Abstract
Microbial physiological processes are intimately involved in nutrient cycling. However, it remains unclear to what extent microbial diversity or community composition is important for determining the rates of ecosystem-scale functions. There are many examples of positive correlations between microbial diversity and ecosystem function, but how microbial communities ‘map' onto ecosystem functions remain unresolved. This uncertainty limits our ability to predict and manage crucial microbially mediated processes such as nutrient losses and greenhouse gas emissions. To overcome this challenge, we propose integrating traditional biodiversity–ecosystem function research with ideas from genotype–phenotype mapping in organisms. We identify two insights from genotype–phenotype mapping that could be useful for microbial biodiversity–ecosystem function studies: the concept of searching ‘agnostically' for markers of ecosystem function and controlling for population stratification to identify microorganisms uniquely associated with ecosystem function. We illustrate the potential for these approaches to elucidate microbial biodiversity–ecosystem function relationships by analysing a subset of published data measuring methane oxidation rates from tropical soils. We assert that combining the approaches of traditional biodiversity–ecosystem function research with ideas from genotype–phenotype mapping will generate novel hypotheses about how complex microbial communities drive ecosystem function and help scientists predict and manage changes to ecosystem functions resulting from human activities.
This article is part of the theme issue ‘Conceptual challenges in microbial community ecology’.
Keywords: microbial ecology, biodiversity–ecosystem function, structure–function relationships, microbial processes, microbial community composition, biogeochemical cycles
1. Introduction
Ecology is broadly focused on understanding biodiversity and how that biodiversity shapes the ecosystems that humans depend on. Many ecosystem processes essential to all of life are mediated by microorganisms and therefore understanding the relationship between microbial biodiversity and ecosystem function is important [1,2]. Certain ecosystem functions are correlated with microbial diversity, indicating that we should be able to determine what aspects of microbial biodiversity influence ecosystem function. However, attempts to describe that mapping have borne little fruit [3,4]. We argue that to overcome this challenge we should look to other successful attempts at mapping biological variation onto higher order processes. In particular, population genetics and the process of genotype–phenotype mapping provide a number of potentially useful insights. For example, genotype–phenotype mapping often makes few assumptions about the nature of the map, i.e. it is ‘agnostic.' In addition, population geneticists have developed rigorous methods for reducing potentially confounding relationships such as geographic structuring of populations. Finally, the ultimate goal of genotype–phenotype mapping is to identify the unique contribution of genotype to phenotype separately from other drivers of phenotypic variation such as the environment. Inspired by this, we suggest a reframe of the question, ‘is microbial biodiversity related to ecosystem function?' to, ‘what is the unique contribution of the microbiome to ecosystem function independent of the environment?'
(a). There is evidence that microbial biodiversity matters for ecosystem function
There is increasing evidence for microbial biodiversity–ecosystem function relationships. For example, there are positive correlations between microbial diversity and ecosystem multifunctionality for a variety of ecosystems and most major lineages of microorganisms [5–7]. Adding microbial diversity or abundance to ecosystem models can in some cases improve model accuracy [4]. Contrived communities that vary in richness and communities created through sequential dilution or varying filter size to generate differences in diversity can also exhibit differences in ecosystem function [8–11]. Finally, reciprocal transplant and common garden experiments that manipulate the connection between community composition and environment reveal differences in ecosystem function for communities of distinct origins [12–14]. Given these relationships, many investigators have now moved on to the challenge of identifying the aspects of microbial biodiversity (e.g. specific taxa, genes, functional groups, etc.) that influence a given ecosystem function; however, this has proven especially challenging.
(b). The mapping between microbial biodiversity and ecosystem function has been elusive
Most studies that attempt to identify the aspects of microbial biodiversity that influence a given ecosystem function focus on ‘functional' gene or transcript abundance. In this case, qPCR or shotgun metagenomic sequencing is used to estimate the abundance of a gene or transcript that is a putative marker for a microbial process (and thus a marker for the functional group that performs that process). For example, the gene mcrA, which encodes a subunit of the enzyme that performs the final step in methanogenesis, is commonly used as a marker for methanogenesis and for the methanogen functional group. Other examples include pmoA and methanotrophy, nifH and nitrification, and nosZ and denitrification. It is often hypothesized that the abundance of these markers is predictive of the rate of the associated processes (for example, it is hypothesized that the abundance of mcrA is related to the rate of methanogenesis).
Some ecosystem functions in certain ecosystems can be predicted from the abundance or transcriptional activity of genetic markers for those functions. For example, soil methane production and consumption can under some circumstances be predicted from the genetic markers mcrA and pmoA [15–17]. However, for most ecosystem functions, the abundance of a functional gene or transcript is rarely positively correlated with the rate of the corresponding process [3]. The cases where there is a positive correlation tend to be restricted to agricultural ecosystems and certain functions within the nitrogen cycle [3]. In general, including aspects of microbial biodiversity (e.g. functional gene abundance or diversity) improves models of ecosystem function less than one-third of the time and increases variance explained by an average of only eight percentage points over environmental variables [4].
