Short abstract
Objectives
Bioinformatics analysis revealed a potential interaction between long noncoding (lnc)RNA TUG1 (taurine-upregulated gene 1) and microRNA (miR)-27a. miR-27a can promote sepsis by upregulating tumor necrosis factor-α (TNF-α). Our objective was to study the roles of TUG1 in sepsis.
Methods
Plasma levels of TUG1 in patients with sepsis and in healthy controls were measured by quantitative PCR assay. The IntaRNA program was used to predict potential interactions between TUG1 mRNA and miR-27a. The interaction between TUG1 and miR-27a was further explored by transfecting TUG1 expression vector or miR-27a mimic into AC16 human cardiomyocytes, and apoptosis was evaluated by cell apoptosis assay.
Results
TUG1 was downregulated in patients with sepsis. TUG1 was able to directly interact with miR-27a, but overexpression of TUG1 or miR-27a failed to affect the expression of each other. In contrast, TUG1 overexpression led to decreased levels of TNF-α, whereas miR-27a overexpression increased TNF-α in cardiomyocytes. Cell apoptosis analysis showed that TNF-α and miR-27a overexpression promoted apoptosis of cardiomyocytes induced by lipopolysaccharide. TUG1 overexpression had the opposite effect and attenuated the effects of TNF-α and miR-27a overexpression.
Conclusion
We conclude that TUG1 is downregulated in sepsis and may act as a molecular “sponge” of miR-27a to downregulate TNF-α.
Keywords: Sepsis, survival, taurine-upregulated gene 1 (TUG1), miR-27a, tumor necrosis factor-α (TNF-α), long noncoding RNA
Introduction
Sepsis is caused by dysregulated immune and systemic inflammatory responses to microbial infections that cause organ injury.1 Sepsis has a mortality rate of 15% to 25%. In addition, the long-term course of sepsis results in immune suppression, prolonged inflammation, lean tissue wasting, and systemic organ injury.1,2 With the development of novel diagnostic and therapeutic approaches, such as early diagnostic markers, rapid effective delivery of antibiotics, and fluid resuscitation, the overall short-term survival of sepsis has improved significantly.3–5 However, patients who survive sepsis still have a high risk of mortality and long-term functional and cognitive deficits.6 Therefore, novel therapies are needed.
Sepsis is a type of inflammatory disease and its development requires the involvement of multiple inflammatory factors, such as interleukin-3 and tumor necrosis factor-α (TNF-α).7,8 Inhibition of the production of proinflammatory cytokines is a promising approach for the treatment of sepsis.9 Recent studies on the pathogenesis of sepsis revealed the participation of noncoding RNAs, such as microRNAs and long (>200 nt) noncoding RNAs (lncRNAs).10,11 For instance, miR-27a upregulates the gene encoding TNF-α (TNFA) to relieve the inflammation of sepsis.12 Our preliminary bioinformatics analysis revealed the potential interaction between miR-27a and lncRNA TUG1 (taurine-upregulated gene 1); this interaction of miR-27a and TUG1 has a protective role in cardiomyocyte ischemia reperfusion.13 This study was therefore carried out to explore possible interactions between miR-27a and TUG1 in sepsis.
Materials and methods
Participants and plasma
The present study enrolled 70 patients with sepsis (46 men and 24 women, ranging from 35 to 66 years, 50.3 ± 6.5 years) as well as 70 healthy controls (46 men and 24 women, ranging from 36 to 66 years, 50.5 ± 6.2 years) at the First Affiliated Hospital of Kunming Medical University from May 2017 to May 2019. Blood was extracted from each participant on the day of admission. Blood samples were centrifuged in EDTA tubes at room temperature at 1200 × g for 10 minutes to separate plasma. Plasma samples were immediately frozen in liquid nitrogen and stored in a liquid nitrogen sink before use. This study was approved by the review board of Ethics Committee of the First Affiliated Hospital of Kunming Medical University. All participants were informed of the principle of this study and they all signed informed consent.
