Abstract
Phylosymbiosis, where similarities in host-associated microbial communities recapitulate the phylogeny of their hosts, is a newly recognized yet pervasive pattern in the field of host–microbe interactions. While phylosymbiosis has been documented across many systems, we still have a poor understanding of the mechanisms that underlie this emergent pattern. Host selection of the microbiome is a widely cited mechanism, yet other basic ecological and evolutionary processes (dispersal, drift and diversification) may also be at play. This paper discusses the roles that each of these processes and their interactions may play in yielding phylosymbiotic signals across hosts. Finally, this paper will identify open questions and methods that are required to better understand the relative contributions of these basic processes to phylosymbiosis. Given that phylosymbiosis has been shown to relate to functional components of host fitness, understanding the processes that contribute to these patterns will be important for our understanding of the ecology and evolution of host–microbe interactions.
This article is part of the theme issue ‘Conceptual challenges in microbial community ecology’.
Keywords: community assembly, community ecology, host–microbe interactions, microbiome
1. Introduction
Recent advances in the field of host–microbe interactions have demonstrated that macroorganisms are largely colonized by communities of microbes that have the potential to influence host phenotypes [1]. Understanding the forces that drive the structure of these communities is of great interest to microbial ecologists and evolutionary biologists. Studies in numerous systems show that the compositions of host-associated microbial communities are shaped by a complex set of host and environmental factors, including host genotype, species and ontogeny, as well as diet, habitat, geographical location and anthropogenic disturbance [2–6]. However, a thorough understanding of the forces that shape host-associated microbial communities is still needed.
One rather consistent driver of microbial community structuring across numerous host clades is evolutionary history. Specifically, phylosymbiosis, or the congruence between the evolutionary history of various host species and the community structures of their associated microbiomes, has been observed in numerous animal clades, as well as in some plant-associated communities [7–9]. In other words, as host genetic differences increase over time, differences in the structure of host-associated microbial communities will also increase [7]. Detailed descriptions of phylosymbiosis and how it is measured have been reviewed elsewhere [10]. Importantly, phylosymbiosis is different from co-diversification in that the microbiome dendrogram is of microbial community structures, not of specific microbes (though this can be the case and will be discussed more below). A handful of studies also demonstrate that patterns of phylosymbiosis correlate with functional effects on the hosts, such that hosts inoculated with microbial communities from different host species exhibit decreased performance and fitness, suggesting that these relationships may be acted upon by natural selection. For example, Peromyscus mice inoculated with the microbial communities from more distantly related hosts suffer decreases in their ability to digest food material [7], and Nasonia wasps inoculated with interspecific microbiomes exhibit lower survival than those inoculated with intraspecific microbial communities [11]. Moreover, hybridization of host species leads to a breakdown in phylosymbiotic communities, which may lead to negative consequences for host health and survival [12,13]. Despite the ecological and evolutionary importance of phylosymbiosis, we still have an extremely limited understanding of the mechanisms that underlie this pattern.
To date, it is largely assumed that differential selection of microbes by the host drives variation in microbial communities [8]. That is, hosts may be exposed to the same pool of potential microbial colonizers, but then filter and select certain microbes to persist as symbionts. However, theoretical studies have suggested that for some host-associated habitats, such as the vertebrate gut, host selection may not fully explain the observed patterns [8]. Therefore, other forces may contribute to the resulting patterns of phylosymbiosis across host species. This paper will discuss other basic ecological and evolutionary processes that underlie community assembly: dispersal, selection, drift and diversification [14,15], and how each may contribute to patterns of phylosymbiosis. Potential interactions between these processes, such as priority effects, will also be discussed. Finally, this paper will address future directions needed to enhance our understanding of phylosymbiosis, and how ecological and evolutionary theories of community assembly may need to be adapted to understand these processes. For the purposes of this paper, I will largely focus on gut microbial communities, as these systems have been best studied mechanistically and experimentally. Similar processes may occur on other body sites or in plants, and will be discussed briefly throughout the paper.
2. Microbial community ecology
Insights into the structure of microbial communities, including host-associated communities, were significantly advanced with the development of 16S rRNA gene sequencing technologies. These inventories provide thorough snapshots into the structure of a microbial community in a given sample. With these data, researchers have begun to apply classical ecological theory to host-associated microbial communities [16]. For example, one of community ecology's few ‘laws’ is the species–area relationship, such that larger islands should contain more species of macroorganisms [17]. Similar relationships have been found in the gut microbiome, where larger animals harbour more diverse microbial communities [18]. It is possible that ecological and evolutionary processes such as dispersal, selection, drift and diversification may underlie observed patterns of phylosymbiosis. These processes will first be discussed in isolation, but often these processes will occur simultaneously and have the potential to interact with one another. Therefore, interactions between processes (such as priority effects) will also be discussed.
