Abstract
Background: Patients with sepsis exhibit significant long-term immunosuppressive sequelae. Monocyte dysfunction is a hallmark of this damage. Circulating exosomes are an important mediator of the systemic signaling events that occur during the septic response; thus, we sought to characterize the contribution of circulating exosomes to the inflammatory process induced during sepsis
Methods: Monocyte-derived exosomes were isolated from cultured monocytes from healthy adult donors via stimulation with lipopolysaccharide (LPS) or phosphate-buffered saline (PBS). The proteome was determined by capillary-liquid chromatography–nanospray tandem mass spectrometry (capillary–LC/NT/MS). Using pathway analysis, proteomic networks of exosomes derived from LPS-stimulated monocytes were compared with those isolated from patients with surgical sepsis. Naïve monocytes were then treated with these exosomes and stimulated with LPS to determine the effects on recipient-cell immune function.
Results: Proteomic analysis demonstrated 18 differentially expressed proteins (17 down-regulated, one up-regulated) in sepsis-derived exosomes, with 15 differentially expressed proteins (14 down-regulated, one up-regulated) in the LPS-stimulated exosomes. Functional enrichment analysis demonstrated several down-regulated processes, including localization, biogenesis, and metabolic and cellular processes in addition to immune system processes. In LPS-stimulated macrophages, similar down-regulated processes were seen, including metabolic and cellular processes, as well as the response to stimulus. Cells treated with sepsis-derived exosomes or exosomes from LPS-stimulated monocytes demonstrated significant reductions in tumor necrosis factor (TNF)-α generation in response to LPS stimulation.
Conclusions: Proteomic analysis of sepsis-derived exosomes and LPS-stimulated, macrophage-derived exosomes exhibited down-regulation of several important protein networks, including the immune response. In addition, human monocytes treated with exosomes from patients with sepsis or LPS-stimulated monocytes demonstrated significant reductions in TNF-α generation in response to LPS stimulation. These data suggest the contribution of circulating exosomes to systemic signaling and immunomodulation during sepsis.
Keywords: anti-inflammation, compensatory anti-inflammatory response syndrome (CARS), exosomes, immunosuppression, proteomics, sepsis, systemic inflammatory response syndrome (SIRS)
The septic process represents a disease manifested by sequential release of opposing pro- and anti-inflammatory mediators [1–3]. These responses are not mutually exclusive and represent a continuum of immune regulation that evolves from acute inflammation to either a counter-inflammatory resolution or persistent immunoparalysis. The magnitude of both the pro- and anti-inflammatory or immunoparalysis phases correlates with death [4,5]. Deaths within the first 30 days carry a bimodal distribution consisting of early acute deaths followed by delayed deaths [6]. Patients who survive the initial septic insult exhibit significant morbidity and more long-term deaths [6]. These patients can exhibit a characteristic set of symptoms referred to as “persistent inflammatory, immunosuppressed catabolic syndrome” (PICS) [7,8]. Patients with PICS show dramatically reduced survival for at least five years after their septic insult compared with age- and co-morbidity-matched control patients, suggesting a persistent and potentially permanent alteration in the function of these patients [9]. This “post-sepsis immunosuppression” is manifested by significant reductions in the ability to generate pro-inflammatory cytokines in response to inflammatory stimuli [1,10,11].
Monocyte dysfunction has long been recognized as a hallmark of this immunosuppression [12,13]. These cells demonstrate significant reductions in expression of tumor necrosis factor TNF-α, interleukin (IL)-6, and human leukocyte antigen HLA-DR [14,15]. In patients with sepsis, <5% of isolated monocytes manifest a normal response to exogenous inflammatory stimuli, suggesting a profound systemic effect [13,15]. A key piece of missing information is how this immunosuppressive response is amplified systemically.
Exosomes are small, biologically active fragments of cells that are produced in a regulated manner and circulate in the blood [16,17]. They are manufactured in response to many stimuli, from many cell types, and contain proteins and nucleic acid, including small regulatory RNA molecules (miRNAs), which all are important regulators of recipient cell function [18–21]. During sepsis, several cellular targets are engaged by circulating exosomes that lead to worsening immunosuppression, including down-regulation of Toll-like receptor (TLR)4 expression, increased signal transducers and activators of transcription (STAT)3 signaling, and reduced nuclear factor (NF)-Kβ signaling [21,22]. These data suggest an anti-inflammatory role for circulating exosomes during sepsis.
The aim of the current study was to examine the functional role of circulating exosomes and their protein cargo in the transmittance of this systemic signal of immunosuppression. Accordingly, we generated exosomes from human monocytes in response to lipopolysaccharide (LPS) stimulation and performed proteomic analysis to identify robust gene networks that might be targeted by these exosomes. We compared these protein networks with exosomes isolated from patients with sepsis to identify clinical consistency within our in vitro model. In order to assess the effect of these exosomes on recipient-cell immune function, we treated naïve isolated monocytes with exosomes from these LPS-stimulated monocytes and sepsis-derived exosomes and subjected these cells to additional LPS stimulation. We measured TNF-α concentrations in response to LPS stimulation as an indicator of immunosuppression. Our pathway analysis data showed that sepsis is associated with changes in immunodeficiency signaling, acute-phase signaling, and coagulopathy, suggesting that circulating exosomes and, specifically, monocyte-derived exosomes promote immunosuppressive signaling in recipient cells. Additionally, we saw significant reductions in TNF-α expression in response to LPS stimulation in cells conditioned with exosomes from LPS-treated monocytes or patients with sepsis.
Materials and Methods
In vitro exosome generation
Human peripheral blood mononuclear cell (PBMC) isolation was performed using fresh blood leukocyte source packs (American Red Cross, Columbus, OH) through Ficoll-Hypaque density gradient centrifugation (GE Healthcare). Monocytes were selected using magnetic beads conjugated with CD14 antibody (Miltenyi Biotec, Auburn, CA). These monocytes were suspended overnight in Roswell Park Memorial Institute (RPMI) 1640 medium at a density of 4 × 106 cells/mL in 12-well culture plates in a humidified atmosphere (5% CO2, 37°C). These cells were stimulated for 12 hours with 6 ng/mL of LPS or an equal volume of phosphate-buffered saline (PBS) as an in vitro model of sepsis. Media were collected from these cells, and exosomes were isolated for proteomic analysis. Briefly, 63 mcL of Exoquick Exosome Precipitation Solution was added to 250 mcL of medium collected from LPS- or PBS-treated cells. This mixture was then incubated on a roller at 4°C for two hours and subjected to centrifugation at 1,500 × g for 30 minutes. Supernatant liquid was aspirated, and the pellet was subjected to centrifugation at 1,500 × g for 5 minutes. The remaining supernatant liquid was removed, and the exosome pellet was used for protein isolation and mass spectrometry.
