Abstract
Recent theory and experiments have reported a reproducible tendency for the coexistence of microbial species under controlled environmental conditions. This observation has been explained in the context of competition for resources and metabolic complementarity given that, in microbial communities (MCs), many excreted by-products of metabolism may also be resources. MCs therefore play a key role in promoting their own stability and in shaping the niches of the constituent taxa. We suggest that an intermediate level of organization between the species and the community level may be pervasive, where tightly knit metabolic interactions create discrete consortia that are stably maintained. We call these units Metabolically Cohesive Consortia (MeCoCos) and we discuss the environmental context in which we expect their formation, and the ecological and evolutionary consequences of their existence. We argue that the ability to identify MeCoCos would open new avenues to link the species-, community- and ecosystem-level properties, with consequences for our understanding of microbial ecology and evolution, and an improved ability to predict ecosystem functioning in the wild.
This article is part of the theme issue ‘Conceptual challenges in microbial community ecology’.
Keywords: microbial ecology, ecosystem functioning, metabolism, functional groups
1. Alternative community states and ecosystem functioning
A common feature of microbes is their ability to modify their environment by releasing extracellular enzymes, by secreting antibiotics and signalling molecules, and by sequestering nutrients or releasing by-products of metabolism. Through their metabolic activity, microbial communities (MCs)1 can therefore have substantial impacts on human health and ecosystem functioning in natural environments [1–3]. The notion of MC functioning that we consider focuses on ecosystem-level processes that operate at temporal scales that are much longer than bacterial generation times. For instance, the carbon cycle in the biosphere happens at geological time-scales, and is contingent on microbial activity [3]. A second feature we consider is that functioning is not performed by a single species (e.g. a single pathogen) or explained by a single-specific process (e.g. formation of a biofilm) [4]. We are interested in broad functions that are the consequence of alternative MC states, which require considering the combined action of multiple taxa, rather than narrow functions, that are the consequence of a single action or metabolic process [5]. Examples of broad functions are human diseases like inflammatory bowel disease or obesity, for which there is evidence that the consequences of MC functions (here health and disease states occurring over the scale of a human lifetime) are unlikely to be explained by a single pathogen but through whole MC states [6].
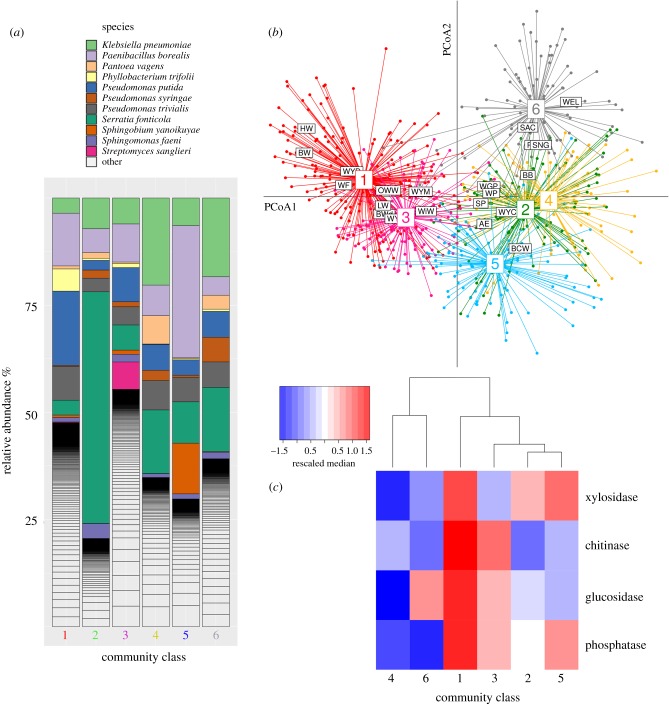
A growing body of research has found persistent MC states likely driven by environmental conditions, including, for example, the controversial discovery of human enterotypes [7,8]. As an example, we illustrate in figure 1a the clustering of more than 700 communities sampled from rainwater-filled puddles formed at the base of beech trees into just six distinct community classes. Pascual-García & Bell [9] showed that there was significant spatial autocorrelation, which may be explained by stochastic processes and dispersal limitation (figure 1b). Nonetheless, the six classes contained communities that were often distant in space (more than 100 km), suggesting community similarity was at least partly driven by local environmental conditions. Comparable large-scale efforts, including Tara Oceans [10] and the Human Microbiome Project [11], have also identified distinct community classes when the same kind of environment (marine, human gut) is sampled across multiple locations or individuals.
Figure 1.
Community-level classes. (a) Approximately 700 natural MCs sampled from tree-holes were classified into six classes. The bar-plot shows the relative abundance of the most representative species (operational taxonomic units (OTUs) at more than 97% sequence similarity) in each class. (b) Projection of the similarity of the communities into the first two principal coordinates of a principal coordinate analysis. Significant spatial autocorrelation was found, which is apparent when the centroid of each sampling location (each site identified by a three-letter code) is superimposed on the ordination, showing that sampling sites are associated with specific community classes. However, the classes (labelled 1 to 6) yield a more economical classification with comparable significance, with the constituent communities often sampled from different locations. (c) In addition, communities contained in these classes have distinct functions and metagenomic repertoires. The functional differences among the classes are illustrated in the heatmap, which indicates the median capacity of communities from each class to degrade a set of common substrates, showing divergent functional ‘signatures’ for each of the community classes. Overall, the data suggest that local environmental conditions occurring in different locations, rather than neutral evolution with dispersal limitation, shape these communities. More details in [9].