(a). Dispersal
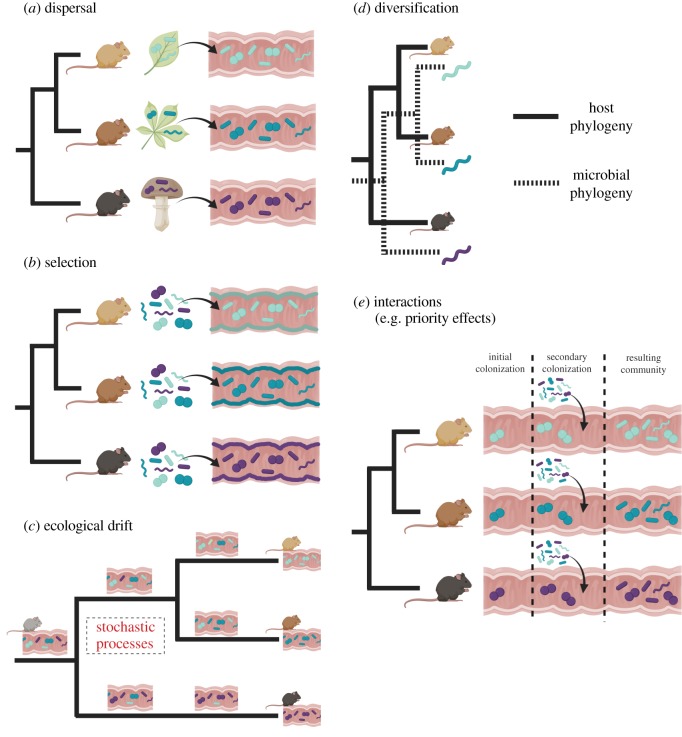
Dispersal, or the movement of microbes between environments, may underlie patterns of phylosymbiosis if species differentially encounter microbes as part of their natural history (figure 1a). For example, if dietary strategies have diverged across species, they may be inoculated with differential communities from food items. Indeed, controlled experimental studies have demonstrated that environmental sources of microbes can be extremely important for inoculation and maintenance of host-associated microbial communities. For example, stinkbugs [19] and bobtail squid [20] acquire specific symbionts from the environment every generation. Moreover, salamanders housed in sterile containers lose a significant proportion of their skin microbiome compared with those housed in containers with soil [21]. Additionally, feeding Drosophila flies on sterile media causes them to lose members of their gut microbial community [22]. Such dispersal effects have also been suggested to exist in the wild, where allopatric animal populations exhibit lower overlap in gut microbial community structure compared with sympatric animal populations, and this overlap decreases with geographical distance [23]. Intraspecific phylosymbiosis has been reported across populations of pikas (Ochotona princeps), which exhibit distinct host genetic structure over geographical scales [24]. While host genetics could play a role (discussed more below), it could be that dispersal limitations across geographical scales underlie the phylosymbiotic pattern observed across these populations.
Figure 1.
Ecological and evolutionary processes that may contribute to observed patterns of phylosymbiosis. Here, colours of microbes represent different species, and shapes represent different functional guilds. (a) Disperal—host species may come into contact with distinct microbial communities as part of their natural history or transmission from conspecifics. (b) Selection—hosts may exert selective forces that allow certain microbial members to colonize, while others are excluded from associating with the host. (c) Ecological drift—stochastic processes may result in the gain or loss of microbial members over evolutionary time. (d) Diversification—microbes may co-speciate with their host species. (e) Interactions—here, priority effects are depicted. Here, assume that there is no host selection occurring (hence the lack of coloured epithelial layers as depicted in (b)). Hosts may be initially colonized with a vertically transmitted microbe that has co-speciated with the host. These initial colonizing microbes may then exert subsequent selective forces through habitat modification or microbe–microbe interactions to only allow certain microbes to colonize, yielding phylosymbiotic patterns. (Online version in colour.)
Dispersal may not be such a passive process as environmental or trophic acquisition. There may be microbial and host adaptations to facilitate dispersal-based symbiosis. For example, environmental microbes that have been experimentally evolved to be associated with the zebrafish gut exhibit increased motility and host colonization rates [25]. Moreover, transmission between conspecifics would represent a type of dispersal. Many animals exhibit distinct adaptations to promote vertical microbial transmission between generations [26]. For example, female stinkbugs (Megacopta punctatissima) place ‘symbiont capsules’ near their eggs, and nymphs engage in a wandering behaviour to increase acquisition of these microbial symbionts [27]. Further, animals may acquire microbes horizontally through social interactions between conspecifics [28]. In mice, gut microbial taxa that are aerotolerant are more likely to be transmitted horizontally across individuals [29]. These microbial and host adaptations may help to promote phylosymbiotic patterns across host species if hosts only acquire microbes through vertical and horizontal transmission from conspecifics. However, it should be noted that in the absence of other interacting forces (selection, drift or diversification), it seems unlikely that dispersal by transmission alone would yield phylosymbiotic patterns across organisms. When clades of animal hosts are reared under common environments (presumably removing dispersal barriers), they still harbour host-specific microbial communities [7,30], suggesting that other factors are at play.
