Abstract
Photosynthesis has become a major trait of interest for cereal yield improvement as breeders appear to have reached the theoretical genetic limit for harvest index, the mass of grain as a proportion of crop biomass. Yield improvements afforded by the adoption of green revolution dwarfing genes to wheat and rice are becoming exhausted, and improvements in biomass and radiation use efficiency are now sought in these crops. Exploring genetic diversity in photosynthesis is now possible using high-throughput techniques, and low-cost genotyping facilitates discovery of the genetic architecture underlying this variation. Photosynthetic traits have been shown to be highly heritable, and significant variation is present for these traits in available germplasm. This offers hope that breeding for improved photosynthesis and radiation use efficiency in cereal crops is tractable and a useful shorter term adjunct to genetic and genome engineering to boost yield potential.
Keywords: CO2 assimilation, electron transport, grain yield, radiation use efficiency, Rubisco
Photosynthetic performance is a key target for yield improvement in cereal crops. Evidence for genetic variation in photosynthesis is reviewed along with new approaches to measure these traits.
Introduction: supply, demand, and the challenges ahead
The challenges of sustainably supplying sufficient food to a burgeoning world population, predicted to exceed 9 billion by 2050, have been well documented (FAO 2009; Crist et al., 2017). Apart from the rise in global consumption, we are faced with diminishing arable land through degradation, changes in food preferences as living standards rise across Asia and Africa, and, importantly, the largely negative impacts of climate change and extreme weather events on agricultural production (Dawson et al., 2016). However, focusing on the needs of global agricultural production out to 2050 runs the risk of complacency in our research and crop breeding goals as it is likely that a major imbalance between supply and demand in the global cereal grain market, such as was seen in 2008, will occur long before 30 years have passed (FAO: The state of food insecurity in the world 2011; http://www.fao.org/3/i2330e/i2330e04.pdf). Indeed, global reserves of cereal grain are again low as a proportion of annual global demand and may be precipitously so (http://www.fao.org/worldfoodsituation/csdb/en/).
Recent climate modelling reported by the International Panel on Climate Change (IPCC) is now geographically fine grained enough to allow us to predict not only global increases in average temperature and the impact on agriculture, but also how this maps onto major cereal-producing regions globally (Elbehri et al., 2017; Arneth et al., 2019). Models also predict increases in the frequency of catastrophic weather events and how these will translate into crop losses through heat stress, drought, flood, frost, etc. (Arneth et al., 2019). The impact of climate change on wheat yields from 1990 to 2015 was recently modelled for Australia (Hochman et al., 2017) and found to have been responsible for a 27% decline in yield potential in rainfed environments, despite significant advances in yield resilience made by breeders during this period. If this effect accelerates and is exacerbated by extreme weather events, the current 1% annual improvement in wheat yield potential (Fischer et al., 2014) will not even keep pace with these detrimental effects over the next decade in dry, hot environments, not only in the developing world but also in our major cereal grain-exporting regions. A more than doubling of our annual yield progress is required in the case of all the globally important cereals, and the search for new ‘frontier traits’ to achieve this is now a major breeding focus (Furbank et al., 2019a).
The ‘green revolution’ and improving photosynthesis
The spectacular yield improvements seen in cereals in the 1960s and 1970s and the high annual increases in yield potential in wheat and rice achieved in the subsequent decades has recently declined (Parry et al., 2011; Furbank et al., 2019a). It is now widely accepted that the breeding strategies of the green revolution based on improvements in harvest index and grain number and largely driven by adoption of dwarfing genes (Fig. 1) have reached a plateau.
