Abstract
Diffusion magnetic resonance (MR) tractography represents a novel opportunity to investigate conserved and deviant developmental programs between humans and other species such as mice. To that end, we acquired high angular resolution diffusion MR scans of mice [embryonic day (E) 10.5 to postnatal week 4] and human brains [gestational week (GW) 17–30] at successive stages of fetal development to investigate potential evolutionary changes in radial organization and emerging pathways between humans and mice. We compare radial glial development as well as commissural development (e.g., corpus callosum), primarily because our findings can be integrated with previous work. We also compare corpus callosal growth trajectories across primates (i.e., humans and rhesus macaques) and rodents (i.e., mice). One major finding is that the developing cortex of humans is predominated by pathways likely associated with a radial glial organization at GW 17–20, which is not as evident in age-matched mice (E 16.5, 17.5). Another finding is that, early in development, the corpus callosum follows a similar developmental timetable in primates (i.e., macaques and humans) as in mice. However, the corpus callosum grows for an extended period of time in primates compared with rodents. Taken together, these findings highlight deviant developmental programs underlying the emergence of cortical pathways in the human brain.
Keywords: brain pathways, callosal, cingulum, diffusion MRI, fetus, human, mouse, tractography
Introduction
Diffusion magnetic resonance imaging (MRI) is a high-throughput and noninvasive method that can be used to investigate neural pathways in adulthood, as well as their development (Mori and Zhang 2006; Qiu et al. 2015). Questions focused on the development and evolution of connectivity patterns and cell migration have traditionally been addressed with tract-tracers or carbocyanine tracers. Although these methods have revealed key developmental processes in the brain, they are time-consuming and invasive and suffer from a number of technical limitations (Nudo and Masterton 1989, 1990; Chen et al. 2006; Heilingoetter and Jensen 2016). Moreover, carbocyanine tracers (e.g., DiI) are one of the few methods available for study of migration and establishment of pathways in humans (Burkhalter et al. 1993; Konstantinidou et al. 1995; Hevner 2000; Tardif and Clarke 2001). As a result, we have been able to only glimpse at the developmental processes dictating evolutionary changes in patterns of brain connections in the human lineage.
Diffusion imaging has revealed important developmental processes in humans such as radial glial pathways in the developing cortex (Takahashi et al. 2012; Kolasinski et al. 2013; Xu et al. 2014; Miyazaki et al. 2016; Vasung et al. 2017; Das and Takahashi 2018), tangential migratory routes (Takahashi et al. 2012; Kolasinski et al. 2013; Miyazaki et al. 2016; Wilkinson et al. 2017), and emerging axonal pathways coursing through the white matter of the brain (e.g., Vasung et al. 2010; Takahashi et al. 2012). We here focus on the developmental timeline of radial glia and commissural fibers in humans and mice to identify conserved and deviant developmental programs giving rise to evolutionary changes in the structure and connectivity patterns in the human brain. Among the commissural fibers available for study, we focus especially on the developmental timeline of corpus callosum maturation in humans and mice because the corpus callosum is clearly homologous across rodents and primates, which is not true for many other cortical association pathways (e.g., uncinate fasciculus and arcuate fasciculus; Schmahmann et al. 2007; Charvet et al. 2017b). The corpus callosum is a white matter fiber pathway integral for interhemispheric cortical communication, which evolved with the emergence of placental mammals and is disproportionately expanded in primates compared with many other eutherian mammals (Katz et al. 1983; Aboitiz and Montiel 2003; Bloom and Hynd 2005; Manger et al. 2010; Caminiti et al. 2013). Diffusion MR tractography provides a 3D perspective with which to compare development of radial fibers and emerging axonal pathways across the entire cortex of mammalian species.
A complex process orchestrates cell production in the human brain. In humans, neurogenesis across the six-layered cortex (called the isocortex) is thought to begin at around embryonic day E 30 (Bystron et al. 2006) and ceases around birth (270 days after gestation; Zecevic et al. 2005; Malik et al. 2013; Charvet et al. 2017a), whereas isocortical neurogenesis in mice begins on E 10 to last only about 8 days (Caviness et al. 2003, 2009). As cells exit the proliferative zone to become neurons, they migrate towards the cortical plate (CP) along radial glia (Rakic 1971, 1972, 1974, 2002, 2003a; Varon and Somjen 1979; Patel et al. 1994; Noctor et al. 2001; Rash and Richards 2001; deAzevedo et al. 2003; Gertz et al. 2014; Nowakowski et al. 2016). The first neurons to extend axons are called pioneers. Similar to radial glial cells, which guide the migration of newly born neurons, pioneer axons act as scaffolds guiding the extension of other axons (Supér et al. 1998; Nishikimi et al. 2013). Pioneer axons guide the development of cortical neurons projecting subcortically and cortically, including neurons projecting to the contralateral hemisphere (McConnell et al. 1989; De Carlos and O’leary 1992; Koester and O’Leary 1994; Rash and Richards 2001; Paul et al. 2007). As more and more cells exit the cell cycle, the progenitor pool, the radial organization of the developing human fetal cortex wane, and the white matter of the developing cortex become increasingly replaced with long-range projecting pathways as assessed from diffusion MRI (Takahashi et al. 2012). The aim of the present study is to compare the timetable of “scaffolds” such as radial glial fibers and pioneer axons between species. Although “pioneer” might imply a guidance function, we define pioneer exclusively temporally, and we focus on the temporal sequence of fibers emerging across the hemisphere of the developing cortex. Another aim of the present study is to compare the timing of cortical association pathway maturation (i.e., corpus callosum) between human and mice to investigate evolutionary changes in developmental processes giving rise to variation in cortico-cortical association pathways between species.
It is well known that primate isocortical neurogenesis is unusual compared with rodents (Rakic 1995; Zecevic et al. 2005; Kriegstein et al. 2006; Martínez-Cerdeño et al. 2006). After controlling for differences in developmental schedules, isocortical neuronal production extends for an unusually long time in humans and macaques compared with mice (Clancy et al. 2001; Workman et al. 2013; Cahalane et al. 2014; Charvet et al. 2017a). The extension in the duration of isocortical neurogenesis leads to an amplification of upper layer neurons (i.e., layer II–IV) and a concomitant expansion of long-range cortico-cortical association pathways in adult primates (Charvet et al. 2015, 2017a, 2019; Krienen et al. 2016). Given these data, it may be expected that there are differences in radial glial organization between humans and mice during neurogenesis because an extension in the duration of cortical neurogenesis should entail an extended period in which fibers are radially organized to allow newly born neurons to migrate from the progenitor pool to the CP. Extending the duration of neurogenesis might also lead to major changes in cortical circuitry by potentially altering the timing of initial axon extension of pioneer neurons and/or extending the duration over which cortical association fibers grow. Many of the callosally projecting neurons are situated in the upper layers (i.e., layer II–IV) (Isseroff et al. 1984; Meissirel et al. 1991; Greig et al. 2013), which have undergone a dramatic expansion in primates (Charvet et al. 2017a, 2019), and a is concomitant increased expression of select genes encoding ion channels, neurofilament, and synaptic proteins in upper layers (Zeng et al. 2012; Krienen et al. 2016). In addition, callosally projecting neurons express a suite of genes, some of which are differentially expressed in macaques compared with mice (Fame et al. 2011, 2016a). These studies highlight major modifications in callosal organization between primates and rodents. Thus, we can use diffusion MR tractography to investigate deviations in developmental processes giving rise to the evolution of cortical association pathways in the human lineage.
Considerable work has been conducted on radial glial development, pioneer, as well as callosal development in model organisms such as mice, rats, monkeys, and cats (Wise and Jones 1976; Marin-Padilla 1978; Innocenti 1981; O’Leary et al. 1981; Ivy and Killackey 1982; Silver et al. 1982; Luskin and Shatz 1985; Berbel and Innocenti 1988; Mrzljak et al. 1988, 1990; Voigt 1989; LaMantia and Rakic 1990; Lent et al. 1990; Aggoun-Zouaoui and Innocenti 1994; Tessier-Lavigne and Goodman 1996; Richards et al. 1997; Shu and Richards 2001; Richards 2002; Niquille et al. 2009; Unni et al. 2012; Gobius et al. 2016; Fame et al. 2016b), as well as humans (Rakic and Yakovlev 1968; Kostovic and Krmpotic 1976; Kostovic and Rakic 1990; Marín-Padilla 1992; Kier and Truwit 1996; DeAzevedo et al. 1997; Huang et al. 2006; Rados et al. 2006; Ren et al. 2006; Huang et al. 2009). Yet, we still know very little about the basic developmental timeline of radial glia, pioneer development, and cortical association pathways in humans and which developmental processes are conserved or variant between humans and other species. Understanding basic developmental differences between humans and model organisms is an essential enterprise in order to relate findings from model organisms to humans and to identify developmental mechanisms that underlie the neural specializations responsible for sensory and cognitive specializations leading to the emergence of the human brain (Clancy et al. 2001; Ward and Vallender 2012; Perlman 2016; Charvet et al. 2017b).
Among the various diffusion MR methods amenable to reconstruct pathways in three dimensions, high-angular resolution diffusion imaging (HARDI) tractography enables the identification of complex crossing tissue coherence in the brain (Tuch et al. 2003), as well as immature brains (e.g., Takahashi et al. 2010, 2011, 2012). It is typically more challenging to identify cell extensions whether they are from radial glia or from bundles of axons because there is a surplus of unmyelinated fibers during fetal brain development. We here use HARDI to track the development of fibers because it is theoretically superior to diffusion tensor imaging (DTI) and permits the study of pathways guiding the development of neural pathways (e.g., radial glia and pioneer axons) in the human brain.
Materials and Methods
We first discuss sample acquisition, scanning parameters, and tractography to compare the developmental time course of fibers from diffusion MR tractography in humans (n = 5) and in mice (n = 5; Figs 1–3). We then discuss how we measured and compared corpus callosum growth trajectories in humans, macaques, and mice (Fig. 4). Corpus callosum scans for humans (Shi et al. 2011; Cohen et al. 2016; Khan et al. 2018a, 2018b), macaques (Young et al. 2017), and mice (Chuang et al. 2011) were acquired from multiple sources. Scanning parameters and sample size, as well sources are listed in Supplementary Table 1.
Figure 1.
