Summary Paragraph
The neuromodulator melatonin synchronizes circadian rhythms and related physiological functions via actions at two G protein-coupled receptors: MT1 and MT2. Circadian release of high nighttime levels of melatonin from the pineal gland activates melatonin receptors in the suprachiasmatic nucleus of the hypothalamus, synchronizing physiology and behavior to the light-dark cycle1–4. The two receptors are established drug targets for aligning circadian phase in disorders of sleep5,6 and depression7,1–4,8,9. Despite their importance, few if any in vivo active MT1 selective ligands have been reported2,8,10–12, hampering both the understanding of circadian biology and the development of targeted therapeutics. We docked over 150 million virtual molecules against an MT1 crystal structure, prioritizing structural fit and chemical novelty. Thirty-eight high-ranking molecules were synthesized and tested, revealing ligands in the 470 pM to 6 μM range. Structure-based optimization led to two selective MT1 inverse agonists, topologically unrelated to previously explored chemotypes, that were tested in mouse models of circadian behavior. Unexpectedly, the MT1-selective inverse agonists advanced the phase of the mouse circadian clock by 1.3–1.5 hrs when given at subjective dusk, an agonist-like effect eliminated in MT1- but not in MT2-knockout mice. This study illustrates opportunities for modulating melatonin receptor biology via MT1-selective ligands, and for the discovery of new, in vivo-active chemotypes from structure-based screens of diverse, ultra-large libraries.
Ultra-large library docking for new melatonin receptor ligands.
The recent determination of the MT1 and MT2 receptor crystal structures13,14 afforded us the opportunity to seek new chemotypes with new functions, including MT1-selective ligands, by computational docking of an ultra-large make-on-demand library15, seeking molecules that complemented the main ligand binding (orthosteric) site of the receptor. Given the similar MT1 and MT2 sites, where 20 of 21 residues are identical, and the challenges of docking for selectivity16, we sought to prioritize new, high-ranking chemotypes from the docking screen, unrelated to known melatonin receptor ligands, expecting these to differentially interact with the two melatonin receptor types17–19.
We docked over 150 million “lead-like” molecules, characterized by favorable physical properties, from ZINC (http://zinc15.docking.org)15,20. These largely make-on-demand molecules have not been previously synthesized, but are usually accessible by two component reactions. Use of complex building blocks in these reactions biases toward diverse, structurally interesting molecules15,20. Each library molecule was sampled in an average of over 1.6 million poses (orientations x conformations) in the MT1 orthosteric site13 by DOCK3.721, more than 72 trillion complexes for the library overall, scoring each for physical complementarity to the receptor site21. Seeking diversity, the top 300,000 scoring molecules were clustered by topological similarity, resulting in 65,323 clusters, and those that were similar to known MT1 and MT2 ligands from ChEMBL2322 were eliminated (see Methods) (Fig. 1, Extended Data Table 1).
Figure 1. Large library docking finds novel, potent melatonin receptor ligands.

a, Docking for new melatonin receptor chemotypes from the make-on-demand library. b, Docked pose of ‘0207, an hMT1/hMT2 non-selective agonist with low nanomolar activity. c, Docked pose of ‘5999, an MT2-selective inverse agonist. In b-c, the crystallographic geometry of 2-phenylmelatonin is shown in transparent blue, for context. d, The initial 15 docking actives are shown, highlighting groups that correspond to melatonin’s acetamide side chain (blue) and its 5-methoxy-indole (red) in their docked poses and receptor interactions. Shaded molecules are inverse agonists.
The best scoring molecules from each of the top 10,000 clusters were inspected for engagement with residues that recognize ligands in the MT1 crystal structure13,14, and for new polar partners in the MT1 site. In the docked complexes, these included hydrogen bonds with Q181ECL2, N1624.60, T178ECL2, N2556.52, and with the backbone atoms of A1584.56, G1043.29, and F179ECL2. Conformationally strained molecules and those with unsatisfied hydrogen bond donors were deprioritized23. Within the best-scoring clusters, all members were inspected and the one that best fit these criteria was prioritized. Ultimately, 40 molecules with ranks ranging from 16 to 246,721, or the top 0.00001% to top 0.1% of the over 150 million docked, were selected for de novo synthesis and testing. Of the 38 molecules successfully synthesized (a 95% fulfillment rate), 15 had activity at either or both of the human MT1 and MT2 receptors in functional assays (Extended Data Table 1, Fig. 1), a hit rate of 39% (number-active/number-physically-tested).
In vitro pharmacology reveals new chemotypes with multiple functions.
These active molecules included both agonists and inverse agonists, consistent with the emphasis on chemotype novelty (Extended Data Table 1, Fig. 1). This novelty is supported quantitatively by their low topological similarity to known melatonin receptor ligands24, and visually by comparison of the new ligands to their closest analogs among the knowns (Extended Data Table 1). The different chemotypes often engaged the same residues that recognize 2-phenylmelatonin in the crystal structures. Examples include the hydrogen-bond interactions with N1624.60 made by the methoxy group of 2-phenylmelatonin, but in the docked models by esters (ZINC92585174), pyridines (ZINC151209032), and benzodioxoles (ZINC301472854). Similarly, while 2-phenylmelatonin stacks an indole with F179ECL2, the docked ligands stack benzoxazines (ZINC482850041), thiophenes (ZINC419113878), and furans (ZINC433313647). While 2-phenylmelatonin hydrogen bonds with Q181ECL2 via its acetamide, the docked ligands use esters or even pyridines (Fig. 1). The new ligands also dock to interact with new residues, including hydrogen bonds with T178ECL2, N2556.52, A1584.56, G1043.29, and F179ECL2 (Fig. 1b,c, Extended Data Fig. 3a–d).
Consistent with docking against an agonist-bound MT1 structure, four of the new ligands were MT1-selective agonists (Extended Data Fig. 1a,b), with EC50 values in the 2 to 6 μM range, and without detectable MT2 activity up to 30 μM: ‘3878, ‘9032, ZINC353044322, and ZINC182731037. Strikingly, ZINC159050207, although non-selective between the receptor types, is a 1 nM MT1 agonist, among the most potent molecules found directly from a docking screen25–30 (Extended Data Table 1, Fig. 1b, Extended Data Fig 1c,d). Admittedly, many ligands were just as active at the MT2 receptor, or even selective for it (Extended Data Table 1, Extended Data Fig 1). Thus, whereas the initial docking against the MT1 structure found new, potent chemotypes, and some of these were type selective, they were just as likely to prefer the MT2 type as the MT1 type. This attests to both the strengths and weaknesses of chemotype novelty as a strategy for compound prioritization, and to the need for further optimization.
We sought to improve twelve of these chemotype families, selecting analogs from the make-on-demand library. Several thousand such were docked into the MT1 site (Extended Data Table 2) (see Methods). Of the 131 synthesized and tested, 94 analogs had activity at either or both MT1 or MT2 melatonin receptors at concentrations ≤ 10 μM (Extended Data Table 2, Supplementary Table 1, Extended Data Fig. 2); of the twelve chemotype families, five saw improved potency. While this structure-based analoging could often find more potent ligands, their efficacy, selectivity, and bias were sensitive to small structural changes (Extended Data Fig. 3).
We were particularly interested in type-selective ligands with in vivo efficacy, as these are unreported in the field. We investigated two MT1-selective inverse agonists, ZINC555417447 and ZINC157673384, and a selective MT2 agonist, ZINC128734226 (from here on referred to as UCSF7447, UCSF3384 and UCSF4226, respectively), for their affinities (Fig. 2, Supplementary Data 4), in vitro signaling, pharmacokinetics (Extended Data Table 3), selectivity on mouse as well as the human receptors (hMT1 and hMT2) (Fig. 2, Supplementary Data 3 and 4), and for their efficacies in mouse models of circadian behavior (Fig. 3, Extended Data Figs. 4–5, Extended Data Fig. 7). As expected, UCSF7447 and UCSF3384 competed for 2-[125I]-iodomelatonin binding with higher affinity for the hMT1 receptors. Ki values in the absence of GTP, 304 nM and 938 nM, respectively, were improved by uncoupling G protein from the receptor by GTP addition, with Ki values improving to 7.5 nM and 63 nM, respectively, supporting their status as inverse agonists (Fig. 2a–b, Supplementary Data 3 and Extended Data Fig. 6). Both UCSF7447 and UCSF3384 increased basal cAMP, also as expected for inverse agonists, with EC50 values of 41 and 21 nM at hMT1, selectivity for hMT1 over hMT2 of 53- and 31-fold, and hMT1 inverse agonist efficacies of 62% and 47%, respectively (Fig. 2c–d, Extended Data Fig. 6). The third molecule, UCSF4226 was an hMT2-selective agonist with an MT2/MT1 selectivity of 54 in 2-[125I]-iodomelatonin binding assays and a selectivity of 91 in BRET assays; in isoproterenol-stimulated cAMP inhibition, the agonist had an EC50 of 7.1 nM at hMT2, a value closely matched by an EC50 of 6.3 nM in BRET assays (Supplementary Data 4). Upon intravenous administration in mice, the three molecules were CNS permeable, with brain/plasma ratios ranging from 1.4 to 3.0. Plasma half-lives ranged from 0.27 to 0.32 hours (Extended Data Table 3), similar to melatonin2. Against mouse MT1 and MT2 receptors (mMT1, mMT2) in vitro, the selectivity of the two inverse agonists improved over the human receptors being over 158 and over 100 times more selective for the mMT1 receptor to increase basal cAMP with no activity observed against the mMT2 receptor up to 10 μM for either compound (Fig. 2e–f; Supplementary Data 3). Conversely, while the agonist UCSF4226 lost little activity on the mouse receptor, its selectivity for the mMT2 receptor was much diminished (Supplementary Data 4). Accordingly, we moved forward to mouse in vivo experiments with the two selective MT1 inverse agonists.
Fig. 2. Affinity, efficacy, and potency of MT1-selective inverse agonists on human (h) and mouse (m) MT1 and MT2 receptors.