(b). Selection
Selection, or differential filtering of a microbial pool, is currently the leading hypothesis to explain patterns of phylosymbiosis (figure 1b; [8]). Studies have been conducted on several clades of animals reared under controlled experimental settings to explicitly remove potential environmental, sex and endosymbiont factors that could underlie or obscure patterns of phylosymbiosis [7]. Indeed, significant degrees of phylosymbiosis were detected in all animal clades, suggesting that there are genetic underpinnings that structure host-associated microbial communities [7]. Other studies have inoculated numerous germ-free animals of different genotypes or species with the same pool of microbes and found that these host genotypes or species select for distinct microbial communities [31,32]. Together, these studies demonstrate that there are selective processes occurring in the animal gut.
What, then, might be the mechanisms of selection to filter and sculpt microbial communities? A likely candidate seems to be the immune system, as these processes recognize self from non-self and filter out pathogenic microbes. In Hydra, species-specific expression profiles of antimicrobial peptides (AMPs) contribute significantly to phylosymbiotic community structures, such that transgenic Hydra that are deficient in AMPs fail to select for particular microbial members [33]. The adaptive immune system contributes to the filtering of environmental microbes to determine the community of the zebrafish gut, though dispersal seems to overwhelm this effect [34–36]. Numerous intraspecific genome-wide association studies between animal genomes and gut microbial community structure have identified immune genes as likely candidates [37–39]. Even components of the plant immune system (e.g. salicylic acid) have been shown to be important in determining the community structure of the root microbiome [40].
There may also be other physiological mechanisms by which hosts select for certain microbial members. For example, the gut is lined with mucins and glycans that can vary in their structure across species [41]. Mice that lack the α1–2 fucosyltransferase gene, which codes for an enzyme that fucosylates glycans on the intestinal epithelium, harbour microbial communities distinct from those of wild-type mice [42]. Host-encoded digestive enzymes can also alter the nutritional environment of the gut, and act to select for certain microbes in the gut [43,44]. Even host-encoded microRNAs, which normally regulate host gene expression, can influence bacterial gene expression and act to sculpt community structure [45,46]. Importantly, these studies have all been conducted within a single host species, and so we still lack an understanding of whether these mechanisms could underlie variation in microbial community structure across host species.
Much research on the selection of the microbiome is rather host-centric, and it should be recognized that microbes may also play a role in ‘choosing’ which hosts to colonize. Microbes may select hosts where their replication rates may be higher, and thus they gain a colonization (and/or transmission) advantage over other microbes [11]. Many pathogenic microbes recognize specific host receptors to determine host specificity [47], and similar processes may occur for non-pathogenic bacteria. Though, some specificity may be combinations of host and microbial factors. For example, the specificity of plant–Rhizobium mutualisms is driven by coordination of host-specific secretion of chemical signals and microbial-specific recognition of these chemical signals [48]. Similarly, it may be challenging to determine whether microbes capable of binding to specific glycans on the gut lining would represent selection by the host or by the microbe. Disentangling the directions and interactions associated with selection (hosts selecting microbes, microbes choosing hosts or a combination) represents a challenging but important gap in our understanding of phylosymbiosis and host–microbe interactions.
(c). Ecological drift
Stochastic or random processes may cause some microbes (especially ones with low abundances) to become extinct. The term ‘ecological drift’ is used to describe the random fluctuations in species abundances over time, and is somewhat analogous to the concept of genetic drift, or random variation in allele frequencies across generations. Processes of ecological drift could result in distinct microbial communities across host species, as these communities have been separated from one another over evolutionary time (figure 1c). Ecological drift generally occurs in communities where diversity and the strength of deterministic processes are low [49]. Given that the gut is typically densely populated and diverse, it seems unlikely that ecological drift in isolation would contribute largely to interspecies differences across hosts that are phylosymbiotic. Indeed, theoretical studies suggest that stochastic processes are unlikely to contribute to microbiome community structure given that selective forces by the host or competitive forces between microbes are typically considerable and suppress the effects of ecological drift [50]. Some experimental studies suggest that stochastic processes may contribute to determining the structure of the phyllosphere microbiome [51], and underlie the well-known ‘cage-effects’ that exist in studies of the mouse gut microbiome [52]. A recent study also suggests that ecological drift likely occurs in gut communities, but after initial environmental selection of microbes [53]. Thus, ecological drift does likely occur in host-associated microbial communities, but is unlikely to be the sole determinant of phylosymbiosis across host species. Instead, interactions between ecological drift and other processes (dispersal, diversification and selection) may be important, and will be discussed below. Overall, our understanding of ecological drift in microbial communities is limited within a host species, and even more so for how these processes could play out over evolution and host speciation.