Studies that have identified community classes have also found that the classes are associated with distinct functional profiles. For the example in figure 1, the six classes had differing functional capacities in terms of their capacity to degrade substrates and their growth efficiencies. The result is consistent with the idea that there is a degree of functional redundancy within community classes (i.e. changes to composition within classes result in similar functional profiles), but that large changes to composition (among classes) alter functioning. Metabolic functions are therefore redundant within community types, but disparate community types reflect disparate functional capacities. Further research has shown that the classes also have differentiated genetic repertoires, pointing towards a relationship between ecological conditions and bacterial traits, and rejecting the hypothesis that communities are stochastically assembled or are functionally equivalent. Similar observations have been found in a study of marine systems, which also found community types with differentiated functional profiles associated with seasonal changes [12].
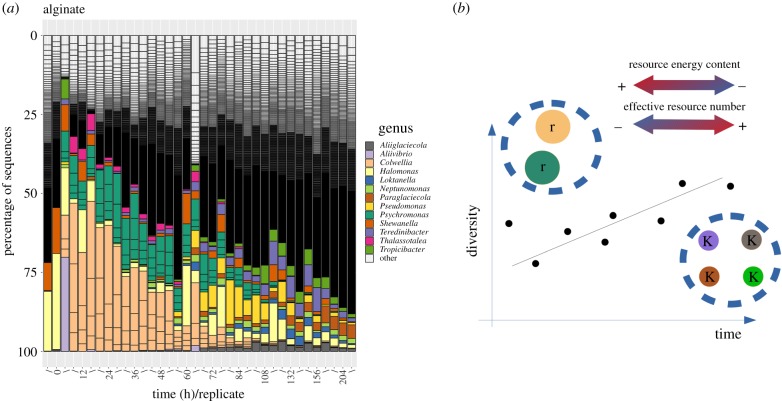
To illustrate how functionally divergent community classes (figure 2) might arise, we show the dynamics of marine bacterial communities colonizing synthetic particles of alginate [13]. We identified the exact sequence variants (ESVs) [14] in each sample, and assigned each ESV to a genus. In the figure, all the ESVs are represented and the genera to which the 20 most abundant ESVs belong are indicated (colours), with the remaining genera shown in dark grey. One remarkable feature of this experiment is the high reproducibility of the trajectories at the genus level (three independent replicates per time-point) despite colonization by a diverse natural community from which we would expect a somewhat stochastic assembly. At the starting stages of the colonization of the particles, a few operational taxonomic units (OTUs) dominate the community, possibly specialized on the breakdown of alginate. At an intermediate stage, around 60–84 h, there is a clear transition in the communities, likely mediated by the release of by-products that are acquired by secondary consumers. As a consequence, there is an increase in the number of microniches and a consistent increase in the diversity of the community. Contrary to the prediction of stochastic community assembly that would result from widespread functional redundancy, these empirical studies imply that broad community classes are determined by environmental conditions, leading to reproducible trajectories of community assembly, and to functional profiles that reflect the metabolisms of the community members.
Figure 2.
Diversity and resources properties in an ecological succession. (a) Population dynamics of natural marine assemblage MCs on synthetic particles of alginate. Each bar represents the relative abundance in the corresponding sample, labelled by time-point, with the 15 most abundant genera highlighted and the remainder shown as ‘others’. Three replicates per time-point are shown. (b) Illustration of the expected diversity increase through time as a function of the energy content and the effective number of resources that results from the degradation of resources. r-strategists will be observed at earlier times where the resources are abundant and rich in energy, while K-strategists should be expected at later times, where the resources are more heterogeneous, lower in energy and scarce (see Discussion).
2. Microbial environmental modifications create metabolically cohesive consortia
These observations require a broad framework to understand the convergence of communities over space (figure 1) and time (figure 2) and how those community dynamics map onto the observed functional dynamics. We follow Tilman’s seminal consumer–resources models, which predict that the outcome of two species competition for a single resource will be determined by the ability of each species to deplete the resource to the lowest concentration when grown in isolation. When both species are grown in co-culture, it is expected that the one depleting the resource to a minimum in isolation will exclude the other in co-culture [15]. However, the resources used by bacteria and other microbes are generally not depleted in this manner. Instead, the ‘winner’ releases metabolic by-products that can be used by other populations, which provides the opportunity for coexistence if an outcompeted species is able to exploit this new niche [16,17]. This reasoning can be extended to a large number of species and resources. Following from Tilman’s model, one hypothesis that emerges from this observation is that the combination of species that deplete resources to a minimum concentration will systematically dominate the community. A corollary is that a community assembled in this manner will optimize functions associated with community-level metabolism, as has been shown for methanogenic communities [18].
We call this kind of community organization a Metabolically Cohesive Consortium (MeCoCo), which is a type of consortium that exhibits a positive feedback loop, where the consortium engineers the environment by both creating and using resources, resulting in stable and reproducible community dynamics. Members of MeCoCos minimize competition within the consortium by specializing on particular resource components, and exclude resource generalists by lowering resource abundances through their combined action. We conjecture that the formation of MeCoCos may be common in natural communities, and would be a parsimonious explanation for the observation of clustered community compositions at broad scales (figure 1), and the predictable successional trajectories exhibited in figure 2. This dynamical formation of reproducible modules is supported by consumer–resource models that have reported the systematic formation of stable communities starting from random assemblages [19–23] due to resource partitioning, and even without the release of metabolic by-products [24].
We illustrate how this idea might be supported using results from a study investigating a consortium found in the production of kefir and wine formed by Saccharomyces cerevisiae and two Lactobacillus species (LAB: L. lactis and L. plantarum). The two LAB species are auxotrophs of some amino-acids that are provided by S. cerevisiae in nitrogen-rich environments [25]. In addition, when the carbon source is lactose, the inability of S. cerevisiae to grow on this carbon source is compensated by a supply of usable carbon sources from L. lactis [25]. Therefore, a mutualistic relationship between these species makes the consortium robust against fluctuations in the available amino-acids or carbon sources in the environment. This occurs through a niche created from overflown metabolism [25], which may rapidly result in a stable consortium. Therefore, niche self-construction would be an expected ecological consequence of the collective reduction of environmental (i.e. resource) fluctuations.