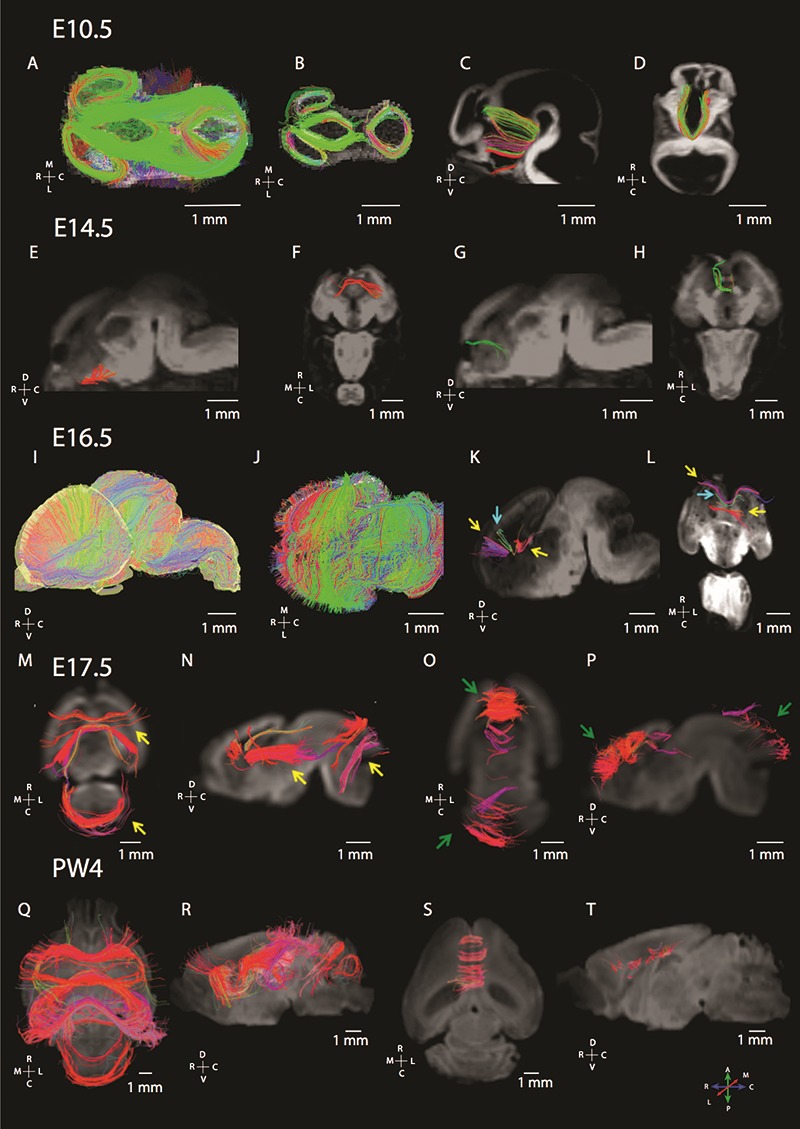
Maturation of commissural fibers as assessed from diffusion MR tractography in mice at various ages of development (E 10.5, E 14.5, E 16.5, E 17.5, and postnatal week 4: PW 4). (A) At E 10.5 (A–D), the dorsal view of brain fibers from whole brain tractography (A) and horizontal slice filters (B) shows that many of the pathways course across the anterior to posterior direction in the dorsal telencephalon as well as in the diencephalon. We identify pathways coursing through the midline in the ventral telencephalon by placing ROIs to identify commissural fibers (C, D). (B) At E 14.5 (E–H), we observe callosal pathways as well as putative pioneer axons. (C) At E 16.5 (I, L), we observe pathways coursing across the medial to lateral axes through the developing cortex. (I) Whole brain tractography through the mouse brain shows that the telencephalon primarily contains fibers coursing medial to lateral. The dorsal view of a horizontal slice through the telencephalon also highlights fibers coursing through the dorsal to ventral and medial to lateral direction within the developing cortex. (K, L) Placing ROI to identify commissural fibers identifies callosal and putative pioneer pathways. Yellow arrows in 16.5E denote putative pioneer axons, while blue arrows denote callosal pathways. At E 17.5 (M–P), horizontal (M, O) and sagittal views (N, P) highlight callosal and putative pioneer pathways coursing through the dorsal midline. ROIs were used to identify callosal and pioneer pathways. At postnatal week 4 (Q–T), ROIs highlighting commissural fibers show callosal fibers as well as the hippocampal commissure fibers coursing across the medial to lateral direction within the caudal cortex. Yellow arrows show the development of fibers in the anterior and posterior brain regions, while green arrows highlight the development of short fibers in the anterior and posterior brain regions. The direction of fibers is color-coded with a map of these color-codes located in the lower left panel. An orientation map is adjacent to each examined plane of section. The following abbreviations are used: R: rostral; C: caudal; M: medial; L: lateral; D: dorsal; and V: ventral.
Figure 3.
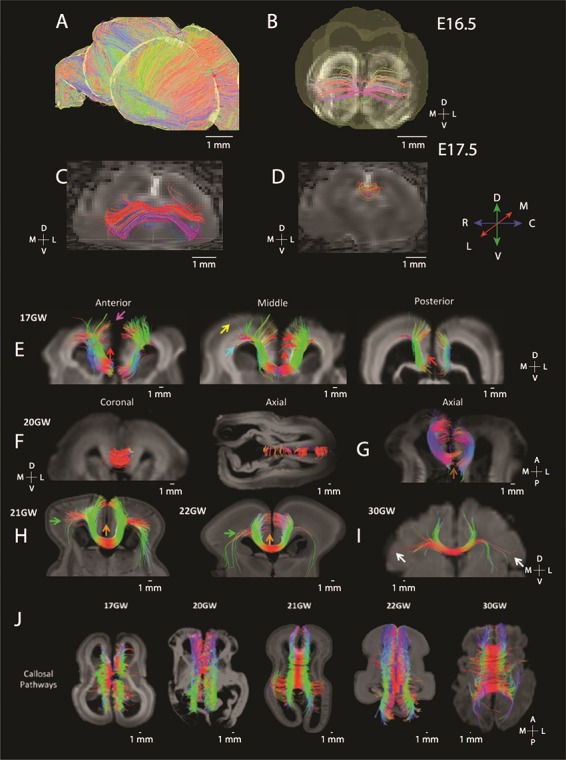
Diffusion MR tractography captures the maturation of the corpus callosum in mice (E 16.5: A, B; E 17.5: C, D) as well as human brain from GW 17 to 30 (E–J). (A) ROIs were used to highlight brain commissural and pioneer fibers in humans and in mice. Sagittal slices set through the E 16.5 mouse brain show that fibers are principally organized across the medial to lateral axis. (B) ROIs set through the dorsal midline highlight callosal fibers at this age at E 16.5 (B) and at E 17.5 (C). The corpus callosum is more prominent at E 17.5 than at E 16.5. At E 17.5, more fibers are evident in the rostral cortex (C) than in more caudal regions (D). At GW 17 in humans (E), putative pioneer and callosal pathways are also observed across the anterior to posterior axis with more prominent callosal fibers evident in anterior regions than in posterior regions. (E) The yellow arrow highlights callosal pathways from the lateral brain regions, and the pink arrow shows callosal pathways from the upper medial wall. (E) The blue arrow shows callosal pathways from the ventricular region, and red arrows show possible residual pioneer axons at the cingulate cortex. The corpus callosum gains increasing prominence with age (F–I). (H) At GW 21, callosal pathways are evident and from the upper medial wall and lateral brain regions (green arrow). (H) Callosal pathways from the upper medial wall, cingulate cortex, and lateral brain regions (green arrow) at GW 21 and GW 22. (F) At GW 30, we observe callosal pathways from the cingulate cortex, upper medial walls, and lateral walls of the brain (white arrows). (J) Dorsal views of horizontal slices through the corpus callosum from GW 17 to GW 30 show that the corpus callosum becomes increasingly prominent with age.
Figure 4.
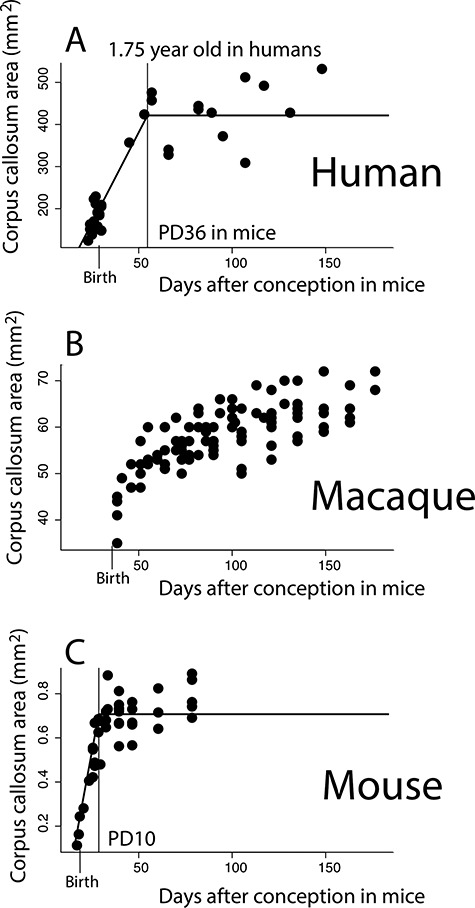
The growth of the corpus callosum is protracted in humans (A) and macaques (B) compared with mice (C). In order to control for variation in the length of developmental schedules, age in humans and macaques is mapped onto mouse age in days after conception according to the translating time website (Clancy et al. 2001; Workman et al. 2013). (A) the area of the corpus callosum reaches a plateau in growth between 1 and 2 years of age in humans (i.e., 1.75 years after birth). (B) The corpus callosum shows sustained growth in macaques across the examined ages and does not plateau. (C) The corpus callosum reaches a plateau in growth at 28.5 days after conception in mice, which is 10 days after birth in mice. The corpus callosum grows for much longer than expected in humans and macaques when human and macaque age is mapped onto that of mice. Vertical bars identify when the corpus callosum ceases to grow, as well as birth.
Sample Acquisition for Diffusion MR Tractography
Mice
Five intact mouse brains at E 10.5, 14.5, 16.5, 17.5, and 4 postnatal weeks (PW) were obtained from ongoing developmental or genetic research projects at Boston Children’s Hospital (BCH) and Massachusetts General Hospital (MGH) under approved protocols at the hospitals. Mice were deeply anesthetized (xylanzine/ketamine 100 mg/mL, 60 mg/kg) and either sacrificed by transcardial perfusion with 0.9% saline followed by 4% paraformaldehyde (PFA) (>postnatal day 5) or decapitated and immersion-fixed (<postnatal day 3). Fetal brains were immersion-fixed after the removal of the skull. Postnatal animals were perfused with 4% PFA followed by immersion fixation for at least 2 weeks in 4% PFA. Brains were placed in Fomblin for imaging.
Humans
A total of five brains at GW 17, 20, 21, 22, and 30 were imaged for diffusion MR tractography. Samples from GW 17–22 were scanned ex-vivo, whereas the GW 30 individual was scanned in vivo. Postmortem brains were obtained from the Department of Pathology, Brigham and Women’s Hospital (BWH, Boston, MA, USA) and the Allen Institute Brain Bank (AIBB, Seattle, WA, USA) with full parental consent. The known primary causes of death were complications from prematurity. The brains were grossly normal, and standard autopsy examinations of all brains undergoing postmortem HARDI revealed no or minimal pathologic abnormalities at the macroscopic level. All brains utilized for this study were analyzed under institutional review board-approved protocols.
Tissue Preparation for HARDI in Humans
At the time of autopsy, all brains were immersion-fixed, and the brains from the BWH were stored in 4% PFA, and the brains from the AIBB were stored in 4% periodate-lysine-paraformaldehyde (PLP). During MR image acquisition, the brains from the Brigham and Women’s Hospital were placed in Fomblin solution (Ausimont; e.g., Takahashi et al. 2012) and the brains from the AIBB were placed in 4% PLP. These different kinds of solutions in which the brains from different institutes were placed tend to change the background contrast (i.e., a dark background outside of the brain using Fomblin and bright background using PLP), but they do not specifically change diffusion properties [e.g., fractional anisotropy (FA) and apparent diffusion coherence (ADC)] within the brain.
