Abstract
Two series of rhodanine-3-acetic and rhodanine-3-propionic acids derivatives having benzylidene and cinnamylidene substituents with additional electron donating and withdrawing groups at the C-5 position, were synthesised. The structures of the obtained derivatives were confirmed by spectroscopic methods and their lipophilicity was screened. The crystal structures were determined for selected compounds. The antibacterial activity of the derivatives was depended on the type of carboxyalkyl group in the N-3 position and on the type of the substituent in the C-5 position. The derivatives of rhodanine-3-propionic acid demonstrated the highest activity against Gram-positive bacteria. However, none of tested derivatives showed activity against Gram-negative bacteria and yeast. We believe that the presence of the N,N-diethylamine group in the aromatic system and the number of carbon atoms in the carboxyalkyl group is more significant for the biological activity than the fact that the benzylidene or cinnamylidene substituent was present at the C-5 position.
Keywords: Rhodanine, Rhodanine-3-carboxyalkyl acids, Antibacterial activity, Antimicrobial activity, Thiazolidine-4-one
1. Introduction
The rapid growth of hospital-acquired infections caused by the bacteria strains resistant to the contemporary antibiotics is one of the greatest challenges of modern antibiotic therapy. Multiple drug-resistant microorganisms, mainly from the Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa strains and the Enterobacteriaceae family, are a major threat to patients (Nowakowicz-Dębek et al., 2016, Santajit and Indrawattana, 2016). The resistance of bacteria and yeast to antimicrobial agents is also a significant economic issue. Data gathered by physicians and epidemiologists allows to conclude that one of the reasons for drug resistance among bacteria is the excessive and irresponsible use of antibiotic therapy (Harris et al., 2002).
Many scientific centres have been conducting the research on discovering the new compounds that will successfully constrain the growth of pathogenic microorganisms. One of the substance groups studied are derivatives having 2-sulfanylidene-1,3-thiazolidin-4-one core, commonly known as rhodanine (Fig. 1).
Fig. 1.
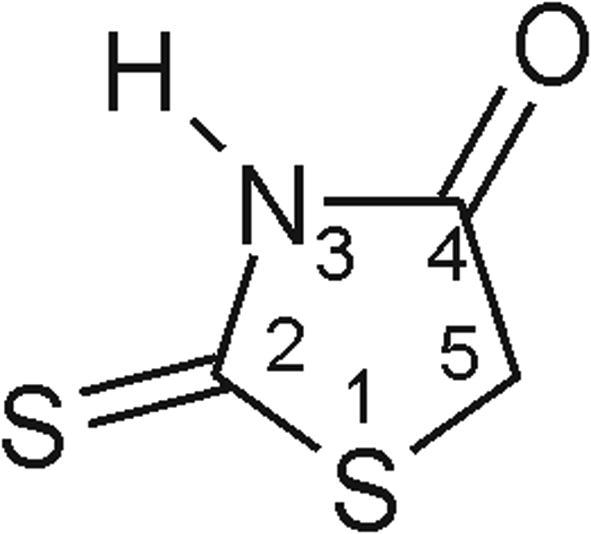
Structure of 2-sulfanylidene-1,3-thiazolidin-4-one (rhodanine).
Rhodanine derivatives have a broad spectrum of biological activity demonstrating antibacterial, antifungal, antiviral, antiparasitic, antidiabetic, antineoplastic and anti-inflammatory activity (Kaminsky et al., 2017).
There is a hope that rhodanine derivatives having a carboxyalkyl acid fragment at the N-3 position and arylidene substituents enriched with additional electron donating and withdrawing function groups at the C-5 position, could be used as drugs.
Shingate et al. studied the 5-benzylidene derivatives of rhodanine-3-acetic acid and its activity against Mycobacterium tuberculosis and Mycobactierium bovis (Subhedar et al., 2016). Several of the compounds investigated showed good activity against both tested bacterial strains.
Ravi and Sundaram examined the antibacterial properties of 5-benzylidene-rhodanine 3-propionic acid (Sundaram and Ravi, 2013a). The compounds they obtained demonstrated moderate properties against Gram-positive and Gram-negative bacteria. Derivatives of rhodanine-3-acetic acid were also used by Lesyk et al., who obtained the derivatives of 3-O-acyloleanolic acids, that demonstrated antineoplastic activity against nine types of cancer (Kaminsky et al., 2012).
Antibacterial and antineoplastic activities were also examined for the 5-benzylidene derivatives of rhodanine-3-isopropionic acid (Sundaram and Ravi, 2015b). Tested by them compounds with NO2 group in the ortho position of the aromatic ring system exhibited good antibacterial activity against MRSA. The research conducted by Miao et al. (2013) and Patel et al. (2013) teams demonstrated that the antibacterial activity of rhodanine-3-acetic derivatives was higher when there was a hydrophobic arylidene group having additional electron withdrawing substituents at the C-5 position.
Based on the subject matter literature, we decided to continue our research upon antibacterial activity of rhodanine-3-carboxyalkyl acids derivatives with various substituents at the C-5 position. In our previous research (Tejchman et al., 2017), we synthesized a series of carboxyalkylrhodanine acids derivatives having an acetic, propionic, butyric or hexanoic acid moiety at the N-3 position. The synthesised compounds underwent the Knoevenagel condensation with 4-diethylaminobenzoic, 4-dibutylaminobenzoic and 4-diphenylaminobenzoic aldehydes. We screened the compound series for their antibacterial activity against selected Gram-positive and Gram-negative bacteria strains. The examined derivatives demonstrated good activity against Gram-positive bacteria and low against Gram-negative ones. During our research, we concluded that the number of carbon atoms in the linker between carboxylic group and the N-3 nitrogen atom has little influence on the antibacterial activity of the investigated compounds. Among the examined group, derivatives having a shorter carbon linker at N-3 had better activity against Gram-positive bacteria than those with longer linker between the carboxylic group and the nitrogen atom. It was observed that the antibacterial activity of the investigated compounds depended on the size of the amino group at the 4′ position in the benzylidene substituent.
Based on our results and the corresponding literature references, we decided to synthesise two series of rhodanine-3-acetic and rhodanine-3-propionic acids derivatives. The first one had a benzylidene substituent with electron donating or withdrawing groups in the aromatic ring. The other had a cinnamylidene substituent with analogous groups in the aromatic ring at the C-5 position.
The compounds used for the investigation are presented in Table 1.
Table 1.
Chemical structures of the investigated compounds. (The compounds marked with an asterisk were previously examined by other teams. The scope of research has been described in the further part of the text).
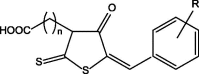 |
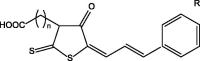 |
||||
|---|---|---|---|---|---|
| n | R | n | R | ||
| 4a* | 1 | 2-NO2 | 12a | 1 | 2-NO2 |
| 4b* | 1 | 3-NO2 | 12b | 1 | 4-NO2 |
| 4c* | 1 | 4-NO2 | 13a | 2 | 2-NO2 |
| 5a* | 2 | 2-NO2 | 13b | 2 | 4-NO2 |
| 5b* | 2 | 3-NO2 | 14a | 1 | 4-N(CH3)2 |
| 5c* | 2 | 4-NO2 | 14b | 1 | 4-N(C2H5)2 |
| 6a* | 1 | 4-N(CH3)2 | 15a | 2 | 4-N(CH3)2 |
| 6b* | 1 | 4-N(C2H5)2 | 15b | 2 | 4-N(C2H5)2 |
| 7a* | 2 | 4-N(CH3)2 | 16 | 1 | 4-Cl |
| 7b* | 2 | 4-N(C2H5)2 | 17 | 2 | 4-Cl |
| 8a | 1 | 2-OCH3 | |||
| 8b | 1 | 3-OCH3 | |||
| 8c* | 1 | 4-OCH3 | |||
| 9a* | 2 | 2-OCH3 | |||
| 9b | 2 | 3-OCH3 | |||
| 9c | 2 | 4-OCH3 | |||
| 10a | 1 | 2-Cl | |||
| 10b* | 1 | 3-Cl | |||
| 10c* | 1 | 4-Cl | |||
| 11a* | 2 | 2-Cl | |||
| 11b* | 2 | 3-Cl | |||
| 11c* | 2 | 4-Cl | |||
2. Experimental part
2.1. Materials and methods
All reagents for the synthesis of rhodanine derivatives were purchased from Sigma-Aldrich and used without subsequent purification.
Melting point (uncorrected) was determined on the Boetius apparatus. The IR spectrum was recorded with Jasco FT IR-670 Plus spectrophotometer in the KBr disk.
Mass spectrometry analysis was made using AmaZon ETD (Bremen, Germany) equipped with standard electrospray ion source and ion trap analyzer. Samples were dissolved in the mixture of methanol and chloroform (1:1, v/v) with addition of 0.1% formic acid (v/v). Usually, concentration of the sample in solution did not exceed 1 nM. Samples were introduced into a mass spectrometer using a syringe pump with a flow rate of 2 μl/min. Basic settings of the mass spectrometer were as follows: scan range 100–700 m/z, ion polarity: positive, heated capillary temperature: 280 °C, capillary voltage: −4500 V, ion trap was set to accumulate ca. 300 000 ions for a single cycle.
Fragmentation was done after manual ion selection with an isolation window set to 4 Da. Low energy collision induced dissociation (low energy CID) was used as a fragmentation technique with the collision energy of 1 eV. Typically for ion traps, fragment ion cut-off was set to 27% of the nominal m/z ratio of selected parent ion. Fragmentation spectra were acquired for about 20 s (equal to ca. 20 spectra) and accumulated.
The NMR spectra were obtained in CDCl3 on the Bruker Avance III HD spectrometer operating at 400.17 MHz (1H) and 100.62 MHz (13C); the chemical shifts (ppm) were referenced to lock out the signal of the solvent, and J was expressed in Hz.
