Abstract
Phospholipid Phosphatase-Related Protein Type 1 (PLPPR1) is a six-transmembrane protein that belongs to the family of plasticity-related gene proteins, which is a novel brain-specific subclass of the lipid phosphate phosphatase superfamily. PLPPR1–5 have prominent roles in synapse formation and axonal pathfinding. We found that PLPPR1 overexpression in the mouse neuroblastoma cell line (Neuro2a) results in increase in cell adhesion and reduced cell migration. During migration, these cells leave behind long fibrous looking extensions of the plasma membrane causing a peculiar phenotype. Cells expressing PLPPR1 showed decreased actin turnover and decreased disassembly of focal adhesions. PLPPR1 also reduced active Rac1, and expressing dominant negative Rac1 produced a similar phenotype to overexpression of PLPPR1. The PLPPR1-induced phenotype of long fibers was reversed by introducing constitutively active Rac1. In summary, we show that PLPPR1 decreases active Rac1 levels that leads to cascade of events which increases cell adhesion.
Keywords: extracellular matrix, cytoskeleton, cell motility, chondroitin sulfate proteoglycans, Neuro2a
1. Introduction
The Phospholipid Phosphatase Related (PLPPR) proteins (PLPPR1 – PLPPR5, previously referred to as Plasticity Related Gene (PRG) proteins, are enriched in the brain with the highest expression levels during development [1]. The first protein of this family to be discovered, PLPPR4, was found in sprouting axons following hippocampal deafferentation, implying a role in neuronal plasticity [2]. The expression of PLPPR1 mRNA correlates with sprouting corticospinal axons after injury [3] and neuronal remodeling in the hippocampus after kainic acid treatment [4]. Decreased PLPPR1 mRNA has been associated with dysregulated neuronal migration [5, 6], suggesting a role for these proteins in neuronal migration, neurogenesis and axon growth after injury.
Overexpression of either PLPPR1, PLPPR4 or PLPPR5 in cultured cells causes dramatic changes in cell phenotype, mainly characterized by multiple, long actin-rich protrusions [4, 7–10]. We have shown that these proteins associate with each other, and that co-expression of PLPPR proteins vastly enhances this phenotype [7]. When PLPPR proteins are expressed in neuronal cells, they produce neuritic extensions [11] and enhance dendritic spine formation [12].
How the PLPPR proteins modulate cytoskeletal dynamics to cause these changes is an open question. While these proteins are structurally related to the lipid phosphatases, there is contradictory evidence as to whether the PLPPRs have any lipid phosphatase activity [2, 13]. It has been reported that PLPPR4 expression increased the expression of integrin β1 in cells [10], and that neuronal sprouting after PLPPR1 expression requires RasGRF activity [9]. However, other reports indicate that the morphological changes following PLPPR1 expression are not mediated through cdc42 or the Arp2/3 complex [8].
Here we report that PLPPR1 expression leads to increased cell adhesion and changes in cytoskeletal dynamics that result in decreased cell migration. This enables cells overexpressing PLPPR1 to overcome the anti-adhesive actions of chondroitin sulfate proteoglycans (CSPGs). We show that these effects of PLPPR1 are due to modulation of Rac1 GTPase activity. These results establish a novel signaling pathway for the PLPPR family of proteins.
2. Material and methods
2.1. Plasmid construction
PLPPR1 was prepared as described previously [7]. Constitutively active Rac1 G12V (CA Rac1) and dominant negative Rac1 T17N (DN Rac1) were subcloned into XhoI/BamHI sites of pC1 (Clontech, Mountain view, CA). mApple-FTR-940 (F-tractin) was originally from Michael Davidson’s laboratory (plasmid # 54902, Addgene, Cambridge, MA). mApple-paxillin was kindly provided by Dr. Clare Waterman (National Institutes of Health, Bethesda, MD). A construct expressing C-terminal HA-tagged PLPPR1 (PLPPR1–HA) was a generous gift from Dr. Andrew Morris (University of Kentucky, Lexington, KY) [8].
2.2. Cell culture and transfection
Neuro2a cells were maintained in DMEM supplemented with penicillin/streptomycin antibiotics and 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA). Twenty-four hours after plating, cells were transfected with either pEGFP or EGFP-PLPPR1, using Avalanche Omni® transfection reagent (EZ Biosystems, College Park, MD).
All cell culture plates (Corning, Corning NY), 6-well (3506), 12-well (3512), 24-well (3527), 48-well (3548), MatTek dishes (P35G-1.5–14-C, MatTek Corporation, Ashland, MA) and Lab-Tek™ culture II chamber slides, (155382, ThermoFisher Scientific, Waltham, MA) were coated with 10 μg/ml fibronectin (Millipore, Billerica, MA), unless otherwise indicated, and incubated overnight at 4°C.
2.3. Immunocytochemistry
Transfected Neuro2a cells were fixed in 4% paraformaldehyde (PFA), 48 h after plating and 24 h after transfection. Cells were briefly rinsed in PBS and incubated in blocking buffer (0.3% Triton X-100 in PBS, supplemented with 10% normal goat serum, NGS) for 1 h and then with primary antibody diluted in 0.3% Triton X-100 in PBS, supplemented with 2.5% NGS. Cells were washed in PBS and then incubated with secondary antibody diluted in 0.3% Triton X-100 in PBS, supplemented with 2.5% NGS. See Table 1 for dilutions and source of antibodies. Cells were then washed in PBS and then mounted with DAPI diluted in Fluoromount (Sigma-Aldrich). All phalloidin (ThermoFisher Scientific) staining was as previously described [7].
Table 1.
Antibodies Used
| Primary Antibodies | Dilution | Source |
| Chicken anti-GFP | 1:2000 | Abcam (ab13970) |
| Anti-p34-Arc/ARPC2 | 1:100 | Sigma Aldrich (07–227) |
| Mouse anti-Rac1 | 1:500 | Cytoskeleton (ARC03) |
| Mouse anti-β-actin | 1:4000 | Sigma Aldrich (A1978) |
| Rabbit anti-phospho-paxillin (Tyr31) | 1:1000 | ThermoFisher Scientific (44–720G) |
| Mouse anti-paxillin | 1:1000 | BD Biosciences (612405) |
| Rabbit anti-phospho FAK (Tyr576) | 1:500 | Epitomics (1700–1) |
| Rabbit anti-FAK | 1:500 | Epitomics (2103–1) |
| Secondary Antibodies | Dilution | Source |
| Alexa Fluor 568, Goat anti-Mouse IgG | 1:1000 | Molecular Probes (A-11019) |
| Alexa Fluor 488, Goat anti-Chicken | 1:1000 | ThermoFisher Scientific (A-11039) |
| Goat anti-Chicken HRP | 1:2000 | Abcam (ab6877) |
| Veriblot for IP Detection Reagent (HRP) | 1:4000 | Abcam (ab131366) |
| Donkey anti-Rabbit IgG HRP-linked F(ab’)2 fragment | 1:2000 | Amersham-GE (NA9310) |
| Sheep anti-Mouse IgG HRP-linked F(ab’)2 fragment | 1:2000 or 1:4000 | Amersham-GE (NA9340) |
| Texas Red-X phalloidin | 1:500 | ThermoFisher Scientific (T7471) |
For STED microscopy, cells were mounted in ProLong™ Diamond mounting media (ThermoFisher Scientific) and imaged on a Leica SP8 STED 3X/Confocal Microscope using 100×/1.4 N.A. oil immersion objective lens (Leica Microsystems, Buffalo Grove, IL). All other imaging was performed on a Zeiss 780 LSM (Carl Zeiss, Thornwood, NY) confocal microscope using a 63×/1.4 N.A. oil immersion objective lens.
2.4. Live cell imaging
All live cell imaging was conducted on microscopes equipped with a heated stage at 37°C and 5% CO2 on either a Nikon A1R microscope (Nikon Instruments Inc., Melville, NY), a Zeiss 780 LSM confocal microscope or an Delta Vision OMX microscope (GE, Marlborough, MA).
2.5. Cell migration assay
Neuro2a cells transfected with either pEGFP or EGFP-PLPPR1were plated in Lab-Tek culture chamber slides coated with fibronectin. Time lapse images were acquired for 10 h at the rate of 1 frame/10 min on a Nikon A1R microscope using a 10×/0.45 N.A. objective lens. Cell migration was analyzed by tracking each transfected cell, using the middle of each cell body as the point of reference. The distance travelled was determined using NIS Elements software (Nikon Instruments Inc.).
2.6. Cell adhesion assay
For the detachment assay, Neuro2a cells were plated on a 12-well plate coated with fibronectin at a density of 2.0 × 105 cells per well and transfected 24 h after plating with either pEGFP or EGFP-PLPPR1. After another 24 h, cells were briefly washed with DMEM culture media to remove serum and subsequently treated with Trypsin-EDTA diluted in DMEM (1:10) with increasing time intervals. Cells were then rinsed with PBS, fixed with 4% PFA and stained with DAPI. 10 × 10 tiled images were acquired on a Zeiss 780 LSM confocal microscope using a 10×/0.45 N.A. dry objective lens. The number of nuclei per field were counted using ImageJ.
