Abstract
Increasing evidence indicates the pivotal role of long noncoding RNAs in a variety of cancers, but there is limited focus on the link between long noncoding RNAs and gestational choriocarcinoma. This study aimed to examine the role of long noncoding RNA OGFRP1 in JEG-3 and JAR cells. Small interfering RNA was used to downregulate long noncoding RNA OGFRP1 level. Cell proliferation was measured by cell counting kit-8 and clone formation assays. Cell cycle and apoptosis were analyzed by flow cytometry. Cell invasion was examined by transwell assay. Protein expression was determined by Western blot. A double-effect inhibitor (BEZ235) that inhibits AKT and mTOR phosphorylation was used as a positive control. Knockdown of long noncoding RNA OGFRP1 significantly inhibited the proliferation of JEG-3 and JAR cells. Knockdown of long noncoding RNA OGFRP1 induced cell cycle arrest in G1 phase and apoptosis. On the other hand, knockdown of long noncoding RNA OGFRP1 inhibited the invasion of JEG-3 and JAR cells. Finally, knockdown of long noncoding RNA OGFRP1 resulted in the inactivation of AKT/mTOR signaling pathway. In addition, knockdown of long noncoding RNA OGFRP1 caused changes in the expression of intracellular cell cycle–related proteins and apoptosis-related proteins, including downregulation of CDK4, CDK6, Cyclin D1, Nusap1, and Bcl2 protein expression and upregulation of Bax protein expression. In conclusion, we found that downregulation of long noncoding RNA OGFRP1 inhibited cell proliferation, cell cycle progression, and invasion of JEG-3 and JAR cells and induced apoptosis through AKT/mTOR pathway. This study extends the understanding of the function of long noncoding RNA OGFRP1 in tumorigenesis, and these findings may be important for developing a potential therapeutic target for gestational choriocarcinoma therapy.
Keywords: lncRNA OGFRP1, gestational choriocarcinoma, AKT/mTOR signaling pathway
Introduction
Gestational choriocarcinoma (GC) derived from placental villus trophoblastic cells is a malignant and rare gestational trophoblastic neoplasia.1 Gestational choriocarcinoma is highly invasive and proliferative and can spread widely and rapidly to the liver, lungs, kidneys, brain, and vagina due to its affinity for the hematogenous route.2,3 It is highly sensitive to chemotherapy and surgical resection improves the cure rate.4,5 The current overall cure rate for GC is about 80%.6 However, a small but significant proportion of patients exhibit resistance to chemotherapy and are unable to survive or have an increased risk of hysterectomy and fertility loss due to metastasis that occurs primarily in the early stages and tumor recurrence.7-9 Moreover, due to severe side effects, the application of chemotherapy in clinical treatment has also been seriously hindered.10 Therefore, it is very necessary to develop new therapies or strategies for the treatment of GC.
Long noncoding RNAs (lncRNAs) are a class of transcripts that are more than 200 nucleotides in length and do not have protein coding ability.11 In recent years, lncRNAs have received extensive attention as potential and crucial regulators of a variety of biological processes.12-15 A growing body of evidence also indicates the pivotal role of lncRNAs in a variety of cancers,16,17 but there is limited focus on the link between lncRNAs and GC. The first lncRNA found to be associated with the progression of GC is H19,18 and a recent study has also shown that H19 is associated with GC resistance.1 Knockout of H19 reduces drug resistance in resistant GC cells.1 Since then, as far as we know, only LINC00261,19 LINC00629,20 and MEG3 21 have been discovered as tumor suppressor genes, inhibiting invasion and proliferation of GC cells and inducing apoptosis. MALAT1 is identified as a cancer-promoting gene that promotes GC proliferation in vivo and in vitro.22
Long noncoding RNA OGFRP1 (lncOGFRP1), a novel lncRNA, is found to be involved in the regulation of autophagy in human coronary artery endothelial cells (HCAECs).23 It has also been identified to regulate proliferation and invasion of hepatocellular carcinoma (HCC) cell lines, although the effects are different in different HCC cell lines.24 However, lncOGFRP1 may still have other features that require annotation.