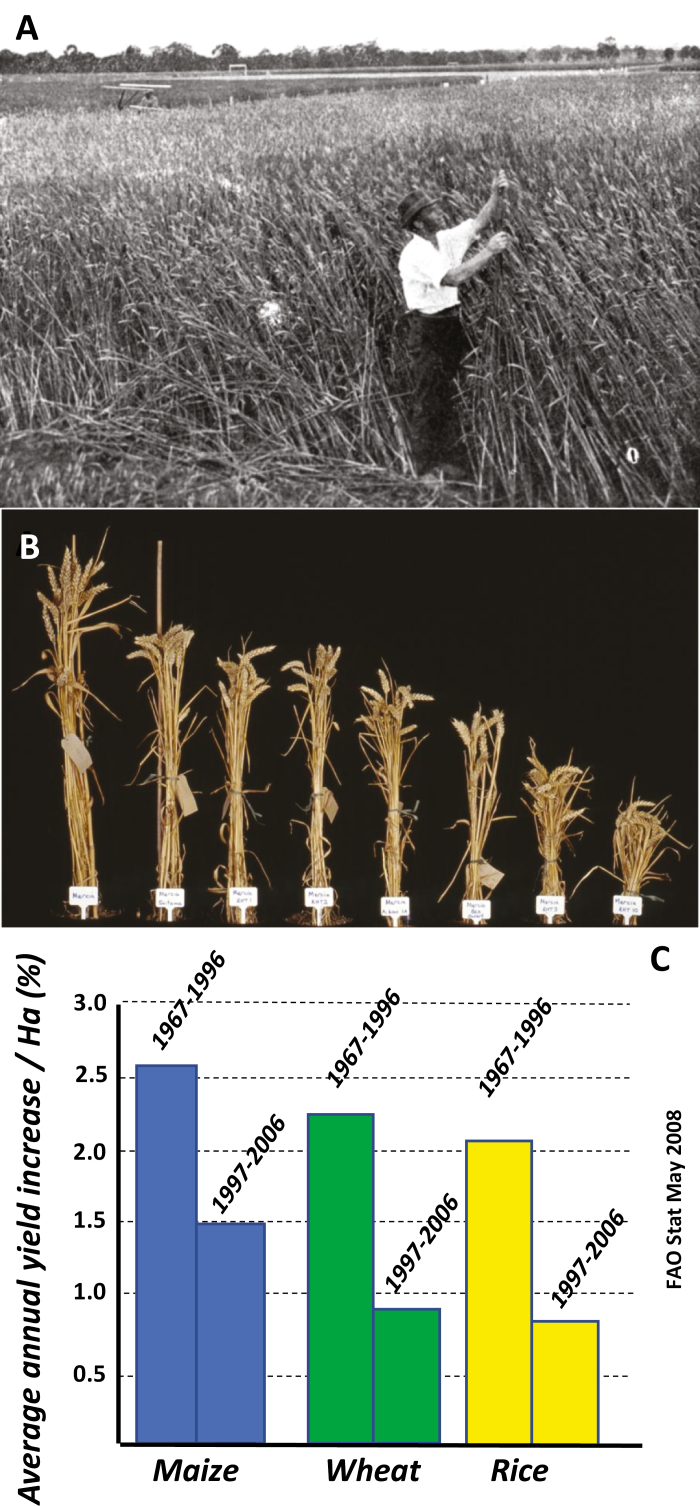
Fig. 1.
The Green Revolution and global cereal yields. (A) A farmer examining a pre-semi-dwarf wheat crop in 1915 (source: ‘Wheat growing in Australia’ McCarron, Bird & Co, Melbourne, Australia; 1915). (B) Effect of various dwarfing genes on plant stature in near isogenic lines of wheat (sourced from the John Innes Centre image archives). (C) Declining annual yield progress from breeding in the three major cereal crops prior to the 2008 food crisis (FAO, 2009).
Genetic potential for harvest index in many elite wheat and rice genotypes has reached the theoretical limit of ~0.6 (60% of the plant biomass is harvestable grain; Foulkes et al., 2011). Since the yield equation comprises only harvest index and biomass as factors, there has recently been a major focus on improving wheat and rice biomass without sacrificing harvest index (Parry et al., 2011; Furbank et al., 2019a), most readily achievable by improvements in radiation use efficiency (RUE) via increased photosynthetic capacity and efficiency. Since the 2008 food crisis, many hundreds of millions of dollars has now been invested in research to improve photosynthetic performance in model plants and crops, by both transgenic and non-transgenic approaches.
Candidate gene engineering approaches
Photosynthesis is one of the most intensively studied biochemical processes in plants. Decades of biochemistry and biophysics of mechanisms and processes have been followed by gene suppression work in transgenic plants to ‘titrate’ out levels of key enzymes and determine the limitations to photosynthetic flux afforded by these steps in C3 and C4 photosynthesis (e.g. Hudson et al., 1992; Furbank et al., 1996; Price et al., 1998; Harrison et al., 2001; von Caemmerer et al., 2005). Modelling has been used extensively to elucidate these limitations to flux under a range of environmental conditions (von Caemmerer, 2000). The contribution of the leaf-level models derived from Farquhar et al. (1980) in guiding these experiments has been invaluable in providing a quantitative, mathematical lens with which to examine impacts of gene expression, enzyme activity, and protein level on leaf photosynthetic physiology. Together with systems models (Zhu et al., 2007) and incorporated into field crop simulations (Wu et al., 2016, 2019; Yin et al., 2017), modelling continues to guide our strategies for candidate gene selection for manipulation and transgenic plant analysis. Targets for engineering-based approaches have included Rubisco improvement by directed evolution (Wilson et al., 2018), increased expression of Rubisco (Salesse-Smith et al., 2018), reduction of photorespiration by CO2-concentrating mechanisms (reviewed in von Caemmerer et al., 2012; Long et al., 2016) and photorespiratory bypasses (South et al., 2019; Shen et al., 2019), overexpression of enzymes important in regeneration of ribulose-1,5-bisphosphate (RuBP) such as sedoheptulose-bisphosphatase (Lefebvre et al., 2005; Driever et al., 2017), modification of photoprotection (Kromdijk et al., 2016), and overexpression of the thylakoid cytochrome b6f complex for ATP production (Simkin et al., 2017; Ermakova et al., 2019a). Modification of mesophyll conductance and stomatal conductance to provide better access of CO2 to Rubisco has also been explored as an engineering target using proposed CO2 porins (reviewed in Groszmann et al., 2017; Condon, 2020).