Protein extraction and digestion
The exosome pellets were resuspended in a volume of 100 mcL (50 mcL for control cells) of 50 mM ammonium bicarbonate (ABC) lysis buffer containing 0.1% Rapigest (Waters Corporation, Milford, MA), followed by sonication twice for 10 seconds and incubation for one hour at room temperature. Extracts were clarified at 13,000 rpm in a microcentrifuge, and the concentration was calculated by standard Bradford assay. The extract was digested with trypsin (protein:enzyme 30:1) at a final concentration of 0.1% Rapigest. Briefly, 5 mcL of 50 mM ABC containing dithiothreitol 5 mcg/mcL was added followed by incubation at 65°C for 15 minutes. Then 5 mcL of 50 mM ABC buffer containing iodoacetamide 15 mcg/mcL was added and incubated for 15 minutes before the addition of sequencing-grade trypsin digestion, which was performed overnight at 37°C. The following day, trifluoroacetic acid (0.5% final concentration) was added and kept at 37°C for 30 minutes to precipitate the Rapigest. The sample was clarified at 13,000 rpm for five minutes in a microcentrifuge, dried in a Vacufuge, and reconstituted to a volume of 20 mcL of inacetic acid (50 mM). Nanodrop (A280nm) was used to calculate the peptide concentration.
Orbitrap fusion proteomics
Protein identification and measurement were performed via ionization by the Orbitrap Fusion mass spectrometer according to the manufacturer protocols (Fisher Thermo Scientific, Waltham, MA) [23]. The injected peptides were separated using a nano column (EASY-SprayTM, PepmapTM RSLC, C18 3 mc 100A, 75 mcm × 150mm; Thermo Scientific). Samples were desalted with 0.1% formic acid for five minutes. Mobile phases A and B were 0.1% formic acid and acetonitrile, respectively. Gradients for phase B were 2%–35% for more than 220 minutes, 35%–55% over 50 minutes, 55%–90% over five minutes, and 90% over five minutes, which then were equilibrated at 2% for 20 minutes. Data were obtained at a spray voltage of 1.7kV, with a capillary temperature of 275°C. The scan was recorded in FT mode, between 400–1600 m/z, with the resolution set at 120,000 and a ACG target of 2 × 105 to determine the amino acid sequences of the most prominent spectral peaks. The collision-induced dissociation (CID; 2+–3+) and escalating density training (ETD; 4+–6+) methods were used to assess MSn spectra, with the CID fragmentation energy set at 35%.
Sequence information was processed as previously described [24]. Briefly, mgf files were analyzed using Mascot Daemon (Martix Science 2.3.2, Boston MA). We then searched this against the Uniprot Human Database (version 12032015). Mass accuracy was set at 10 ppm and fragment mass tolerance at 0.5 Da. Several variable modifications, including oxidation, deamidation, and carbamidomethylation, were considered. Four missed cleavages were permitted. In order to determine the false discovery rate (FDR), we searched a decoy database, which allows us to filter peptides. We set the significance threshold at p < 0.05 with bold red peptides required for valid identification. We then set the FDR to <1% with a minimum of two significant peptides to be considered valid.
We performed label-free quantitation using the spectral count approach. We then compared the number of NT/MS spectra identified from the same protein in multiple LC/NT/MS datasets in order to provide relative quantification. The Student t-test was performed by scaffold to evaluate whether the folder change for certain proteins was significant (p < 0.05).
Patient information
All patients or their surrogates provided written informed consent. Patient samples were obtained in accordance with Institutional Review Board No. 2010H0281 at The Ohio State University Wexner Medical Center. Control patients included critically ill, non-septic patients (n = 4). These patients were defined as those in the intensive care unit requiring intubation, but without infection or signs of systemic inflammation (SIRS criteria). Patients with sepsis were defined as those presenting with an infection and signs of organ dysfunction in accordance with the Third International Consensus Definitions for Sepsis and Septic Shock (n = 4) [25]. The patients used for this investigation all had sepsis with a Sequential Organ Failure Assessment (SOFA) score >8.
Exosome isolation
Total plasma exosomes were isolated from patients immediately after plasma collection at day 1 of diagnosis. Approximately 8 mL of blood was collected in ethylene diamine tetraacetic acid vials, and the plasma was isolated by layering blood over Ficoll-Paque (GE Healthcare Cat. No. 17-5442-03) followed by centrifugation at 2,000 rpm for 20 minutes at room temperature. Exosomes from plasma and culture were isolated using Exoquick Exosome Precipitation reagent (System Biosciences Inc., Mountain View, CA). A one-fourth volume of Exoquick solution was mixed with plasma samples followed by overnight incubation at 4°C. They were then centrifuged at 1,500 × g for 30 minutes. The supernatant liquid was aspirated, and the pellet was centrifuged at 1,500 × g for 5 minutes. The remaining supernatant liquid was removed. The exosome pellet was resuspended in 500 mcL of sterile PBS. Exosome size and concentration were confirmed using the Nanosight NS300 (Malvern Panalytical Technologies, Egham, Surrey, UK).
Monocyte immunosuppression testing
Human monocytes were obtained from healthy donors via CD 14 bead selection as described above. Monocytes were cultured to confluence in RPMI 1640 medium and treated with either sepsis-derived exosomes or exosomes from LPS-stimulated monocytes, healthy patients, or non-stimulated monocytes. Samples were incubated for 12 hours, then the medium was washed off and replaced with fresh RPMI 1640. The cell cultures were then challenged with LPS (6 ng/mL) for four hours to test the degree of immunosuppression present as described in the literature [26]. Concentrations of TNF-α were assessed using a commercial immunoassay as a measure of immune function (Bio-Rad, Cat. No. 171-BK55MR2). All standards, quality controls, and samples were analyzed in duplicate. The lower limit of detection for TNF-α was 0.9 pg/mL.