2.7. Interference reflection microscopy
Neuro2a cells transfected with either pEGFP or EGFP-PLPPR1 and 24 h post-transfection were fixed with 4% PFA for 15 min. Images were acquired with a Zeiss 780 LSM confocal microscope using a 63×/1.4 N.A. oil immersion objective lens using procedures outlined in Barr, et al. [14]. Single-frame TIFF images were extracted from TIFF stacks and processed using ImageJ. The area of each cell was traced from bright field images to obtain an ROI (region of interest). The image was then rescaled to an 8-bit scale of 0–255 based on pixel intensity values that fell within histogram bins exceeding 5% of the mean histogram bin height. A band pass filter was applied based on convolution with a 41 × 41 pixel kernel. The histogram was then stretched again based on a 5% cut-off of the mean values. A binary threshold was fixed by selecting pixel intensities above 90 on the 0 – 255 scale. The image was then inverted and the number of pixels above threshold was divided by the area of the ROI to determine intensity. A color map ‘Gem’ was used to display higher intensity pixels as blue and lower intensity as orange which represent stronger and weaker adhesions respectively. For quantification, the area of each cell was traced from bright field images to obtain the region of interest. A threshold was applied to the IRM image of the same cell and the number of pixels above threshold was divided by the ROI to calculate the area of cell attachment.
To measure the size of focal adhesion (FA) contacts, Neuro2a cells were plated on fibronectin coated MatTek dishes at a density of 2 × 104 cells and transfected with either pEGFP or EGFP-PLPPR1. Cells were fixed and stained with anti-paxillin antibody. Images were acquired using TIRF microscopy on an inverted OMX Delta Vision microscope system using a 60×/1.49 N.A. TIRF objective lens. Counting of FAs in single cells (n = 10 cells) was done after noise removal by thresholding and applying a size constraint to FAs using ImageJ software [15].
2.8. Actin turnover analyzed by FRAP
Neuro2a cells, plated on glass-bottom MatTek dishes coated with fibronectin, were co-transfected with F-tractin (mApple-FTR-940) and either pEGFP or EGFP-PLPPR1. FRAP experiments were conducted on a Nikon A1R microscope using a 60×/1.4 N.A. oil objective lens. A ROI was selected from the leading edge of transfected cells and a 564 nm laser at 20% intensity was used as a pre-bleaching signal for F-tractin. The ROI was subsequently photobleached by three lasers at 458, 564 and 633 nm, all set at 100% intensity. The postbleaching acquisition was carried out by sacquiring 60 frames at a rate of 1 frame/0.6 s. The fluorescence intensity was determined at every time point and automatically generated by NIS Elements software. Images were corrected for photobleaching and the fluorescence intensity of the ROI for each frame was normalized to the average initial intensity of pre-bleaching frames and to the area of the bleached cell. The fluorescence intensity for the pre-bleaching frames was calculated as the average fluorescence intensity in the first 5 frames, Fpre. The value, F0, was designated as the average fluorescence intensity of the 8th frame after the photobleaching phase that occurred in the 6th and 7th frames. Individual plots for each experiment was fitted to a model function. The fluorescence recovery of F-tractin was best fitted by a single-exponential function (double exponential function did not significantly improve the quality of the fit) using Matlab, revealing the presence of mobile actin fractions with different recovery kinetics. The following model was applied:
Where: A is the percentage of mobile fraction of actin and B is the time constant of fluorescence recovery. A and B were obtained from the fitted curve. The mean ± SEM was calculated for the percentage of fluorescence recovery and the time constant for each condition. The final graphs show the fluorescence recovery, mobile fraction of monomeric actin and t1/2 which indicates the time it took for F-actin polymerization.
2.9. Actin turnover Retrograde flow + kymograph
Neuro2a cells were plated on glass-bottom MatTek dishes coated with fibronectin and co-transfected with F-tractin (mApple-FTR-940) and either pEGFP or EGFP-PLPPR1. Cells were imaged using Airyscan microscopy on a Zeiss 880 confocal microscope with a heated incubator stage. Time lapse movies were taken at 1 frame per 15 seconds with 5 Z-stacks that were maximum projected based on intensity. Time-lapse series allowed the identification of actin rich lamellipodia. A one pixel-wide line crossing the edge of the cell perpendicularly at the location of the lamellipodia was traced. This line records actin dynamics relative to the direction of the F-actin in the lamellipodia. A kymograph was generated by ImageJ from frame 1–1000 in the leading edge on the lamellipodia and the slope of the angles were plotted on the kymograph for the time it took for F-actin molecules to retract back into the cell body.
2.10. Analysis of focal adhesion dynamics
Time lapse TIRF images were collected every 60 seconds for 1 hour. Global assessment of focal adhesion dynamics was performed using “The Focal Adhesion Analysis Server” (FAAS) which is a web based tool for analyzing focal adhesion dynamics for automated detection, tracking, and quantification of adhesion structures in living cells [16]. This is a unique tool that allows the assessment of adhesion dynamics in a comprehensive, unbiased manner. A total of 15 cells per transfection condition were analyzed. Dynamics of newly formed adhesions in growing ends of cells was determined by this software. The analysis pipeline for FAAS extracts and quantifies a wide range of properties. For our experimental purposes, measurements were generated for the size of each focal adhesion, the rate of assembly and disassembly and the time it took to achieve each of these phases.
2.11. Immunoblot assay
Neuro2a cells seeded at a density of 4 × 105 cells/well in a 6-well culture dish were transfected with either pEGFP or EGFP-PLPPR1. Media was removed from all cells and rinsed once with warmed PBS. Cell lysates were prepared in 2× SDS cell lysis buffer and clarified by centrifugation. Protein quantitation was performed using either the Ionic Detergent Compatibility Reagent for Pierce™ 660nm Protein Assay Reagent (ThermoFisher Scientific) or the BCA method. Equal amount of proteins (20 μg/lane) was separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). Membranes were blocked for 1 h in 5% nonfat dried milk in either 0.1% Tween20 in PBS (PBST) or 0.1% Tween20 in 10 mM Tris–HCl pH 7.5 (TBST) or blocked in 5% BSA in TBST. Membranes were incubated with primary antibodies overnight at 4°C, washed in either PBST or TBST and subsequently incubated with secondary antibody for 30 min. Antigen–antibody complexes were detected with KPL LumiGlo chemiluminescent substrate (Seracare, Milford, MA). See Table 1 for source and dilutions for all primary and secondary antibodies used.
2.12. Statistical analysis
All analyses were performed using GraphPad Prism (GraphPad Software, La Jolla, CA) version 7. Data were analyzed by unpaired Student’s t tests, one-way ANOVA or two-way ANOVA with Welch’s correction or Tukey’s post-hoc multiple comparison test to determine significance. Normality of the distribution of the data was tested with Kolmogorov–Smirnov normality tests using the column statistics function of GraphPad Software. All tests were two-tailed with significance indicated as follows: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Unless otherwise specified, values represent the means ± SEM. All experiments were repeated independently at least three times.
3. Results
3.1. PLPPR1 reduces cell migration and increases cell adhesion
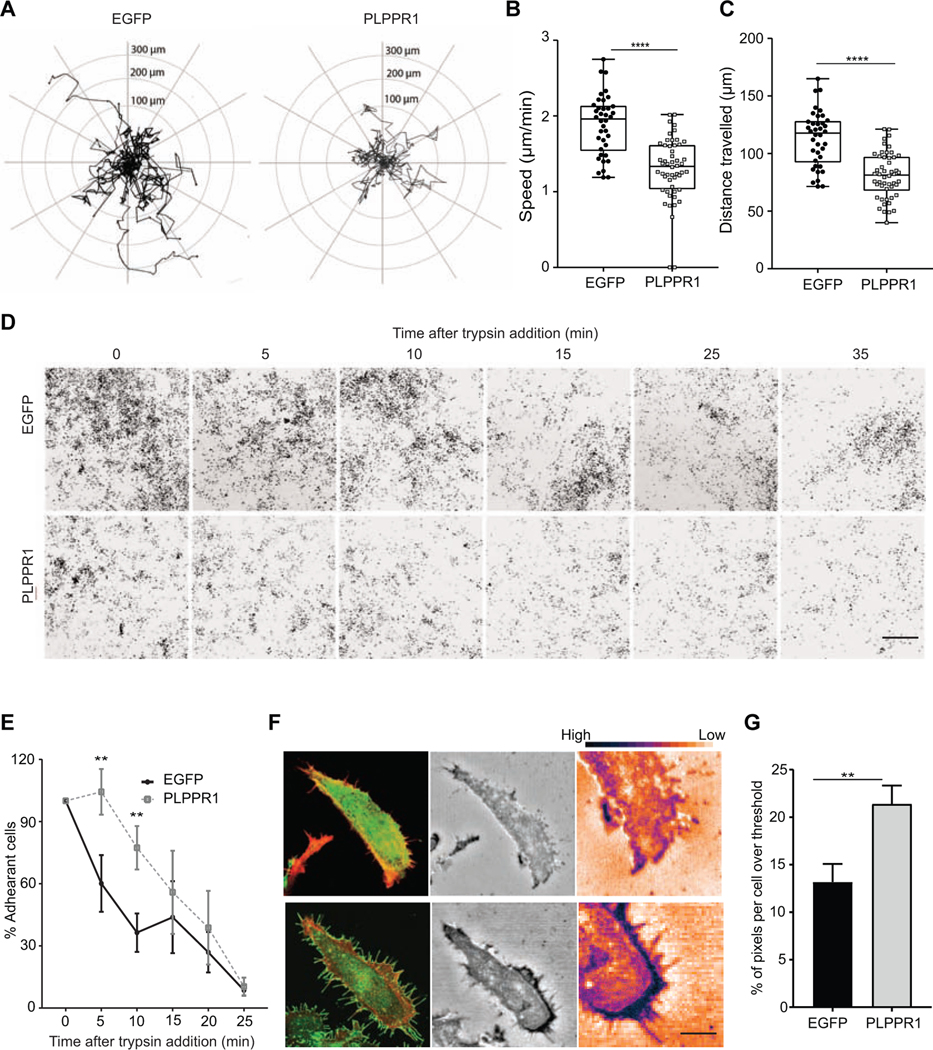
Overexpression of PLPPR1 induces membrane protrusions in several different cell types [4, 7–9, 11], implying a change in the cytoskeleton. Many cellular processes, including migration and adhesion, are governed by dynamic changes to the cytoskeleton. To assess the potential role of PLPPR1 on these processes, we expressed either EGFP or EGFP-PLPPR1 (henceforth mentioned as PLPPR1) in Neuro2a cells, which do not express detectable levels of PLPPR1 mRNA {data from [17, 18] accessible at the NCBI GEO database}, plated on a uniform fibronectin substrate. Live cell imaging was used to observe the migratory trajectories of individual cells (Fig. 1A). Both the speed (Fig. 1B) and total distance travelled (Fig. 1C) by migrating cells expressing PLPPR1 were significantly reduced as compared to cells expressing EGFP.
