Abstract
Chronological age remains an imperfect measure of accumulated physiological stress. Biological measures of aging may provide key advantages, allowing scientists focusing on age-related functional changes to use metrics derived from epigenetic factors like DNA methylation (DNAm), which could provide greater precision. Here we investigated the relationship between methylation-based age and an essential cognitive function that is known to exhibit age-related decline: selective attention. We found that DNAm-age predicted selective attention abilities and fully mediated the relationship between selective attention and chronological age. Using neuroimaging with magnetoencephalography, we found that gamma activity in the anterior cingulate was robustly predicted by DNAm-derived biological age, revealing the neural dynamics underlying this DNAm age-related cognitive decline. Anterior cingulate gamma activity also significantly predicted behavior on the selective attention task, indicating its functional relevance. These findings suggest that DNAm age may be a better predictor of cognitive and brain aging than more traditional chronological metrics.
Keywords: biological age, gamma oscillations, magnetoencephalography, methylation
Introduction
Increases in chronological age have been associated with gradual declines in numerous cognitive processes, including motor control (Mattay et al. 2002), memory (Nyberg et al. 2012), and attention (Verhaeghen and Cerella 2002). These detrimental changes are often mild and can be difficult to distinguish with high specificity from early stages of pathological cognitive decline. Thus, an enhanced understanding of the nature and genesis of age-related cognitive decline would be of high utility. Complicating this matter, chronological measures of aging have been criticized as an incomplete representation of the level of accumulated physiological stress that a person has undergone (Hayflick 2007; Mitnitski et al. 2013). Anecdotally, this rings true: the idiom of a person looking “good for their age” is a common one. Empirically, this is supported by the highly variable rates of mortality and morbidity as a function of chronological age, as well as by the ability of biological markers of aging to provide enhanced prediction of mortality and morbidity in late life (Mitnitski et al. 2002; Levine 2013; Marioni et al. 2015a). Despite this, uncertainty remains regarding the proper definition and measurement of biological markers of aging, as well as the relationship between these measurements and age-related cognitive decline.
In recent years, the accumulation of epigenetic changes across the lifespan has been found to powerfully predict a number of age-related complications. This was largely enabled by new methods of analyzing and interpreting DNA methylation (DNAm) patterns across a large number of CpG (i.e., 5′-cytosine-phosphate-guanine-3′) sites that can be used to predict the chronological age of a person with a high degree of accuracy (Hannum et al. 2013; Horvath 2013; Marioni et al. 2015a; Gross et al. 2016; Field et al. 2018; Horvath and Raj 2018). Discrepancies between these methylation-based assessments of age (DNAm age) and chronological age can be used to provide an index of “accelerated/decelerated” biological aging. Such measures of relative biological age advancement (ΔAge) are highly heritable (Horvath 2013) and have already been tied to health conditions such as cancer, diabetes, and human immunodeficiency virus (HIV), and may also help to predict the onset of other age-related disorders (Bell et al. 2010; Day et al. 2013; Hannum et al. 2013; Levine et al. 2015a; Gross et al. 2016). Robust links have even been drawn between ΔAge and increased all-cause mortality in later life (Marioni et al. 2015a; Christiansen et al. 2016), further supporting the role of these markers as a valuable and potentially more precise metric in the field of aging.
Though increasingly utilized in other areas, it remains unclear whether methylome-derived measures of aging are effective predictors of cognitive and brain decline. Previous research has found that ΔAge can predict domain-general cognitive fitness in later life (Marioni et al. 2015b), and that the epigenetic profiles at a number of differentially methylated regions are associated with cognitive impairment as measured by the Montreal Cognitive Assessment (Nasreddine et al. 2005; Chouliaras et al. 2018; MoCA). This makes sense, as age-related changes in DNAm have been identified in post-mortem tissue in a number of brain regions essential to “higher-order” cognitive processes (Hernandez et al. 2011; Horvath et al. 2012; Horvath 2013). Additionally, earlier work in the field of neuroepigenetics suggested a role for DNAm in memory formation and learning ability (Liu et al. 2009; Day and Sweatt 2010; Levine et al. 2015b). Coupled with recent reports of covariance between DNAm measures of aging and structural neuroimaging measures of brain integrity (Raina et al. 2017; Chouliaras et al. 2018), these findings indicate a general link between DNAm and the neural underpinnings of cognition. However, few studies have independently investigated the relationship between DNAm profiles and specific cognitive functions (Wiers 2012; Nikolova and Hariri 2015; Lancaster et al. 2018), and none has examined whether measures of ΔAge independently predict the neural dynamics underlying cognitive processes that are known to be affected by chronological age. Key among these processes is selective attention (Hasher and Zacks 1988; Plude and Doussard-Roosevelt 1989; Hasher et al. 1991; McDowd and Filion 1992; Maylor and Lavie 1998; Geerligs et al. 2014), which is the ability to preferentially allocate neural resources to task-relevant stimuli, while simultaneously limiting resources to irrelevant features of the environment. The inhibition of interfering information is of key relevance to aging, as impairments in this domain have been directly linked to reductions in specific functional abilities that also decline with age (Jefferson et al. 2006).
In this study, we combine 2 established methylome-based measures of biological aging (Hannum et al. 2013; Horvath 2013; Gross et al. 2016) with high-density, task-based magnetoencephalography (MEG) to characterize the relationship between biological aging, behavioral performance on a flanker selective attention task (Eriksen and Eriksen 1974; McDermott et al. 2017), and the oscillatory neural dynamics serving this performance in a large group of cognitively healthy adults. We find that methylome-based biological age not only predicts selective attention abilities in our participants, but also fully mediates the relationship between chronological age and selective attention performance. Further, we find that changes found in the DNAm-determined biological age (the residual difference between biological and chronological age) strongly covaries with high-frequency (i.e., gamma) oscillatory neural responses in the anterior cingulate cortex during a selective attention task. Finally, we determine that neural activity in the anterior cingulate uniquely predicts behavioral selective attention abilities, supporting the functional importance of this neural signature in behavioral performance and age-related cognitive decline.