Data analysis
Proteome Discoverer 1.4 was used to analyze the data as described in our previous report [27]. To compare proteins packaged in the exosomes derived from individuals with sepsis, the proteins obtained were subjected to Hierarchical Clustering and heatmap visualization software from Genesis (http://genome.tugraz.at/). In addition, PANTHER (http://pantherdb.org/) was used for gene ontology analysis and broad characterization of proteins packaged in sepsis exosomes. Search tool for the retrieval of interacting genes/proteins (STRING) (v.10.5) was used for analyzing protein–protein interactions enriched in the dataset using medium confidence (0.4) (http://string-db.org/).
IPA analysis
The Ingenuity Pathway Analysis (IPA®, QIAGEN, Redwood City, CA; www.qiagen.com/ingenuity) is a database that predicts biological networks enriched in the dataset based on the existing literature. We used this bioinformatics tool to generate a functional network of proteins that were down-regulated in exosomes derived from critically ill individuals or cultured primary monocytes exposed to LPS.
Results
Exosome characterization
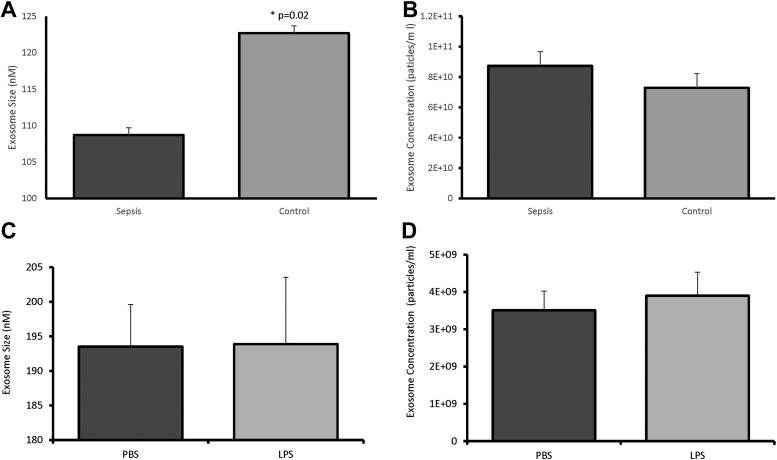
Initial characterization of exosomes isolated from the plasma of patients with sepsis or critically ill patients without sepsis demonstrated a difference in average size (108.7 ± 22.4nm versus 122.7 ± 20.2nm, respectively; p = 0.02; Fig. 1A), but no difference was seen in concentration (8.73 ± 4.9 × 1010 versus 7.29 ± 4.49 × 1010 particles/mL; p = 0.37; Fig. 1B). Exosomes generated from LPS-stimulated monocytes did not differ in size (193.9 ± 21.6nm versus 193.5 ± 10.6nm; p = 0.97; Fig. 1C) or concentration (3.51 ± 0.9 × 109 particles/ml versus 3.9 ± 1.4 × 109 particles/mL; p = 0.69, respectively; Fig. 1D) compared with exosomes derived from monocytes treated with PBS. In addition to size and concentration characterization, proteomic analysis identified significant amounts of the cytosolic protein actin in all monocyte samples without significant differences in expression (fold change 0.68; p = 0.33; Table 1). Actin also was identified in patient samples without a significant difference between patients with and without sepsis (fold change 8.0; p = 0.351; Table 2).
FIG. 1.
Size and concentration of exosomes derived from patients with sepsis (A), critically ill controls (B), and LPS-stimulated (C) and PBS-treated (D) monocytes. In exosomes derived from patients with sepsis, there was a significant decrease in size (A) compared with control patients (108.7 ± 22.4nm vs. 122.7 ± 20.2nm; p = 0.02), but no difference was seen in concentration (B) (8.73 ± 4.9 × 1010 vs. 7.29 ± 4.49 × 1010 particles/mL; p = 0.37). Exosomes generated from LPS-stimulated monocytes did not differ in size (C) (193.9 ± 21.6nm vs. 193.5 ± 10.6nm, p = 0.97) or concentration (D) (3.51 ± 0.9 × 109 particles/mL vs. 3.9 ± 1.4 × 109 particles/mL; p = 0.69)
Table 1.
Changes in Protein Expression in Lipopolysaccharide (LPS) vs. Phosphate-Buffered Saline (PBS)-Treated Monocytes
| Protein | Fold Change (LPS/PBS) | P Value |
|---|---|---|
| Lactotransferrin | 0.20 | 0.007 |
| Prothrombin | 0.43 | 0.008 |
| Phosphoglycerate kinase 1 | 0.35 | 0.010 |
| Vitronectin | 0.40 | 0.013 |
| Thrombospondin-4 | 0.33 | 0.016 |
| Transforming growth factor-β-induced protein | 1.80 | 0.016 |
| Alpha-2-macroglobulin | 0.69 | 0.024 |
| Inter-α-trypsin inhibitor heavy chain H3 | 0.61 | 0.025 |
| Heat shock protein HSP 90-α | 0.10 | 0.033 |
| Apolipoprotein B-100 | 0.31 | 0.033 |
| Filamin-A | 0.07 | 0.041 |
| Coagulation factor V | 0.38 | 0.041 |
| Glyceraldehyde-3-phosphate dehydrogenase | 0.32 | 0.043 |
| Thrombospondin-1 | 0.32 | 0.046 |
| Complement C3 | 0.38 | 0.049 |
| Albumin | 1.63 | 0.163 |
| Actin | 0.68 | 0.330 |
Fifteen proteins were differentially expressed in LPS-stimulated monocytes, one of which, transforming growth factor-β induced (TGFBI), was up-regulated.
Table 2.
Changes in Protein Expression in Sepsis vs. Control Patients
| Protein | Fold Change (Sepsis/Control) | P Value |
|---|---|---|
| Histidine-rich glycoprotein | 0.43 | 0.001 |
| Immunoglobulin heavy constant γ 2 | 0.50 | 0.003 |
| Immunoglobulin heavy constant γ 4 | 0.44 | 0.005 |
| Ig λ-2 chain C regions | 0.49 | 0.006 |
| Immunoglobulin heavy constant γ 1 | 0.49 | 0.012 |
| Immunoglobulin heavy constant γ 3 | 0.51 | 0.014 |
| Complement C4-A | 0.64 | 0.016 |
| Coagulation factor XI | 0.61 | 0.016 |
| Kallikrein | 0.15 | 0.018 |
| Complement C4-B | 0.67 | 0.020 |
| C-reactive protein | 1.53 | 0.024 |
| Immunoglobulin κ variable 1-33 | 0.50 | 0.024 |
| Immunoglobulin λ-like polypeptide 5 | 0.33 | 0.028 |
| Clusterin | 0.48 | 0.031 |
| Immunoglobulin heavy constant αj 1 | 0.64 | 0.039 |
| Complement C3 | 0.48 | 0.040 |
| Selenoprotein P | 0.20 | 0.047 |
| Plasminogen | 0.47 | 0.048 |
| Albumin | 0.95 | 0.812 |
| Actin | 8.00 | 0.351 |
Eighteen proteins were differentially expressed when patients with sepsis were compared with control patients. Of the 18 proteins, 17 were down-regulated and one, C-reactive protein, was up-regulated.