Figure 1. Overexpressing PLPPR1 deceased cell migration and increases cell adhesion.
(A) Cell trajectories during migration were evaluated in Neuro2a cells transfected with EGFP (left) or PLPPR1 (right), using the middle of each cell body as a point of reference. (B) Speed of migrating cells and (C) distance travelled were determined using NIS Elements software. EGFP, n= 38 cells; PLPPR1, n = 52 cells. Data represent mean ± SEM. p-values were calculated using Student’s t-test with Welch’s correction, ****p < 0.0001. Experiment was conducted in triplicate. (D) Representative images of Neuro2a cells transfected with either EGFP (top) or PLPPR1 (bottom) and subjected to trypsin treatment over time. Scale bar, 100 μm. (E) Quantification of the number of adherent cells resistant to detachment was determined as a percentage of total unwashed transfected cells. Data represents mean ± SEM. p-values were calculated using two-way ANOVA repeated measures with Tukey’s posthoc analysis. *p < 0.05, **p < 0.01. Experiment was conducted in triplicate. (F) (Left) Representative confocal images of cells expressing either EGFP (top) or PLPPR1 (bottom). (Center) IRM images of the respective cells Scale bar = 10 μm and white boxes to highlight the inset Scale bar = 2 μm. (Right) A color map ‘Gem’ from ImageJ is used to display higher intensity pixels as blue and lower intensity as orange which represent stronger and weaker adhesions respectively. (G) Quantification of average area of cell attachment to the fibronectin substrate. EGFP, n = 14 cells; PLPPR1, n = 13 cells. Data represent mean ± SEM. p-values were calculated using Student’s t-test with Welch’s correction. **p < 0.01.
Cell adhesion is a major determinant for cell migration. We therefore performed a cell detachment assay to test the strength of cells to remain attached to the substrate. For this detachment assay, Neuro2a cells expressing either EGFP or PLPPR1 were plated on a fibronectin substrate and grown overnight. After 24 hours, their resistance to detachment by trypsin/EDTA was measured. Our results show that cells expressing PLPPR1 were both more adherent and resistant to detachment by Trypsin/EDTA (Fig. 1 D, E)
To examine the intricacies of adhesion in cells expressing PLPPR1, we used Interference Reflection Microscopy (IRM) to visualize and quantify the interface between the ventral cell surface and the fibronectin substrate coated on glass. Cell membranes in close contact with the substrate produce more interference, detected as darker pixels, while membranes further from the substrate have less interference, detected as lighter pixels. IRM showed that the PLPPR1-expressing cells had stronger adhesion around the outer periphery (Fig. 1F). The average area of attachment to the fibronectin substrate was also significantly greater in cells expressing PLPPR1, suggesting a stronger attachment compared to cells expressing EGFP (Fig. 1G). These observations confirm our results that show increased attachment in cells expressing PLPPR1.
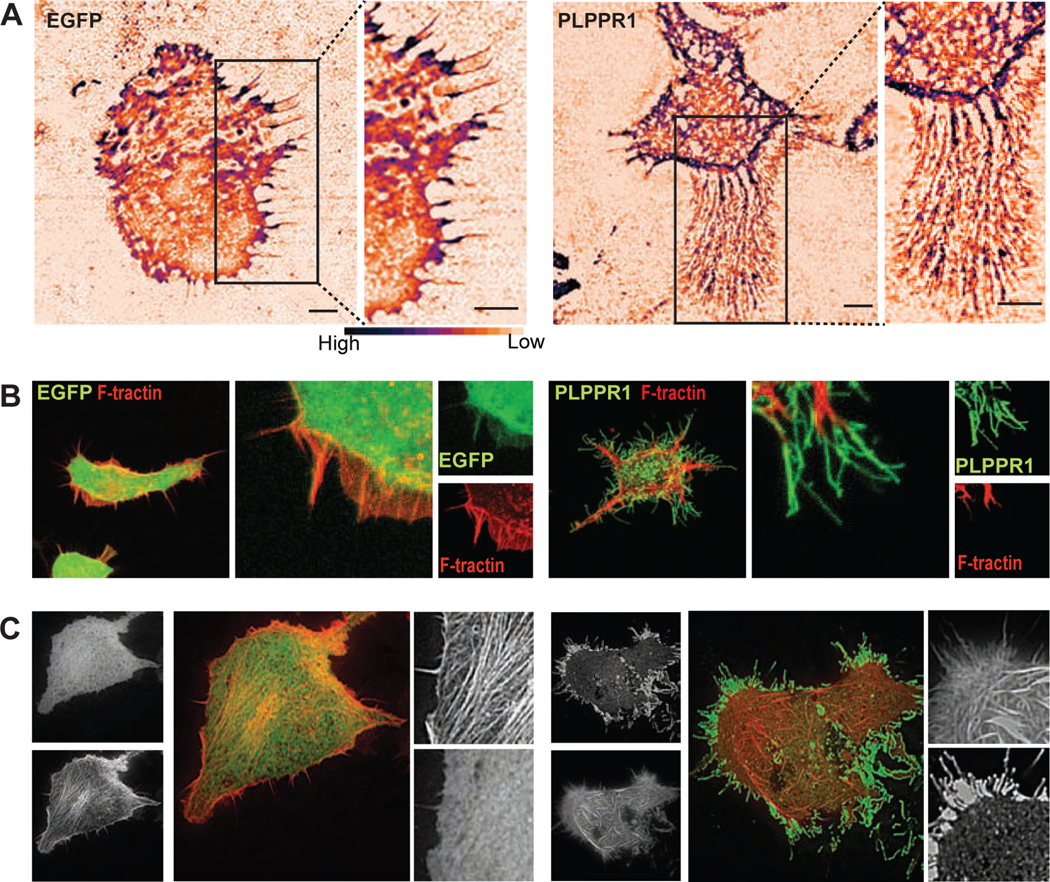
Using live-cell IRM, we observed the formation of a trail of fibrous structures as PLPPR1-expressing cells migrate on the glass substrate (Movie S1, Fig. 2A). These fibrous trails were formed in response to cell movement, and were five to 10 times longer than retraction fibers, suggesting that these “trailing fibers” may be the result of the increased cell attachment. To determine the nature of these “trailing fibers”, we co-expressed mApple-FTR-940 (F-tractin, a cellular probe for filamentous actin) with either EGFP or PLPPR1 in Neuro2a cells. Confocal images showed no expression of F-tractin in these “trailing fibers” (Fig. 2B). The presence of such “trailing fibers”, devoid of cytoskeletal components, have been attributed to active migration in cancer cells [19, 20]. The lack of actin implies they are not protrusions or retraction fibers but may instead be remnants of cell membrane that, due to the PLPPR1-induced increase in cell adhesion, could not detach readily from the fibronectin substrate during migration.
Figure 2. Cells overexpressing PLPPR1 produce “trailing fibers”.
(A) Representative IRM images of Neuro2a cells transfected with either EGFP (left) or PLPPR1 (right) Scale bar, 5μm. Inset is a higher magnification of boxed area showing trailing fibers in cells overexpressing PLPPR1. Scale bar, 5 μm. (B) Representative images of cells co-expressing F-tractin (FTR-940) with either EGFP (left) or PLPPR1 (right) showing membrane protrusions which are devoid of actin filaments. Scale bar, 5 μm. Inset is a higher magnification image of the boxed area. (C) Actin stress fibers in Neuro2a cells co-transfected with F-tractin (FTR-940) and EGFP (left) or PLPPR1 (right). Smaller left panels depict the whole cell in GFP or F-tractin Scale bar, 10 μm. Smaller right panels are insets of higher magnification of boxed area. Scale bar, 2 μm. All experiments were conducted in triplicate.