Materials and Methods
Participants
Sixty-eight cognitively healthy, HIV-negative adults (Mage = 44.71; SD = 15.01; range = 22–72 years; 35 males; 57 right-handed) were included from a large, ongoing study of aging in HIV-infected and uninfected adults (NIH Study: MH103220) that includes collection of whole-methylome and task-based MEG data. Exclusion criteria included any medical illness affecting central nervous system function, any neurological or psychiatric disorder, history of head trauma, current substance abuse, any non-removable metal implants that would adversely affect MEG data acquisition, and incomplete methylome and/or MEG data. All participants had normal or corrected-to-normal vision, and completed a comprehensive neuropsychological battery and a demographics questionnaire (see Supplementary Table 1). The Institutional Review Board at the University of Nebraska Medical Center reviewed and approved this investigation. Written informed consent was obtained from each participant following detailed description of the study. All participants completed the same experimental protocol.
Neuropsychological Testing
Participants completed a standardized battery of neuropsychological assessments, with raw scores for each participant being converted to demographically adjusted z-scores using published normative data (Heaton et al. 2004). This battery assessed multiple functional domains, including fine motor (grooved pegboard), speed of processing (trailmaking A, digit symbol, Stroop color), attention (symbol search, Stroop word), and executive functioning (verbal fluency, semantic fluency, Stroop interference, and trailmaking B). Composite scores for each of these functional domains were computed by averaging the standardized z-scores from the assessment sets comprising the domain. Participants scoring below the population mean (i.e., zero) by more than 2 standard deviations on 2 or more functional domains were suspected to be cognitively impaired and were not included in the study.
Blood Draw and Methylation Analysis
All of the methylation metrics described below, including the Horvath and Hannum models of DNAm age, were computed on the entire data set from the large (N > 180) study of aging mentioned above and reported in previous publications (Lew et al. 2018; Spooner et al. 2018; Wiesman et al. 2018; Spooner et al. 2019; Wiesman and Wilson 2019). Whole-blood samples were collected by blood draw from each participant as closely as possible to their MEG scan date, and this time difference was used as a covariate of no interest in all analyses involving the 2 different types of measures. Importantly, all statistically significant findings held regardless of whether this covariate was partialed-out, indicating that the relative advancement of biological versus chronological age remained stable over the period of time between data collections. The DNA sample collection, methylation analysis, and epigenetic age estimation closely followed the pipeline established in earlier work (Gross et al. 2016).
Briefly, DNA was purified from whole-blood samples using BD Vacutainer EDTA collection tubes and DNeasy blood and tissue extraction kits (QIAGEN). Methylation analysis was performed using Infinium HumanMethylation450 BeadChip Kits (Illumina). Following hybridization, BeadChips were scanned using the Illumina HiScan System. All data were processed through the Minfi R processing pipeline (Aryee et al. 2014). Methylome data were downloaded from Hannum (Hannum et al. 2013) and EPIC (Riboli et al. 2002) (GEO: GSE40279 and GSE51032), and we processed these data together along with methylation data generated from the larger study mentioned above. Beta values were extracted and quantile normalized using Minfi; cell counts were estimated using estimateCellComposition and resulting normalized beta values were adjusted for cell types (Houseman et al. 2012; Gross et al. 2016). All data was then normalized using a modified BMIQ procedure provided by Horvath (Horvath 2013). The gold standard was set to the median beta observed in the Hannum study (Hannum et al. 2013).
For all analyses reported in the main text of this manuscript, the “consensus model” of methylation age was used, which combines both the Horvath (Horvath 2013) and Hannum (Hannum et al. 2013) methods of prediction and has been previously found to provide more substantial predictive capacity than either model in isolation (Gross et al. 2016). In addition, we also computed all of our main analyses using the Hannum and Horvath model data individually to ensure that none of our primary findings were specific to either method, as shown in the Supplementary Material. Also, similarly to the analysis reported in Gross et al. (2016), the residuals from a regression of the consensus metric of biological age on chronological age were used to represent the acceleration/deceleration of biological age relative to chronological age (ΔAge).
MEG Experimental Paradigm and Behavioral Data Analysis
Participants performed an arrow-based version of the Eriksen flanker task (Eriksen and Eriksen 1974; McDermott et al. 2017; Heinrichs-Graham et al. 2018) while seated in a nonmagnetic chair within the magnetically shielded room. Each trial began with a fixation that was presented for an interval of 1450–1550 ms. A row of 5 arrows was then presented for 2500 ms and participants were instructed to indicate with their right hand whether the middle arrow was pointing to the left (index finger) or right (middle finger). The 200 total trials were pseudo-randomized and equally split between congruent and incongruent conditions, with left and right pointing arrows being equally represented in the 2 conditions (Fig. 1). Total MEG recording time was about 14 min. Reaction time (RT) data were extracted from each trial, and trial-wise outliers were excluded based on a fixed threshold of ±2.5 SD from the mean per participant. To provide a behavioral measure of selective attention function, the average reaction times per participant for the incongruent and congruent conditions were subtracted, to produce an RT “flanker effect” for each participant. At this stage, the behavioral data from 3 participant-level outliers was excluded based upon a fixed threshold of ±2.5 SD from the group mean.
Figure 1 .
