Abstract
The messenger RNA (mRNA) 3′ untranslated regions (3′ UTRs), as cis-regulated elements bound by microRNAs (miRNAs), affect their gene translation. However, the role of the trans-regulation of 3′ UTRs during heart dysfunction remains elusive. Compared with administration of angiogenic factor with G-patch and forkhead-associate domains 1 (Aggf1), ectopic expression of Aggf1 with its 3′ UTR significantly suppressed cardiac dysfunction in angiotensin II-infused mice, with upregulated expression of both Aggf1 and myeloid cell leukemia 1 (Mcl1). Along their 3′ UTRs, Mcl1 and Aggf1 mRNAs share binding sites for the same miRNAs, including miR-105, miR-101, and miR-93. We demonstrated that the protein-coding Mcl1 and Aggf1 mRNAs communicate and co-regulate each other’s expression through competition for these three miRNAs that target both transcripts via their 3′ UTRs. Our results indicate that Aggf1 3′ UTR, as a trans-regulatory element, accelerates the cardioprotective role of Aggf1 in response to hypertensive conditions by elevating Mcl1 expression. Our work broadens the scope of gene therapy targets and provides a new insight into gene therapy strategies involving 3′ UTRs.
Keywords: AGGF1, MCL1, 3’UTR, cardiac dysfunction
3′ UTRs act as trans-regulatory elements and play a cardioprotective role in angiotensin II (AngII)-induced heart dysfunction in mice. Overexpression of Aggf1 3′ UTR suppressed the reduction in AGGF1 and MCL1 protein expression in response to AngII, because ectopic 3′ UTRs blocked the interaction between microRNAs and Aggf1 and Mcl1 mRNAs.
Introduction
Hypertensive heart disease is responsible for considerable morbidity and mortality and, therefore, is a major health problem.1 In the course of this condition, hypertensive cardiac dysfunction caused by cardiac inflammation and fibrosis leads to left ventricular hypertrophy, systolic and diastolic dysfunction, and even heart failure.1, 2, 3, 4 However, the molecular mechanisms of hypertensive heart disease remain unclear, and therapeutic strategies for cardiac dysfunction are not effective.
Regulation of gene expression by multiple mechanisms is a key molecular biology phenomenon. Post-transcriptional regulation controls gene expression at the RNA level and is an intermediate level of expression control between epigenetic regulation of transcription and post-translational modifications. MicroRNA (miRNA)-mediated regulation of gene expression is a major method of inhibiting translation and/or promote degradation of messenger RNA (mRNA) encoding the target protein. 3′ UTR non-coding sequences act as cis-regulatory elements and can be recognized and bounded with miRNAs, resulting in the modulation of their mRNA stability and protein expression. However, the function for 3′ UTRs in hypertensive cardiac dysfunction has not been clarified.
The angiogenic factor with G-patch and forkhead-associated domains 1 gene, AGGF1, was discovered during genetic screening for congenital vascular Klippel-Trenaunay syndrome.5 Administration of the AGGF1 protein blocked cardiac apoptosis in ischemia-reperfused heart by inhibiting the nuclear factor κB signaling pathway.6 Our previous studies found that recombinant AGGF1 protein increased the survival rate after myocardial infarction in a mouse model.7 In addition, exogenous AGGF1 blunted cardiac fibrosis in mouse models of isoproterenol-induced hypertrophy and transverse aortic constriction-induced hypertrophy with pressure overload, preventing the development of heart dysfunction and cardiac remodeling.8 Additionally, myeloid cell leukemia 1 (MCL1) is an anti-apoptotic Bcl2 family protein highly expressed in the myocardium.9 MCL1 plays an essential role in myocardial homeostasis and autophagy.9 Mcl1 deficiency leads to impaired autophagy, mitochondrial dysfunction, and lethal heart failure.10,11 Although the cardioprotective role of AGGF1 had been established, whether the AGGF1 3′ UTR non-coding sequence is involved in cardiac dysfunction after angiotensin II (AngII) infusion or not is not clear.
In this study, we showed that administration of the full-length Aggf1 construct that incorporates both the complete protein-coding sequence and its 3′ UTR could recover heart function due to the upregulation of the Mcl1 expression level, compared to constructs containing only the Aggf1 coding sequence. We propose that this novel regulatory mechanism could be utilized as a novel gene therapy approach for cardiovascular and other diseases.
Results
Overexpression of Full-Length Aggf1 Attenuates Cardiac Dysfunction in AngII-Infused Mice
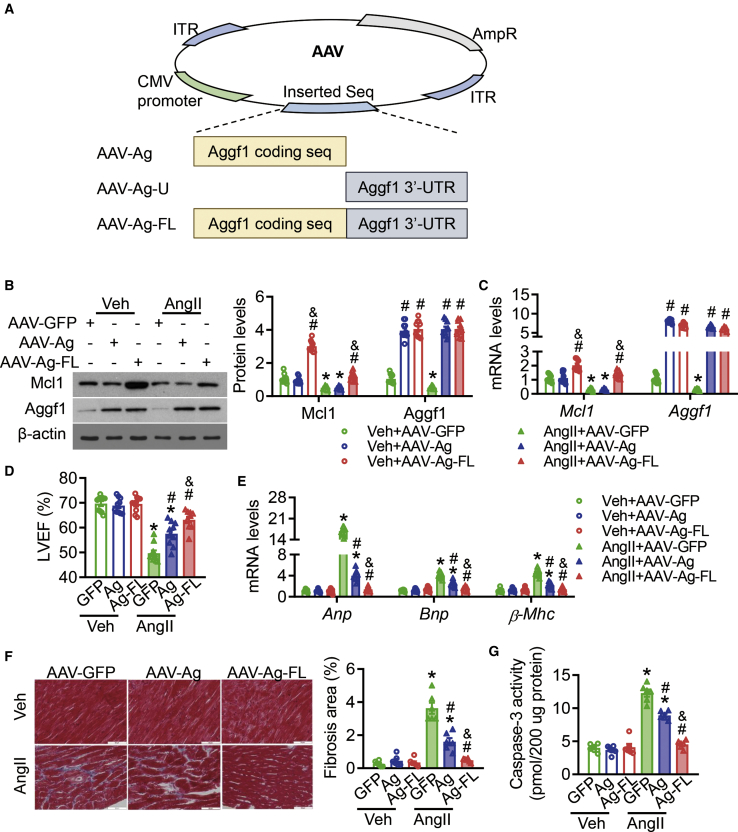
To explore whether the cardio-protective effect of AGGF1 would be on cardiac dysfunction, we used an adeno-associated virus (AAV) serotype 9 (AAV9) delivery system in mice with AngII infusion. We generated two AAVs expressing ectopic AGGF1, including AAV-Ag-FL, which carried a full-length mouse Aggf1 coding sequence and its 3′ UTR, and AAV-Ag (with an Aggf1 coding sequence) (Figure 1A). In AngII-infused mice, AAV-Ag induced higher expression levels of Aggf1 than the negative control AAV-GFP (with GFP tag), and the increased Aggf1 level was observed after AAV-Ag-FL administration (Figures 1B and 1C). AAV-Ag-FL administration induced Mcl1 expression, whereas AAV-Ag did not (Figures 1B and 1C). Echocardiography showed that AAV-Ag was associated with significantly higher left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) values than those observed after AAV-GFP injection in AngII-infused mice (Figures 1D, S1A, and S1B; Table S1). AAV-Ag-FL administration more effectively prevented the reduction of LVEF and LVFS in AngII-infused mice compared to the effects of AAV-GFP and AAV-Ag (Figures 1D, S1A, and S1B; Table S1).
Figure 1.
The Full Length of Aggf1 Administration Plays a Cardio-Protective Role in AngII-Infused Mouse
(A) Depiction of the AAVs with different inserted parts of Aggf1 mRNA. The cartoon depicts the main cassettes along the AAV backbone (top), including cytomegalovirus (CMV) promoter, inverted terminal repeats (ITRs), and ampicillin resistance (AmpR). Different parts of Aggf1 mRNA are inserted, generating different AAVs (bottom). (B–G) 8-week-old mice were infused with vehicle or AngII. One week after mini-pump implantation, the mice were injected intravenously with AAV-GFP, AAV-Ag, or AAV-Ag-FL once per week for 5 weeks (n = 10 each group). (B and C) Western blot (B) and real-time PCR (C) analyses for Aggf1 and Mcl1 expression were performed with heart tissues from mice. (D and E) LVEF levels (D) and the mRNA levels of Anp, Bnp, and β-Mhc (E) are indicated in these six groups. (F and G) Representative images of Masson staining of heart tissues (F) and the cardiac caspase-3 activities (G) are shown. Statistical analysis was carried out using one-way ANOVA with Tukey’s test. *p < 0.05 versus vehicle; #p < 0.05 versus AAV-GFP; &p < 0.05 versus AAV-Ag.
