Abstract
We present a testable hypothesis related to an origin of life on land in which fluctuating volcanic hot spring pools play a central role. The hypothesis is based on experimental evidence that lipid-encapsulated polymers can be synthesized by cycles of hydration and dehydration to form protocells. Drawing on metaphors from the bootstrapping of a simple computer operating system, we show how protocells cycling through wet, dry, and moist phases will subject polymers to combinatorial selection and draw structural and catalytic functions out of initially random sequences, including structural stabilization, pore formation, and primitive metabolic activity. We propose that protocells aggregating into a hydrogel in the intermediate moist phase of wet-dry cycles represent a primitive progenote system. Progenote populations can undergo selection and distribution, construct niches in new environments, and enable a sharing network effect that can collectively evolve them into the first microbial communities. Laboratory and field experiments testing the first steps of the scenario are summarized. The scenario is then placed in a geological setting on the early Earth to suggest a plausible pathway from life's origin in chemically optimal freshwater hot spring pools to the emergence of microbial communities tolerant to more extreme conditions in dilute lakes and salty conditions in marine environments. A continuity is observed for biogenesis beginning with simple protocell aggregates, through the transitional form of the progenote, to robust microbial mats that leave the fossil imprints of stromatolites so representative in the rock record. A roadmap to future testing of the hypothesis is presented. We compare the oceanic vent with land-based pool scenarios for an origin of life and explore their implications for subsequent evolution to multicellular life such as plants. We conclude by utilizing the hypothesis to posit where life might also have emerged in habitats such as Mars or Saturn's icy moon Enceladus.
“To postulate one fortuitously catalyzed reaction, perhaps catalyzed by a metal ion, might be reasonable, but to postulate a suite of them is to appeal to magic.”
—Leslie Orgel
Keywords: Origin of life, Prebiotic chemistry, Hydrothermal systems, Protocells, Progenotes, Microbial communities
1. Introduction
One of the authors knew Leslie Orgel quite well, and the clarity of his thinking has guided our research in many ways. It would be an enjoyable exercise to debate the point Leslie made in the quote above (Orgel, 2000). Sadly, this is not possible, but if it were, we would remind him of our conversations in which he proposed that using methods of combinatorial chemistry would be an efficient way for origins of life research to advance, to which we would heartily agree! However, we would also state that it is a way for self-assembled chemical systems to advance in a cyclic process toward the complexity required for life to begin. A primary point to be made in this review is that instead of single reactions in individual test tubes, we are proposing a novel hypothesis that life began through a combinatorial selection among vast numbers of microscopic test tubes, each containing a varying composition of potential reactants and products. An iterative process drawing from interacting sets of these variations would in fact be able to constitute a suite of catalytic reactions through selection, driven solely by physical and chemical forces without the need to appeal to magic.
Here we analyze one such scenario from this perspective, which goes beyond simple chemical reactions approaching equilibrium to a more complex, systems-chemistry approach (Dyson, 1999; Pross, 2012; Damer and Deamer, 2015). A computer science metaphor of the bootstrapping of a chemical operating system is used. The scenario is further advanced by grounding it in a prebiotically plausible geological landscape of subaerial hydrothermal fields associated with volcanic land masses (Deamer et al., 2006; Damer, 2016). We review experimental testing of the hypothesis in the laboratory and in the field, and then integrate the evidence with computational models and the geological conditions supporting the microbial populations that produced stromatolites, the oldest and most ubiquitous fossil evidence for life on Earth. We then propose a roadmap of avenues to test the hypothesis, consider the implications of a hot spring origin versus the alternative oceanic hydrothermal vent scenario for the evolution of complex life on Earth, and conclude by offering some new insights to the question of how life might arise on other worlds.
2. Section I
2.1. Considering life's origin through the metaphors of computer science
The first question we address concerns a mechanism by which selection and evolution can begin. In his book Life: Its Nature, Origin and Development, Oparin (1962) asked whether life could in fact be understood as a mechanism. He even considered cybernetics, which is pertinent to the thought experiment we describe next. Oparin decided that the properties of life went far beyond mechanical explanations:
“Of course, we may, and should try to understand the physical and chemical basis of the various vital phenomena by means of the construction and study of models which will reproduce the same phenomena as occur in organisms but in a simplified form. In doing so, however, we must always remember that we are dealing with models and not confuse them with living things.”
Keeping Oparin's caveat in mind, we note that computational approaches have gone far beyond the primitive cybernetics he envisaged. Computer-based software tools and metaphors have infiltrated all branches of the scientific enterprise. From the perspective of computer science, the intracellular processes of life have been aptly compared with the functioning of a computer operating system (Bray, 2011; Pang and Maslov, 2013). It is an interesting and perhaps enlightening exercise to compare in silico programs to the operating system of life. Rather than patterns of electrons flowing in silicon circuits, the programs of the cell are embedded in monomer sequences of biopolymers. Base sequences in DNA are linear data stores, analogous to a read-only memory, which are transcribed into base sequences in messenger RNA, then loaded into ribosomes, which generate application programs fashioned out of proteins. Like an operating system's message queues and protocols, a finely tuned chemical network of signaling and feedback enables a cell to execute specific protein programs in a highly regulated manner.
This comparison brings us to the question we are addressing here: Can a chemical computer emerge spontaneously on a sterile yet habitable planet such as the early Earth? The answer is obviously yes, because life did begin, but the process by which this occurred remains a fundamental problem in biology. We argue that by analogy to the development of computers, the hardware of the first forms of life is represented by organic molecules that become self-organized into supramolecular structures (Lehn, 2002) capable of capturing free energy available in the environment and used it to drive polymerization and growth. The programs of life spontaneously developed when initially random systems of polymeric molecules underwent cycles of selection and amplification to express functions within the system of self-assembling hardware.
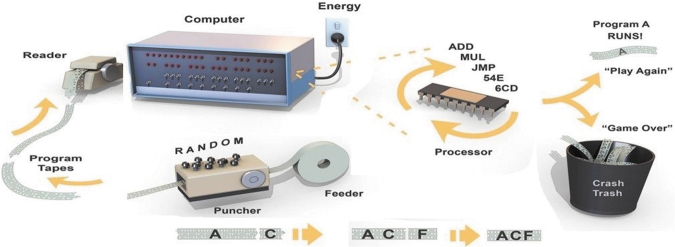
However, how it is possible for programs to emerge in the absence of a programmer? Figure 1 illustrates how this can work by referring to early computers that used holes punched in a paper tape as a way to code instructions using binary bits. The goal is to spontaneously generate a program that will turn on a series of light-emitting diodes (LEDs) in a front panel attached to the computer. At the outset, the tape puncher is linked to a random number generator. The punched tapes are then read one at a time into a simple computer that uses electrical energy to read each tape and then execute the purely random instructions through a primitive processor. Most programs “crash” in that they switch on no LEDs and so are discarded. However, if by chance a rare random program (such as “A” in the example) happens to light up at least one LED, it is then automatically selected and fed back into the puncher. The puncher stores the program “A” and punches it out multiple times, adding new random instructions “B,” “C,” and “D” onto extended sections of tape. The next cycle runs the new programs A-B, A-C, A-D; and A-C is similarly selected to go forward into a new generation of punched programs. Over time a further extended program (A-C-F) will emerge, which operates an even larger number of LEDs on the front panel. This method of programming is inefficient, but given enough time it will create a program that switches on all of the LEDs.
FIG. 1.
Metaphor for how a computer program can be developed without a programmer.
To summarize the four obvious steps, (1) a source of energy (an electrical current) drives a process that punches random holes in a tape, feeds the tape into our simple computer, and operates the computer's CPU. At some point, one set of holes happens to cause an LED to light up and that set is (2) selected, (3) replicated, and (4) extended, and functional instructions are thereby drawn out of a population of randomly punched tapes. Another chance sequence of holes in an elongated tape causes a second LED to light up, and so on, until all of the LEDs are operating. A fifth often overlooked step is essential, which is a feedback loop between the LED status and the punching apparatus, a testing subroutine that instructs the system to save, incorporate, and amplify any chance program that happens to turn on an LED. This simple test could be called combinatorial selection and can enable programming without the need for a programmer, suggesting how similar steps driving combinatorial chemistry could lead to an origin of life. In a chemical system, the energy sources are hot water and sunlight, the paper tapes are polymers such as nucleic acids or proteins, and the holes are chance patterns in monomer sequences. The programs are collections of interacting polymers that undergo selection and amplification if they happen to have functions that increase survivability of those same polymers over time. The collections are contained within membrane-bounded compartments and referred to as protocells.
Using this metaphor of self-writing computer code allows a reframing of the origin of life as a testable hypothesis that incorporates the biophysics of self-assembly, the chemistry of polymerization, and the geophysical properties associated with volcanic land masses on the early Earth. Although this reframing provides a new perspective for life's origin, it also raises many questions. First, we must define an operating protocell. The sole measure of operability of a microbial cell is that encapsulated systems of catalytic polymers use nutrients and energy to undergo growth by polymerization, supporting an eventual division into viable daughter cells. Akin to the punched paper tapes, the system we propose generates immense numbers of microscopic protocells, each different in composition from all the rest, and these protocell populations must then undergo a primitive version of Darwinian selection we have termed combinatorial selection, at the molecular and supramolecular level.
Here is the set of questions we explore in this review:
How can the earliest protocells composed of primitive, self-assembled membranes encapsulate cargoes of random polymers?
How can functional chemical “programs” spontaneously emerge by selection and begin to operate?
What environmental stresses provide selective factors and how can polymer programs be extended so that protocells can survive these factors and propagate sets of polymers into subsequent generations?
Are there prebiotically plausible natural environments in which protocells can emerge and then undergo evolution through a primitive form of selection before the emergence of life?
How can such populations of primitive protocells eventually evolve into the self-sustaining and self-reproducing forms that comprise living systems?
We first address the question of a plausible environment that is conducive to the emergence of protocells. Darwin (1871) provided a clue in an insightful sentence written in a letter to his friend J.D. Hooker:
“But if (& oh what a big if) we could conceive in some warm little pond with all sorts of ammonia & phosphoric salts,—light, heat, electricity & etc. present, that a protein compound was chemically formed, ready to undergo still more complex changes [..]”
Most readers are probably familiar with this quote, but we reiterate it here because Darwin's intuition of a warm little pond makes perfect sense. In contrast to the dilution that inevitably would occur in a global salty ocean, potential organic reactants can accumulate and be concentrated sufficiently for reactions to occur in small bodies of fresh water on volcanic land masses emerging from the ocean (Pearce et al., 2017; Ranjan et al., 2019). Darwin also proposed that a “protein compound” must first form, “ready to undergo still more complex changes.” Although he did not know that proteins were linear chains composed of linked amino acids, Darwin intuitively understood that proteins were important, and that they could somehow be synthesized by an unspecified source of energy. Darwin also hinted that the proteins must be able to undergo evolutionary cycles generating more complex changes, which today is described as an away-from-equilibrium chemical system.
This brings us to a conundrum known as the “water problem” (Benner et al., 2012). Proteins and the other key polymers of life, RNA and DNA, must function in an aqueous medium, but in the absence of continuous synthesis, they undergo hydrolytic decomposition at varying rates depending on temperature and pH. In life today, polymers are continuously synthesized and repaired by enzyme-catalyzed condensation reactions driven by adenosine triphosphate (ATP) as an energy source. However, there were no enzymes or ATP available on the prebiotic Earth, so how could the first polymers form? It has long been understood that condensation reactions can occur if monomers undergo dehydration at elevated temperatures (Fox and Harada, 1958; Lohrmann and Orgel, 1971; Lahav et al., 1978). However, a single cycle might lead to a few oligomers, but these would begin to undergo hydrolysis upon rehydration. We propose that not just a single cycle but instead continuous wet-dry cycles would allow polymers to exist in a steady state and grow more complex as solutes in a prebiotic warm little pond (Hargrave et al., 2018). As long as the rate of synthesis exceeds the rate of hydrolysis, multiple cycles operating in conjunction with a kinetic trap will inevitably produce an accumulation of polymers (Higgs, 2016). In other words, a kinetic trap emerges in a reaction or a series of linked reactions if the rate of synthesis exceeds the rate of a back reaction (Ross and Deamer, 2016). All life on Earth incorporates a kinetic trap because the rate of synthesis of biopolymers by condensation reactions exceeds the rate of their hydrolysis.
One possible site for the emergence of a kinetic trap would be mineral surfaces where fluctuating water levels cause cycles of wetting and drying that localize concentrated, dry solutes. Fast wet-dry cycles happening in seconds to minutes occur when geysers splash water on surrounding hot rocks, medium duration cycles measured in minutes to hours occur with the rise and fall of pool levels caused by regularly fluctuating hot springs, and slower cycles measured in hours to days would be associated with complete evaporation of smaller pools followed by refilling during precipitation.
Volcanoes emerging through a global ocean would be the original land masses on the Hadean Earth (Van Kranendonk, 2010; Bada and Korenaga, 2018), analogous to Hawaii and Iceland today. If we were to travel 4 billion years back in time to visit volcanic landscapes on the Hadean Earth (Fig. 2), we would see abundant hydrothermal fields with multiple hot spring systems replenished by precipitation evaporating from the surrounding ocean. The distilled fresh water would percolate into hot rocks and then circulate back to the surface as springs and geysers. Hydrothermal fields provide sources of heat and chemical energy to drive polymerization reactions in films of concentrated organic solutes that form on mineral surfaces during repeated cycles of wetting and drying (inset A). Inset B shows an example of such a pool on Mount Mutnovsky, an active volcano in Kamchatka, Russia (Kompanichenko et al., 2015). The recent discovery of a 3.5-billion-year-old hot spring setting on a volcanic plateau that formed in the absence of direct plate tectonic involvement is a good analog for an early Earth volcanic landscape emerging from a global ocean just 500 million years after the approximate time frame we are proposing for life's origins (Pearce et al., 2018; Van Kranendonk et al., 2020).
FIG. 2.
An artist's conception of a geyser-driven Hadean volcanic hot spring system in which cycles of evaporation and rehydration can occur. Inset A shows a ring of dried solutes on the mineral surfaces at the edge of a fluctuating pool. Inset B shows a boiling pool associated with a hot spring site on Mount Mutnovsky in Kamchatka, Russia. (Art credit Ryan Norkus; Photo credit Tony Hoffman.)
In summary, the primary polymers of life today—proteins and nucleic acids—are synthesized enzymatically by condensation reactions. The chemical energy of ATP is used to remove the equivalent of water molecules from the amine and carboxyl groups on amino acids to form peptide bonds, or the phosphate and ribose hydroxyl groups on nucleotides to form ester bonds. A certain degree of caution should be exercised when envisaging prebiotic versions of condensation reactions driving polymerization processes. For instance, Krishnamurthy (2018) wrote: “Coincidentally, it turns out that this ester-to-amide mechanism is part of extant biology, with more evolved molecules, as exemplified by (a) the peptidyl-transfer reaction within the confines of the ribosome and (b) the thioester-amide exchange equivalent in the nonribosomal peptide synthesis pathway. Whether this is a ‘preservation’ of a primordial reaction mechanism or a case of convergence due to chemical contingency is an open-ended debate.”