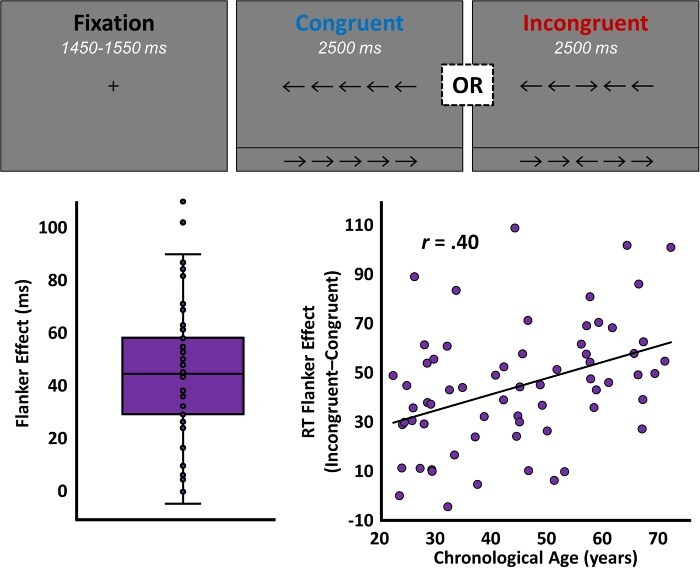
Selective attention task design and behavior. The single-trial layout of the flanker task is shown above, in which participants viewed a centrally presented crosshair for 1500 ms and then were presented with 1 of the 4 arrow arrays shown. Upon presentation of the arrow array, participants were instructed to indicate the direction of the central arrow, regardless of congruency. The significant effect of congruency (i.e., the flanker effect) on reaction time (P < 0.001) is illustrated by the box-and-whisker plot on the bottom left. The scatterplot on the bottom right shows the relationship between chronological age (x-axis, in years) and the effect of congruency on reaction time (y-axis, in milliseconds), with the line of best fit and correlation coefficient for this relationship overlaid.
MEG Data Acquisition
Four head-position indicator (HPI) coils were attached to the participant’s head and localized, together with the 3 fiducial points and scalp surface, using a 3D digitizer (Fastrak 3SF0002, Polhemus Navigator Sciences, Colchester, VT, USA). Once the participant was positioned for MEG recording, an electric current with a unique frequency label (e.g., 322 Hz) was fed to each of the coils. This induced a measurable magnetic field and allowed each coil to be localized in reference to the sensors throughout the recording session. All recordings were conducted in a one-layer magnetically shielded room with active shielding engaged for environmental noise compensation. Neuromagnetic responses were sampled continuously at 1 kHz with an acquisition bandwidth of 0.1–330 Hz using a 306-sensor Elekta MEG system (Helsinki, Finland) equipped with 204 planar gradiometers and 102 magnetometers. Participants were monitored during data acquisition via real-time audio-video feeds from inside the shielded room. Importantly, since head movement differences as a function of age are a concern in neuroimaging, we correlated head movement parameters (i.e., individual participant mean, median, and range of motion over the course of the task) with consensus predicted age, chronological age, and ΔAge, and found no significant covariance between any of these metrics (all P’s > 0.15). Further, to ensure that movement parameters were not having any specific effect on our main neural measure of interest, we correlated our head movement parameters with the flanker effect on gamma oscillations in the anterior cingulate cortex and again found no indication of a relationship (all P’s > 0.40).
MEG Preprocessing, Time-Frequency Transformation, and Sensor-Level Statistics
Each MEG data set was individually corrected for head motion, aligned to the starting head position for each participant, and subjected to noise reduction using the signal space separation method with a temporal extension (tSSS; Taulu and Simola 2006). The tSSS and movement compensation parameters were as follows: buffer length = 6 s, subspace correlation threshold = 0.95, HPI extraction window = 0.2 s, HPI sliding-window step = 0.01 s, and HPI goodness-of-fit = 0.90. Only data from the gradiometers was used for further analysis. Cardiac and blink artifacts were removed from the data using signal-space projection, which was subsequently accounted for during source reconstruction (Uusitalo and Ilmoniemi 1997). The continuous magnetic time series was then divided into 1500 ms epochs, with the baseline extending from −450 to −50 ms prior to the onset of the probe stimuli. Epochs containing artifacts were rejected using a fixed threshold method, supplemented with visual inspection. An average amplitude threshold of 1037.13 (SD = 200.67) fT and an average gradient threshold of 234.85 (SD = 88.60) fT/s was used to reject artifacts. Across the group, an average of 174.12 (SD = 10.29) trials per participant were used for further analysis. Importantly, none of our statistical comparisons were compromised by relevant differences in trial number, as the number of accepted trials did not differ across congruency conditions (P > 0.60), and the difference in trials between conditions did not covary with any measures of chronological age, biological age, or ΔAge (all P’s > 0.30).
The artifact-free epochs were next transformed into the time-frequency domain using complex demodulation, and the resulting spectral power estimations per sensor were averaged over trials to generate time-frequency plots of mean spectral density. These sensor-level data were normalized by each respective bin’s baseline power, which was calculated as the mean power during the −450 to −50 ms time period. The specific time-frequency windows used for subsequent source imaging were determined by statistical analysis of the sensor-level spectrograms across all conditions and the entire array of gradiometers. Each data point in the spectrogram was initially evaluated using a mass univariate approach based on the general linear model. To reduce the risk of false-positive results while maintaining reasonable sensitivity, a two-stage procedure was followed to control for type 1 error. In the first stage, one-sample t-tests were conducted on each data point and the output spectrogram of t-values was thresholded at P < 0.05 to define time-frequency bins containing potentially significant oscillatory deviations across all participants. In stage two, the time-frequency bins that survived the threshold were clustered with temporally and/or spectrally neighboring bins that were also above the threshold (P < 0.05), and a cluster value was derived by summing all of the t-values of all data points in the cluster. Nonparametric permutation testing was then used to derive a distribution of cluster values and the significance level of the observed clusters (from stage one) were tested directly using this distribution (Ernst 2004; Maris and Oostenveld 2007). For each comparison, at least 10 000 permutations were computed to build a distribution of cluster values. Based on these analyses, the time-frequency windows that contained significant oscillatory events across all participants were subjected to a beamforming analysis.
Structural MRI Processing and MEG Coregistration
Given the digitization of fiducials, head coils, and the scalp surface performed prior to the recordings (see “MEG Data Acquisition”), each participant’s MEG data could be transformed into a common coordinate system and co-registered with their individual structural T1-weighted MRI data. This was done prior to source-space analysis using BESA MRI (Version 2.0). Structural MRI data were aligned parallel to the anterior and posterior commissures and transformed into standardized space. MRI data were acquired with a Philips Achieva 3 T X-series scanner using an 8-channel head coil and a 3D fast-field echo sequence with the following parameters: TR: 8.09 ms, TE: 3.70 ms, field of view: 24 cm, slice thickness: 1 mm with no gap, in-plane resolution: 1.0 × 1.0 mm. Following source-space analysis (i.e., beamforming), each participant’s 4.0 × 4.0 × 4.0 mm functional images were also transformed into standardized space using the transform that was previously applied to the structural MRI volume and spatially resampled.