Scanners
Different scanner systems were used to accommodate brains of various sizes (same as in e.g., Takahashi et al. 2012; Xu et al. 2014; Miyazaki et al. 2016; Wilkinson et al. 2017). Magnetic resonance (MR) coils that best fit each brain sample were used to ensure optimal imaging. Mouse specimens as well as human specimens from BWH [2 fetal specimens (GW 17, 20)] were imaged with a 9.4T Bruker Biospec MR system. Ex-vivo specimens from the AIBB were imaged with a 3 T Siemens MR system [2 fetal specimens (GW 21, 22)] at the A. A. Martinos Center, MGH, Boston, MA, USA, because they did not fit in the 4.7T bore. In order to obtain the best signal-to-noise ratio and highest spatial resolution, we used custom-made MR coils with one channel on the 4.7T and 3T systems (Takahashi et al. 2012; Kolasinski et al. 2013; Xu et al. 2014). The in vivo subject (GW 30) from BCH was imaged on a 3T Siemens MR system at BCH, Boston, MA, USA, when the individual was asleep and not anesthetized. At a 3T scanner, the use of high b values such as 8000 s/mm2 is problematic because it causes artifacts in that the tractography appears inconsistent with previous findings from tract-tracers. We reason that the issue of different acquisition systems is not significant for tractography because both systems yielded similar patterns.
Scanning Protocols
Scanning protocols are listed in Supplementary Table 1. For the human brains at GW 21 and GW 22 from BWH and for mouse specimens, a 3D diffusion-weighted spin-echo echo-planar imaging sequence was used. Sixty diffusion-weighted measurements (with the strength of the diffusion weighting, b = 8000 s/mm2) and one nondiffusion-weighted measurement (no diffusion weighting or b = 0 s/mm2) were acquired with δ = 12.0 ms and Δ = 24.2 ms. For the human brains at GW 21 & 22 from AIBB, diffusion-weighted data were acquired over two averages using a steady-state free-precession sequence with TR/TE = 24.82/18.76 ms, α = 60°, and the spatial resolution was 400 × 400 × 400 μm. Diffusion weighting was isotropically distributed along 44 directions (b = 730 s/mm2) with 4 b = 0 images, using a custom-built, single channel, 2-turn solenoid coil (6.86 cm inner diameter, 11.68 cm length). The solenoid design minimizes the distance between the copper turns of the coil and the sample, yielding a higher signal-to-noise ratio with each turn and improved signal uniformity compared with a phased array receive coil design. We determined the highest spatial resolution for each brain specimen with an acceptable signal-to-noise ratio of more than 130 for diffusion MR tractography.
Reconstruction and Identification of Diffusion MR Tractography
We reconstructed white matter tracts in each brain with the Diffusion Toolkit and TrackVis (http://trackvis.org). We used a FACT algorithm for diffusion MR tractography (Mori et al. 1999), as in previous publications (for humans, Takahashi et al. 2010, 2011, 2012; Schmahmann et al. 2007; D’Arceuil et al. 2008; Das and Takahashi 2018; for mice, Rosen et al. 2013; Lodato et al. 2014; Fame et al. 2016a; Kanamaru et al. 2017), which connects fibers using a local maximum or maxima. The FACT algorithm is limited in its ability to resolve crossing pathways when used with the traditional DTI technique because one simply connects the direction of the principal eigenvector on a tensor to produce the DTI tractography pathways. This feature is a recognized limitation of DTI (Mori et al. 1999). Hence, in the current study, we used HARDI, which can theoretically detect multiple local maxima on an orientation distribution function (ODF). Using each local maxima on an ODF, we applied the streamline algorithm with Q ball to initiate and continue tractography (Tuch et al. 2003), thus enabling us to identify crossing pathways within a voxel.
Diffusion MR trajectories were propagated by consistently pursuing the orientation vector of least curvature. We terminated tracking when the angle between two consecutive orientation vectors was greater than the given threshold (40°), or when the fibers extended outside of the brain surface. For visualization purposes, the color-coding of tractography connections was based on a standard red–green–blue (RGB) code using directions of the middle segment of each fiber. In some cases, this RGB code relies on the direction of every fiber segment. In other cases, the RGB code applies to the average direction of fibers.
Regions of Interest to Select Fibers
We first extracted fibers from whole brain tractography, and we used slices or hand-drawn regions of interest (ROIs) to highlight pathways of interest in humans and mice. Whole brain tractography captures fibers coursing through the whole brain and permits to detect how overall organization of fibers might vary between species. For whole brain tractography and slice filters, we had a minimum length threshold set to about 1 mm for a visualization purpose. A single slice filter (either set along the horizontal or sagittal direction) permits viewing fibers coursing through the slice of interest. To study fibers in contact with the medial walls of the developing cortex, multiple sagittal slice filters of varying thickness with “either end” restrictions were used. Thickness of slice filters and placement were dependent on which pathway we aimed to capture during development (e.g., corpus callosum and anterior commissure). No threshold as to minimum or maximum FA was used to include or exclude fibers. We did not constrain the ROIs to the developing white or gray matter to highlight select commissural fibers (e.g., corpus callosum and anterior commissure) and track their developmental time course in both species. ROIs were variably placed to highlight commissural fibers of interest. By changing the size of the ROIs, we ensured not to miss pathways of interest.
Variation in Corpus Callosum Area Between Primates and Rodents
It has been previously reported that the corpus callosum is expanded in primates compared with other mammals (Manger et al. 2010). We aimed to confirm that the corpus callosum is indeed expanded in primates compared with rodents. To that end, we use structural MRI and atlases to measure the corpus callosum area from midsagittal sections in small-brained monkeys and large-brained rodents in order to control for variation in overall brain size. We supplement these data with previously published data (see Supplementary Table 1; Manger et al. 2010; Yuasa et al. 2010; Charvet et al. 2015, 2019; Schilling et al. 2017). We use phylogeny-generalized-least-squares statistics (PGLS) to obtain phylogenetically controlled slopes of the logged values of the area of corpus callosum versus the logged values of brain size (Pagel 1999; Freckleton et al. 2000). We test whether taxonomy (i.e., primate versus rodent) accounts for a significant percentage of the variation in corpus callosum area. These phylogenetically controlled generalized linear models were performed via restricted estimated maximum likelihood with the software program R and the library package ape (Pagel 1999; Freckleton et al. 2000). The phylogeny, which includes branch lengths for these species, was taken from Bininda-Emonds et al. 2007.
After controlling for variation in brain size, primates possess a larger corpus callosum compared with rodents (see Supplementary Table 1). A PGLS model with brain size and taxonomy are both significant independent variables accounting for 98.2% of the variance in corpus callosum area (F = 348; R2 = 0.98.1; P = 1.137 × 10−10; λ = 0) and an Akaike information criterion (AIC) score of −25.51. A PGLS model that uses brain size alone to predict corpus callosum area only accounts for 96.8% of the variance (F = 393.3; P = 1.539 × e−10, λ = 0), with an AIC score model of −18.50. The addition of taxonomy as an independent variable in the PGLS model results in a lower AIC score than the model without taxonomy as an independent variable. These findings confirm that primates possess a larger corpus callosum than rodents when controlled for brain size.
Growth Trajectories of the Human, Macaque, and Mouse Corpus Callosum Area
Comparative analyses of growth trajectories can pinpoint what developmental programs might account for variation in cortical association pathways in humans versus mice. To quantitatively investigate whether the corpus callosum growth follows a similar trajectory across primates and rodents, we measured the area of the corpus callosum from midsagittal sections of FA images from mice, macaques, and humans at several stages of development (see Supplementary Tables 1 and 2). We here select FA images from diffusion MR scans, rather than structural MR scans, as we had done when comparing the corpus callosum of adult primates with those of rodents. We select FA images to track the development of the corpus callosum because the corpus callosum is easily identifiable from its bright white appearance from midsagittal sections at successive stages of development and across species (Fig. 5). The FA images range from E 17 to postnatal day 60 in mice (n = 42), from GW 24 to 18 years of age in humans (n = 37), and from two PW to 36 months after birth in macaques (n = 102).
Figure 5.
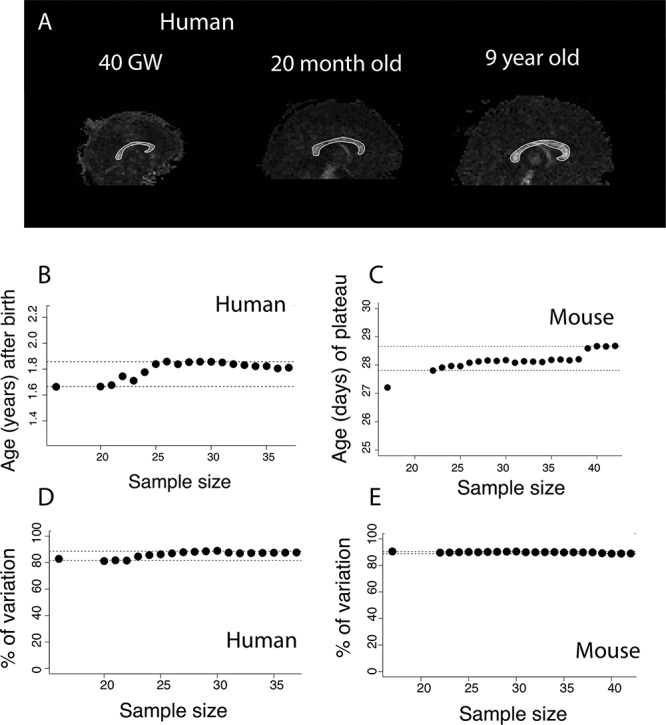
(A) Midsagittal planes through FA images of human brains at GW 40, 20 months, and 9 years of age show that the corpus callosum has a bright white appearance in contrast to surrounding brain regions. We have contoured the area of the corpus callosum in white as examples of corpus callosum measurements. To assess whether corpus callosum measurements are robust to variation in sample size, we subsampled the number of individuals from which the corpus callosum area was measured in humans and in mice. We select nonlinear regression models that account for a significant percentage of the variance. Identified age in which the corpus callosum ceases to grow ranges between 1.6 and about 1.8 years of age in humans (B) and between postconception day 27 and 29 in mice (C). Regardless of sample size, our model accounts for more than 80% of the variance in humans (D) and in mice (E). Horizontal dashed lines represent 95% CI from the age in which the corpus callosum ceases to grow in humans (B) and in mice (C), and the percentage of variance explained in humans (D) and in mice (E).
Multiple datasets were used to capture corpus callosum areas in humans (see Supplementary Table 1; Shi et al. 2011; Cohen et al. 2016; Khan et al. 2018a, 2018b; Fig. 4). To corroborate our findings of corpus callosum growth trajectories in humans, we also consider corpus callosum growth trajectories from a previously published dataset that relied on structural MRI scans to capture the growth of the corpus callosum in humans (n = 86). Ages range from shortly after birth to 25 years of age (Sakai et al. 2017). The FA images of mice are made available by John Hopkins University and the human brain scans were previously published (Chuang et al. 2011). The FA images of macaques are from the UNC-Wisconsin Neurodevelopment Rhesus Database (Young et al. 2017). We manually measured the area of the corpus callosum in mice, rhesus macaques, and humans from midsagittal sections at different ages (Fig. 5A). We excluded one outlier for macaques because the corpus callosum measurement from FA images was inconsistent with that obtained from the T1 structural MR.