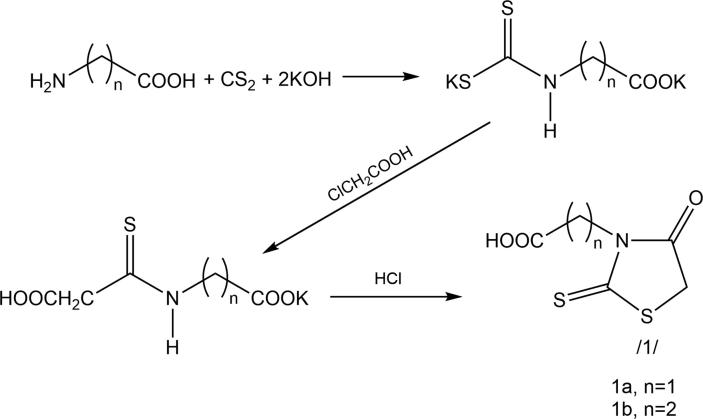
2.2. General procedure of rhodanine-3-alkanoic acids synthesis
The solution of 11.22 g (0.2 mol) potassium hydroxide in 50 cm3 of water was added to the suspension of 0.1 mol of the appropriate amino acid (aminoethanoic acid and 3-aminopropanoic acid). The resulting solution was cooled to 5–10 °C and 7.6 g (0.1 mol) carbon disulfide was added. The content of the flask was mixed at 5–10 °C for 6 h. The cooling bath was removed and mixing was continued at room temperature for 24 h.
The solution of 9.45 g (0.1 mol) chloroacetic acid in 50 cm3 water was added to the resulting solution. The solution was mixed for 7 h at the temperature below 15 °C. Next, the solution of 60 cm3 hydrochloric acid in 100 cm3 of water was added to the flask content. The resulting mixture was heated to 90 °C and kept at that temperature for 20 min. After cooling, a sediment was received, which was drained and crystallized from water.
2.3. General procedure of rhodanine-3-alkanoic acids condensation with aldehydes
0.005 mol of appropriate rhodanine-3-alkanoic acid, 5 g molecular sieves 4A, 25 cm3 isopropyl alcohol, 0.0055 mol appropriate aldehyde and 2.53 g (0.025 mol) triethylamine were put into a flask. The mixture was heated under a reflux condenser for 5 h in nitrogen. After heating was finished, the solution was filtered hot. The permeate was cooled and 50 cm3 of 2 M hydrochloric acid solution was added. The resulting sediment was filtered using Büchner funnel and crystallized from isopropyl alcohol or glacial acetic acid.
/4a/ 5-(2′-nitrobenzylidene)-rhodanine-3-acetic acid (Subhedar et al., 2016, Hardej et al., 2010), m.p. 194–196 °C, yield 55.1%, log 1/kw 2.340, MS [M+1]+ 325, IR cm−1: 1729.83 C O, 1714.41 C O conj. 1600.63 C C exo., 1333.53 C—N, 1198.54 C S.
/4b/ 5-(3′-nitrobenzylidene)-rhodanine-3-acetic acid (Hardej et al., 2010), m.p. 260–262 °C, yield 84.7%, log 1/kw 2.391, MS [M+1]+ 325, IR cm−1: 1761.65 C O, 1714.41 C O conj., 1601.59 C C exo. 1327.75 C—N 1195.65 C S.
/4c/ 5-(4′-nitrobenzylidene)-rhodanine-3-acetic acid (Subhedar et al., 2016, Hardej et al., 2010, Tanouchi et al., 1985), m.p. 269–271 °C, yield 64.5%, log 1/kw 2.114, MS [M+1]+ 325, IR cm−1: 1712.48 C O, 1604.48 C C exo., 1325.82 C—N, 1186.97 C S.
/5a/ 5-(2′-nitrobenzylidene)-rhodanine-3-propionic acid, m.p. 193–196 °C, yield 59.9%, log 1/kw 2.613, MS [M+1]+ 339, IR cm−1: 1721.16 C O, 1702.84 C O conj., 1603.52 C C exo., 1180.22 C S, 1H NMR (400 MHz, DMSO), ppm, 13.55 (br. s, 1H, HOOC—), 8.23 (d, J = 8.20 Hz, 1H, Ar—H), 8.05 (s, 1H, CH CH—Ar), 7.91 (t, 1H, Ar—H), 7.75 (d, J = 9.04 Hz, 1H, Ar—H), 7.23 (d, J = 9.04 Hz, 1H, Ar—H), 4.25 (t, 2H, HOOC—CH2—CH2—), 2.66 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.35 (S C—S), 172.19 (HOOC—), 166.41 (N—C O), 143.31 (Ar—NO2), 135.18 (C CH—Ar), 131.90 (S—C CH—), 129.21 (Ar—C), 129.13 (Ar—C), 127.57 (Ar—C), 126.09 (Ar—C), 40.20 (HOOC—CH2—CH2—), 31.22 (HOOC—CH2—CH2—).
/5b/ 5-(3′-nitrobenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 145–147 °C, yield 65.0%, log 1/kw 2.761, MS [M+1]+ 339, IR cm−1: 1714.41 C O, 1604.48 C C exo., 1334.50 C—N, 1163.83 C S.
/5c/ 5-(4′-nitrobenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 248–250 °C, yield 81.2%, log 1/kw 2.922, MS [M+1]+ 339, IR cm−1: 1705.73 C O, 1692.23 C O conj., 1606.41 C C exo., 1318.11 C—N, 1175.54 C S.
/6a/ 5-(4′-N,N-dimethylaminobenzylidene)-rhodanine-3-acetic acid (Subhedar et al., 2016, Tanouchi et al., 1985), m.p. 276–278 °C, yield 77.4%, log 1/kw 3.511, MS [M+1]+ 323, IR cm−1: 1717.30 C O, 1703.80 C O conj., 1615.09 C C exo., 1321.96 C—N, 1185.04 C S.
/6b/ 5-(4′-N,N-diethylaminobenzylidene)-rhodanine-3-acetic acid (Tejchman et al., 2017), m.p. 239–241 °C, yield 43.5%, log 1/kw 3.402, MS [M+1]+ 351, IR cm−1: 1719.30 C O, 1699.93 C O conj., 1612.21 C C exo., 1322.95 C—N, 1185.10 C S.
/7a/ 5-(4′-N,N-dimethylaminobenzylidene)-rhodanine-3-propionic acid (Tanouchi et al.,1985), m.p. 256–257 °C, yield 40.2%, log 1/kw 3.097, MS [M+1]+ 337, IR cm−1: 1714.41 C O, 1698.98 C O conj., 1616.07 C C exo., 1338.36 C—N, 1166.72 C S.
/7b/ 5-(4′-N,N-dimethylaminobenzylidene)-rhodanine-3-propionic acid (Tejchman et al., 2017), m.p. 201–203 °C, yield 63.5%, log 1/kw 3.921, MS [M+1]+ 365, IR cm−1: 1716.21 C O, 1700.94 C O conj., 1610.27 C C exo., 1331.61 C—N, 1199.51 C S.
/8a/ 5-(2′-methoxybenzylidene)-rhodanine-3-acetic acid, m.p. 220–221 °C, yield 76.8%, log 1/kw 2.478, MS [M+1]+ 310, IR cm−1: 1731.76 C O, 1711.51 C O conj., 1590.99 C C exe., 1325.82 C—N, 1195.65 C S, 1H NMR (400 MHz, DMSO), ppm, 13.45 (s, 1H, HOOC—), 7.99 (s, 1H, C CH—Ar), 7.57–7.11 (m, 4H Ar—H), 4.74 (t, 2H, HOOC—CH2—), 3.93 (s, 3H, —CH3), 13C NMR (101 MHz, DMSO) ppm, 194.27 (S C—S), 167.78 (HOOC—), 166.95 (N—C O), 158.66 (Ar—OCH3), 134.02 ( CH—Ar), 130.86 (S—C CH—), 129.81 (Ar—C) 122.32 (Ar—C), 121.78 (Ar—C), 121.66 (Ar—C), 112.53 (Ar—C), 56.28 (—O—CH3), 45.47 (HOOC—CH2—).
/8b/ 5-(3′-methoxybenzylidene)-rhodanine-3-acetic acid, m.p. 193–195 °C, yield 72.5%, log 1/kw 2.391, MS [M+1]+ 310, IR cm−1: 1722.12 C O, 1708.62 C O conj., 1602.56 C C exe., 1328.71 C—N, 1201.43 C S, 1H NMR (400 MHz, DMSO), ppm, 13.45 (s, 1H, HOOC—), 7.89 (s, 1H, C CH—Ar), 7.50 (t, 1H, Ar—H) 7.25 (m, 2H, Ar—H), 7.13 (d, J = 7.95, 1H, Ar—H), 4.23 (t, 2H, HOOC—CH2—), 3.92 (s, 3H, —CH3), 13C NMR (101 MHz, DMSO) ppm, 193.72 (S C—S), 167.72 (HOOC—), 166.77 (N—C O), 160.25 (Ar—OCH3), 134.64 (Ar—C), 134.42 ( CH—Ar), 131.17 (Ar—C), 122.70 (S—C CH—), 118.45 (Ar—C), 117.75 (Ar—C) 116.37 (Ar—C), 55.85 (—O—CH3), 45.51 (HOOC—CH2—).
/8c/ 5-(4′-methoxybenzylidene)-rhodanine-3-acetic acid (Subhedar et al., 2016, Kaminsky et al., 2012, Tanouchi et al., 1985), m.p. 247–249 °C, yield 79.6%, log 1/kw 2.506, MS [M+1]+ 310, IR cm−1: 1719.23 C O, 1703.30 C O conj., 1588.09 C C exo., 1322.93 C—N, 1173.47 C S.
/9a/ 5-(2′-methoxybenzylidene)-rhodanine-3-propionic acid, m.p. 163–165 °C, yield 71.9%, log 1/kw 2.890, MS [M+1]+ 324, IR cm−1: 1711.51 C O, 1702.53 C O conj., 1588.09 C C exo., 1333.53 C—N, 1174.44 C S, 1H NMR (400 MHz, DMSO), ppm, 12.50 (s, 1H, HOOC—), 7.94 (s, 1H, C CH—Ar), 7.18 (d, J = 8.48, 1H, Ar—H), 7.12 (t, 1H, Ar—H), 4.23 (t, 2H, HOOC—CH2—CH2—), 3.92 (s, 3H, —CH3), 2.64 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 194.24 (S C—S), 172.21 (HOOC—), 167.05 (N—C O), 158.62 (Ar—OCH3), 133.74 (Ar—C), 130.52 (S—C CH—), 128.70 (Ar—C) 122.79 ( CH—Ar), 121.81 (Ar—C), 121.75 (Ar—C), 112.53 (Ar—C), 56.40 (—O—CH3), 31.28 (HOOC—CH2—CH2—), 21.51 (HOOC—CH2—CH2—).
