Abstract
N6-methyladenosine (m6A) is a dynamic RNA modification that regulates various aspects of RNA metabolism and has been implicated in many biological processes and transitions. m6A is highly abundant in the brain; however, only recently has the role of m6A in brain development been a focus. The machinery that controls m6A is critically important for proper neurodevelopment, and the precise mechanisms by which m6A regulates these processes are starting to emerge. However, the role of m6A in neurodegenerative and neuropsychiatric diseases still requires much elucidation. This review discusses and summarizes the current body of knowledge surrounding the function of the m6A modification in regulating normal brain development, neurodegenerative diseases and outlines possible future directions.
Keywords: : epitranscriptomics, N6-methyladenosine, neurodegenerative disease, neurodevelopment, neurogenesis
Tight regulation of the nervous system is required to ensure proper function, and predictably, anomalies during brain development have been associated with neurological disorders. Indeed, several neural and non-neural cell types must be generated at the required numbers, in the appropriate location, at a specific time for proper brain development. Furthermore, accurate synaptic connections between neurons and efficient communication between distinct cell populations are essential for the proper function of the brain. Indeed, many studies have provided a greater understanding into human neurodevelopmental processes [1–5]. Clearly, such an orchestrated, precise developmental plan requires a multilayered genetic blueprint to maintain proper development. To establish the correct RNAs and proteins at each stage of neurogenesis, dynamic, rapid changes in gene expression are required. To accomplish this there needs to be precise, time-sensitive mechanisms in place in order to maintain proper brain development. Both epigenetic and transcriptional mechanisms have been shown to control neurogenesis. For example, DNA methylation and histone modification-mediated gene regulation are crucial for neural cell differentiation [6–9]. Furthermore, transcriptional regulation has also been shown to have a vital role in regulating neurogenesis [10–12]. Transcriptional control is exerted by transcription factors that regulate processes such as cell cycle exit, loss of progenitor properties and acquisition of neuronal features [13–17]. In addition to epigenetic and transcriptional control, epitranscriptomic regulation could potentially afford yet another layer of regulation.
The most abundant epitranscriptomic mark is the N6-methyladenosine (m6A) modification. This modification is dynamic and is deposited onto mRNA by ‘writer’ methyltransferases (METTL3/METTL14) [18–20] in complex with the METTL3 adaptor, WTAP [21], and other associated proteins, KIAA1429 [22] and RBM15/15B [23]. ‘Eraser’ enzymes (FTO, ALKBH5) remove the modification [24,25], and its function is mediated by proteins that specifically recognize it, ‘reader’ proteins (e.g., YTHDF 1, 2, 3) [26–28]. m6A has been implicated in mRNA stability, translation, splicing and miRNA processing [27–31]. Depending on the specific context, specific reader proteins will recognize the m6A mark and mediate its function (for a more detailed review see Shi H, Wei J, He C [32]; Figure 1). Furthermore, as m6A is a dynamic modification, mRNAs can be rapidly marked and unmarked, allowing for tight, fast regulation of mRNAs. Altogether, it is conceivable that m6A could exert regulatory control over a biological process. This review focuses on the role of m6A in neurodevelopmental pathways (see Table 1 for a summary) and neurodegenerative disease (see Table 2 for a summary).
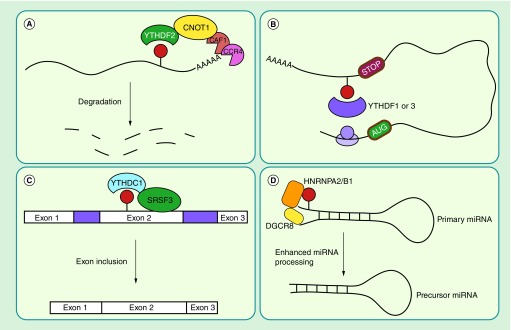
Figure 1. . N6-methyladenosine affects many aspects of RNA metabolism.