In this study, we researched the role of lncOGFRP1 in GC cell line JEG-3 and JAR. Small interfering RNA (siRNA) specific for lncOGFRP1 was synthesized and used to downregulate lncOGFRP1 levels. The effects of lncOGFRP1 knockdown on proliferation, invasion, cell cycle, and apoptosis of JEG-3 and JAR cells were examined. The molecular mechanism by which lncOGFRP1 affected the biological function of JEG-3 and JAR cells had also been explored.
Materials and Methods
Cell Culture
The human CC cell lines JEG-3 and JAR purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were laboratory preserved. The cells were cultured in Dulbecco's modified Eagle medium (DMEM; Gibco, Grand Island, New York) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37 C with 5% CO2.
Transfection
The siRNA targeting lncOGFRP1 and the negative control siRNA were designed and synthesized by the Ruibo (Guangzhou, China). Transfection of all siRNA was performed by Lipofectamine 2000 (Invitrogen, Carlsbad, California).
Quantitative Real-Time Polymerase Chain Reaction
Total messenger RNA from cells was extracted using TRizol reagent (Invitrogen) and reverse transcribed to complementary DNAs by PrimeScript Reverse Transcription Reagent Kit (TaKaRa, Dalian, China). Quantitative real-time polymerase chain reaction was performed using SYBR PremixEx Taq II (TaKaRa) with ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). The levels of lncOGFRP1 were calculated with the 2−ΔΔCt method and normalized to β-actin.
Cell Counting Kit-8 Assay
5 × 103 transfected cells were seeded in each well of a 96-well plate. Cell proliferation was monitored by the cell counting kit-8 (CCK-8) assay every 24 hours. Ten microliter of CCK-8 solution (Beyotime Biotechnology, Shanghai, China) was added to each well and incubated at 37 C for 2 hours. The absorbance at 450 nm was measured by a microplate reader.
Colony-Forming Assay
Two hundred transfected cells were normally cultured in each well of a 6-well plate. After 2 weeks, cells were fixed with methanol and stained with 0.1% crystal violet. Visible colonies were counted and pictured.
Flow Cytometry Detection for Cell Cycle
After 48 hours of transfection, supernatants were transferred from culture dishes into collecting tubes, and cells were trypsinized (without EDTA). All cells were collected by centrifugation and resuspended in cold phosphate-buffered solution (PBS). Subsequently, cells were fixed with chilled 70% ethanol at 4°C for 24 hours. Fixed cells were washed twice with PBS, incubated with RNaseA for 30 minutes, and stained with propidium oxide for another 30 minutes in dark by Cell Cycle Analysis Kit (Beyotime Biotechnology). Cell cycle progression was immediately detected using a flow cytometer and analyzed using FCS Express 4 software (De NovoSoftware, Los Angeles, California).
Flow Cytometry Detection for Apoptosis
After 48 hours of transfection, cells were detached using trypsin-EDTA. Then, cells were collected by centrifugation and resuspended in Annexin V binding buffer. Subsequently, 100 μL of the cell suspension was double stained with 5 μL V-fluorescein isothiocyanate fluoresceine isothiocyanate (V-FITC) and 5 μL propidium iodide (BD Biosciences, Franklin Lakes, New Jersey) for 15 minutes at room temperature in the dark. Then, 400 μL of 1× binding buffer was added. Cell apoptosis was measured by BD FACS Canto II (BD Biosciences) and analyzed using FlowJo software (version 7.6.5).
Transwell Invasion Assay
The invasion ability of JEG-3 cells was evaluated using the 24-well Matrigel-coated Transwell chambers (BD Biosciences). The upper chamber was filled with 1 × 105 cells/mL in FBS-free DMEM, and the lower chamber was filled with 600 μL of DMEM containing 10% FBS. After incubation at 37°C for 24 hours, cells on the upper chambers were removed, and invaded cells on the lower surface of the chambers were fixed in 100% methanol and stained with 0.1% crystal violet. Invaded cell was counted and photographed under a microscope (×200 magnification, Nikon TE2000).