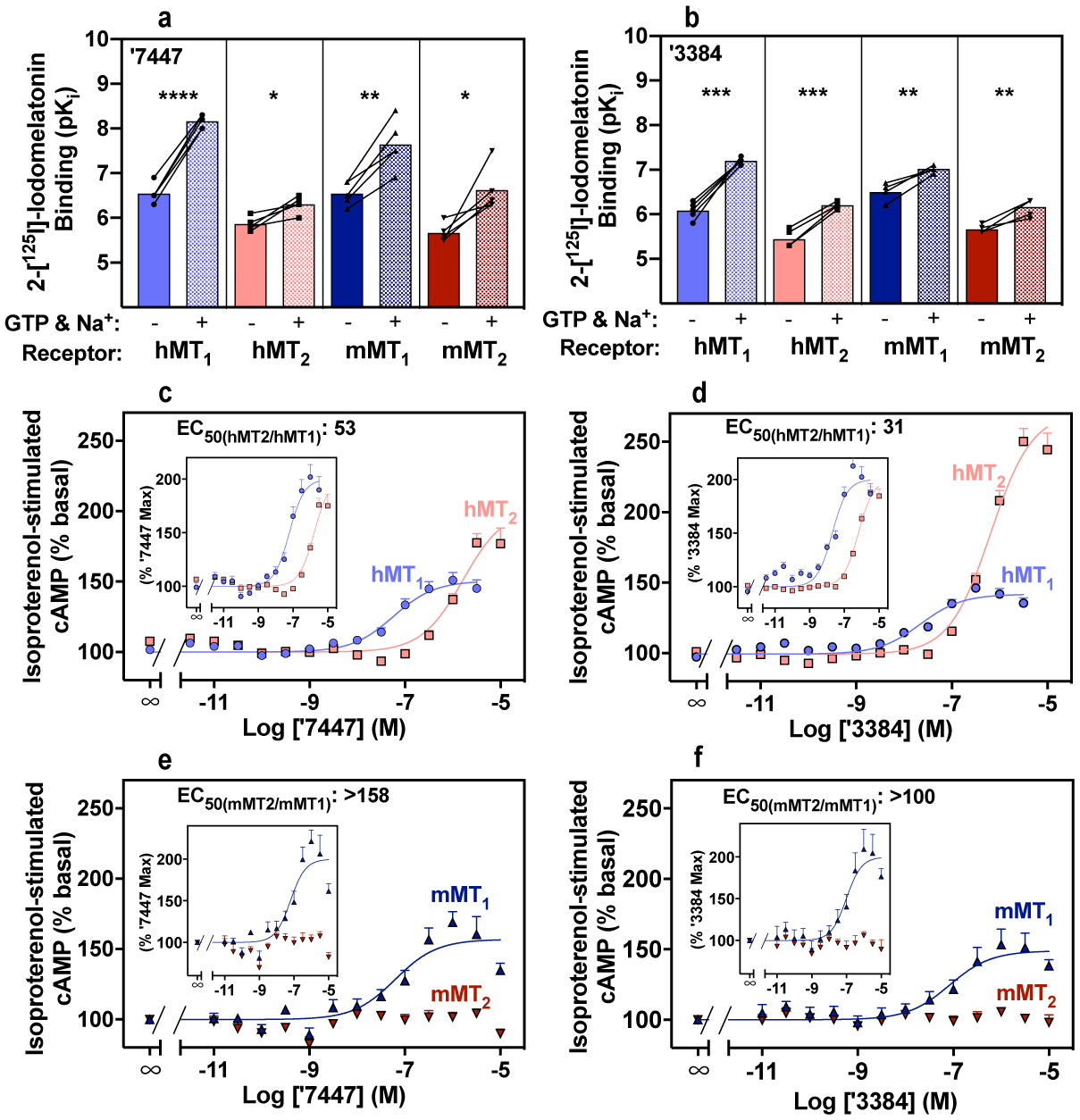
(a,b) Affinity (pKi) of inverse agonists ‘7447 (a) and ‘3384 (b) by 2-[125I]-iodomelatonin competition for hMT1, hMT2, mMT1, and mMT2 receptors stably expressed in CHO cells. Binding was measured in the absence and presence of 100 μM GTP, 1 mM EDTA.Na2, and 150 mM NaCl. GTP uncouples G proteins from melatonin receptors promoting inactive conformations46 and higher affinity for inverse agonists; thus, the solid bars show higher affinity than the paired checker bars. Connected symbols represent pKi values of individual determinations run in parallel. Ki values were derived from competition binding curves (see Supplementary Data Fig. 3). Bars represent the averages of five independent determinations. Statistical significance between pKi averages were calculated by two-tailed paired student t test (t, df and P values under described under Data Analysis in Methods).
*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 when compared with corresponding pKi averages values derived in the absence of GTP.
(c - f) Concentration-response curves on hMT1, hMT2, mMT1, and mMT2 receptors transiently-expressed in HEK cells, monitoring isoproterenol-stimulated cAMP production with ‘7447 c: hMT1 pEC50: 7.39 ± 0.10, Emax: −62 ± 13%, n = 8; hMT2 pEC50: 5.66 ± 0.10, Emax: −84 ± 9%, n = 8, and e: mMT1 pEC50: 7.20 ± 0.17, Emax: -56 ± 5 %, n = 5; mMT2 pEC50: n/d, n=5, Emax: n/d, n = 5) and d: ‘3384: hMT1pEC50: 7.68 ± 0.09, Emax: −47 ± 12%, n = 13; hMT2 pEC50: 6.18 ± 0.04, Emax: −153 ± 14 %, n = 12; and f: mMT1 pEC50: 7.00 ± 0.22, Emax: -49 ± 3 %, n = 5; and mMT2 pEC50: n/d, Emax: n/d, n = 5) treatment. Data for ‘7447 and ‘3384 was normalized to isoproterenol-stimulated basal activity. Inset graphs represent data normalized to maximal ligand effect.
Data represent mean ± s.e.m. from the indicated number (n) of biologically independent experiments run in triplicate.
UCSF7447 (‘7447); UCSF3384 (‘3384)
Figure 3. In vivo, MT1-selective inverse agonists decelerate re-entrainment rate (a-c) and phase advance circadian activity when administered at dusk (CT 10) (d-f).

a - b, Inverse agonists ‘3384 and ‘7447 decelerate re-entrainment rate [a, VEH vs ‘7447 (30 μg/mouse); mixed-effect two-way repeated measures ANOVA (treatment x time interaction: F16,735 = 3.39 P = 8.20 × 10−6], and increase number of days to re-entrainment after 6 h advance of dark onset in the “east-bound jet-lag” paradigm [b, VEH vs. MLT, ‘3384, and ‘7447 (30 μg/mouse) or LUZ (300 μg/mouse); one-way ANOVA (F4,92 = 16.97 P = 1.86 × 10−10)]. c, Inverse agonist ‘7447 targets MT1 receptors to increase number of days to re-entrainment [VEH (white) vs. ‘7447 (blue; 30 μg/mouse); two-way ANOVA (treatment: F1,120 = 24.82 P = 2.14 × 10−6, genotype: F2,120 = 23.44 P = 2.55 × 10−9)]. d, Inverse agonists ‘3384 and ‘7447 phase advance circadian wheel activity onset in constant dark at CT 10 (dusk), resembling agonist melatonin [left: VEH vs. MLT or ‘7447 (0.9 μg/mouse); one-way ANOVA (F2,26 = 13.60 P = 9.08 × 10−5); center: VEH vs. MLT, ‘3384 or ‘7447 (30 μg/mouse); one-way ANOVA (F3,52 = 32.05 P = 7.15 × 10−12); right: VEH vs LUZ (300 μg/mouse); two-tailed unpaired students t test (t = 0.92 df = 7 P = 0.39)]. e, The phase advance of wheel activity onset by ‘7447 is mediated via the MT1 receptor at CT 10 (dusk) [VEH (white) vs. ‘7447 (blue; 30 μg/mouse); two-way ANOVA (treatment x genotype interaction: F2,49 = 4.46 P = 0.0166)]. f, Inverse agonist ‘7447, unlike melatonin, did not phase delay in constant dark at CT 2 (dawn) [VEH (white) vs. ‘7447 (blue; 30 μg/mouse); two-way ANOVA (treatment x genotype interaction: F2,49 = 0.384 P = 0.684)]. Panel f has 1 value not shown due to scale, but is included in the analysis (value = 0.91 h). Data shown represent mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 for comparisons to WT VEH. &P < 0. 001 for comparisons to MT2KO VEH. Post-test analysis used Sidak’s (a), Tukey’s (c, e, f), or Dunnet’s (b & d; all P < 0.05). Details for all statistical analyses and reporting of n values for each condition (depicted as scatter dot plots where appropriate) are found in Methods (Statistics & Reproducibility). Vehicle (VEH), melatonin (MLT), luzindole (LUZ), UCSF7447 (‘7447), UCSF3384 (‘3384). All treatments were given via s.c. injection.
In vivo pharmacology reveals new MT1-selective activities. We first examined the in vivo activity of the two MT1-selective inverse agonists in a mouse model of re-entrainment. In this “east-bound jet-lag” model, mice are subjected to an abrupt six-hour advance of the light-dark cycle and treated at the new dark onset for three consecutive days to assess re-entrainment rate. At 30 μg/mouse, the agonist melatonin accelerates re-entrainment to the new cycle, consistent with its use in the treatment of east-bound human jet-lag (Fig. 3b). Conversely, the prototypical non-selective antagonist/inverse agonist luzindole, administered at 300 μg/mouse, decelerates re-entrainment, measured by the number of days to adapt to the new dark onset, as expected for an inverse agonist43,31,32,33. The selective MT1 inverse agonists UCSF7447 and UCSF3384, dosed 30 μg/mouse (about 1 mg/Kg), also decelerated re-entrainment (Fig. 3a,b, Extended Data Fig. 4c,d,g), phenocopying luzindole (encouragingly, at a 10-fold lower dose).
Superficially, the shared effect of decelerating re-entrainment by UCSF7447, UCSF3384 (Fig. 3a–c) and luzindole33 might seem expected, as they all share the same function as melatonin receptor antagonists/inverse-agonists. However, luzindole is MT1/MT2 non-selective, unlike UCSF7447 and UCSF3384. Their phenocopying of luzindole suggests that deceleration of re-entrainment by all three molecules—slowing “jet-lag” accommodation—is mediated via the MT1 receptor alone. Supporting this, the effect of UCSF7447 was eliminated in an MT1KO mouse (Fig. 3c, Extended Data Fig. 4d,i), but not in an MT2KO mouse, where its effect was actually increased, adding to the deceleration afforded by deletion of the MT2 receptor alone (Fig. 3c, Extended Data Fig. 4i,k,m,n).,
The effect of the MT1-selective inverse agonists on circadian phase was even more unexpected. Here, we measured their effects on circadian phase by monitoring the running wheel activity onset of freely running mice in constant darkness34–36 and administering them at subjective dusk (circadian time 10, CT 10). Both inverse agonists phase-advanced circadian wheel running rhythm onset, an effect characteristic of melatonin, the endogenous, non-selective agonist, and of non-selective agonist drugs like ramelteon37 and agomelatine9,38 (Fig. 3d–f, Extended Data Fig. 5b–d). Whereas MT1-selective inverse agonists have few if any precedents in vivo, we would have ordinarily expected the opposite effect of the agonist39,40, delaying rather than advancing circadian phase. Instead, UCSF7447 advanced the onset of activity by approximately 1 hour at 0.9 μg/mouse (about 0.03 mg/Kg), an effect similar to that of melatonin at its ED50 (0.72 μg/mouse)34 (Fig. 3d). At a higher dose (30 μg/mouse, about 1 mg/Kg), both UCSF7447 and UCSF3384 advanced the onset of running wheel activity with an amplitude similar to melatonin34 at this circadian time (CT 10). Intriguingly, whereas melatonin and ramelteon advance phase when dosed at dusk (CT 10), and delay phase when given at dawn (CT 2)35–37,41, UCSF7447 did not affect phase at dawn (Fig. 3f, Extended Data Figure 5i,r,s), only working at dusk (Extended Data Fig. 7a).
The phenocopying of the non-selective agonist melatonin by the MT1-selective inverse agonists, in shifting circadian phase, motivated us to investigate mechanism of action and the role of off-targets. Accordingly, both molecules, as well as the hMT2-selective agonist UCSF4226, were tested against a panel of common off-targets (Supplementary Data 1). By radioligand competition, no activity was seen up to a concentration of 10 μM for the new ligands. Against a panel of 318 GPCRs, activity was observed for only seven receptors when screened at a single concentration, none of which replicated in full concentration-response (Supplementary Data 2). Consistent with activity via the MT1 receptor, the advance in the onset of running wheel activity at dusk (CT 10) by UCSF7447 was eliminated in MT1KO mice but not in MT2KO mice (Fig. 3e, Extended Data Fig 5a–f). These observations suggest that the MT1-selective inverse agonists UCSF7447 and UCSF3384 are not only potent, with effects on phase shift for UCSF7447 at 0.9 μg/mouse (about 0.03 mg/Kg) (Fig 3c) and efficacies resembling the long-established reagent luzindole in the jet-lag model at 10-fold lower doses, but that their unexpected activity in circadian phase is via the MT1 receptor. We note that the lack of precedence for this behavior reflects a lack of MT1 selective inverse agonists to probe for it, something addressed by this study.