To confirm the purity of the monocyte-derived exosome samples, albumin concentrations were assessed. There was no change in the expression of albumin in exosomes derived from monocytes stimulated with LPS compared with exosomes derived from monocytes treated with PBS (fold change 1.63; p = 0.163; Table 1). There also was no difference in expression of albumin in patients with sepsis compared with control patients (fold change 0.95; p = 0.812; Table 2).
Protein detection
In the exosome samples from patients with sepsis (n = 4), a total of 198 proteins were detected. Eighteen proteins were expressed differentially when patients with sepsis were compared with control patients. Of the 18 proteins, 17 were down-regulated and one, C-reactive protein, was up-regulated (Table 2). Exosomes generated from human monocytes treated in vitro with LPS exhibited similar characteristics, albeit with some differences in protein profiles. In sum, 192 proteins were detected, of which 14 were down-regulated and one, transforming growth factor β induced (TGFBI), was up-regulated compared with PBS-treated monocytes (Table 1).
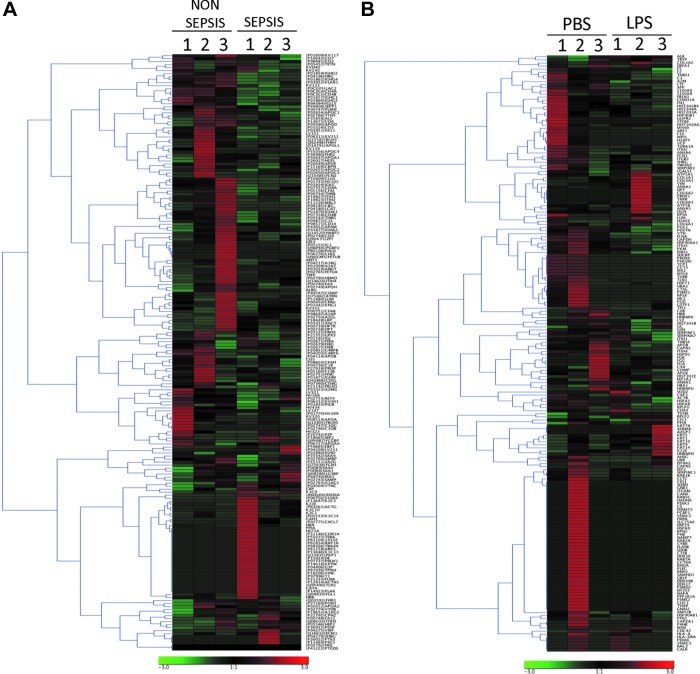
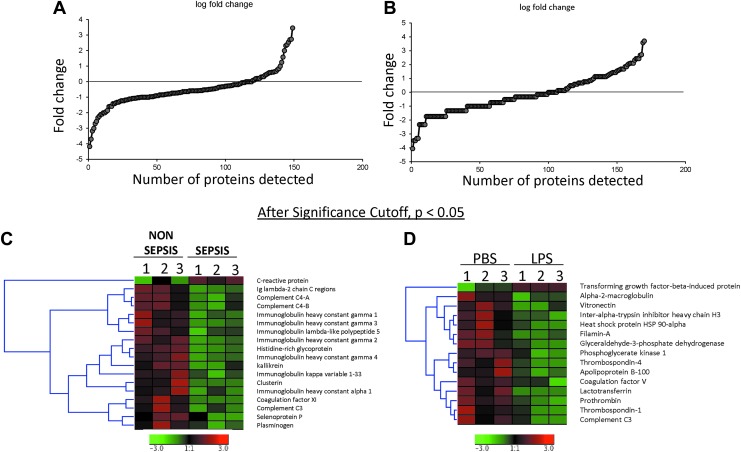
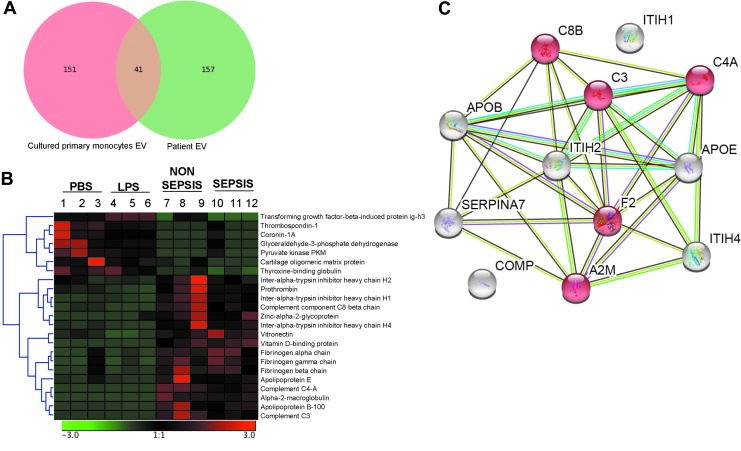
Proteins obtained through LC/NT/MS were subjected to clustering analysis using Genesis software (Fig. 2). Of all the proteins detected, the majority were found in the log ratio of a ±2 range (Fig. 3A, B). After applying a significance cutoff of <0.05, heat map data were generated (Fig. 3C, D). The amount of overlap of the proteins expressed in exosomes from human monocytes and patients with sepsis was then compared. A total of 41 proteins were detected across all groups (Fig. 4A), of which 23 were expressed differentially between human monocytes and cells from patients with sepsis (Fig. 4B). After fold change analysis, 12 proteins were found to be down-regulated in the exosomes of septic patients as well as in monocytes exposed to LPS (Fig. 4C). String analysis demonstrated these proteins belong to the complement and coagulation cascades (Fig. 4C).
FIG. 2.
