Abstract
Objectives:
The aim of the present study was to evaluate the clinical relevance of mutations in tumor suppressor genes using whole-exome sequencing data from centenarians and young healthy individuals.
Methods:
Two pools, one of centenarians and one of young individuals, were constructed and whole-exome sequencing was performed. We examined the whole-exome sequencing data of Bulgarian individuals for carriership of tumor suppressor gene variants.
Results:
Of all variants annotated in both pools, 5080 (0.06%) are variants in tumor suppressor genes but only 46 show significant difference in allele frequencies between the two studied groups. Four variants (0.004%) are pathogenic/risk factors according to single nucleotide polymorphism database: rs1566734 in PTPRJ, rs861539 in XRCC3, rs203462 in AKAP10, and rs486907 in RNASEL.
Discussion:
Based on their high minor allele frequencies and presence in the centenarian group, we could reclassify them from pathogenic/risk factors to benign. Our study shows that centenarian exomes can be used for re-evaluating the clinically uncertain variants.
Keywords: tumor suppressor genes, whole-exome sequencing, centenarians, Bulgarian, reevaluation of clinical significance
Introduction
Tumor suppressor genes (TSGs) are part of the cell cycle regulation mechanism. Mutations in TSGs have long been associated with malignant disorders and often lead to the onset of neoplasms in different organs and systems. Germline genetic disturbances in TSGs could be transmitted and predispose their carriers to inherited forms of cancer. Additional somatic mutations might trigger cell transformation and cancer development.
The introduction of next generation sequencing technology has made it possible many new variants in TSGs to be discovered and their role in different malignant disorders evaluated.
Online publicly available databases have compiled lists of TSGs with germline and somatic mutations associated with oncogenesis. Interpreting the clinical significance of variants in TSGs is however complicated, and databases with information about the functional significance of variants are often incomplete and based on contradictory evidence. For most variants no meta-analyses are performed, which hinders risk estimation and clarification of their role as driver mutations. Yet, such information is very important in medical genetic counseling.
Centenarians are individuals who presumably carry protective variants against risk factors in the environment and are most likely to carry fully functional TSGs. Also, the frequency of risk alleles in TSGs in centenarians should be lower compared to the frequency in the general population. Centenarian exomes/genomes could thus be considered “golden standard” for healthy longevity, making them appropriate for studying the functional significance of variants in TSGs.
The aim of this study is to determine the carriership of risk/pathogenic TSG alleles in healthy Bulgarian individuals and to evaluate their clinical relevance using centenarian exomes. Whole-exome sequencing (WES) data were obtained from pools of individual DNA (pool-seq), an approach which has repeatedly been shown to provide reliable allele frequency estimates.1
Methods
Ethics Statement
The study was approved by the Ethics committee of the Medical University No. 1030/08.03.2017. It is conducted in accordance with national and international legislation for conducting research with participation of human subjects. Each participant in the study received information about the aims of the project and signed written informed consent. Interviews conducted with the individuals gathered information about their health status and medical history for major age-related diseases.
Sample Collection, DNA Isolation, and Pool Preparation
Blood or buccal swap samples were collected from 93 healthy Bulgarian individuals. The individuals have been divided into two groups according to their age: 32 centenarians aged 100 to 106 years and 61 healthy individuals aged 18 to 30 years. Whole genome DNA has been extracted using phenol-chloroform extraction protocol, and equimolar amounts of DNA from each individual were used to construct the pools.
Whole-Exome Sequencing
The DNA pools were whole-exome sequenced (BGI Genomic Services, China) at ×250 coverage which is required for pool-seq sequencing to ensure that alleles with low frequency are also detected. The obtained .vcf files were annotated using the web-based service wANNOVAR.2 Following the “best practice” recommendations for pool-seq data, we performed robust filtering on variant calling: number of individuals per pool >30, genotype quality >99, mapping quality >60, number of reads per minor allele frequency >2, total depth of coverage—above 30.1,2 The total number of variants annotated in both pools after applying these filters was 89 810 (72 791 in both pools, 8253 in centenarian pool only, and 8766 in control pool only).