Additionally, although AAV-Ag administration significantly reduced the heart weight/body weight (HW/BW) and heart weight/tibia length (HW/TL) ratios in AngII-infused mice (Figure S1C), administration of AAV-Ag-FL was associated with a reversal of the HW/BW and HW/TL ratios in AngII-infused mice (Figure S1C). Similarly, AAV-Ag administration significantly repressed Anp, Bnp, and β-Mhc mRNA levels after AngII infusion (Figure 1E), whereas AAV-Ag-FL had an even stronger effect on the levels of these mRNAs (Figure 1E). Neither AAV-Ag administration nor AAV-Ag-FL administration affected the heart rate or blood pressure regardless of AngII infusion (Figures S2A and S2B). These data indicated that although AAV-Ag effectively attenuated cardiac dysfunction in AngII-infused mice, an even more potent effect was achieved by AAV-Ag-FL.
In order to confirm the efficacy of AAV-Ag and AAV-Ag-FL administration, Masson staining was performed on heart sections. AAV-Ag significantly repressed cardiac fibrosis compared to the effect of AAV-GFP in AngII-infused mice (Figure 1F), whereas cardiac fibrosis was diminished in the hearts of animals injected with AAV-Ag-FL (Figure 1F). Similarly, AAV-Ag and AAV-Ag-FL administration inhibited the AngII-induced increase in ColIa2, ColIVa1, and Ctgf mRNA levels (Figure S3A). Increases in Mcp1, Tnfα, and Il6 mRNA levels in the hearts of AngII-infused animals were prevented by AAV-Ag and AAV-Ag-FL administration, indicating that AGGF1 repressed the cardiac inflammatory response to AngII infusion (Figure S3B). Importantly, AAV-Ag-FL more strongly suppressed the increases in ColIa2, ColIVa1, Ctgf, Mcp1, Tnfα, and Il6 mRNA levels than did AAV-Ag, indicating that AAV-Ag-FL had a more potent anti-inflammation effect than AAV-Ag (Figures S3A and S3B).
To verify the protective effects of AAV-Ag and AAV-Ag-FL on AngII-induced hypertensive cardiac dysfunction, apoptosis in cardiac tissue was analyzed by TUNEL staining. AAV-Ag administration repressed the AngII-induced increase in TUNEL-positive cells, whereas AAV-Ag-FL administration diminished the TUNEL-positive signal entirely (Figure S4). Consistent with TUNEL-staining results, AAV-Ag attenuated the increased activity of caspase-3 after AngII infusion, whereas AAV-Ag-FL reversed the increase in caspase-3 activity (Figure 1G). Together, these results suggested that both AAV-Ag and AAV-Ag-FL administration successfully inhibited cardiac apoptosis and prevented cardiac dysfunction and the progression of cardiac dysfunction in AngII-infused mice. Furthermore, AAV-Ag-FL demonstrated a more powerful cardiac protective effect than AAV-Ag.
Downregulation of Mcl1 and Aggf1 Expression in Heart Tissues from AngII-infused Mice
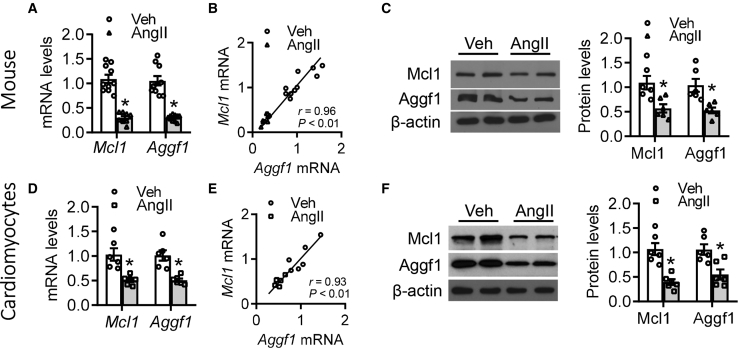
To clearly demonstrate that the reduction of Mcl1 and Aggf1 expression is associated with the severity of hypertensive cardiac dysfunction, a mouse model was generated by AngII infusion. Notably, Mcl1 and Aggf1 mRNA levels in the heart tissue of AngII-infused mice were lower than those in vehicle-treated hearts (Figure 2A). Aggf1 mRNA levels were positively correlated with Mcl1 mRNA levels in both vehicle and AngII-treated mice (Figure 2B). Accordingly, Mcl1 and Aggf1 protein levels were lower in the hearts of AngII-treated mice than in the hearts of vehicle-treated mice (Figure 2C). Thus, in vivo experiments in a mouse model of cardiac dysfunction showed that Mcl1 and Aggf1 levels were downregulated in the heart in response to AngII infusion.
Figure 2.
Hypertension Induces the Reduction of Aggf1 and Mcl1 in Heart Tissues
(A–C) 8-week-old mice were infused with vehicle or AngII (n = 10 each group). (A) Real-time PCR analysis of Aggf1 mRNA levels in hearts. (B) The association between Aggf1 and Mcl1 mRNA levels in heart tissues from mice. (C) Western blot analysis for Aggf1 expression in heart tissues from vehicle/AngII-infused mice. Statistical analysis was carried out by a Student’s t test. *p < 0.05 versus vehicle. (D–F) Purified mouse neocardiomyocytes from wild-type mice were incubated with vehicle or AngII for 24 h. (D) Real-time PCR analysis for Aggf1 and Mcl1 mRNA levels. (E) The association between Aggf1 and Mcl1 mRNA levels in mouse neocardiomyocytes. (F) Western blot analysis for Aggf1 and Mcl1 protein levels after treatment. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05.
Reduction in Mcl1 and Aggf1 Expression Levels in Cultured Neocardiomyocytes following AngII Infusion
Although reduction in MCL1 and AGGF1 expression levels was observed in heart samples following AngII infusion, we first set up an in vitro system using isolated mouse neocardiomyocytes that were incubated with AngII or vehicle. Cultured cardiomyocytes exposed to AngII had lower Mcl1 and Aggf1 mRNA levels than those treated with vehicle (Figure 2D). Furthermore, a correlation between Mcl1 and Aggf1 mRNA levels was also observed in cultured cardiomyocytes (Figure 2E). Similarly, the levels of Mcl1 and Aggf1 proteins were also decreased after incubation with AngII (Figure 2F). These results indicated that AngII treatment was associated with lower levels of Mcl1 and Aggf1 expression in cardiac tissue both in vivo and in vitro.
Reduction of Mcl1 Expression by Aggf1 Knockdown
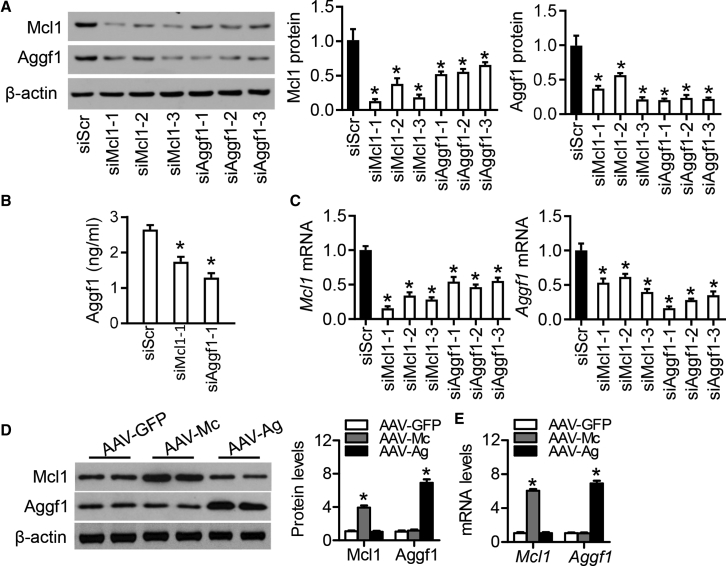
Additionally, exposure to three small interfering RNAs (siRNAs) that targeted Aggf1 protein also reduced the levels of Mcl1 protein (Figure 3A) and secretory Aggf1 (Figure 3B). After Aggf1 knockdown, Mcl1 and Aggf1 mRNA levels were decreased (Figure 3C), and Mcl1 and Aggf1 mRNA levels were positively correlated (Figure S5). However, there was no change in Mcl1 expression after Aggf1 overexpression (AAV-Ag), whereas ectopic expression of Aggf1 was substantially increased at both the mRNA and protein levels (Figures 3D and 3E). These data showed that Mcl1 is not downstream of Aggf1.