2. Genotype–phenotype mapping as a source of inspiration
In the approaches described above, microbial ecologists often use microbiome data to infer taxonomic composition, essentially creating species lists from data such as 16S rRNA marker genes or shotgun metagenomes. Interpreting microbiome data in this way has allowed us to use approaches from biodiversity–ecosystem function research (which are often focused on taxonomic or functional groups), but it has generally not been useful for creating more detailed descriptions of the relationship between microbial biodiversity and ecosystem function. But this approach is not the only way we could determine the relationship between a complex set of highly variable data and an aggregate function.
This kind of ‘many-to-one’ mapping is analogous to the challenge of identifying the genetic basis of complex traits in organismal populations. In such ‘genotype–phenotype' mapping studies, a population exhibits variation in a phenotype (e.g. height or disease state) as well as variation in potentially thousands of single-nucleotide polymorphisms (SNPs). To identify the genetic basis for a trait, investigators sample from this population and correlate phenotype with genotype. While some phenotypes (e.g. the propensity for diseases such as Parkinson's) are controlled by a single locus [18,19], most traits depend on a large number of genes that control variation in phenotype [20,21]. In addition, there is often no a priori expectation about which regions of the genome control that trait so we must search for genetic markers throughout the genome. If a marker is significantly correlated with the phenotype of interest, this either indicates it is inside a gene with a direct or indirect effect on phenotype or that it is in linkage disequilibrium (i.e. non-random association between two alleles) with a causal gene.
There are a number of parallels between this challenge faced by organismal biologists and that facing microbial community ecologists. They both involve large numbers of statistical comparisons. Both are attempting to identify causal relationships that are potentially confounded by complex patterns of covariation. There is often no strong expectation about which entities (i.e. which genomic regions or which microbial genes or lineages) are most likely to be causally related to phenotype or function, and thus ‘agnostic' approaches are needed. For some ecosystem functions, it is possible that a single taxon could substantially influence its rate. For example, methane flux from permafrost in Sweden may be controlled by a single taxon [17]. But for most ecosystem functions, there could be many taxa of small effect that contribute to the rate of ecosystem function. Finally, both ultimately require manipulation (of genes or taxa) to establish causation.
(a). The importance of a taxonomically ‘agnostic’ approach
Most microbial biodiversity–ecosystem function research up to this point has used an approach analogous to that used by plant ecologists studying biodiversity–ecosystem function relationships. This approach is to measure or manipulate the diversity of a taxonomic group (e.g. plants) and look for an association with the function performed by that group (e.g. primary productivity). We can think of plants as a ‘functional group,' i.e. a group of taxa united by their ability to perform a particular ecosystem function. For microbes, estimating functional group abundance can be much more challenging. From a small number of cultured isolates, we have a provisional understanding of which microbes might be involved in some ecosystem functions. By sequencing the genomes of these isolates, we have identified genetic markers for certain functions, which we call ‘functional genes.' But most microbial taxa remain uncultured and we do not know the function of most microbial taxa detected in environmental samples [22,23]. In addition, there have been recent discoveries of functions in unexpected taxonomic groups, for example methanogenesis by fungi and cyanobacteria, a function previously considered restricted to archaea [24,25].
As stated earlier, these functional markers are not correlated with their corresponding ecosystem function in most ecosystems and for most processes. In addition, they provide little explanatory power to the models of ecosystem function. Because of this, it might be prudent to look more agnostically at microbial communities to identify taxa, groups of taxa or genes that are important for predicting the rates of ecosystem functions rather than assuming that the genetic markers we have provisionally identified for a given function represent the most likely taxa or genes involved. This agnostic approach is analogous to the approach of many genotype–phenotype mapping studies (e.g. genome-wide association studies), which often look for associations between a phenotype and loci anywhere in the genome.
Beyond finding new physiologies in unexpected lineages, there are other reasons for looking agnostically. In the case of microbial functions, it may be that the organism that performs a function is not the limiting factor for the rate of that function. For example, the rate of soil-to-atmosphere methane flux could be limited by methanogens or methanotrophs or the balance of the two. However, it could also be limited by the bacteria that produce the fermentative byproducts that methanogens use as substrates. Or there could be indirect limitation by organisms that liberate nitrogen or phosphorus into mineral forms. In other words, the influence of microbial communities on the rate of ecosystem function could represent a complex metabolic network much like the regulation of gene expression in organisms that partially determines their phenotype. These broader patterns of biodiversity–ecosystem function relationships would be invisible to any study that solely focuses on the most relevant functional group without considering the possible influence of other taxa.
(b). Controlling for population stratification
It is widely accepted that organisms, including microorganisms, exhibit population stratification due to geographic and environmental separation [26,27]. This can lead to spurious associations between phenotypes and genetic markers that are at high frequency in isolated sub-populations. Association studies generally control for population stratification by accounting for shared ancestry among organisms in a population when modelling the connection between genotype and phenotype. Typically, microbial biodiversity–ecosystem function studies do not account for population stratification (i.e. community similarity among ecosystems), although there are some exceptions [28–30]. Community similarity (the community analogue of shared ancestry among organisms) is not as tightly linked to geography or environment as is shared ancestry. Therefore, it could be useful to account for these separately in microbial studies, particularly if we are interested in quantifying the effect of microbial communities on ecosystem function independent of these other factors.