The likelihood that a MeCoCo forms will depend on many factors. For instance, a systematic formation of modules with reproducible composition is a result predicted for low immigration rates [21]. Thus, any process that homogenizes ecosystems, and in so doing increases immigration rates into local communities, would tend to disrupt MeCoCos that develop. We would therefore predict MeCoCos would be more likely in spatially heterogeneous ecosystems with low immigration (e.g. soil), and would not be as prevalent in well-mixed systems (e.g. aquatic environments). Many other factors may also influence the importance of MeCoCos as a level of organization. For example, it has been recently shown with simulations that stable consortia often emerge in complex communities containing large population sizes or high mutation rates [26], in which evolutionary events in the population occur at a more rapid rate than the time required for populations to equilibrate. Once established, these consortia can then spread to adjacent areas by outcompeting resident communities. A future challenge is to use theory, experiments and observations to identify the conditions under which MeCoCos are likely to form, and to model their spatial and temporal dynamics.
3. Empirical evidence of MeCoCos
There is a large body of literature reporting the stable coexistence of species pairs through cross-feeding, including details of the metabolic [27] and physiological mechanisms [28,29]. In vitro experiments using larger consortia have also generated metabolic co-dependencies using genetically modified strains [30] and through the generation of auxotrophic strains [31]. This work demonstrates the capacity to engineer simple communities with metabolic dependencies under laboratory conditions.
Aside from culture-based approaches, there is evidence of stable metabolic consortia created in natural environments. These include controlled biotechnological settings such as anaerobic reactors [18], generation of biofuel [32] and degradation of pesticides [33]. Examples in natural environments are less common. Examples include dental plaque consortia containing 18 genera [34], consortia composed of phototrophic green sulfur bacteria and chemotrophic bacteria such as Chlorochromatium aggregatum [35], and anaerobic food chains in methanogenic environments [36].
A more generic analysis was conducted by Freilich et al. [37] and Zelezniak et al. [38], which found metabolic co-dependencies among co-occurring taxa. Zelezniak et al. [38] first searched for genomes that matched the 16S sequences, and the genome sequences were used to characterize the metabolic capacity of each population. They then used flux-balance models to infer metabolic co-dependencies, finding increasing levels of co-dependencies within co-occurring phylotypes, but increasing levels of inferred competition (shared metabolic requirements) among phylotypes that did not co-occur. Freilich et al. [37] explored metabolic interactions using flux-balance models, finding that many taxa had the capacity for cooperative interactions. When these interactions were contrasted with patterns of co-occurrence in natural samples, they found a significantly higher number of uni-directional ‘cooperative loops’ (metabolic exchanges) in co-occurring taxa, suggesting the existence of whole communities engaged in indirect mutualistic interactions via the gradual degradation of complex resources. This research therefore pioneered the MeCoCo idea, providing some of the first empirical data indicating that MeCoCos may often emerge in complex communities.
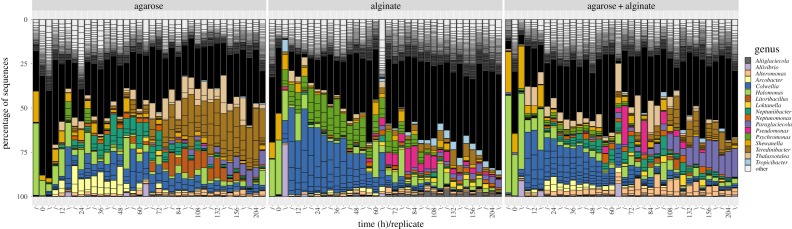
We illustrate the prominence of MeCoCos by expanding the alginate-particle experiment shown in figure 2 to also include particles made of agarose or a mixture of agarose and alginate (figure 3, [13]). We observed highly reproducible dynamics in all three conditions, with a shift in the composition at intermediate times, and partitioning of the time-series into two or three groups of distinct species likely as a result of an abrupt transition of the underlying substrate. Despite the different substrates, this consistent dynamic occurred in all three experiments but with different community members, suggesting a tight relationship between the substrate and the communities. Moreover, the abundances of the most representative species on particles composed of a mix of alginate and agarose (figure 3c) experienced dynamics that could be approximated by a simple linear combination of their dynamics on the single-substrate particles (figure 3a,b) [13]. The communities occupying the particles were composed of just a few genera, which would be candidate MeCoCos in this dynamic environment, with most of the genera involved in metabolizing previously degraded substrates. Similarly, in controlled environments, experiments analysing the growth of natural communities on single-carbon sources reported a systematic convergence to the same steady states at the family level [39].
Figure 3.
Population dynamics in single- and mixed-resource environments. Microbial communities were allowed to colonize particles composed of alginate (a), agarose (b) or a mixture of the two (c). The community dynamics in all three conditions exhibit a transition at intermediate times, in which the dynamics are split into well-differentiated communities. Although the community composition on the two substrates differs, the remarkable modularity of the experiments allows us to predict the relative abundance for the most abundant members as a linear combination of the relative abundances of the pure substrates [13].
In some of the best-known examples of metabolic dependencies, such as the dental plaque ecosystem, it is necessary for community members to be in close proximity, or even in direct contact [27]. Therefore, an open question is whether such metabolic exchanges can be maintained in more dynamic or open environments [40]. The consistent regularities in community composition observed in the analysis of large datasets (figures 1–3), along with the observed examples of inferred metabolic co-dependencies across a wide range of environments [37,38], provide support for the existence of MeCoCos in many habitats. Taken together, the results suggest that such close physical proximity is not a requirement for MeCoCos, but that metabolic exchanges can occur at a distance. However, physical distance between MeCoCo members can clearly hinder the emergence of MeCoCos if the metabolic products produced by one MeCoCo member become too dilute as they diffuses away from the producer [41]. An interesting question is whether physical proximity evolves in older MeCoCos, for which there has been a greater opportunity for selection to operate, leading to the prediction that metabolic inter-dependencies become increasingly intimate over time.