Cardiomyocytes and transient transfections
Human cardiomyocyte cell line AC16 (Sigma-Aldrich, St. Louis, MO, USA) was used in this study. Cardiomyocyte growth medium (ScienCell Research, Carlsbad, CA, USA) containing 10% fetal bovine serum was used to culture AC16 cells at 37°C under 95% humidity and 5% CO2. Cells were harvested at 75% to 85% confluence to perform subsequent transient transfections.
TUG1 and TNFA expression vectors were constructed using pcDNA3.1 vector (Sigma-Aldrich) as backbone. Negative control (NC) miRNA and miR-27a mimic were synthesized by GenePharma (Shanghai, China). AC16 cells were harvested and counted and then 106 cells were transfected with 10 nM vector or 45 nM short interfering (si)RNA using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The NC cells were transfected with empty pcDNA3.1 vector or NC miRNA. Control (C) cells were cells that did not undergo transfection.
Dual luciferase reporter assay
The full length TUG1 cDNA was cloned into pGL3 Luciferase Reporter Vector (Promega, Madison, WI, USA) to make the TUG1 vector. AC16 cells were transfected with TUG1 vector + NC miRNA (NC group) or TUG1 vector + miR-27a mimic (miR-27a group). Cells were cultivated at 37°C under 95% humidity and 5% CO2 for 48 hours, and then relative luciferase activity was measured using a dual luciferase reporter assay kit (Promega). Renilla luciferase was used to normalize firefly luciferase activity.
RNA extractions and PCR assays
Plasma (0.2 mL) or 105 cells were mixed with 1 mL of Trizol reagent (Invitrogen, Carlsbad, CA, USA) to extract total RNAs. For the lipopolysaccharide (LPS) treatment, AC16 cells were incubated with LPS at 0, 100, 200, or 300 ng/mL for 48 hours before RNA isolation. All RNA samples were digested with DNA eraser (Takara Bio, Kyoto, Japan) to completely remove genomic DNA. QuantiTect reverse transcription kit (Qiagen, Valencia, CA, USA) was used to perform reverse transcription with RNA samples as template, followed by real-time quantitative (q)PCR performed using QuantiTect SYBR Green PCR Kit (Qiagen) with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as endogenous control to measure the expression levels of TUG1 and TNFA mRNA. A PureLink miRNA Isolation Kit (Thermo Fisher Scientific) was used to extract miRNAs, followed by the measurement of expression levels of miR-27a with U6 as endogenous control using All-in-One miRNA qRT-PCR Detection Kit (Genecopoeia, Rockville, MD, USA). All PCR reactions were repeated 3 times, and fold changes of gene expression were calculated using the 2−ΔΔCt method.
Protein extraction and western blot
Phosphate-buffered saline was used to wash AC16 cells twice. Then, 105 AC16 cells were mixed with 1 mL of radioimmunoprecipitation assay (RIPA) solution (Sigma-Aldrich) to prepare cell lysates. Protein concentrations were measured using the bicinchoninic acid (BCA) assay (Sigma-Aldrich), followed by protein denaturation in boiling water for 10 minutes. Protein molecules were separated using a 10% sodium dodecyl sulfate-PAGE gel, followed by gel transfer to polyvinylidene difluoride membranes. Blocking was performed using PBS containing 5% nonfat milk for 2 hours at room temperature. After that, membranes were first incubated with rabbit primary antibodies to GAPDH (ab9845, Abcam, Cambridge, UK) and TNF-α (ab9635, Abcam) for 12 hours at 4°C. Then, membranes were further incubated with secondary antibodies of horseradish peroxidase (HRP)-conjugated goat anti-rabbit (IgG; ab6721, Abcam) for 2 hours at room temperature. Signals were then produced using ECL chemiluminescence kit (Pierce Biotechnology, Rockford, IL, USA), and ImageJ v1.48 software (National Institutes of Health, Bethesda, MD, USA) was used to normalize signals.