All these approaches have also been recently reviewed (Simkin et al., 2019) and are the focus of major national and international consortium efforts (e.g. ARC Centre of Excellence for Translational Photosynthesis, https://photosynthesis.org.au/; C4 Rice Project, https://c4rice.com/; and Realising Improvements in Photosynthetic Efficiency or RIPE, https://ripe.illinois.edu/). However, while modelling predicts that these modifications alone and in combination could have quantum effects on photosynthetic performance in crops, as yet only a few of these genetically modified traits have been tested in the field in cereals (e.g. Shen et al., 2019) or indeed in any crops apart from tobacco. Recently, interpretation of phenotypes and field performance of transgenic tobacco has also been controversial (Evans, 2019; Fischer et al., 2019). Given the challenges of field translation of genetically modified photosynthetic traits, is there significant genetic variation in photosynthesis in our existing cereal germplasm collections to breed for large increases in biomass and yield? Are these traits heritable and stable and what are the genetics underpinning such variation?
Genome to phenome: mining allelic variation
The plummeting costs of genome and transcriptome sequencing means that burgeoning collections of cereal germplasm can be cheaply genotyped, and in many cases high-quality genome re-sequence data are available (Wang et al., 2018; Langridge and Waugh, 2019; Milner et al., 2019). Coupled with the high level of domain knowledge available to identify candidate genes in photosynthetic improvement, this provides an opportunity to mine allelic variation in photosynthetic genes (Furbank et al., 2019a, b; Fig. 2) and determine their importance. Several initiatives have been established across the globe in wheat, rice, and barley to capture the genetic diversity of both current and historic/heirloom varieties and wild relatives or genome donors in the case of wheat. In wheat, most notable of these are the CIMMYT activities under the Seeds of Discovery program (https://seedsofdiscovery.org/) and the efforts of the Leibniz Institute for Crop Breeding (IPK) to carry out Illumina sequencing of a large proportion of their barley and wheat germplasm collection (https://bridge.ipk-gatersleben.de).
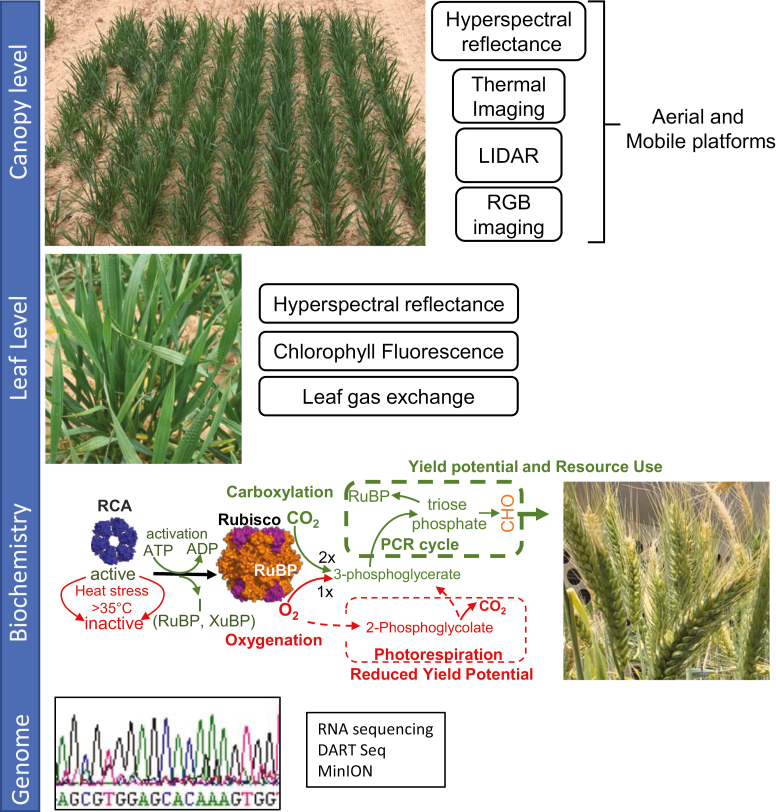
Fig. 2.
