Short abstract
Objective
Serum proteomic analysis of tuberculosis (TB) antigens to identify biomarkers enabling discrimination of active TB (ATB) from latent TB infection (LTBI).
Methods
Serum samples from patients with ATB, individuals with LTBI and healthy controls (HCs) were used to probe proteome microarrays. Based on signal intensities of IgG and IgM antibodies, 100 TB proteins were selected for fabrication of mini-protein microarrays, which were then used to screen 204 serum samples.
Results
Proteome microarray analyses showed that 58 IgG or IgM specific antibodies were significantly more abundant in ATB patients than in individuals with LTBI or HCs. Serological evaluation of mini-protein microarrays demonstrated that average levels of 15 specific antibodies were higher in ATB patients than in individuals with LTBI or HCs. This combination of 15 TB serum biomarkers had a sensitivity of 85.4% and specificity of 90.3% in discriminating ATB from LTBI.
Conclusion
Combinations of serum biomarkers can offer improved diagnostic performance in discriminating ATB from LTBI. Five biomarkers (MT1560.1, Rv0049, Rv0270, Rv1597 and Rv3480c) associated with ATB induced stronger IgM responses in these patients.
Keywords: Mycobacterium tuberculosis, active TB, latent TB, proteome microarray, serum biomarkers, IgG, IgM
Introduction
Tuberculosis (TB), a life-threatening infectious disease, caused about 1.2 million deaths among human immunodeficiency virus (HIV)-negative individuals worldwide in 2018. Over 65% of the estimated 10 million new TB cases occurred in eight high-TB burden, low/middle-income developing countries: India, China, Indonesia, the Philippines, Pakistan, Nigeria, Bangladesh and South Africa.1 Latent TB infection (LTBI) affects about 1.7 billion people worldwide and progresses to active TB (ATB) in about 5% to 10 of infected individuals during their lifetimes.1 Therefore, development of new diagnostics and serological biomarkers to identify LTBI and its risk of progression to ATB is a promising public health strategy to prevent new infections.1–3
Interferon gamma release assays (IGRAs) are nonmolecular technologies endorsed by the World Health Organization for diagnosis of LTBI. However, these assays lack sufficient diagnostic power to reliably discriminate LTBI from ATB.4–6 The diagnostic accuracy and cost-effectiveness of two IGRAs, T-SPOT.TB and QuantiFERON GOLD In-Tube (QFT-GIT), were compared in a recent UK study.6 T-SPOT.TB was more sensitive than QFT-GIT (82.3% versus 67.3%) and both assays were similarly specific (82.6% versus 80.4%). However, both assays were insufficiently sensitive to rule out ATB in routine clinical practice.6 Thus, new biomarkers able to discriminate LTBI from ATB are urgently needed.
Proteome microarrays containing 4,099 TB proteins were previously probed with sera from 561 suspected TB cases, and 13 proteins were found to be significantly associated with ATB.7 Recently, additional serum proteomic approaches have been used to identify new candidate biomarkers for distinguishing LTBI from ATB, for monitoring disease progression from LTBI to ATB in nonhuman primates and in TB patients, and for investigating serum IgG and IgM antibodies against TB antigens in ATB patients.8–10 In this study, we aimed to evaluate serum responses to Mycobacterium tuberculosis antigens as potential biomarkers for discriminating ATB from LTBI using proteome microarrays containing 4,262 TB antigens and mini-protein microarrays containing 100 selected TB proteins.
Materials and methods
Collection of serum samples from participants
This study was approved by the ethics committee of the Affiliated Hospital of Zunyi Medical University and was conducted in accordance with the principles laid out in the Declaration of Helsinki. Written informed consent for the use of samples was obtained from all participants. At the time of drawing blood samples, ATB patients had been treated for less than 2 weeks following a diagnosis of TB confirmed by the presence of acid-fast bacilli on sputum smears and positive M. tuberculosis cultures. The T-SPOT.TB kit (Oxford Immunotec, Abingdon, UK) was used to identify individuals with LTBI. Individuals with LTBI and healthy controls (HCs) had neither clinical symptoms nor abnormal chest radiographic findings associated with ATB. Patients with HIV and patients taking immunosuppressant drugs were excluded. All participants were Bacille Calmette–Guérin (BCG) vaccinated. Peripheral venous blood (5 mL) was drawn from each of the participants (ATB patients, individuals with LTBI and HCs) and sera were obtained by centrifugation at 3,000 ×g for 10 minutes. Serum samples were aliquoted (1 mL each) into 1.5 mL microcentrifuge tubes and stored at −80°C until use.
Proteome and mini-protein microarrays
Proteome microarrays containing 4,262 TB proteins were purchased from BCBIO (Guangzhou, China). The proteome microarrays were described in detail by Cao et al.8 Based on the screening results from TB proteome microarrays, 100 distinguishing TB proteins were specifically selected for custom fabrication of mini-protein microarrays.
Serological screening of proteome and mini-protein microarrays
Proteome and mini-protein microarrays were blocked in blocking buffer (Tris-buffered saline, pH 7.4, containing 10% bovine serum albumin) for 2 hours at room temperature (25°C) with agitation. Two hundred microliters of serum samples, diluted 1:200 in phosphate-buffered saline (PBS), pH 7.4, containing 0.05% Tween-20 (PBST), were added to proteome or mini-protein microarrays and incubated at 4°C overnight. After washing three times for 5 minutes with PBST, proteome and mini-protein microarrays were incubated in the dark with Cy3-conjugated goat anti-human IgG and Cy5-conjugated goat anti-human IgM (Jackson Laboratory, PA, USA; both diluted 1:1,000) for 45 minutes at room temperature (25°C). The proteome and mini-protein microarrays were washed at room temperature (25°C) three times with PBST and twice with double-distilled water in the dark. Finally, proteome and mini-protein microarrays were dried in a SlideWasher (CapitalBio, Beijing, China) at room temperature (25°C).