Hierarchical cluster images illustrating proteins quantified in exosomes obtained from septic patients (A) and critically ill subjects (B) and cultured primary monocytes exposed to lipopolysaccharide or phosphate-buffered saline created using Genesis software. Protein expression data were obtained using liquid chromatography–nanospray tandem mass spectrometry. A scale representing fold change is at the bottom.
FIG. 3.
Log ratio distribution of all quantitated proteins present in exosomes obtained from septic patients (A) and critically ill subjects (B) and cultured primary monocytes exposed to lipopolysaccharide (LPS) or phosphate-buffered saline (PBS). Hierarchical cluster images illustrating proteins up-regulated or down-regulated in the exosomes obtained from septic patients (C) and critically ill subjects (D) and cultured primary monocytes exposed to LPS or PBS created using Genesis software.
FIG. 4.
Protein expression. (A) Vein diagram illustrating proteins detected in exosomes obtained from cultured primary monocytes exposed to lipopolysaccharide or phosphate-buffered saline (red) and septic patients compared with critically ill subjects (green). Protein expression data were obtained using liquid chromatography–nanospray tandem mass spectrometry. A total of 41 proteins were detected in both analyses. (B) Hierarchical clustering analysis of significant proteins (one-way analysis of variance; p < 0.05) detected both in the exosomes obtained from cultured primary monocytes exposed to lipopolysaccharide (LPS) or phosphate-buffered saline (left) and septic patients compared with critically ill subjects (right). (C) STRING version 10.5 was used to construct protein–protein interaction network of common, significantly down-regulated proteins in exosomes obtained from septic patients compared with cultured primary monocytes exposed to LPS. Line or strings indicate protein interactions.
Enrichment Analysis
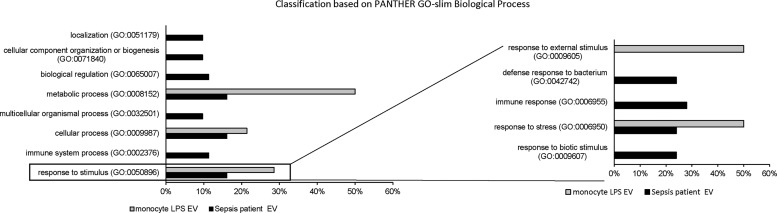
To investigate the differences at a functional level, we performed functional enrichment analysis using PANTHER-GO. In patients with sepsis, down-regulated processes included localization (GO:0051179; 9.7%) biogenesis (GO:0071840; 9.7%), metabolic (GO:0008152; 16.1%) and cellular processes (GO:0009987; 16.1%), as well as immune system processes (GO:0002376; 11.3%) and response to stimuli (GO:0050896; 16.1%) (Fig. 5). Such diversity of biological processes was not seen in vitro in endotoxin-stimulated monocytes. Many (71%) of down-regulated proteins belonged to metabolic (GO:0008152; 50%) and cellular processes (GO:0009987; 21.4%). The only other pathways in which the proteins were enriched in was response to stimulus (GO:0050896; 28.6%) (Fig. 5). When analyzing the response to stimulus pathways, in patients with sepsis, these down-regulated pathways were further subdivided to include the defensive response to bacteria, stress, and biotic stimuli and the immune response (Fig. 5). Similarly, exosomes from endotoxin-stimulated monocytes demonstrated down-regulation of pathways that included the response to external stimuli and to stress (Fig. 5). Together, these pathway analyses suggest an immunosuppressive role for exosomes.
FIG. 5.
Bar chart comparing GO classification of down-regulated proteins in sepsis-patient exosomes with lipopolysaccharide-treated cultured monocytes on the basis of biological process. Response to stimulus process was further sub-classified. The bar graphs represent percentage of each functional group and total number of proteins in each group.
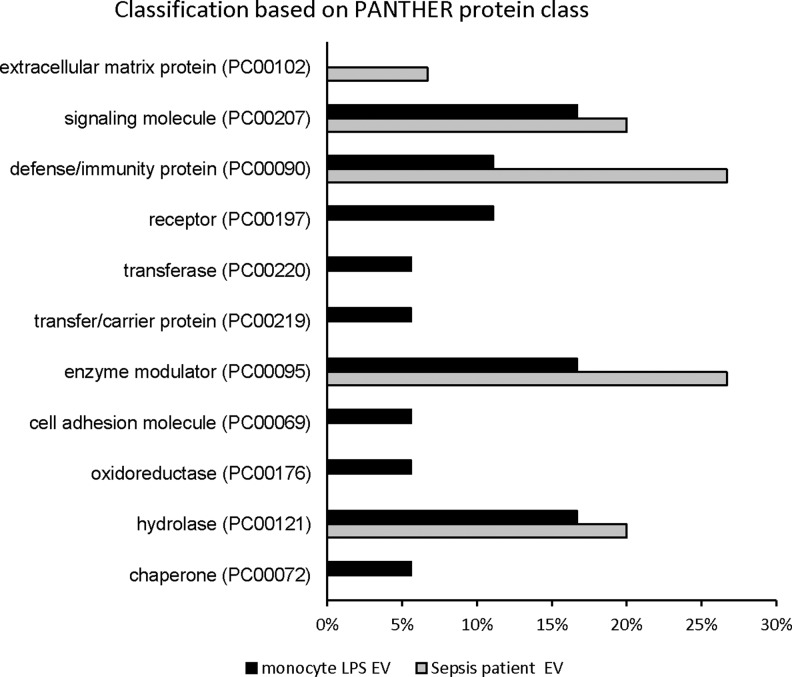
When analyzing protein classes that were down-regulated, several patterns emerged. In patients with sepsis, the majority of the down-regulated proteins were extracellular matrix proteins (PC00102), signaling molecules (PC00207), defense/immunity proteins (PC00090), enzyme modulators (PC00095), and hydrolases (PC00121) (Fig. 6). In LPS-stimulated monocyte-derived exosomes, the down-regulated protein classes included signaling molecules (PC00207), defense/immunity proteins (PC00090), receptors (PC00197), transferases (PC00220), transfer/carrier proteins (PC00219), enzyme modulators (PC00095), cell adhesion molecules (PC00069), oxidoreductases (PC00176), hydrolases (PC00121), and chaperones (PC00072) (Fig. 6). In a fashion similar to biological processes, defense and immunity proteins were identified as significantly down-regulated in the protein class GO classification. Cellular component analysis of these significantly different proteins using STRING confirmed that the majority of them were indeed packaged as a cargo in extracellular vesicles (Fig. 7). Functional enrichment of the down-regulated proteins using the KEGG pathway demonstrated that they belong to the complement and coagulation cascades in both patient- and in vitro sepsis-derived exosomes.