/9b/ 5-(3′-methoxybenzylidene)-rhodanine-3-propionic acid, m.p. 148–150 °C, yield 73.7%, log 1/kw 2.810, MS [M+1]+ 324, IR cm−1: 1710.55 C O, 1696.09 C O conj., 1601.59 C C exe., 1335.46 C—N, 1174.44 C S, 1H NMR (400 MHz, DMSO), ppm, 12.40 (s, 1H, HOOC—), 7.82 (s, 1H, C CH—Ar), 7.49 (t, 1H, Ar—H), 7.23 (t, 2H, Ar—H), 7.13 (d, J = 7.00, 1H, Ar—H), 4.24 (t, 2H, HOOC—CH2—CH2—), 3.83 (s, 3H, —OCH3), 2.65 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.77 (S C—S), 172.46 (HOOC—), 167.12 (N—C O), 160.23 (Ar—OCH3), 134.80 (C CH—Ar), 133.38 (Ar—C), 131.15 (Ar—C), 123.29 (Ar—C), 122.92 (S—C CH—), 117.47 (Ar—C), 116.27 (Ar—C), 55.88 (—O—CH3), 31.25 (HOOC—CH2—CH2—),21.52 (HOOC—CH2—CH2—).
/9c/ 5-(4′-methoxybenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 226–229 °C, yield 58.7%, log 1/kw 2.981, MS [M+1]+ 324, IR cm−1: 1705.73 C O, 1694.16 C O conj., 1588.04 C C exe., 1338.36 C—N, 1169.62 C S.
/10a/ 5-(2′-chlorobenzylidene)-rhodanine-3-acetic acid (Tanouchi et al.,1985), m.p. 236–238 °C, yield 52.8%, log 1/kw 3.049, MS [M+1]+ 315, IR cm−1: 1720.09 C O przeg., 1712.48 C O conj., 1594.84 C C exo., 1321.00 C—N, 1203.36 C S.
/10b/ 5-(3′-chlorobenzylidene)-rhodanine-3-acetic acid (Tanouchi et al.,1985), m.p. 225–228 °C, yield 83.9%, log 1/kw 3.171, MS [M+1]+ 315, IR cm−1: 1735.62 C O, 1713.44 C O conj., 1602.56 C C exo., 1327.75 C—N, 1193.72 C S.
/10c/ 5-(4′-chlorobenzylidene)-rhodanine-3-acetic acid (Subhedar et al., 2016, Tanouchi et al., 1985), m.p. 270–271 °C, yield 78.9%, log 1/kw 3.962, MS [M+1]+ 315, IR cm−1: 1728.87 C O, 1711.51 C O conj., 1599.66 C C exo., 1331.61 C—N, 1185.04 C S.
/11a/ 5-(2′-chlorobenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 144–146 °C, yield 87.8%, log 1/kw 3.567, MS [M+1]+ 329, IR cm−1: 1710.55 C O, 1699.94 C O conj., 1599.66 C C exo., 1328.71 C—N, 1176.36 C S.
/11b/ 5-(3′-chlorobenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 193–195 °C, yield 80.8%, log 1/kw 3.267, MS [M+1]+ 329, IR cm−1: 1711.51 C O, 1698.98 C O conj., 1604.48 C C exo., 1314.25 C—N, 1172.51 C S.
/11c/ 5-(4′-log 1/kw 3.472, chlorobenzylidene)-rhodanine-3-propionic acid (Sundaram and Ravi, 2013), m.p. 246–248 °C, yield 83.5%, MS [M+1]+ − 329, IR cm−1: 1705.73 C O, 1695.12 C O conj., 1601.59 C C exo., 1338.36 C—N, 1171.54 C S.
/12a/ 5-(2′-nitrocinnamylidene)-rhodanine-3-acetic acid, m.p. 241–242 °C, yield 61.9%, log 1/kw 2.974, MS [M+1]+ 351, IR cm−1: 1751.05 C O, 1715.37 C O conj., 1604.48 C C exo., 1579.41 C C—C C, 1341.25 C—N, 1196.51 C S, 1H NMR (400 MHz, DMSO), ppm, 13.43 (br. s, 1H, HOOC—), 8.08 (d, J = 7.68 Hz, 1H, CH—CH CH—Ar), 8.02 (d, J = 7.83 Hz, 1H, CH—CH CH—Ar), 7.78 (t, 1H, Ar—H), 7.68–7.62 (m, 3H, CH—CH CH—Ar), 7.19 (dd, J = 7.57 Hz, J = 14.24 Hz, 1H, CH—CH CH—Ar), 4.72 (s, 2H, HOOC—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.42 (S C—S), 167.36 (HOOC—), 166.13 (N—C O), 149.74 (Ar—NO2), 139.26 ( CH—CH CH—Ar), 133.96 ( CH—CH CH—Ar), 133.65 ( CH—CH CH—Ar), 131.07 (Ar—C), 130.31 (Ar—C) 129.38 (Ar—C) 128.26 (Ar—C), 126.24 (Ar—C), 125.09 (S—C CH—), 45.43 (HOOC—CH2—).
/12b/ 5-(4′-nitrocinnamylidene)-rhodanine-3-acetic acid, m.p. 261–263 °C, yield 92%, log 1/kw 2.632, MS [M+1]+ 351, IR cm−1: 1713.44 C O, 1609.31 C C exo., 1595.81 C C—C C, 1327.75 C—N, 1192.76 C S, 1H NMR (400 MHz, DMSO), ppm, 13.38 (s, 1H, HOOC—), 8.24 (d, J = 8.80 Hz, 2H, Ar—H), 7.96 (d, J = 8.82 Hz, 2H, Ar—H), 7.59 (d, J = 11.40 Hz, 1H, CH—CH CH—Ar), 7.51 (d, J = 15.17, 1H, CH—CH CH—Ar), 7.36 (dd, J = 11.45, J = 15.11, 1H, CH—CH CH—Ar), 4.71 (s, 2H, HOOC—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.33 (S C—S), 167.75 (HOOC—), 166.16 (N—C O), 147.97 ((Ar—NO2), 143.05 (CH—CH CH—Ar), 142.27 (CH—CH CH—Ar), 133.53 (CH—CH CH—Ar), 129.51 (Ar—C), 128.10 (Ar—C), 126.39 (Ar—C), 124.51 (S—C CH—), 45.41 (HOOC—CH2—).
/13a/ 5-(2′-nitrocinnamylidene)-rhodanine-3-propionic acid, m.p. 198–201 °C, yield 58.6%, log 1/kw 3.269, MS [M+1]+ 365, IR cm−1: 1724.05 C O, 1707.66 C O conj., 1606.41 C C exo., 1583.27 C C—C C, 1344.14 C—N, 1176.36 C S, 1H NMR (400 MHz, DMSO), ppm, 13.38 (s, 1H, HOOC—), 8.15 (d, J = 7.82, 1H, CH—CH CH—Ar), 7.72 (t, 1H, Ar—H), 7.70–7.65 (m, 3H, Ar—H), 7.32 (dd, J = 7.58, J = 14.86, 1H, CH—CH CH—Ar), 4.35 (t, 2H, HOOC—CH2—CH2—), 2.70 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 192.54 (S C—S), 171.30 (HOOC—), 166.38 (N—C O), 151.20 ((Ar—NO2), 136.15 (CH—CH CH—Ar), 135.21 (CH—CH CH—Ar), 132.55 (CH—CH CH—Ar), 131.10 (Ar—C), 129.55 (Ar—C), 128.70 (S—C CH—), 128.17 (Ar—C), 127.62 (Ar—C), 125.25 (Ar—C), 43.38 (HOOC—CH2—CH2—), 32.25 (HOOC—CH2—CH2—).
/13b/ 5-(4′-nitrocinnamylidene)-rhodanine-3-propionic acid, m.p. 244–245 °C, yield 56.3%, log 1/kw 3.742, MS [M+1]+ 365, IR cm−1: 1703.80 C O, 1698.06 C O conj., 1596.77 C C—C C, 1320.10 C—N, 1159.97 C S, 1H NMR (400 MHz, DMSO), ppm, 12.50 (br. s, 1H, HOOC—), 8.26 (d, J = 8.85 Hz, 2H, Ar—H), 7.96 (d, J = 8.84 Hz, 2H, Ar—H), 7.55(d, J = 11.41 Hz 1H, CH—CH CH—Ar), 7.52 (d, J = 15.09 Hz, 1H, CH—CH CH—Ar), 7.37 (dd, J = 11.49 Hz, J = 15.06 Hz, 1H, CH—CH CH—Ar), 4.21 (t, 2H, HOOC—CH2—CH2—), 2.62 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.44 (S C—S), 172.20 (HOOC—), 166.54 (N—C O), 147.94 ((Ar—NO2), 142.46 (CH—CH CH—Ar), 142.39 (CH—CH CH—Ar), 132.56 (CH—CH CH—Ar), 129.45 (Ar—C), 128.24 (S—C CH—), 127.14 (Ar—C), 124.55 (Ar—C), 40.95 (HOOC—CH2—CH2—), 31.25 (HOOC—CH2—CH2—).
/14a/ 5-(4′-N,N-dimethylaminocinnamylidene)-rhodanine-3-acetic acid, m.p. 273–274 °C, yield 44.0%, log 1/kw 3.398, MS [M+1]+ 349, IR cm−1: 1736.58 C O, 1697.05 C O conj., 1609.62 C C exo., 1563.99 C C—C C, 1321.90 C—N, 1160.94 C S, 1H NMR (400 MHz, DMSO), ppm, 13.35 (br.s, 1H, HOOC—), 7.59–7.56 (m, 3H, CH—CH CH—Ar), 7.34 (d, J = 14.76 Hz, 1H, CH—CH CH—Ar), 6.83 (dd, J = 11.81 Hz, J = 14.79 Hz, 1H, CH—CH CH—Ar), 6.74 (d, J = 8.93 Hz, 2H, Ar—H), 4.69 (s, 2H, HOOC—CH2—),), 3.02 (s, 6H, -N(CH3)2, 13C NMR (101 MHz, DMSO) ppm, 192.85 (S C—S), 167.94 (HOOC—), 166.03 (N—C O), 152.34 (Ar—N(CH3)2), 148.60 ( CH—CH CH—Ar), 136.73 (Ar—C), 130.93 (Ar—C), 123.49 ( CH—CH CH—Ar), 118.62 ( CH—CH CH—Ar), 118.50 (S—C CH—), 112.35 (Ar—C), 45.32 (HOOC—CH2—N ), 40.17 (-N(CH3)2).