4. Ecological consequences of MeCoCos
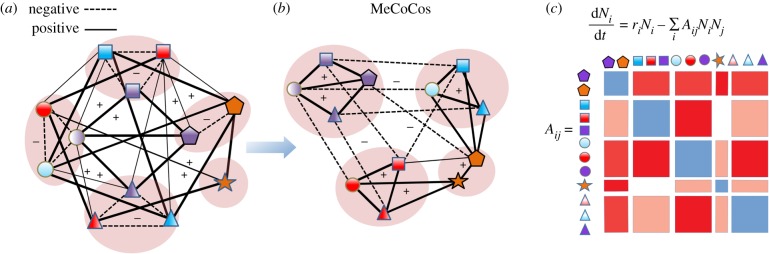
An important corollary to the MeCoCo hypothesis is the prediction that MeCoCos are stable against invasions because no other species would be able to acquire nutrients at a sufficiently high rate to coexist with the established members. This conjecture is supported by recent results that highlight the importance of metabolic interactions in shaping stable communities [42–44]. If MeCoCos are stable structures that resist invasion, it would be possible to understand microbial ecosystems in terms of MeCoCo building blocks. To illustrate this point, we outline a hypothetical example in which we consider groups of species with similar metabolic capabilities (figure 4a), which we call functional groups because this is the definition of function that we have adopted here. In this example, we would typically expect competitive interactions within these functional groups (represented in the networks with dashed lines) because members occupy similar metabolic niches [48]. Similarly, we may expect that metabolites are traded between different functional groups, leading to commensal or mutualistic interactions (solid lines in the network). With this picture in mind, we would expect that competing species would tend to segregate while those cooperating would tend to aggregate. We present an alternative configuration of the same network, where taxa have been arranged into groups according to their positive interactions. This rearrangement allows us to observe another level of organization (figure 4b). In this visualization, there are positive intra-specific interactions within MeCoCos, while negative interactions (e.g. competition) occur between MeCoCos. Although we believe that metabolic interactions are critical, other kinds of interactions may also be accommodated. For instance, bacteria may also be organized into cooperative groups as a result of their collective antibiotic resistance, while antibiotic-mediated antagonisms may occur between populations [49].
Figure 4.
MeCoCos as a level of organization. (a) The symbols represent different species, where the same shape represents similar metabolic capabilities (functionally redundant groups), which would lead to competitive interaction (dotted lines) owing to a high niche overlap. On the other hand, members of different functional groups may engage in commensal or mutualistic relationships, driven by metabolic complementarity (solid lines). (b) A rearrangement of the left network leads to a new representation, in which members related through complementary functions tend to co-occur, forming MeCoCos, which constitute an intermediate level of organization between the species and the community levels. Understanding community-level dynamics may thus be simplified to understanding how MeCoCos compete (figure 5). (c) The interaction matrix corresponding to the networks has a block structure that makes it feasible to build population dynamics models as in macroscopic systems (e.g. [45–47]). Blue blocks represent competitive interactions, and red blocks represent mutualistic interactions arising from metabolic complementarity.
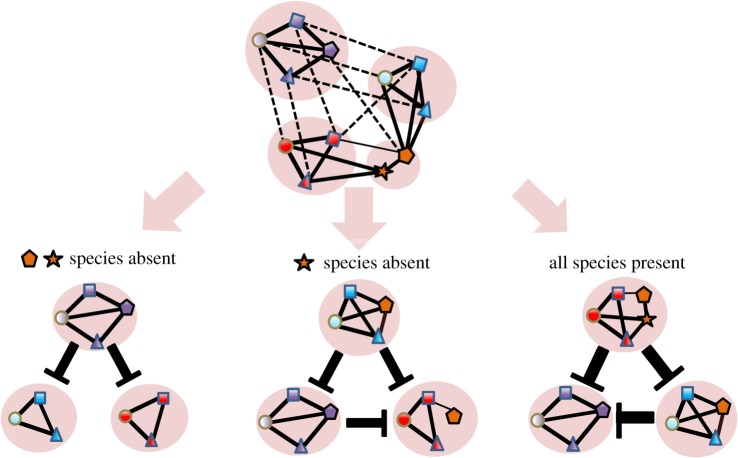
The specific members of MeCoCos are likely context-dependent [6], with some metabolic dependencies only occurring in particular environments. For example, some metabolic interactions may only occur when the appropriate complex substrates are available [37]. As a result, the MeCoCo that prevails in competition with other MeCoCos will depend on the environmental conditions. However, in predicting which MeCoCo prevails we would need to understand the behaviour of the MeCoCos as a whole, and not of every single constituent species. Interestingly, rare species may play a particularly prominent role in MeCoCos, since they may act as ‘metabolic switches’, facilitating the formation of specialized metabolic pathways under certain conditions (figure 5). In general, the framework proposed would therefore move the emphasis from species to MeCoCos. In figure 4c, we show how the MeCoCo structure leads, in terms of an interaction matrix, to a block structure. This would allow us to simplify these systems to reasonable levels of complexity as is done using macroscopic systems [45], for example, dividing ecosystems into mutualistic interactions between plants and their pollinators and competitive interactions within plant and pollinator functional groups [46,47].
Figure 5.
Competition between MeCoCos. Illustration of how changes in species composition alter which MeCoCo becomes dominant. The dominant community is the one that depletes resources to a minimum concentration, which we assume is related to the number of realized metabolic (complementary) links (solid lines connecting species). The orange species may be understood as community-level ‘metabolic switches’ that determine the outcome of the dynamics.