Cell apoptosis assay
AC16 cells were harvested 24 hours after transfection and incubated with cardiomyocyte medium with 10% fetal bovine serum containing 300 g/mL LPS for 48 hours. Then, the medium was aspirated and cells were resuspended in chilled PBS. The fluorescein isothiocyanate (FITC) annexin V Apoptosis Detection Kit with propidium iodide (PI) (BioLegend, San Diego, CA, USA) was used to stain the cells according to the protocol from BioLegend. Finally, apoptotic cells were detected by flow cytometry (FACSCalibur; Becton-Dickinson, Franklin Lakes, NJ, USA).
Statistical analysis
All experiments were performed in three independent biological replicates. Mean values of the data were processed by Graphpad Prism 6 software (GraphPad Inc., San Diego, CA, USA). Differences between two groups were explored using an unpaired t-test. Explorations of differences among multiple groups were performed by one-way ANOVA and Tukey test. A P-value < 0.05 was considered to indicate statistical significance.
Results
Plasma TUG1 was downregulated in sepsis
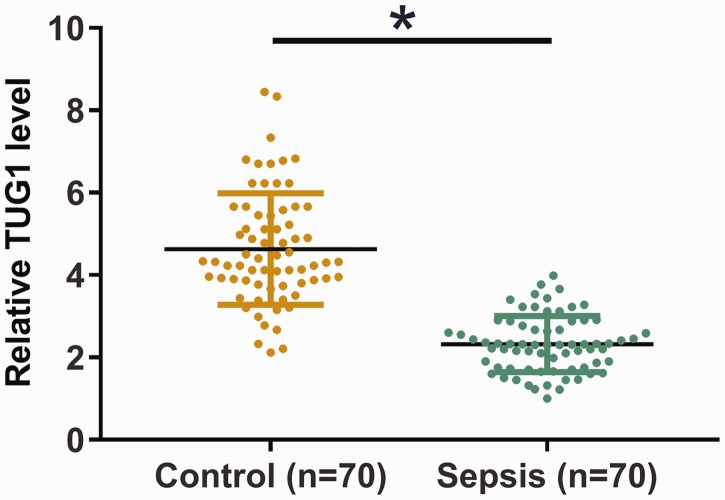
Plasma levels of TUG1 in patients with sepsis (n = 70) and in healthy controls (n = 70) were measured by qPCR. Plasma levels of TUG1 were significantly lower in the sepsis group than in the control group (Figure 1, P < 0.05), indicating the involvement of TUG1 in sepsis.
Figure 1.
Plasma TUG1 is downregulated in sepsis. Plasma levels of TUG1 in sepsis patients (n = 70) and healthy controls (n = 70) were measured using qPCR. Data were compared between two groups by an unpaired t-test. The PCR was repeated three times and data are expressed as mean values. The bars represent the mean value of the two groups, the whiskers represent the standard deviations of all data in each group, and the data points indicate the exact value of each sample in the groups. *P < 0.05. TUG1, taurine-upregulated gene 1; qPCR, real-time quantitative PCR.
TUG1 directly interacted with miR-27a
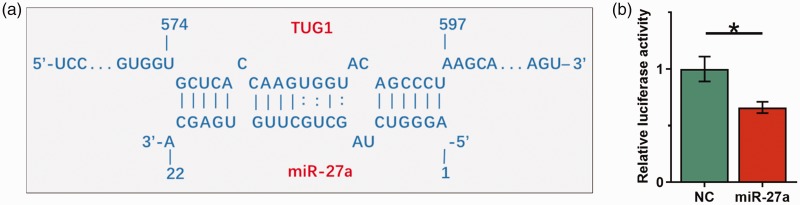
IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) was used to predict potential interactions between TUG1 and miR-27a. We observed that TUG1 and miR-27a may interact through base pairing (Figure 2a). The dual luciferase reporter assay was performed by transfecting TUG1 vector + NC miRNA (NC group) or TUG1 vector + miR-27a mimic (miR-27a group) into AC16 cells. Significantly lower relative luciferase activity was observed in the miR-27a group than in the NC group (Figure 2b, P < 0.05).