Genome-wide association studies correct for stratification using a variety of methods. Generally, they ignore the underlying environmental and spatial distance between samples and instead use shared ancestry as a proxy for local selection and assortative mating. A common approach is to perform a regression of phenotype and shared ancestry (computed as the first one or more principal components of a genotype matrix) and then use the residuals from this model as the values for phenotype in a subsequent regression using the genotypes directly [31]. This principal component correction is designed to test the effect of individual genes after removing the effect of shared ancestry among individuals. Another approach, employed in our example, is variance component modelling (or mixed modelling, hierarchical modelling, etc.), where genotypic similarity is included as a covariate in the model to control for stratification while testing the genotype–phenotype connection [32].
If we control for covariates such as community similarity, geographic distance or environmental similarity, it changes the nature of our question. For example, if we test the correlation between the relative abundance of a taxon and the rate of methane flux, we are asking ‘is this taxon correlated with methane flux?' If we find a significant result, that may be because variation in the abundance of that organism directly or indirectly contributes to methane flux. However, it might also be that that organism lives only in ecosystems that happen to have a high rate of methane flux. In this scenario, we are unable to distinguish between these possibilities. However, if we add environmental variables or environmental similarity as a covariate in our model, we can ask, ‘Is this taxon uniquely associated with function in a way that it is independent of the environment?' By ‘uniquely associated', we mean those taxa associated with the function irrespective of environmental conditions, local community structure or spatial proximity. This slight reframing of the question could be especially rewarding for microbial biodiversity–ecosystem function research, particularly as it relates to incorporating microbial community data into ecosystem models. Finally, it is interesting in its own right to understand whether microbial communities are selected by the underlying environmental conditions to produce a particular rate of ecosystem function or whether community variation has functional consequences independent of the environment.
3. An example: high-affinity methane oxidation
To illustrate the ideas outlined above, we reanalysed a subset of previously published data from a paper that has demonstrated one successful approach for applying genotype–phenotype mapping to microbial communities [28]. In our reanalysis, we do not intend to challenge the conclusions of that paper, but instead we want to demonstrate how to perform this type of study for microbial ecologists unfamiliar with association studies. A full description of the study design, samples and data generation can be found in that article. Briefly, these data were gathered from intact soil cores taken from diverse ecosystems of the Congo Basin in Gabon, Africa. Cores were incubated in the laboratory under different concentrations of methane to identify the rates of specific methane cycling pathways. In this example, we analyse data from just one of these pathways, high-affinity methane oxidation (the oxidation of atmospheric concentrations of methane), which we will refer to simply as ‘methane oxidation'. In addition, we only include amplicon sequences from the DNA-inferred community and not the RNA-inferred community, both of which are presented in the original paper [28]. The data we analysed include methane oxidation rates, amplicon sequence variants (ASVs) generated using the ‘DADA2’ pipeline and inferred from unique 16S rRNA gene sequences [33], pmoA abundance estimates (via qPCR), and four environmental covariates (soil moisture, bulk density, carbon and nitrogen).
Analyses were conducted in the ‘R’ statistical environment using the ‘phyloseq’ package [34,35]. The relative abundances of ASVs were corrected using the variance stabilizing transformation from ‘DESeq2’ [36,37]. We first tested the correlation between ecosystem function and typical measures of microbial community structure: functional gene abundance and community richness, which were estimated using the ‘breakaway’ package [38]. We then tested covariation between community structure (estimated as Bray–Curtis distance using ‘vegan’), environmental variation (Euclidean distance) and geographic distance (Euclidean distance) using Mantel tests [39,40]. Finally, we identified taxa that were significantly associated with function independent of the environment by fitting variance component models using ‘varComp’ to test the relationship between relative abundance of each ASV and methane oxidation rate [32,41]. To illustrate how including different covariates (environmental, geographic and community) can result in different conclusions about which taxa are associated with function, we fitted this model with and without random effects variance components for environmental similarity, geographic site ID and Bray–Curtis similarity. Significant taxa were determined by controlling the false discovery rate at q-value < 0.05 [42]. Figures were created using ‘ggplot2’ [43]. All raw data and scripts required to recreate this analysis are available in the electronic supplementary material.