Databases of TSG and Variants
The Tumor Suppressor Gene Database (TSGene 2.0) is an online comprehensive resource for data based on pan-cancer analysis of TSGs and contains a list of 1217 TSGs (1018 protein-coding and 199 non-coding genes).3
We examined the WES data of Bulgarian individuals for the presence and frequency of variants in TSGs from this database.
Additionally, we compiled a list of 30 161 variants in 248 genes from the following publicly available TSG databases:
Results
TSG Database
The TSG database contains a list of 1217 TSGs. The obtained WES data from Bulgarian samples was inspected for the presence and frequency of variants from the genes in this database.
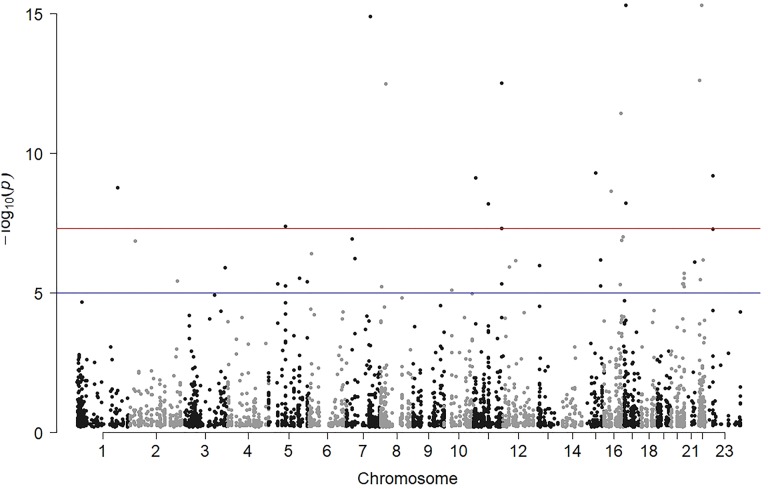
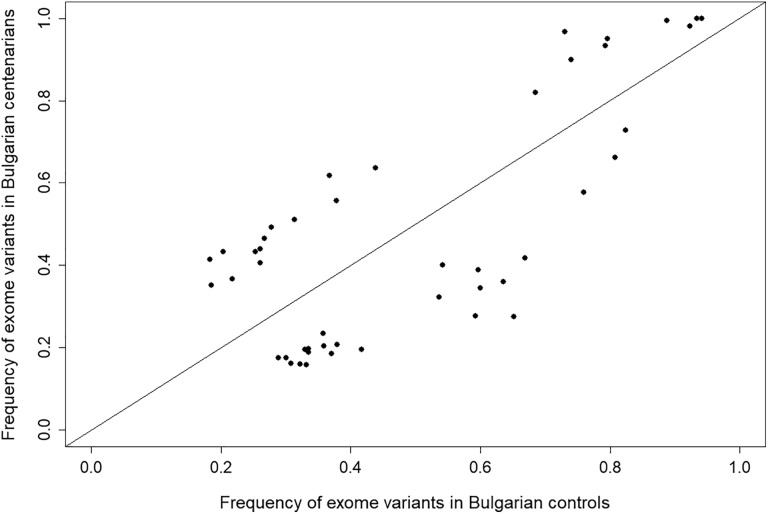
Altogether 5042 variants in 851 TSGs—4092 variants in both pools, 424 variants in centenarians only, and 526 in controls only—were found in the Bulgarian WES data. For the 4092 variants found in both pools, we used Fisher exact test to determine the significance of allele frequency differences between the two pools (Figure 1).The frequencies of variants with significant difference (false discovery rate > 1.0 × 10−5) in allele frequency, altogether 46 variants, are plotted on Figure 2.