As noted earlier, heating dry amino acids at elevated temperatures was among the first and most obvious energy sources to be tested as a way to drive polymerization. Over ensuing years, dry heat was abandoned because the yields were minimal and products were too short to be able to fold into catalytically active structures such as ribozymes. Furthermore, the bonds formed were often not related to the kinds of bonds used in biology and there was a concern that intractable “tars and asphalts” were the most likely products (Benner et al., 2012). Therefore, researchers went on to explore condensing agents, activated mononucleotides, and mineral surfaces as a way to generate longer polymers.
We do not intend to exclude other versions of activated polymerization or possible involvement of condensing agents, but it seems appropriate to focus first on polymerization that does not require activated monomers or condensing agents. Wet-dry cycles in fresh water would be abundant in the prebiotic environment as long as volcanic land masses had emerged from the global ocean, and a surprising amount of chemical energy is introduced by concentrating potential reactants in dilute solutions as they evaporate to form dry films on mineral surfaces (Ross and Deamer, 2016). Furthermore, wet-dry cycles pump mixtures of reactants toward ever increasing complexity as products accumulate in kinetic traps. Such cycles have the additional advantage that if amphiphilic compounds are present in the mixture of reactants, encapsulation of polymers in membranous compartments occurs as a system goes through cycles of hydration and dehydration.
We conclude that hot springs and fluctuating freshwater pools are plausible candidates for prebiotic sites supporting the assembly of protocells, defined as encapsulated systems of random polymers forming in numerous pools over many millions of years. As described in the next section, protocell populations can be exposed to a variety of environmental stresses. Most protocells are likely to be unstable and their components will be dispersed, but a few will have properties that enhance their robustness. These will be selected in a process representing the first step in a pre-Darwinian version of evolution.
3. Section II
3.1. Comparative environments for an origin of life
Alternatives to membranous compartments have been proposed as settings for prebiotic chemistry. These include montmorillonite clay surfaces catalyzing polymerization of activated mononucleotides, compartments based on liquid/liquid phase separation such as coacervate droplets, droplets of hydrocarbon oils present as emulsions, and compartments composed of oligonucleotide origami or amphiphilic peptides. See Fishkis (2007) and Joyce and Szostak (2018) for reviews of these approaches. Szostak (2016) proposed an alternative to wet-dry cycling for the formation and replication of RNA polymers within membranous vesicles by heating and cooling in hot spring pools, possibly involving an ice-covered surface. Mayer et al. (2018) investigated phase transitions of super- and subcritical CO2 boundaries in deep tectonic fault zones as potential drivers of peptide synthesis and evolution. Perhaps most influential over the past 30 years has been the proposal that life began in seawater at hydrothermal vents (Corliss et al., 1981; Russell et al., 1993, 2014 Russell and Hall, 1997; Martin and Russell, 2003, 2007; Kelley et al., 2005; Lane and Martin, 2012; Barge et al., 2015, 2017). It was suggested that rock cavities (Baaske et al., 2007) or mineral gels (Westall et al., 2018) could concentrate solutes and serve as compartments to counteract the dispersive environment of vent fluid flow. In earlier publications, we and others have compared freshwater and seawater environments (Mulkidjanian et al., 2012; Deamer and Damer, 2017; Milshteyn et al., 2018) and comment further in this section.
3.2. Cycling freshwater pools as an optimal environment for the assembly and selection of protocells on an evolutionary pathway to life
The hypothesis we are presenting here has a number of advantages in common with other proposals, and also has some novel and possibly essential additional features. A key strength is the empirical foundation and weight of evidence that emerged over the past decade (see Deamer et al., 2019).
Amphiphilic compounds are present in carbonaceous meteorites and can assemble into membranous vesicles (Deamer, 1985; Deamer and Pashley, 1989; Naraoka et al., 1999; Deamer et al. 2002; Deamer and Dworkin, 2005). Amphiphiles are also synthesized by photochemical reactions (Dworkin et al., 2001) and are products of Fischer- Tropsch type reactions that may occur in volcanic geochemistry (McCollom et al., 1999; Rushdi and Simoneit, 2006). These results suggest that membranous compartments were available in the prebiotic environment and could accumulate in subaerial pools.
Recent investigations of a 3.5 billion-year-old, anoxic hot spring setting from the Pilbara Craton in Western Australia show that its hydrothermal veins and compositionally varied pools and springs had the capacity to mobilize and concentrate many of the elements required for prebiotic chemistry, including C, H, N, O, P, S, B, Zn, Mn, K, and others (Van Kranendonk et al., 2020).
As described earlier, biological polymers such as oligonucleotides and oligopeptides are products of condensation reactions and can be synthesized when their solutions are exposed to multiple wet-dry cycles in acidic fresh water (Rajamani et al., 2008; Toppozini et al., 2013; da Silva et al., 2014; DeGuzman et al., 2014; Forsythe et al., 2015; Rodriguez-Garcia et al., 2015; Himbert et al., 2016; Misuraca et al., 2017; Yu et al., 2017). Mineral surfaces and peptides may play roles such as nonenzymatic polymerization in early steps in life's origins and bear further investigation (Dalai and Sahai, 2018; Kaddour et al., 2018).
If lipids are present during a wet-dry cycle, polymers are encapsulated to form protocells (Deamer and Barchfeld, 1982; Apel et al., 2002).
Other forms of self-assembly may support key processes in prebiotic chemistry such as the emergence of homochirality or the purification of polymers (Budin and Szostak, 2010).
The results are not only confined to laboratory conditions but have also been demonstrated in hot spring analogs of the early Earth (Milshteyn et al., 2018; Deamer et al., 2019).
Pools on land have access to multiple potential sources of biologically relevant organic compounds. Three possibilities include the following: extraterrestrial infall in the form of meteorites, interplanetary dust particles, and impacting comets (Chyba and Sagan, 1992; Ehrenfreund and Sephton, 2006; Callahan et al., 2011); atmospheric synthesis of compounds such as formaldehyde and hydrogen cyanide that react to produce amino acids, nucleobases, and sugars (Parker et al., 2011; Bada and Korenaga, 2018; Wu and Sutherland, 2019); and geochemical synthesis of similar organic compounds as volcanic gases pass through hot minerals (Cody et al., 2000). The rates of meteoritic infall 4 billion years ago delivered a much larger fraction of extraterrestrial material than today, including substantial amounts of organic compounds (Pearce et al., 2017). Because oxygen was absent, organic compounds accumulated over longer time spans than they would in today's oxidizing atmosphere. Whatever the source, if the organic compounds happened to fall and be dispersed into the oceans, they would form a very dilute solution. In contrast, those same compounds falling on or synthesized at volcanic land masses exposed to the atmosphere would become concentrated on the mineral surfaces and then would be flushed into pools connected to hydrothermal systems, enabling their participation in prebiotically significant reactions.
We assume that subaerial hydrothermal fields on the early Earth would resemble similar volcanic sites today. Carbon dioxide and sulfur dioxide are major components of volcanic gases and are weak acids when dissolved in water. Therefore, the circulating water in hydrothermal fields is moderately acidic, with pH ranging from 2 to 5. The concentrations of common cations such as sodium, potassium, calcium, and magnesium are in the millimolar range, with accompanying anions of sulfate, chloride, and silicate (Deamer et al., 2019). The water is not stagnant, but instead circulates and undergoes multiple cycles of evaporation to a dry state in response to heat and low relative humidity, followed by rehydration from precipitation or periodic hot spring and geyser activity. The cycling frequency is as short as minutes when caused by geyser splashing, hours due to fluctuating levels of pools fed by pulses from hot springs, or days to weeks in cycles of evaporation followed by precipitation.
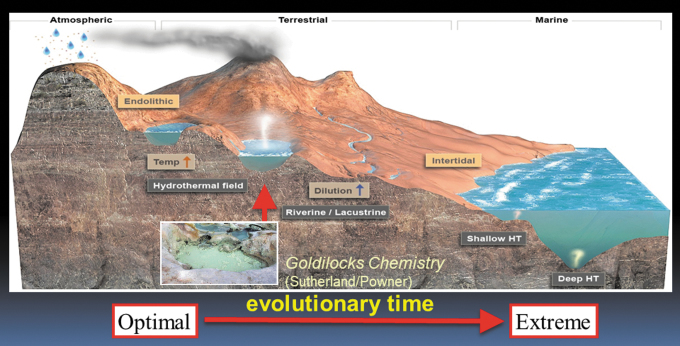
Figure 3 shows a local scale illustration of geological structures related to volcanism and associated hydrothermal fields. Salty seawater is distilled by evaporation and falls as precipitation on terrestrial volcanic land masses where it forms hydrothermal fields associated with hot springs, small pools, and geyser activity. The inset shows a small boiling pool at Bumpass Hell on Mt. Lassen, California, which is an example of dilute solutions that can undergo wet-dry cycles. Small, freshwater pools like this could provide the “Goldilocks” conditions for prebiotic chemistry suggested by Powner and Sutherland (2011). These conditions incorporate variable chemical compositions and pH, mesothermic temperatures (Bada and Lazcano, 2002), access to extraterrestrial and atmospheric exogenous delivery of organic compounds, and an ability to undergo wet-dry cycles that support condensation reactions and encapsulate polymers. A downward flow of water driven by gravity would carry populations of protocells and early living cells on a journey through evolutionary time into new pools with novel conditions. They would adapt to dilute rivers and lakes feeding into salty intertidal zones, the marine shoreline, and the open ocean. The varying pH and chemical composition of water bodies during this downward migration would select populations that are robust to dilution and salinity, and much later be able to tolerate the extreme conditions of submarine hydrothermal vents. This scenario is treated in greater detail in Section IV.
FIG. 3.
A local scale depiction of a volcanic land mass interacting with freshwater and saltwater conditions (inset: pool in Bumpass Hell on Mount Lassen California). Credit: Bruce Damer and Ryan Norkus.
3.3. Critique of an origin of life at submarine hydrothermal vents
For hydrothermal vents to become plausible sites conducive for life's origin, the hypothesis must be experimentally tested, or in Popper's words (1963), be capable of being falsified. One such test proposed for the vent scenario is that carbon dioxide can be reduced to simple organic solutes such as formic acid in a vent environment (Herschy et al., 2014; Sojo et al., 2019). The experimental evidence and thermodynamic models of the vent hypothesis have recently been challenged and these critiques should be addressed (Jackson, 2016, 2017; Wächtershäuser, 2016; Ross, 2018). One such model proposed by Lane and Martin (2012) suggested that a pH gradient would be present across mineral membranes of alkaline vents because the vent fluid is at a high pH compared with a lower acidic pH assumed for the early ocean. They posit that if a primitive version of an ATP synthase was embedded in the mineral membrane, the energy of the gradient could be used as a chemiosmotic energy source just as it is in life today. The membranes of bacteria, mitochondria, and chloroplasts can use the energy of a proton gradient equivalent to 3 pH units (a 1000-fold gradient), but only if the ATP synthase operates within a membrane that is 5 nm thick and relatively impermeable to proton flux. All the energy of the proton gradient is concentrated in that 5 nm thickness. However, the pH gradient of 3 pH units between alkaline vent fluid and an acidic ocean is proposed to lie across a mineral membrane three orders of magnitude thicker than a lipid bilayer, with dimensions of micrometers rather than nanometers. There would be almost no energy available to an ATP synthase embedded in such a thick membrane (Gogarten and Deamer, 2016). By analogy, a turbine and generator can produce electricity if it is in a waterfall 1 m thick and 100 m high, but if it is in a river three orders of magnitude longer (1 km) that falls through the same distance of 100 m, the energy available will hardly be sufficient to move the turbine blades.
Milshteyn et al. (2019) described an alternative source of chemiosmotic energy that can develop across lipid bilayer membranes 5 nm thick. If an electron acceptor (ferricyanide) is present inside lipid vesicles and an electron donor such as a reduced sulfur compound is outside, quinones present in the Murchison meteorite can pick up electrons and protons from the donor, transport them by diffusion across the bilayer, and deliver the electrons to the ferricyanide in the vesicles. The protons are released in the process, producing an acidic interior and substantial gradients over 2 pH units. An important goal for future research is to discover a process by which chemiosmotic energy can be used to synthesize the biochemical compounds necessary for primitive metabolism.
Hypotheses for an origin of life must also propose that a cell-sized compartment is able to maintain sufficient concentrations of reactants so that metabolic reactions can be initiated. Proponents of the vent hypothesis have suggested mineral gels as an alternative compartment for prebiotic chemistry (Westall et al., 2018). While interesting chemistry has been performed in hydrogels (Trevors et al., 2005; Gorrell et al., 2017), the products of reactions become trapped in the amorphous mineral matrix, whereas a gel composed of lipid vesicles can encapsulate distinct polymer sets that can interact with one another. Mineral cavities within vent rock formations have also been proposed to serve as compartments for primitive forms of life (Lane and Martin, 2012; Russell et al., 2014; Weiss et al., 2016). A recent report demonstrated that certain selected lipid mixtures could assemble into vesicles at 70C in the alkaline water of “white smoker” vents (Jordan et al., 2019). Monnard and Deamer (2002) also showed that specialized lipid mixtures could form stable membranes in seawater at ordinary temperatures and pH ranges. However, a plausible prebiotic source of such mixtures remains uncertain. As a general rule, the much higher concentrations of ionic solutes composing seawater inhibit self-assembly of membranous structures and encapsulation of polymers. This is not a problem for self-assembly of membranes in hot spring water supplied by precipitation with its much lower concentrations of ionic solutes (Milshteyn et al., 2018). Lastly, the water activity within a submerged mineral cavity, gel or vesicle will be at equilibrium with the surrounding ocean bulk. This presents a significant thermodynamic hurdle because in aqueous solutions condensation reactions leading to polymer synthesis would require chemically activated monomers such as the nucleoside triphosphates that drive biological metabolism or the imidazole esters of mononucleotides used in the laboratory (Inoue and Orgel, 1982; Ferris, 2006). So far, a plausible prebiotic source of activated monomers has not been established experimentally. Due to the aforementioned water problem, should any catalytic polymer, let alone one so complex as a primitive ATP synthase, be formed by chance in a vent environment, without the constant repair and resynthesis by the enzymes of biology, it would soon be disassembled by hydrolytic decomposition.