Figure 2.
TUG1 directly interacted with miR-27a. IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) was used to predict the potential interaction between TUG1 and miR-27a (a). Dual luciferase reporter assay was performed by transfecting AC16 cells with TUG1 vector + NC miRNA (NC group) or TUG1 vector + miR-27a mimic (miR-27a group) (b). Data were compared by unpaired t-test. Experiments were repeated three times and mean values are shown; error bars represent standard deviations. *P < 0.05. TUG1, taurine-upregulated gene 1; AC16, human cardiomyocyte cell line; NC, negative control.
TUG1 overexpression led to decreased TNFA expression
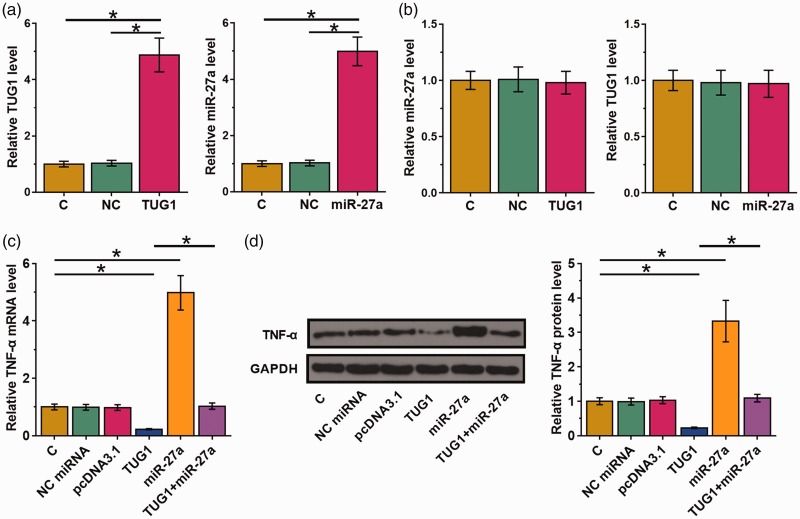
The interaction between TUG1 and miR-27a was further explored by transfecting TUG1 expression vector or miR-27a mimic into AC16 cells. Overexpression of TUG1 and miR-27a was confirmed by qPCR at 24 hours post-transfection (Figure 3a, P < 0.05). Compared with the C and NC groups, overexpression of TUG1 or miR-27a failed to affect the expression of each other (Figure 3b). The function of miR-27a in inflammatory responses is mainly mediated by upregulation of TNFA. Therefore, effects of TUG1 and miR-27a on the expression of TNFA in AC16 cells were analyzed by qPCR and western blot at the mRNA (Figure 3c) and protein (Figure 3d) levels, respectively. We observed that miR-27a overexpression led to upregulation of TNFA (P < 0.05). TUG1 overexpression showed the opposite and reduced the effects of miR-27a overexpression (P < 0.05).
Figure 3.
TUG1 overexpression led to decreased TNFA mRNA expression. The interaction between TUG1 and miR-27a was further explored by transfecting TUG1 expression vector or miR-27a mimic into AC16 cells. Overexpression of TUG1 and miR-27a was confirmed by qPCR at 24 hours post-transfection (a). The effect of TUG1 or miR-27a overexpression on the expression of each other was also analyzed by qPCR at 24 hours post-transfection (b). Effects of TUG1 and miR-27a on the expression of TNFA in AC16 cells were analyzed by qPCR and western blot at the mRNA (c) and protein (d) levels, respectively. Experiments were repeated three times and mean values are shown; error bars represent standard deviations. *P < 0.05. TNFA, tumor necrosis factor-α gene; TUG1, taurine-upregulated gene 1; AC16, human cardiomyocyte cell line; qPCR, real-time quantitative PCR.