Discussion
From a large library docking screen emerged multiple new chemotypes for melatonin receptors (Fig. 1), with new signaling and new pharmacology. Three features of this study merit emphasis. First, docking a library of over 150 million diverse, make-on-demand molecules found ligands topologically unrelated to known melatonin receptor ligands, with picomolar and nanomolar activity on the melatonin receptors. Second, the chemical novelty of these molecules translated functionally, conferring melatonin receptor type selectivity. Whereas the deceleration of re-entrainment (jet-lag model) by the new inverse agonists resembled that of the classic non-selective antagonist/inverse agonist luzindole, their high selectivity for the MT1 receptor, and the chemical-genetic epistasis in the MT1KO mouse, convincingly implicates the MT1 receptor in this response. Unexpectedly, the new inverse agonists conferred an agonist-like effect in circadian phase shift experiments when administered at dusk, perhaps suggesting previously unknown signaling control for the MT1 receptor in the SCN, which has known time of day dependent receptor mediated signaling pathways42.Third, these are the first MT1-selective inverse agonists active in vivo, with efficacy at doses as low as 0.9 μg/mouse in circadian phase shift. Their efficacy in modulating time-dependent circadian entrainment supports their potential as leads towards therapeutics in conditions and diseases affected by alterations in phase5–7,43.
Certain caveats bear airing. While we sought MT1-selective ligands, we found ligands for both melatonin receptor types, reflecting their conserved orthosteric sites. Indeed, rather than adopting a structure-based strategy for type selectivity, we simply focused on chemical novelty among the high-ranking docked molecules15,17. While the 39% docking hit rate was high, and the hits were potent, this likely reflects a site that is unusually well-suited to ligand binding: it is small, solvent-occluded, and largely hydrophobic. These high hit rates and potencies may not always translate to other targets44,45.
The key observations of this work should nevertheless be clear. From a structure-based screen of a diverse, 150 million compound virtual library sprang 15 new chemical scaffolds, topologically unrelated to known melatonin receptor ligands and synthesized de novo for this project. From their chemical novelty emerged new activities, including inverse agonists and ligands with melatonin receptor type-selectivity. The potency, brain exposure, and selectivity of these new ligands enable one to begin to disentangle the physiological role of the MT1 receptor. Accordingly, we are making the MT1-selective inverse agonist UCSF7447, and the hMT2 selective agonist UCSF4226, openly available to the community, as probe pairs coupled with a close analog that has no measurable activity on the melatonin receptors (Extended Data Table 4). We note that only a small fraction of even the highest-ranking chemotypes from the docking were tested here; it is likely that hundreds-of-thousands of melatonin receptor ligands, representing tens-of-thousands of new chemotypes15, remain to be discovered from the make-on-demand library, which continues to grow (http://zinc15.docking.org). This study suggests that not only potent ligands may be revealed by docking such a library, but also that the new chemotypes explored can illuminate new in vivo pharmacology.
Online Methods
Molecular docking
The MT1 receptor bearing nine thermostabilizing point mutations, as determined crystallographically13, was used in the docking calculations. To prepare the structure for docking, atoms of the co-crystallized ligand, 2-phenylmelatonin, were used to seed the matching sphere calculation in the orthosteric site; these spheres represent favorable positions for individual ligand atoms to dock; overall 45 spheres were used. DOCK3.7 orients flexibases of pre-calculated ligand conformations into the orthosteric site by overlaying atoms of each library molecule onto these matching spheres. The receptor structure was protonated by REDUCE47 and assigned AMBER united atom charges48. For residues N1624.60 and Q181ECL2, the partial atomic charges of the side chain amide was increased without changing residue net charge, as previously49. The volume of the low protein dielectric, which defines the boundary between solute and solvent in Poisson-Boltzmann electrostatic calculations, was extended out 1.9 Å from the protein surface using spheres calculated by SPHGEN. Scoring grids were pre-calculated by CHEMGRID for AMBER van der Waals potential, QNIFFT50 for Poisson-Boltzmann-based electrostatic potentials, and SOLVMAP51 for ligand desolvation.
The resulting potential grids and ligand matching parameters were evaluated for their ability to enrich known MT1 ligands over property-matched decoys. Decoys share the same physical properties as known ligands but are topologically dissimilar and so unlikely to bind. Thirty-one known MT1 melatonin receptor ligands, both agonists and antagonists, were extracted from the IUPHAR database52, and 1550 property-matched decoys were generated using the DUD-E pipeline. Docking success was judged on the ability to enrich the known ligands over the decoys by docking rank, using adjusted logAUC; this is widely done in the field. We also ensured that molecules with extreme physical properties were not enriched, as can happen when only counter-screening against property-matched decoys. In particular, we wanted to ensure that neutral molecules were enriched over charged ones. The docking parameters were also judged on how well they reproduced the known ligands’ expected binding modes and their ability to hydrogen-bond with N1624.60 and Q181ECL2.
The “lead-like” subset of ZINC15 (http://zinc15.docking.org), characterized by favorable physical properties (e.g., with calculated octanol-water partition coefficients (cLopP) ≤3.5, and with molecular weights ≤350), was then docked against the MT1 orthosteric site, using DOCK3.721. This library contained over 150 million molecules, mostly make-on-demand from the Enamine REAL set15. Of these, over 135 million molecules successfully docked, with over 36 million receiving a favorable score (<0 kcal/mol). An average of 3,445 orientations were calculated for each, and for each orientation, an average of 485 conformations were sampled. A simplex minimizer was used for rigid-body minimization on the best-scored pose for each ligand. Overall, about 72 trillion complexes were sampled and scored. The calculation time was 45,020 core hours, or 1.25 calendar days on 1,500 cores.
To reduce redundancy of the best-ranking docked molecules, the top 300,000 ranked molecules were clustered by ECFP4-based Tanimoto coefficient (Tc) of 0.5, and the best-scoring member was used to represent the cluster. The resulting 65,323 clusters were filtered for novelty by calculating ECFP4-based Tcs against >1,100 MT1 and MT2 receptor ligands from the CHEMBL2322 database. Molecules with Tc ≥0.38 to known MT1/MT2 ligands were not further pursued.
After filtering for novelty, the docked poses of the best-scoring members of each cluster were filtered by the proximity of their polar moieties to N1624.60 or Q181ECL2, and manually inspected for favorable geometry and interactions. Of the best-scoring molecules so prioritized, all members of its cluster within the top 300,000 molecules were also inspected, and sometimes one of these was chosen if they exhibited more favorable poses or chemical properties. Ultimately, forty compounds were chosen for testing, thirty-eight of which were successfully synthesized. To our knowledge, none of these compounds has been previously available and we are unaware of reports of them being previously synthesized.
Make-on-demand synthesis
Compounds were synthesized using 72,000 qualified in stock building blocks and 130 well-characterized, two component reactions at Enamine. Historically, molecules have been synthesized in three to four weeks with an 85% fulfilment rate; in this project delivery time was six weeks, but with a 95% fulfilment rate for the 40 molecules prioritized from the initial docking screen. Each reaction is tested for conditions including temperatures, completion time, and mixing53. Typically, compounds are made in parallel by combining reagents and solvents in a single vial in the appropriate conditions to allow the reaction to proceed to completion. The product-containing vial is filtered by centrifugation into a second vial to remove precipitate and the solvent is evaporated under reduced pressure; the product is then purified by HPLC. Identity and purity are assessed by LC/MS and, as appropriate, 1H NMR (Supplementary Data 2 & 7). All compounds were shipped 90% pure or better, and the main three compounds UCSF7447, UCSF3384 and UCSF4226 were independently confirmed to be ≥95% pure by LC/MS in secondary confirmation analyses at a second lab (Supplementary Data 5). Details on synthesis and analyses are provided in Supplementary Data 6.
Structure-based ligand optimization
After experimental testing (below), 12 of the 15 active ligands from docking were prioritized for optimization, representing a range of activities and type selectivity (Extended Data Table 2 and Supplementary Table 1). Several thousand analogs of these ligands, each bearing the same scaffold as the parent molecule and with Tc <0.38 to annotated melatonin receptor ligands, were selected from the ZINC database and docked to the MT1 binding site, again using DOCK3.7. The resulting docked poses were manually evaluated for interactions with N1624.60 or Q181ECL2, and 132 analogs were selected for de novo synthesis at Enamine, in two iterations. Of these, 131 were successfully synthesized, a >99% fulfillment rate.
Cell Culture
HEK293T cells were maintained with complete Dulbecco’s modified Eagle’s medium (DMEM), supplemented by 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin G and 100 μg/ml streptomycin. Cells were maintained at 37°C in the presence of 5% CO2.
Tango arrestin recruitment assay
MT1 and MT2 Tango constructs were designed and assays were performed as previously described54. Briefly, HTLA cells stably expressing TEV protease fused ß-arrestin (kindly provided by Dr. Richard Axel) and tTA dependent luciferase reporter gene were transfected with MT1 or MT2 Tango construct. The next day, transfected cells were seeded into poly-L-lysine coated 384-well white clear bottom cell culture plates with DMEM containing 1% dialyzed FBS at a density of 20,000 cells per well in 40 μl for another six hours. Drug solution was prepared in the same media used for cell plating at 5X final concentration and 10 μl per well was added for overnight incubation. The next day, media and drug solutions were discarded and loaded with 20 μl per well of Bright-Glo reagent (Promega). Plates were incubated for 20 mins in the dark followed by being counting using SpectraMax luminescence reader (Molecular Device). Data were analyzed using GraphPad Prism 6.0.
cAMP assay
MT1 and MT2 receptors were tested using Promega’s split luciferase based GloSensor cAMP biosensor technology. HEK293T cells were plated in 15 cm cell culture dish (at a ~50% cell confluency) with DMEM supplemented with 10% dialyzed FBS, 2 mM L-glutamine, 100 units/ml penicillin G and 100 μg/ml streptomycin for 4–6 hour. Then, cells were co-transfected with 8 μg of construct which encodes either MT1 or MT2 (de-Tango-ized constructs) and 8 μg of Glosensor DNA. Next day, transfected cells were seeded into poly-L-lysine coated 384-well white clear bottom cell culture plates with complete DMEM supplemented with 1% dialyzed FBS at a density of 20,000 cells per well for another 24 h. The next day, cell medium was discarded and loaded with 20 μl of assay buffer (1× HBSS, 20 mM HEPES, pH 7.4, 0.1% BSA). To measure agonist activity of MT1 or MT2 receptor, 10 μl of test compound solution at 3X final concentration was added for 15 minutes followed by addition of 10 μl of luciferin/isoproterenol mixture (at a final concentration of 4 mM and 200 nM respectively) for another 15 mins for luminescence quantification. Then, plates were counted using SpectraMax luminescence reader (Molecular Device). Data were analyzed using GraphPad Prism 8 (Graphpad Software Inc., San Diego, CA).
Log(Emax/EC50) calculation and ligand bias quantification
The ΔLog(Emax/EC50) was calculated with melatonin as a reference agonist for G protein and ß-arrestin pathway, and the ΔΔLog(Emax/EC50) was calculated between two pathways for each ligand55, as were corresponding bias plots56. The bias factor is unitless and defined as 10ΔΔLog(Emax/EC50).