5. Evolutionary consequences of MeCoCos
The large number of metabolic genes found in the prokaryotic flexible genome is consistent with a picture in which metabolic trading is widespread in natural environments [50,51]. Under the MeCoCo hypothesis, the genomic regions that are critical for the maintenance of the species in the consortium will be subjected to a strong selective pressure. Conversely, those genes associated with functions that are covered by other species will experience reduced selective pressures, akin to that observed in symbiotic or parasitic species, leading to a reduction in genome size. For these organisms, low effective population sizes (Ne) would favour genetic drift and hence the appearance of genomic structures that are not purged by selection, like pseudogenes, whereas such features would be purged for populations in MeCoCos with high Ne. In general, we expect species with high Ne would be more likely to be part of MeCoCos because they gain a more immediate selective advantage by rapidly shedding unnecessary parts of their genome.
Widespread loss of genes has also been observed in oligotrophic environments, like the open ocean, where ‘streamlining theory’ explains this loss by pointing to the necessity of these species to be efficient consumers in environments with low nutrient availability [52]. Under these conditions, there is a strong selective pressure to maintain an efficient core metabolism and to lose genes that are not immediately necessary [52]. Under such ‘streamlined’ conditions, it is noteworthy that genes associated with inter-specific interactions are maintained [53], providing further evidence for the central role of metabolic interactions under oligotrophic conditions.
We expect the genetic signature associated with MeCoCos to lie between the scenarios observed for parasites/symbionts and oligotrophic species. This is the prediction made by a related hypothesis, the Black Queen Hypothesis (BQH), which posits loss of genes due to leaky metabolic products frequently present in MCs [54]. For the BQH, the loss of genes is because metabolic products (e.g. metabolites) are often public goods. Consequently, purifying selection leads to a loss of the metabolic pathways needed for the biosynthesis of these metabolites in taxa that can simply take them up from the environment. However, the reduction in genome size would not necessarily be as extreme as the reduction predicted by streamlining theory, and would only involve loci that exploit the leaky metabolites of other species in the environment.
In the case of MeCoCos, we expect a dynamic eco-evolutionary process in which there is selection not only to lose genes that are redundant within the MeCoCo, but also to promote genes that maintain the consortium through positive selection. Therefore, we expect both auxotrophic species, and also the selection of genes related with other functions that may lie beyond metabolism, including signalling to identify partner species and chemotaxis, to stay in close proximity [27].
6. Discussion
In this article, we were interested in developing our understanding of community-level functioning in MCs. There is a need to focus on the relationship between the complexity of the community (the number and identity of the species and their interactions) and community-level metabolism. However, instead of approaching this relationship from the bottom up by building detailed models of each species that may fail in predicting community-level function if high-order interactions are present in the system [55], we follow the lead of recent studies (e.g. [56]) in advocating top-down approaches, in which natural communities are isolated and manipulated under controlled conditions.
An important property of MCs is their stability over space and time because we expect stable communities will result in stable functions. However, functional redundancy among microbial species results in complex relationships among the community composition, the community metabolome, and ecosystem functioning. To address this complexity, we advocate experiments that monitor functioning while directly manipulating community and ecosystem properties [57,58] and while also characterizing the mechanistic basis of MC functions [43,59]. The patterns that emerge from such experiments suggest that the ability of bacterial communities to modify their environment may give rise to MeCoCos through complementary syntrophic interactions. These patterns emerge despite the coarseness of amplicon-based surveys, which may not provide a full picture of microbial diversity in these communities.
The formation of MeCoCos results in increased control of the existing resources by members of the consortium, which promotes community and functional stability. We predict that MeCoCos deplete resources to a minimum concentration and would therefore be more stable against invasions. The importance of structural stability has been emphasized in macroscopic organisms in the context of mutualistic systems [46,47,60], and has recently been applied to microbial populations [22,23,61]. We predict MeCoCos exhibit high levels of structural stability such that fluctuations in external resource inputs would be buffered by species producing the resource or its derivatives. The prominence of MeCoCos should depend on the environmental context. Since the MeCoCo hypothesis is formulated around the degradation of resources and exchanges of metabolites, such predictions would revolve around classifying environments according to the qualitative properties of the resources. We propose three resource axes, which are not fully independent:
-
Axis 1:
Resource diversity, which we define here as the effective number of resources. ‘Effective’ is used here to distinguish resources in terms of their molecular content. For instance, an environment containing chitin and cellulose would have a lower effective number of resources than an environment containing chitin and phosphoric acid because the former pair of molecules have the more similar composition.
-
Axis 2:
Resource abundance. Environments may be oligotrophic or eutrophic (i.e. the resource abundance and the stage of degradation, particularly the compounds, degradation products, and energy that can be retrieved from each unit of effective resource).
-
Axis 3:
Resource heterogeneity. The structure of the environment (well-mixed versus spatially structured) and the degree of turnover of resources (e.g. due to high immigration rates or pulses of resources).
Predicting whether MeCoCos are important in a particular environment will depend on its position in ‘resource space’ across these three axes. For example, in resource-rich, homogeneous environments, we expect MeCoCos would be formed by only a few ‘r-strategist’ taxa that dominate resource exploitation [62,63] (illustrated in figure 2b). Without resource replenishment, we expect environments to become more energetically depauperate over time owing to the degradation of resources, but that the degradation process would also produce a higher effective number of resources. Under these conditions, we expect microbial resource ‘markets’ [64] to be more sophisticated, where density-dependent processes such as competition for resources select for specialist ‘K-strategists’ with mutualistic or commensal interactions that reduce competition. This prediction is aligned with recent simulations showing that the likelihood of engaging in syntrophic interactions increases in resource-poor environments [65]. In another example of stressful environment, toxic metal fluids, it was shown that facilitation leads to coexistence of four species, while reducing toxicity or adding nutrients increased competition [66], since a more habitable environment favours r-strategies. In environments with pulses of resource and of migrants, like the gut, we broadly expect neutral community dynamics owing to the high species turnover, but a significant departure from neutrality for taxa that are a subset of established species, as was found in the zebrafish gut [67]. Such environments would therefore be composed of a core MeCoCo that is continuously out-competing immigrant populations.