Genome to phenome and back: identification of photosynthetic traits for integration into breeding programmes or gene technologies. Analysis of photosynthetic CO2 assimilation from the canopy and leaf level can be achieved through rapid phenotyping techniques (see Furbank et al., 2019a, b). These techniques enable rapid determination of photosynthetic parameters that help select germplasm for detailed analyses. At the canopy level, LIDAR is used for non-destructive biomass determination, drones or unmanned aerial vehicles are used for imaging crop canopies which can include RGB cameras for crops coverage, and thermal imaging is used for canopy temperature, which can be utilized for screening germplasm for differences in water use efficiency. At the leaf level, tools such as hyperspectral reflectance can be used to estimate electron transport capacity and Vcmax, in addition to leaf N and leaf mass per area (Silva-Perez et al., 2018). Tools such as MultispecQ (Kuhlgert et al., 2016) and SPAD provide surrogates for leaf N content, with the former measuring electron transport and non-photochemical dissipation of incoming light energy. Determining the underpinning biochemistry and gene sequence diversity is requisite to deploy traits crucial for improving CO2 assimilation.
In rice, the International Rice Research Institute (IRRI) have developed a large sequenced diversity panel of indica and japonica rice genotypes with diverse pedigrees and geographic origins: the 3K population, currently being extended to include 10 000 entries (Wang et al., 2018; http://snp-seek.irri.org/). Maize germplasm collections and genotyping data are perhaps the best developed and have been utilized for many years (reviewed in Romay, 2018). In sorghum, the first C4 grass to undergo full genome assembly, genetic resources are also building, with genetic material dating from pre-domestication to current elite lines (Mace et al., 2013; Boyles et al., 2019).
While the genetic resources now exist in a variety of crops to examine allelic variation in all candidate genes/proteins known to be important in controlling photosynthetic flux, Rubisco has been the focus of most efforts to date.
Exploring the Triticeae tribe for variation in Rubisco catalysis
CO2 assimilation within key C3 crops is often limited by the catalytic activity of Rubisco (von Caemmerer, 2000; reviewed in Sharwood, 2017). Rubisco is a bifunctional enzyme that can either fix substrate CO2 or oxygen to substrate RuBP (Sharwood, 2017). Carboxylation of RuBP is the productive reaction of Rubisco that results in formation of 3-phosphoglycerate that is used for the synthesis of carbohydrate backbones, which are then utilized for plant growth and productive yield, whereas oxygenation is unfavourable because of the production of 2-phosphoglycolate that must be recycled through the photorespiratory pathway (Bauwe et al., 2010). This process consumes energy and releases previously fixed CO2 (Sharkey, 1988). Therefore, Rubisco is regarded as an inefficient catalyst, with a catalytic cycle of 2–3 carboxylations per second, a meagre catalytic efficiency, and poor specificity for CO2 (Sharwood, 2017). In addition, Rubisco requires the assistance of Rubisco activase (RCA) that is required to maintain and modulate enzymatic activity through metabolic repair of the activity that removes inhibitory sugar phosphates (Mueller-Cajar, 2017). These include the misfiring product xylulose bisphosphate (XUBP), the nocturnal inhibitor carboxyarabinitol-1-phosphate (CA1P), and substrate (Parry et al., 2008). Surprisingly, RCA is thermolabile within C3 crops, with activity dropping significantly as temperatures exceed 35 °C (Crafts-Brandner and Salvucci, 2000). To circumvent Rubisco catalytic inefficiencies, terrestrial plants devote significant amounts of leaf nitrogen (N) to synthesize large amounts of Rubisco to ensure appropriate CO2 assimilation. However, this has burdened agriculture with the high use of N fertilizers to ensure sufficient Rubisco is synthesized for assimilating carbon for growth and yield. Therefore, Rubisco and RCA are two primary targets for improving CO2 assimilation by ameliorating catalysis to improve both photosynthetic N use efficiency and photosynthetic water use efficiency.