Data collection and analysis
Fluorescent signals were measured at 532 nm and 635 nm using a GenePix® Professional 4200A Microarray Scanner (Molecular Devices/Axon Instruments, Foster City, CA, USA). Signal intensities for IgG and IgM binding to proteome and mini-protein microarrays were measured and analyzed using GenePix Pro 6.0 Microarray Acquisition and Analysis software (Axon Instruments). Methods for protein microarray analyses were essentially as described previously by Cao et al.8 Briefly, samples producing high background signals were eliminated and the Limma package for R was used to normalize signal intensities. The F635 median fluorescence intensity represented IgM signal strength and the B532 median fluorescence intensity represented IgG signal strength. The ratios of F635 an B532 intensities between ATB and LTBI/HC samples were calculated as fold changes.
Evaluation of biomarkers for diagnosis of ATB
The diagnostic performance of each TB protein was examined by calculating the areas under the receiver operating characteristic (ROC) curves (AUCs) along with their 95% confidence intervals (CIs) as described by Cao et al.8 The cutoff level for each biomarker was determined by calculating the maximum Youden’s index.11 The methods for analysis of biomarker combinations, evaluation of models, and prediction of protein–protein association networks were also described previously by Cao et al.8 We selected 58 TB biomarkers as factors to construct the logistic regression model, which included serological data for 100 ATB patients, 60 individuals with LTBI and 44 HCs.
Statistical analysis
Hierarchical cluster analysis was performed using R statistical software (www.r-project.org) using log-transformed intensity values. Sensitivity and specificity were calculated as follows: sensitivity = number of true positives/(number of true positives + number of false negatives) and specificity = number of true negatives/(number of false positives +number of true negatives). Differences between groups were assessed using t-tests or one-way analysis of variance followed by Bonferroni correction using GraphPad Prism version 6.0 (Graphpad Software Inc., La Jolla, CA, USA). Values of P < 0.05 were considered statistically significant.
Results
Study participants
A total of 204 participants were enrolled in this study, including 100 patients with ATB (54 women and 46 men), 60 individuals with LTBI (all women) and 44 HCs (36 women and 8 men). The median ages for these three groups of participants were 45.4 ± 17 years (ATB), 42.1 ± 10.4 years (LTBI) and 46.4 ± 13.1 years (HC). Patients were diagnosed with ATB via the presence of acid-fast bacilli in sputum smears followed by positive sputum cultures for M. tuberculosis. All individuals with LTBI had positive results of T-SPOT.TB tests, whereas all HCs tested negative.
Biomarkers identified by serological screening of proteome microarrays
In the initial study, nine proteome microarrays were divided into three groups. Each proteome microarray was incubated with one of the following serum samples. For the ATB group (including sera from 16 TB patients), one serum sample was used for the first microarray, a pool of five serum samples was used for the second microarray, and a pool of ten serum samples was used for the third microarray. For the LTBI group (including sera from nine individuals), one serum sample, a pool of three serum samples, and a pool of five serum samples were used for each of the three proteome microarrays. For the HC group (including seven sera), one serum sample and two pools of three serum samples each were used for three proteome microarrays. The rationale for using a pool of three or more serum samples for each microarray was to maximize the probability of detecting signals.
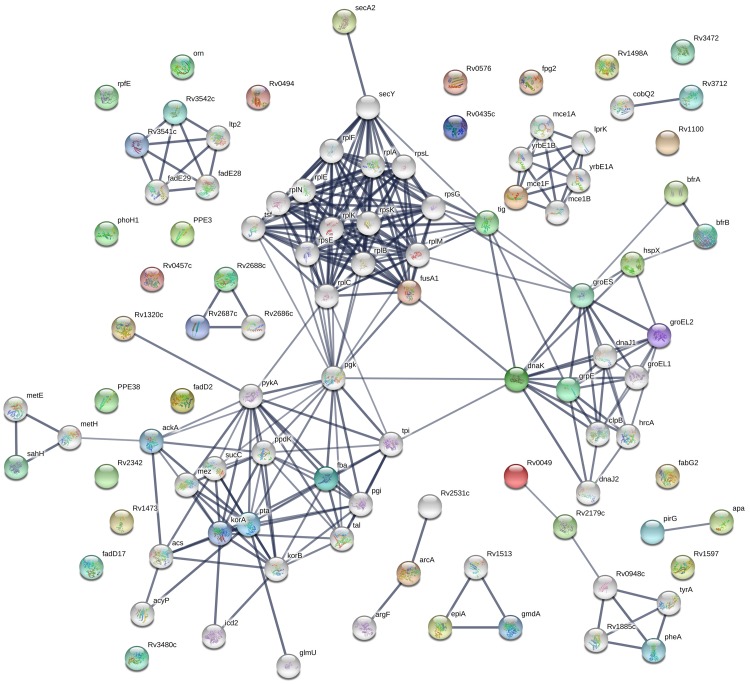
The average levels of 31 IgG and 27 IgM antibodies specific for 46 TB antigens were significantly higher in the ATB group than in the LTBI and HC groups (Table 1). These TB antigens were categorized into eight functional groups: intermediary metabolism and respiration (17, 29.31%), cell wall and cell processes (10, 17.24%), conserved hypothetical proteins (10, 17.24%), virulence, detoxification and adaptation (8, 13.79%), lipid metabolism (5, 8.62%), Pro-Pro-Glu (3, 5.17%), regulatory proteins (3, 5.17%), and information pathways (2, 3.45%). Predicted interactions of these TB antigens with other proteins were analyzed using the STRING 10.0 database (Figure 1).