MEG Source Imaging and Statistics
Cortical networks were imaged using dynamic imaging of coherent sources (Gross et al. 2001), which applies spatial filters to time-frequency sensor data in order to calculate voxel-wise source power for the entire brain volume. The single images are derived from the cross spectral densities of all combinations of MEG gradiometers averaged over the time-frequency range of interest, and the solution of the forward problem for each location on a grid specified by input voxel space. For this analysis, the forward solution was computed using a spherically symmetric model (Sarvas 1987). Following convention, we computed noise-normalized, source power per voxel in each participant using active (i.e., task) and passive (i.e., baseline) periods of equal duration and bandwidth. Such images are typically referred to as pseudo-t maps, with units (pseudo-t) that reflect noise-normalized power differences (i.e., active vs. passive) per voxel. This generated participant-level pseudo-t maps for each time-frequency-specific response identified in the sensor-level cluster-based permutation analysis.
These whole-brain images per time-frequency response were computed for each condition (i.e., congruent and incongruent) and then subtracted within each participant to generate maps representing the effect of congruency (i.e., the “flanker” effect). This has the net effect of retaining the neural differences resultant of selective attention load while nulling the responses underlying lower-level visual and perceptual processing. To examine the effects of biological age measures on the neural dynamics serving selective attention, the whole-brain congruency effect maps were then regressed on linear measures of DNAm age while controlling for the effect of chronological age, with a two-step process to control for multiple comparisons. First, to examine potentially significant 3-dimensional clusters of covariance, the statistical output maps of this analysis were thresholded with a voxel-wise significance cutoff of P < 0.005 and cluster-size cutoff of k > 500. Second, the voxel-wise threshold was increased to a very stringent familywise error-corrected significance cutoff of P < 0.05 using second-level analysis in SPM12. MEG pre-processing and imaging used the Brain Electrical Source Analysis (BESA version 6.0) software. Whole-brain regressions used SPM12, and all other statistical analyses relating behavioral performance, neuroimaging metrics, and aging measures were performed using R. Bootstrapping was performed to derive 90% (one-tailed) confidence intervals for indirect effects for the hypothesized mediations using the robmed package in R. Note that one-tailed tests were appropriate here, as only 1 direction of indirect effect would be interpretable for each of the mediations performed. All other statistical tests were two-tailed, unless otherwise stated.
Results
Methylation Age Predicts Selective Attention Performance
Sixty-eight adult participants each completed 200 trials of our selective attention task (Fig. 1, top) while undergoing high-density MEG. Please see Supplementary Table 1 for comprehensive sample characteristics. Participants performed generally well (meanRT = 664.12, SDRT = 139.42; mean%Correct = 97.87, SD%Correct = 4.31) and, as is typically reported, a significant effect of congruency was found on RT (t(67) = 9.88, P < 0.001), such that participants were significantly slower to respond on incongruent compared to congruent trials (Fig. 1, bottom left). This congruency or “flanker effect,” which is representative of selective attention function, was predicted by both chronological (r(63) = 0.40, P = 0.001; Fig. 1, bottom right) and methylation-based biological (r(63) = 0.44, P < 0.001; Figure 2, right) measures of aging. Intriguingly, methylation-based biological age fully mediated the relationship between chronological age and selective attention ability (ΔR = 0.46, indirect effect = 0.93, 95% CIlower = 0.05), indicating that biological age is responsible for the observed effect of chronological age on selective attention function. Finally, as hypothesized, biological age predicted selective attention ability above and beyond the effect of chronological age (r(63) = 0.22, P = 0.041, one-tailed test), such that increased biological age, relative to chronological age, predicted worse performance on the task.
Figure 2 .
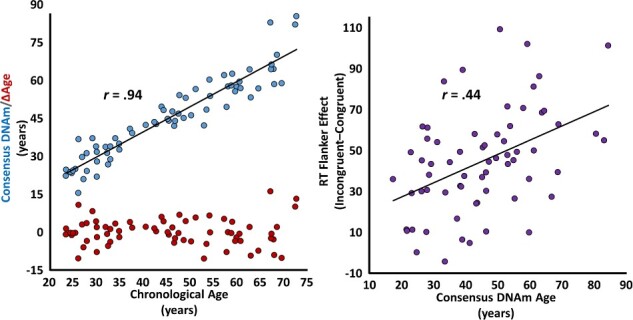
Methylation-based biological age predicts chronological age and the behavioral flanker effect. The scatterplot on the left represents the relationship between chronological age (x-axis, in years) and methylation-based biological age (y-axis, blue, in years). The line of best fit and effect size for this relationship are overlaid. On the same plot, each participant’s biological age acceleration/deceleration (y-axis, red, in years) is indicated below in red. The scatterplot on the right represents the relationship between methylation-based biological age (x-axis, in years) and the effect of congruency (i.e., the flanker effect) on reaction time (y-axis, in milliseconds), with the line of best fit and correlation coefficient for this relationship overlaid.