We do agree that mineral gels could have played a protective and structural role for the first protocell populations arising in hot spring pools, just as they do within microbial mats growing in Yellowstone National Park or Rotorua in New Zealand today. Silica gels also form hard mineral sinters that have harbored and preserved evidence for life throughout the fossil record (Handley et al., 2008; Campbell et al., 2015; Havig and Hamilton, 2019). The alternative offered here is that microscopic membranous compartments, rather than minerals, served as a primary organizing matrix to initiate the first steps along the pathway to the origin of life. We also note that the downhill direction for life originating in freshwater pools on land and then adapting to salty seawater is opposite to the assumption that life began deep in the ocean and only later found its way uphill into fresh water on land. In our view, submarine hydrothermal vents represent a later adaptation for extremophilic microbial life that can thrive in conditions vastly different from the clement pools where life can begin. As we demonstrate next, it is the cycling of systems of polymers through distinctive dry, wet, and moist phases that can drive molecular evolution along the path to cellular life, a process that is not available in a continuously immersed environment.
3.4. The central role of cycling
Energy-driven cycles are central to life's ability to maintain itself in a far from equilibrium state against the trend toward ever increasing entropy. Therefore, it is reasonable to consider the possibility that life's origin also depended on cycles. Unlike an ocean vent setting, surface pools are subject to a rich variety of environmental cycles. If a mixture of amphiphiles, for instance, monocarboxylic acids simulating those delivered by meteorites, is added to a hot spring pool, they form microscopic membrane-bounded compartments (Deamer et al., 2006; Milshteyn et al., 2018). These would be mixed with other organic solutes, particularly amino acids and nucleobases delivered by meteoritic infall or synthesized by geochemical processes. An important point is that these potential monomers are not passive solutes, but instead are involved in a variety of chemical reactions that increase their chemical complexity. For instance, Becker et al. (2018, 2019) established cycling reaction pathways by which all four nucleobases, nucleosides and nucleotides can be synthesized in “one pot” systems through wet-dry cycling. Nam et al. (2018) and Powner et al. (2009) described surface conditions in which nucleosides and nucleotides can also be nonenzymatically synthesized. These recent discoveries support the proposal that small freshwater pools are conducive for the assembly of polymers and protocells in the initial steps toward primitive life.
Figure 4 illustrates how protocells can form and respond to cycles of dehydration followed by rehydration in a pool subjected to regular drying and refilling. The process assumes that a variety of polymers are being continuously synthesized from monomers such as amino acids and nucleotides, and that the system incorporates an indefinite series of cycles that pump the components away from equilibrium toward a steady state in which the polymers are maintained in a kinetic trap. Significantly, cycling also drives a series of natural experiments that undergo combinatorial selection in the form of encapsulated polymers. We refer to this process as “coupled phases” (Damer and Deamer, 2015; Deamer, 2019) because during each cycle, polymers are synthesized and transferred or “coupled” between three distinct phases that involve the self-assembly properties of amphiphilic compounds (Monnard and Deamer, 2002): a multilamellar phase characterized by reduced water activity, a hydrated phase in which encapsulated polymer mixtures are budded off into protocell compartments into an aqueous bulk phase, and an intermediate hydrogel (or gel) phase in which surviving protocells and concentrated solutes form a moist aggregate before drying and fusing back into the multilamellar phase. It is important to note that these polymers remain enclosed within various states of lipid encapsulation analogous to solutions circulating within laboratory glassware.
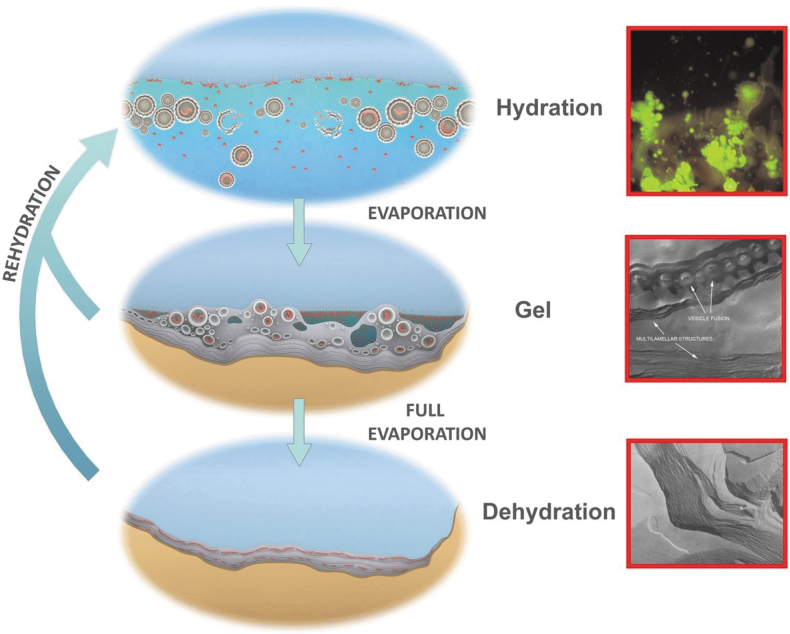
FIG. 4.
Natural drying and rewetting cycles in a small pool supporting three phases of membranous encapsulation: a hydrated phase (top), inset: protocells containing DNA budding out of a dried mixture of DNA and phospholipid, stained with acridine orange; a gel phase (center), inset: freeze-fracture image of a lipid hydrogel with ∼50% water by weight showing lipid vesicles fusing into lamellae; and a dehydrated phase (bottom), inset: freeze-fracture image of anhydrous lipid lamellae of phosphatidylcholine. Micrographs credit: Deamer. (Image adapted from Damer, 2016.)
Each of the three phases subjects sets of polymers to different forms of combinatorial selection. Figure 5 illustrates these forms through the cycling of polymer populations within the kinetic trap of the three distinct phases. Kinetic traps are defined as a steady state in which polymers accumulate because the initial rate of synthesis exceeds the rate of hydrolysis or other decomposition processes (Ross and Deamer, 2016). Polymers are first subjected to selection through competition for monomers during their synthesis in the dehydrated lamellae (1). During hydrous immersion (2), polymers are tested for their ability to stabilize their membranous enclosures (3). Protocell compartments surviving immersion (4) are delivered into a gel aggregate (5) as the pool dries down. The gel aggregate enables protocell-to-protocell interaction (Chen et al., 2004; Adamala and Szostak, 2013a, 2013b) through a form of network selection and niche construction (Odling-Smee et al., 2003). The gel phase supports gradients of increasing concentration during drying, which would also promote the emergence of autocatalytic metabolic cycles (Hordijk et al., 2019). Protocells fuse (6) depositing their polymer cargoes back into the two-dimensional lamellar volumes for resynthesis. The lamellae are not completely dry and immobile but remain in a fluid state so reactants can diffuse within the planes between bilayers. Monomers transition from a dilute, disorganized state in aqueous solution to a highly concentrated and organized state in the multilamellar lipid matrix. Because the water activity is low, ester bond formation is promoted so that polymers of nucleic acids are synthesized.
FIG. 5.
An integrated three-phase scenario showing how membranes and polymers can self-assemble and coevolve driven by fluctuating hydrothermal conditions on the prebiotic Earth.
4. Section III
4.1. Stepwise emergence of functional systems of polymers through three coupled phases
“We may envisage that such a process of molecular self-organization includes many random events which do not have any instructed functional significance. What really matters is how certain such random effects are able to feed back to their origin and thus become themselves the cause of some amplified action.” Eigen, 1971
Section II laid the foundation of experimental and observational results and their significance. Section III is necessarily speculative because a freshwater origin has implications that have not previously been explored. The above quote from Eigen (1971) proposed the emergence, from a random background, of functional molecular systems able to amplify some action and by implication their own continued presence. We have proposed that such a system can start, before the existence of genes, through simple effects such as protocell stabilization. The early part of the scenario relies on a form of phenotypic inheritance similar to the “compositional genome” described by Segre et al. (2001), in which properties are inherited according to their composition rather than information stored in a polymer sequence.
What are the heritable properties of protocells capable of surviving successive cycles? These properties must emerge from a synergy between the components of the system, which are the polymers and the surrounding membrane. All of the processes up to this point have been driven by self-assembly and a very simple source of energy—the chemical potential of dehydration that drives ester and peptide bond synthesis so that monomers can form polymers. The prebiotic synthesis of nucleic acids must have been nonenzymatic and would lead to complex mixtures of variable length strands having random sequences of nucleotides. How can these possibly begin to incorporate catalytic functions? One answer is found in SELEX, the Systematic Evolution of Ligands by EXponential enrichment, an experimental procedure that allows extraction of oligomers with a desired binding affinity for a given molecular target from an initially random pool of oligonucleotides (Djordjevic, 2007). Evolution of catalytic RNA from mixtures of random sequences was demonstrated by Bartel and Szostak (1993) and Horning and Joyce (2016), but these were laboratory experiments using purified nucleoside triphosphates as a source of energy, and the reactions were catalyzed by multiple enzymes. It is possible, however, that similar selective processes could have discovered catalytic functions within vast numbers of self-assembled, encapsulated random polymers on the prebiotic Earth. What is needed now is a similar demonstration that systems of encapsulated polymers can also evolve useful functions such as pore formation and a primitive metabolism that supports growth by catalyzing the formation of copies of additional useful polymers. The following discussion describes what we believe could be fruitful directions for future research.
Walker et al. (2012) used computer models to show how sequences of functional polymers can become established in a pre-existing pool of nonfunctional sequences and monomers. We develop this further by proposing a mechanism for the emergence of specific functional polymers by cycles that incorporate a kinetic trap described in Section II. Cycling encapsulated polymers promotes selection between molecular systems at the level of the protocell while reducing loss of products due to diffusion. The system that would emerge through such a large number of natural combinatorial experiments could plausibly evolve the structures and functions necessary for the transition to cellular life.
Returning to the computing metaphor for the origin of life, there is another concept that is useful: the bootstrap. A bootstrap is an initial small program loaded by a computer, which when executed activates hardware that loads subsequent programs. Those programs in turn begin to start up services and peripherals, such as a network or data stores. The programs must establish a handshake that is coordinated by shared messages. The emergence of large numbers of simple protocells housing random sets of polymers that assemble in a cycling pool provides an opportunity for one to be selected as a bootstrap. The criterion for that first bootstrap is simple: that the polymers contained within the membrane-bounded compartment act in concert with the membrane itself to stabilize the system and prevent the loss of the polymers due to hydrolytic effects or dispersion into the surrounding bulk solution. Therefore, the first selective factor is that protocells are stabilized long enough to return their cargoes into subsequent cycles of synthesis and testing. The stabilizing polymers are analogous to the cytoskeletal proteins that strengthen the membranes of living cells. We refer to these as S-polymers and propose that they constitute the earliest bootstrap for life. With the S-polymers present, the stabilized system of encapsulated polymers is maintained and provides the platform of a kinetic trap so that other functional polymers can emerge.
A second essential function of polymers in the earliest steps toward life would be pore formation in boundary membranes to have access to potential nutrients available in the surrounding medium. Living cells today have evolved enzymes and channels that transport nutrients and maintain ion concentration gradients, but much simpler peptides can also produce pores. One example is the antibiotic peptide gramicidin, composed of just 15 amino acids. When added to membrane-bounded vesicles called liposomes, gramicidin partitions into the lipid bilayers and two molecules assemble into transient transmembrane channels capable of transporting sodium and potassium ions. Pore formation in protocells is important for two reasons. First, the pores allow nutrient solutes to diffuse into the intracellular volume so that primitive metabolism can be supported. They also have a protective function by allowing ionic solutes to equilibrate across the membrane. As evaporation occurs during a wet-dry cycle, the initial modest concentrations of the various ionic solutes of hydrothermal fields increase a 100-fold or more as most of the water is lost to the atmosphere. When freshwater returns, a large osmotic gradient develops that can cause membranous compartments to swell in volume and release encapsulated components. Selectively permeable protocells can survive into future generations because pores permit equilibration of otherwise destructive osmotic gradients. An interesting corollary is that protocells able to survive the osmotic effects of wet-dry cycles will be preadapted to the stresses they experience later in salty seawater. The same pores that ameliorate osmotic stress will also permit increasingly concentrated nutrients to enter protocells during a dry-down. Nutrients combined with sources of energy entering the crowded interiors of aggregated protocells in the gel phase set up the possibility for the emergence of autocatalyzed metabolic cycles (Saha et al., 2014; Kee and Monnard, 2016; Hordijk et al., 2019; Kauffman, 2019).
The above scenario provides a platform available to experimental testing. Indeed, the first steps of this scenario have been confirmed: the stabilizing effect of initially random polymers on protocells allows elongation and potential amplification of those polymers (Rajamani et al., 2008; da Silva et al., 2014; DeGuzman et al., 2014; Himbert et al., 2016; Deamer et al., 2019). While the emergence of pores or metabolic cycles has not yet been demonstrated, pore-forming polymers could be observed in future experiments carried out through multiple cycles under a number of conditions. In fact, Vlassov et al. (2001) demonstrated that selection and amplification of certain RNA species lead to ion-conducting defects in lipid bilayers. Because protocells with encapsulated polymers vary in size, composition, and contents, a natural version of combinatorial chemistry becomes possible in which each protocell can be considered to be an experiment in which relatively stable molecular systems will be preferentially selected, as suggested by Adamala and Szostak (2013a). We emphasize that the synthesis, amplification, and selection of encapsulated polymers within populations of increasingly robust protocells is the sole driver of this process.
4.2. Selection and evolution of functional polymers
Laboratory studies of mononucleotide polymerization have shown that each wet-dry cycle yields polymeric products in the range of several percent of the monomers present (DeGuzman et al., 2014; da Silva et al., 2014), and these become encapsulated in lipid vesicles. We are not specifying their composition, but if the solutes include monomers such as nucleotides and amino acids, the polymers would resemble RNA and peptides, as well as possible complexes of RNA with peptides (Fishkis, 2007). It is not difficult to guess what the additional functional properties of the polymers will be, because through hindsight they would be precursors to functional polymers in cells today. However, what order of emergence might these polymers take?