TUG1 inhibits LPS-induced apoptosis of AC16 cells through TNF-α and miR-27a
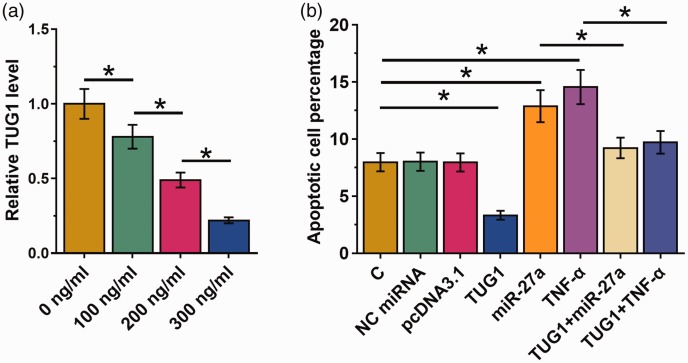
AC16 cells were first incubated with LPS at 0, 100, 200 or 300 ng/mL for 48 hours, and then TUG1 expression was measured by qPCR assay. The LPS treatment downregulated TUG1 expression in a dose-dependent manner (Figure 4A, P < 0.05). A cell apoptosis assay was performed to analyze the effects of transfection on the apoptosis of AC16 cells. Cell apoptosis analysis showed that TNFA and miR-27a overexpression promoted the apoptosis of cardiomyocytes induced by LPS. TUG1 overexpression had the opposite effect and attenuated the effects of TNFA and miR-27a overexpression (Figure 4b, P < 0.05).
Figure 4.
TUG1 inhibits LPS-induced apoptosis of AC16 cells through TNFA and miR-27a. AC16 cells were first incubated with LPS at doses of 0, 100, 200 or 300 ng/mL for 48 hours, followed by the measurement of TUG1 expression by qPCR assay (a). Cell apoptosis assay was performed to analyze the effects of transfections on the apoptosis of AC16 cells (b). Experiments were repeated three times and mean values are shown; error bars represent standard deviations. *P < 0.05. LPS, lipopolysaccharide; TNFA, tumor necrosis factor-α; TUG1, taurine-upregulated gene 1; AC16, human cardiomyocyte cell line; qPCR, real-time quantitative PCR.
Discussion
This study mainly investigated the roles of TUG1 in sepsis. We found that TUG1 was downregulated in sepsis and may regulate the expression of TNFA through miR-27a to inhibit the apoptosis of cardiomyocytes induced by LPS.
The development of sepsis is accompanied by changes in expression levels of a large number of lncRNAs.14 Some differentially expressed lncRNAs have been shown to play critical roles in the development of sepsis.15,16 For instance, lncRNA MALAT1 interacts with the p38 MAPK/NFκB signaling pathway and miR-125b to regulate cardiac dysfunction and inflammation induced by sepsis.15 LncRNA H19 regulates the expression of miR-874 by competing with aquaporin 1 to participate in the development of LPS-induced sepsis.16 In this study, we reported for the first time the downregulation of TUG1 in sepsis, indicating its possible involvement in this disease.
The heart is one of the most commonly affected organs in sepsis, and heart failure caused by sepsis leads to an unacceptably high mortality rate.17 In this study, we observed a decreased rate of LPS-induced apoptosis of cardiomyocytes after TUG1 overexpression, indicating a potential protective role of TUG1 in sepsis-induced apoptosis of cardiomyocytes. Therefore, TUG1 overexpression may serve as a potential therapeutic target for the treatment of sepsis. However, clinical studies are needed to further test the clinical application of TUG1. In addition, sepsis induces dysregulation of multiple organs. More studies are needed to analyze the protective roles of TUG1 in other organs of sepsis patients.