Table 1.
Features of TB antigens identified as putative biomarkers by serological screening of proteome microarrays.
| Locus tag-Ab type | ATB/LTBI+HC | P value | Gene symbol | Gene product description |
|---|---|---|---|---|
| Cell wall and cell processes (10, 17.24%) | ||||
| MT3503-IgM | 1.024 | 0.0031 | Rv3395A | Probable membrane protein |
| Rv0435c-IgM | 1.040 | 0.0273 | Rv0435c | ATPase |
| Rv1473-IgM | 1.034 | 0.0421 | Rv1473 | Macrolide ABC transporter ATP-binding protein |
| Rv1821-IgG | 1.015 | 0.0097 | secA2 | Accessory Sec system translocase SecA2 |
| Rv1860-IgG | 1.144 | 0 | apa | Alanine and proline-rich secreted protein Apa |
| Rv1860-IgM | 1.110 | 0 | apa | Alanine and proline-rich secreted protein Apa |
| Rv2450c-IgM | 1.051 | 0.0284 | rpfE | Resuscitation-promoting factor RpfE |
| Rv2462c-IgG | 1.019 | 0.0024 | tig | Trigger factor |
| Rv2688c-IgM | 1.056 | 0.0081 | Rv2688c | Antibiotic ABC transporter ATP-binding protein |
| Rv3810-IgG | 1.045 | 0.0064 | pirG | Cell surface protein |
| Conserved hypothetical proteins (10, 17.24%) | ||||
| MT1547-IgG | 1.033 | 0.0075 | MT1547 | Hypothetical protein |
| Rv0049-IgG | 1.014 | 0.037 | Rv0049 | Hypothetical protein |
| Rv0049-IgM | 1.046 | 0 | Rv0049 | Hypothetical protein |
| Rv1100-IgG | 1.040 | 0.038 | Rv1100 | Hypothetical protein |
| Rv1100-IgM | 1.092 | 0 | Rv1100 | Hypothetical protein |
| Rv1597-IgM | 1.066 | 0.0012 | Rv1597 | Hypothetical protein |
| Rv2342-IgG | 1.029 | 0.0406 | Rv2342 | Hypothetical protein |
| Rv3472-IgG | 1.026 | 0.0241 | Rv3472 | Hypothetical protein |
| Rv3542c-IgG | 1.027 | 0.0004 | Rv3542c | Hypothetical protein |
| Rv3542c-IgM | 1.027 | 0.0292 | Rv3542c | Hypothetical protein |
| Information pathways (2, 3.45%) | ||||
| Rv0684-IgM | 1.005 | 0.0115 | fusA1 | Elongation factor G |
| Rv0944-IgM | 1.053 | 0.0218 | Rv0944 | Formamidopyrimidine-DNA glycosylase |
| Intermediary metabolism and respiration (17, 29.31%) | ||||
| MT1560.1-IgG | 1.040 | 0.0368 | MT1560.1 | Hypothetical protein |
| Rv0363c-IgG | 1.025 | 0.0002 | fba | Fructose-bisphosphate aldolase |
| Rv0408-IgM | 1.025 | 0.0072 | Pta | Phosphate acetyltransferase |
| Rv0457c-IgG | 1.019 | 0.0342 | Rv0457c | Peptidase |
| Rv1001-IgM | 1.049 | 0.0016 | arcA | Arginine deiminase |
| Rv1320c-IgG | 1.027 | 0.0204 | Rv1320c | Adenylate cyclase |
| Rv1876-IgG | 1.030 | 0.019 | bfrA | Bacterioferritin BfrA |
| Rv1876-IgM | 1.083 | 0 | bfrA | Bacterioferritin BfrA |
| Rv2179c-IgG | 1.007 | 0.0031 | Rv2179c | 3′-5′ exoribonuclease |
| Rv2368c-IgG | 1.028 | 0.0064 | phoH1 | Phosphate starvation-inducible protein PhoH |
| Rv2368c-IgM | 1.022 | 0.0033 | phoH1 | Phosphate starvation-inducible protein PhoH |
| Rv2511-IgG | 1.036 | 0.0007 | orn | Oligoribonuclease |
| Rv3248c-IgG | 1.046 | 0.0001 | sahH | Adenosylhomocysteinase |
| Rv3248c-IgM | 1.024 | 0.0092 | sahH | Adenosylhomocysteinase |
| Rv3712-IgG | 1.018 | 0.0083 | Rv3712 | Ligase |
| Rv3838c-IgG | 1.019 | 0.0414 | pheA | Prephnate dehydratase |
| Rv3841-IgG | 1.022 | 0.0121 | bfrB | Bacterioferritin BfrB |