(a). Results and discussion
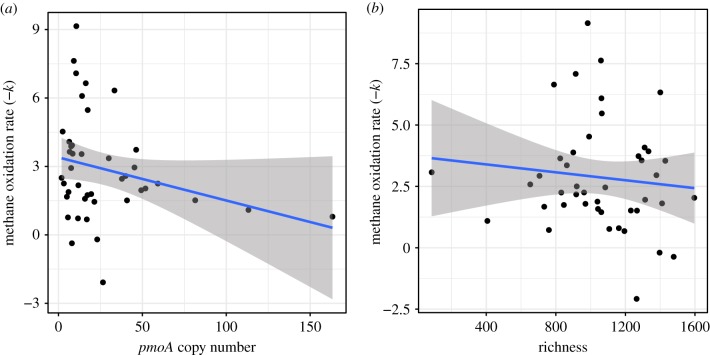
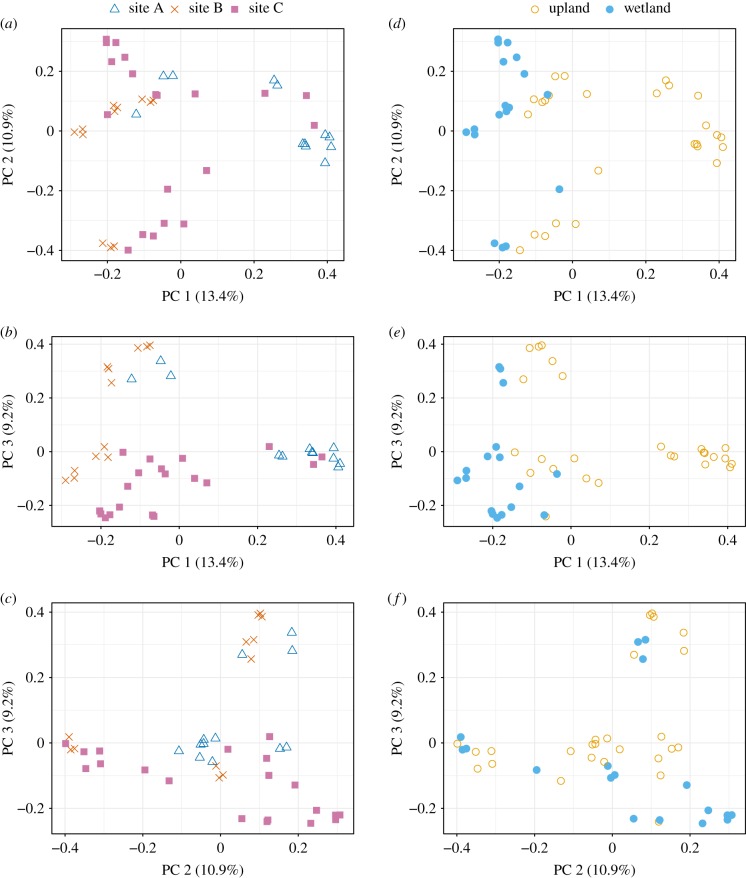
Microbial biodiversity–ecosystem function studies typically test functional group abundance or community richness as it relates to ecosystem function. In our case, methane oxidation rate was not significantly correlated with pmoA gene abundance or 16S rRNA gene-based taxonomic richness (table 1 and figure 1). To demonstrate that the covariance structure of the data might alter our conclusions about which taxa regulate ecosystem function, we tested collinearity between each pair of distance matrices for community, environment and geography. We found a moderate and significant correlation between community composition and environmental variation, geography and community composition, and geography and environmental variation (table 2 and figure 2). To visualize this population stratification, principal coordinate plots show that beta diversity of samples separated by site ID and by ecosystem type (wetland or upland; figure 2), which indicates substantial spatial and environmental structuring of microbial populations. This suggests that the presence or abundance of certain taxa will be elevated in specific ecosystems. In this case, high-affinity methane oxidation is typically greater in upland ecosystems than in wetland ecosystems and so any taxa differentially abundant in uplands will tend to be correlated with methane oxidation regardless of their involvement in that process. It is necessary to control for this stratification to rigorously identify associations between taxa and function.
Table 1.
Estimates for the linear relationship between methane oxidation rate and two measures of microbial community structure: pmoA functional gene abundance and ASV richness.
| term | estimate | s.e. | t-statistic | p-value |
|---|---|---|---|---|
| pmoA copy number | 0.019 | 0.011 | 1.705 | 0.096 |
| richness | 0.001 | 0.001 | 0.694 | 0.491 |
Figure 1.
Methane oxidation rate is not correlated with functional gene abundance or ASV richness. Correlations between methane oxidation rate and (a) abundance of the functional gene pmoA (n = 42), and (b) ASV richness (n = 44). Lines represent the ordinary least squares regression lines with standard errors. (Online version in colour.)
Table 2.
Mantel tests for each pair of dissimilarity matrices. Community distance matrix was based on Bray–Curtis distance while both environment and geography distance matrices were based on Euclidean distance. p-values were determined by permutation test with 999 permutations.
| terms | Mantel statistics (r) | 95% upper quantile of permutations | p-value |
|---|---|---|---|
| community ∼ geography | 0.353 | 0.056 | 0.001 |
| community ∼ environment | 0.474 | 0.109 | 0.001 |
| geography ∼ environment | 0.241 | 0.055 | 0.001 |
Figure 2.
Microbial community composition is spatially and environmentally structured. Principal coordinate plots of Bray–Curtis distance representing the first three axes of community composition. In (a–c) points are identified by site ID, and in (d–f) points are identified by wetland or upland ecosystem. All four environmental covariates separate strongly by wetland/upland. Axis length is proportional to variance explained as indicated in parentheses. PC, principal coordinate. (Online version in colour.)
