for the canine samples, and these probabilities were compared with sample age, anatomical location, tumor grade, tumor purity, and mutation rate (Figure 4).
Immunohistochemistry
Staining
Hematoxylin & Eosin staining was used to classify glioma grade and lineage. The immunohistochemistry panel included those antibodies that have been documented to work in canine tissues and include myeloid microglia/macrophages (IBA1), monocytes (CD14) and their M2 skew subtype (CD163), and lymphoid T cells (CD3) and B cells (CD79a). Slides with 5um sections, were deparaffinized and rehydrated in a dry incubator (60°C for 1 hour), xylene, and histological grade ethanol. Antigen retrieval was performed using citrate buffer and a pressure cooker at 120°C23 for 12 minutes. Quenching for endogenous peroxidase was performed with 3% H2O2 for 15 minutes at room temperature. Non-specific binding was minimized using ready-to-use protein blocker (Dako) applied for 15 minutes before the application of the primary antibody overnight at 4°C. All the washing was done using 1x T-PBS buffer mixed with 0.1% Tween 20. The biotinylated secondary antibody was applied for 30 minutes at room temperature followed by three washes with buffer for 5 minutes each. Color development was performed using the DAKO DAB kit and color change was monitored until an appropriate detectable level was achieved (10–60 sec depending on the antibody). Slides were counterstained with hematoxylin (25 seconds) and bluing buffer, then rehydrated and cover-slipped with long lasting mounting solution. The immunohistochemistry quantification were done blindly relative to the tumor pathology. Scanning and tissue segmentation - Scanning and analysis were performed using the PerkinElmer Vectra Automated Quantitative Pathology Imaging System and the inForm Cell Analysis software (ver 2.4). Slides were scanned twice on low- and high-power fields as follows: the first scan was of the whole slide on low power field (10x) for manual tissue segmentation to identify three tumor regions/categories as necrotic center, tumor and invasive edge under the neuropathologist’s supervision/direction. For each region, every fourth field was imaged (25%) on high-power field (20x) and resulted in 21 to 274 fields per slide, which varies according to the size of the tissue and presence or absence of necrosis. For the training set, heterogeneous fields were randomly selected to include tissue, non-tissue and damaged areas. Hematoxylin and DAB was used to identify the nuclei. Positive and negative cells were distinguished visually and three optical densities (OD) thresholds were set accordingly. The thresholds allowed 4-bin (0 = negative, +1 = weak positive, +2 = intermediate, +3 = strong positive) sorting of cells depending on the positivity and its intensity. The intermediate positivity threshold was calculated as the midpoint after setting the lower and higher threshold. The algorithm of the training set was applied for all the high-power fields captured. The results were inspected and the nonspecific and defective fields were removed before compiling the dataset. The same process was applied for all seven markers (Figure 5).
CIBERSORT Based Expression Analysis
Processed RNA-seq expression matrices from canine (n=40; 25 HGG, 14 LGG, 1 unknown grade), adult (n=703; 529 LGG, 174 GBM), and pediatric glioma (n=92; 42 LGG, 50 HGG) were each run as separate jobs into the CIBERSORT webserver (https://cibersort.stanford.edu) and processed in relative mode using the following parameters: Signature Genes: LM22 CIBERSORT default, Permutations run: 100, Quantile normalization disabled (Newman et al., 2015). The resulting cellular fraction tables were then collapsed from 22 cell types into 11 based on lineage, using groupings from a prior publication (Gentles et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using R version 3.6.1. Statistical details for analyses are described in the respective sub-section under the Method Details section above and summarized in figure legends. p value of <0.05 were considered statistically significant. No statistical methods were used to predetermine study sample size.