Figure 3.
Reduction of Aggf1 or Mcl1 Affects Both Aggf1 and Mcl1 Expression
(A–C) The isolated mouse neocardiomyocytes were transfected with scramble siRNA, different siMcl1, or siAggf1 for 36 h. Western blot analysis for Aggf1 and Mcl1 protein levels (A) and ELISA assays for secretory Aggf1 levels (B) were performed after siRNA transfection. (C) Real-time PCR assay analyzed Aggf1 and Mcl1 mRNA levels in cardiomyocytes. Statistical analysis was carried out with a Student’s t test. n = 3. *p < 0.05 versus siScr. (D and E) The isolated mouse neocardiomyocytes were incubated with AAV-GFP, AAV-Mc, or AAV-Ag. Western blot (D) and real-time PCR (E) analyses were performed for Aggf1 and Mcl1 expression after AAV treatment. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05 versus AAV-GFP.
Regulation of Gene Expression by 3′ UTRs as Trans-Regulatory Elements
Our data demonstrated that Aggf1 knockdown caused a reduction in Mcl1 expression but that Aggf1 overexpression did not affect Mcl1 expression. Furthermore, whereas Aggf1 expression was decreased by Mcl1 knockdown, no change in Aggf1 expression after Mcl1 overexpression was observed. Similar results were obtained in experiments with different siRNAs, which allowed excluding off-target effects of siRNAs (Figures 3C and S5). The difference between the gene knockdown and overexpression consequences could be due to differential involvement of the 3′ UTR and 5′ UTR of the mRNA. Recently, it was noted that competing endogenous RNAs (ceRNAs) bind miRNAs via miRNA binding sites (miRNA recognition elements; MREs) and thereby de-repress the genes targeted by corresponding miRNAs.
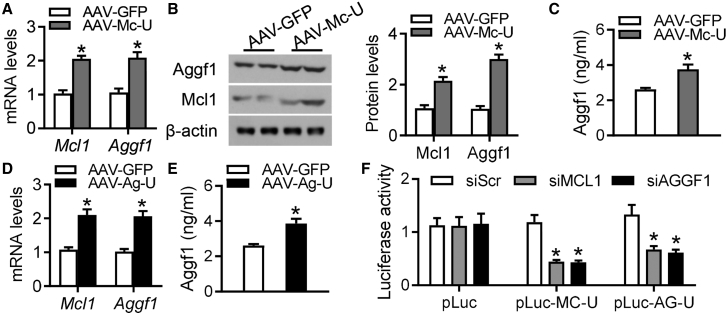
In order to determine whether crosstalk among 3′ UTRs affected gene expression changes in our experiments, we first generated AAV-Mc-U, which incorporated only the mouse Mcl1 3′ UTR, and AAV-Ag-U, which contained the 3′ UTR of mouse Aggf1 mRNA. Ectopic expression of the Mcl1 3′ UTR via AAV-Mc-U in mouse neocardiomyocytes increased the mRNA levels of both Mcl1 and Aggf1 (Figure 4A). Western blot analyses showed that AAV-Mc-U also increased Mcl1 and Aggf1 protein levels (Figure 4B), which was accompanied by elevated levels of secreted Aggf1, compared to those after AAV-GFP incubation (Figure 4C). Additionally, mouse Aggf1 3′ UTR overexpression in neocardiomyocytes via incubation with AAV-Ag-U increased the expression of Mcl1 and Aggf1 at both the mRNA and protein levels compared to those following AAV-GFP treatment (Figures 4D and S6). Furthermore, elevated levels of secreted Aggf1 were detected after AAV-Ag-U treatment (Figure 4E). These results showed that Mcl1 and Aggf1 expression levels were regulated by the 3′ UTRs of Mcl1 and Aggf1 mRNAs in mouse neocardiomyocytes.
Figure 4.
Regulation of Gene Expression via Trans-Elemental 3′ UTRs
(A–C) The purified mouse neocardiomyocytes were pre-cultured with AAV-GFP or AAV-Mc-U. Real-time PCR (A) and western blot (B) analyses were performed for Aggf1 and Mcl1 expression after treatment. (C) The secretory Aggf1 levels were detected by ELISA assays. n = 6. *p < 0.05 versus AAV-GFP. (D and E) The purified mouse neocardiomyocytes were pre-cultured with AAV-GFP or AAV-Ag-U. Real-time PCR analyses for Aggf1 and Mcl1 mRNA levels (D) and ELISA assays for secretory Aggf1 levels (E) were performed. n = 6. *p < 0.05 versus AAV-GFP. (F) The luciferase activities of pLuc-AG-U and pLuc-MC-U were measured in the deficiency of MCL1 or AGGF1 in HEK293 cells. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05 versus siScr.
Regulation of MCL1 and AGGF1 Expression Levels by Their 3′ UTRs
In order to further confirm that the decreases in MCL1 and AGGF1 expression levels are mutually interdependent, we generated the pLuc-MC-U plasmid containing the human MCL1 3′ UTR and performed luciferase activity assays in HEK293 cells, which revealed that AGGF1 deficiency resulted in a suppressed luciferase activity of pLuc-MC-U compared with that detected in the presence of siScr (Figure 4F). Similarly, the reduction of MCL1 led to a lower luciferase activity of pLuc-AG-U compared with that observed with siScr (Figure 4F). The knockdown efficiency on either AGGF1 or MCL1 was detected (Figures S7A and S7B). These results suggested that the decrease in AGGF1 reduced MCL1 expression via the 3′ UTR of MCL1 mRNA, whereas the attenuated expression of AGGF1 was caused by MCL1 knockdown via the 3′ UTR of AGGF1 mRNA.
Our data showed that the ectopic expression of the Aggf1 3′ UTR induced an increase in Aggf1, and then the increase in Aggf1 induced the increased Mcl1 expression via another pathway not dependent on Aggf1 3′ UTR. In order to examine this possibility, an AGGF1-deficient HEK293 cell strain (AG-Cas9) was generated with the CRISPR/Cas9 method and then was treated with AAV-Ag-U or AAV-GFP. The expression of AGGF1 and MCL1 at the protein and mRNA levels was increased in wild-type (WT) HEK293 cells (Figures S8A and S8B). The increased level of MCL1 was also observed in the AGGF1-deficient HEK293 cells compared with WT cells (Figures S8A and S8B). Upon AGGF1 deficiency, the ectopic expression of the Aggf1 3′ UTR induced the increase in MCL1 at both the protein and mRNA levels, indicating that the Aggf1 3′ UTR regulates MCL1 expression via an AGGF1-independent pathway (Figures S8A and S8B).
Identification of miRNAs Interacting with Mcl1 and Aggf1 3′ UTRs
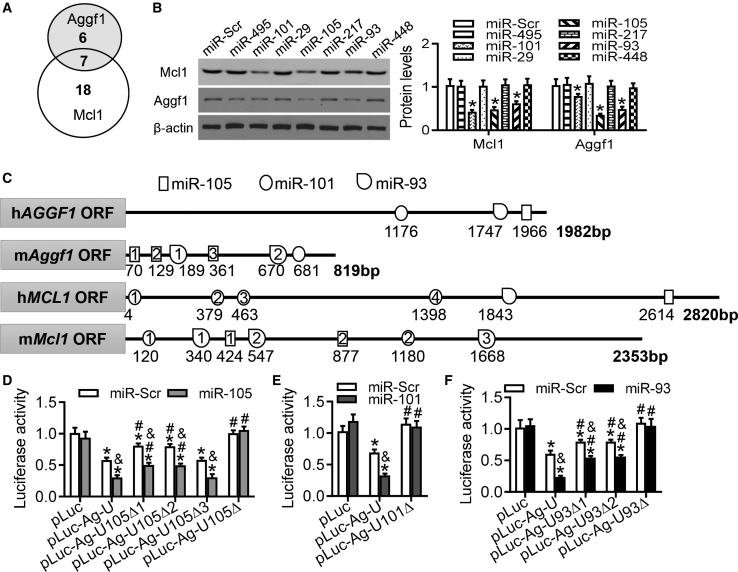
To elucidate which miRNAs, targeting Mcl1 and Aggf1, mediated the crosstalk, we sought to identify miRNAs that were predicted to bind the 3′ UTRs of mouse Mcl1 and Aggf1 using the starBase system (v.2.0). There were seven miRNAs identified for Mcl1 and Aggf1 (Figure 5A), including miR-495, miR-101, miR-29, miR-105, miR-217, miR-93, and miR-448 (Figure 5A).