Gelatin Zymography
The 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel containing 0.5 mg/mL gelatin was prepared. Three microgram of protein was added to the SDS-PAGE gel and electrophoretically separated. Subsequently, the gel was incubated twice in the eluate (2.5% Triton X-100, 50 mM Tris–HCl, 5 mmol/L CaCl2, pH 7.6) for 40 minutes and washed twice with a rinse solution (50 mM Tris-HCl, 5 mmol/L CaCl2, pH 7.6) for 20 minutes. Then, gel was placed in an incubation solution (50 mM Tris–HCl, 5 mmol/L CaCl2, 0.02% Brij-35, pH 7.6) at 37°C for 4 hours. After incubation, the gel was stained with 0.25% Coomassie Brilliant Blue and destained in 7.5% acetic acid containing 20% methanol. Matrix metallopeptidase 9 activity was indicated by a white band on the blue gel.
Western Blot
Total proteins from cells were dissolved with RIPA buffer and quantified with BCA Protein Assay Kit (Tiangen, Beijing, China). Fifteen microgram of proteins were loaded onto a 10% SDS-PAGE gel and then transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% fat-free skim milk for 1 hour at room temperature, PVDF membranes were incubated with primary antibodies at 4°C overnight and subsequently incubated with secondary antibody at room temperature for 2 hours. Immunoreactive bands were developed using enhanced chemiluminescence kit (Amersham, Little Chalfont, United Kingdom).
Anti-mTOR (1:1000, 20657-1-AP), anti-p-mTOR (1:1000, Ser2448, 66888-1-Ig), anti-AKT (1:1000, 10176-2-AP), and anti-p-AKT (1:1000, Ser473, 66444-1-Ig) were purchased from Proteintech (Manchester, United Kingdom). Anti-CDK4 (1:1000, ab108357), anti-CDK6 (1:1000, ab124821), anti-Nusap1 (1:1000, ab169083), anti-Cyclin D1 (1:1000, ab16663), anti-Bax (1:1000, ab32503), anti-Bcl2 (1:1000, ab32124), and anti-GAPDH (1:1000, ab181602) were purchased from Abcam (Cambridge, Massachusetts).
Statistical Analysis
Each experiment was repeated at least 3 times. Statistical analysis was performed using SPSS 22.0 (SPSS, IBM, Beijing, China) and GraphPad Prism 6 (GraphPad, San Diego, California). Data were presented as the mean ± standard deviation. Statistical analyses were performed with the Student t test between 2 groups or with 1-way analysis of variance among multiple groups. A P < .05 was considered statistically significant.
Results
Knockdown of lncOGFRP1 Inhibits the Viability of JEG-3 and JAR Cells
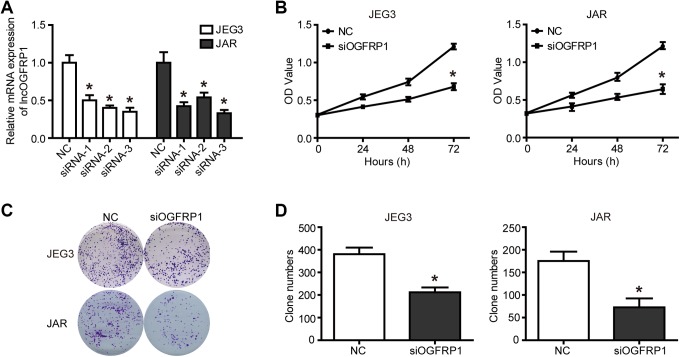
In order to reveal the role of lncOGFRP1 in JEG-3 and JAR cells, loss-of-function assays were performed and lncOGFRP1 was targeted using siRNA (Figure 1A, P < .05). Small interfering RNA-3 was selected for subsequent experiments. Cell counting kit-8 assay showed that lncOGFRP1 knockdown significantly inhibited the proliferation of JEG-3 and JAR cells (Figure 1B, P < .05). In addition, colony-forming assay revealed that the number of colony cells decreased when lncOGFRP1 level was downregulated (Figure 1C and D, P < .05).
Figure 1.
Knockdown of lncOGFRP1 inhibits the viability of JEG-3 and JAR cells. After siRNA, targeting lncOGFRP1 (siOGFRP1) was transfected into JEG-3 and JAR cells, (A) lncOGFRP1 levels were decreased, as detected by qRT-PCR, (B) cell proliferation was inhibited, as examined by CCK-8 assays, (C and D) clonality was impeded, as evaluated by clone formation assay. *P < .05. CCK-8 indicates cell counting kit-8; lncOGFRP1, long noncoding RNA OGFRP1; qRT-PCR, quantitative real-time polymerase chain reaction; siRNA, small interfering RNA.