DATA AND CODE AVAILABILITY
Sequencing data generated during this study is available in the Binary Alignment Map (BAM) format at the NCBI SRA database with the BioProject accession ID PRJNA579792 [URL: https://dataview.ncbi.nlm.nih.gov/object/PRJNA579792]. Software code used to generate figures is available at https://github.com/TheJacksonLaboratory/canineglioma and documented at the URL, https://canineglioma.verhaaklab.com.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Canine Glioma Patient Samples | This paper | Detailed under Table S1 |
| Critical Commercial Assays | ||
| AllPrep DNA/RNA Mini Kit | Qiagen | N/A |
| AllPrep DNA/RNA FFPE Kit | Qiagen | N/A |
| KAPA Hyper Prep Kit (Illumina) | KAPA Biosystems/Roche | N/A |
| SeqCap EZ Canine Exome Custom Design | Roche Nimblegen | canine 140702_canFam3_exomeplus_BB_EZ_HX1 probe set |
| Nimblegen SepCap EZ Kit | Roche Nimblegen | N/A |
| KAPA Stranded mRNA-Seq kit | KAPA Biosystems/Roche | N/A |
| Premium RRBS Kit | Diagenode | N/A |
| Deposited Data | ||
| DNA sequencing data - WGS and Exome | This paper | NCBI SRA Accession ID: PRJNA579792 |
| RNA sequencing data | This paper | NCBI SRA Accession ID: PRJNA579792 |
| RRBS sequencing data | This paper | NCBI SRA Accession ID: PRJNA579792 |
| Software and Algorithms | ||
| bwa | v0.7.15-r1140 | http://bio-bwa.sourceforge.net/ |
| Genome Analysis ToolKit (GATK) | v4.0.8.1 | https://software.broadinstitute.org/gatk/ |
| Qualimap | v2.2.1 | http://qualimap.bioinfo.cipf.es/ |
| fastp | v0.19.5 | https://github.com/OpenGene/fastp |
| kallisto | v0.45.0 | https://pachterlab.github.io/kallisto |
| sleuth | v0.30.0 | https://pachterlab.github.io/sleuth |
| FastQC | v0.11.7 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| TrimGalore | v0.5.0 | https://github.com/FelixKrueger/TrimGalore |
| Bismark Bisulfite Mapper | v0.19.1 | https://github.com/FelixKrueger/Bismark |
| Bowtie2 | v2.2.3 | N/A |
| NGSCheckMate | v1.0.0 | https://github.com/parklab/NGSCheckMate |
| Mutect2 - GATK4 | v4.0.8.1 | https://software.broadinstitute.org/gatk/ |
| VarScan2 | v2.4.2 | https://github.com/dkoboldt/varscan |
| LoFreq | v2.1.3.1 | https://github.com/CSB5/lofreq |
| SomaticSeq | v3.1.0 | https://github.com/bioinform/somaticseq |
| Variant Effect Predictor (VEP) | v91 | https://github.com/Ensembl/ensembl-vep |
| dNdScv | 0.0.1.0 | https://github.com/im3sanger/dndscv |
| MuSiC2 | v0.2 | https://github.com/ding-lab/MuSiC2 |
| GISTIC2 | v2.0.22 | ftp://ftp.broadinstitute.org/pub/GISTIC2.0/GISTICDocumentation_standalone.htm |
| HMMCopy | v1.22.0 | http://bioconductor.org/packages/release/bioc/html/HMMcopy.html |
| TitanCNA | v1.19.1 | https://github.com/gavinha/TitanCNA |
| Snakemake | v5.2.1 | https://snakemake.readthedocs.io/en/stable/ |
| flowr | v0.9.10 | https://github.com/sahilseth/flowr |
| NMF R package | v0.21.0 | https://cran.r-project.org/web/packages/NMF |
| Entropy R package | v1.2.1 | https://cran.r-project.org/web/packages/entropy |
| outliers R package | v0.14 | https://cran.r-project.org/web/packages/outliers/ |
| MutationalPattern R package | v1.6.2 | https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html |
| Palimpsest R package | v1.0.0 | https://github.com/FunGeST/Palimpsest |
| BradleyTerryScalable R package | 0.1.0.9000 | https://cran.r-project.org/web/packages/BradleyTerryScalable/vignettes/BradleyTerryScalable.html |
| DNANexus app for St Jude Cloud data analysis | v1.1.6 (This paper) | https://dxapp.verhaaklab.com/dnanexus_ngsapp |
| CIBERSORT | webserver | https://cibersort.stanford.edu |
Significance.