In a recent study, Wang et al. reported that miR-27a was overexpressed in sepsis and can upregulate the expression of TNFA to promote the development of sepsis.12 Consistent with this, we observed upregulated TNFA expression in AC16 cells after miR-27a overexpression. In addition, we showed that TNFA and miR-27a interacted in the regulation of LPS-induced apoptosis of AC16 cells.
Interestingly, miR-27a is predicted to interact with TUG1, whereas overexpression of miR-27a failed to affect expression of TUG1. Therefore, TUG1 is unlikely to be a target of miR-27a. Instead, TUG1 overexpression led to decreased expression of TNFA. Therefore, TUG1 may sponge miR-27a to downregulate TNFA.
In conclusion, TUG1 is downregulated in sepsis and may sponge miR-27a to downregulate expression of TNFA, thereby inhibiting apoptosis of AC16 cells induced by LPS.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
We are grateful for financial support from Yunnan provincial health commission medical discipline leader training project (D201609).
ORCID iD
Jiang Duan https://orcid.org/0000-0002-5037-6415
References
- 1.Hotchkiss RS, Moldawer LL, Opal SM, et al. Sepsis and septic shock. Nat Rev Dis Primers 2016; 30: 16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 2013; 13: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotts JE, Matthay MA. Sepsis: pathophysiology and clinical management. BMJ 2016; 353: i1585. [DOI] [PubMed] [Google Scholar]
- 4.Polat G, Ugan RA, Cadirci E, et al. Sepsis and septic shock: current treatment strategies and new approaches. Eurasian J Med 2017; 49: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour CW, Gesten F, Prescott HC, et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med 2017; 376: 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Bahr V, Hultman J, Eksborg S, et al. Long-term survival in adults treated with extracorporeal membrane oxygenation for respiratory failure and sepsis. Crit Care Med 2017; 45: 164–170. [DOI] [PubMed] [Google Scholar]
- 7.Deng M, Loughran PA, Zhang L, et al. Shedding of the tumor necrosis factor (TNF) receptor from the surface of hepatocytes during sepsis limits inflammation through cGMP signaling. Sci Signal 2015; 8: ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber GF, Chousterman BG, He S, et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 2015; 347: 1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steeland S, Van Ryckeghem S, Vandewalle J, et al. Simultaneous inhibition of tumor necrosis factor receptor 1 and matrix metalloproteinase 8 completely protects against acute inflammation and sepsis. Crit Care Med 2018; 46: e67–e75. [DOI] [PubMed] [Google Scholar]
- 10.Ho J, Chan H, Wong SH, et al. The involvement of regulatory non-coding RNAs in sepsis: a systematic review. Crit Care 2016; 20: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meydan C, Bekenstein U, Soreq H. Molecular regulatory pathways link sepsis with metabolic syndrome: non-coding RNA elements underlying the sepsis/metabolic cross-talk. Front Mol Neurosci 2018; 11: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Ruan Z, Mao Y, et al. miR-27a is up regulated and promotes inflammatory response in sepsis. Cell Immunol 2014; 290: 190–195. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, Dong Z, Gao H. LncRNA TUG1 protects against cardiomyocyte ischaemia reperfusion injury by inhibiting HMGB1. Artif Cells Nanomed Biotechnol 2019; 47: 3511–3516. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrina D, Severino P, Barbeiro H, et al. Insights into the function of long noncoding RNAs in sepsis revealed by gene co-expression network analysis. Noncoding RNA 2017; 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Wang X, Yan X, et al. LncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFκB. Int Immunopharmacol 2018; 55: 69–76. [DOI] [PubMed] [Google Scholar]
- 16.Fang Y, Hu J, Wang Z, et al. LncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomed Pharmacother 2018; 105: 1183–1191. [DOI] [PubMed] [Google Scholar]
- 17.Arfaras-Melainis A, Polyzogopoulou E, Triposkiadis F, et al. Heart failure and sepsis: practical recommendations for the optimal management. Heart Fail Rev 2019; 21. doi: 10.1007/s10741-019-09816-y. [DOI] [PubMed] [Google Scholar]