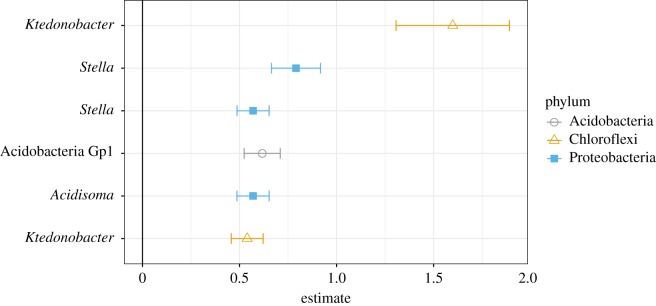
To demonstrate this approach, we tested the effect of the relative abundance of each ASV on methane oxidation rate while controlling for different sets of covariates including environment, geography and community. After controlling the false discovery rate, 460 unique ASVs were identified as significantly correlated with function when no covariates were included in the model. We found the different numbers of taxa significantly associated with methane oxidation depending on which covariates were included in the model (table 3). Each of these sets of taxa represents different versions of the biodiversity–ecosystem function mapping question. For example, by attempting to control for environmental variation statistically, we can identify taxa whose traits may contribute to variation in function that is independent of environmental conditions. Similarly, by controlling for geographic distance among samples, we can reduce the likelihood that the taxa we identify are only related to function because of an association with unmeasured environmental variation that is spatially structured or because of differences in dispersal history among sites. In the model that controlled for all three covariates (community, environment and geography), only six ASVs were significantly correlated with methane oxidation rate (figure 3). These taxa could be useful indicators of methane oxidation rate across space and different ecosystems. Researchers could elaborate on these findings using targeted cultivation and manipulative experiments to further understand their contribution to methane oxidation.
Table 3.
Number of significant taxa after including each set of covariates in a variance component model. ‘removed' and ‘added' columns are relative to the no-covariate model. Significance was determined by controlling the false discovery rate at q-values < 0.05.
| term(s) | removed | added | significant |
|---|---|---|---|
| none | 0 | 0 | 460 |
| geo | 338 | 21 | 143 |
| com | 460 | 0 | 0 |
| env | 281 | 1 | 180 |
| geo + com | 458 | 0 | 2 |
| geo + env | 377 | 13 | 96 |
| com + env | 447 | 0 | 13 |
| geo + com + env | 454 | 0 | 6 |
Figure 3.
Taxa associated with methane oxidation rates after controlling for geographic location, environmental similarity and community composition. Points are estimates for the linear relationship between the relative abundance of a single ASV and methane oxidation rate with standard errors from variance component models including similarity matrices as covariates for community and environment and site ID for geographic location. Amplicon sequence variants are labelled at the finest resolution available: genus for all except the Group 1 Acidobacteria. Points are identified by phylum. Significant taxa were determined by controlling the false discovery rate at q-value < 0.05. (Online version in colour.)
Notably, these six taxa fall into three genera and one class with cultured representatives that are not known to consume methane [44–47]. These taxa could be related to ecosystem function in multiple ways. The most interesting possibility is that each of these taxa is statistically related because it is causally connected to the function. This could be direct––for example, an organism that consumes methane––or indirect––for example, an organism that regulates substrates necessary for methane oxidizers. Alternatively, a significant association could occur for non-causal reasons. For example, any organism that tends to be in high abundance where methane oxidation rates are high would be correlated with methane oxidation, even if it has no causal relationship. This could be because such an organism is favoured under the same conditions that favour methane oxidation. Such covariation can drive associations that are not causal, but the effects of covariation would have been reduced by controlling for covariates in our tests.
4. Caveats and future directions
Once taxa have been identified with an association test (such as the one we outline above), there are multiple ways they could be used for future study. One approach common in genetics, particularly for markers of genetic disorders, is to generate a polygenic score based on the summed effect of many genes on a phenotype of interest, such as the probability of developing a disorder. A similar aggregate bioindicator could be generated for ecosystems that would summarize the probability of the rate or occurrence of a particular ecosystem function. This would be accomplished by measuring the abundance of the taxa identified in an association study and determining their association with the rate of an ecosystem function. Alternatively, the identified taxa could be incorporated into a structural equation model in an attempt to better understand the individual effects and interactions among taxa as they contribute to the rate of ecosystem function [48]. This might give an indication of the relative importance of different taxa as compared with other factors, such as environmental variables, and also identify underlying latent variables that explain variation in ecosystem function.
Ultimately, the relationships identified in any comparative mapping study must be verified. For genotype–phenotype studies in organisms, there are multiple ways that this verification is accomplished. In some cases, organisms can be artificially selected for a particular phenotype (e.g. through experimental evolution in an environment that favours the phenotype of interest) and the genetic changes that occur in response to selection can be compared with those identified via mapping studies. An analogous approach for microbial biodiversity–ecosystem function studies would be to apply artificial ecosystem selection (sensu [49]) on a given function and compare the taxa (or genes) that change in response to selection with those identified via a comparative approach (such as the one illustrated in our example).