Diffuse gliomas are the most common malignant brain tumors, with high-grade tumors carrying a dismal prognosis. Preclinical models have proven themselves as poor predictors of clinical efficacy. Spontaneous glioma in dogs provides an attractive alternative model because of their comparable tumor microenvironment and tumor life history. We determined the similarities and differences between human and canine gliomas through genomic profiling, and leveraged our datasets to identify conserved somatic drivers, mutational processes, and temporal ordering of somatic glioma events across species. Canine gliomas resemble human gliomas at (epi-)genetic levels and are more reminiscent of pediatric than adult disease, thus rationalizing sporadic canine glioma as a preclinical model tailored to measuring treatment efficacies in patients with canine or human glioma.
Highlights.
Genomic, epigenomic, and transcriptomic characterization of sporadic glioma in dogs
Somatic alterations in canine glioma converge with human glioma drivers
Canine glioma resemble pediatric human glioma by mutation rate and DNA methylation
Microenvironment similarity between canine and human pediatric and adult glioma
ACKNOWLEDGMENTS
This work is supported by grants from the National Institutes of Health (NIH): Cancer Center Support grants P30CA16672 and P30CA034196 and Cancer Center Support Grant Supplement 3P30CA016672-41S7 (A.B.H.); R01 CA190121 (R.G.W.V.), R01 CA120813 (A.B.H.); P0 1CA207206 (J.H.R.); an unrestricted grant from Agilent Technologies (R.G.W.V.); and philanthropic support from Mr. Herb Simmons (A.B.H.). E.K. is recipient of an MD-Fellowship by the Boehringer Ingelheim Fonds and is supported by the German Academic Scholarship Foundation. F.S.V. is supported by a postdoctoral fellowship from The Jane Coffin Childs Memorial Fund for Medical Research. F.P.B. is supported by the JAX Scholar program and K99 CA226387. K.C.J. is the recipient of an American Cancer Society Fellowship (130984-PF-17-141-01-DMC). This work was partially supported (A.K.L., C.M.) by the Intramural Program of the National Cancer Institute, NIH (Z01-BC006161). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
DECLARATION OF INTERESTS
R.G.W.V. declares equity in Boundless Bio, Inc. A.B.H. receives royalties and milestone payments for licensed intellectual property from Celldex Therapeutics, research grant support from Merck, and is a scientific board member for Caris Life Sciences. The other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.ccell.2020.01.004.
REFERENCES
- Addissie S, and Klingemann H (2018). Cellular immunotherapy of canine cancer. Vet. Sci. 5, 10.3390/vetsci5040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktipis CA, Boddy AM, Gatenby RA, Brown JS, and Maley CC (2013). Life history trade-offs in cancer evolution. Nat. Rev. Cancer 13, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, Caldas C, Califano A, Doherty M, Elsner M, et al. (2015). Toward understanding and exploiting tumor heterogeneity. Nat. Med. 21, 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermueller C, Lee HJ, Reik W, and Stegle O (2017). DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 18, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage P, and Doll R (1954). The age distribution of cancer and a multistage theory of carcinogenesis. Br. J. Cancer 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. (2018). Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, and Cantley LC (2018). The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel FP, Wesseling P, and Verhaak RGW (2018). Reconstructing the molecular life history of gliomas. Acta Neuropathol. 135, 649–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank HM, Sheltzer JM, Meehl CM, and Amon A (2015). Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol. Biol. Cell 26, 1440–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiotis VA, and Charest A (2018). Immunotherapies for malignant glioma. Oncogene 37, 1121–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. (2013). The somatic genomic landscape of glioblastoma. Cell 155, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckx BJ, Hitte C, Coopman F, Verhoeven GE, De Keulenaer S, De Meester E, Derrien T, Alfoldi J, Lindblad-Toh K, Bosmans T, et al. (2015). Improved canine exome designs, featuring ncRNAs and increased coverage of protein coding genes. Sci. Rep. 5, 12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NF, Carter TJ, Ottaviani D, and Mulholland P (2018). Harnessing the immune system in glioblastoma. Br. J. Cancer 119, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buque A, and Galluzzi L (2018). Modeling tumor immunology and immunotherapy in mice. Trends Cancer 4, 599–601. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. (2015). Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, et al. (2018). DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. (2016). Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, and Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, and Elledge SJ (2017). Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker B, Davis BW, Rimbault M, Long AH, Karlins E, Jagannathan V, Reiman R, Parker HG, Drögemüller C, Corneveaux JJ, et al. (2015a). Comparison against 186 canid whole-genome sequences reveals survival strategies of an ancient clonally transmissible canine tumor. Genome Res. 25, 1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker B, Parker HG, Dhawan D, Kwon EM, Karlins E, Davis BW, Ramos-Vara JA, Bonney PL, McNiel EA, Knapp DW, and Ostrander EA (2015b). Homologous mutation to human BRAF V600E is common in naturally occurring canine bladder cancer—evidence for a relevant model system and urine-based diagnostic test. Mol. Cancer Res. 13, 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, Mooney TB, Callaway MB, Dooling D, Mardis ER, et al. (2012). MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J (2017). Connecting cancer to its causes requires incorporation of effects on tissue microenvironments. Cancer Res. 77, 6065–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, and Morris Q (2015). PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 16, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PJ, York D, Higgins RJ, LeCouteur RA, Joshi N, and Bannasch D (2016). Chromosomal aberrations in canine gliomas define candidate genes and common pathways in dogs and humans. J. Neuropathol. Exp. Neurol. 75, 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang LT, Afshar PT, Chhibber A, Mohiyuddin M, Fan Y, Mu JC, Gibeling G, Barr S, Asadi NB, Gerstein MB, et al. (2015). An ensemble approach to accurately detect somatic mutations using SomaticSeq. Genome Biol. 16, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleshner I, and Chernett NL (1997). A wellness model for the geriatric population. Home Care Provid 2, 321–323. [DOI] [PubMed] [Google Scholar]
- Fortunato A, Boddy A, Mallo D, Aktipis A, Maley CC, and Pepper JW (2017). Natural selection in cancer biology: from molecular snowflakes to trait hallmarks. Cold Spring Harb. Perspect. Med. 7, 10.1101/cshperspect.a029652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D, Schwenzer H, Marino G, Butcher LM, Pollara G, Kriston-Vizi J, Venturini C, Austin R, de Castro KF, et al. (2018). Molecular signatures of regression of the canine transmissible venereal tumor. Cancer Cell 33, 620–633.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Mitchell TJ, Rubanova Y, Anur P, Rosebrock D, Yu K, et al. (2017). The evolutionary history of 2,658 cancers. bioRxiv. 10.1101/161562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, et al. (2018). The landscape of genomic alterations across childhood cancers. Nature 555, 321–327. [DOI] [PubMed] [Google Scholar]
- Ha G, Roth A, Khattra J, Ho J, Yap D, Prentice LM, Melnyk N, McPherson A, Bashashati A, Laks E, et al. (2014). TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 24, 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis KM, Cichowski K, and Elledge SJ (2019). Tissue-specificity in cancer: the rule, not the exception. Science 363, 1150–1151. [DOI] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hendricks WPD, Zismann V, Sivaprakasam K, Legendre C, Poorman K, Tembe W, Perdigones N, Kiefer J, Liang W, DeLuca V, et al. (2018). Somatic inactivating PTPRJ mutations and dysregulated pathways identified in canine malignant melanoma by integrated comparative genomic analysis. PLoS Genet. 14, e1007589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, Ma J, Edmonson MN, Hedlund EK, Rusch MC, et al. (2014). The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 5, 3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, Garcia-Alonso L, Brammeld JS, Martincorena I, Wille DR, McDermott U, and Saez-Rodriguez J (2018). Pathway-based dissection of the genomic heterogeneity of cancer hallmarks’ acquisition with SLAPenrich. Sci. Rep. 8, 6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. (2017). Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121. [DOI] [PubMed] [Google Scholar]