In addition to understanding the conditions under which MeCoCos are formed, there is also a need for the development of methods that identify MeCoCos in nature. At the coarsest level, we predict that species belonging to the same MeCoCo should systematically co-occur because certain community configurations would be the most successful in shaping their own niche [43]. Co-occurrence patterns might, therefore, provide a straightforward route to identifying MeCoCos. More refined methods could develop null models to detect MeCoCos. For example, MeCoCos could be identified within subsets of taxa that depart from neutrality.
If MeCoCos are widespread, there would be a need to integrate this idea into the broader evolutionary literature that discusses levels of selection. If species and their interactions can be understood as portions of metabolic processes that together complete more complex metabolisms [68], the metaphor of a bacterial community as a ‘supra-organism’ is compelling [69]. There is likely little need to re-open the debate around the existence of group selection [70,71] since the formation and maintenance of MeCoCos could be explained by selection at the population level based on eco-evolutionary feedbacks [26]. However, there are parallels with emerging views of the evolution of multicellularity, since the formation of stable consortiums has been suggested as an intermediate state for the formation of multicellular life [35,72,73]. If MeCoCos are viewed as a level of selection, the concept would be useful for developing methods for artificial selection of desirable properties at the community level [74], and in predicting outcome of community coalescent events, such as community transplantations [24].
This study was motivated by the need to simplify the extraordinary complexity of microbial life that has been discovered in natural systems so that we can predict and control ecosystem functioning. The idea that much microbial life lives off metabolic by-products produced by other microbes is as old as modern microbiology, including some of the earliest examples of how microbial ecosystems operate Winogradsky columns. We believe a refinement of this view is warranted—that functions are ultimately driven by self-selecting consortia, driven by the way in which microbes exploit and modify their surrounding environment. Identification of MeCoCos using the wealth of genomic data currently being generated would pave the way for both a simpler and more mechanistic understanding of the link between microbial dynamics and the functioning of ecosystems.
7. Methods
Data used in the figures were obtained from publicly available data used in the following studies: [5] (figure 1) and [13] (figures 2 and 3). Bar-plots were generated rarefying samples to 1000 reads and represented with the R package phyloseq [75,76]. For figure 1, the classes, β-diversity distance matrix and GPS locations of the samples were retrieved from [9], and we performed a principal coordinate analysis (PCoA) of the samples with the R function dudi.pco, from package ade4 [77]. Results are represented projecting the samples into the first two PCoA coordinates and computing the centroids of the clusters defined by both the sampling sites and the community classes. Functions represented in figure 1 quantify the median of the log-transformed exoenzymatic activities of communities belonging to the β-diversity classes. Medians were rescaled by rows to make functions comparable, and the classes were clustered with complete linkage computing the Euclidean distance. Results were represented with the function heatmap.2 of the R package gplots [78].
Endnote
For simplicity, in this article, we use the term ‘bacteria’ referring to members of the Bacteria and Archaea kingdoms, while ‘microbial’ may also include Eukarya.
Data accessibility
This article has no additional data.
Authors' contributions
A.P.-G.: Conceived the idea, conducted the analyses, wrote the original manuscript. S.B.: Contributed to the writing. T.B.: Contributed to the writing and helped develop the ideas.
Competing interests
We declare we have no competing interest.
Funding
The research was funded by a European Research Councilstarting grant (no. 311399-Redundancy) awarded to T.B. and by the Simons Collaboration: Principles of Microbial Ecosystems (PriME), award number 542381, to S.B. T.B. was also funded by a Royal Society University Research Fellowship.
References
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. ( 10.1038/nrmicro1978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becerra-Castro C, Lopes AR, Vaz-Moreira I, Silva EF, Manaia CM, Nunes OC. 2015. Wastewater reuse in irrigation: a microbiological perspective on implications in soil fertility and human and environmental health. Environ. Int. 75, 117–135. ( 10.1016/j.envint.2014.11.001) [DOI] [PubMed] [Google Scholar]
- 3.Falkowski PG. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. ( 10.1126/science.281.5374.200) [DOI] [PubMed] [Google Scholar]
- 4.Chicurel M. 2000. Slimebusters. Nature 408, 284–286. ( 10.1038/35042737) [DOI] [PubMed] [Google Scholar]
- 5.Rivett DW, Bell T. 2018. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat. Microbiol. 3, 767 ( 10.1038/s41564-018-0180-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenblum S, Turnbaugh PJ, Borenstein E. 2012. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl Acad. Sci. USA 109, 594–599. ( 10.1073/pnas.1116053109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9, e1002863 ( 10.1371/journal.pcbi.1002863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ. et al. 2008. A core gut microbiome in obese and lean twins. Nature 457, 480–484. ( 10.1038/nature07540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascual-García A, Bell T. In press. Community-level signatures of ecological succession in natural bacterial communities. Nat. Commun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunagawa S. et al. 2015. Structure and function of the global ocean microbiome. Science 348, 1261359 ( 10.1126/science.1261359) [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The Human Microbiome Project. Nature 449, 804 ( 10.1038/nature06244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galand PE, Pereira O, Hochart C, Auguet JC, Debroas D. 2018. A strong link between marine microbial community composition and function challenges the idea of functional redundancy. ISME J. 12, 2470 ( 10.1038/s41396-018-0158-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, Pascual-García A, Cordero OX. 2019. Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, 1528–1535. ( 10.1016/j.cub.2019.03.047) [DOI] [PubMed] [Google Scholar]