Accelerated Biological Aging Predicts the Neural Dynamics of Selective Attention
Chronological age covaried with methylome-predicted biological age (r(66) = 0.94, P < 0.001; Fig. 2, left, blue); however, a reasonable amount of variability remained in the residuals of this relationship (SDresiduals = 5.39; Fig. 2, left, red). Since these residuals (ΔAge) represent the standardized deviation of biological age from chronological age, they are commonly used as a measure of “accelerated/decelerated aging”. Notably, we ran these same analyses using both the Hannum and Horvath models of DNAm age separately, and the results remained nearly identical (Supplementary Fig. 1), with the consensus of these models providing the most robust estimator of chronological age. Before relating DNAm age directly to neural activity, we first had to identify the significant neural responses during our selective attention task. Significant oscillatory responses were observed in 4 distinct time-frequency windows (Fig. 3). These included a strong increase or synchronization in the theta (4–8 Hz) range over medial posterior and lateral anterior cortices extending temporally from stimulus onset to 250 ms post-stimulus onset. In a spatially overlapping group of MEG sensors, there was also a broadband gamma (62–72 Hz) increase, which ranged temporally from 200–325 ms post-stimulus onset. In addition, there was a slightly more lateral posterior decrease or desynchronization response in the alpha (8–16 Hz) band, which stretched from 200 to 600 ms post-stimulus onset. Finally, in medial central sensors, a strong decrease in the beta (18–26 Hz) band was observed from 300 to 550 ms post-stimulus onset. See Supplementary Figs 2–4 for a visualization of the spectral, temporal, and spatial extents of each of these clusters in sensor space. To identify the anatomical basis of these 4 oscillatory responses, we performed source reconstruction on each per congruency condition. The conditional volumetric maps for each participant where then subtracted (i.e., incongruent-congruent) to generate 4 spectrally specific whole-brain maps reflecting selective attention-related neural activity per participant. Importantly, the beta response was found to originate from the hand-knob region of the precentral gyrus contralateral to movement, indicating a motor-related origin. As the focus of this study was the neural dynamics relating to selective attention function, we did not examine this response further.
Figure 3 .
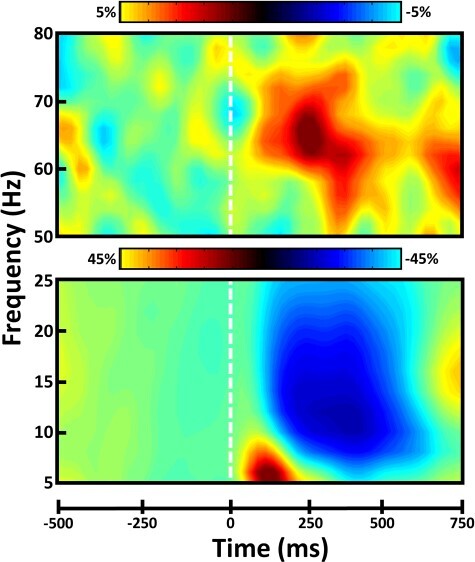
Spectral and temporal definitions of neural responses to the selective attention task. Sensor-level spectrograms showing the time-frequency representations of neural responses identified through the cluster-based permutation analysis across the entire array of gradiometers. Time (in ms) is shown on the x-axis, frequency (in Hz) is denoted on the y-axis, and the dashed white line at 0 ms indicates the onset of the flanker stimuli. The color scale bar for percent change from baseline is displayed above each plot. Each spectrogram represents group-averaged data from 1 gradiometer sensor that was representative of the neural response. The beta motor response is not shown.
Next, we probed whether the spectrally specific neural dynamics serving selective attention were predicted by biological age, above and beyond the effect of chronological age. Put another way, we examined the covariance between ΔAge and these neural dynamics. Interestingly, ΔAge predicted selective attention-related oscillations only in the gamma range. Regressions of ΔAge on whole-brain selective attention-related neural activity in the gamma band revealed significant clusters in the anterior cingulate (rpeak = 0.57; peak coordinates: x = 2, y = −2, z = 41), left superior parietal (rpeak = 0.47; peak coordinates: x = −14, y = −80, z = 45), and left primary visual cortices (rpeak = 0.44; peak coordinates: x = −7, y = −84, z = −4). In all 3 of these regions, greater ΔAge predicted a greater effect of congruency (Supplementary Fig. 5), indicating increased neural resources were needed for selective attention processing. In other words, greater biological age relative to chronological age was associated with greater selective attention-related brain activity in the gamma range in these 3 regions. Although all 3 regions exhibited significant relationships with relative age advancement at their peaks, only the anterior cingulate cluster survived stringent family-wise error correction. Again, these same analyses were run using the Hannum and Horvath DNAm age predictions separately and produced very similar results (Supplementary Fig. 6). To determine the importance of anterior cingulate gamma activity in selective attention performance, we next correlated the neural effect of congruency in this region with the behavioral flanker effect, which reflects selective attention function. We found that these variables were significantly related, such that greater congruency effects on gamma-range neural activity in this region were associated with greater congruency effects on behavior (r(59) = 0.30; P = 0.020; Fig. 4). Importantly, the relationship between ΔAge and selective attention function was mediated by congruency effects on gamma activity in the anterior cingulate (ΔR = 0.17, indirect effect = 0.79, 95% CIlower = 0.14). Lastly, whole-brain selective attention-related neural activity did not covary significantly with ΔAge in the theta or alpha bands.
Figure 4 .
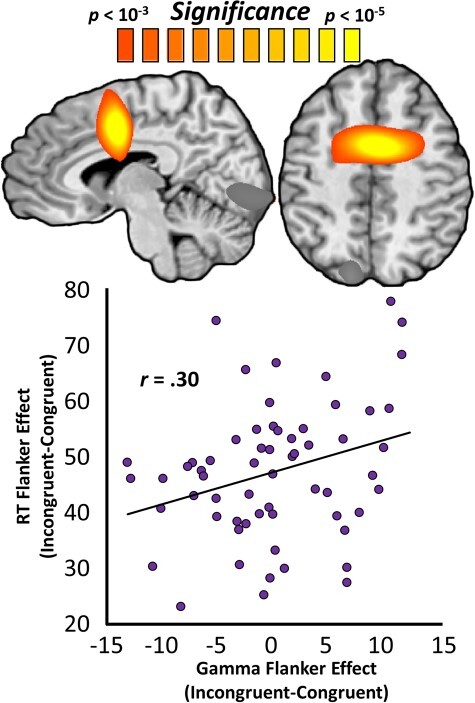
DNAm-predicted anterior cingulate gamma oscillations predict the behavioral flanker effect. Whole-brain maps are voxel-wise regressions of gamma-range congruency maps on biological age advancement and represent the anterior cingulate cluster that survived stringent multiple comparisons correction. The color scale-bar (above maps) indicates the statistical significance of the test at each voxel, with warmer colors representing more robust relationships. The scatterplot below represents the relationship between the congruency effect on gamma oscillations (x-axis, in units of pseudo-t) at the peak shown above and the effect of stimulus congruency on reaction time (y-axis, in milliseconds), with the line of best fit and effect size overlaid.