/14b/ 5-(4′-N,N-diethylaminocinnamylidene)-rhodanine-3-acetic acid, m.p. 263–265 °C, yield 51.2%, log 1/kw 4.152, MS [M+1]+ 377, IR cm−1: 1713.44 C O, 1693.19 C O conj., 1611.23 C C exo., 1588.09 C C—C C, 1322.93 C—N, 1195.65 C S, 1H NMR (400 MHz, DMSO), ppm, 13.39 (s, 1H, HOOC—), 7.62–7.49 (m, 3H, CH—CH CH—Ar), 7.34 (d, J = 14.76 Hz, 1H, CH—CH CH—Ar), 6.78 (dd, J = 11.89 Hz, J = 14.58 Hz, 1H, CH—CH CH—Ar), 6.70 (d, J = 8.80 Hz, 2H, Ar—H), 4.68 (s, 2H, HOOC—CH2—), 3.42 (q, 4H, —N(CH2CH3)2, 1.12 (t, 6H, —N(CH2CH3)2), 13C NMR (101 MHz, DMSO) ppm, 192.72 (S C—S), 167.94 (HOOC—), 165.93 (N—C O), 149.96 (Ar—C), 148.70 ( CH—CH CH—Ar), 136.87 ( CH—CH CH—Ar), 131.34 (Ar—C), 122.80 ( CH—CH CH—Ar), 118.07 (S—C CH—), 117.95 (Ar—C), 111.82 (Ar—C), 45.31 (—N(CH2CH3)2), 44.36 (HOOC—CH2—N ), 12.96 (—N(CH2CH3)2).
/15a/ 5-(4′-N,N-dimethylaminocinnamylidene)-rhodanine-3-propionic acid, m.p. 244–245 °C, yield 46.4%, log 1/kw 3.711, MS [M+1]+ 363, IR cm−1: 1734.66 C O, 1697.05 C O conj., 1612.20 C C exo., 1593.88 C C—C C, 1326.79 C—N, 1159.97 C S, 1H NMR (400 MHz, DMSO), ppm, 12.49 (br.s, 1H, HOOC—), 7.55 (d, J = 8.93 Hz, 2H, Ar—H), 7.50 (d, J = 11.61, 1H, CH—CH CH—Ar), 6.78 (dd, J = 11.81 Hz, J = 14.88 Hz, 1H, CH—CH CH—Ar), 7.34 (d, J = 14.76 Hz, 1H, CH—CH CH—Ar), 4.19 (t, 2H, HOOC—CH2—CH2—), 3.03 (s, 6H, —N(CH3)2, 2.60 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 192.76 (S C—S), 172.23 (HOOC—), 166.35 (N—C O), 152.24 (Ar—N(CH3)2), 147.56 ( CH—CH CH—Ar), 130.77 (Ar—C), 129.15 (Ar—C), 123.54 ( CH—CH CH—Ar), 119.14 ( CH—CH CH—Ar), 118.69 (S—C CH—), 112.34 (Ar—C), 40.15 (HOOC—CH2—CH2—N ), 39.79 (—N(CH3)2), 31.35 (HOOC—CH2—CH2—N ).
/15b/ 5-(4′-N,N-diethylaminocinnamylidene)-rhodanine-3-propionic acid, m.p. 242–245 °C, yield 56.1%, log 1/kw 4.551, MS [M+1]+ 391, IR cm−1: 1713.44 C O, 1703.80 C O conj., 1611.20 C C exo., 1563.02 C C—C C, 1324.75 C—N, 1193.72 C S, 1H NMR (400 MHz, DMSO), ppm, 12.49 (br.s, 1H, HOOC—), 7.54–7.74 (m, 3H, CH—CH CH—Ar), 7.29 (d, J = 14.76 Hz, 1H, CH—CH CH—Ar), 6.78 (s, 1H, CH—CH CH—Ar), 6.75 (d, J = 2.96 Hz, 2H, Ar—H), 4.19 (t, 2H, HOOC—CH2—CH2—),), 3.42 (q, 4H, —N(CH2CH3)2, 1.12 (t, 6H, —N(CH2CH3)2, 13C NMR (101 MHz, DMSO) ppm, 192.67 (S C—S), 172.23 (HOOC—), 166.33 (N—C O), 149.85 (Ar—C), 148.10 ( CH—CH CH—Ar), 136.12 ( CH—CH CH—Ar), 131.19 (Ar—C), 120.85 ( CH—CH CH—Ar), 118.61 (Ar—C), 118.16 (S—C CH—), 111.84 (Ar—C), 111.82 (Ar—C), 44.34 (—N(CH2CH3)2), 40.35 (HOOC—CH2—CH2—N ), 31.36 (HOOC—CH2—CH2—N ), 12.96 (—N(CH2CH3)2).
/16/ 5-(4′-chlorocinnamylidene)-rhodanine-3-acetic acid, m.p. 262–265 °C, yield 63.0%, log 1/kw 3.984, MS [M+1]+ 341, IR cm−1: 1715.37 C O, 1703.80 C O conj., 1604.48 C C exo., 1572.65 C C—C C, 1331.61 C—N, 1165.76 C S, 1H NMR (400 MHz, DMSO), ppm, 13.45 (s, 1H, HOOC—), 7.59 (d, J = 8.32 Hz, 2H, Ar—H), 7.50 (d, J = 11.44 Hz, 1H, CH—CH CH—Ar), 7.43 (t, 3H, CH—CH CH—Ar), 7.18 (dd, J = 11.69 Hz, J = 14.89 Hz, 1H, CH—CH CH—Ar), 4.70 (s, 2H, HOOC—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.39 (S C—S), 167.80 (HOOC—), 166.18 (N—C O), 144.79 (CH—CH CH—Ar), 135.12 ((Ar—Cl), 134.89 (Ar—C), 134.46 (CH—CH CH—Ar), 130.34 (Ar—C), a29.51 (Ar—C), 124.92 (CH—CH CH—Ar), 124.17 (S—C CH—), 45.42 (HOOC—CH2—).
/17/ 5-(4′-chlorocinnamylidene)-rhodanine-3-propionic acid, m.p. 258–261 °C, yield 72.3%, log 1/kw 4.501, MS [M+1]+ 355, IR cm−1: 1715.37 C O, 1701.87 C O conj., 1603.52 C C exo., 1590.77 C C—C C, 1334.50 C—N, 1159.97 C S, 1H NMR (400 MHz, DMSO), ppm, 12.50 (s, 1H, HOOC—), 7.74 (d, J = 8.55 Hz, 2H, Ar—H), 7.50 (t, 3H, CH—CH CH—Ar), 7.39 (d, J = 15.17 Hz, 1H, CH—CH CH—Ar), 7.14 (dd, J = 11.53 Hz, J = 15.12 Hz, 1H, CH—CH CH—Ar), 4.20 (t, 2H, HOOC—CH2—CH2—), 2.62 (t, 2H, HOOC—CH2—CH2—), 13C NMR (101 MHz, DMSO) ppm, 193.40 (S C—S), 172.21 (HOOC—), 166.51 (N—C O), 144.17 (CH—CH CH—Ar), 134.97 ((Ar—Cl), 134.94 (CH—CH CH—Ar), 130.25 (Ar—C), 124.86 (CH—CH CH—Ar), 124.81 (S—C CH—), 121.78 (Ar—C), 40.91 (HOOC—CH2—CH2—), 31.27 (HOOC—CH2—CH2—).
2.4. Antibacterial activity in vitro assay
All target compounds were screened for antibacterial and antifungal activities by micro-dilution broth method using Mueller-Hinton broth and Mueller-Hinton broth with 5% lysed sheep blood for growth of non-fastidious and fastidious bacteria, respectively, or Mueller-Hinton broth with 2% glucose for growth of fungi. Minimal inhibitory concentration (MIC) of tested compounds were evaluated for the panel of reference microorganism from American Type Culture Collection (ATCC), including Gram-negative bacteria (S. typhimurium ATCC 14028, E. coli ATCC 25922, P. mirabilis ATCC 12453, K. pneumoniae ATCC 13883, P. aeruginosa ATCC 9027), Gram-positive bacteria (S. aureus ATCC 25923, S. aureus ATCC 6538, S. aureus ATCC 43300, S. epidermidis ATCC 12228, M. luteus ATCC 10240, B. subtilis ATCC 6633, B. cereus ATCC 10876, S. pyogenes ATCC 19615, S. pneumoniae ATCC 49619, S. mutans ATCC 25175) and fungi (C. albicans ATCC 102231, C. albicans ATCC 2091, C. parapsilosis ATCC 22019, C. glabrata ATCC 90030, C. krusei ATCC 14243). A detailed procedure for the determination of MIC value was presented in earlier publication (Tejchman et al., 2017).
2.5. Measurement of lipophilicity
The chromatographic measurements were carried out using Shimadzu LC-20A liquid chromatographic system and a computer software LC Solution Version 1.24 to control the hardware and store the chromatographic data. Measurements were performed applying a column packed with Eurospher II 100-3-C18A (Knauer; 12.5 cm × 4 mm i.d.) using methanol-water mixtures as mobile phases. Volume fraction of the organic modifier varied from 0.9 to 0.4 with increment 0.1. The mobile phase flow-rate was 1 mL min−1. All measurements were made at 20 °C. The test compounds were separately dissolved (10−3 mg mL−1) in the organic modifier and detected using UV–VIS detector. For calculations, average values of the retention factors from at least three experimental replicates were taken. The dead time was evaluated from uracil peak.
The descriptor of lipophilicity or hydrophobicity (log kw value) was determined on the basis of a series of isocratic measurements at different concentrations of the organic modifier (methanol) in a two-component mobile phase and linear extrapolation of the relationship between log k and the volume fraction (φ) of the methanol to its zero content (to 100% of water). The log kw values determined by the extrapolation methods depended to a high degree on the range of the organic modifier concentrations, at which measurements and then extrapolation were made (Kwietniewski, 2010). In this case, the concentration range (φ) from 0.4 to 0.9 was used for all examined substances.
2.6. X-ray analysis
Crystals suitable for an X-ray structure analysis were obtained from a mixture of ethanol and dimethylformamide for 15b and from a mixture of methanol and dimethyl sulfoxide for 17, by slow evaporation of the solvent at room temperature.