Figure 5.
Characterization of miRNAs in the 3′ UTRs of Aggf1 and Mcl1 mRNAs
(A) Summarization of predicted MREs in the 3′ UTRs of Aggf1 and Mcl1. (B) Western blot analysis for Aggf1 and Mcl1 expression was performed after overexpression of different miRNA candidates in purified mouse neocardiomyocytes. *p < 0.05 versus miR-Scr. (C) Bioinformatic analysis of the Aggf1 and Mcl1 mRNA sequences identified binding sites for miR-105, miR-101, and miR-93 at the 3′ UTRs. (D) HEK293 cells were transfected with different luciferase plasmids and miR-Scr or miR-105. The luciferase activities were measured. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr. (E) HEK293 cells were transfected with different luciferase plasmids and miR-Scr or miR-101. The luciferase activities were measured. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr. (F) The luciferase activities were measured in HEK293 cells transfected with different luciferase plasmids and miR-Scr or miR-93. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05 versus pLuc; #p < 0.05 versus pLuc-Ag-U; &p < 0.05 versus miR-Scr.
To determine whether these miRNA candidates affected the expression of Mcl1 and Aggf1, mouse neocardiomyocytes were transfected with each of these seven miRNAs. Both western blot and real-time PCR analyses showed that treatments with miR-101, miR-105, or miR-93 decreased Mcl1 and Aggf1 expression at both the mRNA and protein levels (Figures 5B and S9).
Previous microarray analyses of miRNAs in heart tissues from mice infused with AngII for 7 days showed lower levels of miR-29, miR-448, and miR-495 and higher levels of miR-101, miR-105, and miR-93 compared to those in control animals.12 The levels of all seven candidate miRNAs for Mcl1 and Aggf1 were determined in heart tissues from AngII- and vehicle-infused mice. We found increased expression levels of miR-101, miR-105, and miR-93 in AngII-treated animals (Figure S10). Additionally, upregulation of miR-101, miR-105, and mir-93 was observed after AngII treatment in mouse neocardiomyocytes (Figure S11).
In order to further confirm that these miRNAs, indeed, regulate expression levels of Mcl1 and Aggf1 genes, mouse neocardiomyocytes were transfected with a combination of miR-101, miR-105, and miR-93 (Figures S12A and S12B). This treatment induced a dramatic reduction in Mcl1 and Aggf1 gene expression levels compared with the effects of individual miRNAs (Figures S12A and S12B).
Characterization of miRNA Binding Sites of the Aggf1 3′ UTR
3′ UTRs have multiple miRNA binding sites. In order to elucidate how miRNAs regulate Aggf1 expression, 3′ UTRs of human and mouse AGGF1-encoding genes were analyzed using a TargetScan prediction algorithm, miRDB prediction software, and miRSearch v.3.0 (QIAGEN, Hilden, Germany). In the human AGGF1 3′ UTR, the candidate binding region is at 1,966–1,972 bp for miR-105, at 1,176–1,181 bp for miR-101, and at 1,747–1,752 bp for miR-93 (Figure 5C). In the mouse Aggf1 3′ UTR, the candidate binding regions for miR-105 are at 70–74 bp, 129–133 bp, and 361–365 bp. For miR-101, one site is at 681–685 bp, whereas for miR-93, the two candidate binding regions are located at 189–192 bp and 670–673 bp (Figure 5C).
The luciferase activity of pLuc-Ag-U, which contained the whole 3′ UTR of the mouse Aggf1 gene, was decreased compared with that of pLuc. It is possible that the endogenous miRNAs cause the reduction of luciferase activities (Figure 5D). Overexpression of miR-105 deteriorated the reduction of luciferase activities, compared with miR-Scr (Figure 5D). However, the declined luciferase activities were recovered in the cells expressing pLuc-Ag-U105Δ1 (harboring a deletion of the 70- to 74-bp region) or pLuc-Ag-U105Δ2 (harboring a deletion of the 129- to 133-bp region), not in the cells with pLuc-Ag-U105Δ3 (harboring a deletion of the 361- to 365-bp region) (Figure 5D). Additionally, a reduction of luciferase activities of pLuc-Ag-U105Δ1 and pLuc-Ag-U105Δ2 was observed after miR-105 overexpression, compared with miR-Scr (Figure 5D). Deletion of both 70- to 74-bp and 129- to 133-bp sites (pLuc-Ag-U105Δ) did not affect luciferase activities compared with pLuc (Figure 5D), indicating that miR-105 recognized the mouse Aggf1 mRNA 3′ UTR at the 70- to 74-bp and 129- to 133-bp regions.
Overexpression of miR-101 also attenuated pLuc-Ag-U luciferase activity, and this effect was sensitive to the deletion of the 681- to 685-bp site (Figure 5E). Additionally, miR-93 overexpression caused a significant reduction of pLuc-Ag-U luciferase activity (Figure 5F), which was rescued when pLuc-Ag-U93Δ1 and pLuc-Ag-U93Δ2 constructs were used that harbored deletions of the 189- to 192-bp and 670- to 673-bp sites, respectively (Figure 5F). The reduced luciferase activity was also restored to the normal level by a combined deletion of these two sites (pLuc-Ag-U93Δ) in both miR-Scr- and miR-93-transfected cells (Figure 5F). Moreover, the reduction of luciferase activity in miR-93-overexpressing cells, compared to control cells, disappeared after these two sites were deleted (pLuc-Ag-U93Δ) (Figure 5F). These data demonstrate that miR-93 recognized the mouse Aggf1 mRNA 3′ UTR at 189- to 192-bp and 670- to 673-bp regions.
Characterization of miRNA Binding Sites of the Mcl1 3′ UTR
There are multiple candidate binding sites for miRNAs in the 3′ UTRs of human and mouse genes encoding Mcl1 (Figure 5C). In the mouse Mcl1 3′ UTR, two candidate regions for miR-105 were identified at 424–429 bp and 877–882 bp. miR-101 presumably binds the Mcl1 3′ UTR at the 120- to 125-bp and 1,180- to 1,185-bp sites, whereas the three miR-93-binding candidate regions are located at 340–344 bp, 547–551 bp, and 1,668–1,672 bp (Figure 5C).
Overexpression of miR-105 caused a significant reduction in luciferase activity of pLuc-Mc-U, which contained the whole mouse Mcl1 3′ UTR (Figure S13A). The decrease in luciferase activity caused by miR-105 was rescued when pLuc-Mc-U105Δ1 and pLuc-Mc-U105Δ2 constructs were utilized that carried deletions of the 424- to 429-bp and the 877- to 882-bp sites, respectively (Figure S13A), indicating that miR-105 likely affected Mcl1 mRNA by interaction with both the 424- to 429-bp and 877- to 882-bp sites.
The luciferase activity of pLuc-Mc-U was also decreased by miR-101 overexpression, whereas luciferase activity of pLuc-Mc-U101Δ1 harboring a deletion of the 3′ UTR 120- to 125-bp site was resistant to miR-101 treatment (Figure S13B). Meanwhile, the decrease in luciferase activity after miR-101 overexpression was unaffected by deletion of the 1,180- to 1,185-bp site in pLuc-Mc-U101Δ2, compared to pLuc-Mc-U101 (Figure S13B), indicating that miR-101 recognized the mouse Mcl1 mRNA 3′ UTR at the 120- to 125-bp site.
A significant reduction of pLuc-Mc-U luciferase activity was also noted after miR-93 overexpression (Figure S13C), whereas the activity was rescued when pLuc-Mc-U93Δ1 and pLuc-Mc-U93Δ3 constructs were used, which had deletions of the 3′ UTR 340- to 344-bp and 1,668- to 1,672-bp sites, respectively (Figure S13C). However, the effect of miR-93 overexpression on luciferase activity was not changed with the deletion of the 340- to 344-bp and 1,668- to 1,672-bp sites (pLuc-Mc-U93Δ) (Figure S13C). These data suggest that Mcl1 translation is regulated by miR-93 binding to the 3′ UTR 340- to 344-bp and 1,668- to 1,672-bp sites.