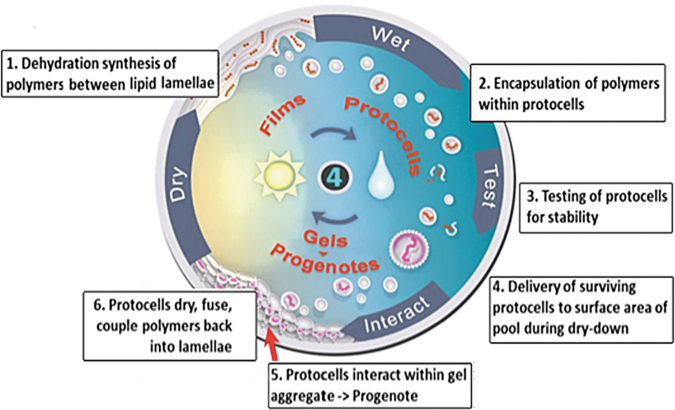
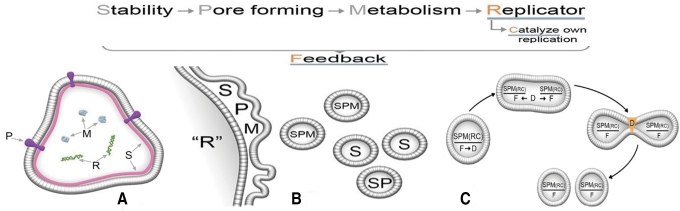
Figure 6 summarizes the stepwise emergence of functions. The text across the top indicates a proposed order of emergence, with the purely physical property of stabilization being the initial bootstrap followed by pore formation (P), metabolic cycles (M), which then provides a platform for a replicator (R) able to use a catalyst (C). At some point, a feedback function (F) emerges to control the rates of these reactions. Inset A depicts a cartoon representation of a model protocell having S, P, M, and R functions operating in tandem. Inset B illustrates how protocells containing random sets of these polymers bud off from lamellae during rehydration. The budding process acts as a natural mechanism for a protocell reproductive function (R). Selective pressures acting on the population through the three coupled phases would select for the most general stabilizing factors, and so, protocells with more functions working in tandem are likely to emerge and establish coherent sets of functional polymers. Not shown but implied is an informational (I) polymer that enables the replication of specific sets of functional polymers. When it first emerges and how it operates are subject to speculation, but perhaps it begins as a template-based system (Olasagasti et al., 2011; Adamala and Szostak, 2013a, 2013b). Finally, inset C shows (clockwise) a protocell with all functions SPM(RC) under the control of feedback (F) and then collectively operated on by a set of D-polymers able to duplicate minimally viable sets of functional polymers. By minimally viable we mean that when another D-polymer enables the division of protocells into two daughter compartments, they are viable and able to grow and divide again. This division marks the transition to lines of living cells. The D-polymer enabling division may have resembled a primitive version of the FtsZ protein that forms a contractile ring for cell division in bacteria (Erickson et al., 2010). In summary, the above scenario steps beyond serial approaches to the origin of life sometimes characterized as “metabolism first” or “genetics first” by proposing a complex mixture in which both metabolic and genetic functions coemerge within a cycling, lipid-encapsulated polymolecular system (Lazcano, 2010).
FIG. 6.
A cartoon representation of the emergence of functions within cycling protocell populations. The top of the figure represents a simplified order of emergence of these functions, executed by a combination of polymers. Figure (A) is a model early protocell with S (stabilizing) factors and P (pore forming) polymers. At some point, polymers that can function as catalysts must emerge, and the two illustrated here are metabolic (M), capable of catalyzing growth, and (R) self-replication. Figure (B) depicts budding protocells capturing various combinations of these polymers, which affect their survivability. Figure (C) is an abstract representation of a protocell containing a more evolved system of growing polymers controlled by feedback networks (F). This system triggers a polymer mechanism D that can duplicate minimally viable sets of these polymers and deliver them by division into daughter cells. If these daughter cells contain fully functional polymer systems and are able to grow and divide again, they form the first lines of living cells.
5. Section IV
5.1. Integrating the chemical, combinatorial, and adaptive scenario in a geological setting
Any scenario describing the origin of life must first escape the bounds of a working hypothesis into laboratory testing followed by further testing in natural conditions. For instance, laboratory simulations have inspired experiments in volcanic hydrothermal fields in Kamchatka, Russia, Bumpass Hell at Lassen National Park, California, Yellowstone National Park in the United States, and most recently in Rotorua, New Zealand. The results of these and the Rotorua study are reported by Deamer et al. (2019). With some confidence we can now integrate the chemical, combinatorial, and adaptive aspects of our scenario within a macroscale environment meant to represent a Hadean landscape.
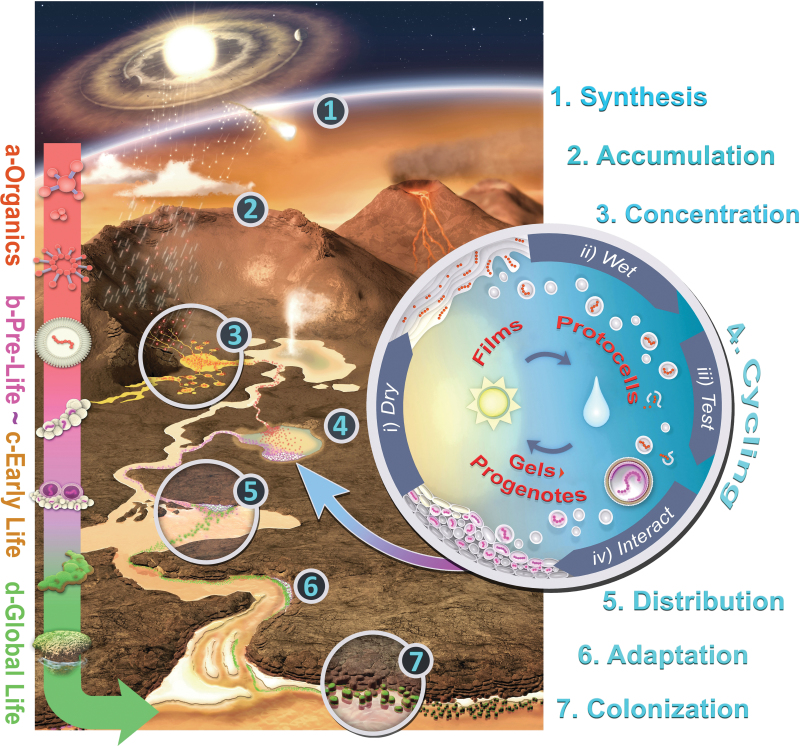
Figure 7 depicts this landscape, and the seven specific stages we propose can lead to the origin of the first microbial communities. Each stage has a number, and we proceed through them one by one to describe how our scenario, in which cycling hot spring pools play a central role, integrates physical, chemical, and combinatorial processes within a prebiotic geological landscape.
FIG. 7.
Integrating a prebiotic Hadean geological landscape with the chemistry of life's origins in hydrothermal fields and its subsequent adaptive pathways into early microbial communities. Credit: Bruce Damer and Ryan Norkus. (Image adapted from an earlier version in Damer, 2016.)
5.1.1. Synthesis
The setting is the Hadean Earth, ∼4 billion years ago by which time a global ocean has formed, with emerging volcanic land masses (Pearce et al., 2018). The solar system is a dusty place with its accretion disc still being swept by young planets and large asteroids that experience numerous collisions. These dust particles are coated with a thin layer of ice containing organic compounds synthesized earlier in their history, along with organic compounds adhering to their interior porous mineral matrix. This organic material was delivered to Earth's surface as abundant accretionary infall that continues at a reduced level today (Pearce et al., 2017). Atmospheric photochemistry and volcanic geochemistry also contributed organic compounds to the available organic inventory (Parker et al., 2011; Wu and Sutherland, 2019). Separation of organic meteoritic compounds such as amphiphiles and monomers will begin as soon as dust particles and meteorites come into contact with acidic atmospheric moisture (Deamer, 1985; Tuck, 2002) delivering larger organic complexes designated “(a)” in Fig. 7 on the left side bar of chemical complexity.
5.1.2. Accumulation
If this material falls into the ocean or larger bodies of water on land, it will be dispersed into a dilute solution that cannot support chemical reactions. However, if these organic compounds accumulate on volcanic land masses, they become available to participate in cycles of significant chemical reactions. The initial collection points are mineral surfaces similar to those present in volcanic sites today. For instance, rough or vesicular surfaces of basaltic lavas can collect solutions. Small pools also form when clay particles or precipitated silicate minerals seal otherwise porous surfaces. The mineral edges of pools, which may have moist sediments, can preferentially capture windblown dust laden with these compounds. Hydration driven by precipitation, dew, humidity changes, or deliquescence (Campbell et al., 2019) can then release organics and begin to promote the condensation reactions described in Section I.
5.1.3. Concentration
Assuming that amphiphilic compounds are present in the mixture, the reactants will be encapsulated and concentrated within vesicular compartments. Because hydrothermal field pools are interconnected, chemically complex, and vary widely in temperature and pH, products formed in one pool can mix with different compounds formed in neighboring pools (Van Kranendonk et al., 2020), and so, chemical processing is not limited to a single pool having one set of conditions. Examples of such pools are found today at hydrothermal fields in Iceland, on Mount Mutnovsky in Kamchatka, Russia, Mount Lassen and Yellowstone National Parks in the United States, and Rotorua, New Zealand. The association of these products within membranous structures keeps them in a concentrated state regardless of the hydration and dilution of the pool and sets the stage for combinatorial selection to occur in a cycling environment.
5.1.4. Cycling
Hydrothermal pools undergo wet-dry cycles resulting from precipitation and fluctuating water levels driven by active hot springs and geysers. The large circle (introduced first in Fig. 5) is set within one such pool that has a “bathtub ring” of dry solutes around the mineral edge. Polymers are synthesized by condensation reactions occurring within these dried films, which form a multilamellar matrix (i). During the hydration phase (ii), vesicles bud off, encapsulating systems of polymers to form protocells (iii). A partially hydrated gel phase of crowded protocells (iv) forms an interactive aggregate within an increasingly concentrated solution and then undergoes drying and fusion into a multilamellar matrix (Fig. 7).
This process generates random sets of polymers captured in vesicles to form vast numbers of protocells. Frequent cycling of these populations initiates the combinatorial selection process that drives chemical evolution and enables the emergence of ever more robust protocells. As the protocells continue to undergo the stresses of cycling, most will be disrupted, their components leaking out or dispersing through disrupted membranes, but a rare few are likely to contain polymers that enhance their survival.
Some protocells are successful natural experiments that survive testing during the hydrated phase, but they cannot express catalytic functions nor live independent lives. For the system to evolve, it must engage in a network in which properties and innovations of molecular selection are shared. Here the gel phase of the cycle can play a role, because the close contacts and fusion events within the aggregate allow sharing of polymers that have beneficial functions, particularly the ability to create useful metabolic products and catalyze polymerization and replication. At this point, the system has reached a form of “Pre-Life” (“(b)” in Fig. 7 on the left side bar). If this seems implausible, it is worth recalling research results from Phil Holliger's group, in which it was reported that ribozymes evolved in the laboratory by selection and amplification were able to catalyze the polymerization of RNA molecules nearly 200 nucleotides in length (Wochner et al., 2011). Similar results were reported more recently by Horning and Joyce (2016). The products of selection that cycling systems generate can lead to the stepwise emergence of increasingly functional polymers described in Section III.
The aggregated protocells in the gel phase resemble the progenote concept described by Woese and Fox (1977) and Woese (1998). Progenotes represent a transitional form between simple chemical systems and the more complex biochemical functions of biology. It is interesting that progenote evolution bears a certain analogy to booting up a computer's operating system in which services start up, share, and support higher level applications. The programs of life are polymer systems selected to perform primitive versions of biological functions: capture nutrients and chemical energy from the environment, metabolically alter the nutrients into useful compounds, grow by polymerization of monomers, and catalyze replication of certain of those polymers. Eventually these systems no longer depend on the scaffold of hydration/dehydration cycles for the chemical potential driving polymer synthesis. Instead, individual protocells will begin to utilize polymer catalysts to perform these tasks and must therefore pass on complete molecular tool sets that include primitive genetic instructions to guide their synthesis. The evolutionary chasm to life will be crossed when protocells begin to divide into daughter cells, each receiving viable sets of these catalysts and genetic instructions. Speciation and specialization of living cells then cross the “Darwinian Threshold” described by Woese (2002). The tangled roots of horizontal sharing through communal progenotes now support the growth of multiple interwoven branches as the tree of life continues to evolve. By 3.5 billion years ago, a common ancestor has emerged with a genetic code and translation system, leaving behind telltale signatures of stromatolites and microfossils preserved so well in ancient marine, lakeshore, and freshwater hot spring settings in the Pilbara region of Western Australia (Walter et al., 1980; Van Kranendonk et al., 2008, 2020; Djokic et al., 2017, 2020).
5.1.5. Distribution
Any hypothesis of the origin of life must propose a pathway for distribution. If the cyclic processes described above occurred in an isolated pool in which a robust progenote population happened to emerge, there would be no way for it to escape this potentially short-lived pool and continue to evolve and adapt to new niches. Interconnected hot spring pools on mountainous volcanic landscapes have the obvious advantage that there is a continuous downhill flow from pool to pool driven by gravity, so that protocell populations can spread to other pools with distinct characteristics related to temperature, pH, ionic content, and mineral composition. There they will be subjected to additional stresses, driving them uphill in complexity, ultimately to the division of protocells into living cells and the emergence of the primitive, early microbial communities. Dehydrated films can also undergo windborne distribution, a mechanism still used today by microbial and algal mat communities (Parker et al., 1982; Neuman et al., 1996). These films are an effective preservation and dispersal mechanism for systems of polymers that would otherwise be degraded by long-term exposure to hydrolysis. Seeds and spores are blown by wind and preserve their genetic polymers against water activity between generations of plants and certain microbial populations today. As protocell and then early living cellular populations become physically distributed across the landscape, they are exposed to a range of conditions that drive increasing robustness to multiple stresses. Innovations developed in one pool can be shared with populations occupying another. This process eventually produces tolerance to the extreme environment of salty seawater, thereby providing access for early life to more extensive but often turbulent tidal marine shorelines.
Such an adaptive gradient is observed in the hot springs of Yellowstone National Park today. Thermophilic microbial communities that are chemotrophic use the energy and chemicals available in the upwelling high-temperature spring, while further downhill in the cooler and more dilute outflow, photosynthetic organisms capture sunlight as an energy source. Beginning with the first generation of stabilizing polymers, the stepwise evolution of more complex polymer systems would continue to depend upon a partnership of polymers with the membranous boundaries of protocells. For instance, Groen et al. (2012) reported that polycyclic aromatic hydrocarbons (PAHs), components of meteoritic organics, have a stabilizing effect on fatty acid membranes, similar to the effect of another polycyclic compound—cholesterol—on membranes today. Black et al. (2013) showed that adenine has a stabilizing effect on fatty acid membranes, and Cornell et al. (2019) observed similar effects of certain amino acids. For an origin of life on a terrestrial world possessing liquid water, it is the exposed land surfaces that would provide the richest array of compounds, environments, cycling systems, and the greatest range of energy sources (Deamer and Weber, 2010).
Distribution of progenotes into cooler watery environments such as dilute streams or lakes leads to a major selective hurdle. Regular wet-dry cycling is no longer available as an energy source and potential nutrients drop to much lower concentrations. Progenote communities fail until selection discovers some form of photoautotrophy supporting a much more active, energy-coupled metabolism. Much more complex molecular machines would be required at this stage, including a genetic code driving a primitive version of the ribosome, catalyzed duplication of genetic information, and cell division. It therefore seems reasonable to assume that the transition from passive protocells into more active and autonomous living cells would occur as populations become distributed away from the hot spring environment at “(c)” in Fig. 7.