3.2. PLPPR1 increases nascent focal adhesion complexes and decreases FA disassembly.
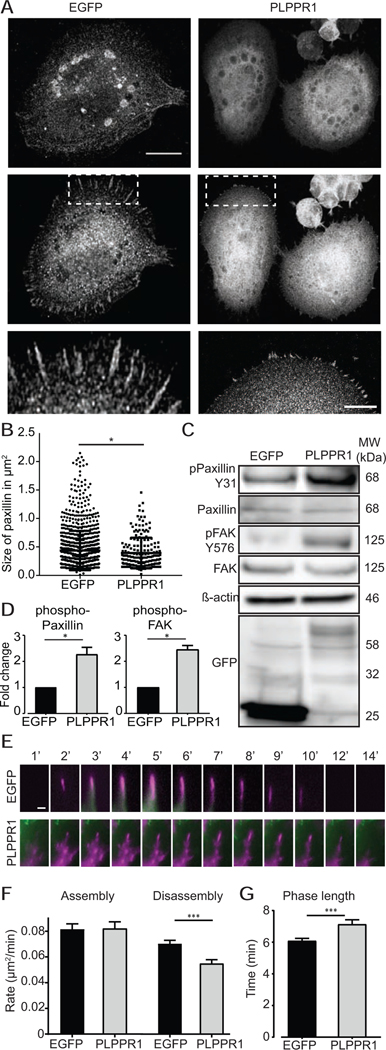
The increased adhesion observed in cells expressing PLPPR1 led us to take a closer look at focal adhesions (FA). FAs are complex plasma membrane-associated proteins that engage with the surrounding extracellular matrix (ECM) and physically connect with the actin cytoskeleton through the recruitment of numerous FA-associated proteins. They operate as mechanosensitive structures that require the recruitment of several proteins in order to change their composition from nascent to mature FAs [21]. This tension-mediated maturation involves the recruitment of paxillin to nascent focal adhesion complexes that are initially small in size, but then mature into focal adhesions as the size of the focal complexes increase and are stabilized [21, 22]. Therefore, cells expressing either EGFP or PLPPR1 were plated on a fibronectin substrate, immunostained for paxillin and F-actin and imaged using TIRF microscopy (Fig. 3A). Measurement of the size of individual FA revealed more nascent paxillin in cells overexpressing PLPPR1 as compared to cells expressing EGFP, suggesting that nascent FAs do not mature (Fig. 3B). In addition, FAs were not observed in the trailing fibers (Movie S2).
Figure 3. Focal adhesions stay in their nascent form with decreased disassembly rates in cells overexpressing PLPPR1.
(A) TIRF microscopy of Neuro2a cells transfected with either EGFP or PLPPR1 and immunostained for paxillin. Scale bar, 10 μm. (B) Mean size of paxillin-containing FAs. Data represent mean ± SEM. p-values were calculated using Student’s t-test with Welch’s correction, *p < 0.05, ****p < 0.0001. (C) Immunoblot analysis of phosphorylated levels of paxillin and FAK in cells expressing either EGFP or PLPPR1. Membranes were stripped and re-probed with anti-GFP antibody and β-actin for loading control. (D) Densitometry analysis of phosphorylated protein versus total protein for paxillin and FAK. All experiments were performed in triplicate. Data represent mean ± SEM. p-values were calculated using Student’s t-test with Welch’s correction, *p < 0.05. (E) Neuro2a cells were co-transfected with m-Apple F-tractin with either EGFP or PLPPR1. Time lapse TIRF microscopy imaged at 1 frame / min for a total of 60 mins. Top 12 panels show the assembly and disassembly of a single Paxillin containing FA in EGFP cell. Bottom 12 panel shows the same for PLPPR1 cell. White asterisks show their appearance and disappearance in the frame Scale bar, 1 μm. (F) Rate of assembly and disassembly of FAs in EGFP and PLPPR1. (G) Phase (of assembly and disassembly) length in minutes per FA. All experiments were conducted in triplicate with each experiment having 15 cells per condition.
Phosphorylation of paxillin on Y31 is crucial for the regulation of paxillin turnover and FA complex maturation [21] as well as dissassmbly [23]. Studies have demonstrated a reduced phospho-paxillin to total paxillin ratio in the transition from nascent to mature FA [24]. Therefore, we assessed phosphorylation levels of paxillin to account for the nascent FAs observed in the PLPPR1-expressing cells. Immunoblotting of Neuro2A cell lysates revealed an increase in phosphorylation of paxillin in cells expressing PLPPR1 (Fig. 3C, D). Paxillin phosphorylation at Y31 is mediated by FAK [25, 26], therefore, we evaluated phosphorylation levels of FAK. Phosphorylation of FAK at Y576 in cells expressing PLPPR1 was increased compared to EGFP-expressing cells (Fig. 3C, D). Altogether, these results suggest that the increased phosphorylation of paxillin and FAK in cells expressing PLPPR1 likely account for the increase in nascent FAs, resulting in increased cell adhesion [27].
Live cell imaging confirmed that focal adhesion complexes in cells co-expressing PLPPR1 and mCherry-paxillin remained nascent compared to the mature FAs observed in cells expressing EGFP (Movie S2). The dynamic measurements for movement of mCherry-paxillin present in each condition as well as how FAs change over time were analyzed using the automated Focal Adhesion Analysis Server (FAAS) [16]. Cells transfected with PLPPR1 and mCherry-paxillin had similar rates of FA assembly but a much lower rate of FA disassembly (Fig. 3E, F). The FA in cells expressing PLPPR1 had longer phase lengths which was due to the slower disassembly (Fig. 3G).
3.3. PLPPR1 overcomes CSPG inhibition
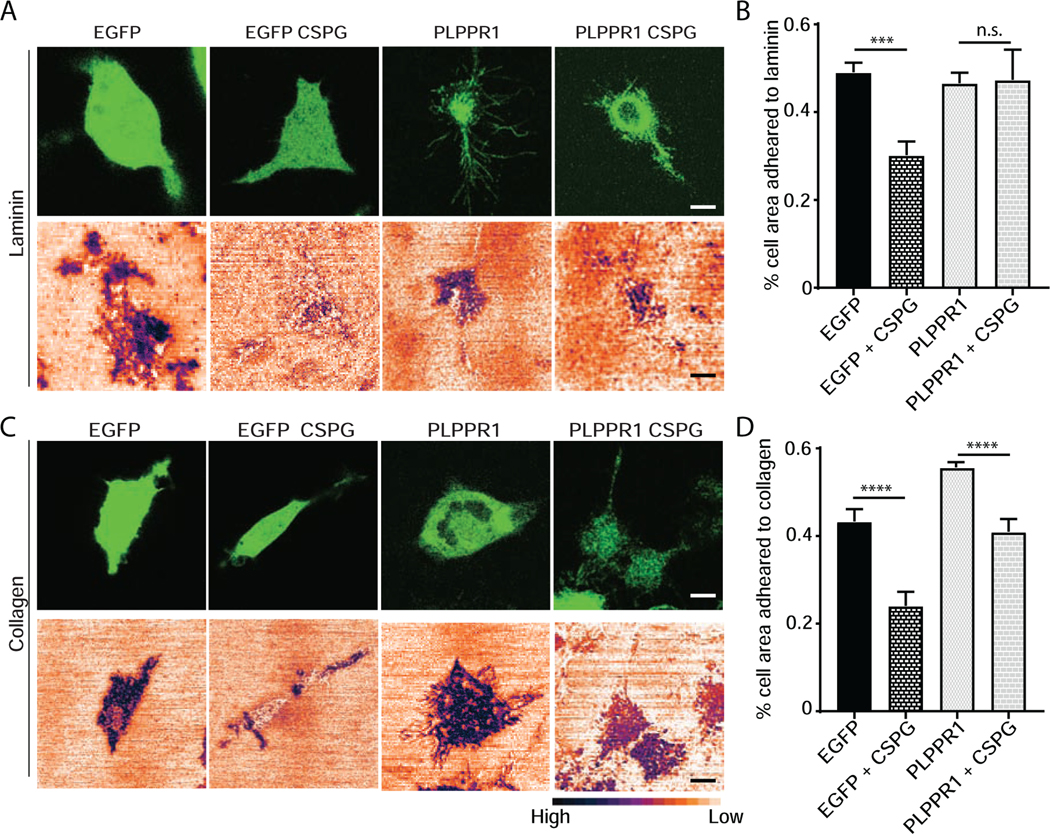
Overexpression of PLPPR1 protein can overcome the inhibitory activity of LPA-induced axon collapse and neurite retraction [9]. Furthermore, PLPPR1 has been demonstrated to promote axon regeneration after spinal cord injury in mice [3]. Chondroitin sulfate proteoglycans (CSPGs) are universally upregulated in the glial scar following injury and, similar to LPA, are potent negative regulators of axon growth by modulating cell adhesion [28–31]. Consequently, we asked if overexpression of PLPPR1 could overcome the anti-adhesive activity of CSPGs on collagen or laminin substrates. Glass-bottom dishes were coated with laminin and laminin/CSPG or collagen and collagen/ CSPG. Neuro2a cells expressing either EGFP or PLPPR1 were plated onto these substrates and 24 hours after transfection the cells were imaged using confocal imaging and IRM. Confocal imaging outlined the entire transfected cell and IRM outlined the ventral area of the cell adhered to the respective substrates. Cells plated onto laminin or collagen showed a high degree of adhesion (Fig. 4A, C), while addition of CSPG to either substrate produced a significant inhibition of adhesion (Fig. 4). In Neuro2a cells expressing PLPPR1, there was no significant difference in adhesion from cells expressing EGFP alone. However, PLPPR1 cells plated onto substrates containing CSPGs had a significant increase in adhesion as compared to cells expressing EGFP (Fig. 4B, D).
Figure 4. PLPPR1 overcomes the inhibition of adhesion by CSPG.
(A) Confocal (top) and IRM (bottom) images of EGFP and PLPPR1-transfected Neuro2a cells plated on laminin or laminin + CSPG. (B) Percentage of ventral surface of cells adhering to laminin or laminin + CSPG in cells expressing EGFP or EGFP-PLPPR1. (C) Confocal (top) and IRM (bottom) images EGFP and EGFP-PLPPR1-transfected Neuro2a cells plated on collagen or collagen + CSPG. (D) Percentage of ventral surface of cells adheting to collagen or collagen + CSPG in EGFP and PLPPR1 cells. All experiments were conducted in triplicate with each experiment having 20 cells per condition. Scale bar, 10 μm.