Discussion
Despite the growing popularity of physiologically-based measures of biological aging, virtually no studies have found significant relationships between these metrics and specific cognitive abilities that are known to covary with chronological age. Herein, we found a robust and behaviorally relevant connection between biological age and the oscillatory neural dynamics serving selective attention function. Specifically, we found that methylation-based measures of biological aging predict a well-established behavioral index of selective attention function and even fully mediate the classic relationship between chronological age and this measure. Further, biological age was found to predict selective attention-related gamma activity in the anterior cingulate cortex, a region that is widely known to be essential to conflict processing and selective attention (Heinze et al. 1994; Weissman et al. 2003; Botvinick et al. 2004; Geerligs et al. 2014; McDermott et al. 2017). Importantly, neural activity in this region was also found to predict behavior on the task, as well as to mediate the novel relationship between biological age and selective attention function, supporting the relevance of this response for selective attentional performance.
Previous studies have reported a relationship between performance on selective attention tasks and chronological aging (Plude and Doussard-Roosevelt 1989; McDowd and Filion 1992; Geerligs et al. 2014), however, whether such performance could be linked to biological aging metrics has remained uncertain. Our finding of strong covariance between biological aging metrics and both behavioral and neural markers of selective attention function provides powerful support for such a link. Importantly, biological aging predicted these neural and behavioral markers “above and beyond” the effects of chronological aging, indicating that DNAm age may provide a more effective model of aging than chronological measures in the context of age-related cognitive decline. Additionally, we identified population-level neuronal dysfunction in the anterior cingulate cortex as an important mediator of these declines.
The anterior cingulate has been found in a number of previous studies to be critically involved in the performance of selective attention tasks (Heinze et al. 1994; Weissman et al. 2003; Botvinick et al. 2004; Weissman et al. 2005; Frühholz et al. 2011; Iannaccone et al. 2015; McDermott et al. 2017), and activity in this region has been previously shown to vary as a function of chronological age (Milham et al. 2002; Geerligs et al. 2014). Further, the anterior cingulate has also been repeatedly linked to the processing of cognitive interference and conflict in the context of similar tasks (Heinze et al. 1994; Milham et al. 2002; Peterson et al. 2002; van Veen and Carter 2002; Bush et al. 2003; Weissman et al. 2003; Botvinick et al. 2004; Weissman et al. 2005; Hanslmayr et al. 2008; Frühholz et al. 2011; Iannaccone et al. 2015; McDermott et al. 2017). Thus, the increased effect of congruency in this region as ΔAge increased likely reflects an amplified effect of interfering stimuli on conflict processing circuitry in physiologically older adults. In other words, as DNAm age accelerates disproportionately with chronological age, fast gamma-range activity in the anterior cingulate may be more affected by the presence of visual distractor stimuli, which in turn interferes with the participant’s ability to inhibit the effect of these distractors on behavior. In short, from these data we can logically conclude that the essential role of the anterior cingulate cortex in selective attention and conflict processing is tied to the physiological impact of epigenetic alterations on the genome. Finally, it should also be noted that these findings were consistent across the 2 models of DNAm age (i.e., the Hannum and Horvath methods), reinforcing the robustness of these effects.
Although our results are of interest, the limitations of this study must be kept in mind. First and foremost, the methylation data from our participants were derived from peripheral blood and not directly from the brain, as the collection of neural tissue from healthy adult brains is not possible. Age-related patterns of CpG methylation have been found to vary across tissue types (Christensen et al. 2009; Thompson et al. 2010). Thus, while the observed relationship between methylation-based measures of biological age and neural activity is statistically robust, a level of extrapolation is necessary to assume that the within-participant variability in methylation patterns across tissue types (i.e., blood and brain tissues) is less than that observed across participants as a function of age. Importantly, recent research supports this extrapolation, as DNAm measures of age have been found to be remarkably consistent across the vast majority of tissue types, with some of the highest concordance observed between brain and blood samples (Horvath et al. 2012; Horvath 2013). Regardless, these results can be interpreted to indicate that, at least at the level of peripheral epigenetic alterations, physiological measures of accumulated biological stress can be used to predict extremely important neurophysiological and cognitive features that also vary across the lifespan. Such an analysis can now be used in studies of neurodegenerative and other disorders that affect cognitive function. It should also be noted that due to the nature of our analysis pipeline and neuroimaging research in general, in which the maximization of signal-to-noise is always a concern, it is likely that these results are not exhaustive in nature. It is possible that only the most robust and consistent link between DNAm, ΔAge, and selective attention-related neural activity are reported here. As new research emerges, more targeted, seed-based analyses of different cortical and sub-cortical areas may be warranted. Finally, it is important to acknowledge that only 1 cognitive faculty (i.e., selective attention) was investigated here. While these findings provide an essential first step, a great deal of future investigation is warranted into the interdependencies between DNAm age, chronological age, other cognitive functions (e.g., stimulus-response interference, working memory, executive function, etc.), and the underlying neural dynamics.
Supplementary Material
Funding
MH103220, MH103220-S2, and MH116782 (T.W.W.), MH062261 (H.S.F.), and AG055332 (A.I.W.) from the National Institutes of Health; grant #1539067 from the National Science Foundation (T.W.W.), and by a Research Support Fund grant from the Nebraska Health System and the University of Nebraska Medical Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Notes
Conflict of Interest: None declared.
Author's Contributions
Conceptualization, H.S.F. and T.W.W.; methodology, H.S.F. and T.W.W.; formal analysis, A.I.W., B.M., and T.W.; resources, H.S.F. and T.W.W.; data curation, M.T.R., J.O., H.S.F., T.I., and T.W.W.; writing—original draft, A.I.W. and T.W.W.; writing—review and editing, A.I.W., M.T.R., J.O., B.M., T.W., T.I., S.S., H.S.F., T.W.W.; visualization, A.I.W.; supervision, H.S.F., T.I., and T.W.W.; funding acquisition, H.S.F. and T.W.W.