| Lipid metabolism (5, 8.62%) | ||||
| Rv0270-IgM | 1.017 | 0.0096 | fadD2 | Fatty-acid-CoA ligase |
| Rv1350-IgG | 1.019 | 0.0168 | fabG2 | 3-oxoacyl-ACP reductase |
| Rv3480c-IgM | 1.049 | 0.0028 | Rv3480c | Diacyglycerol O-acyltransferase |
| Rv3506-IgG | 1.038 | 0.0001 | fadD17 | Long-chain-fatty-acid-CoA ligase FadD17 |
| Rv3506-IgM | 1.047 | 0.0436 | fadD17 | Long-chain-fatty-acid-CoA ligase FadD17 |
| PE/PPE (3, 5.17%) | ||||
| Rv0280-IgM | 1.012 | 0.0187 | PPE3 | PPE family protein PPE3 |
| Rv2352c-IgG | 1.026 | 0.012 | PPE38 | PPE family protein PPE38 |
| Rv2352c-IgM | 1.060 | 0.0002 | PPE38 | PPE family protein PPE38 |
| Regulatory proteins (3, 5.17%) | ||||
| Rv0494-IgG | 1.002 | 0.016 | Rv0494 | HTH-type transcriptional regulator |
| Rv0494-IgM | 1.035 | 0.0033 | Rv0494 | HTH-type transcriptional regulator |
| Rv0576-IgG | 1.019 | 0.039 | Rv0576 | Transcriptional regulator |
| Virulence, detoxification and adaptation (8, 13.79%) | ||||
| Rv0174-IgM | 1.027 | 0.0216 | mce1F | MCE-family protein Mce1F |
| Rv0350-IgG | 1.047 | 0.0462 | dnaK | Chaperone protein DnaK |
| Rv0350-IgM | 1.083 | 0.014 | dnaK | Chaperone protein DnaK |
| Rv0351-IgM | 0.995 | 0.048 | grpE | Stress response protein GrpE |
| Rv0440-IgG | 1.090 | 0.0014 | groEL2 | Molecular chaperone GroEL |
| Rv2031c-IgG | 1.023 | 0.0045 | hspX | Alpha-crystallin |
| Rv2031c-IgM | 1.029 | 0.0027 | hspX | Alpha-crystallin |
| Rv3418c-IgG | 1.019 | 0.0466 | groES | Ahaperonin GroES |
Ab, antibody; ABC: ATP-binding cassette; ACP: acyl-carrier protein; ATB, active TB; ATP: adenosine triphosphate; Co-A: coenzyme A; HC, healthy control; HTH, helix-turn-helix; IgG, immunoglobulin G; IgM, immunoglobulin M; LTBI: latent TB infection; MCE, mammalian cell entry; PE: Pro-Glu; PPE: Pro-Pro-Glu; and TB, tuberculosis.
Figure 1.
STRING analysis of 58 candidate biomarkers identified by serological screening of proteome microarrays containing 4,262 TB proteins. Interaction networks with confidence levels of 0.7 were visualized with edges representing protein-protein associations. Empty nodes, proteins of unknown 3D structure; filled nodes, some 3D structures known or predicted; colored nodes, query proteins and first shell of interactions; white nodes, second shell of interactions; and thicker lines, stronger protein–protein associations.
Biomarkers identified by serological screening of mini-protein microarrays
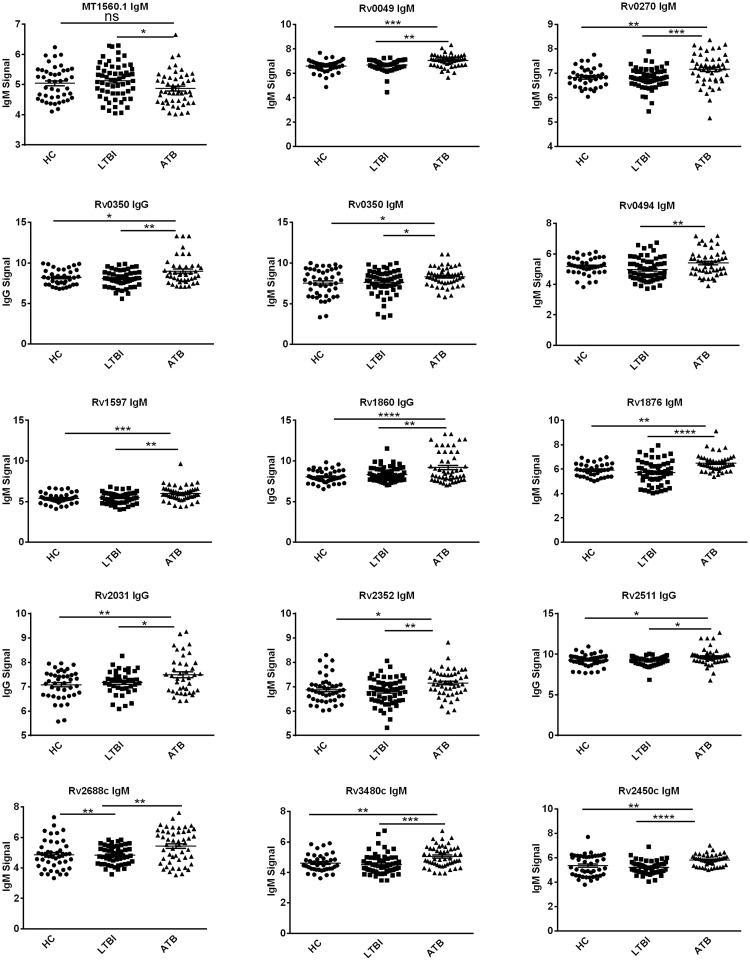
Based on the serum proteomic analysis, we selected 100 distinguishing TB proteins for custom fabrication of mini-protein microarrays. In the subsequent experiment, 204 serum samples were divided into three groups: ATB (100 sera), LTBI (60 sera), and HC (44 sera). All serum samples were incubated separately with mini-protein microarrays, which were processed in the same manner as proteome microarrays. The average signal levels of IgG or IgM specific for 14 TB proteins were higher in the ATB group than in the LTBI and HC groups (P < 0.001) (Figure 2 and Table 2).
Figure 2.