3.4. PLPPR1 decreases the rate of actin polymerization and increases actin retrograde flow
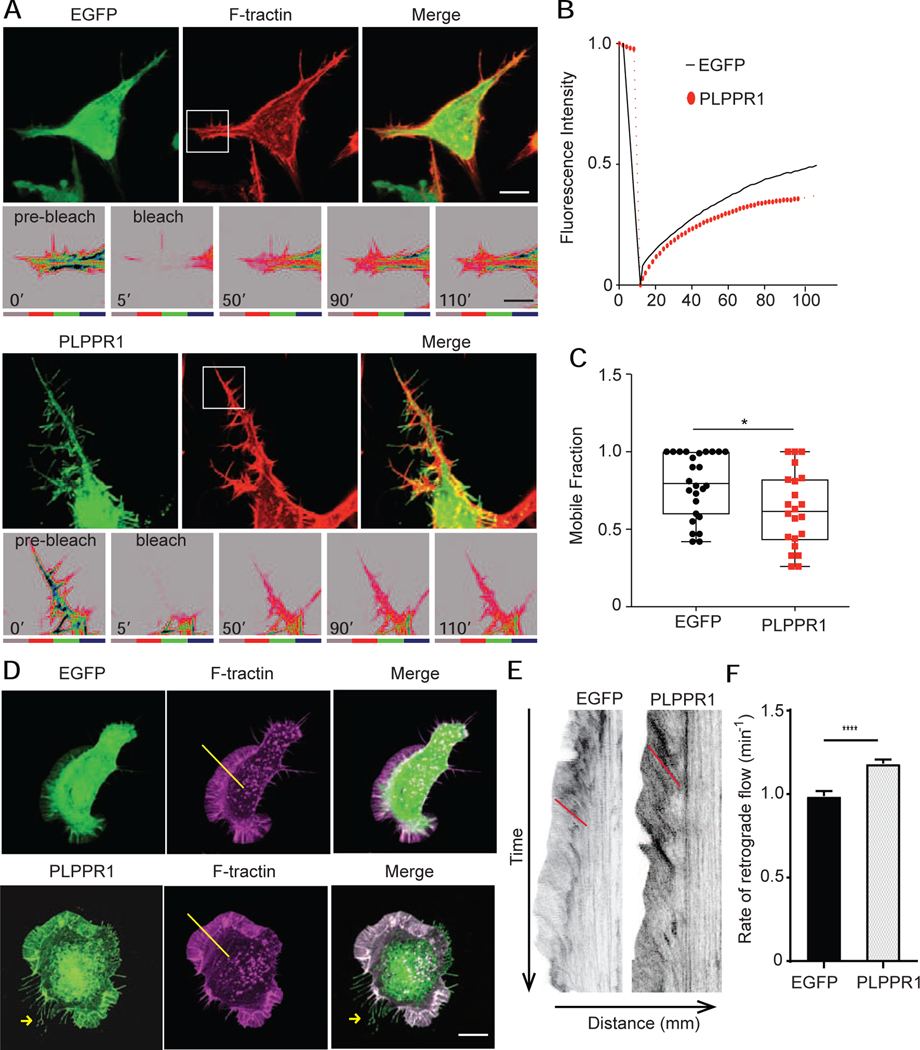
The dynamic organization of the actin cytoskeleton causes changes in cell shape and adhesion strength. This generates tension and creates the pushing and pulling forces that enable cell migration [32, 33]. Given our results showing increased adhesion and decreased migration in cells expressing PLPPR1, we investigated the effect of PLPPR1 on the organization of the actin cytoskeleton. Confocal imaging demonstrated that Neuro2a cells expressing PLPPR1 lack the network of organized linear stress fibers observed in cells expressing EGFP (Fig. 2C). Because actin polymerization and depolymerization are required for stress fiber formation, we evaluated actin turnover by Fluorescence Recovery After Photobleaching (FRAP) measurements in cells co-expressing F-tractin with either EGFP or PLPPR1 (Fig. 4B). The leading edge of the cell was photo-bleached and fluorescence recovery was measured. Results show a decrease in the rate of fluorescence recovery of actin in cells expressing PLPPR1 (Fig. 5A). The mobile fraction of monomeric actin was also decreased in PLPPR1-expressing cells (Fig. 5 B, C).
Figure 5. PLPPR1 expression induces changes in actin polymerization and retrograde flow.
(A) Rate of actin polymerization was assessed in Neuro2a cells co-transfected with F-tractin (FTR-940) and either EGFP (top) or PLPPR1 (bottom) using FRAP and live cell imaging. Scale bar, 10 μm. Bottom insets are the time of pre-bleach, time of bleach and recovery. Intensity of actin is shown in a color gradient, grey being the lowest to blue being the highest Scale bar, 5 μm. (B) Graph showing fluorescence recovery over time in cells transfected with EGFP (black solid line) and cells transfected with PLPPR1 (red dotted line). (C) The mobile fraction of actin monomers was determined as outlined in Materials and Methods. Data represent mean ± SEM. p-values were calculated using Student’s t-test with Welch’s correction. *p < 0.05. (D) Actin retrograde flow was measured in Neuro2a cells co-transfected with F-tractin (FTR-940) and either EGFP (top) or PLPPR1. Cells were imaged at 1 frame per 15 seconds with 5 z-stacks maximum projected for intensity and 500 total time points. The area of lamellipodia (between white arrowheads) was observed for actin retrograde flow. Scale bar, 10 μm. (E) Kymographs showing the actin flow vs. time for a cell transfected with EGFP (left) and PLPPR1 (right). Red lines angles are drawn to indicate the slope of the actin flow. (F) Rates of retrograde flow in EGFP and PLPPR1-expressing cells calculated from slopes of actin tracks. All experiments were conducted in triplicate with each experiment having 10 cells per condition.
In a migrating cell, actin filaments polymerize at the leading edge, usually in the lamellipodia, and flow back into the body of the cell, called retrograde actin flow [34], believed to facilitate cell migration when linked to cell adhesion molecules [35]. It has been suggested to promote forward protrusion via linkage between the lamellipodia and FAs in the clutch model [36]. We therefore compared retrograde actin flow in Neuro2a cells expressing either EGFP or PLPPR1. Neuro2a cells were co-transfected with either EGFP or PLPPR1 with and mAppleFTR-940. To follow the flow of actin from the lamellipodia in a single quantifiable image, kymographs were generated from time lapse movies (Movie S3). Actin retrograde flow was faster in Neuro2a cells expressing PLPPR1than in cells expressing EGFP alone (Fig. 5E, F). The lamellipodia in PLPPR1 also retracted faster due to faster retrograde flow.
3.5. Constitutively active (CA) Rac1 overexpression rescues PLPPR1 induced morphological change
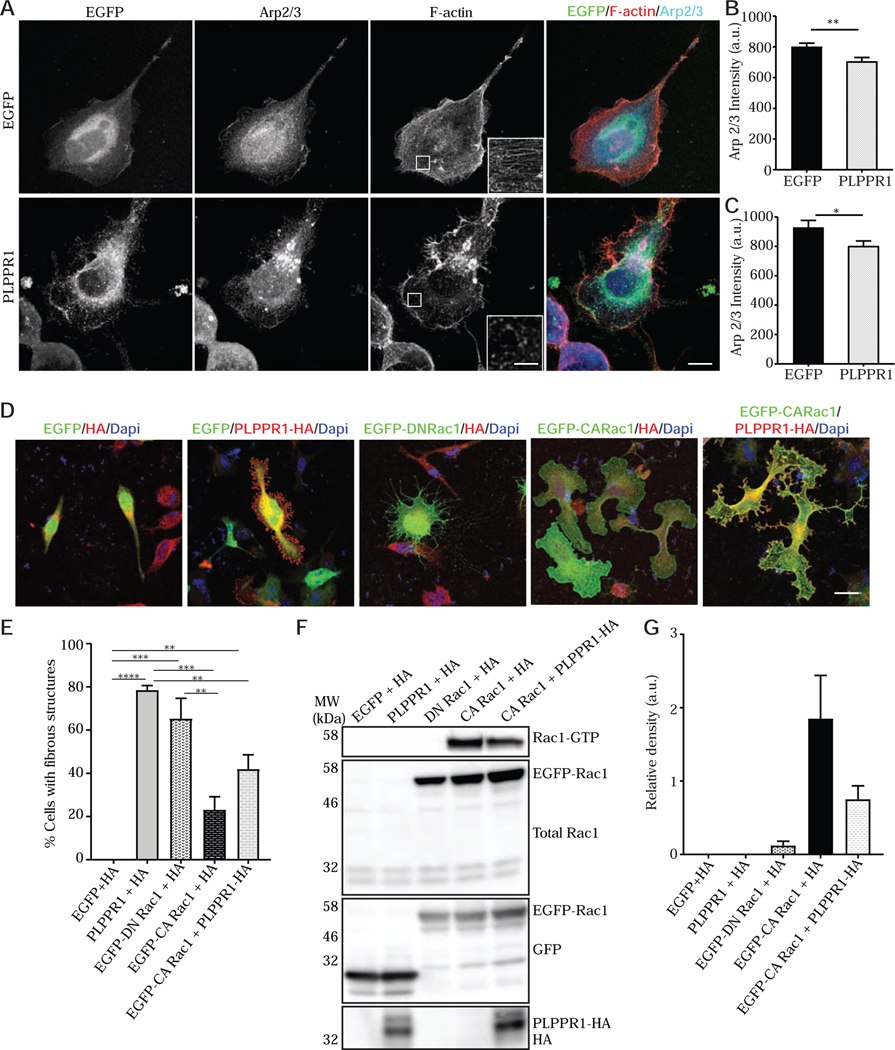
Arp2/3 is an actin nucleating complex that acts downstream of Rac1 and is essential to actin assembly and lamellipodia formation [37–40]. We examined Arp2/3 localization in cells expressing PLPPR1 using STED microscopy. We found that Arp2/3 was localized to the actin-rich lamellipodia and exhibited a diffuse cytoplasmic distribution in cells expressing EGFP. However, the distribution of Arp2/3 was different in Neuro2a cells expressing PLPPR1, which displayed a compact aggregation of Arp2/3 (Fig. 6A), suggesting a disruption in Arp2/3 localization. In addition, total Arp2/3 levels were reduced in PLPPR1 cells (Fig. 6B). When considering only the lamellipodia, the level in Arp2/3 was further reduced in the PLPPR1-expressing cells (Fig.6C). Together, these results suggest a modulatory effect of PLPPR1 on Rac1 activity upstream of Arp2/3.