Involvement of miR-93, miR-101, and miR-105 in the Reduction of Mcl1 and Aggf1 Expression in Response to AngII Treatment
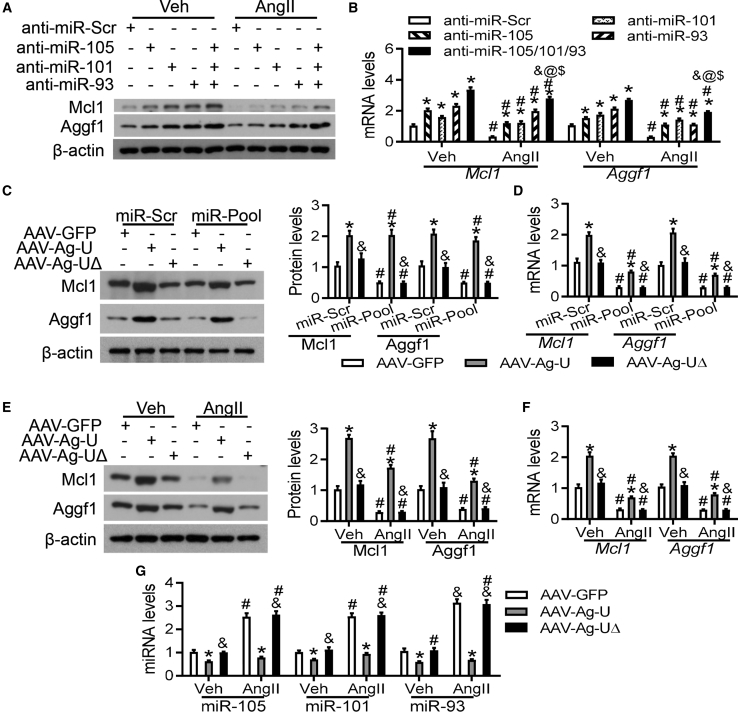
Our data showed that the levels of miR-105, miR-101, and miR-93 were increased in both AngII-induced hearts (Figure S10) and cardiomyocytes (Figure S11). To determine whether decreases in Mcl1 and Aggf1 expression were regulated by AngII via miR-93, miR-101, and miR-105, mouse neocardiomyocytes were transfected with anti-miR-101, anti-miR-105, and anti-miR-93, separately or in combination, and incubated with AngII or vehicle. Both western blot and real-time PCR analysis showed that the reductions in Mcl1 and Aggf1 expression caused by AngII were prevented by all of these treatments (Figures 6A and 6B; Figure S14).
Figure 6.
Aggf1 3′ UTR Prevents the Reduction of Aggf1 and Mcl1
(A and B) Mouse neocardiomyocytes were transfected with anti-miR-101, anti-miR-105, anti-miR-93, or anti-miR-101/105/93, respectively, and then incubated with AngII for 24 h. Western blot (A) and real-time PCR analyses (B) were performed for Aggf1 and Mcl1 expression after treatment. n = 6. *p < 0.05 versus anti-miR-Scr; #p < 0.05 versus vehicle; &p < 0.05 versus anti-miR-105; @p < 0.05 versus anti-miR-101; $p < 0.05 versus anti-miR-93. (C and D) Mouse neocardiomyocytes were incubated with different AAVs and then transfected with miR-Scr or miR-105/101/93. Western blot (C) and real-time PCR (D) analyses were performed for Aggf1 and Mcl1 expression after treatment. n = 6. *p < 0.05 versus AAV-GFP; #p < 0.05 versus miR-Scr; &p < 0.05 versus AAV-Ag-U. (E–G) Mouse neocardiomyocytes were incubated with AAVs and then incubated with vehicle or AngII for 24 h. Western blot (E) and real-time PCR (F) analyses were performed for Aggf1 and Mcl1 expression after treatment. n = 6. (G) The levels of miR-105, miR-101, and miR-93 were measured. Statistical analysis was carried out using one-way ANOVA with Tukey’s test. n = 6. *p < 0.05 versus AAV-GFP; #p < 0.05 versus vehicle; &p < 0.05 versus AAV-Ag-U.
Aggf1 3′ UTR Prevents the Reduction in Mcl1 and Aggf1 Expression upon AngII Treatment
Given that increased expression of Aggf1 following the knockdown of miR-93, miR-101, and miR-105 correlated with increased Mcl1 expression, we predicted that exogenous Aggf1 3′ UTR would be recognized by these miRNAs, which would result in the reduction of their levels. In turn, this would prevent the interactions between these miRNAs and endogenous Mcl1 and Aggf1 3′ UTRs. To this end, we generated a virus that carried the mouse Aggf1 3′ UTR in which MREs were deleted (AAV-Ag-UΔ); namely the 70- to 74-bp and 129- to 133-bp sites (recognized by miR-105), the 681- to 685-bp site (recognized by miR-101), and the 189- to 192-bp and 670- to 673-bp sites (recognized by miR-93). Consistent with our earlier observation, AAV-Ag-U increased expression levels of Mcl1 and Aggf1 compared to those following incubation with AAV-GFP (Figures 4D and 4E), but AAV-Ag-UΔ did not (Figures 6C and 6D). Additionally, exposure to miR-93, miR-101, and miR-105 suppressed Mcl1 and Aggf1 expression compared to that for the vehicle and prevented the increase in Mcl1 and Aggf1 expression caused by AAV-Ag-U (Figures 6C and 6D).
AngII reduced Mcl1 and Aggf1 expression, and this reduction was sensitive to treatment with AAV-Ag-U but not with AAV-Ag-UΔ (Figures 6E and 6F). Furthermore, AAV-Ag-U suppressed the increase in expression levels of miR-105, miR-101, and miR-93 caused by AngII treatment, but AAV-Ag-UΔ did not (Figure 6G). These results showed that exogenous Aggf1 3′ UTR prevented the reduction of Mcl1 and Aggf1 expression upon AngII treatment by blocking the interaction between miR-93, miR-101, and miR-105 and the endogenous 3′ UTRs of the Mcl1 and Aggf1 genes.
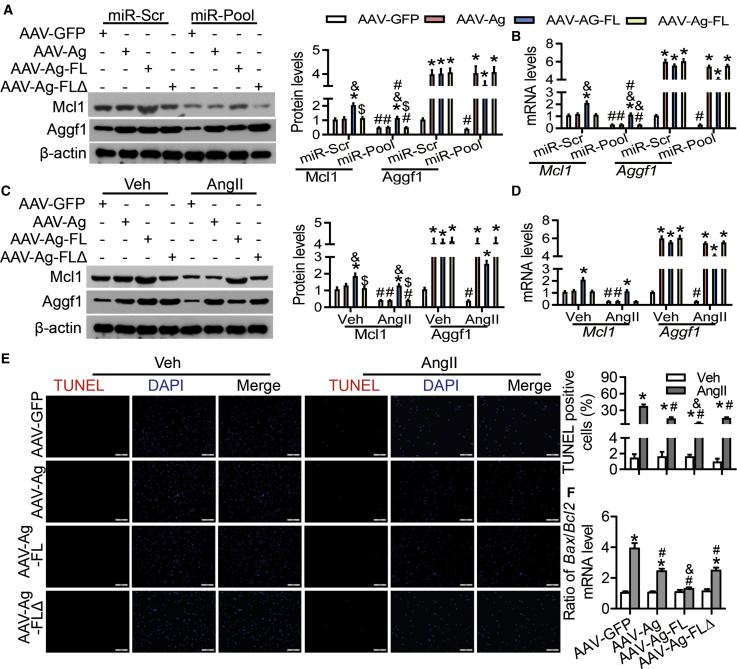
AAV-Ag-FL Prevents the Reduction in Mcl1 and Aggf1 Expression at Both the mRNA and Protein Levels upon AngII Treatment
To examine the regulation of Mcl1 and Aggf1 expression levels by 3′ UTRs, we generated AAV-Ag-FL that carried the full-length mouse Aggf1 coding sequence and its 3′ UTR (Figure 1A). Increased expression levels of Mcl1 and Aggf1 were observed in AAV-Ag-FL-transfected neocardiomyocytes (Figures 7A and 7B). Combined overexpression of miR-101, miR-105, and miR-93 reduced basal Mcl1 and Aggf1 expression as well as the increased expression caused by AAV-Ag-FL (Figures 7A and 7B). We also generated AAV-Ag-FLΔ, which harbored a mouse Aggf1 coding sequence with the 3′ UTR, in which several MREs—such as the 70- to 74-bp and 129- to 133-bp sites (recognized by miR-105), the 681- to 685-bp site (recognized by miR-101), and the 189- to 192-bp and 670- to 673-bp sites (recognized by miR-93)—were deleted. There was no significant difference in Aggf1 expression levels in the cells treated with AAV-Ag and AAV-Ag-FL (Figures 7A and 7B). Furthermore, overexpression of miR-101, miR-105, and miR-93 did not change AGGF1 expression in cells treated with AAV-Ag-FLΔ compared with that observed in the presence of miScr (Figures 7A and 7B). Significantly increased expression of Mcl1 was observed only after AAV-Ag-FL treatment but not in the cells treated with either AAV-Ag or AAV-Ag-FLΔ (Figures 7A and 7B).