5.1.6. Adaptation
Autotrophic microbial colonies ultimately encounter the increasingly brackish water of estuaries and then the high salt and divalent cation concentration of seawater. The microbial communities respond by developing robust means to exchange intracellular sodium chloride for potassium chloride and actively transport toxic divalent calcium ions out of the cells. This adaptation requires complex, energy-dependent transport of ions by membrane-bound enzymes, suggesting that the first organisms able to live in seawater would have advanced far from the initial Goldilocks conditions required at the site of life's origins (Mulkidjanian et al., 2012).
5.1.7. Colonization
As increasingly robust microbial communities arrive in the even harsher tidal marine margins, another set of stresses challenges them to adapt. To respond to increased osmotic stress and shear forces, the microorganisms must develop durable cell walls and an extracellular matrix composed of polymers that strengthen adhesion between individual cells in communal aggregates. The aggregates cling to mineral surfaces of pools and shorelines that are washed by tides 10 times higher than those of today's oceans. The adhesive mucus that allows the cells to remain fixed on mineral surfaces also binds and cements together mineral grains in sediments settling over them. To continue to have access to sunlight, microorganisms composing these microbial mats migrate upward and grow new colonies on top of the cemented substructure. Over many years, multiple layers of microbial films and cemented grains produce the laminations characteristic of stromatolites, which dominate the earliest fossil record and are still forming today in freshwater and shallow marine environments such as the Hamlin Pool at Shark Bay, Western Australia (Papineau et al., 2005). At this stage, life would have gained access not only to aqueous environments on the land but also to the extensive marine margins of volcanic land masses and later, the ocean volume itself, as cyanobacteria colonized the globe “(d)” in Fig. 7.
6. Section V
6.1. Network effects and other evolutionary properties of the progenote
“The demonstration of the existence of a complex, nonenzymatic metabolic cycle, such as the reverse citric acid, would be a major step in research on the origin of life, while demonstration of an evolving family of such cycles would transform the subject.”
—Leslie Orgel
We agree with Orgel's statement that the emergence of closed cycle metabolic reaction networks remains a key unsolved problem for the origin of life (2008). Life depends on highly evolved networks which harvest energy to synthesize products and to coordinate and regulate reactions. In parallel with the initial bootstrap of stable protocells and stepwise selection of functional polymers within the kinetic trap is a necessary emergence of a “network effect”. Computer networks such as the Internet are made up of services based on protocols that enable packet delivery, error correction, dynamic table lookup, and pathway optimization. Each service is simple on its own and often described by a few programmatic steps, yet the emergent network composed entirely of closed cycles attains a high degree of operational robustness and adaptive capability. This capability was specifically designed into the original ARPANET, which grew into today's highly robust and scalable Internet (Leiner et al., 2009). The network effect codifies the old adage “the whole is greater than the sum of its parts.” During the evaporation phase of wet-dry cycles, protocells aggregate into a hydrogel within which a network effect can be initiated. Such gels retain moisture and concentrate nutrients, providing for the selection and sharing of the additional polymer functions such as pores required for solute transport supporting emergent metabolic cycles within protocells. Recent work demonstrated radiolysis reaction networks connecting geochemical reservoirs to the production of carboxylic acids, amino acids, and ribonucleotide precursors (Adam et al., 2019). These networks exhibited hierarchical organization and families of closed loops common to biological systems. Such networks of interacting radicals driven by UV and other radiation sources operating within protocell aggregates could have provided the initial cycles which Orgel sought. Protocells are cell-sized compartments whose interiors enable molecular crowding and when aggregated together they provide an ideal environment for selection through passive forms of protocell competition (Chen et al., 2004; Adamala and Szostak, 2013a). Furthermore, an aggregate would tend to retain the components of disrupted protocells, thereby selecting for surviving protocells that can absorb and conserve the components in a primitive form of heterotrophy.
What follows is a conjecture. However, we believe it has value because a conjecture becomes a working hypothesis if it can be falsified by experiments, several of which are proposed in Section VI. A protocell aggregate can be considered to be a prototypical form of niche construction as described by Odling-Smee et al. (2003). Damer (2016, 2019) extended this concept to prebiotic conditions in which protocell populations form moist gel aggregates during the drying process that protect individual protocells and their polymer cargoes against shear forces, pH extremes, and temperature changes. A gel could also maintain a stable concentration of ionic solutes, thereby protecting protocells against osmotic stress.
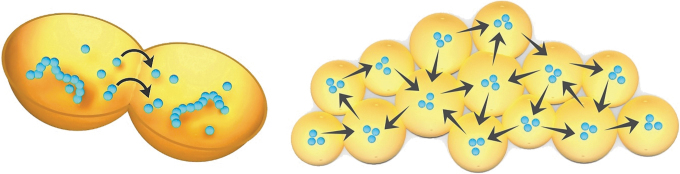
Figure 8 (left) illustrates how the close aggregation of compartments allows metabolic products to diffuse within protocell populations. Such products, and whole complexes of functional polymers, can be mixed throughout the aggregate and thereby benefit the entire population, enabling a mechanism to support inclusive fitness (Hamilton, 1964). This property also models the network effect described earlier and is a primitive version of the horizontal gene sharing characteristic of all microbial communities today (Woese, 2002). As introduced in the previous section, an aggregate of protocells is a candidate progenote originally described by Woese and Fox (1977) and Woese (1998) in which its member protocells are engaged in the collective process of evolving the relationship between phenotype and genotype (Arnoldt et al., 2015). We introduce the name “Progenean” to describe this transitional evolutionary period. There is no rock record left by protocells or their progenote aggregates, so the term cannot specify a geological era but instead refers to the stages of chemical evolution from simple self-assembling protocell populations into living microbial communities. For example, early in the Progenean, consortia of protocells would emerge, which are capable of growth and adaptation through phenotypic properties emerging by simple selection from random sequences. A middle division of the Progenean would begin with the first protogenetic system, perhaps initiated by simple templated replication. The late stages of the Progenean would be characterized by sets of increasingly functional polymers that are duplicated by a primitive replication and translation system and collectively passed on to subsequent generations of protocell populations.
FIG. 8.
Product diffusion between neighboring protocells and a network of interactions set up within protocell aggregates forming in the moist gel phase of a wet-dry cycle.
Throughout this period, passive self-assembly operating on the scaffold of wet-dry cycling is gradually replaced by systems of more efficient, active polymers interacting in networks giving protocells an increasingly coherent biological identity. A key question is how the progenote served as an evolutionary environment to transform simple, passively budding protocells into active, dividing living cells. It is reasonable to assume that the first functional polymers operated most effectively while bound to a protocell's membranous enclosure rather than dispersed within the dilute interior. This binding has been shown to stabilize the membrane against stresses (Black et al., 2013; Mayer et al., 2018), and would also enable sets of polymers to become more crowded on the two-dimensional surface and interact more easily. Recently, Cornell et al. (2019) proposed that this colocalization sets up “a positive feedback loop in which amino acids bind to self-assembled fatty acid membranes, resulting in membrane stabilization and leading to more binding… [and that] high local concentrations of molecular building blocks at the surface of fatty acid membranes may have aided the eventual formation of proteins.” Just as the viable operation of protocells would have been dependent on binding and interactions between enclosures and polymers, the process of their continued evolution would have been dependent upon the protective and nutritive environment of the surrounding progenote aggregate. The long evolutionary climb to cell division would have been a stochastic process driven by innumerable tests and setbacks (Damer, 2011). Failure of a key process within a protocell dwelling separately in solution might result in disruption and dispersal of its entire cargo of polymers, whereas such faults would not be so deleterious if they occurred inside a communal matrix. Challenges arising from unreliable metabolic circuits within a protocell can be mitigated through networks of resupply of materials and energy from other protocells operating in the surrounding aggregate. At some point, evolutionary innovations converge to where protocells are selected, which begin to produce subdivisions of their own compartments, possibly beginning as a simple adaptation to bud and eject vacuoles containing wastes. In the case of a failed compartment division resulting in disruption of the entire protocell, valuable functional polymers would still be retained and reused by other protocells in the surrounding aggregate. The final transition to life could have been facilitated through the chance combination of a fortuitous division of a compartment occurring just after the duplication of two sets of membrane-bound polymers, each pushed into opposite ends of an elongated protocell. In this first example of binary fission, the timed triggering of a transverse division of such a compartment would securely deliver duplicate sets of minimally viable polymers into daughter cells. Through the resulting innovation of cellular reproduction comes a vertical inheritance of traits and the transition out of the Progenean into the era of living microbial communities. As is obvious from the above example, a key evolutionary property of progenotes is that they can support attempts at and failures of risky multistep processes such as reproduction without the loss of genetic material. This communal support system thereby permits the incremental writing of the complex code of life.
The speciation and specialization of increasingly robust living cells would allow communities to scale steeper gradients of environmental stresses and colonize more niches on the landscape. During the Progenean, there would undoubtedly have been innumerable starts and failures, not only of individual protocells but also of entire progenote communities. However, we posit that a landscape of large numbers of interconnected hydrothermal pools continuously driven by regular cycles of energy and supplied by feedstocks of organic material over tens of millions of years had the “combinatorial power” to code the operational kernel of life. Eventually, individual cells would become sufficiently advanced to survive and reproduce freely in solution, detached from their original communal complexes. Wider distribution and the competition for resources of communities and free-living cells would then extend the tangled branches of early life along the path to the cohort of last universal common ancestors (LUCAs).
7. Section VI
7.1. A roadmap for testing the hypothesis
“Origin of Life cannot be discovered; it has to be reinvented.”
—Albert Eschenmoser
We interpret Eschenmoser's statement (2007) that scenarios for an origin of life ought to quickly move beyond paper proposals to become full hypotheses, tested both in the laboratory and in the much more complex environments of natural analogs (Deamer et al., 2019). For any hypothesis, a quick empirical falsification is preferable to endless speculation, yet life is such a complex phenomenon that falsification is not as simple as it can be in physics. Workers in the field must perform multiple experimental tests and use the weight of accumulating evidence to measure the plausibility and explanatory power of any scenario.
Platt's article “Strong Inference” published in Science (1964) advises experimenters to devise alternative hypotheses and a crucial experiment with alternative possible outcomes. Carrying out the crucial experiment to achieve a clean result will exclude one or more of the hypotheses. The following language describes a crucial experiment that can be applied to test any origin of life hypothesis: Confirm or falsify a nonenzymatic process by which catalytic and replicating systems of polymers are produced under plausible prebiotic conditions that also support cycles of combinatorial selection in which encapsulated sets of these polymers emerge and begin to evolve.
Proposals for an origin of life in salty ocean water at hydrothermal vents, or in freshwater at hydrothermal fields can be considered to be alternative hypotheses. We propose the following seven tests that can support or falsify the hot spring hypothesis.
-
1.
A laboratory observation must also work in the natural conditions it is intended to simulate. In the laboratory, we have demonstrated that in the presence of membrane-forming lipids, wet-dry cycles can drive polymerization of mononucleotides, followed by encapsulation of products in membranous compartments upon rehydration. This experiment was recently carried out in a hot spring setting near Rotorua, New Zealand (Deamer et al., 2019). The results were positive, but are preliminary, and the experiments need to be repeated.
-
2.
The wet-dry synthesis of RNA-like polymers recently repeated by others (Mungi and Rajamani, 2015; Mungi et al., 2019) has thus far been limited to the use of canonical mononucleotides of RNA. Other groups have demonstrated polypeptide formation through wet-dry cycling (Forsythe et al., 2015; Rodriguez-Garcia et al., 2015). More complex mixtures of polynucleotides and polypeptides present in protocells represent a next frontier in the testing of this hypothesis. In addition, there may be prebiotically plausible monomers that are close cousins to modern forms, some of which have been demonstrated to lead to polymers of use to biology, such as depsipeptides (Forsythe et al., 2017) and the use of alternate nucleobases (Mungi et al., 2016). Also important is the consideration of the parameters that will limit the participating chemicals' availability due to decomposition or side reactions. A key next step is to observe the selection of the functional polymers described in Section III by carrying out tests 3–7 below. Their presence and activity will be marked by an increased population of protocells in subsequent cycles. Analysis of protocell contents can confirm populations of these polymers and their by-products.
-
3.
Encapsulated polymers must stabilize otherwise fragile membranous compartments having boundaries composed of lipid bilayers. We have proposed (Damer and Deamer, 2015; Deamer, 2019) that the first selective step initiating evolution in a population of protocells is that polymers interact with the membranes in such a way that the protocells become more resistant to environmental stresses such as shear forces. As introduced in Section III, this “S-polymer” is analogous to that of cytoskeletal proteins that strengthen the membranous boundaries of cells today. In the absence of a stabilizing effect, fragile protocells will be continuously disrupted, or simply leak their contents.
-
4.
Another stress associated with wet-dry cycles is that ionic solutes become much more concentrated as water is lost to evaporation. When the dry membranes are rehydrated, a large osmotic gradient is established that causes the membranous compartments to swell as water diffuses across the membranes into the concentrated interior, potentially disrupting protocells by osmotic lysis. The osmotic pressure can be reduced if some of the encapsulated polymers can form ion-conducting defects or pores (“P-polymers”) in the lipid bilayers that allow the concentrated ions to escape. Pores have the additional effect of letting nutrients enter the protocells, which could support the emergence of primitive metabolic cycles (“M-polymers”) utilizing these nutrients. A significant observation by Vlassov et al. (2001) is that RNA oligomers selected by in vitro evolution can produce ion-conducting defects in lipid bilayers. Mayer et al. (2018) have devised and tested a cycling protocell system operating at the phase transition between supercritical and subcritical gaseous CO2 in deep tectonic fault zones. They have observed that oligopeptides polymerize from amino acids and selectively integrate into the bilayer structure of vesicle membranes, thereby facilitating the mutual arrangement of their protection against hydrolysis while stabilizing the surrounding vesicle. Some of these peptides have also increased membrane permeability and protocell durability, effectively expressing the function of P-polymers. A similar detection of the first P-polymers emerging de novo in a wet-dry cycling system of protocells would be a significant milestone in testing the hypothesis.
-
5.
An important next prediction is that random mixtures of encapsulated polymers can evolve catalytic activity supporting primitive forms of metabolism. Bartel and Szostak (1993) demonstrated that 300mers of RNA starting from entirely random sequences can evolve ligase activity by a process of selection and amplification. Similar experiments performed with encapsulated polymers will test the idea that catalytic functions (“C-polymers”) can also emerge when protocells are subjected to selection through hydration/dehydration cycles.
-
6.
At some point, sets of polymers must be reliably replicated to amplify functional polymers that emerge from initial mixtures of random sequences. The first informational “I-polymer” could be a simple templated replication system (Olasagasti et al., 2011; Adamala and Szostak, 2013a, 2013b) or resemble more sophisticated ribozymes as reported by Wochner et al. (2011) and Horning and Joyce (2016). The complexities of cellular functions today suggest that chemical circuits operating in advanced protocells and progenote networks will also necessarily be complex and will be controlled by well-tuned feedback systems mediated by “F-polymers.”