Figure 6. PLPPR1 reduces Arp2/3 level in Neuro2a and constitutively active Rac1 rescues PLPPR1 phenotype.
(A) Cells transfected with either EGFP (top) or PLPPR1 (bottom) were immunostained for Arp2/3 and phalloidin was used to visualize F-actin Scale bar, 10 μm. White arrows show localization of Arp2/3. Inset is a higher magnification of boxed area Scale bar, 2 μm. (B) Arp2/3 levels within the entire cell body of cells expressing either EGFP or PLPPR1. (C) Total Arp2/3 levels within the lamellipodia of cells expressing either EGFP or PLPPR1. (D) Representative confocal images of Neuro2a cells co-expressing EGFP-HA as control, EGFP-PLPPR1-HA which has “trailing fibers”, EGFP-DN Rac1-HA showed “trailing fibers”, EGFP-CA Rac1-HA had rounded large lamellipodia or EGFP-PLPPR1-CA Rac1-HA which resembled control. Scale bar, 10 μm. (E) Percentage of cells expressing the “trailing fiber” phenotype was calculated in each condition of EGFP-HA, EGFP-PLPPR1-HA, EGFP-DN Rac1-HA, EGFP-CA Rac1-HA or EGFP-PLPPR1-CA Rac1-HA. (F) Rac1-GTP was pulled down using PAK-PBD beads from cell lysates prepared from Neuro2a cells co-expressing HA-tag with either EGFP, PLPPR1, EGFP-Rac1T17N or cells co-transfected with PLPPR1 and EGFP-Rac1G12V. Membranes were immunoblotted with Rac1 antibody. Membranes were stripped and reprobed with anti-GFP antibody. (G) Densitometry analysis of phosphorylated protein versus total protein. Data represents mean ± SEM. p-values were calculated using two-way ANOVA with Tukey posthoc analysis, **p < 0.01, ****p < 0.0001. All experiments were conducted in triplicate.
Rac1 activation is essential to lamellipodial formation [41–43]. Given the possibility that PLPPR1 may regulate Rac1 activity, we examined the morphology of cells expressing dominant negative (DN) Rac1 T17N, constitutively active (CA) Rac1 G12V and cells co-expressing PLPPR1 with CA Rac1. We observed that expression of DN Rac1 alone led to the physiology of broken lamellipodia formation of “trailing fibers” during migration similar to those observed in cells expressing PLPPR1 (Fig. 6D). Interestingly, cells co-expressing CA Rac1 with PLPPR1 had intact lamellipodia with no “trailing fibers” (Fig. 6D). We quantified the percentage of cells that displayed trailing fibers and saw a significant reduction in this phenotype when co-expressing CA Rac1 with PLPPR1 as compared to PLPPR1 alone (Fig 6E).
To further explore the effect of PLPPR1 on Rac1 activation, DN Rac1 and CA Rac1 were expressed in Neuro2a cells and active Rac1 bound to GTP was pulled down using PAK-PBD beads. Basal levels of Rac1-GTP were near zero in Neuro2a cells expressing EGFP and DN Rac1, as well as in cells expressing PLPPR1. High levels of active Rac1 were detected in cells expressing CA Rac1, as expected. Interestingly, co-expression of CA Rac1 with PLPPR1 significantly reduced active Rac1 (Fig. 6 F, G). Expression of constitutively active Rac1 leads to lamellipodia formation, presumably via activation of WAVE- and Arp2/3 complexes. Arp2/3 complex activity is required for lamellipodia maintenance during cell movement and migration. Since the CA Rac1 replenishes the Arp2/3 levels in PLPPR1 cells, there is s rescue of the “trailing fiber” phenotype.
4. Discussion
Five members of the PLPPR family have been identified so far, but their distinct roles or mechanisms are not well understood. The expression pattern of the PLPPR proteins during brain development and after injury supports the idea that they have functions in the CNS [11, 44]. Knocking out PLPPR1 affected hippocampal spine density, long term potentiation and spatial memory [10], while a knockout of PLPPR3 altered thalamocortical neuronal guidance [45]. At the cellular level, several members of the family, including PLPPR1 [7] and PLPPR5 [46] induce membrane protrusions when they are expressed in heterologous cells, and we have shown that there are functional interactions between the members of the PLPPR family [7]. However, no mechanism has been identified for these actions of PLPPRs. In this paper, we show that overexpression of PLPPR1 in cultured Neuro2a cells increases cell adhesion through modulation of GTPase signaling. In addition, this increase in cell adhesion is also observed when cells are plated onto CSPGs. We also find that PLPPR1 expression leads to a peculiar morphology of extended retraction fibers during cell migration, likely due to the increase in cell adhesion. Finally, we show an involvement of Rac1 signaling as participating in these changes.
We first demonstrated that expression of PLPPR1 in Neuro2a cells results in decreased migration. In addition, migrating cells expressing PLPPR1 leave long plasma membrane extensions that we have termed “trailing fibers” which are more than 10 times longer than retraction fibers. Cell migration requires both increased adhesion of the substrate at the front of the cell, and the ability of the rear of the cell to detach [47]. Many different cell types have been observed to leave behind membrane fragments at the trailing edge of the cell [48, 49], In fact, regulation of migration speed may be primarily due to the rate of detachment, rather than engagement at the front of the cell [50]. The reduced motility and persistence of these “trailing fibers” in cells expressing PLPPR1 suggest an inability of the cells to detach from the substrate during movement, likely due to the increase in adhesion to the fibronectin substrate.
Cell adhesion is mediated through the engagement and activation of integrins, which then promote the establishment of focal adhesions [51–53]. Studies have shown that PLPPR4 mediates cell adhesion through activation of ß1-integrins through the unique calmodulin-binding domain in its C-terminal tail [10]. While PLPPR1 lacks the key motif in its C-terminal tail, there is evidence that it may associate with ß1-integrins [54]. The nature of this interaction and its role in the observed increase in cell adhesion has yet to be explored. We showed that there were more nascent FAs in PLPPRs and that the disassembly of these FAs was much slower. This reduced disassembly rate might attribute to stronger adhesion interfering with migration.
The maturation of FAs is dependent upon mechanical tension through the actin skeleton [55, 56] engaging a molecular clutch linked to FAs [36]. The fact that many nascent FAs failed to mature suggests less tension is being applied through the actin cytoskeleton, i.e., that the “clutch” is disengaged, preventing motle force. Retrograde actin flow is considered to contribute to forward advance through actin engagement of the clutch [57]. Flow is faster in slower moving or stationary cells as the clutch disengages from FAs [58, 59], consistent with the increase in the rate of retrograde actin flow in cells expressing PLPPR1. Moreover, mature, but not nascent, FAs slow down retrograde actin flow [60]. Tension on FAs also increases the accumulation of disassembly factors [61], which is consistent with the slower turnover of FAs in cells expressing PLPPR1.
Our FRAP experiments demonstrated reduced mobility of actin monomers in these cells. In some cells, retrograde flow appears to regulate FA assembly, which would then translate into regulation of movement [62]. However, we found no change in the rate of FA assembly due to the expression of PLPPR1. This suggests that the migration rate in Neuro2a cells is primarily regulated by the rate of detachment from the substrate.
Several studies have demonstrated that inhibition of Rho GTPases activity prevents the maturation of focal adhesion complexes [52, 63, 64]. Therefore, it is conceivable that the reduced Rac1 activity and the nascent focal adhesion complexes that persist in cells expressing PLPPR1 may account for the increase in cell adhesion and decrease in cell migration. This is consistent with the increase in phosphorylation of paxillin and FAK observed in these cells, given that FAK mediated phosphorylation of paxillin plays a role in the assembly, turnover and stability of focal adhesions [21, 23, 24, 65, 66].
Nucleation of actin filaments by Arp2/3 complex is driven by WAVE (WASP-family verprolin-homologous protein), downstream of Rac1 activation [38, 67]. Localization of Arp2/3 is disrupted in cells overexpressing PLPPR1 and this may be a direct consequence of altered Rac1 activity. We show that expression of DN Rac1 exhibits a similar morphological change as expression of PLPPR1 in Neuro2a cells. Expression of DN Rac1 inhibits GTP loading by sequestering endogenous GEFs and forming non-functional RhoGTPase – RhoGEF complexes. This leads to decreased membrane ruffling and a failure to form proper lamellipodia [42, 68, 69]. The co-expression of PLPPR1 with CA Rac1 rescues the trailing fiber morphology. CA Rac1 blocks the GAP binding domain and inhibits GTPase activity [70]. With an excess of mutated Rac1 fused to GTP, there may be an increased pool of GTP available to other RhoGTPases leading to increased RhoA activity in cells expressing CA Rac1 rescuing the morphology.