Scatter plots of 15 IgG and IgM signals for reactivity against 14 TB proteins in ATB patients, individuals with LTBI and HCs. Each point represents one serum sample and horizontal solid lines indicate the mean signal for each group of serum samples. *P < 0.05; **P < 0.01; ***P < 0.001; ATB, active TB; Control, healthy control; IgG, immunoglobulin G; IgM, immunoglobulin M; LTBI: latent TB infection; ns, not significant; and TB, tuberculosis.
Table 2.
Features of 15 TB antigens identified as putative biomarkers by serological screening of mini-protein microarrays.
| Locus tag-Ab type | ATB/LTBI+HC | P value | Gene symbol | Gene product description |
|---|---|---|---|---|
| MT1560.1-IgM | 1.040 | 0.0368 | MT1560.1 | Hypothetical protein |
| Rv0049-IgM | 1.014 | 0.037 | Rv0049 | Hypothetical protein |
| Rv0270-IgM | 1.017 | 0.0096 | fadD2 | Fatty-acid-CoA ligase FadD2 |
| Rv0350-IgG | 1.047 | 0.0462 | dnaK | Chaperone protein DnaK (HSP70) |
| Rv0350-IgM | 1.083 | 0.014 | dnaK | Chaperone protein DnaK (HSP70) |
| Rv0494-IgM | 1.002 | 0.016 | Rv0494 | HTH-type transcriptional regulator |
| Rv1597-IgM | 1.066 | 0.0012 | Rv1597 | Hypothetical protein |
| Rv1860-IgG | 1.144 | 0 | apa | Alanine and proline rich secreted protein Apa |
| Rv1876-IgM | 1.083 | 0 | bfrA | Bacterioferritin BfrA |
| Rv2031c-IgG | 1.023 | 0.0045 | hspX | Heat shock protein HspX |
| Rv2352c-IgM | 1.060 | 0.0002 | PPE38 | PPE family protein PPE38 |
| Rv2450c-IgM | 1.051 | 0.0284 | rpfE | Resuscitation-promoting factor RpfE |
| Rv2511-IgG | 1.036 | 0.0007 | orn | Oligoribonuclease Orn |
| Rv2688c-IgM | 1.056 | 0.0081 | Rv2688c | Antibiotic ABC transporter ATP-binding protein |
| Rv3480c-IgM | 1.049 | 0.0028 | Rv3480c | Diacylglycerol O-acyltransferase |
Ab, antibody; ABC, ATP-binding cassette; ATB, active TB; ATP, adenosine triphosphate; Co-A: coenzyme A; HC, healthy control; HSP, heat shock protein; IgG, immunoglobulin G; IgM, immunoglobulin M; LTBI: latent TB infection; PPE, Pro-Pro-Glu; and TB, tuberculosis.
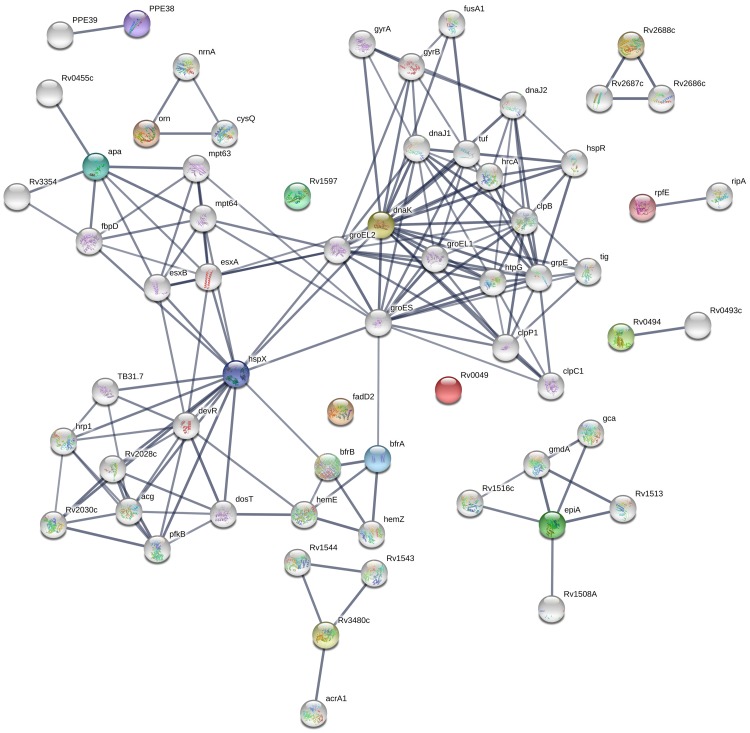
These candidate biomarkers included the chaperone protein DnaK (HSP70), the ATP-binding cassette transporter, and a helix-turn-helix-type transcriptional regulator. Biomarker gene symbols and descriptions are listed in Table 2. Predicted interactions of these TB antigens with other proteins were also analyzed using the STRING 10.0 database (Figure 3). Rv0350 (chaperone protein DnaK), Rv2031 (alpha-crystallin), Rv1860 (alanine and proline-rich secreted protein Apa), and Rv1876 (bacterioferritin BfrA) exhibited strong associations with the highest confidence (combined score > 0.7) (Figure 3), implying that multiple TB antigens may participate collaboratively in pathogen-host interactions.
Figure 3.
STRING analysis of 15 candidate biomarkers identified by serological screening of mini-protein microarrays containing 100 selected TB proteins. Interaction networks with confidence levels of 0.7 were visualized with edges representing protein–protein associations. Empty nodes, proteins of unknown 3D structure; filled nodes, some 3D structures known or predicted; colored nodes, query proteins and first shell of interactions; white nodes, second shell of interactions; and thicker lines, stronger protein-protein associations.