Figure 1.
Manhattan plot of varants in TSGs called in both centenarian and control pools. The horizontal lines correspond to P = 1.0 × 10−5 and 5.0 × 10−7. TSGs indicate tumor suppressor genes.
Figure 2.
Forty-six variants in TSGs showing significant difference (FDR > 1.0 × 10−5) in allele frequency between centenarians and controls. FDR indicates false discovery rate; TSG, tumor suppressor gene.
The variants below the diagonal identity line (n = 24) have significantly higher allele frequency in the control pool. Three of these variants are benign (rs558114 in MUS81, rs741810 in FUS, and rs1057090 in MCPH1), but for the remaining 21 variants no information is reported in dbSNP database. It can be speculated that these variants can potentially have negative impact on longevity, and further analyses on them are needed.
Analysis of all variants called in both pools of the Bulgarian WES data, regardless of their allele frequency differences, revealed only 2 pathogenic/risk alleles, rs1566734 in PTPRJ gene and rs861539 in XRCC3 (Table 1). There were no variants designated as pathogenic or risk alleles among those called in the centenarian pool only (containing 424 variants) or in the control pool only (containing 526 variants).
Table 1.
Risk Variants in TSGs in Bulgarian Healthy Individuals.
| Pool | Ref | Alt | Func | Gene | dbSNP | dbSNP Significance | MAFCent | MAFCont | P Value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| Risk variants in TSGs from TSG database | ||||||||||
| Both | A | C | Exonic | PTPRJ | rs1566734 | Pathogenic | 0.198 | 0.246 | .246 | 0.246 |
| Both | C | T | Exonic | XRCC3 | rs861539 | Risk factor | 0.474 | 0.375 | .089 | 0.201 |
| Risk variants from UniProt and DisGeNet | ||||||||||
| Both | T | C | Exonic | AKAP10 | rs203462 | Risk factor | 0.375 | 0.350 | .491 | 0.466 |
| Both | C | T | Exonic | RNASEL | rs486907 | Risk factor | 0.328 | 0.349 | .420 | 0.442 |
Abbreviations: AKAP10, A-kinase anchoring protein 10; Alt, alternative allele; cent, centenarians; cont, controls; dbSNP, single nucleotide polymorphism database; FDR, false discovery rate adjusted P value; Func, functional consequence; MAF, minor allele frequency; PTPRJ, protein tyrosine phosphatase receptor type J; Ref, referent allele; RNASEL, ribonuclease L; TSGs, tumor suppressor genes; XRCC3, X-ray repair cross complementing 3.
UniProt and DisGeNet Databases
Of the 30 161 variants in TSGs (assembled from UniProt and DisGeNet), 38 were found in the pooled exome sequencing data of Bulgarian individuals.
Two of 38 variants were found as risk factors in the dbSNP (rs203462 in AKAP10 and rs486907 in RNASEL; Table 1).
Among TSG variants found in the centenarian or young individual pools only, none were pathogenic or risk factors.
Discussion
Pathogenic variants in TSGs can be inherited and entail risk for oncogenesis. Different pathogenic alleles predispose to tumor development to a different degree. Assessment of the risk in the healthy carriers of pathogenic alleles is important for medical genetic counseling and in setting up preventive measures for reducing cancer risk.
For this study, we used exome data from Bulgarian centenarians as a tool for evaluation of the clinical effect of germline pathogenic tumor suppressor mutations in healthy carriers.