(A) mRNA stability – YTHDF2 specifically reads m6A marked transcripts localizing the mRNA to P-bodies for degradation. (B) Translation – YTHDF1 and 3 specifically recognize m6A marked transcripts, resulting in enhanced translation through a closed-loop model. (C) Splicing – YTHDC1 is a m6A reader which directs the splicing factor SRSF3 to its target, resulting in exon inclusion. (D) miRNA processing – HNRNPA2/B1, in complex with DGCR8, recognizes primary miRNAs modified by m6A delivering DGCR8 to its target for enhanced miRNA processing.
m6A: N6-methyladenosine.
Table 1. . Role of N6-methyladenosine in neurodevelopment.
| Developmental pathway | Related enzymes and proteins | Role of m6A | Key publications |
|---|---|---|---|
| Cerebellar development | METTL3: KO negatively affects Purkinje cell numbers, laminal structure, dendrite formation and the organization of glial cell fibers | m6A both positively and negatively modulates gene expression of genes important in proper cerebellar development | Ma et al. (2018) [36] Chang et al. (2017) [37] |
| Neurogenesis | METTL14: KO results in a reduction in cortical length and thickness FMRP: KO results extended neuronal progenitor cell cycle FTO: KO leads to decreased brain size and body weight YTHDF2: KO has a strong negative impact on NSPC self-renewal and neuron generation in embryonic neocortex |
m6A marks transcripts for degradation to ensure proper cortical neurogenesis m6A is involved in transcriptional pre-patterning for cortical neurogenesis m6A could regulate the BDNF pathway, and this may be linked to postnatal neurogenesis |
Yoon et al. (2017) [35] Wang et al. (2018) [43] Edens et al. (2019) [44] Zhang et al. (2018) [45] Li, Weng et al. (2017) [46] Li, Zhao et al. (2018) [47] |
| Synaptogenesis | YTHDF1 and 3: KO results in altered spine morphology, dampened excitatory synaptic transmission, and altered cell-surface protein content | m6A marked mRNAs known to function in synaptic plasticity | Merkurjev et al. (2018) [48] Koranda et al. (2018) [49] |
m6A: N6-methyladenosine; NSPC: Neural stem/progenitor cell.
Table 2. . Role of N6-methyladenosine and components of N6-methyladenosine pathways in neurodegenerative and neuropsychiatric diseases.
| Disease | Related enzyme and proteins | Role of m6A | Key publications |
|---|---|---|---|
| Deficit hyperactivity disorder in children, major depressive disorder and Alzheimer’s disease | FTO | No known role | Choudhry et al. (2013) [52] Velders et al. (2012) [53] Milaneschi et al. (2014) [54] Keller et al. (2011) [56] Reitz et al. (2012) [57] |
| Parkinson’s disease | FTO | m6A may affect dopaminergic signaling | Hess et al. (2013) [55] Chen et al. (2019) [62] |
| Autism spectrum disorder | YTHDC2 | No known role | Liu et al. (2016) [58] |
| Stress-related psychiatric disorders | FTO ? Conflicting data as to whether FTO has a positive or negative affect on anxiety and depression |
Altered levels of m6A was observed in depressed patients after glucocorticoid stimulation | Engel et al. (2018) [59] Spychala, Ruther (2019) [60] Sun et al. (2019) [61] |
m6A: N6-methyladenosine.
m6A in brain development
Both gene expression and environment are critical for normal brain development, and clearly, perturbation in either can negatively affect neural outcomes. Brain development is correctly maintained through context-specific dynamic and adaptive processes that are perfectly regulated for proper neural structure and functions [33]. With the dynamic nature of m6A and its multifaceted functionality, m6A adds another layer of regulation during brain development. m6A has been shown to be highly pervasive in the brain, more so in humans compared with mice, and, there is a high level of conservation between the two, suggesting a pivotal role for m6A in brain development [34,35].