Knockdown of lncOGFRP1 Induces Cell Cycle Arrest and Apoptosis
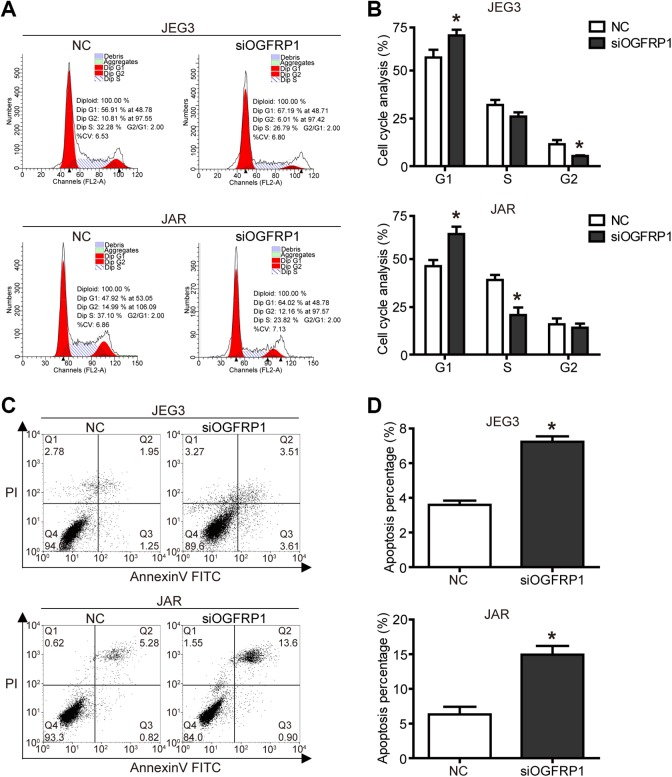
In order to further investigate how knockdown of lncOGFRP1 inhibited JEG-3 and JAR cell proliferation, flow cytometry was used to examine the effect of lncOGFRP1 knockdown on cell cycle and apoptosis. The results of flow cytometry showed that when lncOGFRP1 was knocked down, the proportion of cells in G1 phase was significantly increased, and the proportion of cells in S and G2 phases was decreased (Figure 2A and B, P < .05). In addition, the percentage of apoptosis of JEG-3 and JAR cells was significantly increased when lncOGFRP1 level was downregulated (Figure 2C and D, P < .05). These results indicated that knockdown of lncOGFRP1 inhibited cell proliferation by inducing cell cycle arrest in G1 phase and apoptosis.
Figure 2.
Knockdown of lncOGFRP1 induces cell cycle arrest and apoptosis. After siRNA, targeting lncOGFRP1 (siOGFRP1) was transfected into JEG-3 and JAR cells, (A and B) cell cycle was arrested in G1 phase, as analyzed by flow cytometry, (C and D) apoptosis was induced, as analyzed by flow cytometry. *P < .05. lncOGFRP1 indicates long noncoding RNA OGFRP1; siRNA, small interfering RNA.
Knockdown of lncOGFRP1 Inhibits Invasion of JEG-3 and JAR Cells
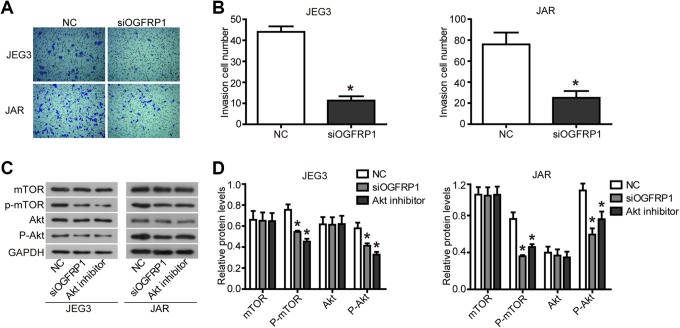
We continued to research the effect of lncOGFRP1 knockdown on invasion ability of JEG-3 and JAR cells. The results of transwell assays showed that when lncOGFRP1 level was downregulated, the number of JEG-3 and JAR cells passing through Matrigel was significantly reduced (Figure 3A and B, P < .05).