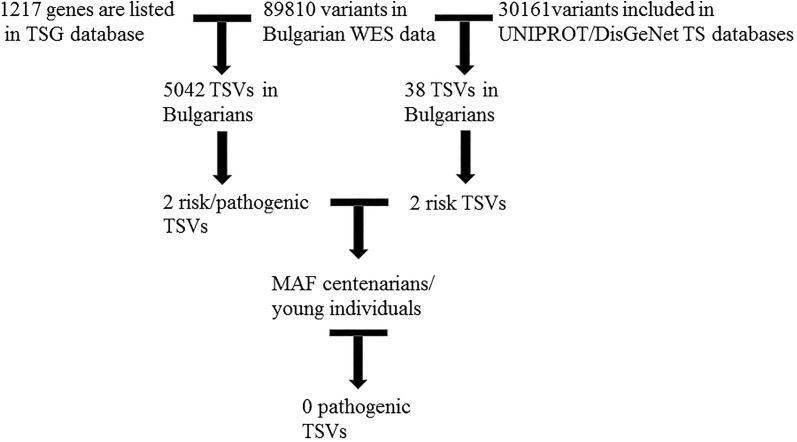
Data for 89 810 variants in 17 133 genes was obtained by performing WES on two DNA pools, one composed of Bulgarian centenarians and one of coethnic young and healthy individuals. The 1217 genes listed in TSGene 2.0 database were screened for variants in our WES data and 5042 were found (see Figure 3, left side of the workflow chart). Two of these are nominated as having clinical relevance in dbSNP database—pathogenic/risk factor (cf Table 1).
Figure 3.
Flowchart for pathogenic (TSVs) in Bulgarian WES data. TSVs indicate tumor suppressor variants; WES, whole-exome sequencing.
From the 30 161 tumor suppressor variants (TSVs) from UniProt/DisGeNet databases, 38 were found in Bulgarian WES data, and 2 of these are proposed by dbSNP as risk factors (Figure 3, right side of the workflow chart).
All in all, from 89 810 variants in Bulgarian healthy individuals—young individuals and centenarians, 5080 are in TSGs (0.06%), and only 4 of these are pathogenic/risk TS variants (0.004%).
None of these 4 variants show significant difference in allele frequencies between the two pools, raising the question about the risk entailed by these variants.
The rs1566734 (Gln276Pro) polymorphism in PTPRJ (protein tyrosine phosphatase receptor type J) is a missense single nucleotide polymorphism, which is classified as pathogenic TSV by DisGeNet and as associated with colon cancer (variant disease-associated ([VDA] score = 0.700), colorectal cancer (VDA score = 0.020), papillary thyroid carcinoma (VDA score = 0.010). Our data show no significant difference in allele frequency of this variant between Bulgarian centenarians and young individuals, and the frequency is relatively high (0.198/0.246, respectively). As this variant is not associated with tumor development in centenarians and could thus be reclassified from pathogenic to benign in Bulgarian population.
This polymorphism in PTPRJ has previously been genotyped in Japanese population and was found that in combination with another variant in the same gene (rs1503185, Arg326Gln) increases the risk for developing lung, head and neck, colorectal, and esophageal cancers.7-10 On the other hand, Toland et al 11 had found that this variant was not associated with increased colorectal cancer risk. Iuliano et al 7 also observed a nonsignificant increased frequency of homozygotes for Gln276Pro polymorphism in papillary thyroid carcinoma cases in 2 distinct Caucasian populations. A meta-analysis has also found this variant not to be associated with increased risk of colorectal cancer.12
The findings for nonassociation of rs1566734 polymorphism are in line with our proposal this variant to be reclassified from pathogenic to benign.
The rs861539 (Thr241Met) polymorphism in the DNA repair XRCC3 (X-ray repair cross-complementing 3) gene is classified as risk factor in DisGeNet database based on repeated findings of association with increased risk of melanoma (VDA score = 0.800), breast carcinoma (VDA score = 0.100), lung cancer (VDA score = 0.100). Our results show that Bulgarian healthy individuals have higher, albeit nonsignificantly different, frequency in the centenarian group compared to the young individuals. This variant thus seems to be benign in Bulgarian population.