Two recent studies have focused on understanding the postnatal m6A landscape in the mouse brain. Ma C, Chang M, Lv H et al. [36] profiled m6A in the mouse cerebellum across four postnatal developmental stages ranging from 7 days to 60 days postbirth (P7, P14, P21 and P60). Chang M, Lv H, Zhang W et al. [37] have provided m6A maps in both the cerebellum and cortex of 2-month old mice. In the first study, they identified a similar number of methylated mRNAs across the four stages and similar methylation patterns across all four stages. Interestingly, they observed a significant number of m6A peaks in start codons, which increased slightly from P7 to P60. A considerable number of m6A peaks were also identified in the coding region and stop codon. To glean insight into m6A function, they identified differential m6A peaks in a pairwise fashion. They noted the presence of 12,452 emerging m6A peaks or ‘ON’ peaks (i.e., peaks present only at a later stage, when compared with an earlier stage), and 11,192 disappearing m6A peaks or ‘OFF’ peaks (i.e., peaks present only at an earlier stage, when compared with a later stage). The distribution of these m6A peaks varied by type: ‘ON’ switches were more likely to be deposited at start codons, whereas ‘OFF’ switches were more observed around stop codons. Importantly, the authors find that the majority of m6A peaks are present at all time points, however, they were also able to identify some temporal specific peaks. Chang M, Lv H, Zhang W et al. [37] also identified common and region specific m6A methylation. The common m6A peaks were typically located at the stop codons and 3′ UTR. However, cerebellum-specific peaks were distributed in the start codons and 3′ UTR, while cortex-specific peaks were mainly concentrated in the coding region. Ma C, Chang M, Lv H et al. [36] reported similar findings. They noted that the distribution of P7-specific m6A peaks were enriched at the stop codon, while P60-specific m6A peaks were enriched at the start codon. Gene ontology analyses revealed that biological processes such as cell cycle, cell division and DNA repair were strong for genes containing P7–P14 OFF switches. However, genes that contain ‘ON’ methylation switches are involved in signal transduction, cell adhesion, learning and synaptic plasticity. Furthermore, they show that the m6A methylation is able to positively and negatively affect gene expression. Thus, m6A is clearly important for cerebellar development by modulating gene expression of cell-fate determining genes. In the same study, localization studies of the five major players in the m6A pathway (METTL3, METTL14, WTAP, ALKBH5 and FTO) revealed that all five proteins were present in the external granule cell layer, Purkinje cell layer and the inner granule cell layer [36]. Knockdown of Mettl3 was shown to negatively affect Purkinje cell numbers, laminal structure, dendrite formation and the organization of glial cell fibers [36]. Interestingly, expression levels of the five proteins decreased when transitioning from P7 to P60, suggesting that m6A has a more crucial role early in cerebellar development. Chang M, Lv H, Zhang W et al. [37] also found that protein levels of the m6A machinery are higher in the cerebellum than in the cortex. Taken together, these findings suggest that m6A is a significant regulator of developmental processes in cerebellum and cortex.
m6A in neurogenesis
During development, neurogenesis results in the large diversity of neurons in the brain. During the process, neural stem cells (NSCs) differentiate at specific times and regions in the brain. In neurogenesis, stem cells have the capacity for self-renewal through cell division, or they can differentiate into specialized cell types, such as neural progenitor cells [38–40]. These progenitor cells themselves then can differentiate into specific types of neurons. Clearly, proper maintenance of neurogenesis is essential for normal neurodevelopment, and perturbation of the NSC self-renewal and neurogenesis at any time during development can directly lead to neurological and psychiatric disorders [41,42]. Two recent papers have demonstrated that m6A exerts some control over the neurogenesis process.