Data for single crystals of 15b and 17 were collected using the Oxford Diffraction SuperNova four circle diffractometer equipped with the Mo (0.71073 Å) Kα for 15b and Cu (1.54184 Å) Kα for 17 radiation source and graphite monochromator. The phase problems were solved by direct methods using SIR-2014 program (Burla et al., 2015) and all non-hydrogen atoms were refined anisotropically using weighted full-matrix least-squares on F2. Refinement and further calculations were carried out using SHELXL program (Sheldrick, 2015). The hydrogen atoms bonded to carbons were included in the structure at idealized positions and were treated using a riding model with Uiso(H) fixed at 1.2 Ueq of C and 1.5 Ueq for methyl groups. Hydrogen atoms attached to oxygen atoms were found from the difference Fourier map and refined without any restraints. For molecular graphics, MERCURY (Macrae et al., 2006) software was used.
15b: C19H22N2O3S2, Mr = 390.50, crystal size = 0.11 × 0.22 × 0.59 mm3, triclinic, space group P, a = 8.3065(4) Å, b = 11.5949(6) Å, c = 12.2345(6) Å, α = 112.675(5)°, β = 98.541(4)°, γ = 108.445(4)°, V = 980.92(9) Å3, Z = 2, T = 130(2) K, 13,135 reflections collected, 4605 unique reflections (Rint = 0.0280), R1 = 0.0464, wR2 = 0.1138 [I > 2σ(I)] and R1 = 0.0659, wR2 = 0.1265 [all data].
17: C15H12ClNO3S2·C2H6OS, Mr = 431.95, crystal size = 0.04 × 0.16 × 1.00 mm3, monoclinic, space group C2/c, a = 41.440(1) Å, b = 4.8420(2) Å, c = 20.0207(5) Å, β = 97.650(3)°, V = 3981.4(2) Å3, Z = 8, T = 130(2) K, 28,073 reflections collected, 3848 unique reflections (Rint = 0.0490), R1 = 0.0384, wR2 = 0.1020 [I > 2σ(I)] and R1 = 0.0416, wR2 = 0.1052 [all data].
CCDC 1877705 and 1877706 contain the supplementary crystallographic data, for 15b and 17, respectively. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
3. Results and discussion
3.1. Chemical synthesis
The synthesis of rhodanine-3-acetic and rhodanine-3-propionic acids was conducted according to the modified procedure proposed by Körner at the beginning of the 20th century (Körner, 1908) (Scheme 1).
Scheme 1.
3-Carboxyalkylrhodanine acids synthesis.
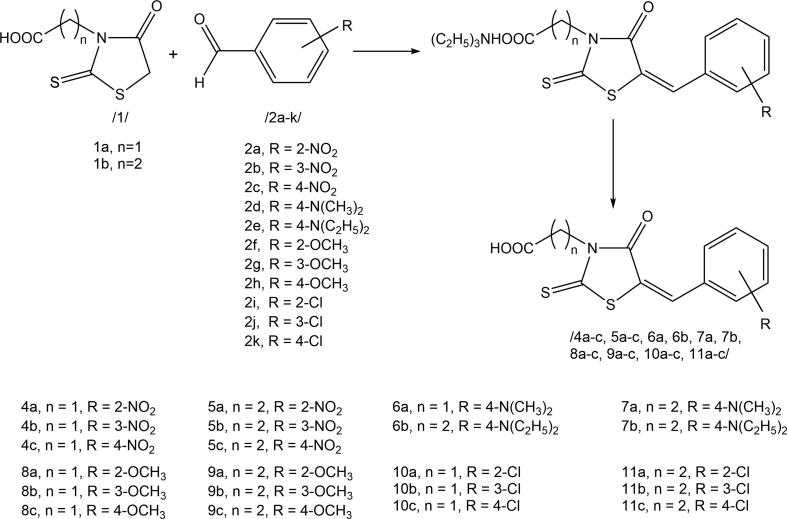
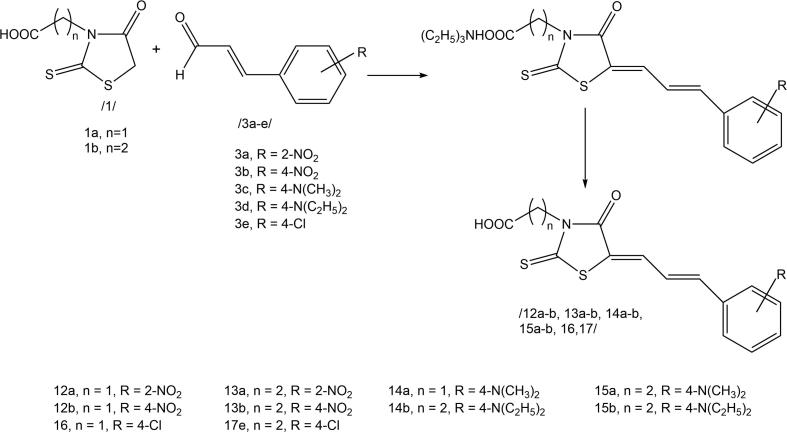
The Knoevenagel condensation of the obtained rhodanine-3-carboxyalkyl acids with benzaldehyde and cinnamaldehyde derivatives was conducted according to the procedure proposed for the analogues of 2-[(5Z)-5-[(E)-2-methyl-3-phenylprop-2-enylidene]-4-oxo-2-sulfanylidene-1,3-thiazolidin-3-yl]acetic acid commonly referred to as epalrestat (Żesławska et al., 2015) (Scheme 2 and Scheme 3).
Scheme 2.
3-Carboxyalkylrhodanine acids condensation with benzaldehyde derivatives.
Scheme 3.
3-Carboxyalkylrhodanine acids condensation with cinnamaldehyde derivatives.
3.2. Antimicrobial activity
The 3-carboxyalkylrhodanine acids derivatives having a cinnamaldehyde fragment at the C-5 position are rarely discussed in the literature on the subject. The best known example of such compound is epalrestat, 2-[(5Z)-5-[(E)-2-methyl-3-phenylprop-2-enylidene]-4-oxo-2-sulfanylidene-1,3-thiazolidin-3-yl]acetic acid, that is an aldose reductase inhibitor used for the treatment of diabetic neuropathy (Hu et al., 2014).
In our current research, we decided to compare the antibacterial activity of rhodanine-3-acetic and rhodanine-3-propionic acids having benzylidene and cinnamylidene groups with additional electron donating or withdrawing groups at the C-5 position. None of the tested compounds influenced effectively against Gram-negative bacteria and yeast (MIC > 1000 mg/L, data not shown in main text. All measured MIC and MBC values are given in the Supplement Materials. Tables S1–S3). Minimal inhibitory concentration (MIC) values of the derivatives against Gram-positive bacteria are included in Table 2, Table 3, Table 4.
Table 2.
Biological activity of derivatives containing a nitro or amino group in the benzylidene substituent.
| Compound |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microorganism |
4a |
4b |
4c |
5a |
5b |
5c |
6a |
6b |
7a |
7b |
| MIC (mg/L) | ||||||||||
| S. aureus ATCC 25923 | 125 | 250 | 125 | 15.6 | 250 | >1000 | 500 | 250 | 500 | 31.25 |
| S. aureus ATCC 6538 | 250 | 250 | 250 | 125 | 125 | >1000 | 500 | 125 | 250 | 31.25 |
| S. aureus ATCC 43300 | 250 | 250 | 250 | 125 | 125 | >1000 | 500 | 125 | 250 | 31.25 |
| S. epidermidis ATCC 12228 | 125 | 500 | 31,3 | 31.3 | 250 | >1000 | >1000 | 250 | 500 | 31.25 |
| M. luteus ATCC 10240 | 250 | 1000 | 125 | 125 | 500 | >1000 | 250 | 62.5 | 1000 | 62.5 |
| B. subtilis ATCC 6633 | 62.5 | 500 | 250 | 15.6 | 62.5 | >1000 | 250 | 62.5 | 500 | 15.6 |
| B. cereus ATCC 10876 | 250 | 250 | 250 | 125 | 250 | >1000 | >1000 | 125 | 250 | 31.25 |
| S. pyogenes ATCC 19615 | 125 | >1000 | 125 | 62.5 | >1000 | >1000 | 250 | 125 | 250 | 62.5 |
| S. pneumoniae ATCC 49619 | 250 | >1000 | 125 | 250 | >1000 | >1000 | 125 | 62.5 | 250 | 62.5 |
| S. mutans ATCC 25175 | 250 | >1000 | 250 | 500 | >1000 | >1000 | 1000 | 250 | 500 | 250 |
Table 3.
Biological activity of derivatives containing a methoxy or chloro group in the benzylidene substituent.
| Microorganism | Compound |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
8a |
8b |
8c |
9a |
9b |
9c |
10a |
10b |
10c |
11a |
11b |
11c |
|
| MIC (mg/L) | ||||||||||||
| S. aureus ATCC 25923 | 1000 | 1000 | 500 | 125 | >1000 | >1000 | 250 | >1000 | 500 | 62.5 | 250 | 125 |
| S. aureus ATCC 6538 | 500 | 500 | 1000 | 125 | >1000 | >1000 | 250 | >1000 | 1000 | 31.25 | 250 | 125 |
| S. aureus ATCC 43300 | 500 | 500 | 1000 | 62.5 | >1000 | >1000 | 250 | >1000 | 500 | 31.25 | 250 | 125 |
| S. epidermidis ATCC 12228 | 1000 | 1000 | 1000 | 1000 | >1000 | 500 | 250 | >1000 | 500 | 31.25 | 500 | 125 |
| M. luteus ATCC 10240 | 250 | 250 | 250 | 1000 | >1000 | >1000 | 250 | >1000 | 250 | 62.5 | 1000 | 500 |
| B. subtilis ATCC 6633 | 500 | 1000 | 500 | 250 | >1000 | >1000 | 250 | >1000 | 500 | 62.5 | 250 | 125 |
| B. cereus ATCC 10876 | 1000 | 1000 | 1000 | 1000 | >1000 | 1000 | 500 | >1000 | 500 | 62.5 | 250 | 125 |
| S. pyogenes ATCC 19615 | 1000 | 500 | >1000 | 250 | >1000 | >1000 | 250 | >1000 | 500 | 62.5 | 1000 | 1000 |
| S. pneumoniae ATCC 49619 | 500 | >1000 | >1000 | 125 | >1000 | >1000 | 500 | >1000 | 500 | 62.5 | 1000 | 1000 |
| S. mutans ATCC 25175 | 1000 | >1000 | >1000 | 1000 | >1000 | >1000 | 500 | >1000 | 1000 | 500 | >1000 | 1000 |
Table 4.