- 14.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639 ( 10.1038/ismej.2017.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilman D. 1982. Resource competition and community structure. Princeton, NJ: Princeton University Press. [PubMed] [Google Scholar]
- 16.Pfeiffer T, Bonhoeffer S. 2004. Evolution of cross-feeding in microbial populations. Am. Nat. 163, E126–E135. ( 10.1086/383593) [DOI] [PubMed] [Google Scholar]
- 17.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, Barraclough TG. 2012. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10, e1001330 ( 10.1371/journal.pbio.1001330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierocinski P. et al. 2017. A single community dominates structure and function of a mixture of multiple methanogenic communities. Curr. Biol. 27, 3390–3395. ( 10.1016/j.cub.2017.09.056) [DOI] [PubMed] [Google Scholar]
- 19.Posfai A, Taillefumier T, Wingreen NS. 2017. Metabolic trade-offs promote diversity in a model ecosystem. Phys. Rev. Lett. 118, 028103 ( 10.1103/PhysRevLett.118.028103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taillefumier T, Posfai A, Meir Y, Wingreen NS. 2017. Microbial consortia at steady supply. eLife 6, e22644 ( 10.7554/eLife.22644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal A, Maslov S. 2018. Diversity, stability, and reproducibility in stochastically assembled microbial ecosystems. Phys. Rev. Lett. 120, 158102 ( 10.1103/PhysRevLett.120.158102) [DOI] [PubMed] [Google Scholar]
- 22.Marsland R III, Cui W, Goldford J, Sanchez A, Korolev K, Mehta P. 2019. Available energy fluxes drive a transition in the diversity, stability, and functional structure of microbial communities. PLoS Comput. Biol. 15, e1006793 ( 10.1371/journal.pcbi.1006793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tikhonov M, Monasson R. 2017. Collective phase in resource competition in a highly diverse ecosystem. Phys. Rev. Lett. 118, 048103 ( 10.1103/PhysRevLett.118.048103) [DOI] [PubMed] [Google Scholar]
- 24.Tikhonov M. 2016. Community-level cohesion without cooperation. eLife 5, e15747 ( 10.7554/eLife.15747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponomarova O. et al. 2017. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345–357. ( 10.1016/j.cels.2017.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotil SE, Vetsigian K. 2018. Emergence of evolutionarily stable communities through eco-evolutionary tunneling. Nature Ecol. Evol. 2, 1644–1653. ( 10.1038/s41559-018-0655-7) [DOI] [PubMed] [Google Scholar]
- 27.D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C. 2018. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488. ( 10.1039/C8NP00009C) [DOI] [PubMed] [Google Scholar]
- 28.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. 2013. Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. ( 10.1111/1574-6976.12019) [DOI] [PubMed] [Google Scholar]
- 29.Schink B, Stams AJ. 2013. Syntrophism among prokaryotes. Berlin, Germany: Springer. [Google Scholar]
- 30.Harcombe WR. et al. 2014. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115. ( 10.1016/j.celrep.2014.03.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mee MT, Collins JJ, Church GM, Wang HH. 2014. Syntrophic exchange in synthetic microbial communities. Proc. Natl Acad. Sci. USA 111, E2149–E2156. ( 10.1073/pnas.1405641111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabaey K, Boon N, Siciliano SD, Verhaege M, Verstraete W. 2004. Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl. Environ. Microbiol. 70, 5373–5382. ( 10.1128/AEM.70.9.5373-5382.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsuyama C, Nakaoka S, Takeuchi Y, Tago K, Hayatsu M, Kato K. 2009. Complementary cooperation between two syntrophic bacteria in pesticide degradation. J. Theor. Biol. 256, 644–654. ( 10.1016/j.jtbi.2008.10.024) [DOI] [PubMed] [Google Scholar]
- 34.Kolenbrander PE, London J. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175, 3247 ( 10.1128/JB.175.11.3247-3252.1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overmann J. 2010. The phototrophic consortium “Chlorochromatium aggregatum” – a model for bacterial heterologous multicellularity. In Recent advances in phototrophic prokaryotes (ed. PC Hallenbeck), pp. 15–29. New York, NY: Springer.
- 36.Stams AJ, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568 ( 10.1038/nrmicro2166) [DOI] [PubMed] [Google Scholar]
- 37.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. 2011. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2, 589 ( 10.1038/ncomms1597) [DOI] [PubMed] [Google Scholar]
- 38.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. 2015. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl Acad. Sci. USA 112, 6449–6454. ( 10.1073/pnas.1421834112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segré D, Mehta P, Sanchez A. 2018. Emergent simplicity in microbial community assembly. Science 361, 469–474. ( 10.1126/science.aat1168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavaliere M, Feng S, Soyer OS, Jiménez JI. 2017. Cooperation in microbial communities and their biotechnological applications. Environ. Microbiol. 19, 2949–2963. ( 10.1111/1462-2920.13767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shou W, Ram S, Vilar JM. 2007. Synthetic cooperation in engineered yeast populations. Proc. Natl Acad. Sci. USA 104, 1877–1882. ( 10.1073/pnas.0610575104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillesland KL, Stahl DA. 2010. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl Acad. Sci. USA 107, 2124–2129. ( 10.1073/pnas.0908456107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medlock GL, Carey MA, McDuffie DG, Mundy MB, Giallourou N, Swann JR, Kolling GL, Papin JA. 2018. Inferring metabolic mechanisms of interaction within a defined gut microbiota. Cell Syst. 7, 245–257. ( 10.1016/j.cels.2018.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445, 533–536. ( 10.1038/nature05514) [DOI] [PubMed] [Google Scholar]