To study the effect of METTL14-mediated m6A on neurogenesis, two prominent studies employed the use of a conditional Mettl14 KO x Nestin-cre mouse model [35,43]. Yoon KJ, Ringeling FR, Vissers C et al. [35] showed that the conditional Mettl14 KO (cKO) in mice produced animals that were visibly smaller in size at P5 compared with littermate controls, and those cKO animals failed to survive past P25. Comparison of the Mettl14 cKO to the WT revealed differences in cortical structure and development. Wang Y, Li Y, Yue M et al. [43] reported that the cKO pups were viable, did not display any obvious morphologic phenotypes and appeared to have normal body weight at P0. However, all Mettl14-cKO mice were dead within the first neonatal week. Wang Y, Li Y, Yue M et al. [43] also observed a reduction in cortical length and thickness, and both studies found an enlargement of the ventricle. Altogether, these results demonstrate a critical function for METTL14 in neurogenesis. Furthermore, Yoon KJ, Ringeling FR, Vissers C et al. [35], observed METTL14-induced m6A depletion resulted in prolonged cell cycle of adult radial glial, whereas neuron production still occurred into postnatal stages: an observation clearly implicating m6A as a significant regulator of cortical neurogenesis [35]. Wang Y, Li Y, Yue M et al. [43] also reported that the Mettl14 cKO resulted in a loss of late neurons, further suggesting a role for METTL14 in cortical neurogenesis. These in vivo findings were further bolstered by performing m6A-seq on E13.5 mouse forebrains (where NSCs are prominent) [35]. These authors found that many transcripts that encode transcription factors, such as Pax6, Sox1, Sox2, Emx2 and Neurog2/Neurogenin 2, are marked with m6A. A gene ontology analysis of m6A marked transcripts yielded enrichment in cell cycle, stem cell and neuronal differentiation pathways. m6A in this context, marks the transcripts for degradation, maintaining proper cortical neurogenesis. Aberrant m6A during the process negatively affects temporal specification and cell cycle progression of neuronal progenitor cells [35]. Further, a recent study uncovered an additional layer of regulation for transcripts involved in neurogenesis. Edens BM, Vissers C, Su J et al. [44] showed that, during neural differentiation, fragile X mental retardation protein (FMRP) can specifically read the m6A modification facilitating the nuclear export of m6A-marked transcripts (Figure 2A). Zhang F, Kang Y, Wang M et al. [45] also showed that mRNA targets of FMRP are enriched for m6A marks. Similar to what was described in the Mettl14 cKO mice, Fmr1 KO mice exhibited an extended neuronal progenitor cell cycle, with neural progenitors still proliferating into postnatal stages. From these studies, it is clear that both FMRP and METTL14 are required for neural progenitor differentiation (Figure 2A & B). At last, transcriptome-wide m6A profiling also showed that m6A has a role in actively suppressing transcription of mRNAs present in adult radial glial, but whose protein product is only functional downstream in the lineage, suggesting that m6A is involved in transcriptional pre-patterning for cortical neurogenesis [35]. The advantage of such an epitranscriptomic post-transcriptional mechanism is that the dynamic m6A mark can suppress translation (e.g., by marking the transcript for decay), but then can be removed rapidly to allow translation to occur whenever the protein is required.
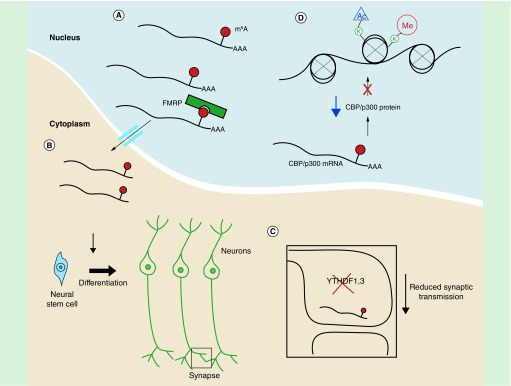
Figure 2. . N6-methyladenosine is critically important in the brain.
(A) FMRP facilitates nuclear export of m6A marked transcripts. (B) m6A promotes decay of these transcripts to maintain proper cortical neurogenesis. (C) YTHDF1 and 3 are required to recognize m6A at synapses to ensure proper synaptic function. (D) m6A results in the decay of the CBP/p300 acetyltransferase mRNA, which in turn results in decreased histone modifications, ultimately regulating gene expression.
m6A: N6-methyladenosine.