(d). Diversification
As hosts speciate over evolutionary time, single or multiple symbionts may also evolve into new species in a manner that is congruent with host phylogeny (figure 1d). Such co-phylogenies are often observed in endosymbionts of invertebrates, which are often transmitted with high fidelity from generation to generation [54]. As mentioned earlier, animals exhibit a wide array of mechanisms to promote vertical transmission of the microbiome from one generation to another [26], and plants can transmit microbial symbionts in seeds [55]. With stable relationships over evolutionary time, it is possible that hosts and microbial species may exhibit co-speciation or co-phylogeny. Though, co-phylogenies can also occur as a result of diversification of microbes onto an existing host clade, a process known as host-shift speciation [56]. As mentioned above, there are many environmental sources of microbial symbionts which may obscure these patterns across microbial taxa, and often only a subset of the microbiome is transmitted across generations [57]. Thus, disentangling the relative importance of diversification in complex and diverse communities, and whether co-phylogenies represent co-speciation or host-shift speciation, remains poorly understood.
Only limited studies have addressed this question using members of complex host-associated microbiomes. One study used a novel sensitivity analysis to demonstrate that patterns of phylosymbiosis observed in ants and apes may be due to recent bacterial evolution, even if the evolving microbial groups that contribute to this pattern make up only a small portion of the overall microbiota [58]. Indeed, detailed phylogenies of individual microbial taxa do exhibit co-phylogeny with their hosts [59,60]. However, as described above, there are numerous examples of horizontal transmission both within and across host species [23,28], suggesting that diversification is unlikely to contribute entirely to observed patterns of phylosymbiosis.
(e). Interactions
The processes described above are unlikely to act in complete isolation to yield phylosymbiosis. Rather, there may be interactions between these processes that result in distinct microbial communities across host species. One example might be priority effects, where dispersal history can change later aspects of selection, drift and diversification [61]. For example, the first microbial colonizers of the gut may alter the gut environment by activating the host immune system or though the production of bioactive compounds that inhibit the growth of other microbial species (figure 1e). Indeed, one study elegantly demonstrated priority effects by inoculating germ-free mice in sequence with two contrasting microbial communities, and showing that the resulting communities inventoried six weeks later were distinct depending on colonization history [62]. Additionally, birds may transmit some microbial members in ovo [63], and experimental administration of bacteria in ovo can alter the community structure of the chicken gut microbiome at 10 days of age [64] More evidence supporting the hypothesis that priority effects act in the assembly of the gut microbiome, as well as a discussion of the mechanisms that could underlie these processes, are excellently reviewed elsewhere [61].
Related to the idea of priority effects is the notion of hub taxa, or microbial species that may have a large number of interactions with other microbes to either promote or prevent colonization, and thus alter overall community structure [65]. Hub taxa are often predicted through network analysis, though the experimental demonstration of their effects should be conducted. Experimental inoculations of Arabidopsis thaliana with putative hub taxa accounted for 15–20% of the variation in the community structure of the phyllosphere microbiome. Further, inoculation of mice with just four isolates of tannin-degrading bacteria significantly altered the community structure and function of the gut microbiome [66].
To date, studies on priority effects and hub taxa have been conducted within a single host species. How might priority effects or hub taxa then drive observed patterns of phylosymbiosis across host species? Again, both plants and animals have mechanisms to transmit microbial symbionts between generations [26,55]. However, it is usually a subset of the microbiota that is transmitted rather than the entire community. It could be that important hub taxa are transmitted early, which then exert priority effects to determine the resulting host-species-specific microbiome. With regards to hub taxa, it could be that hosts exert selection to favour a particular hub microbe, which then sculpts the rest of the microbial community through microbe–microbe interactions. These proposed mechanisms are purely speculative at this point, and require experimental validation in a diversity of systems.
3. Future directions
To date, most studies on phylosymbiosis have investigated concordance between host phylogeny and dendrograms of microbial communities, and potential functional effects of host-specific microbial communities [7–9]. These studies demonstrate the presence of phylosymbiotic patterns and their association with functional effects, but do not elucidate the processes that yield these patterns. Understanding the ecological and evolutionary processes that contribute to microbial community structures across host species will require numerous advances in theory and methodology. The processes discussed above are largely drawn from our understanding of community ecology in macroorganisms and free-living microbes. However, host-associated microbiomes present unique challenges in our understanding of community ecology, such as the high prevalence of horizontal gene transfer among organisms, which may alter aspects of selection, and the difficulty of disentangling host control from other environmental factors such as diet or geography [67]. Therefore, methodological advances and creative experimental design will be required to elucidate the processes that contribute to phylosymbiosis and their relative contributions.