- Jolly C, and Van Loo P (2018). Timing somatic events in the evolution of cancer. Genome Biol. 19, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. (2013). Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 45, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, Breen M, Kitchell B, McNeil E, Modiano JF, et al. (2006). The dog as a cancer model. Nat. Biotechnol. 24, 1065–1066. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Larson DE, and Wilson RK (2013). Using VarScan 2 for germline variant calling and somatic mutation detection. Curr. Protoc. Bioinformatics 44 15.14.1–17. 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler JW, Miller AD, Miller CR, Porter B, Aldape K, Beck J, Brat D, Cornax I, Corps K, Frank C, et al. (2018). A revised diagnostic classification of canine glioma: towards validation of the canine glioma patient as a naturally occurring preclinical model for human glioma. J. Neuropathol. Exp. Neurol. 77, 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, and Rahmann S (2018). Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 34, 3600. [DOI] [PubMed] [Google Scholar]
- Krueger F, and Andrews SR (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc AK, Mazcko C, Brown DE, Koehler JW, Miller AD, Miller CR, Bentley RT, Packer RA, Breen M, Boudreau CE, et al. (2016). Creation of an NCI comparative brain tumor consortium: informing the translation of new knowledge from canine to human brain tumor patients. Neuro Oncol. 18, 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee S, Ouellette S, Park WY, Lee EA, and Park PJ (2017). NGSCheckMate: software for validating sample identity in next-generation sequencing studies within and across data types. Nucleic Acids Res. 45, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. (2011). A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch G, Sivaprakasam K, Zismann V, Perdigones N, Contente-Cuomo T, Nazareno A, Facista S, Wong S, Drenner K, Liang WS, et al. (2019). Identification of recurrent activating HER2 mutations in primary canine pulmonary adenocarcinoma. Clin. Cancer Res. 25, 5866–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, and Ellison DW (2016). The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820. [DOI] [PubMed] [Google Scholar]
- Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, et al. (2017). Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour TA, Lucot K, Konopelski SE, Dickinson PJ, Sturges BK, Vernau KL, Choi S, Stern JA, Thomasy SM, Doring S, et al. (2018). Whole genome variant association across 100 dogs identifies a frame shift mutation in DISHEVELLED 2 which contributes to Robinow-like syndrome in Bulldogs and related screw tail dog breeds. PLoS Genet. 14, e1007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, and Campbell PJ (2017). Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, and Swanton C (2019). Neoantigen quality, not quantity. Sci. Transl. Med. 11, 10.1126/scitranslmed.aax7918. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, and Swanton C (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, and DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, and Cunningham F (2016). The ensembl variant effect predictor. Genome Biol. 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, and Getz G (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC (1976). The clonal evolution of tumor cell populations. Science 194, 23–28. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, and Garcia-Alcalde F (2016). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, and Gilad Y (2011). A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 7, e1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. (2009). An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325, 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. [DOI] [PubMed] [Google Scholar]
- Pollack IF, Agnihotri S, and Broniscer A (2019). Childhood brain tumors: current management, biological insights, and future directions. J. Neurosurg. Pediatr. 23, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RC, and Dou Y (2015). Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthikumar S, Elvers I, Kim J, Arendt ML, Thomas R, Turner-Maier J, Swofford R, Johnson J, Schumacher SE, Alfoldi J, et al. (2018). SETD2 is recurrently mutated in whole-exome sequenced canine osteosarcoma. Cancer Res. 78, 3421–3431. [DOI] [PubMed] [Google Scholar]