A study by Li Z, Weng H, Su R et al. [46] provided evidence that FTO-catalyzed m6A modification is important in neurogenesis, learning and memory. They found FTO in both adult NSCs and neurons. They further showed an increase in expression of FTO from postnatal day 1 to postnatal 8 weeks. To characterize the role of m6A regulated by FTO, the authors generated an m6A map at 2- and 6-week-old, and compared that to RNA-seq data from wild-type and Fto KO mice. They found 363 genes that had altered expression were also marked with m6A. These genes were associated with neuronal development, cell proliferation and migration pathways. Furthermore, they note that brain-derived neurotrophic factor (BDNF) is important for neurogenesis and is regulated by FTO. They found that many players in the BDNF pathway are m6A methylated at both 2 and 6 weeks, and have reduced expression in the Fto KO mice compared with WT. These results suggest that FTO-catalyzed m6A modification could regulate the BDNF pathway, and, in turn, this may affect postnatal neurogenesis. Another study by Li M, Zhao X, Wang W et al. [47] further implicated m6A in neurogenesis by looking at Ythdf2 knockdown mice. They showed that YTHDF2 is highly expressed during the early stage of neural development and Ythdf2−/− mouse embryos at E12.5 and E14.5 were alive but displayed reduced overall cortical thickness, as is seen in Mettl14 cKO mice [47]. In addition, they showed that loss of Ythdf2 has a strong negative impact on neural stem/progenitor cell self-renewal and neuron generation in embryonic neocortex. To gain insight into the molecular mechanism, the authors performed RNA-seq and compared gene expression in Ythdf2−/− and wild-type mice. They found that differentially expressed genes had functions related to axon regulation, synapse assembly and neuron differentiation. Interestingly, these genes, such as, Ddr2, Rnf135, Flrt2, Hlf, Nrp2, Nrxn3 and Ptprd have a marked increase in expression and m6A levels in Ythdf2−/− compared with WT. This study provides evidence that YTHDF2-mediated m6A plays an important role in neurogenesis during embryonic neural development. Critically, m6A has also been implicated as a major regulator of synaptic function. Merkurjev D, Hong WT, Iida K et al. [48] profiled m6A modifications of synaptosomal RNA and discovered 2921 methylated genes comprising the ‘synaptic m6A epitranscriptome’. This epitranscriptome is important in many pathways involved in neurodevelopment and neuropsychiatric diseases. Indeed, they reported that the m6A reader proteins YTHDF1, YTHDF2 and YTHDF3 are enriched in hippocampal dendrites, and the loss of YTHDF1 or YTHDF3 there resulted in altered spine morphology, dampened excitatory synaptic transmission and altered cell-surface protein content (Figure 2C) [48]. Furthermore, mRNAs that are specifically associated with synaptic function were both enriched and differentially methylated in synapses. Altogether, these observations provide evidence that m6A at synapses is required for proper synaptic function. Koranda JL, Dore L, Shi H et al. [49] also suggested a role for m6A in synaptic signaling. They profiled m6A in Mettl14-deleted striatum where they found a correlation between loss of m6A and downregulation of those mRNAs. Interestingly, gene ontology analysis showed that these downregulated mRNAs encode neuron and synapse-specific proteins. More specifically, they found m6A methylation in mRNAs known to function in synaptic plasticity, such as Homer1 and Cdk5r1.
Link between m6A, neurogenesis & histone modifications
By deleting Mettl14 in embryonic NSCs, Wang Y, Li Y, Yue M et al. [43] observed a significant reduction in proliferation, which in turn resulted in their premature differentiation. These findings suggest METTL14 regulates NSC self-renewal. An RNA-seq analysis revealed Mettl14 cKO NSCs displayed an altered gene expression profile compared with the control. Significantly upregulated genes are involved in NSC differentiation, whereas significantly downregulated genes are associated with cell proliferation; the observed phenotype of these cells concurred with the changes in gene expression. The authors only observed a mild correlation between m6A and mRNA levels, suggesting another mechanism was at play here. Interestingly, they observed significantly increased levels of histone modifications (e.g., histone H3 at lysine 27, trimethylation of histone H3 at lysine 4 and trimethylation of histone H3 at lysine 27) in Mettl14 KO NSCs compared with control. Furthermore, by using a chemical inhibitor to block H3K27me3 and H3K27ac, the authors showed that the observed abnormalities of Mettl14 KO NSCs could be rescued. At last, they detected m6A methylation in transcripts that encoded the H3K27 acetyltransferases CBP and p300 in the control sample but the m6A methylation was lost in Mettl14 KO NSCs. Methylation in these two transcripts was directly correlated with mRNA abundance. These findings implicate METTL14 (and m6A) as a specific regulator of histone modifiers, which in turn regulates gene expression (Figure 2D). Chen J, Zhang YC, Huang C et al. [50] also reported similar findings: they showed that m6A is present on the transcript of the histone methyltransferase Ezh2. Loss of Mettl3 resulted in a reduction of EZH2 protein levels and a concomitant decrease in H3K27me3. They also characterized the effect of METTL3 on neurogenesis. Knockdown of Mettl3 resulted in reduced m6A levels in adult NSCs, manifesting itself in the reduced proliferation of adult NSCs, and, furthermore, directed their differentiation toward the glial lineage. Also, the maturation of newborn neurons was affected upon knockdown of Mettl3. Chen J, Zhang YC, Huang C et al. [50] showed that overexpression of Ezh2 could rescue the defects of neurogenesis and neuronal development that were observed following the loss of Mettl3.