In vivo, CSPGs and myelin are known to be major inhibitors of neural regeneration after injury [71]. Cells plated onto substrates of CSPGs have reduced neurite outgrowth [72, 73], likely due to the anti-adhesive actions of CSPGs [31]. We show here that overexpression of PLPPR1 in Neuro2a cells overcomes this anti-adhesive substrate induced by CSPGs. This phenomenon is observed when cells were plated on laminin or collagen. Laminins engage integrins, while CSPGs do not. Even though CSPGs interfere with integrin-mediated adhesion [74], PLPPR1 appears to circumvent that interference, and allowing cells to remain adhered to the substrates. PLPPR1 also has a pro-regenerative effect in vivo after injury [3], perhaps mediated through this same mechanism.
Conclusion
Taken together, our results provide evidence for increased adhesion as a result of PLPPR1 expression. This serves as a potential explanation for the ability of PLPPR1to alter neuronal migration during development [5] and increasing axonal growth after injury [3]. While increased phosphorylation of paxillin and FAK and alteration of Rac1 and Arp2/3 are observed in Neuro2a cells expressing PLPPR1, as our previous proteomic investigations did not reveal an association between PLPPR1 and these proteins [7], the exact signaling pathway between PLPPR1 and these proteins is a subject for future investigations.
Supplementary Material
Acknowledgements
We wish to thank Xufeng Wu and Daniela Malide of the NHLBI Light Microscopy Core facility for their help and guidance and Dr. Clare Waterman for materials provided.
Funding: This work was funded by the NHLBI Intramural Research Program.
Abbreviations:
- PLPPR
Phospholipid Phosphatase Related Protein
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wang WZ, Molnar Z, Dynamic pattern of mRNA expression of plasticity-related gene-3 (PRG-3) in the mouse cerebral cortex during development, Brain Res. Bull. 66 (2005) 454–460. 10.1016/j.brainresbull.2005.05.010 [DOI] [PubMed] [Google Scholar]
- [2].Brauer AU, Savaskan NE, Kuhn H, Prehn S, Ninnemann O, Nitsch R, A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting, Nat. Neurosci. 6 (2003) 572–578. 10.1038/nn1052 [DOI] [PubMed] [Google Scholar]
- [3].Fink KL, Lopez-Giraldez F, Kim IJ, Strittmatter SM, Cafferty WBJ, Identification of intrinsic axon growth modulators for intact CNS neurons after injury, Cell Rep.18 (2017) 2687–2701. 10.1016/j.celrep.2017.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Savaskan NE, Brauer AU, Nitsch R, Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family, Eur. J. Neurosci. 19 (2004) 212–220. 10.1046/j.1460-9568.2003.03078.x [DOI] [PubMed] [Google Scholar]
- [5].Khalaf-Nazzal R, Stouffer MA, Olaso R, Muresan L, Roumegous A, Lavilla V, Carpentier W, Moutkine I, Dumont S, Albaud B, Cagnard N, Roest Crollius H, Francis F, Early born neurons are abnormally positioned in the doublecortin knockout hippocampus, Hum. Mol. Genet. 26 (2017) 90–108. 10.1093/hmg/ddw370 [DOI] [PubMed] [Google Scholar]
- [6].Pfurr S, Chu YH, Bohrer C, Greulich F, Beattie R, Mammadzada K, Hils M, Arnold SJ, Taylor V, Schachtrup K, Uhlenhaut NH, Schachtrup C, The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development, Development 144 (2017) 3917–3931. 10.1242/dev.145698 [DOI] [PubMed] [Google Scholar]
- [7].Yu P, Agbaegbu C, Malide DA, Wu X, Katagiri Y, Hammer JA, Geller HM, Cooperative interactions of LPPR family members in membrane localization and alteration of cellular morphology, J. Cell Sci. 128 (2015) 3210–3222. 10.1242/jcs.169789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sigal YJ, Quintero OA, Cheney RE, Morris AJ, Cdc42 and ARP2/3-independent regulation of filopodia by an integral membrane lipid-phosphatase-related protein, J. Cell Sci. 120 (2007) 340–352. 10.1242/jcs.03335 [DOI] [PubMed] [Google Scholar]
- [9].Broggini T, Schnell L, Ghoochani A, Mateos JM, Buchfelder M, Wiendieck K, Schafer MK, Eyupoglu IY, Savaskan NE, Plasticity Related Gene 3 (PRG3) overcomes myelin-associated growth inhibition and promotes functional recovery after spinal cord injury, Aging (Albany N. Y.) 8 (2016) 2463–2487. 10.18632/aging.101066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu X, Huai J, Endle H, Schluter L, Fan W, Li Y, Richers S, Yurugi H, Rajalingam K, Ji H, Cheng H, Rister B, Horta G, Baumgart J, Berger H, Laube G, Schmitt U, Schmeisser MJ, Boeckers TM, Tenzer S, Vlachos A, Deller T, Nitsch R, Vogt J, PRG-1 regulates synaptic plasticity via intracellular PP2A/β1-integrin signaling, Dev. Cell 38 (2016) 275–290. 10.1016/j.devcel.2016.06.019 [DOI] [PubMed] [Google Scholar]
- [11].Velmans T, Battefeld A, Geist B, Farres AS, Strauss U, Brauer AU, Plasticity-related gene 3 promotes neurite shaft protrusion, BMC Neurosci. 14 (2013) 36 10.1186/1471-2202-14-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coiro P, Stoenica L, Strauss U, Brauer AU, Plasticity-related gene 5 promotes spine formation in murine hippocampal neurons, J. Biol. Chem. 289 (2014) 24956–24970. 10.1074/jbc.M114.597880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McDermott MI, Sigal YJ, Sciorra VA, Morris AJ, Is PRG-1 a new lipid phosphatase?, Nat. Neurosci. 7 (2004) 789; author reply 789–790 [DOI] [PubMed] [Google Scholar]
- [14].Barr VA, Bunnell SC, Interference reflection microscopy, Curr. Protoc. Cell Biol. Chapter 4 (2009) Unit 4 23. 10.1002/0471143030.cb0423s45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat Methods 9 (2012) 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berginski ME, Gomez SM, The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics, F1000Res 2 (2013) 68 10.12688/f1000research.2-68.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].[dataset] Iida K, The RNA-binding protein Sfpq regulates long neuronal genes in transcriptional elongation, geo (2018). GSE60246 [Google Scholar]
- [18].Llorens F, Carulla P, Villa A, Torres JM, Fortes P, Ferrer I, del Rio JA, PrP(C) regulates epidermal growth factor receptor function and cell shape dynamics in Neuro2a cells, J. Neurochem. 127 (2013) 124–138. 10.1111/jnc.12283 [DOI] [PubMed] [Google Scholar]
- [19].Haemmerli G, Strauli P, In vitro motility of cells from human epidermoid carcinomas. A study by phase-contrast and reflection-contrast cinematography, Int. J. Cancer 27 (1981) 603–610. 10.1002/ijc.2910270506 [DOI] [PubMed] [Google Scholar]
- [20].DePasquale JA, Cell matrix adhesions and localization of the vitronectin receptor in MCF-7 human mammary carcinoma cells, Histochem. Cell Biol. 110 (1998) 485–494. 10.1007/s004180050310 [DOI] [PubMed] [Google Scholar]
- [21].Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM, Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation, J. Cell Biol. 188 (2010) 877–890. 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B, Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells, J. Cell Sci. 116 (2003) 4605–4613. 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- [23].Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF, FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly, Nat. Cell Biol. 6 (2004) 154–161. 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- [24].Zaidel-Bar R, Milo R, Kam Z, Geiger B, A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions, J. Cell Sci. 120 (2007) 137–148. 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
- [25].Bellis SL, Miller JT, Turner CE, Characterization of tyrosine phosphorylation of paxillin in vitro by focal adhesion kinase, J. Biol. Chem. 270 (1995) 17437–17441. 10.1074/jbc.270.29.17437 [DOI] [PubMed] [Google Scholar]
- [26].Schaller MD, Parsons JT, pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk, Mol. Cell. Biol. 15 (1995) 2635–2645. 10.1128/mcb.15.5.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL, Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts, J. Cell Biol. 153 (2001) 881–888. 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mckeon RJ, Schreiber RC, Rudge JS, Silver J, Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes, J. Neurosci. 11 (1991) 3398–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang H, Katagiri Y, McCann TE, Unsworth E, Goldsmith P, Yu ZX, Tan F, Santiago L, Mills EM, Wang Y, Symes AJ, Geller HM, Chondroitin-4-sulfation negatively regulates axonal guidance and growth, J. Cell Sci. 121 (2008) 3083–3091. 10.1242/jcs.032649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M, The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth, J. Cell Biol. 125 (1994) 669–680. 10.1083/jcb.125.3.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jin J, Tilve S, Huang Z, Zhou L, Geller HM, Yu P, Effect of chondroitin sulfate proteoglycans on neuronal cell adhesion, spreading and neurite growth in culture, Neural Regen. Res. 13 (2018) 289–297. 10.4103/1673-5374.226398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM, Mechanical integration of actin and adhesion dynamics in cell migration, Annu. Rev. Cell Dev. Biol. 26 (2010) 315–333. 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].van Helvert S, Storm C, Friedl P, Mechanoreciprocity in cell migration, Nat. Cell Biol. 20 (2018) 8–20. 10.1038/s41556-017-0012-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bray D, Surface movements during the growth of single explanted neurons, Proc. Natl. Acad. Sci. U. S. A. 65 (1970) 905–910. 