Diagnostic performance of 15 TB biomarkers
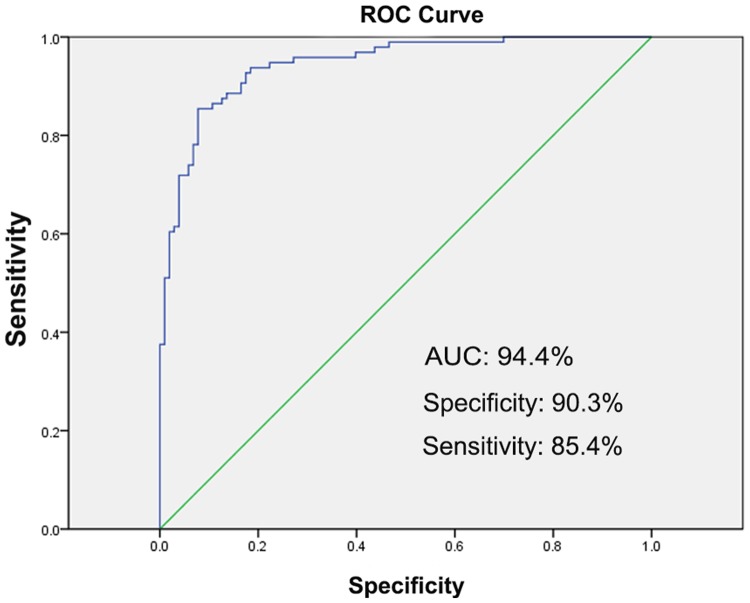
The diagnostic performance of 15 TB biomarkers in discriminating individuals with LTBI and ATB was evaluated using ROC curves. We calculated AUCs, specificities, and sensitivities for each of the 15 TB biomarkers. MT1560.1-IgM, Rv0049-IgM, Rv0270-IgM, Rv0350-IgG, Rv0350-IgM, Rv0494-IgM, Rv1597-IgM, Rv1860-IgG, Rv1876-IgM, Rv2031c-IgG, Rv2352c-IgM, Rv2450c-IgM, Rv2511-IgG, Rv2688c-IgM and Rv3480c-IgM had high AUCs (P < 0.001) (Table 3). These biomarkers could discriminate the antibody responses of individuals with ATB and LTBI with sensitivities ranging from 16.7% (HspX-IgG) to 85.4% (Rv3480c-IgM) and specificities ranging from 36.9% (Rv3480c-IgM) to 99.0% (Apa-IgG) (Table 3). To further assess the diagnostic performance of these 15 TB biomarkers, we combined them using the threshold values determined by maximizing the AUC values. As showed in Figure 4, the sensitivity and specificity of this biomarker combination in discriminating individuals with ATB and LTBI increased to 85.4% and 90.3%, respectively.
Table 3.
Diagnostic parameters of 15 serum biomarkers for discriminating ATB from LTBI.
| Locus tag-Ab type | Genesymbol | AUC (95% CI) | P value | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| MT1560.1-IgG | epiA | 0.62 (0.542–0.698) | <0.001 | 40.6 | 82.5 |
| Rv0049-IgM | Rv0049 | 0.673 (0.598–0.749) | <0.001 | 52.1 | 80.6 |
| Rv0270-IgM | fadD2 | 0.582 (0.502–0.662) | <0.001 | 43.8 | 74.8 |
| Rv0350-IgG | DnaK | 0.56 (0.48–0.641) | <0.001 | 60.4 | 53.4 |
| Rv0350-IgM | DnaK | 0.607 (0.529–0.685) | <0.001 | 77.1 | 44.7 |
| Rv0494-IgM | Rv0494 | 0.566 (0.486–0.646) | <0.001 | 74.0 | 40.8 |
| Rv1597-IgM | Rv1597 | 0.635 (0.558–0.713) | <0.001 | 83.3 | 43.7 |
| Rv1860-IgG | apa | 0.665 (0.588–0.742) | <0.001 | 36.5 | 99.0 |
| Rv1876-IgM | BfrA | 0.64 (0.563–0.717) | <0.001 | 61.5 | 63.1 |
| Rv2031c-IgG | HspX | 0.521 (0.439–0.602) | <0.001 | 16.7 | 94.2 |
| Rv2352c-IgM | PPE38 | 0.729 (0.658–0.799) | <0.001 | 66.7 | 72.8 |
| Rv2450c-IgM | RpfE | 0.61 (0.532–0.689) | <0.001 | 81.2 | 42.7 |
| Rv2511-IgG | Orn | 0.646 (0.569–0.722) | <0.001 | 63.5 | 65.0 |
| Rv2688c-IgM | Rv2688c | 0.577 (0.496–0.658) | <0.001 | 53.1 | 67.0 |
| Rv3480c-IgM | Rv3480c | 0.614 (0.536–0.692) | <0.001 | 85.4 | 36.9 |
95% CI, 95% confidence interval; Ab, antibody; ATB, active TB; AUC, area under the ROC curve; IgG, immunoglobulin G; IgM, immunoglobulin M; LTBI: latent TB infection; ROC, receiver operating characteristic; and TB, tuberculosis.
Figure 4.
ROC curve of the combination of 15 TB antigens as biomarkers for distinguishing individuals with LTBI and ATB. The blue line represents the logistic regression model including the 15 TB antigens, and the green line represents the reference. AUC, area under the ROC curve; ROC, receiver operating characteristic.