The most common way that loci identified in a genotype–phenotype mapping study are verified is through manipulative genetics. The identified loci can be knocked out or over-expressed and the effect on phenotype compared with that predicted from mapping studies. In the case of microbial biodiversity studies, it may be possible to inhibit a particular functional group through the use of specific antimicrobial or chemical inhibitors or using phages that exhibit high host-specificity [50,51], but this is not generally possible. In some cases, we may be able to isolate a microorganism of interest in pure culture and add it back to an ecosystem, transiently increasing its abundance (roughly analogous to ‘overexpressing' a gene). A greater focus on culture-based approaches could increase the success of these kinds of microbial enrichments. Finally, synthetic communities (contrived assemblages of microorganisms) may be the most powerful way to test hypotheses about microbial biodiversity–ecosystem function relationships, but currently these approaches are limited by the small number of taxa that can be routinely cultured from most environments (but see [10] and [52]).
There are a number of limitations to the biodiversity–ecosystem function mapping approach we describe, some in common with organismal mapping studies and others unique. For example, simple linear models such as the variance component model used in this study are typical for genetics studies, but may not be the best way to identify correlations for microbiomes because of the unique challenges of microbial data. Marker gene and metagenome sequences are inherently compositional, reads are often absent from most samples (i.e. they are zero-inflated), and differences in sequencing depth make it difficult to compare relative abundances across samples, challenges that are not faced by population geneticists. We have addressed these challenges using a variance stabilizing transformation, but other models that test differential abundance and differential variance which can control for differences in sequencing depth and are robust to zero-inflation might be more appropriate (e.g. [53]). Clustering reads at higher taxonomic levels could circumvent zero-inflation by providing more continuous variation in taxon abundances across ecosystems. However, this approach introduces biases based on the completeness of taxonomic databases, the accuracy of 16S-based taxonomic assignment, and the removal of reads that lack a taxonomic assignment (although, newer approaches to taxonomic classification might help [54]). Alternatively, decreasing the threshold of sequence similarity to cluster reads without taxonomy could be analogous to aggregating at higher taxonomic levels, but it is uncertain whether these larger aggregates of taxa have any trait conservatism related to function. Here, we chose to test ASVs at the level of the individual read so as not to bias our results in these ways. Finally, we have applied this approach to ASVs inferred from 16S rRNA gene sequences, but any unit of microbiome data such as metagenomic reads or metatranscriptomic mRNA reads could be tested in an association study.
Experimentally, future studies could improve on our example by sampling a more homogeneous set of ecosystems. Our survey includes an especially broad assortment of ecosystems, including grasslands, plantations, forests, peatlands and mineral soil wetlands among others. These ecosystems represent a range of moisture conditions that could regulate the abundance and activity of methane oxidizers and access to methane and oxygen, which methane oxidizers rely on. While this captured substantial variation in methane oxidation rates, sampling from such diverse ecosystems could result in spurious associations between taxa and function. For example, taxa differentially abundant in upland ecosystems that are unrelated to methane oxidation might appear correlated simply as a result of their presence in those ecosystems with high oxidation rates. Future studies could try restricting their search to a more homogeneous population of ecosystems specific to the question at hand.
5. Conclusion
Microbial biodiversity–ecosystem function research has demonstrated positive correlations between diversity and ecosystem function. However, the abundances of microbial functional groups (as currently defined) are often poor predictors of ecosystem function and commonly do not add substantial explanatory power to ecosystem models. Therefore, a new perspective on how to determine the relationship between microbial communities and ecosystem functions is sorely needed. Organismal biologists have over a hundred years of experience identifying relationships between complex sets of highly variable data (genotypes or genome sequences) and aggregate functions (organismal phenotypes). We assert that combining the approaches of traditional biodiversity–ecosystem function research with ideas from genotype–phenotype mapping could provide this new perspective. This integration could not only make underutilized approaches such as covariate modelling and artificial selection more available to microbial ecologists, but also provide instructive examples of how best to conceive of microbial biodiversity–ecosystem function questions. If this integration is successful, it is possible that in the not-so-distant future our field will be able to robustly identify taxa, genes, or even molecules that will allow us to accurately predict the response of ecosystems to environmental change. Doing so will not only generate novel hypotheses about how complex microbial communities drive ecosystem function, but also help scientists predict and manage changes to ecosystem functions resulting from human activities.
Supplementary Material
Acknowledgments
The data re-analysed here were generated with the permission of the Government of Gabon, Centre National de la Recherche Scientifique et Technologique (permit no. AR0035/14/MESRS/CENAREST/CG/CST/CSAR) and the assistance of F. Bivigou, H. Memiaghe, L. Tchignoumba, E. Tobi and I. Akendengue. The University of Oregon, the Gabon–Oregon Transnational Center on Environment and Development, the Smithsonian Conservation Biology Institute and Shell Gabon provided financial and logistical support for the original study re-analysed here. We also thank W. Cresko for insightful discussions regarding population genetics and H. Tavalire for contributions to the statistical analyses.