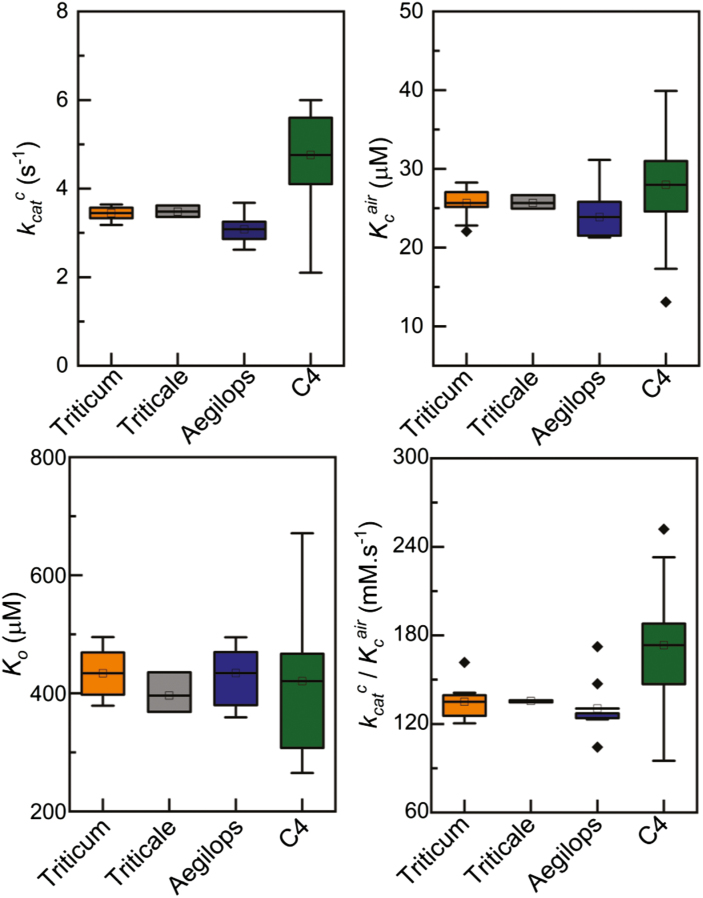
Screens of in vitro Rubisco catalytic parameters among terrestrial plants (C3 and C4) and algae have revealed substantial diversity in Rubisco catalysis and identified versions better suited to current and predicted future climate scenarios (Galmés et al., 2014; Orr et al., 2016; Sharwood et al., 2016a; Young et al., 2016; Heureux et al., 2017). Three catalytic parameters are required for modelling to assess performance. These are: (i) the Michaelis constant for CO2 in air (Kcair); (ii) carboxylation speed (kcatc); and (iii) the specificity for CO2 as opposed to O2 (Sc/o) (Sharwood, 2017). Until recently, little information was known about the diversity of Rubisco catalysis within the Triticeae tribe. Analysis of 25 species with the tribe demonstrated diversity of Rubisco catalysis, with variation observed in kcatc, Kcair, and Ko (the Michaelis constant for oxygen) (Fig. 3A–C; (Prins et al., 2016). Interestingly, Triticum species showed improved carboxylation efficiency at 21% O2 compared with Aegilops relatives (Fig. 3D; Prins et al. (2016). Understanding the source of this change, probably variation in the sequence of Rubisco small subunits, will provide key information to further improve wheat Rubisco catalysis.
Fig. 3.
Exploring Rubisco catalytic diversity within Triticeae and C4 plants. Variation in key Rubisco catalytic parameters from Triticeae and compared with plant C4 Rubisco. Parameters include the carboxylation speed, kcatc, the Michaelis constants for CO2 and O2 and carboxylation efficiency (kcatc/Kcair). Rubisco catalytic data for the Triticeae tribe were replotted from Prins et al. (2016) into box plots alongside C4 Rubisco. Kcair was calculated for Triticeae Rubisco using the formula Kcair (μM)=Kc(1+O/Ko), where Kc and Ko are the Michaelis constants for CO2 and oxygen, respectively, and O is the atmospheric O2 concentration—252 μM. The C4 Rubisco parameters were replotted from Jordan and Ogren (1983); Kubien et al. ( 2008); Whitney et al. (2011); and Sharwood et al. (2016a, b). In each box plot, the black square and the horizontal bar indicate the mean. The lower and upper edge of each box indicate the interquartile (25–75%) range of the values reported. The whiskers extend to 1.5 times the interquartile range.
Further exploration of the Triticeae tribe is required to fully assess diversity in Rubisco catalysis. Identifying catalytic switches in the Rubisco large and small subunits will open up new opportunities for improving wheat CO2 assimilation. However, it is evident in Fig. 3 that C4 Rubisco provides solutions to improve the current wheat forms. The kcatc and carboxylation efficiency (kcatc/Kcair) of C4 Rubisco outperform those of wheat. Comparison of Rubisco specificity is difficult as Prins et al. (2016) used a different technique for measuring Sc/o from that for C4 plants presented. Nevertheless, it is evident that maize Rubisco provides improved CO2 assimilation when compared at 25 °C (Sharwood et al., 2016b).
While substantial catalytic diversity exists within the Rubisco superfamily of enzymes, more interrogation of catalytic diversity is required. Efforts to include Rubisco as a breeding target have been limited due to the labour-intensive measurements of Rubisco catalysis. High-throughput surrogates for Rubisco capacity and catalytic properties measured on intact leaves offer hope of tractable screening methods, and these are discussed in the following sections.