Germ-free systems will likely be imperative to testing how ecological and evolutionary processes contribute to phylosymbiosis. In the past, germ-free techniques have largely been developed for model systems such as mice [68], fruit flies [69] and zebrafish [70]. However, new methods have been developed to generate germ-free Nasonia jewel wasps [71] and sticklebacks [72], allowing host–microbe interactions to be studied in an evolutionary context [11,73]. The generation of additional germ-free systems that can be applied to several species within a host clade will be important for testing mechanisms underlying phylosymbiosis. For example, reciprocal transplant experiments of microbial communities across various germ-free host species could be used to test aspects of selection [32]. Additionally, previous experiments regarding priority effects in the gut have only been conducted on single host species [62]. Thus, establishing germ-free rearing techniques that can be used in several species of closely related host species will open more opportunities to test the processes that drive patterns of phylosymbiosis.
Experimental evolution experiments may also provide powerful systems in which to test the effects of community processes on phylosymbiosis [74]. For example, bank voles (Myodes glareolus) that have undergone repeated artificial selection for the ability to feed on high fibre diets exhibit distinct gut microbial communities compared with randomly bred control lines [75]. Under tightly controlled laboratory settings, the rodent gut microbiome is primarily inherited vertically [29], and so follow up experiments could be conducted to understand how host selection, microbial diversification or ecological drift may have contributed to these results in bank voles. Experimental evolution approaches have also been applied to host-associated microbes [25,76]. One could experimentally evolve microbes to specific hosts and then investigate their performance or success in another host species.
There has also been a call to use cultivated microbes to understand aspects of community assembly [77]. For example, microcosm experiments can help to disentangle aspects of dispersal, priority effects and microbial competition [78,79]. While a longstanding paradigm has been that less than 1% of microbes are culturable, a recent analysis found that approximately 75% of human-associated microbial taxa have close relatives that are culturable (as determined by 16S rRNA similarity; [80]). Further, studies that use a variety of culturing techniques have successfully isolated 50–95% of microbial taxa from human samples [81,82] and 23% of taxa from laboratory rodent samples [83]. Cultured microbes from diverse clades of host species, along with the development of microcosms that replicate the gut environment or experiments in germ-free model systems will help to understand the role of ecological processes in phylosymbiosis.
To truly understand community-level variation, it may be important to track individual strains of microbes, rather than relying solely on the 16S rRNA gene. Further, using the 16S rRNA gene may obscure functional differences between host-specific microbes. Two species of nematodes (Caenorhabditis elegans and C. briggsae) both host strains of Enterobacter cloacae that are indistinguishable at the 16S rRNA gene, yet confer different functional phenotypes to their specific hosts [30]. Strain tracking, or the use of genomic or metagenomic sequencing to track transmission of individual microbial strains, has been used to demonstrate the vertical transmission of some members of the human gut microbiota [84] and horizontal transmission across wild giraffes through social contact [85]. This powerful technique will help to better understand community composition and the processes that underlie phylosymbiosis across host species.
Finally, I argue that these questions should be addressed in a diversity of systems. The field of comparative physiology subscribes to an idea known as Krogh's principle, which states that ‘for such a large number of problems there will be some animal of choice, or a few such animals, on which it can be most conveniently studied’ [86, p. 247]. Such a principle can easily be applied to the field of host–microbe interactions [87]. The strength of phylosymbiotic patterns varies across host clades; patterns are strong and consistent in mammals [24,60,88], unclear in Drosophila flies [7,89], and rare on plant surfaces [8]. Disentangling the ecological processes that contribute to the presence or absence of phylosymbiosis across organisms will greatly inform our understanding of the mechanisms behind these patterns.
4. Conclusion
While host selection or host filtering of microbial communities has been largely assumed to be a dominant contributor to patterns of phylosymbiosis, other ecological and evolutionary processes may also contribute. Aspects of dispersal, drift, diversification and interactions between these processes may also contribute. While these processes have been demonstrated within a single or a few host species, they have rarely been studied in the context of phylosymbiosis across several host species. With advances in sequencing technologies, the development of more experimental systems to investigate host–microbe interactions, including many more germ-free systems, and creative experimental design, the time is ripe to investigate these processes and how they contribute to aspects of ecology and evolution of hosts and host-associated microbial communities.
Acknowledgements
I am grateful to Seth Bordenstein, Samantha Fontaine and two anonymous reviewers for providing comments that improved this paper.