Data accessibility
Raw data and all scripts necessary to recreate the analysis, figures and tables are provided in the electronic supplementary material.
Authors' contributions
A.M. assisted with molecular laboratory work, performed the data analysis and drafted the manuscript; K.M. collected field data, performed molecular laboratory work and critically revised the manuscript; B.B. conceived of the study, designed the study and helped draft the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed herein.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by the National Science Foundation – Dimensions of Biodiversity program (grant no. DEB 14422214), the National Science Foundation Graduate Research Fellowship Program (grant no. DGE 1255832) and the ARCS Foundation Florence and Mike Nudelman Scholarship.
References
- 1.Schimel JP, Gulledge J. 1998. Microbial community structure and global trace gases. Glob. Change Biol. 4, 745–758. ( 10.1046/j.1365-2486.1998.00195.x) [DOI] [Google Scholar]
- 2.Singh BK, Bardgett RD, Smith P, Reay DS. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. ( 10.1038/nrmicro2439) [DOI] [PubMed] [Google Scholar]
- 3.Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB, Nemergut DR, Graham EB, Wallenstein MD. 2015. Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J. 9, 1693–1699. ( 10.1038/ismej.2014.252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham EB, et al. 2016. Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front. Microbiol. 7, 214 ( 10.3389/fmicb.2016.00214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing X, et al. 2015. The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate. Nat. Commun. 6, 8159 ( 10.1038/ncomms9159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK. 2016. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541 ( 10.1038/ncomms10541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado-Baquerizo M, et al. 2017. Circular linkages between soil biodiversity, fertility and plant productivity are limited to topsoil at the continental scale. New Phytol. 215, 1186–1196. ( 10.1111/nph.14634) [DOI] [PubMed] [Google Scholar]
- 8.Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron P-A. 2013. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619. ( 10.1038/ismej.2013.34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagg C, Bender SF, Widmer F, van der Heijden MGA. 2014. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl Acad. Sci. USA 111, 5266–5270. ( 10.1073/pnas.1320054111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnyder E, Bodelier PLE, Hartmann M, Henneberger R, Niklaus PA. 2018. Positive diversity-functioning relationships in model communities of methanotrophic bacteria. Ecology 99, 714–723. ( 10.1002/ecy.2138) [DOI] [PubMed] [Google Scholar]
- 11.Maron P-A, et al. 2018. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 84, e02738-17 ( 10.1128/AEM.02738-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavigelli MA, Robertson GP. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81, 1402–1414. ( 10.1890/0012-9658(2000)081[1402:TFSODC]2.0.CO;2) [DOI] [Google Scholar]
- 13.Strickland MS, Lauber C, Fierer N, Bradford MA. 2009. Testing the functional significance of microbial community composition. Ecology 90, 441–451. ( 10.1890/08-0296.1) [DOI] [PubMed] [Google Scholar]
- 14.Glassman SI, Weihe C, Li J, Albright MBN, Looby CI, Martiny AC, Treseder KK, Allison SD, Martiny JBH. 2018. Decomposition responses to climate depend on microbial community composition. Proc. Natl Acad. Sci. USA 115, 11 994–11 999. ( 10.1073/pnas.1811269115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freitag TE, Prosser JI. 2009. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl. Environ. Microbiol. 75, 6679–6687. ( 10.1128/AEM.01021-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitag TE, Toet S, Ineson P, Prosser JI. 2010. Links between methane flux and transcriptional activities of methanogens and methane oxidizers in a blanket peat bog. FEMS Microbiol. Ecol. 73, 157–165. ( 10.1111/j.1574-6941.2010.00871.x) [DOI] [PubMed] [Google Scholar]
- 17.McCalley CK, et al. 2014. Methane dynamics regulated by microbial community response to permafrost thaw. Nature 514, 478–481. ( 10.1038/nature13798) [DOI] [PubMed] [Google Scholar]
- 18.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. 1989. Identification of the cystic fibrosis gene: genetic analysis. Science 245, 1073–1080. ( 10.1126/science.2570460) [DOI] [PubMed] [Google Scholar]
- 19.MacDonald ME, et al. 1992. The Huntington's disease candidate region exhibits many different haplotypes. Nat. Genet. 1, 99–103. ( 10.1038/ng0592-99) [DOI] [PubMed] [Google Scholar]
- 20.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367. ( 10.1073/pnas.0903103106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reich DE, Lander ES. 2001. On the allelic spectrum of human disease. Trends Genet. 17, 502–510. ( 10.1016/S0168-9525(01)02410-6) [DOI] [PubMed] [Google Scholar]
- 22.Hug LA, et al. 2016. A new view of the tree of life. Nat. Microbiol. 1, 16048 ( 10.1038/nmicrobiol.2016.48) [DOI] [PubMed] [Google Scholar]
- 23.Martiny AC. 2019. High proportions of bacteria are culturable across major biomes. ISME J. 13, 2125–2128. ( 10.1038/s41396-019-0410-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bižić-Ionescu M, Klintzsch T, Ionescu D, Hindiyeh MY, Günthel M, Muro-Pastor AM, Eckert W, Keppler F, Grossart H-P. 2019. Widespread methane formation by Cyanobacteria in aquatic and terrestrial ecosystems. bioRxiv 398958 ( 10.1101/398958) [DOI] [PMC free article] [PubMed]
- 25.Lenhart K, et al. 2012. Evidence for methane production by saprotrophic fungi. Nat. Commun. 3, 1046 ( 10.1038/ncomms2049) [DOI] [PubMed] [Google Scholar]
- 26.Wright S. 1943. Isolation by distance. Genetics 28, 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martiny JBH, et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. ( 10.1038/nrmicro1341) [DOI] [PubMed] [Google Scholar]
- 28.Meyer KM, Hopple AM, Klein AM, Morris AH, Bridgham S, Bohannan BJM. 2019. Community structure ecosystem function relationships in the Congo Basin methane cycle depend on the physiological scale of function. bioRxiv 639989 ( 10.1101/639989) [DOI]
- 29.Qin J, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. ( 10.1038/nature11450) [DOI] [PubMed] [Google Scholar]
- 30.Lloyd-Price J, et al. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. ( 10.1038/s41586-019-1237-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. ( 10.1038/ng1847) [DOI] [PubMed] [Google Scholar]
- 32.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354. ( 10.1038/ng.548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. ( 10.1038/ismej.2017.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMurdie PJ, Holmes S. 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 ( 10.1371/journal.pone.0061217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See http://www.R-project.org/.