Genetic diversity in cereal crop photosynthesis
High-throughput direct measurement of carbon assimilation presents a large technical challenge, even using so-called ‘rapid’ gas analysis techniques (e.g. Stinziano et al., 2017). Exploring genetic diversity for photosynthetic traits in germplasm collections and large structured mapping populations of cereals has long been considered a laborious and almost an intractable task (Parry et al., 2011; Furbank et al., 2019a, b). Single point measurements of assimilation rate on flag leaves of spring and winter wheat have been seen to have both a positive and negative correlation with yield and year of release (Murthy and Singh, 1979; Evans, 1993; Reynolds et al., 1994, 2000; Fischer et al., 1998; Fischer et al., 2010; Sadras et al., 2012; Gaju et al., 2016; Tang et al., 2017). There is of course the complication in such studies of which leaf and at which developmental stage to measure and how to compare germplasm with vastly different phenology (Parry et al., 2011). Obtaining gas exchange data in the field is slow, with stomatal limitation often complicating the measurement (Condon et al., 2004; Feng et al., 2018). While even more time-consuming than assimilation measured at ambient CO2, CO2 response curves or ‘A versus Ci’ curves potentially provide a better estimate of photosynthetic traits as they allow extraction of estimates of Rubisco amount and kinetic efficiency (Vcmax) and electron transport capacity (J) using the modelling frameworks discussed above (Farquhar et al., 1980; Silva-Perez et al., 2018, 2019). Given the time constraints for measurement, a limited number of studies have been done with diversity panels of wheat using gas exchange, with few of these done in the field or incorporating leaf structural traits.
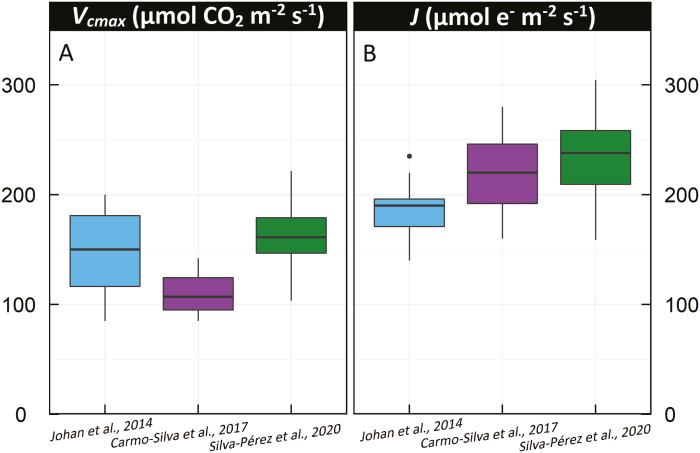
Figure 4 summarizes the most recent of these published data (Driever et al., 2014; Jahan et al., 2014; Carmo-Silva et al., 2017; Silva-Perez et al., 2018, 2019). Across elite germplasm (including historic germplasm sets), there appears to be substantial genetic variation in the modelled parameters reflecting Rubisco capacity (Vcmax) and photosynthetic electron transport (J) (Fig. 4). In the larger data sets of Silva-Pérez et al. (2020) and Carmo-Silva et al. (2017), genetic variation in J appeared to be larger than in Vcmax, but this was not the case in the smaller germplasm set of Jahan et al. (2014). Heritability of these gas exchange-derived traits can be quite high (broad-sense heritabilities of 0.31–0.76 have been reported for A in winter wheat grown in the UK; Carmo-Silva et al., 2017) and for spring wheats grown in Mexico and Australia, heritability of modelled parameters Vcmax and J were also as high as 0.7 (Silva-Pérez et al., 2020). These data suggest that photosynthetic traits are genetically robust enough to breed with and that substantial variation is present in elite material and potentially even greater variation in landraces and wild relatives.
Fig. 4.
Genetic variation in wheat for (A) Rubisco activity (Vcmax) and (B) electron transport rate (J). Vcmax and J for 11 wheat genotypes measured in young plants grown in a controlled-environment growth cabinet (Jahan et al., 2014), 64 winter wheat genotypes grown in the field in the UK (Carmo-Silva et al., 2017), and 74 spring wheat genotypes measured in a glasshouse (only high N treatment shown) and field in Australia and Mexico (Silva-Perez et al., 2018, 2020).
In rice, several studies have been carried out on diverse germplasm to investigate genetic variation in A from gas exchange. In a collection of 20 diverse japonica and indica genotypes, light-saturated A at ambient CO2 concentration varied by >40% from lowest to highest genotypes (Jahn et al., 2011). However, low heritability of A observed in this study (0.17) is indicative of the challenges in using a single point assimilate rate to find the genetic architecture underlying photosynthetic traits.