References
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium dna methylation microarrays. Bioinformatics. 30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. 2010. Genome-wide dna methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinick MM, Cohen JD, Carter CS. 2004. Conflict monitoring and anterior cingulate cortex: an update. Trends Cogn Sci. 8(12):539–546. [DOI] [PubMed] [Google Scholar]
- Bush G, Shin LM, Holmes J, Rosen BR, Vogt BA. 2003. The multi-source interference task: validation study with fmri in individual subjects. Mol Psychiatry. 8(1):60–70. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Pishva E, Haapakoski R, Zsoldos E, Mahmood A, Filippini N, Burrage J, Mill J, Kivimäki M, Lunnon Ket al. 2018. Peripheral dna methylation, cognitive decline and brain aging: pilot findings from the Whitehall ii imaging study. Epigenomics. 10(5):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno Ret al. 2009. Aging and environmental exposures alter tissue-specific dna methylation dependent upon cpg island context. PLoS Genet. 5(8):e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. 2016. Dna methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 15(1):149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2010. Dna methylation and memory formation. Nat Neurosci. 13(11):1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, Myers RM, Absher D. 2013. Differential dna methylation with age displays both common and dynamic features across human tissues that are influenced by cpg landscape. Genome Biol. 14(9):R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen BA, Eriksen CW. 1974. Effects of noise letters upon the identification of a target letter in a nonsearch task. Perception & Psychophysics. 16(1):143–149. [Google Scholar]
- Ernst MD. 2004. Permutation methods: a basis for exact inference. Statistical Science. 19(4):676–685. [Google Scholar]
- Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. 2018. Dna methylation clocks in aging: categories, causes, and consequences. Mol Cell. 71(6):882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühholz S, Godde B, Finke M, Herrmann M. 2011. Spatio-temporal brain dynamics in a combined stimulus-stimulus and stimulus-response conflict task. Neuroimage. 54(1):622–634. [DOI] [PubMed] [Google Scholar]
- Geerligs L, Saliasi E, Maurits NM, Renken RJ, Lorist MM. 2014. Brain mechanisms underlying the effects of aging on different aspects of selective attention. Neuroimage. 91:52–62. [DOI] [PubMed] [Google Scholar]
- Gross AM, Jaeger PA, Kreisberg JF, Licon K, Jepsen KL, Khosroheidari M, Morsey BM, Swindells S, Shen H, Ng CTet al. 2016. Methylome-wide analysis of chronic hiv infection reveals five-year increase in biological age and epigenetic targeting of hla. Mol Cell. 62(2):157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Kujala J, Hamalainen M, Timmermann L, Schnitzler A, Salmelin R. 2001. Dynamic imaging of coherent sources: studying neural interactions in the human brain. Proc Natl Acad Sci U S A. 98(2):694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Yet al. 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 49(2):359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanslmayr S, Pastötter B, Bäuml KH, Gruber S, Wimber M, Klimesch W. 2008. The electrophysiological dynamics of interference during the stroop task. J Cogn Neurosci. 20(2):215–225. [DOI] [PubMed] [Google Scholar]
- Hasher L, Stoltzfus ER, Zacks RT, Rypma B. 1991. Age and inhibition. J Exp Psychol Learn Mem Cogn. 17(1):163–169. [DOI] [PubMed] [Google Scholar]
- Hasher L, Zacks RT. 1988. Working memory, comprehension, and aging: a review and a new view. In: Psychology of learning and motivation. Cambridge (MA): Academic Press Inc., pp. 193–225. [Google Scholar]
- Hayflick L. 2007. Biological aging is no longer an unsolved problem. Ann N Y Acad Sci. 1100:1–13. [DOI] [PubMed] [Google Scholar]
- Heaton RK, Miller SW, Taylor MJ, Grant I. 2004. Revised comprehensive norms for an expanded halstead-reitan battery: demographically adjusted neuropsychological norms for african american and caucasian adults. Psychological Assessment Resources.
- Heinrichs-Graham E, Hoburg JM, Wilson TW. 2018. The peak frequency of motor-related gamma oscillations is modulated by response competition. Neuroimage. 165:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze HJ, Mangun GR, Burchert W, Hinrichs H, Scholz M, Münte TF, Gös A, Scherg M, Johannes S, Hundeshagen H. 1994. Combined spatial and temporal imaging of brain activity during visual selective attention in humans. Nature. 372(6506):543–546. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJet al. 2011. Distinct dna methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 20(6):1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. Dna methylation age of human tissues and cell types. Genome Biol. 14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Raj K. 2018. Dna methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 19(6):371–384. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA. 2012. Aging effects on dna methylation modules in human brain and blood tissue. Genome Biol. 13(10):R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. 2012. Dna methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannaccone R, Hauser TU, Staempfli P, Walitza S, Brandeis D, Brem S. 2015. Conflict monitoring and error processing: new insights from simultaneous eeg-fmri. Neuroimage. 105:395–407. [DOI] [PubMed] [Google Scholar]
- Jefferson AL, Paul RH, Ozonoff A, Cohen RA. 2006. Evaluating elements of executive functioning as predictors of instrumental activities of daily living (iadls). Arch Clin Neuropsychol. 21(4):311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster K, Morris JP, Connelly JJ. 2018. Neuroimaging epigenetics: challenges and recommendations for best practices. Neuroscience. 370:88–100. [DOI] [PubMed] [Google Scholar]