- 45.Bastolla U, Lässig M, Manrubia SC, Valleriani A. 2005. Biodiversity in model ecosystems. I: Coexistence conditions for competing species. J. Theor. Biol. 235, 521–530. ( 10.1016/j.jtbi.2005.02.005) [DOI] [PubMed] [Google Scholar]
- 46.Bastolla U, Fortuna MA, Pascual-García A, Ferrera A, Luque B, Bascompte J. 2009. The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature 458, 1018–1020. ( 10.1038/nature07950) [DOI] [PubMed] [Google Scholar]
- 47.Pascual-García A, Bastolla U. 2017. Mutualism supports biodiversity when the direct competition is weak. Nat. Commun. 8, 14326 ( 10.1038/ncomms14326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meszéna G, Gyllenberg M, Pásztor L, Metz JA. 2006. Competitive exclusion and limiting similarity: a unified theory. Theor. Popul. Biol. 69, 68–87. ( 10.1016/j.tpb.2005.07.001) [DOI] [PubMed] [Google Scholar]
- 49.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. 2012. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231. ( 10.1126/science.1219385) [DOI] [PubMed] [Google Scholar]
- 50.Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263 ( 10.1038/nrmicro3218) [DOI] [PubMed] [Google Scholar]
- 51.Goyal A. 2018. Metabolic adaptations underlying genome flexibility in prokaryotes. PLoS Genet. 14, e1007763 ( 10.1371/journal.pgen.1007763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giovannoni SJ, Thrash JC, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J. 8, 1553 ( 10.1038/ismej.2014.60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia SL, Buck M, Hamilton JJ, Wurzbacher C, Grossart HP, McMahon KD, Eiler A. 2018. Model communities hint at promiscuous metabolic linkages between ubiquitous free-living freshwater bacteria. mSphere 3, e00202-18. ( 10.1128/mSphere.00202-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, 3390–3395. ( 10.1128/mBio.00036-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez-Gorostiaga A, Bajić D, Osborne ML, Poyatos JF, Sanchez A. 2018. High-order interactions dominate the functional landscape of microbial consortia. bioRxiv 333534 ( 10.1101/333534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X, Polz MF, Alm EJ. 2019. Interactions in self-assembled microbial communities saturate with diversity. ISME J. 13, 1602 ( 10.1038/s41396-019-0356-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gravel D, Bell T, Barbera C, Bouvier T, Pommier T, Venail P, Mouquet N. 2011. Experimental niche evolution alters the strength of the diversity–productivity relationship. Nature 469, 89–92. ( 10.1038/nature09592) [DOI] [PubMed] [Google Scholar]
- 58.Langenheder S, Bulling MT, Solan M, Prosser JI. 2010. Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PLoS ONE 5, e10834 ( 10.1371/journal.pone.0010834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnyder E, Bodelier PL, Hartmann M, Henneberger R, Niklaus PA. 2018. Positive diversity-functioning relationships in model communities of methanotrophic bacteria. Ecology 99, 714–723. ( 10.1002/ecy.2138) [DOI] [PubMed] [Google Scholar]
- 60.Rohr RP, Saavedra S, Bascompte J. 2014. On the structural stability of mutualistic systems. Science 345, 1253497 ( 10.1126/science.1253497) [DOI] [PubMed] [Google Scholar]
- 61.Butler S, O'Dwyer JP. 2018. Stability criteria for complex microbial communities. Nat. Commun. 9, 2970 ( 10.1038/s41467-018-05308-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gadgil M, Solbrig OT. 1972. The concept of r- and K-selection: evidence from wild flowers and some theoretical considerations. Am. Nat. 106, 14–31. ( 10.1086/282748) [DOI] [Google Scholar]
- 63.Andrews JH, Harris RF. 1986. r- and K-selection and microbial ecology. In Advances in microbial ecology (ed. KC Marshall), pp. 99–147. Berlin, Germany: Springer.
- 64.Werner GDA, et al. 2014. Evolution of microbial markets. Proc. Natl Acad. Sci. USA 111, 1237–1244. ( 10.1073/pnas.1315980111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pacheco AR, Moel M, Segrè D. 2019. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103 ( 10.1038/s41467-018-07946-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piccardi P, Vessman B, Mitri S. 2019. Toxicity drives facilitation between 4 bacterial species. Proc. Natl Acad. Sci. USA 116, 15 979–15 984. ( 10.1073/pnas.1906172116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. 2016. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664. ( 10.1038/ismej.2015.142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Braakman R, Smith E. 2013. The compositional and evolutionary logic of metabolism. Phys. Biol. 10, 011001 ( 10.1088/1478-3975/10/1/011001) [DOI] [PubMed] [Google Scholar]
- 69.Borenstein E. 2012. Computational systems biology and in silico modeling of the human microbiome. Brief. Bioinform. 13, 769–780. ( 10.1093/bib/bbs022) [DOI] [PubMed] [Google Scholar]
- 70.Mayr E. 1997. The objects of selection. Proc. Natl Acad. Sci. USA 94, 2091–2094. ( 10.1073/pnas.94.6.2091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okasha S. 2001. Why won’t the group selection controversy go away? Br. J. Philos. Sci. 52, 25–50. ( 10.1093/bjps/52.1.25) [DOI] [Google Scholar]
- 72.Moreira D, López-García P. 1998. Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J. Mol. Evol. 47, 517–530. ( 10.1007/PL00006408) [DOI] [PubMed] [Google Scholar]
- 73.Pfeiffer T, Bonhoeffer S. 2003. An evolutionary scenario for the transition to undifferentiated multicellularity. Proc. Natl Acad. Sci. USA 100, 1095–1098. ( 10.1073/pnas.0335420100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arias-Sanchez FI, Vessman B, Mitri S. 2019. Artificially selecting microbial communities: if we can breed dogs, why not microbiomes?. PLoS Biol. 17, e3000356 ( 10.1371/journal.pbio.3000356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R Core Team 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/.
- 76.McMurdie PJ, Holmes S. 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 ( 10.1371/journal.pone.0061217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, JSSv022i04 ( 10.18637/jss.v022.i04) [DOI] [Google Scholar]
- 78.Warnes GR. et al. 2007. gplots: Various programming tools for plotting data. See https://CRAN.R-project.org/package=gplots (accessed 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.