Figure 3.
Knockdown of lncOGFRP1 inhibits invasion and the activation of AKT/mTOR signaling pathway in JEG-3 cells. After siRNA, targeting lncOGFRP1 (siOGFRP1) was transfected into JEG-3 and JAR cells, (A and B) the invasion ability was hindered, as determined by transwell assay, (C and D) the expression of key proteins in the AKT/mTOR signaling pathway was regulated, as detected by Western blot. *P < .05. lncOGFRP1 indicates long noncoding RNA OGFRP1; siRNA, small interfering RNA.
Knockdown of lncOGFRP1 Inhibits the Activation of AKT/mTOR Signaling Pathway
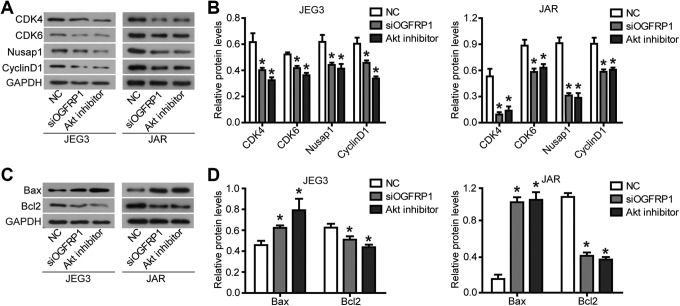
Finally, we explored the specific molecular mechanisms by which lncOGFRP1 promoted the malignant features of JEG-3 and JAR cells. As shown in Figure 3C and D, knockdown of lncOGFRP1 resulted in decreased levels of phosphorylation of AKT and mTOR, but no effect on AKT and mTOR protein levels. In addition, knockdown of lncOGFRP1 caused changes in the expression of intracellular cell cycle–related proteins and apoptosis-related proteins, including downregulation of CDK4, CDK6, Cyclin D1, Nusap1, and Bcl2 protein expression and upregulation of Bax protein expression (Figure 4). These results were consistent with the previous results of flow cytometry to detect the effect of lncOGFRP1 knockdown on cell cycle and apoptosis. In addition, a double-effect inhibitor (BEZ235) that inhibits AKT and mTOR phosphorylation was used as a positive control (Figure 3C and D), and the same result was obtained (Figure 4). These results indicated that lncOGFRP1 may affected the biological function of JEG-3 and JAR cells by affecting the activation of AKT signaling pathway.
Figure 4.
Knockdown of lncOGFRP1 affects the expression of cell cycle–related proteins and apoptosis-related proteins. After siRNA, targeting lncOGFRP1 (siOGFRP1) was transfected into JEG-3 and JAR cells, cell cycle–related proteins (A and B) and apoptosis-related proteins (C and D) were regulated, as examined by Western blot. A double-effect inhibitor (BEZ235) that inhibits AKT and mTOR phosphorylation (AKT inhibitor) was used as a positive control to treat JEG3 and JAR cells, and the same result was obtained. *P < .05. lncOGFRP1 indicates long noncoding RNA OGFRP1; siRNA, small interfering RNA.
Discussion
Combined treatment regimen is extremely effective in the cure of GC.25 However, the latest statistics show that approximately 10% to 30% of patients with GC still have incomplete response to first-line multidrug chemotherapy.26,27 In addition, GC has extensive propagation transferability.28 There is, therefore, an urgent need to find new markers that could serve as early diagnostic and therapeutic targets for GC.
Knockdown of lncOGFRP1 was previously reported to inhibit Hep3B cell proliferation, cell cycle progression, migration, and invasion and to induce apoptosis.24 However, it has no significant effect on these biological functions of another cell line HepG2.24 In addition, a study has shown that lncOGFRP1 participates not only in the proliferation, cell cycle, migration, invasion, and apoptosis of HCAECs but also in autophagy.23 However, the role of lncOGFRP1 in other cell or tissue types remains unknown. In this study, we showed the role of lncOGFRP1 in JEG-3 and JAR cells.