Statistical analyses used to compare the growth of the corpus callosum between species were performed with the software programming language R. We tested whether a linear plateau would fit the data in each species with the library package easynls (model = 3; linear plateau). We subsampled the number of individuals from which the corpus callosum was measured and repeated the same analyses to ensure that the identified ages in which the corpus callosum ceases to grow in both species are not driven by outliers and that the sample size is sufficient to detect when the corpus callosum ceases to grow. We tested for a nonlinear regression (model 3) with the log 10-transformed values for corpus callosum area and the log-based 10 values for age expressed in days after conception (Fig. 5B–E). Tested samples range from 3 to total sample size obtained for each species. For each sample size, we tested for nonlinear regressions 10 times and we only consider ages in which the corpus callosum ceases to grow from models that detect a significant cessation of corpus callosum growth (P < 0.05). We extract the 95% confidence interval (CI) from these data to identify the range of possible ages as well as the percentage of variance explained by the models.
We assess whether the corpus callosum growth is extended in primates compared with rodents after controlling for variation in developmental schedules. The timing of developmental transformations (i.e., events) can be used to identify corresponding ages across species (Clancy et al. 2001). Controlling for variation in developmental schedules permits identifying whether certain development processes occur for an unusually long or short period of time in humans (Finlay and Darlington 1995; Clancy et al. 2001, 2007; Nagarajan and Clancy 2008; Workman et al. 2013; Charvet et al. 2017a; Charvet and Finlay 2018). Because the translating time resource extends up to 2 years of age in humans and its equivalent in other species, we extrapolated ages at later time points as we had done previously (Charvet et al. 2019). After controlling for variation in developmental schedules between species, we assess whether the corpus callosum ceases to grow later than expected in humans and macaques relative to mice.
Results
We describe the overall organization of pathways in the forebrain of humans and mice during development with a particular focus on radial and emerging commissural fibers. We also observe that diffusion MR tractography identifies pioneer axons originating from the medial wall of the developing cerebral cortex in both humans and in mice.
Radial Glia Organization of the Cortex in Humans and Mice
We explore the development of fiber organization in developing mouse brains from E 10.5 to PW 4 (Fig. 1, see Supplementary Fig. 2) and in humans from GW 17 to 30 (Figs 2 and 3). In mice, many of the fibers are observed coursing across the medial to lateral direction in the forebrain throughout development (Figs 1 and 2). According to the translating time model, a mouse at E 16.5 is roughly equivalent to humans at GW 12–16 and a mouse at E 17.5 is equivalent to humans at GW 14–18 (Fig. 2). We therefore compare mice at E 16.5–17.5 with humans at GW 17–21. We use whole brain tractography and coronal slice filters meant to capture fibers coursing through coronal slices shown in Figure 2 to identify species differences in the overall pattern of fiber organization in the two species. At GW 17–21, fibers are principally oriented radially with fibers extending from the proliferative pool towards the CP (Fig. 2A–C). These fibers likely represent radial glial cells known to guide the migration of neurons from the proliferative pool towards the CP. At E 16.5 (Fig. 2D,E) as in E 17.5 (Fig. 2F), we observe fibers coursing principally across the medial to lateral direction through the cortex. Some of these fibers likely correspond to thalamo-cortical, cortico-thalamic, or callosally projecting pathways. Thus, diffusion MRI highlights that mice and humans matched for age differ in their cortical organization. Radially organized fibers are evident in the human cortex but this is not the case in mice, presumably because radial fibers dissipate earlier in mice than in humans. Indeed, cortical neurogenesis wanes around E 18 in mice (Caviness et al. 2003, 2009), but cortical neurogenesis is thought to persist beyond GW 20 in humans (Zecevic et al. 2005; Malik et al. 2013; Charvet et al. 2017a).
Figure 2.
Diffusion MR tractography of cortices at GW 17–21 (A–C) and mice at E 16.5–E 17.6 (E, F) shows major differences in the organization of fibers spanning the cortex. In humans, many fibers course radially within the developing cortex. At GW 17 whole brain tractography (A) and 1 mm thick coronal slices through the anterior and posterior regions of the cortex (B) show fibers coursing from the proliferative zones to the CPs (white arrowheads). A similar situation is observed at GW 21 (C) where coronal slices set to capture fibers coursing through these slices identify radial fibers spanning the proliferative pool and the CP in the anterior and posterior cortex (white arrowheads). In contrast, diffusion MR scans of mice at E 16.5 (D, E) and at E 17.5 (F) show fibers principally organized across the medial to lateral axis (white arrowheads). (D) Whole brain tractography of mice show fibers coursing across the medial to lateral and dorsal to ventral direction through the developing cortex. (E) Coronal slices through the rostral and caudal cortex at E 16.5 capture fibers coursing tangential to the cortical surface rather than radially (white arrowheads). (F) At E 17.5, a similar situation is observed in the mouse where fibers are observed coursing across the medial to lateral direction (white arrowheads). Humans at GW 17–21 and mice at E 16.5–17.5 were selected for comparison because they are at roughly equivalent ages of development (Workman et al. 2013). The following abbreviations are used: A: anterior; P: posterior; R: rostral; C: caudal; M: medial, L: lateral; D: dorsal; and V: ventral.
Commissural Development in Mice and Humans
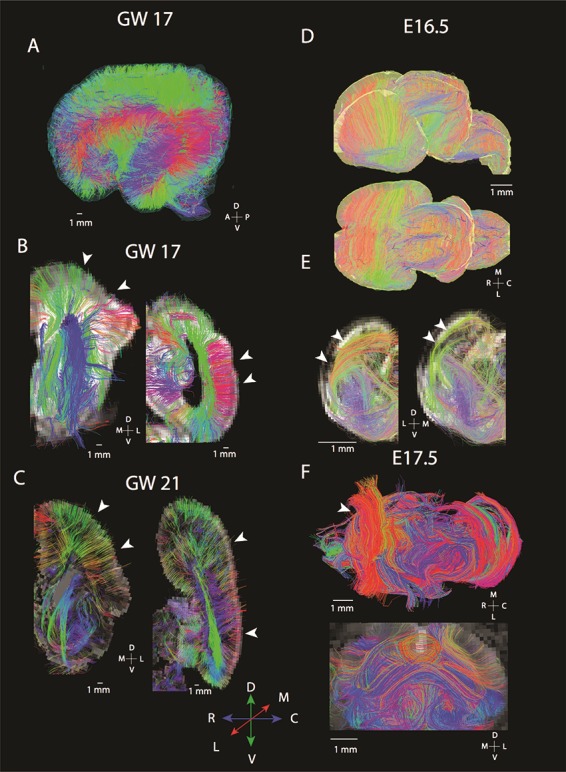
In mice, at the earliest gestational age studied (E 10.5), pathways crossing the midline were observed in anterior and ventral regions of the brain (Fig. 1A–D). We did not observe callosal fibers at this age. At E 14.5 (Fig. 1E–H), we observed short pathways crossing the midline in the presumptive corpus callosum (Fig. 1G,H). These fibers emerge or terminate within the cingulate cortex, and project contralaterally through the dorsal midline of the telencephalon (i.e., the developing corpus callosum). We consider these fibers emerging or terminating within the cingulate cortex to represent pioneer axons coursing through the corpus callosum because they are reminiscent of previously described pioneer axons that act as scaffolds to guide development of other axons crossing the dorsal midline (Koester and O’Leary 1994; Rash and Richards 2001). At E 16.5, fibers course across the medial to lateral direction throughout the developing cortex (Fig. 1I,J). We also observe a population of anteriorly located callosal pathways extending across the midline (Fig. 1K,L, yellow arrows). Some of these fibers originate or terminate within the cingulate cortex (Fig. 1K,L, blue arrows), whereas other fibers originate or terminate from more lateral regions of the developing cerebral cortex. Thus, fibers reminiscent of pioneer axons are first observed emerging or terminating from the cingulate cortex, followed by fibers spanning more lateral regions of the cortex. Given this developmental sequence, neurons emerging from the cingulate cortex may act as a scaffold for later-developing neurons that originate from the lateral regions of the cortex. As development progresses, the corpus callosum becomes increasingly evident at E 17.5 (Fig. 1M–P) and PW 4 (Fig. 1Q–T).
We next compare corpus callosum development in mice (Fig. 3A–D) and humans (Fig. 3E–J) matched for age. In mice, the corpus callosum is evident at E 16.5 and E 17.5 with pathways extending across the medial to lateral axis (Fig. 3A–D). At E 17.5, a larger callosal structure is evident in the rostral cortex compared with the caudal cortex (Fig. 3C,D). In humans, at GW 17 (Fig. 3E, pink arrow), at GW 20 (Fig. 3G), and through GW 22 (Fig. 3H) and GW 30 (Fig. 3I), we observe some pathways extending from the superior medial wall of the developing cortex projecting towards the contralateral hemisphere through the corpus callosum. Interestingly, other fibers arising from more lateral regions of the cortex also project towards the corpus callosum at GW 17 (Fig. 3E, yellow arrow), at GW 21–22 (Fig. 3H, green arrow), and at GW 30 (Fig. 3I, white arrow). Some of these fibers course within the subventricular and ventricular zones (Fig. 3E, blue arrow). A few fibers extend into the gray matter of the medial wall of the developing cortex at GW 17 (Fig. 3E, red arrows), GW 21, and GW 22 (Fig. 3H, orange arrows). We also observe variation in the corpus callosal structure across the anterior to posterior axis with seemingly larger callosal structures located in the frontal cortex than in more posterior regions of the cortex at GW 17 (Fig. 3E). This variation in the corpus callosum structure across the anterior to posterior axis of the cortex observed in the GW 17 human is reminiscent of those observed in mice at E 17.5. Thus, in humans as in mice, there is a tendency for more anterior callosal pathways to be expanded than in posterior regions. As development progresses, the corpus callosum increasingly extends across the medial to lateral axis in both species.
Corpus Callosal Growth Trajectories Across Species
To test whether there are deviations in the timing of corpus callosum development between humans and mice, we measured the area of the corpus callosum at successive ages across primates (i.e., humans and macaques) and mice (Figs 4 and 5). Ages in humans and macaques are mapped onto mouse age to control for variation in developmental schedules across species. We use the translating time model to find corresponding ages, which relies on the timing of developmental transformations to find corresponding ages across species (Clancy et al. 2001; Workman et al. 2013; Charvet et al. 2017a; Charvet and Finlay 2018).
In both humans and in mice, the corpus callosum grows and ceases to grow (i.e., plateaus) after birth (Fig. 4). In humans, the corpus callosum grows and reaches a plateau in growth at about 1.75 years of age in humans, which is equivalent to 35 days after conception in mice (easynls fit model = 3; adj R2 = 0.74; P < 0.05; n = 37; Fig. 4A). In macaques, the corpus callosum continues to grow past the predicted ages for those of mice (postnatal day 10) but our nonlinear model does not capture a linear plateau in growth with these data (easynls fit model = 3; n = 102; Fig. 4B). In mice, the corpus callosum reaches a plateau in growth at around 28.5 days after conception, which is 10 days after birth (easynls fit model = 3 adj R2 = 0.89; P < 0.05; n = 42; Fig. 4C).