A study on Taiwanese population has previously found that carriers of this variant, in combination with certain other variants in XRCC group genes, had up to ×10 increased risk of developing oral cancer,13 and that TT genotype was more prevalent in patients with breast cancer.14 Also, a Belgian study found that the T allele of this variant is not a risk allele for breast cancer.15 A meta-analysis16 concludes that this variant is the risk factor for melanoma, but the association is not significant in Caucasians. There was no significant association found between this polymorphism and lung cancer risk17 in a population from northern Spain and prostate cancer in north Indian population.18
The high frequency of this variant in centenarians, who do not have cancer, indicates that this variant is not a risk factor in Bulgarians, adds to the evidence from other studies for no association of this variant with oncogenesis.
The rs203462 in AKAP10 (A-kinase anchoring protein 10) is a missense variant designated as risk factor for breast cancer (VDA score = 0.010) by DisGeNet. Our WES data show that this variant is carried with high frequency in Bulgarian population, and no difference in allele frequencies between the centenarian and young individuals, indicating that the clinical significance of this variant in Bulgarian population is benign.
A study19 from 2007 has shown that this is functional genetic variant, located in the kinase-binding domain of the A-kinase anchoring protein, and is associated with familial breast cancer. However, in line with our results, a study on a Polish population finds no significant differences in genotype or allele distribution of this variant between nonagenarians and newborns.20
The rs486907 in RNASEL (ribonuclease L) is a missense variant, included in DisGeNet as risk factor for prostate cancer (VDA score = 0.100). Our WES data show that this variant is with high and similar frequency in centenarian/young groups, indicating this variant not to be cancer risk factor.
Alvarez-Cubero et al 21 suggest the role of this variant as prognostic marker and predictor of aggressiveness and progression of prostate cancer.21 Analysis of Ashkenazi Jewish descent individuals shows its contribution in early onset and familial forms of prostate cancer.22
Other researchers Shea et al 23 and Robbins et al 24 find no evidence of association between this R462Q polymorphism and prostate cancer risk in case/control analysis of Afro-Caribbeans and Afro-Americans. These results support our proposal for reclassification of this polymorphism from risk factor to benign.
Conclusion
Literature evidence for pathogenic/risk variants is often contradictory and based on single gene sequencing in case–control studies. Single nucleotide polymorphic variants in such studies that have high population frequencies might rather indicate the presence of unknown nearby risk alleles. The limitations of such study design are overcome by the introduction of WGS and WES technologies. Using WES data from Bulgarian centenarians and young individuals, 4 variants could be reclassified from pathogenic/risk factors to benign based on their high minor allele population frequencies and presence in centenarians.
Abbreviations
- dbSNP
single nucleotide polymorphism database
- TSG
tumor suppressor gene
- TSV
tumor suppressor variant
- VDA score
variant disease-associated score
- WES
whole-exome sequencing
Footnotes
Authors’ Note: Lubomir Balabanski, Dimitar Serbezov, Dragomira Nikolova contributed equally to the present study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Bulgarian centenarians project was funded by the National Science Fund of Bulgaria, contract number DN 03/7/18.12.2016, and Bulgarian Ministry of Education and Science under the National Program for Research “Young Scientists and Postdoctoral Students”.