Discussion
We found that levels of 58 specific antibodies were higher in ATB patients than in individuals with LTBI and HCs, suggesting that some of them could potentially be used as new serum biomarkers for discriminating ATB from LTBI. In addition, some of these TB antigens were predicted to have strong interactions with one another and to be involved in various biological processes (Table 1, Figure 1). For instance, GroES was predicted to interact with four proteins (DnaK, GroEL2, GrpE, and HspX) in the same functional group (virulence, detoxification and adaption) as well as with proteins in other functional groups (BrfA, intermediary metabolism and respiration, and Tig, cell wall and cell processes). Through its interactions with Tig and DnaK, GroES could indirectly interact with many other predicted functional partners such as FusA1 (elongation factor G), Pgk (phosphoglycerate kinase), RplC (50S ribosomal protein L3), RplM (50S ribosomal protein L13), RpsG (30S ribosomal protein S7), Tpi (triosephosphate isomerase), SecY (protein translocase subunit) and Tsf (elongation factor EF-Ts) (Figure 1).
In a previous study using the same proteome microarrays containing 4,262 TB proteins, Cao et al. showed that levels of 152 TB antigen-specific IgGs were elevated in patients with ATB compared with individuals with LTBI.8 The reasons for Cao et al. detecting 152 ATB-associated antigens recognized by IgGs while we detected only 31 may relate to the 7- to 13-fold larger sample size of their study (112 ATB, 113 LTBI and 94 HC compared with 16 ATB, 9 LTBI and 7 HC individuals) as well as inter-individual heterogeneity in anti-TB antibody responses.12 Nevertheless, levels of IgGs specific for five TB antigens (Rv0440, Rv0494, Rv2031c, Rv2342 and Rv3418c) were found in both studies to be consistently elevated in ATB patients compared with individuals with LTBI. One of these five TB antigens is a hypothetical protein (Rv2342) and another is a starvation-inducible, lipid-responsive transcriptional regulator (Rv0494).13 The other three antigens are associated with virulence and detoxification (Rv0440, molecular chaperone GroEL; Rv2031c, alpha-crystallin; and Rv3418c, chaperonin GroES), and were previously observed to be consistently over-expressed by TB isolates resistant to amikacin and kanamycin.14 Rv2031c (alpha-crystallin/HspX) was identified as a latent stage-specific antigen15 that could induce a stronger antibody response in patients with pulmonary TB than in individuals with LTBI.16 Rv2031c serves as a master regulator of the HspX operon, modulating the metabolism of Mtb inside host cells and contributing to survival during latency.17 These five TB antigens are good candidates for new TB vaccines because they can induce strong IgG responses in TB patients. Not surprisingly, a DNA vaccine encoding HspX (Rv2031c) was already evaluated for its immunogenicity in mice and its protective efficacy in a guinea pig model in 2005 and 2007.18,19
In the current study, we measured levels of both IgGs and IgMs and detected higher levels of 27 IgM specific antibodies against TB antigens (12 of which were also recognized by IgGs). By contrast, Cao et al only measured levels of IgGs.8 Measuring levels of both IgGs and IgMs should provide more potential serum biomarkers to discriminate ATB from LTBI, since IgG is the most abundant antibody isotype found in all body fluids but IgM is the first antibody to appear in blood and lymph fluid during the response to infection. Consequently, the 12 TB antigens identified in this study (Rv0049, Rv0350, Rv0494, Rv1100, Rv1860, Rv1876, Rv2301c, Rv2352c, Rv2368c, Rv3542, Rv3506c, and Rv3542c) should be further investigated for their roles in inducing both IgG and IgM responses during disease progression from LTBI to ATB, as well as for their potential as novel TB vaccine candidates. Two of these TB antigens (Rv1860 and Rv2031c) were previously found to be associated with ATB.9 In addition, Rv1860 (alanine and proline rich secreted protein Apa) was also found to stimulate interferon gamma-secreting CD4+ and CD8+ T cells,20 to have high specificity and acceptable sensitivity in identifying individuals with LTBI,21 and to induce strong interferon gamma responses in intranasally BCG-vaccinated mice.22
Serological evaluation of mini-protein microarrays showed that average levels of 15 antibodies (11 IgMs and 4 IgGs) specific for 14 TB antigens were higher in ATB patients than in individuals with LTBI and HCs (Table 2). These biomarkers were able to discriminate the antibody responses of individuals with ATB and LTBI with variable sensitivities and specificities. However, using the combination of all 15 biomarkers, the diagnostic performance for discrimination of ATB and LTBI was increased to sensitivity 85.4%, specificity 90.3% and AUC 94.4% (Figure 4). It is worth noting that 11 of 15 biomarkers were recognized by TB-specific IgMs, indicating the importance of detecting IgM responses for diagnosis of ATB. Nevertheless, additional studies will be needed to verify the ability of these 15 biomarkers to discriminate ATB from LTBI using other techniques such as ELISA, western blotting and parallel reaction monitoring methods. Because some of these 15 biomarkers are housekeeping proteins, it will be necessary to assess their cross-reactivity and to eliminate potential false-positive results using sera from patients suffering from other respiratory diseases such as asthma, chronic obstructive pulmonary disease, and pneumonia.
Conclusion
Our serum proteomic study indicated that a combination of 15 TB biomarkers offered improved diagnostic performance for discriminating the antibody responses of individuals with ATB and LTBI. Five new biomarkers (MT1560.1, Rv0049, Rv0270, Rv1597 and Rv3480c) were found to be associated with ATB and could induce stronger IgM antibody responses in these patients.
Acknowledgements
We thank clinical and laboratory staff and graduate students from the Tuberculosis Division of Respiratory and Critical Care Medicine, Affiliated Hospital of Zunyi Medical University, for their assistance in collecting and processing serum samples from patients and healthy controls.
Declaration of conflicting interest
ZP and LC declare no conflicts of interest. HZ is employed by and has shares in Z-BioMed, Inc., which is involved in infectious disease research.