GPCR-ome counter-screen
Screening of compounds in the PRESTO-Tango GPCR-ome was accomplished as previously described54 with several modifications. First, HTLA cells were plated in DMEM with 10% FBS and 10 U/mL penicillin-streptomycin. Next, the cells were transfected using an in-plate PEI method57. PRESTO-Tango receptor DNAs were resuspended in OptiMEM and hybridized with PEI prior to dilution and distribution to 384-well plates and subsequent addition to cells. After overnight incubation, drugs were added to cells without replacement of the medium. The remaining steps of the PRESTO-Tango protocol were followed as previously described. For those six receptors where activity was reduced to less than 0.5 fold of basal (RLU) or for the one receptor where basal signaling was increased greater than 3-fold of basal, assays were repeated in full dose-response. None of the seven confirmed, and we discount the apparent activity seen in the single-point assay.
Inhibition screen
Binding assays were performed by the NIMH Psychoactive Drug Screening program as detailed previously58. Detailed binding assay protocols are available on-line at: https://pdspdb.unc.edu/pdspWeb/content/UNC-CH%20Protocol%20Book.pdf
BRET recruitment assay
To measure G protein recruitment BRET assay, HEK293T cells were co-transfected in a 1:1:1:1 ratio of Gαi3-RLuc, Gβ3, GFP2-Gγ9, and hMT1or hMT2 (de-Tango-ized constructs) respectively. After 24 hours, transfected cells were plated in poly-L-lysine coated 96-well white clear bottom cell culture plates with DMEM containing 1% dialyzed FBS, 100 units/ml Penicillin G, and 100 μg/ml Streptomycin at a density of 40,000 cells in 200 μL per well and incubated overnight. The following day, media was removed and cells were washed once with 100 μL of assay buffer (1X HBSS, 20 mM HEPES, pH 7.4, 0.1% BSA). Then 60 μL of assay buffer was loaded per well followed by addition of 10 μL of the RLuc substrate, Coelenterazine 400a (Nanolight) at 5 μM final concentration for 5 mins. Drug stimulation was performed with the addition of 30 μl of 3X drug dilution of melatonin or UCSF4226 in assay buffer supplemented with 0.01% (w/v) ascorbic acid per well and incubated at RT for another 5 mins. Both luminescence (400 nm) and fluorescent GFP2 emission (515 nm) were read for the plate for 1 second per well using Mithras LB940. The ratio of GFP2/RLuc was calculated per well and analyzed using “log (agonist) vs. response” in Graphpad Prism 8 (Graphpad Software Inc., San Diego, CA).
Radioligand Binding
Reagents and Ligands
2-[125I]-Iodomelatonin (SA: 2,200 ci, 81.4TBq/mmol) was purchased from Perkin Elmer (Shelton, CT, USA). Guanosine 5’-triphosphate sodium salt hydrate (GTP), melatonin and all other chemicals and reagents were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Compound Preparation
For receptor binding studies, UCSF7447 was dissolved in 50% DMSO/50% ethanol for 13 mM stock solution, diluted 1/10 in 100% ethanol then 1/10 again in 50% ethanol/50% Tris-HCl buffer, pH 7.4 25 deg C. Both UCSF3384 and UCSF4226 were dissolved in 100% ethanol for 13 mM stock solutions and then diluted 1/10 in 50% ethanol/50% Tris-HCl buffer, pH 7.4. Further dilutions were done in the same Tris-HCl buffer.
2-[125I]-Iodomelatonin Competition Binding
CHO cells stably expressing FLAG-tagged recombinant hMT1, hMT2, mMT1, or mMT2 melatonin receptors were grown in culture as monolayers in Ham’s F12 media supplemented with fetal calf serum (10%), penicillin (1%; 10,000 I.U/ml)/streptomycin (5%; 10,000 μg/ml) in CO2 at 37°C as described. Cells were grown for 4 days to 90–95% confluence, then washed with PBS (potassium phosphate buffer, 10 mM, pH 7.4), detached with PBS containing 0.25 M sucrose and 1 mM EDTA, and pelleted by centrifugation (1,700 x g, 5 min) as described59. Cell pellets were suspended and homogenized in control buffer (50 mM Tris-HCl, 10 mM MgCl2; pH 7.4 at 25°C) and washed twice by centrifugation (17,000 x g, 15 min) in control or inactive conformation buffer (50 mM Tris-HCl, 10 mM MgCl2, 100 μM GTP, 1 mM EDTA.Na2, 150 mM NaCl, pH 7.4 at 25°C) as described59. 2-[125I]-Iodomelatonin binding affinity was determined on membranes from CHO-hMT1 (9.6 ± 0.3 μg protein/assay; Bmax: 1,154 ± 38 fmol/mg protein, n = 3), CHO-hMT2 (15 ± 1 μg protein/assay; Bmax: 352 ± 19 fmol/mg protein, n = 3), CHO-mMT1 (6.0 + 0.022 μg protein/assay (n=3); Bmax: 1,705 ± 337 fmol/mg protein, n = 3) and CHO-mMT2 (6.4 + 0.7 μg protein/assay (n=3); Bmax: 725 + 93 fmol/mg protein, n = 3) cells. Ligand competition (10 pM to 100 μM) for 2-[125I]-iodomelatonin (104 ± 2 pM, n = 30) binding was performed in control or inactive conformation buffer in a total volume of 0.26 mL as described59. Assays were incubated for 1 hour at 25°C. Bound radioligand was separated from free by rapid vacuum filtration using glass microfiber filters (Whatman, Krackeler Scientific, Inc., Albany NY, USA) saturated in 0.5% polyethylenimine solution. Total radioactivity bound to the filters was determined on a gamma counter59.
Data Analysis
Ki values were calculated from IC50 values using GraphPad PRISM™ 8.0 according to the Cheng-Prusoff equation60: Ki = IC50/(1 + [L]/KD) where L is the concentration of radioligand, KD is the dissociation constant of 2-[125I]-iodomelatonin in control or inactive conformation buffer for the hMT1 (control KD = 116 pM; Inactive KD = 280 pM) and hMT2 receptors (control KD = 80 + 13 pM; GTP KD = 461 + 159 pM), and for mMT1 receptors (control KD = 87 + 6 pM; GTP KD = 201 + 67 pM) (n=3). Affinity shifts induced by G protein uncoupling were measured by subtracting pKi(inactive) from pKi(Control) (ΔpKi) and normalization by melatonin ΔpKi (CHO-hMT1: 1.19; CHO-hMT2: 0.41). Affinity shifts or lack thereof with G protein uncoupling indicate apparent efficacy46 as ligands are classified as agonists (ΔpKi % MLT > 20 %), antagonists (ΔpKi % MLT < 20 %, > -20 %), or inverse agonists (ΔpKi % MLT < -20 %) accordingly. Individual data points were excluded from cell based when meeting the exclusion criteria for the outliers Grubbs test.
Data shown in Fig. 2a and b were analyzed by two-tailed paired student t test.
In-vivo Methods
Animals and Housing
Male and female C3H/HeN (C3H) wild-type (WT), MT1 knockout (MT1KO), and MT2 knockout (MT2KO) mice (average 6.28 months) used in this study were raised in our breeding colony at University at Buffalo. C3H/HeN mice homozygous for the MT1 and MT2 melatonin receptor gene deletion and their WT controls were generated from breeding pairs donated by Dr. S. M. Reppert (University of Massachusetts Medical School, Worcester, MA, USA) and backcrossed with C3H/HeN mice (Harlan, now Envigo, Indianapolis, IN, USA) for at least seven generation as described in detail61. Genotype was confirmed using tail samples at the end of each experiment and was verified periodically during the tenure of the colony. The strains of mice in our breeding colony were re-derived periodically by backcrossing with WT mice to reduce genetic drift.
Mice were group housed (3 – 5 per cage) with corncob bedding in polycarbonate translucent cages (30 × 19 cm) and maintained in a 14/10 light-dark (LD) cycle (Zeitgeber time 0 or ZT 0 corresponds to lights on and ZT 14 to lights off) in temperature and humidity controlled rooms with ad libitum access to food and water in the Laboratory Animal Facility at the University at Buffalo. Light levels were 200 – 300 lux at the level of the cage. Treatments and animal care performed in the dark were under a dim red safelight (15 watts, Kodak 1A filter) with illuminance of less than 3 lux35. All experimental procedures using mice were conducted in accordance with guidelines set forth by the National Institutes of Health and approved by the University at Buffalo Institutional Animal Care and Use Committee.
Circadian Rhythm Measurement
Circadian rhythm phase was determined for each mouse using the onset of running wheel activity defined as CT 12 (circadian time 12: onset of wheel activity). Running wheel activity was measured continuously via magnetic microswitches detecting wheel revolutions with a computer equipped with Clocklab data collection software™ (Actimetrics: Wilmette, IL). All actigraphy data was visualized and analyzed using ClockLab™ and MATLAB™ software. All mice were individually housed in cages (33 × 15 cm) equipped with running wheels in light-tight ventilated cabinets with controlled temperature and LD cycles (Phenome Technologies: Skokie, IL). Male and female mice were housed in separate cabinets for all experiments.
Phase Shift
Changes in circadian phase induced by vehicle or drugs administered at various circadian times were assessed in WT, MT1KO, and MT2KO male and female C3H/HeN mice (3 to 8 months) using methods and protocols previously described34,35. Following a period of 14 days in a LD cycle mice were placed in constant dark (DD) beginning at Zeitgeber Time (ZT) 12 (dark onset) (ZT 0 = lights on). Mice were kept in DD (2 – 3 weeks) until a stable free-running phase of running wheel activity rhythm onset was established. Circadian times of treatment were predicted from best fit lines of running wheel activity onsets for of running either pre (7 – 14 days) and post (7 – 14 days) treatment. Treatment times were within a 2-hour window at CT 2 (CT 1 – 3), CT 6 (CT 5 – 7), or CT 10 (CT 9 – 11). Mice were treated (0.1 ml/mouse, s.c.) with vehicle (30% ethanol saline, s.c.) or drugs (melatonin, UCSF3384, UCSF7447, at 0.9 μg and 30 μg/mouse or luzindole at 300 μg/mouse in vehicle) for three consecutive days at the appropriate circadian time under dim red light. Vehicle or drug treatments were repeated for 3 consecutive days at the selected circadian time following the three-pulse treatment protocol described35. Phase shifts were quantified using the best-fit lines for onsets of activity during pre and post treatment periods. Differences are characterized as phase delays (pre-treatment ahead of post treatment best fit line onset) or phase advances (post treatment ahead of pre-treatment best fit line onset) of running wheel activity onset rhythms.
Re-entrainment Experiments
Male and female C3H/HeN WT, MT1KO, and MT2KO mice (3 to 6 months) were maintained under a 12:12 LD cycle for at least 2 weeks prior experimental manipulations to allow stable entrainment to dark onset before advance of the LD cycle. Actigraphy data was recorded as described above and all experimental protocols performed as described62. On the first day of treatment, the dark onset was advanced 6 hours. This resulted in a short night and mice were treated (0.1 ml / mouse s.c.) with vehicle (30% ethanol/70%saline, s.c.) or drugs (melatonin, UCSF3384 or UCSF7447 at 30 μg /mouse, or luzindole 300μg /mouse, in vehicle) for three consecutive days 10 – 30 minutes prior to the new dark onset. Post treatment, mice were given 14 – 20 days to re-entrain running wheel activity onsets to the new dark onset. Using exported running wheel activity onsets from actograms, onset hours advanced each day were determined by subtracting this value each day from the average onset of running wheel activity for 3 days prior to treatment for each mouse. Further, using the data from this calculation combined with visualization of actograms, the number of days to reach stable re-entrainment was determined for each mouse.