Data accessibility
This article has no additional data
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279 ( 10.1038/nrmicro2540) [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486, 222 ( 10.1038/nature11053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amato KR, et al. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344 ( 10.1038/ismej.2013.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trevelline BK, Fontaine SS, Hartup BK, Kohl KD. 2019. Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B 286, 20182448 ( 10.1098/rspb.2018.2448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. 2018. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3, e00097-18 ( 10.1128/mSystems.00097-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MT. J. 2018. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA 115, E1157–E1165. ( 10.1073/pnas.1717617115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim SJ, Bordenstein SR. 2019. An introduction to phylosymbiosis. PeerJ 7, e27879v2 ( 10.7287/peerj.preprints.27879v2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Opstal EJ, Bordenstein SR. 2019. Phylosymbiosis impacts adaptive traits in Nasonia wasps. mBio 10, e00887-19 ( 10.1128/mBio.00887-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brucker RM, Bordenstein SR. 2013. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. ( 10.1126/science.1240659) [DOI] [PubMed] [Google Scholar]
- 13.Wang J, et al. 2015. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat. Commun. 6, 6440 ( 10.1038/ncomms7440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemergut DR, et al. 2013. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. ( 10.1128/MMBR.00051-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adair KL, Douglas AE. 2017. Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 35, 23–29. ( 10.1016/j.mib.2016.11.002) [DOI] [PubMed] [Google Scholar]
- 16.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. ( 10.1126/science.1224203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lomolino MV. 2001. Ecology's most general, yet protean pattern: the species–area relationship. J. Biogeogr. 27, 17–26. ( 10.1046/j.1365-2699.2000.00377.x) [DOI] [Google Scholar]
- 18.Sherrill-Mix S, et al. 2018. Allometry and ecology of the bilaterian gut microbiome. mBio 9, e00319-18 ( 10.1128/mBio.00319-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73, 4308–4316. ( 10.1128/AEM.00067-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 12, e1001783 ( 10.1371/journal.pbio.1001783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loudon A, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN. 2014. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 8, 830 ( 10.1038/ismej.2013.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blum JE, Fischer CN, Miles J, Handelsman J. 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4, e00860-13 ( 10.1128/mbio.00860-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. 2017. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl Acad. Sci. USA 114, 13 768–13 773. ( 10.1073/pnas.1700122114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl KD, Varner J, Wilkening JL, Dearing MD. 2018. Gut microbial communities of American pikas (Ochotona princeps): evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 87, 323–330. ( 10.1111/1365-2656.12692) [DOI] [PubMed] [Google Scholar]
- 25.Robinson CD, Klein HS, Murphy KD, Parthasarathy R, Guillemin K, Bohannan BJ. M. 2018. Experimental bacterial adaptation to the zebrafish gut reveals a primary role for immigration. PLoS Biol. 16, e2006893 ( 10.1371/journal.pbio.2006893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 11, e1001631 ( 10.1371/journal.pbio.1001631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. 2008. Symbiont acquisition alters behaviour of stinkbug nymphs. Biol. Lett. 4, 45–48. ( 10.1098/rsbl.2007.0510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tung J, et al. 2015. Social networks predict gut microbiome composition in wild baboons. eLife 4, e05224 ( 10.7554/eLife.05224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeller AH, Suzuki TA, Phifer-Rixey M, Nachman MW. 2018. Transmission modes of the mammalian gut microbiota. Science 362, 453–457. ( 10.1126/science.aat7164) [DOI] [PubMed] [Google Scholar]
- 30.Berg M, Zhou XY, Shapira M. 2016. Host-specific functional significance of Caenorhabditis gut commensals. Front. Microbiol. 7, 1622 ( 10.3389/fmicb.2016.01622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikaelyan A, Thompson CL, Hofer MJ, Brune A. 2015. Deterministic assembly of complex bacterial communities in guts of germ-free cockroaches. Appl. Environ. Microbiol. 82, 1256–1263. ( 10.1128/AEM.03700-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433. ( 10.1016/j.cell.2006.08.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, Fraune S. 2013. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl Acad. Sci. USA 110, E3730–E3738. ( 10.1073/pnas.1304960110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. M. 2015. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664. ( 10.1038/ismej.2015.142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stagaman K, Burns AR, Guillemin K, Bohannan BJ. M. 2017. The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J. 11, 1630–1639. ( 10.1038/ismej.2017.28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burns AR, Miller EL, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S, Guillemin K, Bohannan BJM. 2017. Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl Acad. Sci. USA 114, 11 181–11 186. ( 10.1073/pnas.1702511114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE. 2016. Cross-species comparisons of host genetic associations with the microbiome. Science 352, 532–535. ( 10.1126/science.aad9379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blekhman R, et al. 2015. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 ( 10.1186/s13059-015-0759-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki TA, Phifer-Rixey M, Mack KL, Sheehan MJ, Lin D, Bi K, Nachman MW. 2019. Host genetic determinants of the gut microbiota of wild mice. Mol. Ecol. 28, 3197–3207. ( 10.1111/mec.14905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lebeis SL, et al. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. ( 10.1126/science.aaa8764) [DOI] [PubMed] [Google Scholar]
- 41.Hooper LV, Gordon JI. 2001. Glycans as legislators of host–microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11, 1R–10R. ( 10.1093/glycob/11.2.1R) [DOI] [PubMed] [Google Scholar]
- 42.Kashyap PC, et al. 2013. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl Acad. Sci. USA 110, 17 059–17 064. ( 10.1073/pnas.1306070110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malo MS, et al. 2010. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 59, 1476–1484. ( 10.1136/gut.2010.211706) [DOI] [PubMed] [Google Scholar]
- 44.Poole AC, et al. 2019. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 25, 553–564. ( 10.1016/j.chom.2019.03.001) [DOI] [PubMed] [Google Scholar]
- 45.Liu S, da Cunha A Pires, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. 2016. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43. ( 10.1016/j.chom.2015.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan C, Burns MB, Subramanian S, Blekhman R. 2018. Interaction between host microRNAs and the gut microbiota in colorectal cancer. mSystems 3, e00205-17 ( 10.1128/mSystems.00205-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan X, Yang Y, Zhang JR. 2014. Molecular basis of host specificity in human pathogenic bacteria. Emerg. Microbes Infect. 3, 1–10. ( 10.1038/emi.2014.23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Liu J, Zhu H. 2018. Genetic and molecular mechanisms underlying symbiotic specificity in legume–rhizobium interactions. Front. Plant Sci. 9, 313 ( 10.3389/fpls.2018.00313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Phil. Trans. R. Soc. B 366, 2351–2363. ( 10.1098/rstb.2011.0063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Q, Wu S, Sukumaran J, Rodrigo A. 2017. Models of microbiome evolution incorporating host and microbial selection. Microbiome 5, 127 ( 10.1186/s40168-017-0343-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5, e00682-13 ( 10.1128/mBio.00682-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCafferty J, Mühlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. 2013. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 7, 2116–2125. ( 10.1038/ismej.2013.106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sieber M, et al. 2019. Neutrality in the metaorganism. PLoS Biol. 17, e3000298 ( 10.1371/journal.pbio.3000298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. ( 10.1146/annurev.genet.41.110306.130119) [DOI] [PubMed] [Google Scholar]
- 55.Truyens S, Weyens N, Cuypers A, Vangronsveld J. 2015. Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. ( 10.1111/1758-2229.12181) [DOI] [Google Scholar]
- 56.de Vienne DM, Refrégier G, López-Villavicencio M, Tellier A, Hood ME, Giraud T. 2013. Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198, 347–385. ( 10.1111/nph.12150) [DOI] [PubMed] [Google Scholar]
- 57.Wang S, Ryan CA, Boyaval P, Dempsey EM, Ross RP, Stanton C. 2019. Maternal vertical transmission affecting early-life microbiota development. Trends Microbiol. 28, 28–45. ( 10.1016/j.tim.2019.07.010) [DOI] [PubMed] [Google Scholar]
- 58.Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. ( 10.1111/mec.12611) [DOI] [PubMed] [Google Scholar]
- 59.Moeller AH, et al. 2016. Cospeciation of gut microbiota with hominids. Science 353, 380–382. ( 10.1126/science.aaf3951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. 2017. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 ( 10.1038/ncomms14319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sprockett D, Fukami T, Relman DA. 2018. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 15, 197 ( 10.1038/nrgastro.2017.173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martínez I, et al. 2018. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521 ( 10.7554/eLife.36521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trevelline BK, MacLeod KJ, Knutie SA, Langkilde T, Kohl KD. 2018. In ovo microbial communities: a potential mechanism for the initial acquisition of gut microbiota among oviparous birds and lizards. Biol. Lett. 14, 20180225 ( 10.1098/rsbl.2018.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson KM, Rodrigues DR, Briggs WN, Duff AF, Chasser KM, Bielke LR. 2019. Evaluation of the impact of in ovo administered bacteria on microbiome of chicks through 10 days of age. Poult. Sci. 98, 5949–5960. ( 10.3382/ps/pez388) [DOI] [PubMed] [Google Scholar]