To ensure that the identified ages in which the corpus callosum ceases are robust to variation in sample size, we subsampled the number of individuals from which the corpus callosum area was measured in humans and mice. We then extracted identified ages in which the corpus callosum significantly ceases to grow with the easynls model 3 package (Fig. 5). We only extracted results from the model if the model accounts for a significant percentage of the variance (P < 0.05), and we identify ages in which the corpus callosum cease to grow in both humans and mice (Fig. 5B,C). The corpus callosum ceases to grow between 1.66 years of age (i.e., 878 days after conception; 5% CI) to 1.85 years of age (i.e., 948 days after conception, 95% CI; n = 16–37 Fig. 5B). In mice, the sample size in which the models account for a significant percentage of the variance ranged between 17 and 42, and the corpus callosum ceases to grow between PD 9.3 (i.e., 27.8 days after conception; 5% CI) and PD 10.2 (i.e., 28.7, 95% CI; Fig. 5C). Regardless of sample size, the nonlinear regression accounts for more than 80% of the variance in humans (Fig. 5D) and in mice (Fig. 5E). Taken together, these observations demonstrate that the identified age in which the corpus callosum ceases to grow in each species is robust to variation in sample size and is not driven by outliers.
To further ensure that our results align with those of others and that the corpus callosum ceases to grow at around 1–2 years of age in humans, we supplement our analysis with a previously published dataset that captures the growth of the corpus callosum in humans (Sakai et al. 2017; see Supplementary Fig. 3). Such an analysis shows that the corpus callosum growth plateaus at 1.87 years of age (easynls fit model = 3; adj R2 = 0.78; P < 0.05; n = 86; see Supplementary Fig. 3). Although it is possible that there is small amount of corpus callosum growth past those ages, these two different datasets both demonstrate that most of the corpus callosum growth ceases around 1–2 years after birth in humans such that the corpus callosum continues to grow for an extended period of time in humans compared with mice.
We next confirm the protracted growth of the corpus callosum statistically, and we select the most recent and largest dataset capturing developmental transformations across rodents and primates, including humans (Charvet et al. 2017b). Neural transformations that occur on 27.5–28.5 days after conception in mice (n = 4), which is when the corpus callosum ceases to grow in mice, occur between GW 27 and about 2.6 months after birth in humans (lower 95% CI: GW 26.5; upper 95% CI: 2.03 months after birth). Yet, in humans, the corpus callosum growth ends at 1.75 years of age, which is past the 95% CI generated from these data. Taken together, these findings demonstrate that the corpus callosum continues to grow for significantly longer than expected in humans compared with mice.
Discussion
In this study, we tracked the development of fibers in the human and mouse cortex. Our findings demonstrate the ability of HARDI to track fiber development in different species. During cortical neurogenesis, fibers of the developing human cortex are primarily radially aligned, but this is not the case in age-matched mice. We focus on the developmental timeline of corpus callosum development, and we find that the corpus callosum grows for an extended period of time in humans compared with mice. We discuss the strengths and limitations of diffusion MR tractography, as well as the potential implications for the observed conserved and deviant developmental programs giving rise to variation in the cortical structure and connectivity patterns in the human lineage.
Diffusion MR Scanning for Basic Development and Health
Although ultrasound remains the predominant modality for evaluating disorders related to pregnancy, fetal MRI has been increasingly used to track normal and abnormal human development (Mitter et al. 2015; Jakab et al. 2017). Diffusion MRI is still in its infancy, and more work is needed to identify what developmental processes can be observed from diffusion MR tractography. HARDI is a new innovative approach to identify developmental programs in the human fetal brain, which can also be used to identify conserved and deviant developmental programs between humans and model organisms such as mice. It is important, however, to acknowledge the reliability of HARDI tractography when reporting diffusion imaging studies. As this method is in its infancy, HARDI does suffer from certain pitfalls (Maier-Hein et al. 2017). Additionally, the low number of specimens available for study and the differences in imaging procedures used to capture tractographies across development and between species make it challenging to draw quantifiable conclusions from HARDI tractography. It is currently difficult to obtain sufficient postmortem fetal brains to resolve this problem. Due to these limitations, we integrate our data with comparative analyses of callosal growth trajectories. However, studies such as these are essential as they identify key developmental processes (e.g., pioneer axons) in the human fetal brain, which should increase the utility of this work for disease detection and prevention.
Development of Radial Organization in Humans and Mice
Radial glia have been identified as an important player in developmental mechanisms accounting for the expanded isocortex of primates in adulthood (Rakic 1978, 1995), and we observe major differences in radial organization across the isocortex of humans and mice at corresponding ages. Fibers coursing across the mouse cortex from E 16.5 to 17.5 are principally organized across the medial to lateral axis. These fibers, which extend across the medial to lateral axes of the developing cortex, likely represent cortico-thalamic or thalamo-cortical pathways. This is in contrast with humans at corresponding ages where fibers in humans (GW 17–20) are primarily organized radially across the isocortex. The radial organization of the developing cortex has previously been characterized with diffusion MR tractography in humans. Radially aligned fibers are evident at GW 17 and begin to diminish around GW 21 with residual fiber pathways still observed at GW 40 (Miyazaki et al. 2016). As development progresses, radial glia as well as other proliferative cells wane as cells exit the proliferative zone, and newly born neurons assume their final position within the cortex (Rakic 1974; Miyata et al. 2001; Zecevic et al. 2005; Takahashi et al. 2012; Malik et al. 2013; Arshad et al. 2015; Charvet et al. 2017a).
The finding that the human fetal cortex shows a predominance of fibers oriented radially compared with that observed in mice can be integrated with known deviations in developmental programs between primates and rodents. Previous work use thymidine-based methods to identify when cortical neurons are generated (i.e., born) in macaques and rodents (Rakic 1974; Bayer and Altman 1990). A comparative analysis of thymidine-based studies showed that cortical neurogenesis is extended in macaques compared with examined rodents (Clancy et al. 2001). It is not the onset of cortical neurogenesis, but rather, it is the end of cortical neurogenesis that is protracted in primates relative to rodents (Clancy et al. 2001; Cahalane et al. 2014). That is, superficially located cortical neurons (i.e., layers II–IV neurons) undergo neurogenesis later in macaques considering the timing of other developmental transformations (Clancy et al. 2001; Workman et al. 2013). The extended duration of cortical neurogenesis is concomitant with an expansion in proliferative cells during cortical neurogenesis (Smart et al. 2002; Fish et al. 2008; Martínez-Cerdeño et al. 2012; Romero and Borell 2015). The extension in the duration of neurogenesis is concomitant with an expansion of upper layer neuron numbers in the primate cortex relative to those of rodents in adulthood (Charvet et al. 2017a). The notion that primates extend the duration of cortical neurogenesis for an unusually long time is further supported by a comparative analysis of cortical proliferative pool trajectories across humans, macaques, and mice (Charvet et al. 2017a). These data show that the growth of the cortical proliferative pool is protracted in humans and macaques compared with mice after controlling for variation in developmental schedules. Therefore, the duration of cortical neurogenesis extends for longer than expected relative to rodents (Charvet et al. 2011).
Extending the duration of neuron production should lead to an extension in the duration in which neurons are produced and migrate to the CP. Accordingly, mice at E 16.5 and E 17.5 should not possess many radially aligned fibers because neurogenesis is largely complete at these ages (Caviness et al. 2003, 2009; Yuzwa et al. 2017; Preissl et al. 2018). Indeed, single-cell-based sequencing methods have shown that the relative number of radial glia drops from approximately 14–0% between E 14 and E 18.5 (Yuzwa et al. 2017; Preissl et al. 2018; Loo et al. 2019). This is in contrast with humans at GW 17–21, which still possess a large proliferative pool and self-renewing radial glia, as well neurons exiting the proliferative pool to migrate towards the CP (Rakic 1974, 2003b; Zecevic et al. 2005; Malik et al. 2013; Arshad et al. 2015; Charvet et al. 2017b). We here assume that these radial fibers observed in humans are radial glia because these tractographies have been confirmed immunohistochemically in humans (Xu et al. 2014). However, the observed species differences in radial organization between humans and mice reported in the present manuscript have not been confirmed immunohistochemically, and several possibilities exist as to why a radial organization predominates the human fetal cortex but not mice at equivalent ages. For instance, the human fetal cortex may be composed of more or larger radial glia compared with age-matched mice. More work is needed to identify the developmental time course of radial glia development in humans and in mice. Nevertheless, the observation that putative radial glia are evident in humans but not in age-matched mice aligns with known deviations in the duration of cortical neuron production between primates and rodents.
Development of Commissural Fibers in Mice and Humans
We track the developmental sequence of corpus callosum development in mice. We observed fibers reminiscent of pioneer neurons emerging or terminating from the cingulate cortex as early as E 14.5. Similar fibers were also observed at E 16.5 (McConnell et al. 1989; Ozaki and Wahlsten 1992, 1998; Koester and O’Leary 1994; Molnár et al. 1998; Rash and Richards 2001; Fig. 1C). Although it is now well known that callosally projecting pioneers emerge from the cingulate cortex, the origin of pioneer neurons crossing through the dorsal midline of the telencephalon (i.e., the presumptive corpus callosum) was the subject of debate in the 1990s. The debate focused on whether pioneer neurons crossing the dorsal midline originate from the cingulate cortex or from more lateral regions of the cortex. The debate was settled with the use of dies, which showed that pioneer neurons have their cell bodies originate in the cingulate cortex rather than more lateral regions of the cortex (Rash and Richards 2001). Since then, more work has tracked the development of pioneer neurons extending axons to the contralateral hemisphere (Chun et al. 1987; Chun and Shatz 1989; McConnell et al. 1989; Antonini and Shatz 1990; Imai and Sakano 2011).
In humans as in mice, we identify putative pioneer neurons during development, which can be imaged as early as GW 17. These data are in line with the work of others who have shown that neurons emerging from the developing cingulate cortex have axons crossing the dorsal midline, which can be identified immunohistochemically in the human fetal brain at these ages (Ren et al. 2006). At GW 17, short fibers originating from the upper medial wall of the developing cortex and, more sparsely, from the cingulate cortex course towards the contralateral hemisphere in humans (Fig. 3E, see pink arrow). The sparse fibers from the cingulate cortex likely correspond to residual pioneer pathways, which are known to originate in the cingulate cortex and cross the midline (Rakic and Yakovlev 1968; Koester and O’Leary 1994; Ren et al. 2006). As development progresses, the corpus callosum becomes increasingly evident in both humans and mice.
Conservation and Variation in the Timing of Corpus Callosum Development
We compare developmental timing of the corpus callosum in mice, macaques, and humans (Schwartz and Goldman-Rakic 1991; Clancy et al. 2007; Workman et al. 2013). According to the translating time model, early callosal developmental milestones such as the emergence of the corpus callosum and myelination onset are conserved in their timing across humans and mice (Workman et al. 2013). In the present study, we do not observe major differences in callosal development between the two species at relatively early stages of its development, although these findings do not necessarily entail that there are no early differences across species. We do, however, find that there are clear differences in the growth trajectories of the corpus callosum between primates and mice such that deviations in the duration of callosal growth account for the expansion of the corpus callosum in primates.