Funding
This study was supported by the National Natural Science Foundation of China (81760003), the Key Project of Chinese National Programs (2013ZX10003007-001-003), and the 2nd 2011 Collaborative Innovation Center for TB Prevention and Cure in Guizhou Province. The sponsors played no role in the collection, analysis, and interpretation of data; in writing the manuscript; or in the decision to submit the article for publication.
ORCID iD
Hong Zhang https://orcid.org/0000-0002-3930-6731
References
- 1.World Health Organization. Global tuberculosis report 2019. Available: http://www.who.int/tb/publications/global_report/en/. Accessed 28 November 2019.
- 2.Walzl G, McNerney R, du Plessis N, et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis 2018; 18: e199–e210. doi: 10.1016/S1473-3099(18)30111-7. [DOI] [PubMed] [Google Scholar]
- 3.LoBue PA, Mermin JH. Latent tuberculosis infection: the final frontier of tuberculosis elimination in the USA. Lancet Infect Dis 2017; 17: e327–e333. doi: 10.1016/S1473-3099(17)30248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pai M, Behr M. Latent Mycobacterium tuberculosis infection and interferon-gamma release assays. Microbiol Spectr 2016; 4: 1–10. doi: 10.1128/microbiolspec.TBTB2-0023-2016. [DOI] [PubMed] [Google Scholar]
- 5.Kumar G, Dagur PK, Singh PK, et al. Serodiagnostic efficacy of Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex, ESAT-6, and CFP-10 in patients with active tuberculosis. Arch Immunol Ther Exp (Warsz) 2010; 58: 57–65. doi: 10.1007/s00005-009-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takwoingi Y, Whitworth H, Rees-Roberts M, et al. Interferon gamma release assays for Diagnostic Evaluation of Active tuberculosis (IDEA): test accuracy study and economic evaluation. Health Technol Assess 2019; 23: 1–152. doi: 10.3310/hta23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunnath-Velayudhan S, Salamon H, Wang HY, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci USA 2010; 107: 14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao SH, Chen YQ, Sun Y, et al. Screening of serum biomarkers for distinguishing between latent and active tuberculosis using proteome microarray. Biomed Environ Sci 2018; 31: 515–526. doi: 10.3967/bes2018.069. [DOI] [PubMed] [Google Scholar]
- 9.Kunnath-Velayudhan S, Davidow AL, Wang HY, et al. Proteome-scale antibody responses and outcome of Mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J Infect Dis 2012; 206: 697–705. doi: 10.1093/infdis/jis421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Cao S, Liu Y, et al. Potential role for Rv2026c- and Rv2421c-specific antibody responses in diagnosing active tuberculosis. Clin Chim Acta 2018; 487: 369–376. doi:10.1016/j.cca.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Unal I. Defining an optimal cut-point value in ROC analysis: an alternative approach. Comput Math Methods Med 2017; 2017: 3762651. doi: 10.1155/2017/3762651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyashchenko K, Colangeli R, Houde M, et al. Heterogeneous antibody responses in tuberculosis. Infect Immun 1998; 66: 3936–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousuf S, Angara R, Vindal V, et al. Rv0494 is a starvation-inducible, auto-regulatory FadR-like regulator from Mycobacterium tuberculosis. Microbiology 2015; 161: 463–476. doi: 10.1099/mic.0.000017. [DOI] [PubMed] [Google Scholar]
- 14.Kumar B, Sharma D, Sharma P, et al. Proteomic analysis of Mycobacterium tuberculosis isolates resistant to kanamycin and amikacin. J Proteomics 2013; 94: 68–77. doi: 10.1016/j.jprot.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Demissie A, Leyten EM, Abebe M, et al. Recognition of stage-specific mycobacterial antigens differentiates between acute and latent infections with Mycobacterium tuberculosis. Clin Vaccine Immunol 2006; 13: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozumi H, Tsujimura K, Yamamura Y, et al. Immunogenicity of dormancy-related antigens in individuals infected with Mycobacterium tuberculosis in Japan. Int J Tuberc Lung Dis 2013; 17: 818–824. doi: 10.5588/ijtld.12.0695. [DOI] [PubMed] [Google Scholar]
- 17.Mushtaq K, Sheikh JA, Amir M, et al. Rv2031c of Mycobacterium tuberculosis: a master regulator of Rv2028-Rv2031 (HspX) operon. Front Microbiol 2015; 6: 351. doi: 10.3389/fmicb.2015.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khera A, Singh R, Shakila H, et al. Elicitation of efficient, protective immune responses by using DNA vaccines against tuberculosis. Vaccine 2005; 23: 5655–5665. [DOI] [PubMed] [Google Scholar]
- 19.Roupie V, Romano M, Zhang L, et al. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect Immun 2007; 75: 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar P, Amara RR, Challu VK, et al. The Apa protein of Mycobacterium tuberculosis stimulates gamma interferon-secreting CD4+ and CD8+ T cells from purified protein derivative-positive individuals and affords protection in a guinea pig model. Infect Immun 2003; 71: 1929–1937. doi:10.1128/IAI.71.4.1929-1937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathakumari B, Prabhavathi M, Anbarasu D, et al. Dynamic IgG antibody response to immunodominant antigens of M. tuberculosis for active TB diagnosis in high endemic settings. Clin Chim Acta 2016; 461: 25–33. doi: 10.1016/j.cca.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 22.Sable SB, Cheruvu M, Nandakumar S, et al. Cellular immune responses to nine Mycobacterium tuberculosis vaccine candidates following intranasal vaccination. PLoS One 2011; 6: e22718. doi: 10.1371/journal.pone.0022718. [DOI] [PMC free article] [PubMed] [Google Scholar]