- 36.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531 ( 10.1371/journal.pcbi.1003531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willis A, Bunge J. 2015. Estimating diversity via frequency ratios. Biometrics 71, 1042–1049. ( 10.1111/biom.12332) [DOI] [PubMed] [Google Scholar]
- 39.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. ( 10.2307/1942268) [DOI] [Google Scholar]
- 40.Oksanen J, et al. 2019. vegan: Community Ecology Package. See https://CRAN.R-project.org/package=vegan.
- 41.Qu L, Guennel T, Marshall SL. 2013. Linear score tests for variance components in linear mixed models and applications to genetic association studies. Biometrics 69, 883–892. ( 10.1111/biom.12095) [DOI] [PubMed] [Google Scholar]
- 42.Storey JD. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498. ( 10.1111/1467-9868.00346) [DOI] [Google Scholar]
- 43.Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K, Yutani H, RStudio . 2019. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. See https://CRAN.R-project.org/package=ggplot2.
- 44.Belova SE, Pankratov TA, Detkova EN, Kaparullina EN, Dedysh SN. 2009. Acidisoma tundrae gen. nov., sp. nov. and Acidisoma sibiricum sp. nov., two acidophilic, psychrotolerant members of the Alphaproteobacteria from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 59, 2283–2290. ( 10.1099/ijs.0.009209-0) [DOI] [PubMed] [Google Scholar]
- 45.Chang Y, et al. 2011. Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21T). Stand. Genomic Sci. 5, 97–111. ( 10.4056/sigs.2114901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domeignoz-Horta LA, DeAngelis KM, Pold G. 2019. Draft genome sequence of Acidobacteria group 1 Acidipila sp. strain EB88, isolated from forest soil. Microbiol. Resour. Announc. 8, e01464-18 ( 10.1128/MRA.01464-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritz I, Strömpl C, Abraham W-R. 2004. Phylogenetic relationships of the genera Stella, Labrys and Angulomicrobium within the ‘Alphaproteobacteria’ and description of Angulomicrobium amanitiforme sp. nov. Int. J. Syst. Evol. Microbiol., 54, 651–657. ( 10.1099/ijs.0.02746-0) [DOI] [PubMed] [Google Scholar]
- 48.Grace JB, Anderson TM, Olff H, Scheiner SM. 2010. On the specification of structural equation models for ecological systems. Ecol. Monogr. 80, 67–87. ( 10.1890/09-0464.1) [DOI] [Google Scholar]
- 49.Swenson W, Wilson DS, Elias R. 2000. Artificial ecosystem selection. Proc. Natl Acad. Sci. USA 97, 9110–9114. ( 10.1073/pnas.150237597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maxson T, Mitchell DA. 2016. Targeted treatment for bacterial infections: prospects for pathogen-specific antibiotics coupled with rapid diagnostics. Tetrahedron 72, 3609–3624. ( 10.1016/j.tet.2015.09.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823. ( 10.3390/v5030806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg M, Koskella B. 2018. Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 28, 2487–2492.e3. ( 10.1016/j.cub.2018.05.085) [DOI] [PubMed] [Google Scholar]
- 53.Martin BD, Witten D, Willis AD. 2019. Modeling microbial abundances and dysbiosis with beta-binomial regression. arXiv 1902.02776v1 [stat.ME]. See https://arxiv.org/abs/1902.02776 .
- 54.Shah N, Meisel JS, Pop M. 2019. Embracing ambiguity in the taxonomic classification of microbiome sequencing data. Front. Genet. 10, 1022 ( 10.3389/fgene.2019.01022) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and all scripts necessary to recreate the analysis, figures and tables are provided in the electronic supplementary material.