In vivo Compound Preparation
All compounds were administered in fixed doses of either 0.9 μg or 30 μg subcutaneously (s.c.) in a volume of 0.1 ml per mouse, which are equivalent to doses of 0.03 or 1 mg/Kg for a 30 g mouse, respectively. Vehicle (VEH) was 30% ethanol/70% saline for all doses. Melatonin, UCSF7447, and UCSF3384 were prepared as stock solutions of 3 mg/mL (100% ethanol) using sonication and vortexing to ensure each drug was dissolved. Subsequently, stock solutions were diluted to 0.3 mg/mL (30 μg/0.1 mL injection) or 0.009 mg/mL (0.9 μg/0.1 mL injection) in vehicle. Luzindole was prepared similarly except the starting stock solution was 30 mg/mL in 100% ethanol and it was administered from a solution of 3 mg/mL (300 μg/0.1 mL injection) in vehicle. Treatment dilutions were prepared just before use under sonication with intermittent vortexing between steps and used within 5 minutes of preparation.
Biostatistics and Reproducibility
All statistical analyses as described in further detail for each experiment were conducted using GraphPad Prism 8™ (La Jolla, CA). For phase shift and re-entrainment experiments we determined statistical power a-priori (α error probability = 0.05) based on data for a known effect size for melatonin in these paradigms (G-power 3.0.10)34,62. Individual actograms of wheel running activity were excluded from analysis based on the exclusion criteria described below, which was completed by at least two individuals blind to treatment before data analysis was started. For re-entrainment actograms exclusion criteria includes: a) low running, sporadic activity, significant missing wheel activity data and/or lack of entrainment prior to treatment; b) entrainment of running activity more than 1 h before or after the “old” or “new dark” onset; c) re-entrainment to new dark onset before administration of the third injection (entrainment to injection). For phase shift actograms exclusion criteria includes: a) low running, sporadic activity, missing wheel activity data and/or lack of free running activity rhythms; b) tau change > 0.3 h; c) at least 2 out of 3 injections occurred outside of the target pre-determined time-range for treatment (CT 1 – 3, 5 – 7, 10 – 12). All data sets were visualized for normality using QQ plots of predicted vs. actual residuals. Actigraphy data was generated for visualization blind to treatment prior to the quantification and statistical analysis stages. Comparisons for Fig. 3a, Extended Data Fig. 4l - m were made by mixed effect two-way repeated measures ANOVA (treatment x time) with Sidak’s post hoc test (P < 0.05). Number of days to re-entrainment was compared via one-way ANOVA or two-way ANOVA for Fig. 3b & Fig. 3c with a Dunnet’s or Tukey’s post hoc test (P < 0.05) respectively. Group comparisons for phase shift in Fig. 3d (left & center) & Extended Data Fig. 7a - c were made by one-way ANOVA (P < 0.05) comparing hours shifted of circadian running wheel activity rhythm onsets (3d left: 3 groups - vehicle, melatonin, UCSF7447; 3d center: 4 groups - vehicle, melatonin, UCSF7447, UCSF3384; 7a - c: 4 groups - vehicle, melatonin, UCSF7447, luzindole) accompanied with post-hoc analyses by Dunnet’s to determine individual group differences compared to vehicle (P < 0.05). Fig. 3d (right) comparisons between vehicle and luzindole were made via a two-tailed unpaired students t test (P < 0.05). Data in Fig. 3e & f were compared via a two-way ANOVA (3 × 2: genotype x treatment) with Tukey’s post hoc analyses (P < 0.05). P values and values for statistical analyses are included in figure legends or listed in Supplementary Table 4 Either the overall interaction or the main effects were reported and interpreted for two-way ANOVAs as appropriate for assumptions of each data set. No sex differences in treatment effects were evident in any data set when assessed via two-way ANOVA or three-way ANOVA where appropriate; therefore, data were pooled between male and female mice for analyses described. The n values represent the number of individual mice per condition or independent biological replicates in each experiment. Each data set represents 2 – 4 independent experiments. The n value for each in vivo experiment is listed below:
Fig. 3. In vivo, MT1-selective inverse agonists decelerate re-entrainment rate and phase-advance circadian activity when administered at dusk (CT 10) in C3H mice both via selective actions at MT1.
3a, vehicle (n = 28 mice#) vs. UCSF7447 (n = 21 mice#)
3b, vehicle (n = 28) vs. melatonin (n = 21), UCSF7447 (n = 21), UCSF3384 (n = 16), or luzindole (n = 11)
3c, WT (n = 28 vehicle; n = 21 UCSF7447), MT1KO (n = 16 vehicle; n = 16 UCSF7447), and MT2KO (n = 20 vehicle; n = 25 UCSF7447)
3d, (left panel) - vehicle (n = 8) vs. melatonin (n = 8) or UCSF7447 (n = 13)
(center panel) - vehicle (n = 15) vs. melatonin (n = 10), UCSF3384 (n = 16), or UCSF7447 (n = 15)
(right panel) - vehicle (n = 6) vs luzindole (n = 3)
3e, WT (n = 9 vehicle; n = 10 UCSF7447), MT1KO (n = 8 vehicle; n = 8 UCSF7447), and MT2KO (n = 11 vehicle; n = 9 UCSF7447)
3f, WT (n = 8 vehicle; n = 8 UCSF7447), MT1KO (n = 6 vehicle; n = 7 UCSF7447), and MT2-KO (n = 10 vehicle; n = 13 UCSF7447)
# n values for multiple comparisons range from 1 – 2 values less depending on day compared due to missing onset data which is accounted for in statistical models as appropriate.
Extended Data Fig. 4 (re-entrainment) - MT1-selective inverse agonists decelerate re-entrainment rate in vivo via MT1 receptors.
4h, C3H WT - vehicle (n = 28 mice#) vs. UCSF3384 (n = 16 mice#).
4i, C3H MT1KO - vehicle (n = 16 mice#) vs. UCSF7447 (n = 16 mice#).
4j, C3H MT2KO - vehicle (n = 21 mice#) vs. UCSF7447 (n = 25 mice).
# n values for multiple comparisons range from 1 – 2 values less depending on day compared due to missing onset data which is accounted for in statistical models as appropriate.
Extended Data Fig. 7. a - c, Differential phase shift profile for UCSF7447 compared to the agonist melatonin and a prototype antagonist luzindole across the circadian cycle.
7a, CT 2 - vehicle (n = 3), melatonin (n = 3), luzindole (n = 6), or UCSF7447 (n = 3)
7b, CT 6 - vehicle (n = 8), melatonin (n = 4), luzindole (n = 9), or UCSF7447 (n = 9)
7c, CT 10 - vehicle (n = 6), melatonin (n = 8), luzindole (n = 3), or UCSF7447 (n = 4)
Pharmacokinetics
Pharmacokinetic experiments were performed by Sai Life Sciences Limited (Hyderabad, India). Plasma pharmacokinetics and brain distribution for UCSF7447, UCSF3384, and UCSF4226 were investigated following a single intravenous dose of 2 mg/kg in nine male C57BL/6 mice. Each compound was formulated in 5% N-methyl pyrrolidone, 5% Solutol HS-15, and 90% normal saline. Blood samples (approximately 60 μL from each of three mice) were collected under light isoflurane anesthesia from retro orbital plexus at 0.08, 0.25, 0.5, 1, 2, 4, 8, 12, and 24 hr. Immediately after collection, plasma was harvested by centrifugation and stored at -70°C until analysis. For blood collected at 0.5, 4, and 24 hr, animals were euthanized with excess CO2 asphyxiation and brain samples were collected and homogenized in ice-cold phosphate buffer saline (pH-7.4). Total homogenate volume was three times the brain weight.
All samples were processed for analysis by protein precipitation using acetonitrile and analyzed with fit-for-purpose LC/MS/MS method (Lower limit of quantification = 2.01 ng/mL for plasma and 6.03 ng/g for brain for UCSF7447, 5.01 ng/mL for plasma and 3.00 ng/g for brain for UCSF3384, 1.01 ng/mL for plasma and 6.09 ng/g for brain for UCSF4226). The non-compartmental analysis module in Phoenix WinNonlin® (Version 7.0) was used to assess the pharmacokinetic parameters. Maximum concentration (Cmax) and time to reach maximum concentration (Tmax) were measured. The areas under the concentration time curve (AUClast and AUCinf) and elimination half-life was calculated by the linear trapezoidal rule. The terminal elimination rate constant, ke, was determined by regression analysis of the linear terminal portion of the log plasma concentration-time curve. The terminal half-life (T1/2) was estimated as 0.693/ke.
Code Availability:
DOCK3.7 is freely available for non-commercial research http://dock.compbio.ucsf.edu/DOCK3.7/. A web-based version is freely available to all at http://blaster.docking.org/.
Data Availability Statement:
Probe pairs (two similar ligands with and without activity) of inverse agonists selective for MT1 and agonists selective for hMT2 are available by arrangement with Sigma (Extended Data Figure 3). The identities of the compounds docked in this study are freely available from the ZINC database, http://zinc15.docking.org, and active compounds may be purchased from Enamine. Figures with associated raw data include: Fig. 1, Extended Data Tables 1&2, Extended Data Figs. 1&2, Extended Data Table 1, for which further data are included in Supplementary Tables 1 (MT1 and MT2 affinities, MT1 DOCK energies/ranks) and 2 (compound purity information); Extended Data Fig. 3, for which bias information is included in Supplementary Table 3; Fig. 2, for which GPCRome screening, concentration-response curves, competition binding, and LC/MS data is included in Supplementary Data 1–5; Supplementary Data 6&7 (synthesis routes and spectra of compounds); Fig. 3, for which further data is included in Extended Data 4–5; Extended Data Fig. 7; Supplementary Table 4. Raw data values and transform data for in vitro cell based assays as well as in vivo data for phase shift and re-entrainment are available for Fig. 2; Extended Data Fig. 6; Fig. 3; Extended Data Fig. 4 (re-entrainment), Extended Data Fig. 5 (phase shift), Extended Data Fig. 7a–c.
Extended Data
Extended Data Fig. 1. Concentration-response curves of initial 15 compounds in cAMP assays.

hMT1- (a,c,e) or hMT2-mediated (b,d,f) inhibition of isoproterenol-stimulated cAMP in HEK cells by melatonin and 15 initial compounds. Data normalized to melatonin response represent mean ± s.e.m. of four biologically independent experiments (n=4) run in triplicate, unless otherwise indicated, which is indicated in parenthesis next to each compound name.
Extended Data Fig. 2. Concentration-response curves of interesting analogs based on initial hits in cAMP assays.

hMT1- (a,c,e) or hMT2-mediated (b,d,f) inhibition of isoproterenol-stimulated cAMP in HEK cells by melatonin and select analogs. Data normalized to melatonin response represent mean ± s.e.m. of four biologically independent experiments (n=4) run in triplicate, unless otherwise indicated, which is indicated in parenthesis next to each compound name.
Extended Data Fig. 3. Small changes in ligand structure have large effects on melatonin receptor activity and selectivity.

a, Docked pose of ‘9032, an MT1-selective direct docking hit. b, Docked pose of ‘1360, a close analog of ‘9032 that switches 2-fold selectivity for MT2 over MT1. c, Docked pose of ‘2780, an analog where MT2 selectivity climbs to 89-fold over MT1. d, Docked pose of ‘2623, which adds a bulkier 2-chloro-3-methylthiophene into a proposed MT2-selective hydrophobic cleft, resulting in a fully MT2-selective agonist without detectable MT1 activity. All docked poses are overlaid onto the crystallographic pose of 2-phenylmelatonin in transparent blue. e, Concentration-response curves the four analogs at MT1 and MT2. Data normalized to melatonin response represent mean ± s.e.m. of four biologically independent experiments (n=4) run in triplicate. f, Bias plots of ‘0041 and ‘6688 relative to melatonin signaling. Mean values (Supplementary Table 3) are presented as solid lines and the 95% confidence interval for the line is shaded. Data are normalized to melatonin response and represent mean ± s.e.m. of three biologically independent experiments (n=3) run in triplicate, except for ‘6688 for Gi activation (n=4).