In present study, siRNA was used to target and downregulate the expression of lncOGFRP1. Cell counting kit-8 and colony-forming assays indicated that knockdown of lncOGFRP1 significantly inhibited the proliferation of JEG-3 and JAR cells. Moreover, the results of flow cytometry showed knockdown of lncOGFRP1 induced cell cycle arrest in G1 phase and apoptosis. On the other hand, transwell assays showed that knockdown of lncOGFRP1 significantly inhibited invasion of JEG-3 and JAR cells. These results demonstrated that lncOGFRP1 may play a cancer-promoting role in GC, consistent with its role in Hep3B cells.
Finally, we found that knockdown of lncOGFRP1 resulted in the inactivation of AKT/mTOR signaling pathway. The AKT/mTOR signaling pathway plays an important role in the progression of the GC. Both H19 and MEG3 affect the malignant phenotype and drug resistance of GC cells by affecting the activation of this pathway.18,21 Long noncoding RNA OGFRP1 also affects the biological functions of Hep3B and HCAECs by affecting the activation of this pathway.23,24 In addition, knockdown of lncOGFRP1 caused changes in the expression of intracellular cell cycle–related proteins and apoptosis-related proteins, including downregulation of CDK4, CDK6, Cyclin D1, Nusap1, and Bcl2 protein expression and upregulation of Bax protein expression. These results were consistent with the previous results of flow cytometry to detect the effect of lncOGFRP1 knockdown on cell cycle and apoptosis.
Conclusion
In conclusion, we found that downregulation of lncOGFRP1 inhibited cell proliferation, cell cycle progression, and invasion of JEG-3 and JAR cells and induced apoptosis through AKT/mTOR pathway. This study extends the understanding of the function of lncOGFRP1 in tumorigenesis, and these findings may be important for developing a potential therapeutic target for GC therapy.
Abbreviations
- CCK-8
cell counting kit-8
- GC
gestational choriocarcinoma
- lncRNAs
long noncoding RNAs
- lncOGFRP1
long noncoding RNA OGFRP1
- HCAECs
human coronary artery endothelial cells
- HCC
hepatocellular carcinoma
- DMEM
Dulbecco's modified Eagle medium
- FBS
fetal bovine serum
- PVDF
polyvinylidene fluoride
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- siRNA
small interfering RNA.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Haiyan Xue  https://orcid.org/0000-0002-9117-9311
https://orcid.org/0000-0002-9117-9311
References
- 1. Yu S, Wu C, Tan Q, Liu H. Long noncoding RNA H19 promotes chemotherapy resistance in choriocarcinoma cells. J Cell Biochem. 2019;120(9):15131–15144. doi:10.1002/jcb.28775. [DOI] [PubMed] [Google Scholar]
- 2. Pires LV, Yi Y, Cheng JC, et al. Lapatinib inhibits amphiregulin-induced BeWo choriocarcinoma cell proliferation by reducing ERK1/2 and AKT signaling pathways. Anticancer Res. 2019;39(5):2377–2383. doi:10.21873/anticanres.13355. [DOI] [PubMed] [Google Scholar]
- 3. Savage P. Clinical observations on chemotherapy curable malignancies: unique genetic events, frozen development and enduring apoptotic potential. BMC Cancer. 2015;15:11 doi:10.1186/s12885-015-1006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lurain JR. Gestational trophoblastic disease II: classification and management of gestational trophoblastic neoplasia. Am J Obstet Gynecol. 2011;204(1):11–18. doi:10.1016/j.ajog.2010.06.072. [DOI] [PubMed] [Google Scholar]
- 5. Lurain JR. Gestational trophoblastic disease I: epidemiology, pathology, clinical presentation and diagnosis of gestational trophoblastic disease, and management of hydatidiform mole. Am J Obstet Gynecol. 2010;203(6):531–539. doi:10.1016/j.ajog.2010.06.073. [DOI] [PubMed] [Google Scholar]
- 6. Seckl MJ, Sebire NJ, Fisher RA, et al. Gestational trophoblastic disease: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(suppl 6):vi39–vi50. doi:10.1093/annonc/mdt345. [DOI] [PubMed] [Google Scholar]