-
7.
Emergent properties of protocells may be observable in isolation but are more likely to be accessible for study in an aggregate of many protocells—a progenote. Time frames for bottom-up de novo “N-progenotes” evolving interesting prebiotic properties could exceed the lifetimes of laboratory scientists and their institutions. It might therefore be advisable to “jump the queue” of random selection through the use of synthetic biology to create “S-progenotes” by introducing designed interacting polymer systems (Wieczorek et al., 2013). These “programmed” protocell populations would then be subjected to cycles of selective pressures designed to test those polymer systems. Two recent examples informing this approach include RNA template copying inside protocells (Adamala and Szostak, 2013b) and vesicles containing an encapsulated enzyme that promotes synthesis of simple fatty acid derivatives stabilized to the Mg2+ required for ribozyme activity and RNA synthesis (Adamala et al., 2016). Continuing to cycle progenotes with the goal of testing whether evolutionary changes can be observed should use carefully crafted selective hurdles that reflect real conditions of hot spring pools and other prebiotically plausible environments. The emergence of “D-polymers” controlling the duplication of sets of polymers and their delivery into daughter cells is probably inaccessible to experimental observation. Such a system may be too complex to be designed top-down by a synthetic biologist and may only be attainable via an accelerated bottom-up process of evolution. Perhaps this could be undertaken through the construction of an automated “genesis engine” based on a miniaturized network of chemical robots similar to those developed by Cronin and coworkers (Caramelli et al., 2018). Given the challenges and costs involved, a “second genesis” in the laboratory, long a theme of science fiction, will likely therefore remain a scientific fiction.
8. Section VII
8.1. Evolutionary implications of the hypothesis
Two alternatives have been proposed as sites for the origin of life. Either life began in the ocean, perhaps using the energy and nutrients available in hydrothermal vents, or life began in hot springs associated with volcanic land masses that emerged from the global ocean over 4 billion years ago. The hot spring sites are analogous to those seen today in Iceland and Hawaii, supplied with fresh water distilled by evaporation from the salty sea and falling as precipitation.
Given the two alternatives—salty seawater or fresh water on land—it is interesting to consider implications for life's early evolution and its long climb to complex forms such as land plants. The settings of these two scenarios are illustrated in Fig. 9. If life began at submarine hydrothermal vents, its metabolism would at first have used the same energy as today's chemotrophic microbial populations. These organisms are anaerobic and do not require oxygen (respiration) or light (photosynthesis). Instead, the energy source involves electron transport from a reduced substrate such as hydrogen or hydrogen sulfide to oxidants such as sulfate, ferric iron, or carbon dioxide as acceptors. The evolutionary path therefore requires that chemotrophic vent life somehow made its way to the ocean surface where light was available, then discovered photosynthesis, and evolved into the cyanobacteria that became a dominant form of life for the next 3 billion years. The switch to photosynthesis must have been necessarily rapid because as soon as the chemotrophic organisms dispersed from the vent environment, they would be starved of source materials and energy in the much more dilute and cooler surrounding ocean (Jackson, 2017). It is possible that life originating in shallow marine hydrothermal vents with some sunlight exposure might have a better chance to develop photosynthesis. A massive global culture of cyanobacteria in the ocean ultimately produced sufficient molecular oxygen so that the atmosphere became oxidizing about 2 billion years ago. At some point, eukaryotes emerged by symbiosis, still in salty seawater, and primitive eukaryotes in shallow tide pools began to form multicellular life resembling algae. Finally, this scenario suggests that around 500 million years ago, algae evolved the capability to survive when exposed to the atmosphere on mineral surfaces and the first land plants emerged from seawater onto sterile continents.
FIG. 9.
Alternative scenarios for an origin of life and adaptive pathways from freshwater hydrothermal field pools or saltwater hydrothermal vents to eukaryotes and land plants.
The alternative that life began in hot springs on land possesses a more direct evolutionary pathway to land plants. Freshwater life would also be chemotrophic at first, depending on organic compounds available in hot spring environments through geochemical synthesis and extraterrestrial sources delivered during late accretion. However, early life was also continuously exposed to sunlight and could develop photosynthesis utilizing primitive pigments such as PAHs as an energy source (Deamer, 1992; Summers and Rodoni, 2015). Dry land exposure to damaging ultraviolet radiation would have been mitigated through hypolithic lifestyles (Havig and Hamilton, 2019) and might also have promoted key prebiotic reactions (Sutherland, 2017). Although salty seawater would have been an extreme environment for relatively fragile freshwater life, as microbial populations dispersed downhill toward the sea, they evolved cell walls with ion pumps to keep salt ions from disrupting intracellular metabolism. The volume of fresh water available was only a tiny fraction of that of ocean water, so when photosynthetic life was finally able to survive in the ocean, it filled a very large ecosystem niche. However, this marine adaptation does not mean that the evolution of freshwater microbial populations ceased. Instead, their evolutionary pathway paralleled that of marine life but was driven by different environmental stresses. One was that hot spring water was continuously undergoing wet-dry cycles causing 100-fold changes in the concentration of ionic solutes, so it was essential to develop cell walls to survive steep osmotic gradients. Another property of periodic wet-dry cycles is that at the origin of life, individual protocells were being brought into close contact during the drying phase of each cycle, forming symbiotic aggregates—progenotes. This enabled the later development of living multicellular consortia of microbial mats whose close collaborative sharing is a plausible preadaptation for the fusion events of endosymbiosis, which gave rise to eukaryotes. In the hot spring hypothesis scenario, subaerial land masses on Earth were never sterile, but instead were continuously inhabited by freshwater versions of the microbial forms that later dominated the ocean. This proposal is supported by evidence of stromatolites in the Pilbara in Western Australia thriving in subaerial hot springs 3.5 billion years ago (Djokic et al., 2017, 2020). Paleosols carrying evidence of life 3.22 billion years ago have also been discovered in a preserved river flood plain in the Barberton region of South Africa (Nabhan et al., 2016). In addition to an abundant supply of meteoritic organics available to the subaerial pools for life to start, continental rock weathering products such as phosphorus crucial to early life and life today are only available in an environment having a triple interface between water, atmosphere, and minerals (Satkoski et al., 2016).
Summarizing the arguments just presented, chemotrophic microbial communities originating in hot springs are continuously exposed to light energy during the day and therefore have an opportunity to develop photosynthetic metabolism. They are also transported downhill toward increasingly saline conditions that force evolutionary adaptations to osmotic stress and turbulence at intertidal zones. These same land-based microbial communities continue to undergo a 3-billion-year evolution to eukaryotic freshwater algae and plants, followed by fungi, and protozoa developing in the shallow marine margins of continents (Peterson et al., 2008). The earliest ancestor of land plants is currently understood to have been a form of freshwater, terrestrial algae (Kenrick and Crane, 1997; Del-Bem, 2018).
In contrast, if the significant thermodynamic barriers for life to begin at marine hydrothermal vents were overcome, additional hurdles would still be encountered. The first would be delivery from the vent environment to the ocean surface where sufficient light energy was available to support the emergence of photosynthesis. The next is the journey into intertidal zones where eukaryotic marine algae and multicellular life would become possible by symbiotic mergers. The final hurdle would be adaptation to atmospheric exposure on mineral surfaces and appearance of the first plants, such as primitive mosses that could use fresh water supplied by precipitation. Given that microbial communities have inhabited land as far back as the rock record can reveal, a direct evolutionary path from life originating in fresh water pools to the first terrestrial plants is perhaps a more plausible alternative than one in which life started at oceanic hydrothermal vents.
8.2. Astrobiology implications of the hypothesis for the origin of life on other worlds
Continuously submerged environments such as hydrothermal vents clearly support highly adapted forms of life on Earth today and might possibly harbor life on ocean worlds such as the icy moons Europa and Enceladus (Fig. 10, left). Plumes containing organic compounds and water emerging through cracks in Enceladus' icy shell were discovered by the Cassini mission (McKay et al., 2008), suggesting that hydrothermal vents might be operating at a rock interface deep in an interior ocean. However, while potentially habitable, due to their submarine setting and lack of wet-dry cycling, these locations may not possess the chemical, thermodynamic, or combinatorial capacity for life to begin (Deamer and Damer, 2017). Mars, on the contrary, has clear evidence of silica deposits (Fig. 10, right), indicative of subaerial hydrothermal systems of a similar age as those discovered in the Pilbara. Surprisingly, this site at Columbia Hills discovered by the Spirit rover also resembles hot spring environments that preserve biosignatures of microbial communities on Earth (Ruff and Farmer, 2016). This discovery can guide future missions to promising places to search for signs of past life on Mars before it lost its surface habitability.
FIG. 10.
Left: Depiction of Enceladus, a moon of Saturn visited by the Cassini mission, which discovered plumes emerging through cracks in the south polar ice possibly indicating oceanic hydrothermal activity. Right: silica nodules discovered at Columbia Hills on Mars by the Spirit rover consistent with an ancient hydrothermal environment (credits: NASA/JPL-Caltech).
8.3. Concluding remarks
The hot spring hypothesis for an origin of life emerged from a fortuitous collaboration across multiple branches of science. Laboratory demonstrations supporting the “RNA world” hypothesis since the 1980s established an exciting direction in our field (Gilbert, 1986). However, despite the surprising catalytic properties of RNA molecules, we contend that the chemistry of life's beginning cannot be reduced to the action of a single molecular species, but instead required the interactions and selection of vast numbers of encapsulated polymer systems. These protocells were initially formed through processes of self-assembly of both the polymers and their compartments and evolved through the engine of a kinetic trap subjecting them to combinatorial selection within cycling pools. This pre-Darwinian system of selection provided a scaffold for the “booting up” of the active molecular systems of life. Metaphors and insights from computer science have also informed the hypothesis by predicting that a collaborative network effect would occur in aggregates of protocells known as progenotes. Experimental work at multiple laboratories and more recently at field analog sites has accumulated a growing body of evidence. Finally, the discovery of 3.5-billion-year-old Archean fossil hot spring stromatolites of the Pilbara region of Western Australia provided key additional pieces to the puzzle.
It possibly required more than half a billion years for the first fragile protocells emerging in hydrothermal field pools to evolve into the robust microbial life that produced the durable fossil evidence of stromatolites. Somewhere along that timeline, the LUCAs appeared with a full complement of the functional polymers of life as we know it today. Note the purposeful use of plural in the previous sentence to indicate that LUCA was not a single cell from which all subsequent life descended, but instead would have been collectively sourced from widespread microbial mat communities that were sharing genetic information. Orgel (1968) and Morowitz (1992) proposed that all living systems follow a principle of continuity in which new structures and systems emerge through variations on similar earlier forms. A final and perhaps the most remarkable implication of this hypothesis is that the earliest form life would have taken was an aggregate of collaborating units rather than competing individuals. Peering back through the lens of the principle of continuity, stromatolites are clearly the work of microbial mat communities. Perhaps those communities trace their ancestry to a simpler form of community, the progenote, which in turn got its start in deeper time as an aggregate of even simpler self-assembled protocells with encapsulated molecular systems. The tangled roots of the tree of life traced by intertwined protocell and progenote evolution may never be unraveled to reveal a clear picture of how life actually started on the Earth. However, the hypothesis proposed here can be tested at several stages in the laboratory and in the field. An accumulating weight of evidence may then lead to understanding how life can start on a habitable planet such as the early Earth.
Acknowledgments
Portions of this review were adapted from Chapters 9 and 10 in Assembling Life, written by D.D. and B.D. and published by Oxford University Press (2019). The authors thank Ram Krishnamurthy and many other colleagues who provided constructive criticism as the hot spring hypothesis was developed, and Ryan Norkus for the artwork illustrating the concepts presented here.
Abbreviations Used
- ATP
adenosine triphosphate
- LEDs
light-emitting diodes
- LUCAs
last universal common ancestors
- PAHs
polycyclic aromatic hydrocarbons
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was funded by the authors.