After controlling for variation in overall developmental schedules across species, the corpus callosum continues to grow for an extended period of time in humans. According to our nonlinear regressions, we find that the corpus callosum ceases to grow between 1 and 2 years of age, which is consistent with the work of others who have shown that the corpus callosum ceases to grow several months to a few years after birth in humans (Clarke et al. 1989; Giedd et al. 1999; Tanaka-Arakawa et al. 2015; Sakai et al. 2017; Vannucci et al. 2017). In mice, the corpus callosum area only grows for a few days, which is also consistent with previous work (Wahlsten 1984; but see Chuang et al. 2011). Although these nonlinear regressions capture when most or all of the corpus callosum ceases to grow, it is possible that there is still some corpus callosum growth passed 1–2 years of age in humans and PD 10 in mice. We found that the macaque corpus callosum grows throughout the examined postnatal ages, and these findings align with previous work tracking the growth of the corpus callosum in macaques (Scott et al. 2016). We have not here identified the underlying developmental processes responsible for variation in callosal growth trajectories between species, and several possibilities exist. The human corpus callosum might prolong the duration with which corpus callosum axons increase in number or axons increase in thickness. Alternatively or in addition, the duration with which axons myelinate may be protracted in humans relative to mice. Although we do not resolve the underlying cellular process responsible for variation in corpus callosal growth trajectories across species, our findings demonstrate that the protracted development of cortico-cortical pathways is linked to their expansion in the primate lineage (Charvet et al. 2017b, 2019).
The corpus callosum is responsible for a variety of complex behaviors, and protracted growth of long-range cortico-cortical pathways such as the corpus callosum may be responsible for protracted behavioral development observed in humans relative to mice. Some behaviors such as weaning are deviant in their timing in humans relative to other species (Hawkes and Finlay 2018). However, very few studies have compared the timing of behavioral development between humans and other species. Integrating data on the timing of long-range cortico-cortical pathways with behavioral milestones may serve to identify the behavioral implications of extending the maturation of long-range cortico-cortical pathways in the primate lineage.
Conclusion
The timing of developmental transformations may be a powerful mechanism through which variation in cortical association pathways emerge. Several hypotheses have focused on the timing of axonal development as a source of evolutionary changes in connectivity patterns (Innocenti 1995; Striedter 2005). It has been proposed that neurons that reach their targets early can outcompete other targets, thereby expanding their representation within the brain (Catania 2001). A previous study showed that the star-nose mole’s fovea matures earlier than adjacent appendages. Thus, early axon extension might favor competing for targets, and their overrepresentation in the mature isocortex (Catania 2001). On the other hand, prolonging the duration of axon extension and refinement might afford more time to outcompete targets, which would accordingly increase the territory of select pathways in the adult brain (Deacon 1990; Striedter 2005). These two seemingly contradictory hypotheses emphasize the importance of timing in generating variation in connections between species. Our study supports the view that an extended duration with which corpus callosal pathways grow accounts for evolutionary changes in their structure in the adult brain. Thus, evolutionary changes in a cortical association pathway emerge by modifying the time with which they develop.
Funding
National Institute of Neurological Disorders and Stroke (grant/award number R03NS091587 to E.T.); Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant/award number R01HD078561, R21HD098606, R21HD069001 to E.T.); National Institute of Mental Health (grant/award number R21MH118739 to E.T.). This research was carried out in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, NIH P41RR14075, a P41 Regional Resource supported by the Biomedical Technology Program of the National Center for Research Resources (NCRR), National Institutes of Health. This work also involved the use of instrumentation supported by the NCRR Shared Instrumentation Grant Program (NIH S10RR023401, S10RR019307, and S10RR023043) and High-End Instrumentation Grant Program (NIH S10RR016811). Some of this work was also funded by an NIGMS grant (5P20GM103653) for research at Delaware State University.
Notes
A developmental series of mouse FA images were obtained as a courtesy of Dr Mori (lbam.med.jhmi) at Johns Hopkins University. Dr Lana Vasung provided comments on a previous version of this manuscript. This study was conducted partly using postmortem human brain specimens from the tissue collection at the Department of Neurobiology at Yale University School of Medicine (supported by grant NIH MH081896), which form a part of the BrainSpan Consortium collection (http://www.brainspan.org). Dr Rebecca D. Folkerth at Brigham and Women’s Hospital provided the other brain specimens. Conflict of Interest: None declared.
Supplementary Material
References
- Aboitiz F, Montiel J. 2003. One hundred million years of interhemispheric communication: the history of the corpus callosum. Braz J Med Biol Res. 36:409–420. [DOI] [PubMed] [Google Scholar]
- Aggoun-Zouaoui D, Innocenti GM. 1994. Juvenile visual callosal axons in kittens display origin and fate related morphology and distribution of arbors. Eur J Neurosci. 6:1846–1863. [DOI] [PubMed] [Google Scholar]
- Antonini A, Shatz CJ. 1990. Relation between putative transmitter phenotypes and connectivity of subplate neurons during cerebral cortical development. Eur J Neurosci. 2:744–761. [DOI] [PubMed] [Google Scholar]
- Arshad A, Vose LR, Vinukonda G, Hu F, Yoshikawa K, Csiszar A, Brumberg JC, Ballabh P. 2015. Extended production of cortical interneurons into the third trimester of human gestation. Cereb Cortex. 26:2242–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Altman J. 1990. Development of layer I and the subplate in the rat neocortex. Exp Neurol. 107:48–62. [DOI] [PubMed] [Google Scholar]
- Berbel P, Innocenti GM. 1988. The development of the corpus callosum in cats: a light and electron-microscopic study. J Comp Neurol. 276:132–156. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. 2007. The delayed rise of present-day mammals. Nature. 446:507–512. [DOI] [PubMed] [Google Scholar]
- Bloom JS, Hynd GW. 2005. The role of the corpus callosum in interhemispheric transfer of information: excitation or inhibition? Neuropsychol Rev. 15:59–71. [DOI] [PubMed] [Google Scholar]
- Burkhalter A, Bernardo KL, Charles V. 1993. Development of local circuits in human visual cortex. J Neurosci. 13:1916–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Rakic P, Molnár Z, Blakemore C. 2006. The first neurons of the human cerebral cortex. Nat Neurosci. 9:880–886. [DOI] [PubMed] [Google Scholar]
- Cahalane DJ, Charvet CJ, Finlay BL. 2014. Modeling local and cross-species neuron number variations in the cerebral cortex as arising from a common mechanism. Proc Natl Acad Sci U S A. 111:17642–17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminiti R, Carducci F, Piervincenzi C, Battaglia-Mayer A, Confalone G, Visco-Comandini F, Pantano P, Innocenti GM. 2013. Diameter, length, speed, and conduction delay of callosal axons in macaque monkeys and humans: comparing data from histology and magnetic resonance imaging diffusion tractography. J Neurosci. 33:14501–14511. doi: 10.1523/JNEUROSCI.0761-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania KC. 2001. Early development of a somatosensory fovea: a head start in the cortical space race? Nat Neurosci. 4:353. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Nowakowski RS, Bhide PG. 2009. Neocortical neurogenesis: morphogenetic gradients and beyond. Trends Neurosci. 32:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness VS, Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. 2003. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb Cortex. 13:592–598. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Cahalane DJ, Finlay BL. 2015. Systematic, cross-cortex variation in neuron numbers in rodents and primates. Cereb Cortex. 25:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Finlay BL. 2018. Comparing adult hippocampal neurogenesis across species: translating time to predict the tempo in humans. Front Neurosci. 12:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Palani A, Kabaria P, Takahashi E. Forthcoming 2019. Evolution of brain connections: integrating diffusion MR tractography with gene expression highlights increased cortico-cortical projections in primates. Cereb Cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Hof PR, Raghanti MA, Van Der Kouwe AJ, Sherwood CC, Takahashi E. 2017a. Combining diffusion magnetic resonance tractography with stereology highlights increased cross-cortical integration in primates. J Comp Neurol. 525:1075–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Šimić G, Kostović I, Knezović V, Vukšić M, Babić Leko M, Finlay BL. 2017b. Coevolution in the timing of GABAergic and pyramidal neuron maturation in primates. Proc R Soc B. 284:20171169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF, Finlay BL. 2011. Evo-devo and brain scaling: candidate developmental mechanisms for variation and constancy in vertebrate brain evolution. Brain Behav Evol. 78:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BK, Miller SM, Mantilla CB, Gross L, Yaszemski MJ, Windebank AJ. 2006. Optimizing conditions and avoiding pitfalls for prolonged axonal tracing with carbocyanine dyes in fixed rat spinal cords. J Neurosci Methods. 154:256–263. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, et al. 2011. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun JJ, Nakamura MJ, Shatz CJ. 1987. Transient cells of the developing mammalian telencephalon are peptide-immunoreactive neurons. Nature. 325:617–620. [DOI] [PubMed] [Google Scholar]
- Chun JJ, Shatz CJ. 1989. Interstitial cells of the adult neocortical white matter are the remnant of the early generated subplate neuron population. J Comp Neurol. 282:555–569. [DOI] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. 2001. Translating developmental time across mammalian species. Neurosci. 105:7–17. [DOI] [PubMed] [Google Scholar]
- Clancy B, Finlay BL, Darlington RB, Anand KJ. 2007. Extrapolating brain development from experimental species to humans. Neurotoxicology. 28:931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S, Kraftsik R, Van Der Loos H, Innocenti GM. 1989. Forms and measures of adult and developing human corpus callosum: is there sexual dimorphism. J Comp Neurol. 280:213–230. [DOI] [PubMed] [Google Scholar]
- Cohen AH, Wang R, Wilkinson M, MacDonald P, Lim AR, Takahashi E. 2016. Development of human white matter fiber pathways: from newborn to adult ages. Int J Dev Neurosci. 50:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arceuil H, Liu C, Levitt P, Thompson B, Kosofsky B, De Crespigny A. 2008. Three-dimensional high-resolution diffusion tensor imaging and tractography of the developing rabbit brain. Dev Neurosci. 30:262–275. [DOI] [PubMed] [Google Scholar]
- Das A, Takahashi E. 2018. Neuronal migration and axonal pathways linked to human fetal insular development revealed by diffusion MR tractography. Cereb Cortex. 28:3555–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon TW. 1990. Rethinking mammalian brain evolution. American Zoologist. 30:629–705. [Google Scholar]
- deAzevedo LC, Fallet C, Moura-Neto V, Daumas-Duport C, Hedin-Pereira C, Lent R. 2003. Cortical radial glial cells in human fetuses: depth-correlated transformation into astrocytes. J Neurobio. 55:288–298. [DOI] [PubMed] [Google Scholar]
- DeAzevedo LC, Hedin-Pereira C, Lent R. 1997. Callosal neurons in the cingulate cortical plate and subplate of human fetuses. J Comp Neurol. 386:60–70. [DOI] [PubMed] [Google Scholar]