Biological activity of derivatives containing a nitro, amino, methoxy or chloro group in the cinnamylidene substituent.
| Microorganism | Compound |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
12a |
12b |
13a |
13b |
14a |
14b |
15a |
15b |
16 |
17 |
|
| MIC (mg/L) | ||||||||||
| S. aureus ATCC 25923 | 250 | 15.6 | 15.6 | 7.8 | 1000 | 125 | 62.5 | 31.25 | 500 | 125 |
| S. aureus ATCC 6538 | 500 | 250 | 125 | 62.5 | 500 | 125 | 15.6 | 31.25 | 250 | 125 |
| S. aureus ATCC 43300 | 500 | 15.6 | 62.5 | 7.8 | 500 | 62.5 | 62.5 | 31.25 | 250 | 125 |
| S. epidermidis ATCC 12228 | 1000 | 3.9 | 31.25 | 1.98 | 500 | 62.5 | 31.25 | 31.25 | 1000 | 125 |
| M. luteus ATCC 10240 | 250 | 250 | 500 | 31.25 | 250 | 62.5 | 125 | 31.25 | 500 | 125 |
| B. subtilis ATCC 6633 | 500 | 1000 | 62.5 | 3.9 | 500 | 62.5 | 15.6 | 31.25 | 250 | 62.5 |
| B. cereus ATCC 10876 | 500 | >1000 | 125 | 62.5 | 500 | 125 | 15.6 | 31.25 | 125 | 125 |
| S. pyogenes ATCC 19615 | 500 | 31.25 | 125 | 62.5 | >1000 | 125 | 62.5 | 125 | 500 | >1000 |
| S. pneumoniae ATCC 49619 | 125 | 125 | 62.5 | 62.5 | 500 | 125 | 31.25 | 62.5 | 250 | >1000 |
| S. mutans ATCC 25175 | 1000 | >1000 | 1000 | 500 | >1000 | 125 | 250 | 125 | 1000 | >1000 |
Some compounds tested by our team have been obtained and tested by other researchers and by us. We have previously studied the antimicrobial activity of compounds 6b and 7b [12]. In this work, both compounds were used to compare the influence of molecular structure on biological activity.
Derivatives 4a, 4b and 4c were examined by Talele et al. They were reported their anti MRSA activity (Hardej et al., 2010). Derivatives: 5a, 5b, 5c, 9a, 11a, 11b and 11c were examined by Sundaram and Ravi. They determined their influence on two Gram-positive bacteria strains (Staphylococcus aureus, Bacillus cereus) and one Gram-negative bacterium strain (Escherichia coli). The activity of the compounds examined was described as moderate (Sundaram and Ravi, 2013).
Derivative 7a was used by the Lesyk team to synthesize more complex systems demonstrating antineoplastic activity (Kaminsky et al., 2012).
Derivatives 4c, 6a and 10c examined towards their activity against tuberculosis by the Shingate team (Subhedar et al., 2016) were described in patent literature by Tanouchi, who reported them as potential aldose reductase inhibitors (Tanouchi et al., 1985).
Compound 8c was described by three research teams mentioned above (Subhedar et al., 2016, Kaminsky et al., 2012, Tanouchi et al., 1985). The derivative 10b was previously described as a potential aldose reductase inhibitor (Tanouchi et al., 1985).
None of the 3-carboxyalkylrhodanine acids derivatives tested in our study demonstrated activity against Gram-negative bacteria and yeast (MIC > 1000 mg/L), similarly to the compounds described in our previous publication (Tejchman et al., 2017). The conclusion was contrary to our anticipations, since we expected that the derivatives having a cinnamylidene substituent at the C-5 position would demonstrate at least moderate activity in suppressing the growth of this type of microorganisms.
The derivatives of rhodanine-3-propionic acid with p-N,N-diethylcinnamylidene and p-N,N-diethylbenzylidene substituents at the C-5 position demonstrated the highest activity against Gram-positive bacteria. Derivative 15b inhibited the growth of Gram-positive bacteria at the concentration of 31.25 mg/L, whereas compound 7b inhibited the growth of Gram-positive bacteria at the concentration between 15.6 and 250 mg/L. We consider that the presence of the N,N-diethylamine group in the aromatic system and the number of carbon atoms in the carboxyalkyl group is more significant for the biological activity than the fact that the benzylidene or cinnamylidene substituent was present at the C-5 position.
The compounds with the benzylidene substituent containing electron withdrawing NO2 group from the ortho position at the C-5 position demonstrated moderate or low antibacterial activity against Gram-positive bacteria (MIC 125–500 mg/L). Compound 5c, demonstrated the highest activity within this group of compounds.
Opposite effect was observed in the case of the 3-α-carboxy-ethyl-5-benzylidene rhodanine derivatives (Sundaram and Ravi, 2015). In the case of 5-benzylidene derivatives having a nitro group in meta and para positions, we did not observe significant antibacterial activity.
The activity of the derivatives having the cinnamylidene substituent with NO2 group at the C-5 position was significantly higher than that of the benzylidene derivatives. We observed that compounds having the NO2 group in the para-position were more active in hindering bacterial growth than those which had nitro group in the ortho- position.
Only compound 11a demonstrated moderate antibacterial activity against Gram-positive bacteria of the tested derivatives with a chlorine atom at the C-5 position substituent. However, none of Gram-negative bacteria strains was susceptible to its activity. Ravi and Sundaram observed that this compound inhibited the growth of E. coli, but at very high concentrations exceeding 320 µg/ml (Sundaram and Ravi, 2013).
Derivatives having a methoxy groups in the substituent at the C-5 position demonstrated no activity against bacteria regardless of the —OCH3 group location in the aromatic ring. We expected that this type of derivatives, due to the presence of an ether bond, would demonstrate activity against Gram-positive bacteria, similarly to the compounds obtained by Miao, (Miao et al., 2013) namely rhodanine-3-acetic acid derivatives having a ((6-benzyloxy)naphthalen-2-yl)methylene moiety at the C-5 position. The derivatives tested by them showed a higher antibacterial activity than those tested by our team, probably due to the larger size of the hydrophobic groups used.
3.3. Lipophilicity
Transport of chemical compounds, that are potential medicine drugs, through lipids membranes is directly connected to their physical and chemical properties. One of the most important parameters that have significant importance for their biological properties is lipophilicity (Rutkowska et al., 2013, Hawrył et al., 2015).
For all investigated by us compounds we have determined the MIC value and log kw that are important descriptors of lipophilicity.
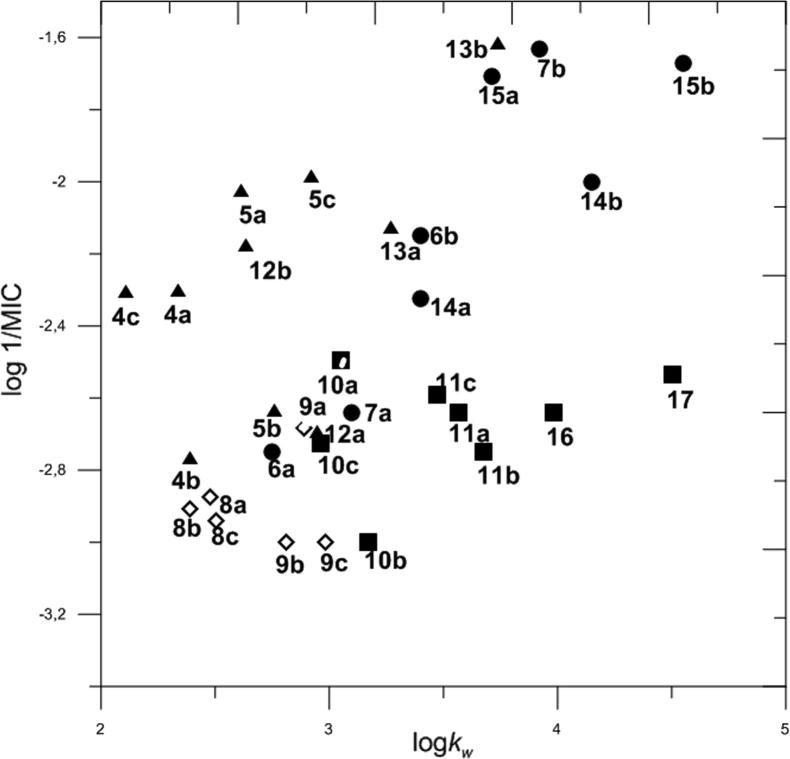
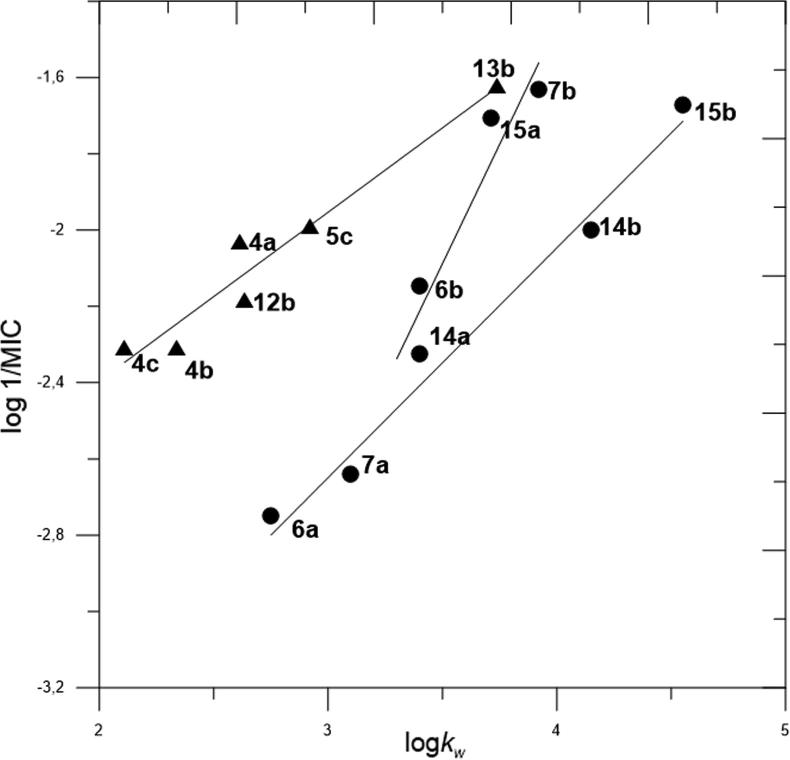
During our research, we took under consideration only the MIC value determined for Gram-positive bacteria, because investigated compounds did not manifest any activity against Gram-negative bacteria and yeast. Graph of antibacterial activity (log 1/MIC) vs. lipophilicity (log kw) allowed to connect the structures of investigated compounds with their biological activity and lipophilicity (Fig. 2).