Extended Data Fig. 4. MT1-selective inverse agonists decelerate re-entrainment rate in vivo via MT1 receptors.

a - e, Representative actograms of running wheel (RW) activity in wild type (WT) C3H/HeN (C3H) mice treated with VEH (a), 30 μg/mouse MLT (b), UCSF7447 (c), UCSF3384 (d), as well as 300 μg/mouse LUZ (e) just prior to the new dark onset (black dots) following an abrupt 6h advance of dark onset in a 12:12 light-dark cycle (gray: dark phase; white: light phase). Compounds were administered once a day for 3 days (see Methods for additional details). Corresponding quantification found in Fig. 3b,c. f - k, Representative actograms of RW activity for VEH [WT (a), MT1KO (c), MT2KO (e)] or inverse agonist ‘7447 [WT (b), MT1KO (d), MT2KO (f)] treated C3H mice following a 6 h advance of dark onset. Mice were kept in a 12:12 light-dark cycle. ‘7447 (30 μg/mouse) was administered for 3 consecutive days just prior to the new dark onset (black dots). l, Inverse agonist ‘3384 decelerates the rate of re-entrainment of RW activity rhythm onset in C3H WT mice. Data expressed in hours advanced each day for VEH vs. ‘3384 (two-way repeated measures ANOVA; treatment x time interaction: F16,647 = 1.99 P = 0.0122). m, Inverse agonist ‘7447 does not modulate the rate of re-entrainment of RW activity rhythm onset in C3H MT1KO mice. Data expressed in hours advanced each day for MT1KO mice treated with VEH vs. ‘7447 (mixed-effect two-way repeated measures ANOVA; treatment x time interaction: F16,474 = 1.44 P =0.117). n, Inverse agonist ‘7447 decelerates the rate of re-entrainment of RW activity rhythm onset in C3H MT2KO mice. Data expressed in hours advanced each day for MT2KO mice treated with VEH vs. ‘7447 (mixed-effect two-way repeated measures ANOVA; treatment x time interaction: F16,683 = 2.57 P = 0.000686). Extension of Fig. 3a - c, Extended Data Fig 3a - d. Data represents mean ± s.e.m. *P < 0.05, **P < 0.01, for multiple comparisons by Tukey’s post test (P < 0.05). Dotted line in j - k refers to the new dark onset. Additional details of all statistical analyses as well as n for each condition can be found in Methods (Statistics & Reproducibility). Vehicle (VEH), melatonin (MLT), luzindole (LUZ), UCSF7447 (‘7447), UCSF3384 (‘3384). All treatments were given via s.c. injection.
Extended Data Fig. 5. MT1-selective inverse agonists phase advance circadian activity at CT 10 via MT1 in vivo.

a - e, Representative actograms of RW activity from individual C3H WT mice kept in constant dark (gray bars) treated with VEH (a), MLT (b), UCSF7447 (c), UCSF3384 (d) or LUZ (e). All treatments were 30 μg/mouse except for LUZ which was 300 μg/mouse as described in Methods. Mice were treated at dusk (CT 10; 2 hours prior to onset of RW activity) for three consecutive days (black dots). Red lines indicate best-fit line of pre-treatment onsets and blue lines indicate best-fit line of post treatment onsets both used for phase shift determinations (see Methods for more details). Corresponding quantification found in Fig. 3d. f - h, Representative actograms of RW activity from individual C3H WT mice kept in constant dark treated with VEH (f), MLT (g), or ‘7447 (h, all treatments 0.9 μg/mouse) at CT 10. Corresponding quantification found in Fig. 3d. i - k, Representative actograms of RW activity from individual C3H WT mice kept in constant dark treated with MLT (i) at CT 2 (10 hours prior to RW onset) or VEH (j) vs. ‘7447(k, all treatments at 30 μg/mouse) at CT 6 (6 hours prior to RW onset). Corresponding quantification found in Extended Data Fig. 7. Extension of Fig. 3d - f. l - q, Representative actograms of running wheel (RW) activity from individual C3H WT (l, m), MT1KO (n, o), and MT2KO (p, q) mice kept in constant dark treated with VEH (white; l, n, p) or UCSF7447(blue; m, o, q; 30 μg/mouse) at CT 10. Corresponding quantification found in Fig. 3e. r - w, Representative actograms of RW activity from individual C3H WT (r, s), MT1KO (t, u), and MT2KO (v, w) mice kept in constant dark treated with VEH (white; r, t, v) or UCSF7447(blue; s, u, w; 30 μg/mouse) at CT 2. Corresponding quantification found in Fig. 3f. Vehicle (VEH), melatonin (MLT), luzindole (LUZ), UCSF7447 (‘7447), UCSF3384 (‘3384). All treatments were given via s.c. injection.
Extended Data Fig. 6. Concentration-response curves and Schild-plots of the inverse agonists ‘7447 and ‘3384 in cAMP assays.
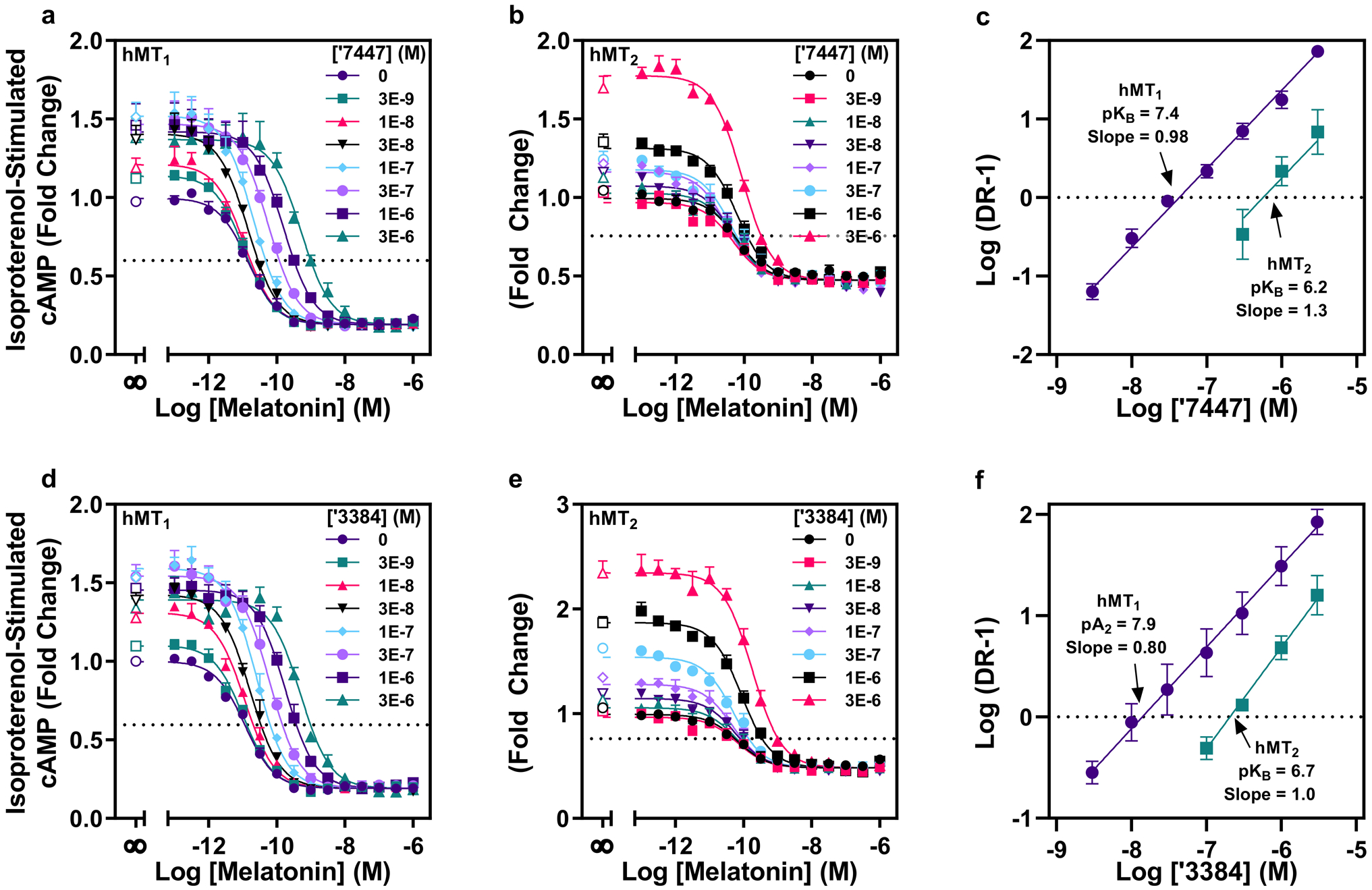
a-d, Modulation of hMT1- (a,d) or hMT2- (b,e) mediated inhibition of isoproterenol-stimulated cAMP in HEK cells by melatonin in the presence of ‘7447 (a,b) or ‘3384 (d,e) over a range of concentrations. Data normalized to effect of isoproterenol alone represent mean ± s.e.m. of three biologically independent experiments (n=3) run in triplicate. c,f. Schild plots depicting competitive antagonism of melatonin by ‘7447 (c) and ‘3384 (f). Schild analysis at hMT1 (purple) and hMT2 (teal) reveal competitive antagonism for ‘7447 (hMT1 pKB: 7.4 ± 0.1, slope: 0.98 ± 0.03; hMT2 pKB: 6.2 ± 0.1, slope: 1.3 ± 0.4) (c), and ‘3384 (hMT1 pA2: 7.9 ± 0.1, slope: 0.80 ± 0.04; hMT2pKB: 6.7 ± 0.1, slope: 1.0 ± 0.1 ) (f). Data represent mean ± s.e.m. of three biologically independent experiments (n=3) run in triplicate. UCSF7447 (‘7447), UCSF3384 (‘3384)
Extended Data Fig. 7. a - c, Differential phase shift profile for inverse agonist ‘7447 compared to the agonist melatonin and a prototype antagonist luzindole across the circadian cycle.

C3H/HeN mice were kept in constant dark and treated with VEH, MLT, LUZ, or ‘7447 (all treatments 30 μg/mouse except for LUZ which was 300 μg/mouse, s.c.). Mice were treated at CT 2, 6, or 10 (10, 6, or 2 hours prior to onset of RW activity) for three consecutive days (see details in Methods). a, CT 2 phase shift data was compared via one-way ANOVA (F3,11 = 28.16 P = 1.85 × 10−5). b, CT 6 phase shift data was compared via one-way ANOVA (F3,26 = 0.61 P = 0.61). c, CT 10 phase shift data was compared via one-way ANOVA (F3,17 = 35.13 P = 1.66 × 10−7). All multiple comparisons made to VEH using Dunnet’s post hoc test (P < 0.05).
Values for MLT & ‘7447 at CT 10 pooled from previous data for comparison to LUZ. Data shown represent mean ± s.e.m. ****P < 0.0001 for comparisons with VEH. Vehicle (VEH), melatonin (MLT), luzindole (LUZ), UCSF7447 (‘7447). All treatments were given via s.c. injection.
Extended Data Table 1.