- De Carlos JA, O’leary DD. 1992. Growth and targeting of subplate axons and establishment of major cortical pathways. J Neurosci. 12:1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame RM, Dehay C, Kennedy H, Macklis JD. 2016a. Subtype-specific genes that characterize subpopulations of callosal projection neurons in mouse identify molecularly homologous populations in macaque cortex. Cereb Cortex. 27:1817–1830. doi: 10.1093/cercor/bhw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame R, MacDonald J, Dunwoodie S, Takahashi E, Macklis J. 2016b. Cited2 regulates neocortical layer II/III generation and somatosensory callosal projection neuron development and connectivity. J Neurosci. 36:6403–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, Macklis JD. 2011. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 34:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. 1995. Linked regularities in the development and evolution of mammalian brains. 268:1578–1584. [DOI] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB. 2008. Making bigger brains-the evolution of neural-progenitor-cell division. J Cell Sci. 121:2783–2793. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. 2000. Phylogenetic analysis and comparative data: a test and review of evidence. Am Nat. 160:712–726. [DOI] [PubMed] [Google Scholar]
- Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. 2014. Diverse behaviors of outer radial glia in developing ferret and human cortex. J Neurosci. 34:2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Rajapakse JC, Vaituzis AC, Liu H, Berry YC, Tobin M, Nelson J, Castellanos FX. 1999. Development of the human corpus callosum during childhood and adolescence: a longitudinal MRI study. Prog Neuropsychopharmacol Biol Psychiatry. 23:571–588. [DOI] [PubMed] [Google Scholar]
- Gobius I, Morcom L, Suárez R, Bunt J, Bukshpun P, Reardon W, Richards LJ. 2016. Astroglial-mediated remodeling of the interhemispheric midline is required for the formation of the corpus callosum. Cell reports. 17:735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. 2013. Molecular logic of neocortical projection neuron specification, development and diversity. Nature Rev Neurosci. 14:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K, Finlay BL. 2018. Mammalian brain development and our grandmothering life history. Physiol Behav. 193:55–68. doi: 10.1016/j.physbeh.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Heilingoetter CL, Jensen MB. 2016. Histological methods for ex vivo axon tracing: a systematic review. Neurol Res. 38:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF. 2000. Development of connections in the human visual system during fetal mid-gestation: a Dil-tracing study. J Neuropathol Exp Neurol. 59:385–392. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang J, Wakana S, Zhang W, Ren T, Richards LJ, Yarowsky P, Donohue P, Graham E, van Zijl PC et al. 2006. White and gray matter development in human fetal, newborn and pediatric brains. Neuroimage. 33:27–38. [DOI] [PubMed] [Google Scholar]
- Huang H, Xue R, Zhang J, Ren T, Richards LJ, Yarowsky P, Miller MI, Mori S. 2009. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. J Neurosci. 29:4263–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Sakano H. 2011. Axon-axon interactions in neuronal circuit assembly: lessons from olfactory map formation. Eur J Neurosci. 34:1647–1654. doi: 10.1111/j.1460-9568.2011.07817.x. [DOI] [PubMed] [Google Scholar]
- Innocenti GM. 1981. Growth and reshaping of axons in the establishment of visual callosal connections. Science. 212:824–827. [DOI] [PubMed] [Google Scholar]
- Innocenti GM. 1995. Exuberant development of connections, and its possible permissive role in cortical evolution. Trends Neurosci. 18:397–402. [DOI] [PubMed] [Google Scholar]
- Isseroff A, Schwartz ML, Dekker JJ, Goldman-Rakic PS. 1984. Columnar organization of callosal and associational projections from rat frontal cortex. Brain Res. 293:213–223. [DOI] [PubMed] [Google Scholar]
- Jakab A, Tuura R, Kellenberger C, Scheer I. 2017. In utero diffusion tensor imaging of the fetal brain: a reproducibility study. Neuroimage Clin. 15:601–612. doi: 10.1016/j.nicl.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy GO, Killackey HP. 1982. Ontogenetic changes in the projections of neocortical neurons. J Neurosci. 2:735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Vasung L, Marami B, Rollins CK, Afacan O, Ortinau CM, Yang E, Warfield SK, Gholipour A. 2018a. Fetal brain growth portrayed by a spatiotemporal diffusion tensor MRI atlas computed from in utero images. NeuroImage. 185:593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Rollins CK, Ortinau CM, Afacan O, Warfield SK, Gholipour A. 2018b. Tract-Specific Group Analysis in Fetal Cohorts Using in utero Diffusion Tensor Imaging (MICCAI: 28-35). Cham: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru Y, Li J, Stewart N, Sidman RL, Takahashi E. 2017. Cerebellar pathways in mouse model of Purkinje cell degeneration detected by high-angular resolution diffusion imaging tractography. Cerebellum. 16:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz MJ, Lasek RJ, Silver J. 1983. Ontophyletics of the nervous system: development of the corpus callosum and evolution of axon tracts. Proc Natl Acad Sci U S A. 80:5936–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kier EL, Truwit CL. 1996. The normal and abnormal genu of the corpus callosum: an evolutionary, embryologic, anatomic, and MR analysis. Am J Neuroradiol. 17:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- Konstantinidou AD, Silos-Santiago I, Flaris N, Snider WD. 1995. Development of the primary afferent projection in human spinal cord. J Comp Neurol. 354:1–12. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Krmpotic J. 1976. Early prenatal ontogenesis of the neuronal connections in the interhemispheric cortex of the human gyrus cinguli. Verh Anat Ges. 70:305–316. [PubMed] [Google Scholar]
- Koester SE, O’Leary DD. 1994. Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J Neurosci. 14:6608–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinski J, Takahashi E, Stevens AA, Benner T, Fischl B, Zöllei L, Grant PE. 2013. Radial and tangential neuronal migration pathways in the human fetal brain: anatomically distinct patterns of diffusion MRI coherence. Neuroimage. 1:412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. 1990. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 297:441–470. [DOI] [PubMed] [Google Scholar]
- Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 113:E469–E478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. 2006. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 7:883–890. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Rakic P. 1990. Axon overproduction and elimination in the corpus callosum of the developing rhesus monkey. J Neurosci. 10:2156–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lent R, Hedin-Pereira C, Menezes JRL, Jhaveri S. 1990. Neurogenesis and development of callosal and intracortical connections in the hamster. Neurosci. 38:21–37. [DOI] [PubMed] [Google Scholar]
- Lodato S, Molyneaux BJ, Zuccaro E, Goff LA, Chen H-H, Yuan W, Meleski A, Takahashi E, Mahony S, Rinn JL et al. 2014. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat Neurosci. 17:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L, Simon JM, Xing L, McCoy ES et al. 2019. Single-cell transcriptomic analysis of mouse neocortical development. Nat Commun. 10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin MB, Shatz CJ. 1985. Studies of the earliest generated cells of the cat’s visual cortex: cogeneration of subplate and marginal zones. J Neurosci. 5:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein K, Neher P, Houde JC, Cote MA, Garyfallidis E, Zhong J, Chamberland M, Yeh FC, Lin YC, Ji Q et al. 2017. Tractography-based connectomes are dominated by false-positive connections. Nat Comm. 8:1349. [Google Scholar]
- Malik S, Vinukonda G, Vose LR, Diamond D, Bhimavarapu BB, Hu F, Zia MT, Hevner R, Zecevic N, Ballabh P. 2013. Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J Neurosci. 33:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manger PR, Hemingway J, Haagensen M, Gilissen E. 2010. Cross-sectional area of the elephant corpus callosum: comparison to other eutherian mammals. Neuroscience. 167:815–824. [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M. 1978. Dual origin of the mammalian neocortex and the evolution of the cortical plate. Anat Embryol. 152:109–126. [DOI] [PubMed] [Google Scholar]
- Marín-Padilla M. 1992. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J Comp Neurol. 321:223–240. [DOI] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. 2012. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One. 7:e30178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Noctor SC, Kriegstein AR. 2006. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb Cortex. 16(Suppl 1):i152–i161. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Ghosh A, Shatz CJ. 1989. Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science. 245:978–982. [DOI] [PubMed] [Google Scholar]
- Meissirel C, Dehay C, Berland M, Kennedy H. 1991. Segregation of callosal and association pathways during development in the visual cortex of the primate. J Neurosci. 11:3297–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C, Prayer D, Brugger PC, Weber M, Kasprian G. 2015. In vivo tractography of fetal association fibers. PLoS One. 10:e0119536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. 2001. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 31:727–741. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Song JW, Takahashi E. 2016. Asymmetry of radial and symmetry of tangential neuronal migration pathways in developing human fetal brains. Front Neuroanat. 10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár Z, Adams R, Blakemore C. 1998. Mechanisms underlying the early establishment of thalamocortical connections in the rat. J Neurosci. 18:5723–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, Van Zijl PC. 1999. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 45:265–269. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J. 2006. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 51:527–539. [DOI] [PubMed] [Google Scholar]
- Mrzljak L, Uylings HBM, Kostovic I, Van Eden CG. 1988. Prenatal development of neurons in the human prefrontal cortex. I. A qualitative Golgi study. J Comp Neurol. 271:355–386. [DOI] [PubMed] [Google Scholar]
- Mrzljak L, Uylings HB, Van Eden CG, Judás M. 1990. Neuronal development in human prefrontal cortex in prenatal and postnatal stages. Prog Brain Res. 85:185–222. [DOI] [PubMed] [Google Scholar]
- Nagarajan R, Clancy B. 2008. Phylogenetic proximity revealed by neurodevelopmental event timings. Neuroinformatics. 6:71–79. doi: 10.1007/s12021-008-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niquille M, Garel S, Mann F, Hornung JP, Otsmane B, Chevalley S, Parras C, Guillemot F, Gaspar P, Yanagawa Y et al. 2009. Transient neuronal populations are required to guide callosal axons: a role for semaphorin 3C. PLoS Biology. 7:e1000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M, Oishi K, Nakajima K. 2013. Axon guidance mechanisms for establishment of callosal connections. Neural Plast. 2013:149060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. 2001. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 409:714–720. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. 2016. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 91:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudo RJ, Masterton RB. 1989. Descending pathways to the spinal cord: II. Quantitative study of the tectospinal tract in 23 mammals. J Comp Neurol. 286:96–119. [DOI] [PubMed] [Google Scholar]
- Nudo RJ, Masterton RB. 1990. Descending pathways to the spinal cord, III: sites of origin of the corticospinal tract. J Comp Neurol. 296:559–583. [DOI] [PubMed] [Google Scholar]
- O’Leary DDM, Stanlield B, Cowan WM. 1981. Evidence that the early postnatal restriction of the cells of origin of the corpus callosal projections is due to the elimination of axon collaterals rather than to the death of neurons. Dev Brain Res. 1:607–617. [DOI] [PubMed] [Google Scholar]