References
- Adam Z., Fahrenbach A.C., Jacobson S.-M., Kacar B., and Zubarev D.Y. (2019) Radiolysis Generates a Complex Organosynthetic Chemical Network. ChemRxiv. Preprint. 10.26434/chemrxiv.10148684.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K. and Szostak J.W. (2013a) Competition between model protocells driven by an encapsulated catalyst. Nat Chem 5:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K. and Szostak J.W. (2013b) Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342:1098–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K., Engelhart A.E., and Szostak J.W. (2016) Collaboration between primitive cell membranes and soluble catalysts. Nat Commun 7:11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel C., Mautner M., and Deamer D.W. (2002) Self-assembled vesicles of monocarboxylic acids and alcohols: conditions for stability and for the encapsulation of biopolymers. BBA Biomembr 1559:1–9 [DOI] [PubMed] [Google Scholar]
- Arnoldt H., Strogatz S., and Timme M. (2015) Toward the Darwinian transition: switching between distributed and speciated states in a simple model of early life. Phys Rev E 92:052909. [DOI] [PubMed] [Google Scholar]
- Baaske P., Weinert F.M., Duhr S., Lemke K.H., Russell M.J., and Braun D. (2007) Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc Natl Acad Sci U S A 104:9346–9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bada J.L. and Korenaga J. (2018) Exposed areas above sea level on Earth >3.5 Gyr ago: implications for prebiotic and primitive biotic chemistry. Life (Basel) 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bada J.L. and Lazcano A. (2002) Some like it hot, but not the first biomolecules. Science 296:1982–1983 [DOI] [PubMed] [Google Scholar]
- Barge L.M., Branscomb E., Brucato J.R., Cardoso S.S.S., and Cartwright J.H.E. (2015) RNA oligomerization in laboratory analogues of alkaline hydrothermal vents. Astrobiology 151:1405–1416 [DOI] [PubMed] [Google Scholar]
- Barge L.M., Branscomb E., Brucato J.R., Cardoso S.S., Cartwright J.H., Danielache S.O., Galante D., Kee T.P., Miguel Y., Mojzsis S.K., Robinson K.J., Russell M.J., Simoncini E., and Sobron P. (2017) Thermodynamics, disequilibrium, evolution: far-from-equilibrium geological and chemical considerations for origin-of-life research. Orig Life Evol Biosph 47:39–56 [DOI] [PubMed] [Google Scholar]
- Bartel D.B. and Szostak J.W. (1993) Isolation of new ribozymes from a large pool of random sequences. Science 261:1411–1418 [DOI] [PubMed] [Google Scholar]
- Becker S., Schneider C., Okamura H., Crisp A., Amatov T., Dejmek M., Carell T. (2018) Wet-dry cycles enable the parallel origin of canonical and non-canonical nucleosides by continuous synthesis. Nat Commun 9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S., Feldmann J., Wiedemann S., Okamura H., Schneider C., Iwan K., Crisp A., Rossa M., Amatov T., and Carell T. (2019) Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science 366: 76. [DOI] [PubMed] [Google Scholar]
- Benner S.A., Kim H.-J., and Carrigan M.A. (2012) Asphalt, water and the prebiotic synthesis of ribose, ribonucleosides and RNA. Acc Chem Res 45:2025–2024 [DOI] [PubMed] [Google Scholar]
- Black R.A., Blosser M.C., Stottrup B.L., Tavaklev R., Deamer D.W., and Keller S.L. (2013) Nucleobases bind to and stabilize aggregates of a prebiotic amphiphile, providing a viable mechanism for the emergence of protocells. Proc Natl Acad Sci U S A 110:13272–13276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. (2011) Wetware: A Computer in Every Living Cell. Yale University Press, New Haven, CT [Google Scholar]
- Budin I. and Szostak J.W. (2010) Expanding roles for diverse physical phenomena during the origin of life. Annu Rev Biophys 39:245–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan M.P., Smith K.E., Cleaves H.J., Ruzicka J., Stern J.C., Glavin D.P., House C.H., and Dworkin J.P. (2011) Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc Natl Acad Sci U S A 108:13995–13998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.A., Lynne B.Y., Handley K.M., Jordan S., Farmer J.D., Guido D.M., Foucher F., Turner S., and Perry R.S. (2015) Tracing biosignature preservation of geothermally silicified microbial textures into the geological record. Astrobiology 15:858–882 [DOI] [PubMed] [Google Scholar]
- Campbell T.D., Febrian R., McCarthy J.T., Kleinschmidt H.E., Forsythe J.G., and Bracher P.J. (2019) Prebiotic condensation through wet–dry cycling regulated by deliquescence. Nat Commun 10: 4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelli D., Salley D., Henson A., Camarasa G.A., Sharabi S., Keenan G., and Cronin L. (2018) Networking chemical robots for reaction multitasking. Nat Commun 9:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.A., Roberts R.W., and Szostak J.W. (2004) The emergence of competition between model protocells. Science 305:1474–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyba C. and Sagan C. (1992) Endogenous production, exogenous delivery and impact-shock synthesis of organic molecules: an inventory for the origins of life. Nature 355:125–132 [DOI] [PubMed] [Google Scholar]
- Cody G.D., Boctor N.Z., Filley T.R., Hazen R.M., Scott J.H., Sharma A., and Yoder H.S. Jr. (2000) Primordial carbonylated iron-sulfur compounds and the synthesis of pyruvate. Science 289:1337–1340 [DOI] [PubMed] [Google Scholar]
- Corliss J.B., Baross J.A., and Hoffman S.E (1981) An hypothesis concerning the relationship between submarine hot springs and the origin of life on Earth. Oceanol Acta 4:59–69 [Google Scholar]
- Cornell C.E., Black R.A., Xue M., Litz H.E., Ramsay A., Gordon M., Mileant A., Cohen Z.R., Williams J.A., Lee K.K., Drobny G.P., and Keller S.L. (2019) Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc Natl Acad Sci U S A 116:17239–17244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva L., Maurel M.C., and Deamer D.W. (2014) Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J Mol Evol 80:86–97 [DOI] [PubMed] [Google Scholar]
- Dalai P. and Sahai N. (2018) Mineral-lipid interactions in the origins of life. Trends Biochem Sci 44:331–341 [DOI] [PubMed] [Google Scholar]
- Damer B.F. (2011) THE EVOGRID: an approach to computational origins of life endeavours (Doctoral dissertation). University College Dublin, Dublin, Ireland. doi:10..13140/RG.2.1.4800.7206. [Google Scholar]
- Damer B.F. (2016) A field trip to the Archaean in search of Darwin's warm little pond. Life (Basel) 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damer B.F. (2019) The Hot Spring Hypothesis for the Origin of Life and the Extended Evolutionary Synthesis. Essay for Extended Evolutionary Synthesis project. Available online at http://extendedevolutionarysynthesis.com/the-hot-spring-hypothesis-for-the-origin-of-life-and-the-extended-evolutionary-synthesis/ (accessed August25, 2019) [Google Scholar]
- Damer B.F. and Deamer D.W. (2015) Coupled phases and combinatorial selection in fluctuating hydrothermal pools: a scenario to guide experimental approaches to the origin of cellular life. Life (Basel) 5:872–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1871) Darwin Correspondence Project, “Letter No. 7471.” Available online at:www.darwinproject.ac.uk/DCP-LETT-7471 (accessed August25, 2019) [Google Scholar]
- Deamer D.W. (1985) Boundary structures are formed by organic compounds of the Murchison carbonaceous chondrite. Nature 317:792–794 [Google Scholar]
- Deamer D.W. (1992) Polycyclic aromatic hydrocarbons: primitive pigment systems in the prebiotic environment. Adv Space Res 12:183–189 [DOI] [PubMed] [Google Scholar]
- Deamer D.W. (2019) Assembling Life. Oxford University Press, Oxford, UK [Google Scholar]
- Deamer D.W. and Barchfeld G.L. (1982) Encapsulation of macromolecules by lipid vesicles under simulated prebiotic conditions. J Mol Evol 18:203–206 [DOI] [PubMed] [Google Scholar]
- Deamer D.W. and Damer B.F. (2017) Can life begin on enceladus? A perspective from hydrothermal chemistry. Astrobiology 17:834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.W. and Dworkin J.P. (2005) Chemistry and physics of primitive membranes. In Prebiotic Chemistry: From Simple Amphiphiles to Protocell Models, edited by P. Walde. Springer, Berlin, pp 1–27 [Google Scholar]
- Deamer D.W. and Pashley R. (1989) Amphiphilic components of the Murchison carbonaceous chondrite: surface properties and membrane formation. Orig Life Evol Biosphere 19:21–38 [DOI] [PubMed] [Google Scholar]
- Deamer D.W. and Weber A.L. (2010) Bioenergetics and life's origins. Cold Spring Harb Perspect Biol 2:a004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.W., Dworkin J.P., Sandford A. B ernstein M.P., and Allamandola L.J. (2002) The first cell membranes. Astrobiology 2:371–382 [DOI] [PubMed] [Google Scholar]
- Deamer D.W., Singaram S., Rajamani S., Kompanichenko V., and Guggenheim S. (2006) Self-assembly processes in the prebiotic environment. Philos Trans R Soc Lond B 361:1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D., Damer B., and Kompanichenko V. (2019) Hydrothermal chemistry and the origin of cellular life. Astrobiology 19:1523–1537 [DOI] [PubMed] [Google Scholar]
- DeGuzman V., Vercoutere W., Shenasa H., and Deamer D.W. (2014) Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J Mol Evol 78:251–262 [DOI] [PubMed] [Google Scholar]
- Del-Bem L.E. (2018) Xyloglucan evolution and the terrestrialization of green plants. New Phytol 219:1150–1153 [DOI] [PubMed] [Google Scholar]
- Djokic T., Van Kranendonk M.J., Campbell K.A., Walter M.R., and Ward C.R. (2020) Earliest signs of life on land preserved in ca. 3.5 Ga hot spring deposits. Nat Commun 8:16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djokic T., Van Kranendonk M.J., Campbell K.A., Havig J., Walter M.R., and Guido D.M. (2020) A reconstructed subaerial hot spring field in the ∼3.5 billion-year-old Dresser Formation, North Pole Dome, Pilbara Craton, Western Australia. Astrobiology 20: 000–000 [DOI] [PubMed] [Google Scholar]
- Djordjevic M. (2007) SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol Eng 24:179–189 [DOI] [PubMed] [Google Scholar]
- Dworkin J.P., Deamer D.W., Sandford S.A., and Allamandola L.J. (2001) Self-assembling amphiphilic molecules: synthesis in simulated interstellar/precometary ices. Proc Natl Acad Sci U S A 98:815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson F. (1999) Origins of Life. Cambridge University Press, Cambridge, UK [Google Scholar]
- Ehrenfreund P. and Sephton M.A. (2006) Carbon molecules in space: from astrochemistry to astrobiology. Faraday Discuss 133:277–288 [DOI] [PubMed] [Google Scholar]
- Eigen M. (1971) Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:465–523 [DOI] [PubMed] [Google Scholar]
- Erickson H.P., Anderson D.E., and Osawa M. (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenmoser A. (2007) The search for the chemistry of life's origin. Tetrahedron 63:12821–12844 [Google Scholar]
- Ferris J.P. (2006) Montmorillonite-catalysed formation of RNA oligomers: the possible role of catalysis in the origins of life. Philos Trans R Soc Lond B Biol Sci 361:1777–1786; discussion 1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkis M. (2007) Steps towards the formation of a protocell: the possible role of short peptides. Orig Life Evol Biosph 37:537–553 [DOI] [PubMed] [Google Scholar]
- Forsythe G., Yu S., Mamajanov I., Grover A., Krishnamurthy R., Fernandez M., and Hud N. (2015) Ester-mediated amide bond formation driven by wet-dry cycles: a possible path to polypeptides on the prebiotic Earth. Angew Chem Int Ed Engl 54:9871–9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe J.G., Petrov A.S., Millar W.C., Yu S.-S., Krishnamurthy R., Grover M.A., Hud N.V., and Fernández F.M. (2017) Surveying the sequence diversity of model prebiotic peptides by mass spectrometry. Proc Natl Acad Sci U S A 114: E7652–E7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox S.W. and Harada K. (1958) Thermal copolymerization of amino acids to a product resembling protein. Science 128:1214. [DOI] [PubMed] [Google Scholar]
- Gilbert W. (1986) The RNA World. Nature 319:618 [Google Scholar]
- Gogarten J.P. and Deamer D. (2016) Is LUCA a thermophilic progenote? Nat Microbiol 1:16229. [DOI] [PubMed] [Google Scholar]
- Gorrell I.B., Henderson T.W., Albdeery K., Savage P.M., and Kee T.P. (2017) Chemical transformations in proto-cytoplasmic media. phosphorus coupling in the silica hydrogel phase. Life (Basel) 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen J., Deamer D.W., Kros A., and Ehrenfreund P. (2012) Polycyclic aromatic hydrocarbons as plausible prebiotic membrane components. Orig Life Evol Biosph 42:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. (1964) The genetical evolution of social behaviour I and II. J Theor Biol 7:1–16, 17–52. [DOI] [PubMed] [Google Scholar]
- Handley K.H., Turner S.J., Campbell K.A., and Mountain B.W. (2008) Silicifying biofilm exopolymers on a hot-spring microstromatolite: templating nanometer-thick laminae. Astrobiology 4:747–770 [DOI] [PubMed] [Google Scholar]
- Hargrave M., Spencer S.K., and Deamer D.W. (2018) Computational models of polymer synthesis driven by dehydration/rehydration cycles: repurination in simulated hydrothermal fields. J Mol Evol 86:501–510 [DOI] [PubMed] [Google Scholar]
- Havig J.R. and Hamilton T.L. (2019) Hypolithic photosynthesis in hydrothermal areas and implications for cryptic oxygen oases on archean continental surfaces. Front Earth Sci 7:15 [Google Scholar]
- Herschy B., Whicher A., Camprubi E., Watson C., Dartnell L., Ward J., Evans J.R.G., and Lane N. (2014) An origin-of-life reactor to simulate alkaline hydrothermal vents. J Mol Evol 79:213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs P. (2016) The effect of limited diffusion and wet-dry cycling on reversible polymerization reactions: implications for pre biotic synthesis of nucleic acids. Life (Basel) 6:pii: E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himbert S., Chapman M., Deamer D.W., and Rheinstadter M.C. (2016) Organization of nucleotides in different environments and the formation of pre-polymers. Sci Rep 6:31285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordijk W., Steel M., and Kauffman S.A. (2019) Molecular diversity required for the formation of autocatalytic sets. Life (Basel) 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning D.P. and Joyce G.F. (2016) Amplification of RNA by an RNA polymerase ribozyme. Proc Natl Acad Sci U S A 113:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T. and Orgel L.E. (1982) Oligomerization of (guanosine 5'-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J Mol Biol 162:201–217 [DOI] [PubMed] [Google Scholar]
- Jackson J.B. (2016) Natural pH gradients in hydrothermal alkali vents were unlikely to have played a role in the origin of life. J Mol Evol 83:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.B. (2017) Ancient living organisms escaping from, or imprisoned in, the vents? Life (Basel) 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S.F., Rammu H., Zheludev I.N., Hartley A.M., Maréchal A., and Lane N. (2019) Promotion of protocell self-assembly from mixed amphiphiles at the origin of life. Nat Ecol Evol 3:1705–1714 [DOI] [PubMed] [Google Scholar]
- Joyce G. and Szostak J.W. (2018) Protocells and RNA self-replication. Cold Spring Harb. Perspect. Biol 10:pii: a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddour H., Gerisioglu S., Dalai P. M iyoshi T., Wesdemiotis C., and Sahai N. (2018) Non-enzymatic polymerization at the mineral-water interface: an insight into adsorption-polymerization relationships and the effects of magnesium. J Phys Chem C 122:29386–29397 [Google Scholar]
- Kauffman S. (2019) A World Beyond Physics: The Emergence and Evolution of Life. Oxford University Press, Oxford, UK [Google Scholar]