Figure 7.
The Cardio-Protective Role of Aggf1 3′ UTR
(A and B) Mouse neocardiomyocytes were incubated with different AAVs and then transfected with miR-Scr or miR-105/101/93. Western blot (A) and real-time PCR (B) analyses were performed for Aggf1 and Mcl1 expression after treatment. Statistical analysis was carried out with a Student’s t test. n = 6. *p < 0.05 versus AAV-GFP; #p < 0.05 versus miR-Scr; &p < 0.05 versus AAV-Ag; $p < 0.05 versus AAV-Ag-FL. (C–F) Mouse neocardiomyocytes were incubated with different AAVs and then treated with vehicle or AngII for 24 h. The protein (C) and mRNA (D) levels were measured. The representative images from TUNEL staining (E) and the ratios for Bax/Bcl2 mRNA levels (F) are shown. Statistical analysis was carried out using one-way ANOVA with Tukey’s test. n = 6. *p < 0.05 versus AAV-GFP; #p < 0.05 versus vehicle; &p < 0.05 versus AAV-Ag; $p < 0.05 versus AAV-Ag-FL.
The reduction in Aggf1 expression after treatment with AngII in neocardiomyocytes was consistent with previous results. During incubation with the vehicle, AAV-Ag induced higher expression of Aggf1 than did AAV-GFP, and high expression of Aggf1 was observed in cells incubated with AAV-Ag-FL and AAV-Ag-FLΔ (Figures 7C and 7D). During incubation with AngII, although a significantly lower Aggf1 expression in neocardiomyocytes was observed in the case of AAV-Ag-FLΔ compared to that achieved after AAV-Ag-FL treatment, there was no significant difference in Aggf1 expression between AAV-Ag and AAV-Ag-FLΔ incubation (Figures 7C and 7D). Additionally, incubation with AAV-Ag-FL elevated Mcl1 expression in both vehicle- and AngII-treated neocardiomyocytes, compared to that observed after exposure to AAV-GFP, whereas treatments with AAV-Ag or AAV-Ag-FLΔ had no effect (Figures 7C and 7D). Similar results were also observed for the secreted Aggf1 levels (Figure S15A). In addition, AAV-Ag-FL suppressed the increase in miR-105, miR-101, and miR-93 expression levels induced by AngII treatment in neocardiomyocytes, whereas AAV-Ag or AAV-Ag-FLΔ did not (Figure S15B).
Overexpression of the Full-Length Aggf1 Prevents AngII-Induced Cardiac Apoptosis
To verify anti-apoptotic effects of AAV-Ag and AAV-Ag-FL in AngII-treated neocardiomyocytes, TUNEL staining was performed, which showed that AAV-Ag treatment inhibited AngII-induced increase in TUNEL-positive (apoptotic) cells, whereas AAV-Ag-FL administration led to the normal level of TUNEL-positive signaling (Figure 7E). However, there was no significant difference in TUNEL-positive cell numbers between treatments with AAV-Ag and AAV-Ag-FLΔ (Figure 7E). Consistent with TUNEL-staining results, the ratio of Bax/Bcl2 mRNA levels was higher in cells that underwent AngII treatment (Figure 7F). AAV-Ag treatment downregulated the increase in this ratio, whereas AAV-Ag-FL exposure reversed the ratio increase back to the normal level (Figure 7F). These results showed that AAV-Ag-FL had more powerful cardioprotective effects than AAV-Ag in mouse neocardiomyocytes in response to AngII-induced stress.
Discussion
In this study, we found that, in AngII-infused hypertensive mice, cardiac function was rescued by the overexpression of a full-length Aggf1 construct that included both the complete coding sequence and its non-coding 3′ UTR, compared with only the Aggf1 coding sequence, due to the upregulation of Mcl1 expression level, indicating that the 3′ UTR non-coding sequences act as trans-regulatory elements and play a cardioprotective role in cardiac dysfunction. Mechanistically, in vitro characterization corroborated Mcl1 as an Aggf1 ceRNA and showed that these genes share 3′ UTRs binding sites for the same miRNAs, including miR-105, miR-101, and miR-93, which were upregulated in response to hypertension/AngII.
We demonstrated that Mcl1 and Aggf1 regulated each other’s expression via their 3′ UTRs. Aggf1 overexpression caused an increase for Aggf1 expression level, whereas overexpression of the full-length Aggf1 constructs that contained the complete coding sequence and its 3′ UTR upregulated Aggf1 and Mcl1 expression both in vivo and in vitro. Furthermore, we directly confirmed that the ectopic expression of either the Aggf1 3′ UTR or the Mcl1 3′ UTR increased Mcl1 and Aggf1 expression. These data clearly showed that 3′ UTRs regulated gene expression. Notably, off-target effects always occur in the process of siRNA interventions. Based on our results, we propose that false-positive off-target effects of siRNAs occur because, with a decrease in the expression of siRNA-targeted genes, the expression levels of their ceRNAs are concomitantly diminished. Targeted mRNA degradation due to siRNA-based intervention may result in increased levels of certain endogenous miRNAs, which, in turn, leads to decreased expression of corresponding ceRNAs or genes that share the same MREs. Interestingly, in AGGF1-deficient HEK293 cells, ectopic expression of the Aggf1 3′ UTR increased MCL1 expression, indicating that upregulation of MCL1 is caused by the Aggf1 3′ UTR via an AGGF1-independent pathway.
It is becoming generally accepted that some ceRNAs regulate the stability of other protein-coding mRNAs through the competitive sequestration of miRNA binding.13,14 It should be noted that dysregulation of the levels of these miRNAs is associated with heart dysfunction and cardiac dysfunction. The expression of miR-101 in the peri-infarct area is decreased at 4 weeks after coronary artery ligation in rats.15 Because of the reduction of Mcl1 and Aggf1 expression levels in response to AngII/hypertension, increased levels of miRNAs such as miR-105, miR-101, and miR-93 were observed. Deficiency in these three miRNAs repressed the reduction of Mcl1 and Aggf1 expression caused by AngII treatment. These results provide further support to our hypothesis that protein-coding mRNAs communicate and co-regulate each other through the competition for miRNAs that target both transcripts.14,16,17 Importantly, as regulation through miRNA sequestration is solely based on MREs, our findings ascribe a predictable and protein-coding-independent function to mRNA molecules.17
Currently, although different chemical modifications have been developed for anti-miRNAs to optimize their biological properties and make them more suitable, their clinical application for treatment is facing tremendous challenges, i.e., non-specific biodistribution and inefficient endocytosis by target cells. Additionally, AAV 3′ UTRs achieve efficient and sustained expression, containing multiple miRNA-binding sites, and affecting multiple relative miRNA levels. Furthermore, in addition to the serotypes of AAV delivery systems, tissue-specific promoters can be considered to maximize ectopic expression in certain cell types. In this manner, the AAV-3′ UTR is a potential clinical strategy for the treatment of cardiac dysfunction.
We showed that the downregulation of MCL1 and AGGF1 expression levels in the hypertensive heart occurs via the upregulation of miR-105, miR-101, and miR-93. Furthermore, we demonstrated that protein-coding Mcl1 and Aggf1 mRNAs communicate and co-regulate each other through competition for those miRNAs that target both transcripts. Given that 3′ UTRs of both Mcl1 and Aggf1 mRNAs appear to exhibit trans-regulatory effects on the regulation of gene expression, we hypothesize that Aggf1 non-coding 3′ UTR sequences may accelerate the cardio-protective role of Aggf1 in AngII-induced cardiac dysfunction. We propose that this novel regulatory mechanism could be utilized as a novel gene therapy approach for cardiovascular and other diseases.