- Schneider G, Schmidt-Supprian M, Rad R, and Saur D (2017). Tissuespecific tumorigenesis: context matters. Nat. Rev. Cancer 17, 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A, and ImmGen C (2013). Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. U S A 110, 2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde J, Bayard Q, Imbeaud S, Hirsch TZ, Liu F, Renault V, Zucman-Rossi J, and Letouze E (2018). Palimpsest: an R package for studying mutational and structural variant signatures along clonal evolution in cancer. Bioinformatics 34, 3380–3381. [DOI] [PubMed] [Google Scholar]
- Snyder JM, Shofer FS, Van Winkle TJ, and Massicotte C (2006). Canine intracranial primary neoplasia: 173 cases (1986–2003). J. Vet. Intern. Med. 20, 669–675. [DOI] [PubMed] [Google Scholar]
- Song RB, Vite CH, Bradley CW, and Cross JR (2013). Postmortem evaluation of 435 cases of intracranial neoplasia in dogs and relationship of neoplasm with breed, age, and body weight. J. Vet. Intern. Med. 27, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Stearns SC (1992). The Evolution of Life Histories (Oxford University Press; ). [Google Scholar]
- Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et al. (2014). Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 14, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targa A, and Rancati G (2018). Cancer: a CINful evolution. Curr. Opin. Cell Biol. 52, 136–144. [DOI] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. (2019). COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al. (2018). Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, vonHoldt B, Horvath S, and Pellegrini M (2017). An epigenetic aging clock for dogs and wolves. Aging (Albany NY) 9, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schiffman JD, and Boddy AM (2017). Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr. Opin. Genet. Dev. 42, 40–47. [DOI] [PubMed] [Google Scholar]
- Truve K, Dickinson P, Xiong A, York D, Jayashankar K, Pielberg G, Koltookian M, Muren E, Fuxelius HH, Weishaupt H, et al. (2016). Utilizing the dog genome in the search for novel candidate genes involved in glioma development-genome wide association mapping followed by targeted massive parallel sequencing identifies a strongly associated locus. PLoS Genet. 12, e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varn FS, Wang Y, Mullins DW, Fiering S, and Cheng C (2017). Systematic pan-cancer analysis reveals immune cell interactions in the tumor microenvironment. Cancer Res. 77, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S, and Swanton C (2016). Tumor evolutionary principles: how intratumor heterogeneity influences cancer treatment and outcome. Am. Soc. Clin. Oncol. Educ. Book 35, e141–e149. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Birkbak NJ, and Swanton C (2017). Constraints in cancer evolution. Biochem. Soc. Trans. 45, 1–13. [DOI] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. (2015). Enhancer evolution across 20 mammalian species. Cell 160, 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. (2017). Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Chen P, Gupta P, Ott M, Zamler D, Kassab C, Bhat KP, Curran MA, de Groot JF, and Heimberger AB (2019). Immune biology of glioma associated macrophages and microglia: functional and therapeutic implications. Neuro Oncol. 10.1093/neuonc/noz212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ, Werner B, Graham TA, and Sottoriva A (2016). Functional versus non-functional intratumor heterogeneity in cancer. Mol. Cell. Oncol. 3, e1162897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, and Nagarajan N (2012). LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, Levy R, Jimenez-Sanchez A, Trabish S, Lee JS, et al. (2019). UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 179, 219–235.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, van der Weyden L, Schott CR, Foote A, Constantino-Casas F, Smith S, Dobson JM, Murchison EP, Wu H, Yeh I, et al. (2019). Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat. Commun. 10, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. (2013). Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 45, 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated during this study is available in the Binary Alignment Map (BAM) format at the NCBI SRA database with the BioProject accession ID PRJNA579792 [URL: https://dataview.ncbi.nlm.nih.gov/object/PRJNA579792]. Software code used to generate figures is available at https://github.com/TheJacksonLaboratory/canineglioma and documented at the URL, https://canineglioma.verhaaklab.com.