- Levine ME. 2013. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 68(6):667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. 2015a. Dna methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY). 7(9):690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S. 2015b. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and alzheimer’s disease related cognitive functioning. Aging (Albany NY). 7(12):1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew BJ, McDermott TJ, Wiesman AI, O’Neill J, Mills MS, Robertson KR, Fox HS, Swindells S, Wilson TW. 2018. Neural dynamics of selective attention deficits in hiv-associated neurocognitive disorder. Neurology. 91(20):e1860–e1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. 2009. Dna methylation impacts on learning and memory in aging. Neurobiol Aging. 30(4):549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SRet al. 2015a. Dna methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie Aet al. 2015b. The epigenetic clock is correlated with physical and cognitive fitness in the lothian birth cohort 1936. Int J Epidemiol. 44(4):1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris E, Oostenveld R. 2007. Nonparametric statistical testing of eeg- and meg-data. J Neurosci Methods. 164(1):177–190. [DOI] [PubMed] [Google Scholar]
- Mattay VS, Fera F, Tessitore A, Hariri AR, Das S, Callicott JH, Weinberger DR. 2002. Neurophysiological correlates of age-related changes in human motor function. Neurology. 58(4):630–635. [DOI] [PubMed] [Google Scholar]
- Maylor EA, Lavie N. 1998. The influence of perceptual load on age differences in selective attention. Psychol Aging. 13(4):563–573. [DOI] [PubMed] [Google Scholar]
- McDermott TJ, Wiesman AI, Proskovec AL, Heinrichs-Graham E, Wilson TW. 2017. Spatiotemporal oscillatory dynamics of visual selective attention during a flanker task. Neuroimage. 156:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowd JM, Filion DL. 1992. Aging, selective attention, and inhibitory processes: a psychophysiological approach. Psychol Aging. 7(1):65–71. [DOI] [PubMed] [Google Scholar]
- Milham MP, Erickson KI, Banich MT, Kramer AF, Webb A, Wszalek T, Cohen NJ. 2002. Attentional control in the aging brain: insights from an fmri study of the stroop task. Brain Cogn. 49(3):277–296. [DOI] [PubMed] [Google Scholar]
- Mitnitski A, Song X, Rockwood K. 2013. Assessing biological aging: the origin of deficit accumulation. Biogerontology. 14(6):709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnitski AB, Graham JE, Mogilner AJ, Rockwood K. 2002. Frailty, fitness and late-life mortality in relation to chronological and biological age. BMC Geriatr. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H. 2005. The Montreal cognitive assessment, Moca: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 53(4):695–699. [DOI] [PubMed] [Google Scholar]
- Nikolova YS, Hariri AR. 2015. Can we observe epigenetic effects on human brain function? Trends Cogn Sci. 19(7):366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg L, Lövdén M, Riklund K, Lindenberger U, Bäckman L. 2012. Memory aging and brain maintenance. Trends Cogn Sci. 16(5):292–305. [DOI] [PubMed] [Google Scholar]
- Peterson BS, Kane MJ, Alexander GM, Lacadie C, Skudlarski P, Leung HC, May J, Gore JC. 2002. An event-related functional mri study comparing interference effects in the simon and stroop tasks. Brain Res Cogn Brain Res. 13(3):427–440. [DOI] [PubMed] [Google Scholar]
- Plude DJ, Doussard-Roosevelt JA. 1989. Aging, selective attention, and feature integration. Psychol Aging. 4(1):98–105. [DOI] [PubMed] [Google Scholar]
- Raina A, Zhao X, Grove ML, Bressler J, Gottesman RF, Guan W, Pankow JS, Boerwinkle E, Mosley TH, Fornage M. 2017. Cerebral white matter hyperintensities on mri and acceleration of epigenetic aging: the atherosclerosis risk in communities study. Clin Epigenetics. 9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondière UR, Hémon B, Casagrande C, Vignat Jet al. 2002. European prospective investigation into cancer and nutrition (epic): study populations and data collection. Public Health Nutr. 5(6B):1113–1124. [DOI] [PubMed] [Google Scholar]
- Sarvas J. 1987. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Phys Med Biol. 32(1):11–22. [DOI] [PubMed] [Google Scholar]
- Spooner RK, Wiesman AI, Mills MS, O’Neill J, Robertson KR, Fox HS, Swindells S, Wilson TW. 2018. Aberrant oscillatory dynamics during somatosensory processing in hiv-infected adults. Neuroimage Clin. 20:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner RK, Wiesman AI, Proskovec AL, Heinrichs-Graham E, Wilson TW. 2019. Rhythmic spontaneous activity mediates the age-related decline in somatosensory function. Cereb Cortex. 29(2):680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulu S, Simola J. 2006. Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements. Phys Med Biol 51:1759. [DOI] [PubMed] [Google Scholar]
- Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, Barzilai N. 2010. Tissue-specific dysregulation of dna methylation in aging. Aging Cell. 9(4):506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo MA, Ilmoniemi RJ. 1997. Signal-space projection method for separating meg or eeg into components. Med Biol Eng Comput. 35(2):135–140. [DOI] [PubMed] [Google Scholar]
- van Veen V, Carter CS. 2002. The anterior cingulate as a conflict monitor: Fmri and erp studies. Physiol Behav. 77(4–5):477–482. [DOI] [PubMed] [Google Scholar]
- Verhaeghen P, Cerella J. 2002. Aging, executive control, and attention: a review of meta-analyses. Neurosci Biobehav Rev. 26(7):849–857. [DOI] [PubMed] [Google Scholar]
- Weissman DH, Giesbrecht B, Song AW, Mangun GR, Woldorff MG. 2003. Conflict monitoring in the human anterior cingulate cortex during selective attention to global and local object features. Neuroimage. 19(4):1361–1368. [DOI] [PubMed] [Google Scholar]
- Weissman DH, Gopalakrishnan A, Hazlett CJ, Woldorff MG. 2005. Dorsal anterior cingulate cortex resolves conflict from distracting stimuli by boosting attention toward relevant events. Cereb Cortex. 15(2):229–237. [DOI] [PubMed] [Google Scholar]
- Wiers CE. 2012. Methylation and the human brain: towards a new discipline of imaging epigenetics. Eur Arch Psychiatry Clin Neurosci. 262(3):271–273. [DOI] [PubMed] [Google Scholar]
- Wiesman AI, O’Neill J, Mills MS, Robertson KR, Fox HS, Swindells S, Wilson TW. 2018. Aberrant occipital dynamics differentiate HIV-infected patients with and without cognitive impairment. Brain. 141(6):1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesman AI, Wilson TW. 2019. The impact of age and sex on the oscillatory dynamics of visuospatial processing. Neuroimage. 185:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