m6A in neurodegenerative & neuropsychiatric disease
Currently, there is little known regarding the specific role, if any, of m6A in neurodegenerative and neuropsychiatric diseases. However, there is strong evidence to suggest a fundamental role for m6A in these diseases. The current knowledge of the role of m6A players and m6A in neurodegenerative and neuropsychiatric disease is summarized in Table 2. As discussed previously, m6A is present in synapses, and aberrant translation at synapses has been associated with autism, fragile X syndrome and other intellectual disorders, suggesting there may be a role for m6A in these diseases. In addition, m6A machinery, but not m6A itself, has been implicated in some neurodegenerative diseases. For example, single nucleotide polymorphisms in FTO have been implicated in many neuropsychiatric diseases [51]. The variants have been linked to attention deficit hyperactivity disorder in children [52,53], major depressive disorder [54] and Alzheimer’s disease [55–57]. Also, mutations in YTHDC2 have been implicated as a risk factor for autism spectrum disorder [58]. Engel M, Eggert C, Kaplick PM et al. [59] found that depressed patients have altered levels of m6A and m6Am (N6, 2’-O-dimethyladenosine) after glucocorticoid stimulation, suggesting that these modifications may also have a role in stress-related psychiatric disorders. The same study also reported no change in anxiety levels were observed in Mettl3 or Fto conditional knockout mice. However, Spychala A, Ruther U [60] did see an increase in anxiety following Fto knockout; whereas, Sun L, Ma L, Zhang H et al. [61] reported that FTO deficiency reduced anxiety and depression-like behaviors. FTO, and indeed m6A, may also be associated with Parkinson’s disease (PD). Hess ME, Hess S, Meyer KD et al. [55] showed that dopaminergic signaling is negatively affected upon the inactivation of FTO. More recently, Chen X, Yu C, Guo M et al. [62] looked more closely into the role of m6A in PD. They modeled the disease in rats and PC12 cells by employing 6-OHDA to selectively destroy dopaminergic neurons. Upon this treatment they found that the m6A modification was reduced in the PC12 cells and in the striatum of the PD rat but not the cortex, hippocampus or midbrain. The authors hypothesized that loss of m6A in the striatum may result from the increased levels of FTO in the midbrain being transmitted to the striatum via the axons of dopaminergic neurons. There are many potential links between m6A, neurodegenerative and neuropsychiatric disease, which may result in attractive targets for therapeutic intervention strategies in patients. For example, the key regulators of the m6A modification (especially the writers and erasers) may act as potential therapeutic targets. Clearly, though, more work is needed to uncover the exact mechanisms by which m6A may affect these diseases, and how this information can be utilized for therapeutics.
Conclusion & future perspective
m6A mRNA methylation is clearly a critical regulator of gene expression in the developing mammalian brain. Further work is needed to fully elucidate the role of m6A in brain development and disease. For example, what is the biological significance of the location of m6A within the mRNA in the context of neural development and neurological disease? How does the position of the m6A affect RNA binding protein binding and how does this contribute to development and disease? This is pertinent because anomalies in RNA processing can result in neurological disease. It is important to understand how m6A affects the RNA binding protein landscape. Also, what cues are required to control the level of m6A, and how does the mark specifically mediate its function in this context? Mechanistically, more work is required to fully appreciate the nuances of regulation by m6A. Emerging long read sequencing technologies such as Nanopore and PacBio will provide us with insights into whether m6A has a preference for a specific mRNA isoform or not. Direct RNA sequencing may also shed light on the different epitranscriptomic marks present on specific mRNAs at a given time. This will indicate if modifications work in concert, starting to provide us with a more comprehensive view of how the epitranscriptome functions. Also, with the advent of single-cell sequencing, studies have started to emerge outlining the view of the transcriptome in each cell type in the brain; this will no doubt be extended to the epitranscriptome as well. This will broaden our understanding of differences in m6A in different neural cell types and the specific impact of m6A on the specific development of particular neural types. Certainly, m6A has an important role in maintaining and modulating multiple processes and pathways in the human brain, the details of which still need to be elucidated. Indeed, a strong emphasis should be placed for determining the role of m6A in the brain.