FIG. 6.
Bar chart comparing GO classification of down-regulated proteins in sepsis patient exosomes with lipopolysaccharide-treated cultured monocytes on the basis of protein class. Bar graphs represent percentage of each functional group and total number of proteins in each group
FIG. 7.
STRING version 10.5 was used to construct protein–protein interaction network of significantly down-regulated proteins in exosomes obtained from septic patients compared with critically ill subjects and cultured primary monocytes exposed to lipopolysaccharide or phosphate-buffered saline. Lines or strings indicate protein interactions. Functional enrichments in networks are mentioned at bottom.
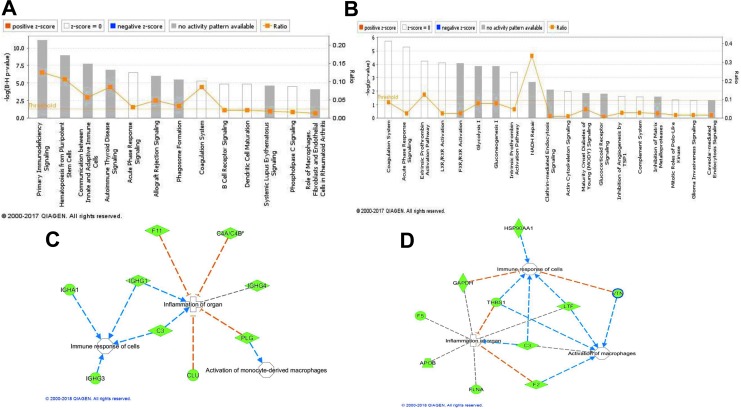
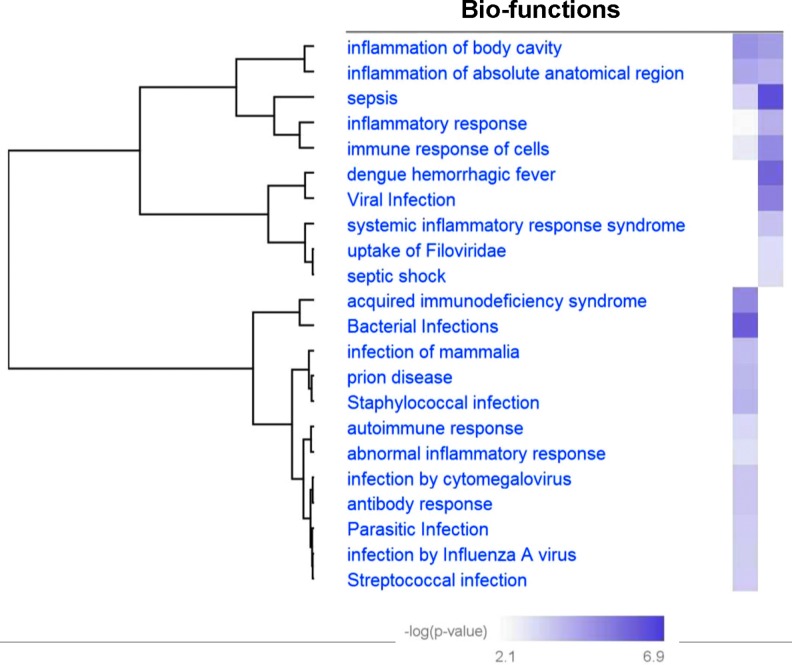
Ingenuity Pathway Analysis was used to delineate the functional significance of down-regulated pathways in sepsis patients and in LPS-induced cultured monocytes. The canonical pathways obtained were compared in these two conditions (Fig. 8A, B). Of interest was the observation that proteins belonging to two canonical pathways, namely, acute-phase response signaling and the coagulation system, were down-regulated in both the above-mentioned conditions (Fig. 8A, B). Furthermore, network analysis using IPA predicted the depletion of important proteins involved in the immune response of cells and activation of macrophages in exosomes of sepsis patients and in LPS-treated monocytes (Fig. 8C, D). The deficiency in the packaging of these proteins in exosomes could be an important indicator or determinant of the inflammation of organs and organ failure under septic condition (Fig. 8C, D, Figure 9).
FIG. 8.
Ingenuity Pathway Analysis (IPA®) of the proteins differentially packaged in exosomes showing canonical pathways obtained in septic patients (A), critically ill subjects (B), and cultured primary monocytes exposed to lipopolysaccharide (LPS) or phosphate-buffered saline (PBS). The analysis showing network proteins and biological functions obtained in septic patients (C), critically ill subjects (D), and cultured primary monocytes exposed to LPS or PBS.
FIG. 9.
Heat map comparing disease and bio-functions of down-regulated proteins. Septic patients compared with critically ill subjects and cultured primary monocytes exposed to lipopolysaccharide or phosphate-buffered saline.
Immunosuppression Testing
Existing data suggest that exosomes derived from patients with sepsis or experimental models of sepsis have immunosuppressive effects on recipient cells [28,29]. In order to test this idea, we treated cultured monocytes with exosomes derived from patient samples (sepsis or controls) and from stimulated monocytes (LPS or PBS). We then re-stimulated these cells with LPS and assessed the concentration of TNF-α as a measure of immunosuppression.
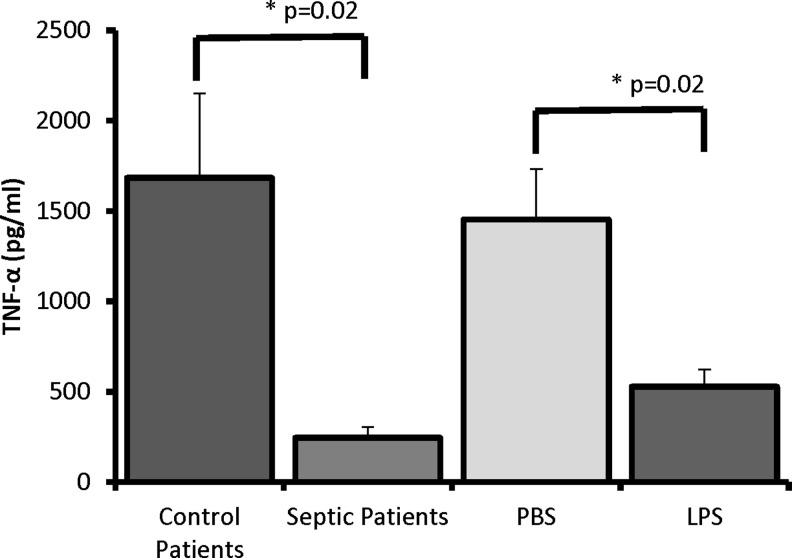
In monocytes treated with exosomes from patients with sepsis, we saw significant decreases in TNF-α expression compared with monocytes treated with exosomes from control patients (245.6 ± 118.7 versus 1684.1 ± 935.1 pg/mL; p = 0.02; Fig. 10). Additionally, monocytes treated with exosomes derived from LPS-stimulated monocytes showed a decreased response to LPS stimulation compared with monocytes treated with exosomes derived from PBS-treated monocytes (528.4 ± 190.6 versus 1452.6 ± 560.1 pg/mL; p = 0.02; Fig. 10).