High-throughput phenomics: accelerating germplasm screening from organs to canopy
Hyperspectral reflectance
As discussed above, high-throughput surrogates for photosynthetic traits traditionally derived from gas exchange are a priority because: (i) the large scale of experiments necessary to screen germplasm diversity sets and mapping populations proves too costly; (ii) time of day and seasonal variation affect trait expression, compressing time available for measurements; and (iii) measurements may need to be made on several leaves/plant organs at different developmental stages in the crop life cycle to obtain a comprehensive analysis. To obtain accurate mapping of photosynthetic traits in a structured population, such as a recombinant inbred set with high-density genetic maps usually requires in excess of 150 lines grown with replication, in the field, preferably across multiple seasons and often multiple locations (Collard et al., 2005). Given that leaf-level measurements can require between two and six replicates per field plot, even at a single developmental stage, this would mean that with 3-fold replication at both the genotype and technical level. a minimum of 1350 individual measurements must be made. If multiple leaf classes at several stages of development are added to the experiment, the phenotyping rapidly becomes unachievable in a reasonable amount of time, particularly if time of day effects are to be avoided. For genome-wide association panels, the size of the experiment may increase to 1000 germplasm entries if gene-level resolution is desired (Ingvarsson and Street, 2011), exacerbating the problem.
Recent advances in machine learning coupled with optical sensing systems at the leaf and canopy level offer a potential solution to this issue. Leaf and canopy spectral reflectance measurements can now be made using affordable visible/near infrared spectrometers or more costly full range visible, NIR, SWIR spectrometers, enabling the collection of many hundreds of wavelengths from below 400 nm to 2500 nm. Predictions of leaf traits from spectral reflectance data are made using machine learning to generate statistical models between every wavelength of reflected light from leaves and the trait of interest measured with traditional methods (termed a ‘training set’). These models are then either validated on another set of germplasm or the training set is divided into a training and validation set for testing (Silva-Perez et al., 2018).
This approach has resulted in prediction of leaf traits related to photosynthesis such as leaf N, phosphorus, mass per area, and photosynthetic traits such as A, J, and Vcmax in plants ranging from trees to annual C3 and C4 crops (e.g. Serbin et al., 2012; Ainsworth et al., 2014; Singh et al., 2015; Yendrek et al., 2017; Silva-Perez et al., 2018). This technique has recently been extended to predictions of respiration rate in wheat (Coast et al., 2019) and field evaluation of transgenic plants with altered photosynthesis (Meacham-Hensold et al., 2019). The attraction of hyperspectral reflectance models is that the collection of spectra can take <20 s per leaf and does not require any equilibration of the leaf in the sensor. There are, however, obstacles to the widespread use of a machine learning to examine genetic variation in crop photosynthetic traits as acquiring the data necessary to build a training set requires many hundreds or even thousands of measurements using the older, slower traditional methods. If training sets are not sufficiently large, containing a wide range of germplasm and even different leaf classes/developmental stages and environments, spurious ‘overfitting’ occurs and the predictive power of models is diminished for leaves which fall outside the panel used to generate the model (see Heckmann et al., 2017; Coast et al., 2019).
Chlorophyll fluorescence
While direct measurement of carbon fixation in high throughput is problematic, estimation of photosynthetic electron transport capacity and related leaf-level efficiencies can be tractable in high throughput using chlorophyll fluorescence techniques without the use of models or proxies (reviewed in Maxwell and Johnson, 2000; Murchie and Lawson, 2013). Compact, commercial pulse amplitude-modulated chlorophyll fluorescence (PAM) systems are available (Cessna et al., 2010; Kuhlgert et al., 2016) which apply a brief saturating flash of light via a high-intensity LED, in the light or dark, allowing calculation of either the photosynthetic electron transport rate (ETR or +) or the intrinsic light-harvesting efficiency of PSII (dark-adapted Fv/Fm). NPQ (non-photochemical quenching, resulting primarily from dissipation of energy as heat) can also be calculated (Maxwell and Johnson, 2000).
Deploying PAM at canopy level in a field crop presents difficulties in both obtaining a uniform saturating flash and in interpreting data from the complex 3D structure of the canopy. For canopy-level measurements, non-imaging fluorescence sensors or light-induced fluorescence transients (LIFTs) which apply a series of ‘flashlets’ to a spot of the canopy up to a few centimetres wide may be useful for field phenotyping (Keller et al., 2019), or imaging sensors for sun-induced fluorescence (SIF) mounted on drones, manned aircraft, or even satellites may be used to generate high-throughput phenotyping data (Zhang et al., 2018; Camino et al., 2019; Furbank et al., 2019b).
Aerial and satellite remote sensing using chlorophyll fluorescence has so far been utilized mainly to study natural vegetation and ecosystems or for horticulture with tree species (Furbank et al., 2019b) and is only recently being used for exploring photosynthetic variation in crops (Camino et al., 2019).