Active molecules from the initial docking screen.
| Compound | Cluster ranka (global rank) | hMT1b pEC50 (% Emax) n | hMT2c pEC50 (% Emax) n | Tcd | Nearest ChEMBL23e MT1/MT2 Ligand |
|---|---|---|---|---|---|
|
ZINC 157665999 |
167 (197) |
4.89±0.38 (63±6) n=3 |
Inverse 7.29±0.16 (Inverse 90±16) n=3 | 0.33 |
 CHEMBL398017 |
 ZINC419113878 |
396 (522) |
5.20±0.08 (84±4) n=4 |
<4.5 n=4 |
0.22 |
 CHEMBL494566 |
 ZINC433313647 |
875 (1242) |
6.81±0.32 (42±2) n=3 |
7.77±0.02 (96±5) n=3 |
0.19 |
 CHEMBL 125226 |
|
ZINC 159050207 |
1559 (2474) |
9.00±0.15 (99±1) n=4 |
8.70±0.25 (83±3) n=4 |
0.24 |
 CHEMBL 1223128 |
 ZINC151209032 |
1981 (3583) |
5.70±0.11 (88±4) n=4 |
<4.5 n=4 |
0.31 |
 CHEMBL394676 |
 ZINC442850041 |
4123 (7872) |
7.91±0.04 (99±3) n=3 |
9.33±0.33 (97 ± 2) n=3 |
0.29 |
CHEMBL344242 |
|
ZINC353044322 |
5764 (28,258) |
5.48±0.05 (87±6) n=4 |
<4.5 n=4 |
0.33 |
 CHEMBL218225 |
 ZINC603324490 |
7612 (53,767) |
Inverse 5.92±0.29 Inverse (37±5) n=3 |
Inverse 6.20±0.08 Inverse (202±30) n=4 |
0.27 |
 CHEMBL3260982 |
 ZINC 182731037 |
7840 (17,095) |
5.30±0.09 (82±2) n=4 |
<4.5 n=4 |
0.29 |
 CHEMBL3612457 |
| ZINC92585174 | 1836 (3010) | 7.80±0.17 (98±1) n=4 |
7.68±0.14 (74±8) n=4 |
0.23 | CHEMBL 1760949 |
| ZINC432154404 | 1849 (3035) | 6.63±0.17 (95±2) n=4 |
7.00±0.17 (74±4) n=4 |
0.27 | CHEMBL 1760956 |
| ZINC664088238 | 2248 (3816) | <5 n=4 |
5.85±0.06 (75±8) n=4 |
0.20 | CHEMBL435032 |
| ZINC576887661 | 4161 (14,292) | 7.10±0.19 (83±0) n=4 | 7.28±0.36 (68±5) n=4 |
0.27 | CHEMBL491605 |
| ZINC301472854 | 5033 (10,022) | 6.03±0.10 (95±5) n=4 |
7.00±0.21 (88±6) n=4 |
0.26 | CHEMBL 115444 |
| ZINC580731466 | 8503 (19,003) | 5.70±0.13 (71 ±3) n=4 |
7.55±0.10 (98±5) n=4 |
0.26 | CHEMBL 115444 |
Cluster rank, Global rank (Methods)
The log half maximal concentration (pEC50) for inhibition of isoproterenol-stimulated cAMP production on hMT1 or hMT2 melatonin receptors transiently expressed in HEK cells. Values in parenthesis represent the percentage of the maximal inhibition normalized to % melatonin response, except for inverse agonists, indicated by (Inverse), where data is normalized to % basal induced by isoproterenol. Data represent mean ± S.E.M. from the indicated number (n) of biologically independent experiments run in triplicate.
ECFP4 Tanimoto coefficient (Tc) to the most similar known MT1 or MT2 ligand in ChEMBL23.
MT1/MT2 ligand in ChEMBL23 most similar to docking active.
Extended Data Table 2.
Some of the potent analogs from initial hits.
| Initial Hita | Analogb | hMT1c pEC50 (% Emax) n | hMT2d pEC50 (% Emax) n |
|---|---|---|---|
 ZINC 157665999 |
 ZINC864032792 |
7.49 ± 0.04 (57 ± 3) n=3 |
Inverse 6.66 ± 0.08 (Inverse 35 ± 5) n=3 |
 ZINC 157665999 |
 ZINC555417447 |
Inverse 7.39 ± 0.10 (Inverse 62 ±13) n=8 |
Inverse 5.66 ±0.10 (Inverse 84 ± 9) n=8 |
 ZINC 157665999 |
 ZINC 157673384 |
Inverse 7.68 ± 0.09 (Inverse 47 ± 12) n=13 |
Inverse 6.18 ± 0.04 (Inverse 153 ±14) n=12 |
 ZINC 157665999 |
ZINC5586789 |
6.81 ± 0.72 (37 ± 8) n=3 |
8.07 ±0.15 (51 ± 3) n=4 |
 ZINC 157665999 |
 ZINC128734226 |
6.83 ± 0.17 (79 ± 3) n=4 |
8.15 ±0.09 (89 ± 3) 77=4 |
 ZINC419113878 |
 ZINC602421874 |
4.70 ±0.11 (51 ± 3) n=4 | 5.35 ± 0.10 (66 ± 7) n=4 |
|
ZINC 159050207 |
ZINC713465976 |
7.75 ± 0.22 (101 ± 0) n=4 |
8.23 ±0.11 (94 ± 3) n=4 |
 ZINC 151209032 |
 ZINC497291360 |
7.05 ± 0.10 (92 ± 2) n=4 |
7.48 ± 0.05 (75 ± 5) n=4 |
 ZINC 151209032 |
 ZINC151192780 |
5.18 ± 0.22 (54 ± 4) n=4 |
7.13 ± 0.12 (95 ± 5) n=4 |
 ZINC 151209032 |
 ZINC485552623 |
<5 n=4 |
5.80 ± 0.06 (107 ±5) n=4 |
 ZINC442850041 |
 ZINC608506688 |
9.78 ± 0.13 (99 ± 1) n=4 |
8.60 ± 0.10 (89 ± 3) n=4 |
|
ZINC301472854 |
 ZINC223593565 |
6.40 ± 0.18 (86 ± 4) n=4 |
6.45 ± 0.20 (58 ± 5) n=4 |
Compound selected directly from the primary docking screen and found to be active on in vitro testing
Analog from initial hit
The log half maximal concentration (pEC50) for inhibition of isoproterenol-stimulated cAMP production on hMT1 or hMT2 melatonin receptors transiently expressed in HEK cells. Values in parenthesis represent the percentage of the maximal inhibition normalized to % melatonin response, except for inverse agonists, indicated by (Inverse), where data is normalized to % basal induced by isoproterenol. Data represent mean ± s.e.m. from the indicated number (n) of biologically independent experiments run in triplicate.
UCSF7447 (‘7447), UCSF3384 (‘3384), UCSF4226 (‘4226)
Extended Data Table 3.
Pharmacokinetics of three melatonin receptor type-selective ligands
| Compound | pIC50 (Emax %) pEC50 (IA) | Cmaxa (ng/ml_) | AUCb (hr*ng/ml_) | T1/2c (hr) | CLd (mL/min/kg) | Vsse | Brain/Plasma ratio |
|---|---|---|---|---|---|---|---|
 ZINC 128734226 MT2 -selective |
pIC50 MT1 – 6.8 (48%) MT2 – 8.2 (80%) |
1922.8 | 282.1 | 0.29 | 117.9 | 1.11 | 1.58 (30’) |
 ZINC555417447 MT1-selective inverse agonist |
pEC50 MT1 – 7.4 (IA) MT2 – 5.8 (IA) |
1948.6 | 494.5 | 0.27 | 67.11 | 1.11 | 3.03 (30’) |
 ZINC157673384 MT,-selective inverse agonist |
pEC50 MT1 – 7.7 (IA) MT2 – 6.2 (IA) |
1299.6 | 563.8 | 0.32 | 58.48 | 1.38 | 1.43 (30’) |
Cmax: Maximum concentration
AUC: Area under plasma concentration-time curve
Half-life
Clearance
Volume of distribution at steady-state
UCSF4226 (‘4226), UCSF7447 (‘7447), UCSF3384 (‘3384)
Extended Data Table 4:
Probe pairs of in vivo tested molecules
| Active Selective Probe (Sigma RefCode) | hMT1 pEC50a (% Emax) n | hMT2 pEC50b (% Emax) n | Inactive analog (Sigma RefCode) | hMT1 pEC50a | hMT2 pEC50b |
|---|---|---|---|---|---|
 ZINC555417447 (SML2751) |
Inverse 7.4 ± 0.10 (Inverse 62 ± 13) n=8 |
Inverse 5.7 ± 0.10 (Inverse 84 ± 9) n=8 |
 ZINC37781618 (SML2752) |
<4.5 n=3 |
<4.5 n=3 |
 ZINC 128734226 (SML2753) |
6.8 ±0.2 (79 ± 3) n=4 |
8.2 ±0.1 (89 ± 3) n=4 |
 Z3670677764 (SML2754) |
<4.5 n=3 |
<4.5 n=3 |
The log half maximal concentration (pEC50) for inhibition of isoproterenol-stimulated cAMP production on hMT1 or hMT2 melatonin receptors transiently expressed in HEK cells. Values in parenthesis represent the percentage of the maximal inhibition normalized to % melatonin response for ‘4226, and to % basal activity for ‘7447. Compounds were tested at concentrations up to 30μM. Data represent mean ± s.e.m. from the indicated number (n) of biologically independent experiments run in triplicate.
UCSF4226 (‘4226), UCSF7447 (‘7447)
Supplementary Material
Acknowledgements.
Supported by the US NIH awards U24DK1169195 (to BLR & BKS), R35GM122481 (to BKS), the NIMH Psychoactive Drug Screening Contract (to BLR), GM133836 (to JJI), ES023684 (to MLD), UL1TR001412 and KL2TR001413 (to the University at Buffalo), PhRMA Foundation Fellowship (73309 to AJJ), Jacobs School of Medicine and Biomedical Sciences unrestricted funds (to MLD), R35GM127086 (to VC), EMBO ALTF 677-2014 (to BS), HFSP long-term fellowship LT000046/2014-L (to LCJ), postdoctoral fellowship from the Swedish Research Council (to LCJ), and the National Science Foundation (NSF) BioXFEL Science and Technology Center 1231306 (to BS & VC). We would like to thank Dr. Gregory Wilding from the Biostatistics, Epidemiology and Research Design (BERD) Core of the Clinical and Translational Science Institute at the University at Buffalo, for statistical advice regarding analyses of in-vivo data.
Footnotes
Competing Financial Interests: B.K.S. and J.J.I. are founders of a company, BlueDolphin LLC, that works in the area of molecular docking. All other authors declare no competing interests.