ORCID iD: Dragomira Nikolova  https://orcid.org/0000-0002-8929-8522
https://orcid.org/0000-0002-8929-8522
References
- 1. Schlotterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat Rev Genet. 2014;15(11):749–763. doi:10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- 2. Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–1566. doi:10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2015;44(D1):D1023–D1031. doi:10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Consortium TU. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47(D1):D506–D515. doi:10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi:10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016;45(D1):D833–D839. doi:10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iuliano R, Palmieri D, He H, et al. Role of PTPRJ genotype in papillary thyroid carcinoma risk. Endocr Relat Cancer. 2010;17(4):1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mita Y, Yasuda Y, Sakai A, et al. Missense polymorphisms of PTPRJ and PTPN13 genes affect susceptibility to a variety of human cancers. J Cancer Res Clin Oncol. 2010;136(2):249–259. doi:10.1007/s00432-009-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shangkuan WC, Lin HC, Chang YT, et al. Risk analysis of colorectal cancer incidence by gene expression analysis. Peer J. 2017;5:e3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei W, Jiang M, Luo L, Li Z, Wang P, Dong WQ. Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. Genet Mol Res. 2013;12(4):6268–6274. doi:10.4238/2013. [DOI] [PubMed] [Google Scholar]
- 11. Toland AE, Rozek LS, Presswala S, Rennert G, Gruber SB. Haplotypes and Colorectal Cancer Risk. Cancer Epidemiol Biomark Prev. 2008;17(10):2782 doi:10.1158/1055-9965.EPI-08-0513. [DOI] [PubMed] [Google Scholar]
- 12. Laczmanska I, Sasiadek MM. Meta-analysis of association between Arg326Gln (rs1503185) and Gln276Pro (rs1566734) polymorphisms of PTPRJ gene and cancer risk. J Appl Genet. 2019;60(1):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yen CY, Liu SY, Chen CH, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37(5):271–277. doi:10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 14. Su CH, Chang WS, Hu PS, et al. Contribution of DNA Double-strand Break Repair Gene XRCC3 Genotypes to Triple-negative Breast Cancer Risk. Cancer Genom Proteom. 2015;12(6):359–367. [PubMed] [Google Scholar]
- 15. Vral A, Willems P, Claes K, Poppe B, Perletti G, Thierens H. Combined effect of polymorphisms in Rad51 and Xrcc3 on breast cancer risk and chromosomal radiosensitivity. Mol Med Rep. 2011;4(5):901–912. doi:10.3892/mmr.2011.523. [DOI] [PubMed] [Google Scholar]
- 16. Fan J, Fan Y, Kang X, Zhao L. XRCC3 T241 M polymorphism and melanoma skin cancer risk: A meta-analysis. Oncol Lett. 2015;9(5):2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162 doi:10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mandal RK, Kapoor R, Mittal RD. Polymorphic variants of DNA repair gene XRCC3 and XRCC7 and risk of prostate cancer: a study from North Indian population. DNA Cell Biol. 2010;29(11):669–674. [DOI] [PubMed] [Google Scholar]
- 19. Wirtenberger M, Schmutzhard J, Hemminki K, et al. The functional genetic variant Ile 646 Val located in the kinase binding domain of the A-kinase anchoring protein 10 is associated with familial breast cancer. Carcinogenesis. 2007;28(2);423–426. [DOI] [PubMed] [Google Scholar]
- 20. Loniewska B, Adler G, Gumprecht J, et al. 1936A-->G (I646V) polymorphism in the AKAP10 gene encoding A-kinase-anchoring protein 10 in very long-lived poles is similar to that in newborns. Exp Aging Res. 2012;38(5):584–592. doi:10.1080/0361073x.2012.726177. [DOI] [PubMed] [Google Scholar]
- 21. Alvarez-Cubero MJ, Martinez-Gonzalez LJ, Saiz M, et al. Prognostic role of genetic biomarkers in clinical progression of prostate cancer. Exp Mol Med. 2015;47(8):e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Agalliu I, Leanza SM, Smith L, et al. Contribution of HPC1 (RNASEL) and HPCX variants to prostate cancer in a founder population. Prostate. 2010;70(15):1716–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shea PR, Ishwad CS, Bunker CH, Patrick AL, Kuller LH, Ferrell RE. RNASEL and RNASEL-inhibitor variation and prostate cancer risk in Afro-Caribbeans. Prostate. 2008;68(4):354–359. [DOI] [PubMed] [Google Scholar]
- 24. Robbins CM, Hernandez W, Ahaghotu C, et al. Association of HPC2/ELAC2 and RNASEL non-synonymous variants with prostate cancer risk in African American familial and sporadic cases. Prostate. 2008;68(16):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]