Materials and Methods
Mouse Model
Male C57BL/6 mice were housed in the controlled environment, with regulation of temperature (22°C ± 1°C) and humidity (55%) and a 12-h:12-h dark-light cycle, and were given unrestricted access to food and water. All animal studies were approved by the Animal Care and Use Committee of Central South University and were carried out in accordance with the principles provided by the NIH Guidelines.
The cardiac dysfunction mouse model was generated by AngII infusion. AngII (1 mg/kg/day, Sigma) or saline vehicle was infused via subcutaneous osmotic minipumps (Alzet, Cupertino, CA, USA) for 6 weeks as described previously.18 Blood pressure was monitored in conscious and calm mice by the non-invasive tail-cuff method using plethysmography (Kent Scientific, Torrington, CT, USA) according to the manufacturer’s instructions.19
Ultrasound Measurement
Cardiac structure and function were dynamically measured in mice by echocardiography with a high-resolution ultrasound system (Vevo 2100, Visual Sonics, Toronto, ON, Canada) as described previously.7 Mice were anesthetized with 1% isoflurane in oxygen, and the depth of anesthesia was confirmed by a stable heart rate (400–500 bpm) and a lack of flexor response to a paw-pinch. Subsequently, mice were fixed on a heated pad to maintain their body temperature at approximately 37°C to avoid interference caused by temperature fluctuation. Heart rate and other physiological parameters were continuously monitored by electrocardiogram (ECG) electrodes. A transducer (MS550D) was placed along the chest to obtain the parasternal long-axis images, and then the probe was rotated to acquire short-axis images at the level of the mid-papillary muscles. To obtain high-quality images, image acquisition was performed while avoiding excessive pressure over the sternum.8
All images were acquired and analyzed by the same blinded investigator according to the American Society of Echocardiography or calculated for three consecutive cardiac cycles and then averaged.
Isolation and Culture of Mouse Neocardiomyocytes
Neonatal mouse cardiomyocytes were isolated and cultured with the Pierce Primary Cardiomyocyte Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, hearts harvested from mice 1–3 days old were digested simultaneously.8 The collected enzyme solution was centrifuged, followed by discarding of the supernatant. Then, the cardiomyocytes were re-suspended in growth medium with 10% serum. After 24 h, fresh growth medium was added to the cells. The cells were cultured for 48 h and then incubated with AAV—either AAV-GFP, AAV-Mc, AAV-Ag, AAV-Mc-FL, AAV-Ag-FL, AAV-Mc-U, or AAV-Ag-U (MOI = 20)—for an additional 12 h.
Measurement of Secretory Aggf1 Protein Levels
In the cellular experiments, the supernatants from the mouse neocardiomyocytes were collected, and the level of Aggf1 in the supernatant was measured with an ELISA kit (LS-C46904, LSBio, Seattle, WA, USA).
Plasmids
The whole 3′ UTR of the mouse Aggf1 gene encoding Aggf1 was amplified by PCR using the following primers: 5′-atg cac tag tag gcc tgt ctg tca cac gg-3′ (forward) and 5′-atg cac gcg tta tgt tta tat cat tta-3′ (reverse). The 3′ UTR of Mcl1 was amplified with the forward primer 5′-atg cac tag tcc ttg tga gtg caa tag-3′ and the reverse primer 5′-atg cac gcg tgg ggg gaa aaa ggt-3′. These PCR products were digested with SpeI and MluI restriction enzymes and subcloned into the pLuc (pMIR-REPORT, as control) plasmid.
The site-deletion experiments were performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent), while multiple-site deletion was performed using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent). The primers were shown in Table S2. These plasmids were verified by DNA sequencing.
Transfections
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS) in a humidified incubator with 5% CO2 at 37°C. HEK293 cells were transfected with siRNAs, miRNA mimics, and plasmids using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Mouse neocardiomyocytes were transfected with siRNA (mouse Aggf1, A-051176-15; mouse Mcl1, A-062229-15; scramble, D-001910-02, Dharmacon, Lafayette, CO, USA), miRNAs (Table S3), or miRNA inhibitors (Table S4) using Nucleofector Kits for Primary Cardiac Cells (Lonza, Allendale, NJ, USA) according to the manufacturer’s instructions.
AAV Preparation
The AAV9 system was modified and adjusted according to the previous report. DNA sequences of the mouse Aggf1 gene encoding the Aggf1 protein with the 3′ UTR of mouse Aggf1 (Aggf1-FL) and mouse Mcl1 gene encoding Mcl1 protein with the 3′ UTR of mouse Mcl1 (Mcl1-FL) were synthesized from GenScript. Subsequently, the sequences were amplified by PCR. The primers were as follows: Aggf1-FL, forward, 5′-atc cac cgg tcg cca cca tgg cct cc-3′ and reverse, 5′-atg cgc ggc cgc tta tgt tta tat ca-3′; Aggf1, forward, 5′-atc cac cgg tcg cca cca tgg cct ccg ag-3′ and reverse, 5′-atg cgc ggc cgc ttt act ctg cag t-3′; and Aggf1-UTR, forward, 5′-atc cac cgg tcg cca cca ggc ctg t-3′ and reverse, 5′-atg cgc ggc cgc tta tgt tta-3′. These PCR products were digested with AgeI and NotI restriction enzymes and subcloned into the AAV9 plasmid.
We amplified the sequences of Mcl1 using the following specific primers: Mcl1-FL, forward, 5′-atc cac cgg tcg cca cca tgt ttg gcc-3′ and reverse, 5′-atg cgc ggc cgc tgg ggg gaa aaa ggt-3′; Mcl1, forward, 5′-atc cac cgg tcg cca cca tgt ttg gc-3′ and reverse, 5′-atg cgc ggc cgc tgg ggg gaa aaa gg-3′; and Mcl1-UTR, forward, 5′-atg cgc ggc cgc tct atc tta tta ga-3′ and reverse, 5′-atg cgc ggc cgc tgg ggg gaa aaa gg-3′. These PCR products were digested with AgeI and NotI restriction enzymes and subcloned into the AAV9 plasmid. The primers for site deletion were shown in Table S5.
AAV9 was packaged by triple plasmid cotransfection in HEK293 cells and purified as described previously.7,20, 21, 22 For in vivo experiments, 1 week after mini-pump implantation, the mice were injected intravenously with AAV-GFP, AAV-Ag, or AAV-Ag-FL once per week for 5 weeks, as described previously.23, 24, 25
Dual Luciferase Assays
HEK293 cells were cultured in 24-well plates. After 24 h, we co-transfected 200 ng of either pLuc (pMIR-REPORT, as control), pLuc-AG-U, or pLuc-MC-U together with 100 nM miRNA mimics or miRNA pools as well as 20 ng pRL-TK vector containing the Renilla luciferase gene (Promega, Madison, WI, USA) using Lipofectamine RNAiMAX according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Twelve hours after transfection, cells were incubated with AAVs for additional 36 h and harvested.
Cells were harvested and lysed using 1× Passive Lysis Buffer (Promega, Madison, WI, USA) and were used for luciferase assays according to the manufacturer’s instructions. Firefly and renilla luciferase activities were measured using the Dual-Glo Luciferase Assay Kit (Promega, Madison, WI, USA). The experiments were repeated at least five times.26
Western Blot Analysis
Cells or tissues were collected in RIPA buffer (Santa Cruz Biotechnology, Dallas, TX, USA), and total protein was quantified with a bicinchoninic acid assay (BCA; Thermo Fisher Scientific, Waltham, MA, USA). 30 μg protein lysate was loaded and separated with 8.5%–12.5% SDS-PAGE gel. After transfer to nitrocellulose membrane (EMD Millipore, Darmstadt, Germany), the membranes were blocked with 5% milk at room temperature for 1 h and then incubated with primary antibodies overnight at 4°C, followed by incubation with the suitable horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Waltham, MA, USA) was used to detect the bands. In all cases, equal loading was confirmed by probing blots against β-actin as an internal control. Primary antibodies and dilutions used were as follows: Mcl1 (1:1,000, # ab32087, Abcam, Cambridge, UK), Aggf1 (1:1,000; #11889-1-AP, Proteintech, Rosemont, IL, USA), and β-actin (1:1,000; Santa Cruz Biotechnology, Dallas, TX, USA).
Real-Time qRT-PCR Assays
Total RNA from cells or tissues was extracted in TRIzol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. Total RNA (2 μg) was reverse transcribed using the ImProm-II Reverse Transcription System (Promega, WI, USA). The primers of mRNA and the Power SYBR Green Master Mix (Life Technologies) were used for real-time PCR assay with the LightCycler1.5 Instrument (Roche, Mannheim, Germany). Each sample has triplicate measurements. U6 small nuclear RNA was used as the housekeeping gene to miRNAs (Table S6), while β-actin was used as the endogenous housekeeping gene to mRNAs (Table S7). Data were analyzed using the −ΔΔCt method, comparing threshold cycles first to housekeeping gene expression and then ΔCt of target genes in controls.