Supplementary Information is available for this paper
References
- 1.Zisapel N New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br J Pharmacol 175, 3190–3199, doi: 10.1111/bph.14116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubocovich ML et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev 62, 343–380, doi: 10.1124/pr.110.002832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J et al. MT1 and MT2 Melatonin Receptors: A Therapeutic Perspective. Annu Rev Pharmacol Toxicol 56, 361–383, doi: 10.1146/annurev-pharmtox-010814-124742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubocovich ML Melatonin receptors: role on sleep and circadian rhythm regulation. Sleep Med 8 Suppl 3, 34–42, doi: 10.1016/j.sleep.2007.10.007 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Mundey K, Benloucif S, Harsanyi K, Dubocovich ML & Zee PC Phase-dependent treatment of delayed sleep phase syndrome with melatonin. Sleep 28, 1271–1278, doi: 10.1093/sleep/28.10.1271 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Rajaratnam SM et al. Melatonin agonist tasimelteon (VEC-162) for transient insomnia after sleep-time shift: two randomised controlled multicentre trials. Lancet 373, 482–491, doi: 10.1016/S0140-6736(08)61812-7 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Lewy AJ et al. The phase shift hypothesis for the circadian component of winter depression. Dialogues Clin Neurosci 9, 291–300 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jockers R et al. Update on melatonin receptors: IUPHAR Review 20. Br J Pharmacol 173, 2702–2725, doi: 10.1111/bph.13536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bodinat C et al. Agomelatine, the first melatonergic antidepressant: discovery, characterization and development. Nat Rev Drug Discov 9, 628–642, doi: 10.1038/nrd3140 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Descamps-Francois C et al. Design and synthesis of naphthalenic dimers as selective MT1 melatoninergic ligands. J Med Chem 46, 1127–1129, doi: 10.1021/jm0255872 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Spadoni G et al. Bivalent ligand approach on N-{2-[(3-methoxyphenyl)methylamino]ethyl}acetamide: synthesis, binding affinity and intrinsic activity for MT(1) and MT(2) melatonin receptors. Bioorg Med Chem 19, 4910–4916, doi: 10.1016/j.bmc.2011.06.063 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Zlotos DP, Riad NM, Osman MB, Dodda BR & Witt-Enderby PA Novel difluoroacetamide analogues of agomelatine and melatonin: probing the melatonin receptors for MT1 selectivity. MedChemComm 6, 1340–1344, doi: 10.1039/C5MD00190K (2015). [DOI] [Google Scholar]
- 13.Stauch B et al. Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288, doi: 10.1038/s41586-019-1141-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson LC et al. XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 569, 289–292, doi: 10.1038/s41586-019-1144-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyu J et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229, doi: 10.1038/s41586-019-0917-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss DR et al. Selectivity Challenges in Docking Screens for GPCR Targets and Antitargets. J Med Chem 61, 6830–6845, doi: 10.1021/acs.jmedchem.8b00718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manglik A et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190, doi: 10.1038/nature19112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang XP et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483, doi: 10.1038/nature15699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansu K et al. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol 13, 529–536, doi: 10.1038/nchembio.2334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sterling T & Irwin JJ ZINC 15--Ligand Discovery for Everyone. J Chem Inf Model 55, 2324–2337, doi: 10.1021/acs.jcim.5b00559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman RG, Carchia M, Sterling T, Irwin JJ & Shoichet BK Ligand pose and orientational sampling in molecular docking. PLoS One 8, e75992, doi: 10.1371/journal.pone.0075992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bento AP et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res 42, D1083–1090, doi: 10.1093/nar/gkt1031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin JJ & Shoichet BK Docking Screens for Novel Ligands Conferring New Biology. J Med Chem 59, 4103–4120, doi: 10.1021/acs.jmedchem.5b02008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchmore SW et al. Application of belief theory to similarity data fusion for use in analog searching and lead hopping. J Chem Inf Model 48, 941–948, doi: 10.1021/ci7004498 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Katritch V et al. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J Med Chem 53, 1799–1809, doi: 10.1021/jm901647p (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Graaf C et al. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H(1) receptor. J Med Chem 54, 8195–8206, doi: 10.1021/jm2011589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannel B et al. Structure-Guided Screening for Functionally Selective D2 Dopamine Receptor Ligands from a Virtual Chemical Library. ACS Chem Biol 12, 2652–2661, doi: 10.1021/acschembio.7b00493 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Kiss R et al. Discovery of novel human histamine H4 receptor ligands by large-scale structure-based virtual screening. J Med Chem 51, 3145–3153, doi: 10.1021/jm7014777 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Congreve M et al. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem 55, 1898–1903, doi: 10.1021/jm201376w (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead CJ et al. Identification of novel adenosine A(2A) receptor antagonists by virtual screening. J Med Chem 55, 1904–1909, doi: 10.1021/jm201455y (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamah-Biassi EB, Stepien I, Hudson RL & Dubocovich ML Effects of the Melatonin Receptor Antagonist (MT2)/Inverse Agonist (MT1) Luzindole on Re-entrainment of Wheel Running Activity and Spontaneous Homecage Behaviors in C3H/HeN Mice. The FASEB Journal 26, 1042.1045–1042.1045, doi: 10.1096/fasebj.26.1_supplement.1042.5 (2012). [DOI] [Google Scholar]
- 32.Dubocovich ML Luzindole (N-0774): a novel melatonin receptor antagonist. J Pharmacol Exp Ther 246, 902–910 (1988). [PubMed] [Google Scholar]
- 33.Browning C, Beresford I, Fraser N & Giles H Pharmacological characterization of human recombinant melatonin mt(1) and MT(2) receptors. Br J Pharmacol 129, 877–886, doi: 10.1038/sj.bjp.0703130 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubocovich ML, Yun K, Al-Ghoul WM, Benloucif S & Masana MI Selective MT2 melatonin receptor antagonists block melatonin-mediated phase advances of circadian rhythms. FASEB J 12, 1211–1220, doi: 10.1096/fasebj.12.12.1211 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Benloucif S & Dubocovich ML Melatonin and light induce phase shifts of circadian activity rhythms in the C3H/HeN mouse. J Biol Rhythms 11, 113–125, doi: 10.1177/074873049601100204 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Burgess HJ, Revell VL, Molina TA & Eastman CI Human phase response curves to three days of daily melatonin: 0.5 mg versus 3.0 mg. J Clin Endocrinol Metab 95, 3325–3331, doi: 10.1210/jc.2009-2590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawashdeh O, Hudson RL, Stepien I & Dubocovich ML Circadian periods of sensitivity for ramelteon on the onset of running-wheel activity and the peak of suprachiasmatic nucleus neuronal firing rhythms in C3H/HeN mice. Chronobiol Int 28, 31–38, doi: 10.3109/07420528.2010.532894 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Reeth O et al. Comparative effects of a melatonin agonist on the circadian system in mice and Syrian hamsters. Brain Res 762, 185–194, doi: 10.1016/s0006-8993(97)00382-x (1997). [DOI] [PubMed] [Google Scholar]
- 39.Ersahin C, Masana MI & Dubocovich ML Constitutively active melatonin MT(1) receptors in male rat caudal arteries. Eur J Pharmacol 439, 171–172 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Soares JM Jr., Masana MI, Ersahin C & Dubocovich ML Functional melatonin receptors in rat ovaries at various stages of the estrous cycle. J Pharmacol Exp Ther 306, 694–702, doi: 10.1124/jpet.103.049916 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Lewy AJ et al. The human phase response curve (PRC) to melatonin is about 12 hours out of phase with the PRC to light. Chronobiol Int 15, 71–83 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Gillette MU & Mitchell JW Signaling in the suprachiasmatic nucleus: selectively responsive and integrative. Cell Tissue Res 309, 99–107, doi: 10.1007/s00441-002-0576-1 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Reid KJ et al. Familial advanced sleep phase syndrome. Arch Neurol 58, 1089–1094, doi: 10.1001/archneur.58.7.1089 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Kufareva I, Gustavsson M, Zheng Y, Stephens BS & Handel TM What Do Structures Tell Us About Chemokine Receptor Function and Antagonism? Annu Rev Biophys 46, 175–198, doi: 10.1146/annurev-biophys-051013-022942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooke RM, Brown AJ, Marshall FH & Mason JS Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug Discov Today 20, 1355–1364, doi: 10.1016/j.drudis.2015.08.003 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Lefkowitz RJ, Mullikin D & Caron MG Regulation of beta-adrenergic receptors by guanyl-5’-yl imidodiphosphate and other purine nucleotides. J Biol Chem 251, 4686–4692 (1976). [PubMed] [Google Scholar]
- 47.Word JM, Lovell SC, Richardson JS & Richardson DC Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285, 1735–1747, doi: 10.1006/jmbi.1998.2401 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Weiner SJ et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. Journal of the American Chemical Society 106, 765–784, doi: 10.1021/ja00315a051 (1984). [DOI] [Google Scholar]
- 49.Carlsson J et al. Structure-based discovery of A2A adenosine receptor ligands. J Med Chem 53, 3748–3755, doi: 10.1021/jm100240h (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher K & Sharp K Electrostatic contributions to heat capacity changes of DNA-ligand binding. Biophys J 75, 769–776, doi: 10.1016/S0006-3495(98)77566-6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mysinger MM & Shoichet BK Rapid context-dependent ligand desolvation in molecular docking. J Chem Inf Model 50, 1561–1573, doi: 10.1021/ci100214a (2010). [DOI] [PubMed] [Google Scholar]
- 52.Southan C et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 44, D1054–1068, doi: 10.1093/nar/gkv1037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolmachev A et al. Expanding Synthesizable Space of Disubstituted 1, 2, 4-Oxadiazoles. ACS combinatorial science 18, 616–624 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Kroeze WK et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369, doi: 10.1038/nsmb.3014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kenakin T, Watson C, Muniz-Medina V, Christopoulos A & Novick S A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci 3, 193–203, doi: 10.1021/cn200111m (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kenakin T Biased Receptor Signaling in Drug Discovery. Pharmacol Rev 71, 267–315, doi: 10.1124/pr.118.016790 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Longo PA, Kavran JM, Kim MS & Leahy DJ Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol 529, 227–240, doi: 10.1016/B978-0-12-418687-3.00018-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Besnard J et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220, doi: 10.1038/nature11691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popovska-Gorevski M, Dubocovich ML & Rajnarayanan RV Carbamate Insecticides Target Human Melatonin Receptors. Chem Res Toxicol 30, 574–582, doi: 10.1021/acs.chemrestox.6b00301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Y & Prusoff WH Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 61.Sumaya IC, Masana MI & Dubocovich ML The antidepressant-like effect of the melatonin receptor ligand luzindole in mice during forced swimming requires expression of MT2 but not MT1 melatonin receptors. J Pineal Res 39, 170–177, doi: 10.1111/j.1600-079X.2005.00233.x (2005). [DOI] [PubMed] [Google Scholar]
- 62.Dubocovich ML, Hudson RL, Sumaya IC, Masana MI & Manna E Effect of MT1 melatonin receptor deletion on melatonin-mediated phase shift of circadian rhythms in the C57BL/6 mouse. J Pineal Res 39, 113–120, doi: 10.1111/j.1600-079X.2005.00230.x (2005) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Probe pairs (two similar ligands with and without activity) of inverse agonists selective for MT1 and agonists selective for hMT2 are available by arrangement with Sigma (Extended Data Figure 3). The identities of the compounds docked in this study are freely available from the ZINC database, http://zinc15.docking.org, and active compounds may be purchased from Enamine. Figures with associated raw data include: Fig. 1, Extended Data Tables 1&2, Extended Data Figs. 1&2, Extended Data Table 1, for which further data are included in Supplementary Tables 1 (MT1 and MT2 affinities, MT1 DOCK energies/ranks) and 2 (compound purity information); Extended Data Fig. 3, for which bias information is included in Supplementary Table 3; Fig. 2, for which GPCRome screening, concentration-response curves, competition binding, and LC/MS data is included in Supplementary Data 1–5; Supplementary Data 6&7 (synthesis routes and spectra of compounds); Fig. 3, for which further data is included in Extended Data 4–5; Extended Data Fig. 7; Supplementary Table 4. Raw data values and transform data for in vitro cell based assays as well as in vivo data for phase shift and re-entrainment are available for Fig. 2; Extended Data Fig. 6; Fig. 3; Extended Data Fig. 4 (re-entrainment), Extended Data Fig. 5 (phase shift), Extended Data Fig. 7a–c.