- Ozaki HS, Wahlsten D. 1992. Prenatal formation of the normal mouse corpus callosum: a quantitative study with carbocyanine dyes. J Comp Neurol. 323:81–90. [DOI] [PubMed] [Google Scholar]
- Ozaki HS, Wahlsten D. 1998. Timing and origin of the first cortical axons to project through the corpus callosum and the subsequent emergence of callosal projection cells in mouse. J Comp Neurol. 400:197–206. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature. 401:877–884. [DOI] [PubMed] [Google Scholar]
- Patel CK, Rodriguez LC, Kuwada JY. 1994. Axonal outgrowth within the abnormal scaffold of brain tracts in a zebrafish mutant. J Neurobiol. 25:345–360. [DOI] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs RJ, Tyszka M, Richards LJ, Mukherjee P, Sherr EH. 2007. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nature Rev Neurosci. 8:287–299. [DOI] [PubMed] [Google Scholar]
- Perlman RL. 2016. Mouse models of human disease an evolutionary perspective. Evol Med Public Health. 2016:170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. 2018. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 21:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Mori S, Miller MI. 2015. Diffusion tensor imaging for understanding brain development in early life. Annu Rev Psychol. 66:853–876. doi: 10.1146/annurev-psych-010814-015340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rados M, Judas M, Kostović I. 2006. In vitro MRI of brain development. Eur J Radiol. 57:187–198. [DOI] [PubMed] [Google Scholar]
- Rakic P. 1971. Neuron–glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electron microscopic study in Macacus rhesus. J Comp Neurol. 141:283–312. [DOI] [PubMed] [Google Scholar]
- Rakic P. 1972. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 145:61–84. [DOI] [PubMed] [Google Scholar]
- Rakic P. 1974. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 183:425–427. [DOI] [PubMed] [Google Scholar]
- Rakic P. 1978. Neuronal migration and contact guidance in primate telencephalon. Postgrad Med J. 54:25–40. [PubMed] [Google Scholar]
- Rakic P. 1995. A small step for the cell—a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18:383–388. [DOI] [PubMed] [Google Scholar]
- Rakic P. 2002. Pre- and post-developmental neurogenesis in primates. Clin Neurosci Res. 2:29–39. [Google Scholar]
- Rakic P. 2003a. Developmental and evolutionary adaptations of cortical radial glia. Cereb Cortex. 13:541–549. [DOI] [PubMed] [Google Scholar]
- Rakic P. 2003b. Elusive radial glial cells: historical and evolutionary perspective. Glia. 43:19–32. [DOI] [PubMed] [Google Scholar]
- Rakic P, Yakovlev PI. 1968. Development of the corpus callosum and cavum septi in man. J Comp Neurol. 132:45–72. [DOI] [PubMed] [Google Scholar]
- Rash BG, Richards LJ. 2001. A role for cingulate pioneering axons in the development of the corpus callosum. J Comp Neurol. 434:147–157. [DOI] [PubMed] [Google Scholar]
- Ren T, Anderson A, Shen W, Huang H, Celine P, Zhang J, Mori S, Kinsman SL, Richards LJ. 2006. Imaging, anatomical, and molecular analysis of callosal formation in the developing human fetal brain. Anat Rec. 288:191–204. [DOI] [PubMed] [Google Scholar]
- Richards LJ, Koester SE, Tuttle R, O'Leary DDM. 1997. Directed growth of early cortical axons is influenced by a chemoattractant released from an intermediate target. J Neurosci. 17:2445–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards LJ. 2002. Axonal pathfinding mechanisms at the cortical midline and in the development of the corpus callosum. Braz J Med Biol Res. 35:1431–1439. [DOI] [PubMed] [Google Scholar]
- Romero DJC, Borell V. 2015. Coevolution of radial glial cells and the cerebral cortex. Glia. 63:1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen GD, Azoulay NG, Griffin EG, Newbury A, Koganti L, Fujisaki N, Takahashi E, Grant PE, Truong DT, Fitch RH et al. 2013. Bilateral subcortical heterotopia with partial callosal agenesis in a mouse mutant. Cereb Cortex. 23:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Komaki Y, Hata J, Okahara J, Okahara N, Inoue T, Mikami A, Matsui M, Oishi K, Sasaki E et al. 2017. Elucidation of developmental patterns of marmoset corpus callosum through a comparative MRI in marmosets, chimpanzees, and humans. Neurosci Res. 122:25–34. [DOI] [PubMed] [Google Scholar]
- Schilling KG, Gao Y, Stepniewska I, Wu TL, Wang F, Landman BA, Gore JC, Chen LM, Anderson AW. 2017. The VALiDATe29 MRI based multi-channel atlas of the squirrel monkey brain. Neuroinformatics. 15:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN, Wang R, Dai G, D’Arceuil HE, de Crespigny AJ, Wedeen VJ. 2007. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 130:630–653. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Goldman-Rakic PS. 1991. Prenatal specification of callosal connections in rhesus monkey. J Comp Neurol. 307:144–162. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Rakic P, Goldman-Rakic PS. 1991. Early phenotype expression of cortical neurons: evidence that a subclass of migrating neurons have callosal axons. Proc Natl Acad Sci U S A. 88:1354–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Grayson D, Fletcher E, Lee A, Bauman MD, Schumann CM, Buonocore MH, Amaral DG. 2016. Longitudinal analysis of the developing rhesus monkey brain using magnetic resonance imaging: birth to adulthood. Brain Struct Funct. 221:2847–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siffredi V, Anderon V, Leventer RJ, Spencer-Smith MM. 2013. Neuropsychological profile of agenesis of the corpus callosum: a systematic review. Dev Neuropsychol. 38:36–57. [DOI] [PubMed] [Google Scholar]
- Silver J, Lorenz SE, Wahlsten D, Coughlin J. 1982. Axonal guidance during development of the great cerebral commissures: descriptive and experimental studies, in vivo, on the role of preformed glial pathways. J Comp Neurol. 210:10–29. [DOI] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. 2011. Infant brain atlases from neonates to 1-and 2-year-olds. PloS one. 6:e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Richards LJ. 2001. Cortical axon guidance by the glial wedge during the development of the corpus callosum. J Neurosci. 21:2749–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supér H, Martinez A, Del Rio JA, Soriano E. 1998. Involvement of distinct pioneer neurons in the formation of layer specific connections in the hippocampus. J Neurosci. 18:4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. 2002. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 12:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF. 2005. Principles of Brain Evolution. Sunderland, MA: Sinauer. [Google Scholar]
- Takahashi E, Dai G, Wang R, Ohki K, Rosen GD, Galaburda AM, Grant PE, Wedeen VJ. 2010. Development of cerebral pathways in cats revealed by diffusion spectrum imaging. Neuroimage. 49:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Dai G, Rosen GD, Wang R, Ohki K, Folkerth RD, Grant EP. 2011. Developing neocortex organization and connectivity in cats revealed by direct correlation of diffusion tractography and histology. Cereb Cortex. 21:200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Folkerth RD, Galaburda AM, Grant PE. 2012. Emerging cerebral connectivity in the human fetal brain: an MR tractography study. Cereb Cortex. 2:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Arakawa MM, Matsui M, Tanaka C, Uematsu A, Uda S, Miura K, Sakai T, Noguchi K. 2015. Developmental changes in the corpus callosum from infancy to early adulthood: a structural magnetic resonance imaging study. PLoS One. 10:e0118760. doi: 10.1371/journal.pone.0118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Bartha AI, Norton ME, Barkovich AJ, Sherr EH, Glenn OA. 2009. Agenesis of the corpus callosum: an MR imaging analysis of associated abnormalities in the fetus. Am J Neuroradiol. 30:257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif E, Clarke S. 2001. Intrinsic connectivity of human auditory areas: a tracing study with DiI. Eur J Neurosci. 13:1045–1050. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. 1996. The molecular biology of axon guidance. Science. 274:1123–1133. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. 2003. Diffusion MRI of complex neural architecture. Neuron. 40:885–895. [DOI] [PubMed] [Google Scholar]
- Unni DK, Piper M, Moldrich RX, Gobius I, Liu S, Fothergill T, Donahoo AL, Baisden JM, Cooper HM, Richards LJ. 2012. Multiple slits regulate the development of midline glial populations and the corpus callosum. Dev Biol. 365:36–49. [DOI] [PubMed] [Google Scholar]
- Varon SS, Somjen GG. 1979. Neuronal–glial interactions. Neurosci Res Prog Bull. 17:27–84. [PubMed] [Google Scholar]
- Vannucci RC, Barron TF, Vannucci SJ. 2017. Development of the corpus callosum: an MRI study. Dev Neurosci. 39:97–106. doi: 10.1159/000453031. [DOI] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov-Milošević N, Pletikos M, Mori S, Kostović I. 2010. Development of axonal pathways in the human fetal fronto-limbic brain: histochemical characterization and diffusion tensor imaging. J Anat. 217:400–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Raguz M, Kostovic I, Takahashi E. 2017. Spatiotemporal relationship of brain pathways during human fetal development using high-angular resolution diffusion MRI and histology. Front Neurosci. 11:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt T. 1989. Development of glial cells in the cerebral wall of ferrets: direct tracing of their transformation from radial glia into astrocytes. J Comp Neurol. 289:74–88. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. 1984. Growth of the mouse corpus callosum. Dev Brain Res. 15:59–67. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, Kane T, Wang R, Takahashi E. 2017. Migration pathways of thalamic neurons and development of thalamocortical connections in humans revealed by diffusion MR tractography. Cereb Cortex. 27:5683–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM,, Vallender EJ. 2012. The resurgence and genetic implications of New World primates in biomedical research. Trends Genet. 28:586–591. doi: 10.1016/j.tig.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SP, Jones EG. 1976. The organization and postnatal development of the commissural projection of the rat somatic sensory cortex. J Comp Neurol. 168:313–344. [DOI] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. 2013. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 33:7368–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Takahashi E, Folkerth RD, Haynes RL, Volpe JJ, Grant PE, Kinney HC. 2014. Radial coherence of diffusion tractography in the cerebral white matter of the human fetus: neuroanatomic insights. Cereb Cortex. 24:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JT, Shi Y, Niethammer M, Grauer M, Coe CL, Lubach GR, Davis B, Budin F, Knickmeyer RC, Alexander AL, Styner MA. 2017. The unc-wisconsin rhesus macaque neurodevelopment database: A structural mri and dti database of early postnatal development. Frontiers in neuroscience. 24:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Kohsaka S, Nakamura K. 2010. Stereotaxic atlas of the marmoset brain: with immunohistochemical architecture and MR images. National Institute of Neuroscience, National Center of Neurology and Psychiatry; Japan. [Google Scholar]
- Yuzwa SA, Borrett MJ, Innes BT, Voronova A, Ketela T, Kaplan DR, Bader GD, Miller FD. 2017. Developmental emergence of adult neural stem cells as revealed by single-cell transcriptional profiling. Cell reports. 21:3970–3986. [DOI] [PubMed] [Google Scholar]
- Zecevic N, Chen Y, Filipovic R. 2005. Contributions of cortical subventricular zone to the development of the human cerebral cortex. J Comp Neurol. 491:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, Smith KA, Ebbert AJ, Swanson B, Kuan L, Page DT, Overly DT, Lein ES, Hawrylycz MJ, Hof PR, Hyde TM, Kleinman JE, Jones AR Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, Smith KA, Ebbert AJ, Swanson B, Kuan L, Page DT, Overly DT, Lein ES, Hawrylycz MJ, Hof PR, Hyde TM, Kleinman JE, Jones AR. 2012. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. 149:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