Executive summary.
N6-methyladenosine (m6A) is known to affect many aspects of RNA metabolism including mRNA stability, translation, splicing and miRNA processing.
m6A is a dynamic modification and is critical in regulating brain development by both positively and negatively modulating gene expression in genes important for proper development.
During brain development, distribution of m6A along genes is dependent on both spatial and temporal cues, with different m6A localization along the gene having a different functional outcome.
m6A marks transcripts for degradation to maintain proper cortical neurogenesis. Aberrant m6A results in problems with temporal affects temporal specification and cell cycle progression of neuronal progenitor cells.
The m6A demethylase, FTO, is important in neurogenesis, learning and memory.
YTHDF2-mediated m6A plays an important role in neurogenesis during embryonic neural development.
m6A methylation is present in mRNAs known to function in synaptic plasticity, such as Homer1 and Cdk5r1.
Aberrant translation at synapses has been associated with autism, fragile X syndrome and other intellectual disorders, thus it is possible that m6A may be play a role in those diseases.
Footnotes
Authors’ contributions
AM Shafik wrote the manuscript. EG Allen and P Jin commented and edited the manuscript.
Financial & competing interests disclosure
This research was supported in part by the NIH (NS079625, NS051630, NS111602, NS097206, HG008935, and MH116441 to P Jin). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
Please disclose any relevant information regarding the ethical conduct of your research. For studies involving data relating to human or animal experimental investigations, appropriate institutional review board approval is required and should be described within the article and in this disclosure, as per the ICMJE recommendations on Protection of Research Participants, and the further recommendations of the International Association of Veterinary Editors’ Consensus Author Guidelines for Animal Use. For those investigators who do not have formal ethics review committees, the principles outlined in the Declaration of Helsinki should be followed. In addition, for investigations involving human subjects, authors should obtain informed consent from the participants involved and include an explanation of how this was obtained in the manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Bae BI, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev. Cell 32(4), 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leone DP, Srinivasan K, Chen B, Alcamo E, Mcconnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 18(1), 28–35 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8(6), 427–437 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: making cortical maps. Trends Neurosci. 32(5), 291–301 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell. Dev. Biol. 30, 465–502 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 61(5 Pt 2), 58R–63R (2007). [DOI] [PubMed] [Google Scholar]
- 7.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11(6), 377–388 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Sun J, Ming GL, Song H. Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur. J. Neurosci. 33(6), 1087–1093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17(9), 537–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martynoga B, Drechsel D, Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb. Perspect. Biol. 4(10) a008359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JA, Ding SL, Sunkin SM. et al. Transcriptional landscape of the prenatal human brain. Nature 508(7495), 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85(1), 27–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan CM, Nyman U, Skotte J. et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell 26(3), 223–236 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Taylor CA, Barnes KM. et al. A Myt1 family transcription factor defines neuronal fate by repressing non-neuronal genes. Elife 8, e46703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu L, Berti L, Masserdotti G. et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 32(9), 3067–3080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rraklli V, Sodersten E, Nyman U, Hagey DW, Holmberg J. Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem Cell Res. 16(1), 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Sansom SN, Griffiths DS, Faedo A. et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 5(6), e1000511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3(11), 1233–1247 (1997). [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Yue Y, Han D. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10(2), 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell. Biol. 16(2), 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 8(6), e1002732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horiuchi K, Kawamura T, Iwanari H. et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288(46), 33292–33302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patil DP, Chen CK, Pickering BF. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537(7620), 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia G, Fu Y, Zhao X. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7(12), 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng GQ, Dahl JA, Niu YM. et al. ALKBH5 Is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49(1), 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi H, Wang X, Lu Z. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27(3), 315–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Lu Z, Gomez A. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481), 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhao BS, Roundtree IA. et al. N-6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161(6), 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature 519(7544), 482–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li A, Chen YS, Ping XL. et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27(3), 444–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18(1), 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74(4), 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol. Rev. 20(4), 327–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominissini D, Moshitch-Moshkovitz S, Schwartz S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397), 201–206 (2012). [DOI] [PubMed] [Google Scholar]; •• The first genome-wide profiling of N6-methyladenosine (m6A) methylation in mammalian system.