- Kee T.P. and Monnard P.-A. (2016) On the emergence of a proto-metabolism and the assembly of early protocells. Elements 12:419–424 [Google Scholar]
- Kelley D.S., Karson J.A., Früh-Green G.L., Yoerger D.R., Shank T.M., Butterfield D.A., Hayes J.M., Schrenk M.O., Olson E.J., Proskurowski G., Jakuba M., Bradley A., Larson B., Ludwig K., Glickson D., Buckman K., Bradley A.S., Brazelton W.J., Roe K., Elend M.J., Delacour A., Bernasconi S.M., Lilley M.D., Baross J.A., Summons R.E., and Sylva S.P. (2005) A Serpentinite-hosted ecosystem: the Lost City Hydrothermal Field. Science 307:1428–1434 [DOI] [PubMed] [Google Scholar]
- Kenrick P. and Crane P.R. (1997) The Origin and Early Diversification of Land Plants. A Cladistic Study. Smithsonian Institution Press, Washington & London [Google Scholar]
- Kompanichenko V.N., Poturay V.A., and Shlufman K.V. (2015) Hydrothermal systems of Kamchatka as the model for prebiotic environment. Orig Life Evol Biosph 45:93–103 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy R. (2018) Life's biological chemistry: a destiny or destination starting from prebiotic chemistry? Chem Eur J 24:16708. [DOI] [PubMed] [Google Scholar]
- Lahav N., White D., and Chang S. (1978) Peptide formation in the prebiotic era: thermal condensation of glycine in fluctuating clay environments. Science 201:67–69 [DOI] [PubMed] [Google Scholar]
- Lane N. and Martin W.F. (2012) The origin of membrane bioenergetics. Cell 151:1405–1416 [DOI] [PubMed] [Google Scholar]
- Lazcano A. (2010) Which way to life? Orig Life Evol Biosph 40:161. [DOI] [PubMed] [Google Scholar]
- Lehn J.M. (2002) Toward complex matter: supramolecular chemistry and self-organization. Proc Natl Acad Sci U S A 99:4763–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiner B.M., Vinton V.G., Clark D.D., Kahn R.E., Kleinrock L., Lynch D.C., Postel J., Roberts L.G., and Wolff S. (2009) A brief history of the internet. SIGCOMM Comput Commun Rev 39:22–31 [Google Scholar]
- Lohrmann R. and Orgel L.E. (1971) Urea-inorganic phosphate mixtures as prebiotic phosphorylating agents. Science 171:490–494 [DOI] [PubMed] [Google Scholar]
- Martin W. and Russell M.J. (2003) On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci 358:59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. and Russell M.J. (2007) On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc Lond B Biol Sci 362:1887–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Schreiber U., Davila M.J., Schmitz O.J., Bronia A., Meyer M., Klein J., and Meckelmann S.W. (2018) Molecular evolution in a peptide-vesicle system. Life (Basel) 8:pii: E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom T.M., Ritter G., and Simoneit B.R. (1999) Lipid synthesis under hydrothermal conditions by Fischer-Tropsch reactions. Orig Life Evol Biosph 29:153–166 [DOI] [PubMed] [Google Scholar]
- McKay C.P., Porco C.C., Altheide T., Davis W.L., and Kral T.A. (2008) The possible origin and persistence of life on Enceladus and detection of biomarkers in the plume. Astrobiology 8:909–919 [DOI] [PubMed] [Google Scholar]
- Milshteyn D., Damer B.F., Havig J., and Deamer D.W. (2018) Amphiphilic compounds assemble into membranous vesicles in hydrothermal hot spring water but not in seawater. Life (Basel) 8:pii: E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milshteyn D., Cooper G., and Deamer D. (2019) Chemiosmotic energy for primitive cellular life: proton gradients are generated across lipid membranes by redox reactions coupled to meteoritic quinones. Sci Rep 9:12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misuraca L., Natali F., da Silva L., Peters J., Demé B., Ollivier J., Seydel T., Laux-Lesourd V., Haertlein M., Zaccai G., Deamer D.W., and Maurel M.C. (2017) Mobility of a mononucleotide within a lipid matrix: a neutron scattering study. Life (Basel) 7:pii: E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard P.-A. and Deamer D.W. (2002) Membrane self-assembly processes: steps toward the first cellular life. Anat Rec 268:196–207 [DOI] [PubMed] [Google Scholar]
- Morowitz H. (1992) Beginnings of Cellular Life: Metabolism Recapitulates Biogenesis. Yale University Press, New Haven and London [Google Scholar]
- Mulkidjanian A., Bychkov A., Dibrova D., Galperin M., and Koonin E. (2012) Origin of first cells at terrestrial, anoxic geothermal fields. Proc Natl Acad Sci U S A 109:E821–E830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungi C.V. and Rajamani S. (2015) Characterization of RNA-like oligomers from lipid-assisted nonenzymatic synthesis: implications for origin of informational molecules on early earth. Life (Basel) 5:65–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungi C.V., Singh S.K., Chugh J., and Rajamani S. (2016) Synthesis of barbituric acid containing nucleotides and their implications for the origin of primitive informational polymers. Phys Chem Chem Phys 18:20144–20152 [DOI] [PubMed] [Google Scholar]
- Mungi C.V., Bapat N.V., Hongo Y., and Rajamani S. (2019) Formation of abasic oligomers in nonenzymatic polymerization of canonical nucleotides. Life (Basel) 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan S., Wiedenbeck M., Milke R., and Heubeck C. (2016) Biogenic overgrowth on detrital pyrite in ca. 3.2 Ga Archean paleosols. Geology 44:763 [Google Scholar]
- Nam I., Nam H.G., and Zare R.N. (2018) Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets. Proc Natl Acad Sci U S A 115:36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naraoka H., Shimoyama A., and Harada K. (1999) Molecular distribution of monocarboxylic acids in Asuka carbonaceous chondrites from Antarctica. Orig Life Evol Biosph 29:187–201 [DOI] [PubMed] [Google Scholar]
- Neuman C.M., Maxwell C.D., and Boulton J.W. (1996) Wind transport of sand surfaces crusted with photoautotrophic microorganisms. CATENA 27:229–247 [Google Scholar]
- Odling-Smee J., Laland K., and Feldman M. (2003) Niche Construction: The Neglected Process in Evolution. Princeton University Press, Princeton and Oxford [Google Scholar]
- Olasagasti F., Kim H., Pourmand N., and Deamer D.W. (2011) Non-enzymatic transfer of sequence information under plausible prebiotic conditions. Biochimie 93:556–561 [DOI] [PubMed] [Google Scholar]
- Oparin A.I. (1962) LIFE: Its Nature, Origin and Development. Academic Press, Inc., New York, p 19 [Google Scholar]
- Orgel L.E. (1968) Evolution of the genetic apparatus. J Mol Biol 38:381–393 [DOI] [PubMed] [Google Scholar]
- Orgel L.E. (2000) Self-organizing biochemical cycles. Proc Natl Acad Sci U S A 97:12503–12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L.E. (2008) The implausibility of metabolic cycles on the prebiotic Earth. PLoS biology 6, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang T.Y. and Maslov S. (2013) Universality of component frequency distributions. Proc Natl Acad Sci U S A 110: 6235–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papineau D., Walker J.J., Mojzsis S.J., and Pace N.R. (2005) Composition and Structure of Microbial Communities from Stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl Environ Microbiol 71: 4822–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B.C., Simmons G.M., Wharton R.A., Seaburg K.G., and Love F.G. (1982) Removal of organic and inorganic matter from Antarctic lakes by aerial escape of bluegreen algal mats. J Phycol 18:72–78 [Google Scholar]
- Parker E.T., Cleaves H.J., Dworkin J.P., Glavin D.P., Callahan M., Aubrey A., Lazcano A., and Bada J.L. (2011) Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proc Natl Acad Sci U S A 108:5526–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce B.K.D., Pudritz R.E., Semenov D.A., and Henning T.K. (2017) Origin of the RNA World: the fate of nucleobases in warm little ponds. Proc Natl Acad Sci U S A 114:11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce B.K.D., Tupper A.S., Pudritz R.E., and Higgs P.G. (2018) Constraining the time interval for the origin of life on earth. Astrobiology 18:343–364 [DOI] [PubMed] [Google Scholar]
- Peterson K.J., Cotton J.A., Gehling J.G., and Pisani D. (2008) The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos Trans R Soc Lond B Biol Sci 363:1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt J.R. (1964) Strong inference. Science 146:347–353 [DOI] [PubMed] [Google Scholar]
- Popper K.R. (1963) Conjectures and Refutations: The Growth of Scientific Knowledge. Routledge, London. ISBN 0-415-04318-2 [Google Scholar]
- Powner M.W. and Sutherland J.D. (2011) Prebiotic chemistry: a new modus operandi. Philos Trans R Soc Lond B Biol Sci 366:2870–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powner M.W., Gerland B., and Sutherland J.D. (2009) Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459:239–242 [DOI] [PubMed] [Google Scholar]
- Pross A. (2012) What Is Life?: How Chemistry Becomes Biology. Oxford University Press, Oxford, UK [Google Scholar]
- Rajamani S., Vlassov A., Benner S., Coombs A., Olasagasti F., and Deamer D.W. (2008) Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig Life Evol Biosph 38:57–74 [DOI] [PubMed] [Google Scholar]
- Ranjan S., Todd Z.R., Rimmer P.B., Sasselov D.D., and Babbin A.R. (2019) Nitrogen oxide concentrations in natural waters on early Earth. Geochem Geophys Geosyst 20:202–2039 [Google Scholar]
- Rodriguez-Garcia M., Suran A.J., Cooper G.J.T., Suarez-Marina I., Hosni Z., Lee M.P., and Cronin L. (2015) Formation of oligopeptides in high yield under simple programmable conditions. Nat Commun 6:8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. (2018) It is neither frankenstein nor a submarine alkaline vent, it is just the second law. Bioessays 40:e1800149. [DOI] [PubMed] [Google Scholar]
- Ross D. and Deamer D.W. (2016) Dry/wet cycling and the thermodynamics and kinetics of prebiotic polymer synthesis. Life (Basel) 6:pii: E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff S. and Farmer J. (2016) Silica deposits on Mars with features resembling hot spring biosignatures at El Tatio in Chile. Nat Commun 7:13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushdi A.I. and Simoneit B.R. (2006) Abiotic condensation synthesis of glyceride lipids and was esters under simulated hydrothermal conditions. Orig Life Evol Biosph 36:93–108 [DOI] [PubMed] [Google Scholar]
- Russell M.J. and Hall A.J. (1997) The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J Geol Soc London 154:377–402 [DOI] [PubMed] [Google Scholar]
- Russell M.J., Daniel R.M., and Hall A. (1993) On the emergence of life via catalytic iron-sulphide membranes. Terra Nova 5:343–347 [Google Scholar]
- Russell M.J., Barge M., Bhartia R., Bocanegra D., Bracher P.J., Branscomb E., Kidd R., McGlynn S., Meier D.H., Nitschke W., Shibuya T., Vance S., White L., and Kanik I. (2014) The drive to life on wet and icy worlds. Astrobiology 14:308–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Pohorille A., and Chen I.A. (2014) Molecular crowding and early evolution. Orig Life Evol Biosph 44:319. [DOI] [PubMed] [Google Scholar]
- Satkoski A.M., Lowe D.R., Beard B.L., Coleman M.L., and Johnson C.M. (2016) A high continental weathering flux into Paleoarchean seawater revealed by strontium isotope analysis of 3.26 Ga barite. Earth Planet Sci Lett 454:28 [Google Scholar]
- Segre D., Deamer D.W., and Lancet D. (2001) The lipid world. Orig Life Evol Biosph 31:119–145 [DOI] [PubMed] [Google Scholar]
- Sojo V., Ohno A., McGlynn S.E., Yamada Y.M., and Nakamura R. (2019) Microfluidic reactors for carbon fixation under ambient-pressure alkaline-hydrothermal-vent conditions. Life (Basel) 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D.P. and Rodoni D. (2015) Vesicle encapsulation of a nonbiological photochemical system capable of reducing NAD+ to NADH. Langmuir 31:10633–10637 [DOI] [PubMed] [Google Scholar]
- Sutherland J.D. (2017) Opinion: studies on the origin of life—the end of the beginning. Nat Rev Chem 1:0012 [Google Scholar]
- Szostak J.W. (2016) On the origin of life. Medicina (B Aires) 76:199–203 [PubMed] [Google Scholar]
- Toppozini L., Dies H., Deamer D.W., and Rheinstädter M.C. (2013) Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: insights into assembly of nucleic acid for primitive life. PLoS One 8:e62810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J.T., Gerald H., and Pollack G.H. (2005) Hypothesis: the origin of life in a hydrogel environment. Prog Biophys Mol Biol 89:1–8 [DOI] [PubMed] [Google Scholar]
- Tuck A. (2002) The role of atmospheric aerosols in the origin of life. Surv Geophys 23:379 [Google Scholar]
- Van Kranendonk M.J. (2010) Two types of Archean continental crust: plume and plate tectonics on early Earth. Am J Sci 2010:1187–1209 [Google Scholar]
- Van Kranendonk M.J., Philippot P., Lepot K., Bodorkos S., and Pirajno F. (2008) Geological setting of Earth's oldest fossils in the ca. 3.5 Ga Dresser Formation, Pilbara Craton, Western Australia. Precambrian Res 167:93–124 [Google Scholar]
- Van Kranendonk M.J., Baumgartner R., Djokic T., Ota T., Steller L., Garbe U., and Nakamura E. (2020) Elements for the origin of life on land: a deep-time perspective from the Pilbara Craton of Western Australia. Astrobiology, in press, doi:10..1089/ast.2019.2107. [DOI] [PubMed] [Google Scholar]
- Vlassov A., Khvorova A., and Yarus M. (2001) Binding and disruption of phospholipid bilayers by supramolecular RNA complexes. Proc Natl Acad Sci U S A 98:7706–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G. (2016) In praise of error. J Mol Evol 82:75–80 [DOI] [PubMed] [Google Scholar]
- Walker S.I., Grover M.A., and Hud N.V. (2012) Universal sequence replication, reversible polymerization and early functional biopolymers: a model for the initiation of prebiotic sequence evolution. PLoS One 7:e34166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Buick R., and Dunlop J. (1980) Stromatolites 3,400–3,500 Myr old from the North Pole area, Western Australia. Nature 284:443–445 [Google Scholar]
- Weiss M.C., Sousa F.L., Mrnjavac N., Neukirchen S., Roettger M., Nelson-Sathi S., and Martin W.F. (2016) The physiology and habitat of the last universal common ancestor. Nat Microbiol 1:16116. [DOI] [PubMed] [Google Scholar]
- Westall F., Hickman-Lewis K., Hinman N., Gautret P., Campbell K.A., Bréhéret J.G., Foucher F., Hubert A., Sorieul S., Dass A.V., Kee T.P., Georgelin T., and Brack A. (2018) A hydrothermal-sedimentary context for the origin of life. Astrobiology 18:259–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R., Wamberg M.C., Albertsen A.N., Löffler P.M.G., and Monnard P.A. (2013) Bottom–up protocell design: gaining insights in the emergence of complex functions. In: Evolutionary Biology: Exobiology and Evolutionary Mechanisms, edited by P. Pontarotti. Springer, Berlin, Heidelberg, pp 81–94 [Google Scholar]
- Wochner A., Attwater J., Coulson A., and Holliger P. (2011) Ribozyme-catalyzed transcription of an active ribozyme. Science 332:209–212 [DOI] [PubMed] [Google Scholar]
- Woese C.R. (1998) The universal ancestor. Proc Natl Acad Sci U S A 12:6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R. (2002) On the evolution of cells. Proc Natl Acad Sci U S A 99:8742–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R. and Fox G.E. (1977) The concept of cellular evolution. J Mol Evol 10:1–6 [DOI] [PubMed] [Google Scholar]
- Wu L.-F. and Sutherland J.D. (2019) Provisioning the origin and early evolution of life. Emerg Top Life Sci ETLS20190011, doi:10..1042/ETLS20190011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.-S., Solano M., Blanchard M.K., Soper-Hopper M., Krishnamurthy R., Fernandez F.M., Hud N.V., Schork F.J., and Grover M.A. (2017) Elongation of model prebiotic proto-peptides by continuous monomer feeding. Macromolecules 50:9286–9294 [Google Scholar]