10.1073/pnas.65.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Forscher P, Smith SJ, Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone, J. Cell Biol. 107 (1988) 1505–1516. 10.1083/jcb.107.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mitchison T, Kirschner M, Cytoskeletal dynamics and nerve growth, Neuron 1 (1988) 761–772. 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- [37].Machesky LM, Hall A, Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization, J. Cell Biol. 138 (1997) 913–926. 10.1083/jcb.138.4.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K, Arp2/3 complex interactions and actin network turnover in lamellipodia, EMBO J. 27 (2008) 982–992. 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE, Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis, Cell 148 (2012) 973–987. 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Machesky LM, Insall RH, Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex, Curr. Biol. 8 (1998) 1347–1356. 10.1016/s0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- [41].Kozma R, Sarner S, Ahmed S, Lim L, Rho family GTPases and neuronal growth cone remodelling: Relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid, Mol. Cell. Biol. 17 (1997) 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ehrlich JS, Hansen MD, Nelson WJ, Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion, Dev. Cell 3 (2002) 259–270. 10.1016/S1534-5807(02)00216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Somanath PR, Byzova TV, 14–3-3b-Rac1-p21 activated kinase signaling regulates Akt1-mediated cytoskeletal organization, lamellipodia formation and fibronectin matrix assembly, J. Cell. Physiol. 218 (2009) 394–404. 10.1002/jcp.21612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brauer AU, Nitsch R, Plasticity-related genes (PRGs/LRPs): a brain-specific class of lysophospholipid-modifying proteins, Biochim. Biophys. Acta 1781 (2008) 595–600. 10.1016/j.bbalip.2008.04.004 [DOI] [PubMed] [Google Scholar]
- [45].Cheng J, Sahani S, Hausrat TJ, Yang JW, Ji H, Schmarowski N, Endle H, Liu X, Li Y, Bottche R, Radyushkin K, Maric HM, Hoerder-Suabedissen A, Molnar Z, Prouvot PH, Trimbuch T, Ninnemann O, Huai J, Fan W, Visentin B, Sabbadini R, Stromgaard K, Stroh A, Luhmann HJ, Kneussel M, Nitsch R, Vogt J, Precise somatotopic thalamocortical axon guidance depends on LPA-mediated PRG-2/radixin signaling Neuron 92 (2016) 126–142. 10.1016/j.neuron.2016.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Broggini T, Nitsch R, Savaskan NE, Plasticity-related gene 5 (PRG5) induces filopodia and neurite growth and impedes lysophosphatidic acid- and nogo-A-mediated axonal retraction, Mol. Biol. Cell 21 (2010) 521–537. 10.1091/mbc.E09-06-0506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lauffenburger DA, Horwitz AF, Cell migration: a physically integrated molecular process, Cell 84 (1996) 359–369. 10.1016/s0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- [48].Regen CM, Horwitz AF, Dynamics of β1 integrin-mediated adhesive contacts in motile fibroblasts, J. Cell Biol. 119 (1992) 1347–1359. 10.1083/jcb.119.5.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Francis JW, Fabi AY, Petty HR, Fluorescence microscopy study of polymorphonuclear leukocyte substrate attached materials, Cell Motil. Cytoskeleton 9 (1988) 1–8. 10.1002/cm.970090102 [DOI] [PubMed] [Google Scholar]
- [50].Chen WT, Mechanism of retraction of the trailing edge during fibroblast movement, J. Cell Biol. 90 (1981) 187–200. 10.1083/jcb.90.1.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nobes CD, Hall A, Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia, Cell 81 (1995) 53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- [52].Sinnett-Smith J, Lunn JA, Leopoldt D, Rozengurt E, Y-27632, an inhibitor of Rho-associated kinases, prevents tyrosine phosphorylation of focal adhesion kinase and paxillin induced by bombesin: dissociation from tyrosine phosphorylation of p130(CAS), Exp. Cell Res. 266 (2001) 292–302. 10.1006/excr.2001.5219 [DOI] [PubMed] [Google Scholar]
- [53].Rottner K, Hall A, Small JV, Interplay between Rac and Rho in the control of substrate contact dynamics, Curr. Biol. 9 (1999) 640–648. 10.1016/s0960-9822(99)80286-3 [DOI] [PubMed] [Google Scholar]
- [54].Yu P, Pisitkun T, Wang G, Wang R, Katagiri Y, Gucek M, Knepper MA, Geller HM, Global analysis of neuronal phosphoproteome regulation by chondroitin sulfate proteoglycans, PLoS One 8 (2013) e59285 10.1371/journal.pone.0059285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD, Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism, J. Cell Biol. 153 (2001) 1175–1186. 10.1083/jcb.153.6.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chrzanowska-Wodnicka M, Burridge K, Rho-stimulated contractility drives the formation of stress fibers and focal adhesions, J. Cell Biol. 133 (1996) 1403–1415. 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lin CH, Forscher P, Growth cone advance is inversely proportional to retrograde F-actin flow, Neuron 14 (1995) 763–771. [DOI] [PubMed] [Google Scholar]
- [58].Henson JH, Svitkina TM, Burns AR, Hughes HE, MacPartland KJ, Nazarian R, Borisy GG, Two components of actin-based retrograde flow in sea urchin coelomocytes, Mol. Biol. Cell 10 (1999) 4075–4090. 10.1091/mbc.10.12.4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang YL, Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling, J. Cell Biol. 101 (1985) 597–602. 10.1083/jcb.101.2.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yamashiro S, Mizuno H, Smith MB, Ryan GL, Kiuchi T, Vavylonis D, Watanabe N, New single-molecule speckle microscopy reveals modification of the retrograde actin flow by focal adhesions at nanometer scales, Mol. Biol. Cell 25 (2014) 1010–1024. 10.1091/mbc.E13-03-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kuo JC, Han X, Hsiao CT, Yates JR 3rd, Waterman CM, Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation, Nat. Cell Biol. 13 (2011) 383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Smilenov LB, Mikhailov A, Pelham RJ, Marcantonio EE, Gundersen GG, Focal adhesion motility revealed in stationary fibroblasts, Science 286 (1999) 1172–1174. 10.1126/science.286.5442.1172 [DOI] [PubMed] [Google Scholar]
- [63].Sawada K, Morishige K, Tahara M, Ikebuchi Y, Kawagishi R, Tasaka K, Murata Y, Lysophosphatidic acid induces focal adhesion assembly through Rho/Rho-associated kinase pathway in human ovarian cancer cells, Gynecol. Oncol. 87 (2002) 252–259. 10.1006/gyno.2002.6831 [DOI] [PubMed] [Google Scholar]
- [64].Burridge K, Guilluy C, Focal adhesions, stress fibers and mechanical tension, Exp. Cell Res. 343 (2016) 14–20. 10.1016/j.yexcr.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Turner CE, Paxillin and focal adhesion signalling, Nat. Cell Biol. 2 (2000) E231–236. 10.1038/35046659 [DOI] [PubMed] [Google Scholar]
- [66].Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD, FAK promotes recruitment of talin to nascent adhesions to control cell motility, J. Cell Biol. 196 (2012) 223–232. 10.1083/jcb.201108078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen B, Chou HT, Brautigam CA, Xing W, Yang S, Henry L, Doolittle LK, Walz T, Rosen MK, Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites, Elife 6 (2017). 10.7554/eLife.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].McCarty OJ, Larson MK, Auger JM, Kalia N, Atkinson BT, Pearce AC, Ruf S, Henderson RB, Tybulewicz VL, Machesky LM, Watson SP, Rac1 is essential for platelet lamellipodia formation and aggregate stability under flow, J. Biol. Chem. 280 (2005) 39474–39484. 10.1074/jbc.M504672200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Debreceni B, Gao Y, Guo F, Zhu K, Jia B, Zheng Y, Mechanisms of guanine nucleotide exchange and Rac-mediated signaling revealed by a dominant negative trio mutant, J. Biol. Chem. 279 (2004) 3777–3786. 10.1074/jbc.M308282200 [DOI] [PubMed] [Google Scholar]
- [70].Zhang ZG, Lambert CA, Servotte S, Chometon G, Eckes B, Krieg T, Lapiere CM, Nusgens BV, Aumailley M, Effects of constitutively active GTPases on fibroblast behavior, Cell. Mol. Life Sci. 63 (2006) 82–91. 10.1007/s00018-005-5416-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fawcett JW, Schwab ME, Montani L, Brazda N, Muller HW, Defeating inhibition of regeneration by scar and myelin components, Handb. Clin. Neurol. 109 (2012) 503–522. 10.1016/B978-0-444-52137-8.00031-0 [DOI] [PubMed] [Google Scholar]
- [72].Carbonetto S, Gruver MM, Turner DC, Nerve fiber growth in cultures of fibronectin, collagen, and glycosaminoglycan substrates, J. Neurosci. 3 (1983) 2324–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Snow DM, Brown EM, Letourneau PC, Growth cone behavior in the presence of soluble chondroitin sulfate proteoglycan (CSPG), compared to behavior on CSPG bound to laminin or fibronectin, Int. J. Dev. Neurosci. 14 (1996) 331–349. 10.1016/0736-5748(96)00017-2 [DOI] [PubMed] [Google Scholar]
- [74].Iida J, Skubitz AP, Furcht LT, Wayner EA, McCarthy JB, Coordinate role for cell surface chondroitin sulfate proteoglycan and α4β1 integrin in mediating melanoma cell adhesion to fibronectin, J. Cell Biol. 118 (1992) 431–444. 10.1083/jcb.118.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.