- 35.Yoon KJ, Ringeling FR, Vissers C. et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171(4), 877.e817–889.e817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• m6A methylation is critically important in regulating cortical neurogenesis in both humans and mice.
- 36.Ma C, Chang M, Lv H. et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19(1), 68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Widespread and dynamic m6A methylation is required for proper cerebellar development.
- 37.Chang M, Lv H, Zhang W. et al. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7(9), 170166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gage FH, Ray J, Fisher LJ. Isolation, characterization, and use of stem cells from the CNS. Annu. Rev. Neurosci. 18, 159–192 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Kilpatrick TJ, Bartlett PF. Cloning and growth of multipotential neural precursors: requirements for proliferation and differentiation. Neuron 10(2), 255–265 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255(5052), 1707–1710 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Christian KM, Song H, Ming GL. Functions and dysfunctions of adult hippocampal neurogenesis. Annu. Rev. Neurosci. 37, 243–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang E, Wen Z, Song H, Christian KM, Ming GL. Adult neurogenesis and psychiatric disorders. Cold Spring Harb. Perspect. Biol. 8(9), a019026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Li Y, Yue M. et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21(2), 195–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• m6A regulates histone modifications affecting gene expression and impacting neural stem cell renewal.
- 44.Edens BM, Vissers C, Su J. et al. FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 28(4), 845.e845–854 e845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Kang Y, Wang M. et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27(22), 3936–3950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Weng H, Su R. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31(1), 127–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, Zhao X, Wang W. et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19(1), 69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • The m6A reader protein Ythdf2 regulates degrades m6A-marked genes involved in neural development-related pathways.
- 48.Merkurjev D, Hong WT, Iida K. et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21(7), 1004–1014 (2018). [DOI] [PubMed] [Google Scholar]; •• This is the first map of m6A in synaptosomal RNA (synaptic m6A epitranscriptome), which has an impact on neurodevelopmental pathways and neuropsychiatric diseases.
- 49.Koranda JL, Dore L, Shi H. et al. Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99(2), 283.e285–292.e285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, Zhang YC, Huang C. et al. m(6)A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics 17(2), 154–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, Yang Y, Sun BF, Zhao YL, Yang YG. FTO and obesity: mechanisms of association. Curr. Diab. Rep. 14(5), 486 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Choudhry Z, Sengupta SM, Grizenko N. et al. Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring) 21(12), E738–E744 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Velders FP, De Wit JE, Jansen PW. et al. FTO at rs9939609, food responsiveness, emotional control and symptoms of ADHD in preschool children. PLoS ONE 7(11), e49131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milaneschi Y, Lamers F, Mbarek H, Hottenga JJ, Boomsma DI, Penninx BW. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry 19(9), 960–962 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Hess ME, Hess S, Meyer KD. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16(8), 1042–1048 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Keller L, Xu W, Wang HX, Winblad B, Fratiglioni L, Graff C. The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J. Alzheimers Dis. 23(3), 461–469 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Reitz C, Tosto G, Mayeux R, Luchsinger JA, Group N-LNFS, Alzheimer’s Disease Neuroimaging I. Genetic variants in the fat and obesity associated (FTO) gene and risk of Alzheimer’s disease. PLoS ONE 7(12), e50354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Shimada T, Otowa T. et al. Genome-wide association study of autism spectrum disorder in the East Asian populations. Autism Res. 9(3), 340–349 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Engel M, Eggert C, Kaplick PM. et al. The role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99(2), 389.e389–403.e389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spychala A, Ruther U. FTO affects hippocampal function by regulation of BDNF processing. PLoS ONE 14(2), e0211937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun L, Ma L, Zhang H. et al. Fto deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 9(3), 721–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Yu C, Guo M. et al. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10(5), 2355–2363 (2019). [DOI] [PubMed] [Google Scholar]