FIG. 10.
Tumor necrosis factor TNF-α expression after lipopolysaccharide (LPS) stimulation in human monocytes previously treated with exosomes from (1) patients with sepsis, (2) patients without sepsis, (3) human monocytes stimulated with LPS, or (4) human monocytes treated with phosphate-buffered saline (PBS). In monocytes treated with exosomes from patients with sepsis, there were decreases in TNF-α expression compared with monocytes treated with exosomes from control patients (245.6 ± 118.7 vs. 1684.1 ± 935.1 pg/mL; p = 0.02). Monocytes treated with exosomes derived from LPS-stimulated monocytes showed a decreased response to LPS stimulation compared with monocytes treated with exosomes derived from PBS-stimulated monocytes (528.4 ± 190.6 vs. 1452.6 ± 560.1 pg/mL; p = 0.02).
Discussion
Using patient samples and in vitro evidence, we were able to identify several key pathways that are altered in sepsis-derived exosomes and in exosomes derived from LPS-stimulated monocytes. Specifically, in surgical patients with sepsis, the down-regulated processes included localization, biogenesis, metabolic and cellular processes, and immune system processes. In LPS-stimulated monocytes, similar down-regulated processes were seen, including metabolic and cellular processes, as well as the response to a stimulus. Within this category, the down-regulated networks were focused on the immune response to stress and bacteria. In both sepsis-derived exosomes and LPS-stimulated exosomes, protein classes involving defense and immunity were significantly down-regulated. Taken together, these data suggest that exosomes produced during sepsis contain protein networks that may have immunoregulatory and possibly immunosuppressive function.
Sepsis-related cellular dysfunction and immunosuppression represent a continuum of systemic aberrations that result in worse outcomes for patients [4-9]. Our data suggest a role for exosome-mediated immune regulation, specifically with monocyte-derived exosomes. Pathway analysis revealed specific targeting of immune system processes, response to stimuli, and immune responses as down-regulated pathways. This is a poorly described function of circulating exosomes during sepsis, with current literature focusing on protein or recipient cell-specific effects of these exosomes [17–22]. This investigation offers insight into a potential role for circulating exosomes in systemic regulation and function during inflammation and sepsis. In our in vitro system, monocytes treated with sepsis-derived exosomes or exosomes produced from monocytes stimulated with LPS demonstrated significant reductions in TNF-α generation in response to additional LPS stimulation. This suggests a regulatory role for circulating exosomes in TLR4 signaling and TNF-α generation.
It is important to note that these data are superficial and not meant to describe the mechanism in detail. This is one of the first published descriptions of the proteomic networks present within these exosomes, and additional investigations are necessary to demonstrate causative functional changes in recipient cells. It may be that through circulating exosomes as a mediator, the immune system projects a systemic regulatory signal that transitions from the pro-inflammatory response to anti-inflammation. One of the Important aspects of immunoregulation is the ability to down-regulate the initial pro-inflammatory phase to hamper some of the deleterious effects of prolonged inflammation. The data from the present study suggest that monocyte-derived exosomes are involved in this process, specifically with TLR4 signaling and TNF-α expression. These data suggest that this regulation may, in part, be mediated by circulating exosomes and the protein milieu they contain.
Through the use of in vitro modeling via endotoxin stimulation and tolerance, these results suggest that stimulated monocytes produce exosomes that carry protein networks with a potentially immunosuppressive pattern of expression. Lipopolysaccharide stimulation has long been used a model of post-sepsis immunosuppression, and the effects are well characterized [30]. However, within this model, LPS stimulation of each individual cell is required for the cell to exhibit a tolerant phenotype. In this study, using multiple proteomic network analyses, we were able to identify monocyte-produced exosomes as an additional possible mediator of this immunosuppressive phenotype. These observations are in line with emerging research that strongly suggests an immunosuppressive role for exosomes during inflammatory states [30–33]. Previous studies have focused on a small number of individual mediators as possible causes of the immunosuppressive effects. The data from the present study expand on this by focusing on the protein cargo as a functional network. This improves our understanding of the immunomodulatory effects of exosomes. Importantly, the functions that were altered in our model system were altered similarly in our pilot investigation of samples from human patients. Although the human data are not meant to be exhaustive or to serve as conclusive evidence, they do offer proof-of-concept corroboration that the observations from our model system potentially occur in vivo. However, more analysis of these data is necessary.
There are several limitations of the current investigation. Because of the scope of proteomic data analysis, a large, heterogeneous patient profile could not be created. There may be differences in age, sex, disease pathology, and other confounders that we cannot account for. It is known that age, among many other characteristics, affects the composition and quantity of circulating exosomes; however, its effect during sepsis is not known [16]. Circulating exosomes have complex function in patients with sepsis. Depending on their cell of origin and cellular target, circulating exosomes can be pro-thrombotic or pro-inflammatory, cause endothelial dysfunction, or have anti-inflammatory effects [28,29,31–33]. These functions may be dictated by the cargo contained in these exosomes; however, this adds considerable complexity, as exosomes carry proteins, RNA, microRNA, and other regulatory components that may be responsible for the effects seen [17,19].