- 65.Banerjee S, Schlaeppi K, van der Heijden MGA. 2018. Keystone taxa as drivers of microbiome structuring and functioning. Nat. Rev. Microbiol. 16, 567–576. ( 10.1038/s41579-018-0024-1) [DOI] [PubMed] [Google Scholar]
- 66.Kohl KD, Stengel A, Dearing MD. 2016. Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environ. Microbiol. 18, 1720–1729. ( 10.1111/1462-2920.12841) [DOI] [PubMed] [Google Scholar]
- 67.Koskella B, Hall LJ, Metcalf CJ. E. 2017. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1, 1606 ( 10.1038/s41559-017-0340-2) [DOI] [PubMed] [Google Scholar]
- 68.Arvidsson C, Hallén A, Bäckhed F. 2012. Generating and analyzing germ-free mice. Curr. Protoc. Mouse Biol. 2, 307–316. ( 10.1002/9780470942390.mo120064) [DOI] [PubMed] [Google Scholar]
- 69.Kietz C, Pollari V, Meinander A. 2018. Generating germ-free Drosophila to study gut-microbe interactions: protocol to rear Drosophila under axenic conditions. Curr. Protoc. Toxicol. 77, e52 ( 10.1002/cptx.52) [DOI] [PubMed] [Google Scholar]
- 70.Melancon E, Gomez De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, Eisen JS, Guillemin K. 2017. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 138, 61–100. ( 10.1016/bs.mcb.2016.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shropshire JD, van Opstal EJ, Bordenstein SR. 2016. An optimized approach to germ-free rearing in the jewel wasp Nasonia. PeerJ 4, e2316 ( 10.7717/peerj.2316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forberg T, Milligan-Myhre K. 2017. Gnotobiotic fish as models to study host–microbe interactions. In Gnotobiotics (eds Schoeb TR, Eaton KA), pp. 369–383. New York, NY: Academic Press. [Google Scholar]
- 73.Milligan-Myhre K, Small CM, Mittge EK, Agarwal M, Currey M, Cresko WA, Guillemin K. 2016. Innate immune responses to gut microbiota differ between oceanic and freshwater threespine stickleback populations. Dis. Models Mech. 9, 187–198. ( 10.1242/dmm.021881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoang KL, Morran LT, Gerardo NM. 2016. Experimental evolution as an underutilized tool for studying beneficial animal–microbe interactions. Front. Microbiol. 7, 1444 ( 10.3389/fmicb.2016.01444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kohl KD, Sadowska E, Rudolf A, Dearing MD, Koteja P. 2016. Experimental evolution on a wild mammal species results in modifications of gut microbial communities. Front. Microbiol. 7, 634 ( 10.3389/fmicb.2016.00634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martino ME, Joncour P, Leenay R, Gervais H, Shah M, Hughes S, Gillet B, Beisel C, Leulier F. 2018. Bacterial adaptation to the host's diet is a key evolutionary force shaping Drosophila-Lactobacillus symbiosis. Cell Host Microbe 24, 109–119. ( 10.1016/j.chom.2018.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolfe BE. 2018. Using cultivated microbial communities to dissect microbiome assembly: challenges, limitations, and the path ahead. mSystems 3, e00161-17 ( 10.1128/mSystems.00161-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Svoboda P, Lindström ES, Osman OA, Langenheder S. 2018. Dispersal timing determines the importance of priority effects in bacterial communities. ISME J. 12, 644–646. ( 10.1038/ismej.2017.180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Friedman J, Higgins LM, Gore J. 2017. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 ( 10.1038/s41559-017-0109) [DOI] [PubMed] [Google Scholar]
- 80.Martiny AC. 2019. High proportions of bacteria are culturable across major biomes. ISME J. 13, 2125–2128. ( 10.1038/s41396-019-0410-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257. ( 10.1073/pnas.1102938108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, Surette MG. 2016. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8, 72 ( 10.1186/s13073-016-0327-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lagkouvardos I, et al. 2016. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 1, 16131 ( 10.1038/nmicrobiol.2016.131) [DOI] [PubMed] [Google Scholar]
- 84.Asnicar F, et al. 2017. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2, e00164-16 ( 10.1128/mSystems.00164-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.VanderWaal KL, Atwill ER, Isbell LA, McCowan B. 2014. Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). J. Anim. Ecol. 83, 406–414. ( 10.1111/1365-2656.12137) [DOI] [PubMed] [Google Scholar]
- 86.Krogh A. 1929. The process of physiology. Am. J. Physiol. 90, 243–251. ( 10.1152/ajplegacy.1929.90.2.243) [DOI] [Google Scholar]
- 87.Kohl KD. 2018. A microbial perspective on the grand challenges in comparative animal physiology. mSystems 3, e00146-17 ( 10.1128/mSystems.00146-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kohl KD, Dearing MD, Bordenstein SR. 2018. Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol. Ecol. 27, 1874–1883. ( 10.1111/mec.14460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinson VG, Douglas AE, Jaenike J. 2017. Community structure of the gut microbiota in sympatric species of wild Drosophila. Ecol. Lett. 20, 629–639. ( 10.1111/ele.12761) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data