Fig. 2.
Dependence of log 1/MIC on log kw for all investigated compounds.
The figure illustrates three groups of investigated compounds showing similar type of dependence between biological activity and lipophilicity. These three groups of compounds are detailed in Fig. 3.
Fig. 3.
Three groups of compounds exhibiting a similar type of dependence of biological activity on lipophilicity.
The first group contains compounds 6a, 7a, 14b and 15b. In this group, much higher activity was shown by derivatives that contain 4′-N,N-diethylaminocinnamylidene substituent than these containing 4′-N,N-dimethylaminobenzylidene substituent at C-5 position. In pairs 6a, 7a and 14b, 15b, higher activity and lipophilicity was shown by rhodanine-3-propionic acid derivatives than by rhodanine-3-acetic acid derivatives.
The second group includes following derivatives: 6b, 7b, 14a and 15a. In this group, activity and lipophilicity of rhodanine-3-acetic acid derivatives was significantly lower than activity of rhodanine-3-propionic acid derivatives. A factor that caused that derivatives containing in their structure an amino substituent did not form one group with similar properties, was probably different distribution of electron density within molecules (Yashin, 1989, Ciura et al., 2017).
The third group includes four derivatives containing nitro group at the para position and two derivatives containing nitro group at the ortho position. They are following compounds: 4c, 5c, 12b, 13b and 4a, 5a. In this group, the highest activity and the highest lipophilicity was shown by compound 13b, while the lowest activity and the lowest lipophilicity was shown by 4c. Compounds 4a and 5a, that contain 2′-nitrobenzylidene substituent at position C-5, showed similar activity as their isomers containing 4′-nitrobenzylidene substituent at the position C-5. Derivatives 12a, 13a, as well as 4b and 5b showed lower biological activity than other compounds containing nitro group at position C-5.
Other investigated compounds did not show clear dependence between their structure and biological activity or lipophilicity.
3.4. X-ray crystallographic studies of 15b and 17
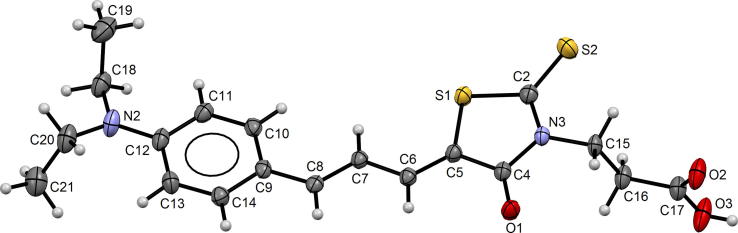
The unit cell consists of two 15b molecules related by the inversion center. The molecular geometry in the crystal structure of 15b with the atom numbering scheme is shown in Fig. 4.
Fig. 4.
Molecular structure of 15b with the atom numbering scheme.
The molecule has two double bonds C5 C6 and C7 C8 and can form four geometric isomers (Z, Z; E, E; Z, E or E, Z). In the crystal structure of 15b the 5Z, 7E isomer is observed. This isomer also occurred in another crystal structure determined earlier, containing rhodanine and aromatic rings connected together with the same chain (Żesławska et al., 2015). In the Cambridge Structural Database (CSD, Version 5.39) (Gromm et al., 2016) there are no other crystal structures with the same chain connecting rhodanine and aromatic rings. The majority of rhodanine derivatives with determined crystal structures, deposited in the CSD, have additional methyl group at the C7 atom and form the same isomer.
The rhodanine and aromatic rings are almost coplanar, the interplanar angle is 10.9(1)°. Two ethyl groups at the N2 atom are directed oppositely to the phenyl ring plane, similar as in other crystal structures of 5-benzylidenerhodanine derivative containing N,N-diethyl substituent at benzylidene moiety (Stawoska et al., 2017, Żesławska et al., 2018). The angle between the plane of thiazolidine ring (S1, C2, N3, C4, C5) and the plane of carboxyl group (C17, O2, O3) is 88.7(2)°. This value is a little higher as in other 3-carboxyethylrhodanine derivatives, 81.2° and 82.0° (Żesławska et al., 2015, Stawoska et al., 2017) and it differs from the values 40.3° and 55.8° observed in the crystal structure of another 3-carboxyethylrhodanine derivative (Żesławska et al., 2018) and 3-carboxyethylrhodanine (Tejchman et al., 2016).
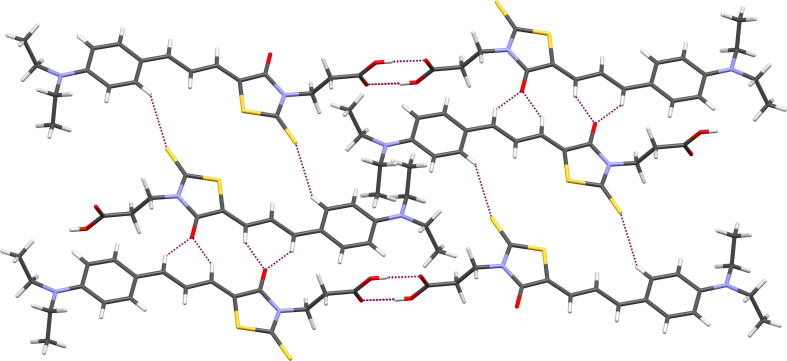
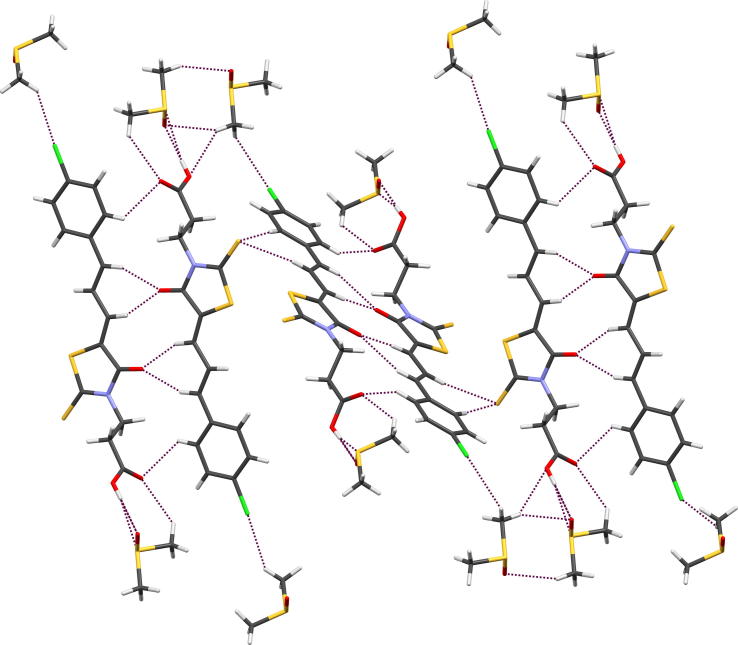
The crystal packing is determined by O—H⋯O intermolecular interactions, created by carboxylic groups of two molecules related by inversion center (Fig. 5). These interactions lead to the formation of dimers, that are joined to each other by C—H⋯O and C—H⋯S contacts. Parameters of mentioned interactions are listed in Table 5. In addition, the layers are observed in the crystal structure. Similar arrangement of 3-carboxyethylrhodanine derivatives was noticed in other crystal structures (Żesławska et al., 2015, Stawoska et al., 2017).
Fig. 5.
Partial packing view of 15b. Dashed lines indicate hydrogen bonds.
Table 5.
Parameters of intermolecular interactions of 15b and 17.
| Compd | D-H⋯A | H⋯A (Å) | D⋯A (Å) | D-H-A (°) | Symmetry codes |
|---|---|---|---|---|---|
| 15b | O3-H3⋯O2 | 1.82(3) | 2.642(2) | 177(3) | −x − 1, −y + 1, −z + 2 |
| C6-H6⋯O1 | 2.43 | 3.282(2) | 149 | −x, −y, −z + 2 | |
| C8-H8⋯O1 | 2.68 | 3.494(2) | 144 | −x, −y, −z + 2 | |
| C10-H10⋯S2 | 2.84 | 3.596(2) | 138 | −x, −y, −z + 1 | |
| C21-H21C⋯O1 | 2.53 | 2.642 | 177(3) | −x + 1, −y, −z + 2 | |
| 17 | O3-H3⋯O4 | 1.87(3) | 2.647(2) | 176(4) | |
| O3-H3⋯S3 | 2.77(3) | 3.478(2) | 152(3) | ||
| C6-H6⋯O1 | 2.52 | 3.399(2) | 154 | −x + 1/2, −y − 3/2, −z | |
| C8-H8⋯O1 | 2.48 | 3.380(2) | 158 | −x + 1/2, −y − 3/2, −z | |
| C7-H7⋯S2 | 2.91 | 3.853(2) | 173 | −x + 1/2, −y, −z + 1/2 | |
| C10-H10⋯S2 | 3.01 | 3.963(2) | 176 | −x + 1/2, −y, −z + 1/2 | |
| C18-H18A⋯O4 | 2.64 | 3.559(4) | 156 | −x, −y, −z | |
| C18-H18B⋯CL1 | 2.92 | 3.738(3) | 142 | x − 1/2, −y − 3/2, z − 1/2 | |
| C18-H18C⋯CL1 | 2.97 | 3.630(3) | 125 | x − 1/2, −y − 1/2, z − 1/2 | |
| C19-H19B⋯O2 | 2.61 | 3.373(4) | 135 | ||
| C19-H19C⋯O4 | 2.52 | 3.262(4) | 132 | x, y + 1, z | |
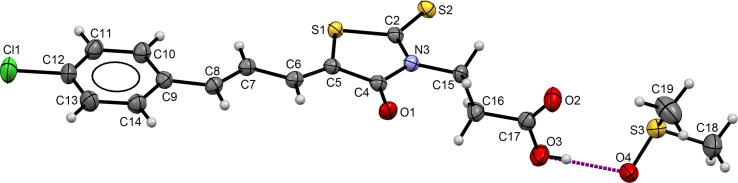
The asymmetric unit consists of one molecule of 17 and one molecule of dimethyl sulfoxide from the solvent (Fig. 6).
Fig. 6.
Molecular structure of 17 with the atom numbering scheme.