This study does not attempt to evaluate the cumulative effect of these regulatory elements. It may be that within exosomes, these layers of regulatory elements are synergistic or counter-regulatory. Additionally, it is unclear whether the packaging of these exosomes actually is meant to serve as a systemic signaling event or if it simply reflects the immunomodulatory changes in the parent cell that produce the exosome. Existing data support a signaling role in many disease processes, including cancer and inflammation; however, detailed analysis of this regulation was not performed in the present investigation [19–22,34]. Although the protein networks identified in this study are predictive, they may not be the only affected networks in vivo; and further characterization is necessary. Despite these limitations, this study offers novel insight and expands the knowledge of the systemic function of exosomes during inflammation and sepsis.
Conclusions
These data suggest that early in the septic process, circulating exosomes, specifically, monocyte-derived exosomes, contain networks of proteins that may mediate an immunomodulatory effect in recipient cells. With future investigation, we aim to elucidate both the in vitro and in vivo mechanisms by which these circulating exosomes regulate the systemic immune response.
Author Disclosure Statement
This work was supported by a Surgical Infection Society Junior Faculty Fellowship as well as the American Association for the Surgery of Trauma Research and Education Foundation Scholarship Award.
References
- 1. Hotchkiss RS, Monnerat G, Payen D. Immunosuppresion in sepsis: A novel understanding of the disorder and a new therapeutic approach. Lancet Inf Dis 2013;13:260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reddy RC, Chen GH, Tekchandani PK, et al. Sepsis-induced immunosuppression: From bad to worse. Immunol Res 2001;24:273–287 [DOI] [PubMed] [Google Scholar]
- 3. Ward NS, Casserly B, Ayala A. The compensatory anti-inflammatory response syndrome (CARS) in critically ill patients. Clin Chest Med 2008;29:617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sasse KC, Nauenberg E, Long A, et al. Long-term survival after intensive care unit admission with sepsis. Crit Car Med 1995;23:1040–1047 [DOI] [PubMed] [Google Scholar]
- 5. Pinsky MR, Vincent JL, Deviere J, et al. Serum cytokine levels in human septic shock: Relation to multiple-system organ failure and mortality. Chest 1993;103:565–575 [DOI] [PubMed] [Google Scholar]
- 6. Daviaud F, Grimaldi D, Dechartres A, et al. Timing and causes of death in septic shock. Ann Intens Care 2015:5(16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gentile LF, Cuenca AG, Efron PA, et al. Persistent inflammation and immunosuppression: A common syndrome and new horizon for surgical intensive care. J Trauma Acute Care Surg 2012;72:1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mira J, Gentile L, Mathias BJ, et al. Sepsis pathophysiology, chronic critical illness, and persistent inflammation–immunesuppression and catabolism syndrome. Crit Care Med 2017;45:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang HE, Szychowski JM, Griffin R, et al. Long-term mortality after community-acquired sepsis: A longitudinal population-based cohort study. BMJ 2014;4:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beeson P. Development of tolerance to typhoid bacterial pyrogen and its abolition by reticulo-endothelial blockade. Proc Soc Exp Biol Med 1946;61:248–250 [DOI] [PubMed] [Google Scholar]
- 11. McCall CE, Yoza BK. Gene silencing in severe systemic inflammation. Am J Respir Crit Care Med 2007;175:763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novakovic B, Habibi E, Wang S, et al. B-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167:1354–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Munoz C, Carlet J, Fitting C, et al. Dysregulation of in vitro cytokine production by monocytes during sepsis. J Clin Invest 1991;88:1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ertel W, Kremer JP, Kenney J, et al. Downregulation of proinflammatory cytokine release in whole blood from septic patients. Blood 1995;85:1341–1347 [PubMed] [Google Scholar]
- 15. Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: Mediators, macrophages, lymphocytes and apoptosis. Shock 1996;6:s27–s38 [PubMed] [Google Scholar]
- 16. Eitan E, Green J, Bodogai M, et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci Rep 2017;7:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meziani F, Delabranche X, Asfar P, Toti F. Bench-to-bedside review: circulating microparticles: A new player in sepsis? Crit Care 2010;14:236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nieuwland R, Berckmans RJ, McGregor S, et al. Cellular origin and procoagulant properties of microparticles in meningococcal sepsis. Blood 2000;95:930–935 [PubMed] [Google Scholar]
- 19. Valadi H, Ekstorm K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659 [DOI] [PubMed] [Google Scholar]
- 20. Li X, Liu L, Yang J, et al. Exosome derived from human cord mesenchymal stem cells mediates miR-181c attenuating burn-induced excessive inflammation. Ebiomedicine 2016;8:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Gu H, Yang L, et al. Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis. Sci Rep 2015;8:13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Connel RM, Kahn D, Gibson WSJ, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010;33:607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tu C, Li J, Shen S, et al. Performance investigation of proteomic identification by HDC/CID fragmentation in combination with high/low-resolution detectors on a tribrid, high-field orbitrap instrument. PLoS One 2016;11(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu H, Rovshan G, Sadygov G, et al. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Ann Chem 2004;76:4193–4201 [DOI] [PubMed] [Google Scholar]
- 25. Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (SEPSIS-3). JAMA 2016;315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Segre E, Fullerton J. Stimulated whole blood cytokine release as a biomarker of immunosuppression in the critically ill: The need for a standardized methodology. Shock 2016;45:490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh K, Pal D, Sinha M, et al. Epigenetic modification of microRNA-200b contributes to diabetic vasculopathy. Mol Ther 2017;25:2689–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monte Real J, Rodriquez L, Ferreira P, et al. Exosomes from patients with septic shock convey miRNAs related to inflammation and cell cycle regulation: New signaling pathways in sepsis? Crit Care 2018;22:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDonald M, Tian Y, Rehman Q, et al. Functional significance of macrophage-derived exosomes in inflammation and pain. Pain 2014;155:1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cavaillon JM, Adib-Conquy M. Bench-to-bedside review: Endotoxin tolerance as a model of leukocyte reprogramming in sepsis. Crit Care 2006;10:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Del Fresno C, Garcia-Rio F, Gomez-Pina V, et al. Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharide-tolerant human monocytes: Demonstration in isolated monocytes from cystic fibrosis patients. J Immunol 2009;182:6494–6507 [DOI] [PubMed] [Google Scholar]
- 32. Domenis R, Cifu A, Quaglia S, et al. Pro inflammatory stimuli enhance the immunosuppressive functions of adipose mesenchymal stem cell-derived exosomes. Sci Rep 2018;8:13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miksa M, Wu R, Dong W, et al. Immature dendritic cell-derived exosomes rescue septic animals via milk fat globule epidermal growth factor VIII. J Immunol 2009;183:5983–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]