Histological Analysis
Hearts were excised from mice, embedded in paraffin, sectioned into 5-μm slices, and stained with Masson’s trichrome. Six regions were randomly studied for each mouse and imaged using a microscope. These images were quantified using the Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA). All measurements were performed in a blinded manner.27
Measurement of Caspase-3 Activity
As described in detail previously, caspase-3 activity in myocardial tissues was analyzed with a caspase-3 fluorescent kit (BIOMOL Research Laboratories).7
TUNEL Assay
Mouse hearts were excised and fixed in optimal cutting temperature (OCT) compound, cut into 6-μm-thick sections, and used for a TUNEL assay using the In Situ Cell Death Detection Kit (Roche Diagnostics, Mannheim, Germany). The images were visualized and captured under a fluorescence microscope. More than eight fields in four different sections were examined for each mouse by a researcher who was blinded to the treatments. The percentage of TUNEL-positive cells of the total number of nuclei as determined by DAPI staining (blue) was analyzed. Heart sections incubated with the labeling solution but without terminal transferase were used as negative controls.7
Statistics
Statistical analyses were performed using Prism 6 (GraphPad) software. All data are expressed as means ± SD. Student’s t test was used for two-group comparisons. For comparisons of more than two groups, one-way ANOVA and one-way ANOVA by Tukey’s test were used for normal distributions and non-normal distributions, respectively. p < 0.05 was considered significant.
Author Contributions
L.D. and Q.L. designed experiments; L.D., C.O., Y.Z., D.L., and S.L. performed experiments; S.L., L.D., C.O., Y.Z., Z.M., and Q.L. analyzed data; S.L. provided statistical support; L.D., S.L., and Q.L. wrote the manuscript.
Conflicts of Interests
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81970414 to Q.L. and 81500720 to L.D.), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJA350001 to Q.L.), and the Health Commission of Hubei Province (WJ2017M248 to C.O.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.02.002.
Supplemental Information
References
- 1.Drazner M.H. The progression of hypertensive heart disease. Circulation. 2011;123:327–334. doi: 10.1161/CIRCULATIONAHA.108.845792. [DOI] [PubMed] [Google Scholar]
- 2.Mann D.L. Inflammatory mediators and the failing heart: past, present, and the foreseeable future. Circ. Res. 2002;91:988–998. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 3.Jia L., Li Y., Xiao C., Du J. Angiotensin II induces inflammation leading to cardiac remodeling. Front. Biosci. 2012;17:221–231. doi: 10.2741/3923. [DOI] [PubMed] [Google Scholar]
- 4.Shahbaz A.U., Sun Y., Bhattacharya S.K., Ahokas R.A., Gerling I.C., McGee J.E., Weber K.T. Fibrosis in hypertensive heart disease: molecular pathways and cardioprotective strategies. J. Hypertens. 2010;28(Suppl 1):S25–S32. doi: 10.1097/01.hjh.0000388491.35836.d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian X.L., Kadaba R., You S.A., Liu M., Timur A.A., Yang L., Chen Q., Szafranski P., Rao S., Wu L. Identification of an angiogenic factor that when mutated causes susceptibility to Klippel-Trenaunay syndrome. Nature. 2004;427:640–645. doi: 10.1038/nature02320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Yang H., Song L., Li N., Han Q.Y., Tian C., Gao E., Du J., Xia Y.L., Li H.H. AGGF1 protects from myocardial ischemia/reperfusion injury by regulating myocardial apoptosis and angiogenesis. Apoptosis. 2014;19:1254–1268. doi: 10.1007/s10495-014-1001-4. [DOI] [PubMed] [Google Scholar]
- 7.Lu Q., Yao Y., Hu Z., Hu C., Song Q., Ye J., Xu C., Wang A.Z., Chen Q., Wang Q.K. Angiogenic factor AGGF1 activates autophagy with an essential role in therapeutic angiogenesis for heart disease. PLoS Biol. 2016;14:e1002529. doi: 10.1371/journal.pbio.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y., Lu Q., Hu Z., Yu Y., Chen Q., Wang Q.K. A non-canonical pathway regulates ER stress signaling and blocks ER stress-induced apoptosis and heart failure. Nat. Commun. 2017;8:133. doi: 10.1038/s41467-017-00171-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas R.L., Gustafsson A.B. MCL1 is critical for mitochondrial function and autophagy in the heart. Autophagy. 2013;9:1902–1903. doi: 10.4161/auto.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas R.L., Roberts D.J., Kubli D.A., Lee Y., Quinsay M.N., Owens J.B., Fischer K.M., Sussman M.A., Miyamoto S., Gustafsson Å.B. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 2013;27:1365–1377. doi: 10.1101/gad.215871.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Bathina M., Lynch J., Koss B., Calabrese C., Frase S., Schuetz J.D., Rehg J.E., Opferman J.T. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 2013;27:1351–1364. doi: 10.1101/gad.215855.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Wang H.X., Li Y.L., Zhang C.C., Zhou C.Y., Wang L., Xia Y.L., Du J., Li H.H. MicroRNA Let-7i negatively regulates cardiac inflammation and fibrosis. Hypertension. 2015;66:776–785. doi: 10.1161/HYPERTENSIONAHA.115.05548. [DOI] [PubMed] [Google Scholar]
- 13.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Z., Sun X., Shan H., Wang N., Wang J., Ren J., Feng S., Xie L., Lu C., Yuan Y. MicroRNA-101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor-β1 pathway. Circulation. 2012;126:840–850. doi: 10.1161/CIRCULATIONAHA.112.094524. [DOI] [PubMed] [Google Scholar]
- 16.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karreth F.A., Tay Y., Perna D., Ala U., Tan S.M., Rust A.G., De Nicola G., Webster K.A., Weiss D., Perez-Mancera P.A. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowley S.D., Gurley S.B., Herrera M.J., Ruiz P., Griffiths R., Kumar A.P., Kim H.S., Smithies O., Le T.H., Coffman T.M. Angiotensin II causes hypertension and cardiac hypertrophy through its receptors in the kidney. Proc. Natl. Acad. Sci. USA. 2006;103:17985–17990. doi: 10.1073/pnas.0605545103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilde E., Aubdool A.A., Thakore P., Baldissera L., Jr., Alawi K.M., Keeble J., Nandi M., Brain S.D. Tail-cuff technique and its influence on central blood pressure in the mouse. J. Am. Heart Assoc. 2017;6:e005204. doi: 10.1161/JAHA.116.005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki K., Fuess S., Storm T.A., Gibson G.A., Mctiernan C.F., Kay M.A., Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bostick B., Ghosh A., Yue Y., Long C., Duan D. Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Ther. 2007;14:1605–1609. doi: 10.1038/sj.gt.3303029. [DOI] [PubMed] [Google Scholar]
- 22.Nakai H., Fuess S., Storm T.A., Muramatsu S., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 24.Li H., Zhang X., Wang F., Zhou L., Yin Z., Fan J., Nie X., Wang P., Fu X.D., Chen C., Wang D.W. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation. 2016;134:734–751. doi: 10.1161/CIRCULATIONAHA.116.023926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F., Chen C.L., Qian J.Q., Yan J.T., Cianflone K., Xiao X., Wang D.W. Long-term modifications of blood pressure in normotensive and spontaneously hypertensive rats by gene delivery of rAAV-mediated cytochrome P450 arachidonic acid hydroxylase. Cell Res. 2005;15:717–724. doi: 10.1038/sj.cr.7290341. [DOI] [PubMed] [Google Scholar]
- 26.Lu Q., Xie Z., Yan C., Ding Y., Ma Z., Wu S., Qiu Y., Cossette S.M., Bordas M., Ramchandran R., Zou M.H. SNRK (sucrose nonfermenting 1-related kinase) promotes angiogenesis in vivo. Arterioscler. Thromb. Vasc. Biol. 2018;38:373–385. doi: 10.1161/ATVBAHA.117.309834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S., Lu Q., Wang Q., Ding Y., Ma Z., Mao X., Huang K., Xie Z., Zou M.H. Binding of FUN14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation. 2017;136:2248–2266. doi: 10.1161/CIRCULATIONAHA.117.030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.