Similar as in 15b the 5Z, 7E isomer in the crystal structure is observed. The fragment of molecule containing rhodanine and aromatic ring, connected by prop-2-enylidene chain, is more planar than in 15b. The interplanar angle between the planes of both rings is 3.6(1)°. Thus the flat fragment was also observed in another crystal structure containing phenyl and rhodanine rings joined together by prop-2-enylidene chain (Żesławska et al., 2015). The angle between the rhodanine ring and carboxyl group is close to the value of this angle presented above, which is equal to 86.5(1)°.
In the crystal lattice, the carboxylic groups are engaged in the hydrogen bonds with DMSO molecules (Fig. 7). Additionally, the C—H⋯O, C—H⋯S and C—H⋯Cl intermolecular interactions were observed. The parameters of these interactions are listed in Table 5.
Fig. 7.
Partial packing view of 17. Dashed lines indicate hydrogen bonds.
4. Conclusions
Among all compounds investigated by us, the highest activity characterized derivatives of rhodanine-3-acetic and rhodanine-3-propionic acids having at the C-5 position cinnamylidene substituent containing a dialkylamino or nitro group in the para position. The size of alkyl groups in the dialkylamine moiety significantly affects the antimicrobial activity.
Derivatives containing a diethylamino group have higher activity than those containing a dimethylamino group. The presence of chlorine atoms or methoxy groups in the arylidene moiety does not increase the antibacterial activity of the tested compounds. The obtained results will allow to plan further research in order to obtain compounds with even higher antibacterial activity.
The crystal structure of more active compound 15b shows the less planarity of 5-cinnamylidenerhodanine fragment in comparison to 17 and the involvement of N,N-diethyl substituent as donor in C—H⋯O intermolecular contacts.
Acknowledgments
Acknowledgements
We are thankful to the Vice-Rector of the Pedagogical University in the Cracow, Professor Mariusz Wołos for the financial support of our project.
Declaration of Competing Interest
The authors declare no conflicts of interest with regard to the publication of this article.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2020.02.002.
Contributor Information
Waldemar Tejchman, Email: waldemar.tejchman@up.krakow.pl.
Izabela Korona-Glowniak, Email: iza.glowniak@umlub.pl.
Ludomir Kwietniewski, Email: mirekkw@o2.pl.
Ewa Żesławska, Email: ewa.zeslawska@up.krakow.pl.
Wojciech Nitek, Email: nitek@chemia.uj.edu.pl.
Piotr Suder, Email: suder.p@gmail.com.
Marek Żylewski, Email: marek.zylewski@jci.pl.
Anna Malm, Email: anna.malm@umlub.pl.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Burla M.C., Caliandro R., Carrozzini B., Cascarano G.L., Cuocci C., Ciazovazzo C., Mallamo M., Mazzone A.G., Polidori G. Crystal structure determination and refinement via SIR2014. J. Appl. Cryst. 2015;48:306–309. doi: 10.1107/S1600576715001132. [DOI] [Google Scholar]
- Ciura K., Rutecka A., Kawczak P., Nowakowska J. Quantitative structure—retention relationship modeling of the retention behavior of selected antipsychotic drugs in normal-phase thin-layer chromatography. JPC-J. Planar Chromat. 2017;30:225–230. doi: 10.1556/1006.2017.30.3.13. [DOI] [Google Scholar]
- Gromm C.R., Bruno I.J., Lightfoot M.P., Ward S.C. The cambridge structural database. Acta Cryst. 2016;B72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardej D., Ashby C.S., Jr., Khadtare N.S., Kulkarni S.S., Singh S., Talele T.T. The synthesis of phenylalanine-derivatived C5-substituted rhodanines and their activity against selected methicillin-resistant Staphylococcus Aureus (MRSA) strains. Eur. J. Med. Chem. 2010;45:5827–5832. doi: 10.1016/j.ejmech.2010.09.045. [DOI] [PubMed] [Google Scholar]
- Harris L.G., Foster S.J., Richards R.G. An introduction to Staphylococcus aureus, and techniques for identifying and quantifying S. aureus adhesins in relation to adhesion to biomaterials: Review. Eur. Cell Mater. 2002;4:39–60. doi: 10.22203/eCM.v004a04. [DOI] [PubMed] [Google Scholar]
- Hawrył H.M., Świeboda R.S., Gawroński M.S., Wójciak-Kosior M.A., Popiołek Ł.P., Kocjan R.B. Determination of lipophilicity of some new 1,2,3-triazole derivatives by RP-HPLC and RP-TLC and calculated methods. Current Chem. Lett. 2015;4:101–110. doi: 10.5267/j.ccl.2015.4.002. [DOI] [Google Scholar]
- Hu X., Li S., Yang G., Liu H., Boden G., Li L. Efficacy and safety of aldose reductase inhibitor for the treatment of diabetic cardiovascular autonomic neuropathy: systematic review and meta-analysis. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0087096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky D., Bednarczyk-Cwynar B., Vasylenko O., Kazakova O., Zimenkovsky B., Zaprutko L., Lesyk R. Synthesis of new potential anticancer agents based on 4-thiazolidinone and oleanane scaffolds. Med. Chem. Res. 2012;21:3568–3580. doi: 10.1007/s00044-011-9893-9. [DOI] [Google Scholar]
- Kaminsky D., Kryshchyshyn A., Lesyk R. 5-Ene-4-thiazolidinones–an efficient tool in medicinal chemistry. Eur. J. Med. Chem. 2017;140:542–594. doi: 10.1016/j.ejmech.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner H. Über einige derivate der dithiocarbamino-essigsäure. Ber. Dtsch. Chem. Ges. 1908;41:1901–1905. doi: 10.1002/cber.19080410265. [DOI] [Google Scholar]
- Kwietniewski L. Determination of solute retention in RPLC with pure water as effluent using a numerical method based on the Ościk's equation”. J. Liquid Chromatogr. Rel. Techn. 2010;33:305–323. doi: 10.1080/10826070903524076. [DOI] [Google Scholar]
- Macrae C.F., Edgington P.R., McCabe P., Pidcock E., Shields G.P., Taylor R., Towler M., van de Streek J. Mercury: visualization and analysis of crystal structures. J. Appl. Cryst. 2006;39:453–457. [Google Scholar]
- Miao J., Zheng C.J., Sun L.P., Song M.X., Xu L.L., Piao H.R. Synthesis and potential antibacterial activity of new rhodanine-3-acetic acid derivatives. Med. Chem. Res. 2013;22:4125–4132. doi: 10.1007/s00044-012-0417-z. [DOI] [Google Scholar]
- Nowakowicz-Dębek B., Wlazło Ł., Kasela M., Ossowski M. Epidemiologia wielolekoopornych szczepów Staphylococcus aureus. Probl. Hig. Epidemiol. 2016;97:106–112. [Google Scholar]
- Patel B.A., Ashby C.R., Jr., Hardej D., Talele T.T. The synthesis and SAR study of phenylalanine-derived (Z)-5-arylmethylidene rhodanines as anti-methicillin-resistant Staphylococcus aureus (MRSA) compounds. Bioorg. Med. Chem. Lett. 2013;23:5523–5527. doi: 10.1016/j.bmcl.2013.08.059. [DOI] [PubMed] [Google Scholar]
- Rutkowska E., Pająk K., Jóźwiak K. Lipophilicity – methods of determination and its role in medicinal chemistry. Acta Pol. Pharm. 2013;70:3–18. [PubMed] [Google Scholar]
- Santajit, S., Indrawattana, N., 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Hindawi Publishing Corporation BioMed Research International, 2016, Article ID 2475067, 8 pages, 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed]
- Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015;C71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawoska I., Tejchman W., Mazuryk O., Lycka A., Nowak-Sliwinska P., Żesławska E., Nitek W., Kania A. Spectral characteristic and preliminary anticancer activity in vitro of selected rhodanine-3-carboxylic acids derivatives. J. Heterocyclic Chem. 2017;54:2889–2897. doi: 10.1002/jhet.2897. [DOI] [Google Scholar]
- Subhedar, D.D., Shaikh, M.H., Navale, L., Yeware, A., Sarkar, D., Shingate, B.B., 2016. [Et3NH][HSO4] catalyzed efficient synthesis of 5-arylidene-rhodanine conjugates an d their antitubercular activity, Res. Chem. Intermed. 42, 6607 – 6626. DOI: 1007/s11164-016-2484-0.
- Sundaram K., Ravi S. Microwave assisted synthesis of 3-α-carboxy ethylrhodanine derivatives and their in vitro antibacterial activity. J. Appl. Pharm. Sci. 2013;3:133–135. doi: 10.7324/JAPS.2013.3725. [DOI] [Google Scholar]
- Sundaram K., Ravi S. Synthesis, antibacterial activity against MRSA, and in vitro cytotoxic activity against HeLa cell lines of novel 3-α-carboxy ethyl-5-benzylidene rhodanine derivatives. Res. Chem. Intermed. 2015;41:1011–1021. doi: 10.1007/s11164-013-1251-8. [DOI] [Google Scholar]
- Tanouchi, T., Kawamura, M., Ajima, A., Mohri, T., Hayashi, M., Terashima, H., Hirata, F., Morimura, T., 1985. Rhodanine derivatives, process for their preparation, and aldose reductase inhibitor containing the rhodanine derivatives as active ingredient. European Patent: EP 0047109 B1.
- Tejchman W., Korona-Głowniak I., Malm A., Żylewski M., Suder P. Antibacterial properties of 5-substituted derivatives of rhodanine-3-carboxyalkyl acids. Med. Chem. Res. 2017;26:1316–1324. doi: 10.1007/s00044-017-1852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejchman W., Skórska-Stania A., Żesławska E. The crystal structures of three rhodanine- 3-carboxylic acids. J. Chem. Cryst. 2016;46:181–187. doi: 10.1007/s10870-016-0644-0. [DOI] [Google Scholar]
- Yashin Y.I. Chromatographic retention parameters and molecular structure. Pure Appl. Chem. 1989;61:2021–2026. [Google Scholar]
- Żesławska E., Nitek W., Tejchman W. The synthesis and crystal structures of the homologues of epalrestat. J. Chem. Crystallogr. 2015;45:151–157. doi: 10.1007/s10870-015-0577-z. [DOI] [Google Scholar]
- Żesławska E., Nitek W., Tejchman W., Handzlik J. Influence of 3-{5-[4-(diethylamino)benzylidene]-rhodanine} propionic acid on the conformation of 5-(4-chlorobenzylidene)-2-(4-methylpiperazin-1-yl)-3H-imidazol-4(5H)-one. J. Acta Cryst. 2018;C74:1427–1433. doi: 10.1107/S2053229618013980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.