- 7. Peng Z, Zhang C, Zhou W, Wu C, Zhang Y. The STAT3/NFIL3 signaling axis-mediated chemotherapy resistance is reversed by Raddeanin a via inducing apoptosis in choriocarcinoma cells. J Cell Physiol. 2018;233(7):5370–5382. doi:10.1002/jcp.26362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berkowitz RS, Goldstein DP. Current management of gestational trophoblastic diseases. Gynecol Oncol. 2009;112(3):654–662. doi:10.1016/j.ygyno.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 9. Shen Y, Yang J, Zhao J, Xiao C, Xu C, Xiang Y. The switch from ER stress-induced apoptosis to autophagy via ROS-mediated JNK/p62 signals: a survival mechanism in methotrexate-resistant choriocarcinoma cells. Exp Cell Res. 2015;334(2):207–218. doi:10.1016/j.yexcr.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 10. Braga A, Campos V, Filho JR, et al. Is chemotherapy always necessary for patients with nonmetastatic gestational trophoblastic neoplasia with histopathological diagnosis of choriocarcinoma? Gynecol Oncol. 2018;148(2):239–246. doi:10.1016/j.ygyno.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 11. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi:10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 14. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 15. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 16. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi:10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 18. Wang JM, Kumar S, van Agthoven A, Kumar P, Pye D, Hunter RD. Irradiation induces up-regulation of E9 protein (CD105) in human vascular endothelial cells. Int J Cancer. 1995;62(6):791–796. doi:10.1002/ijc.2910620624. [DOI] [PubMed] [Google Scholar]
- 19. Wang Y, Xue K, Guan Y, et al. Long noncoding RNA LINC00261 suppresses cell proliferation and invasion and promotes cell apoptosis in human choriocarcinoma. Oncol Res. 2017;25(5):733–742. doi:10.3727/096504016X14772362173376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muys BR, Lorenzi JC, Zanette DL, et al. Placenta-enriched LincRNAs MIR503HG and LINC00629 decrease migration and invasion potential of JEG-3 cell line. PLoS One. 2016;11(3): e0151560 doi:10.1371/journal.pone.0151560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ji L, Li X. Long noncoding RNA MEG3 is a tumor suppressor in choriocarcinoma by upregulation of microRNA-211. J Cell Physiol. 2019;234(12):22911–22920. doi:10.1002/jcp.28853. [DOI] [PubMed] [Google Scholar]
- 22. Shi D, Zhang Y, Lu R, Zhang Y. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother. 2018;97:543–550. doi:10.1016/j.biopha.2017.10.083. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X, Liu J, Gu Y, Sun C, Qu F. Down-regulation of lncRNA OGFRP1 induces autophagy and growth inhibition by AKT/mTOR signaling pathway in HCAECs. Cell Biol Int. 2019;43(2):158–166. doi:10.1002/cbin.11081. [DOI] [PubMed] [Google Scholar]
- 24. Chen W, You J, Zheng Q, Zhu YY. Downregulation of lncRNA OGFRP1 inhibits hepatocellular carcinoma progression by AKT/mTOR and Wnt/beta-catenin signaling pathways. Cancer Manag Res. 2018;10:1817–1826. doi:10.2147/CMAR.S164911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. May T, Goldstein DP, Berkowitz RS. Current chemotherapeutic management of patients with gestational trophoblastic neoplasia. Chemother Res Pract. 2011;2011:806256 doi:10.1155/2011/806256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abrao RA, de Andrade JM, Tiezzi DG, Marana HR, Candido dos Reis FJ, Clagnan WS. Treatment for low-risk gestational trophoblastic disease: comparison of single-agent methotrexate, dactinomycin and combination regimens. Gynecol Oncol. 2008;108(1):149–153 doi:10.1016/j.ygyno.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 27. Lurain JR, Singh DK, Schink JC. Primary treatment of metastatic high-risk gestational trophoblastic neoplasia with EMA-CO chemotherapy. J Reprod Med. 2006;51(10):767–772. [PubMed] [Google Scholar]
- 28. Yousefi Z, Mottaghi M, Rezaei A, Ghasemian S. Abnormal presentation of choriocarcinoma and literature review. Iran J Cancer Prev. 2016;9(2):e4389 doi:10.17795/ijcp-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]