In contrast, leaf-level chlorophyll fluorescence has been used to explore diversity in chloroplast electron transport and, to a limited degree, the genetics behind this variation
Wheat cultivars more tolerant to heat stress have been identified based on higher Fv/Fm from dark-adapted, detached leaves from a diversity panel of 41 lines of different origins (Sharma et al., 2015). Although commercial ‘Imaging PAM’ systems and hand-held devices are available for the estimation of chlorophyll-based parameters, they have been costly and have had limited application to high-throughput field phenotyping.
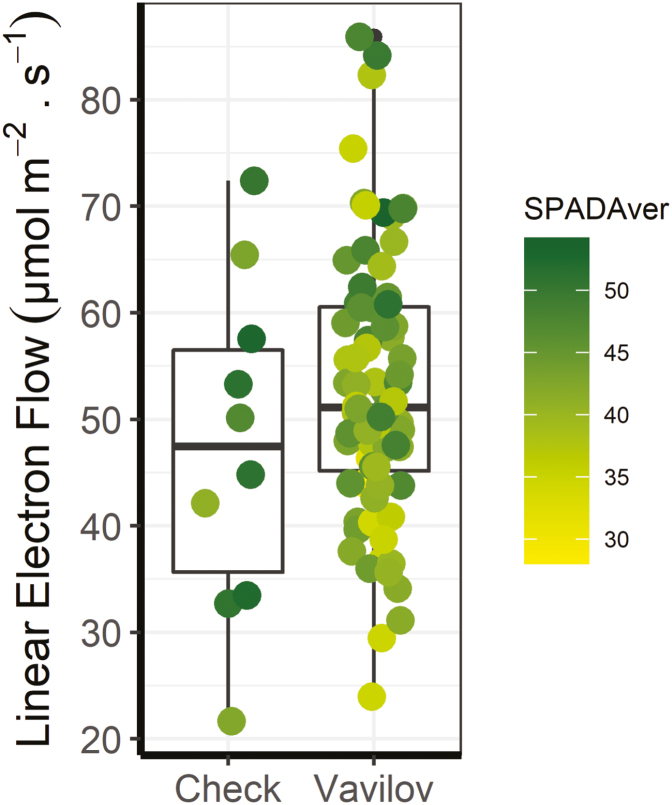
The development of new microelectronics and sensors has led to the design of hand-held instruments amenable for larger screenings. For example, the MultisepQ sensor can rapidly measure several fluorescence-based photosynthetic parameters and other physical leaf traits in field situations at a low cost (Kuhlgert et al., 2016), such as SPAD, linear electron flow (LEF), and PSII quantum yields (Kuhlgert et al., 2016). The MultispeQ sensor has been used successfully to assess the effects of abiotic stresses (i.e. heat and drought) in Phaseolus (Traub et al., 2018) and cowpea (Mwale et al., 2017), and characterization of photosynthetic traits in potato (Prinzenberg et al., 2018) and transgenic tobacco plants with alternative electron transport (Gómez et al., 2018). This system also features an open platform for metadata annotation, robust data acquisition, and easy sharing of information.
To investigate the utility of the MultispeQ sensor in a cereal crop, we screened a subset of the pre-breeding ‘Vavilov’ wheat population representing the genetic diversity of the panel (Riaz et al., 2017) and 10 commercial wheat varieties. A 4-fold difference between the highest and lowest LEF values was detected (GME et al., unpublished). Both Vavilov and commercial checks spread over a similar range of LEF, between 20 µmol e m–2 s–1 and 80 µmol e m–2 s–1. The mean LEF for Vavilov was slightly higher and there were few lines with higher LEF than commercial wheats. In addition, concurrent measurements of SPAD with the same sensor demonstrated a dynamic response of LEF to N content mostly in the Vavilov lines, with commercial varieties presenting higher N. High-throughput screening for leaf photosynthetic-related traits using sensors such as MultispeQ can be useful to identify new diversity in photosynthetic capacity and efficiency in breeding programmes or during surveys of diversity panels at relatively low cost.
Photosynthesis by wheat spikes can provide up to 25% of total grain carbohydrate during grain filling, and this contribution could be higher under stressful conditions. It is anticipated that genetic variability for the spike photosynthesis trait exists, but phenotyping this photosynthetic capacity in this organ is challenging due to their complex structure and to the unknown capacity for re-fixation of carbon respired from the developing grain. The latter makes traditional gas exchange difficult to interpret because it measures net CO2 uptake by the spike. Moreover, and although gas exchange chambers for 3D organs such as spikes may be custom-built (Sanchez-Bragado et al., 2014; Fortineau and Bancal, 2018), screening of individual wheat spike photosynthesis by gas exchange in the field is not practical for high-throughput phenotyping
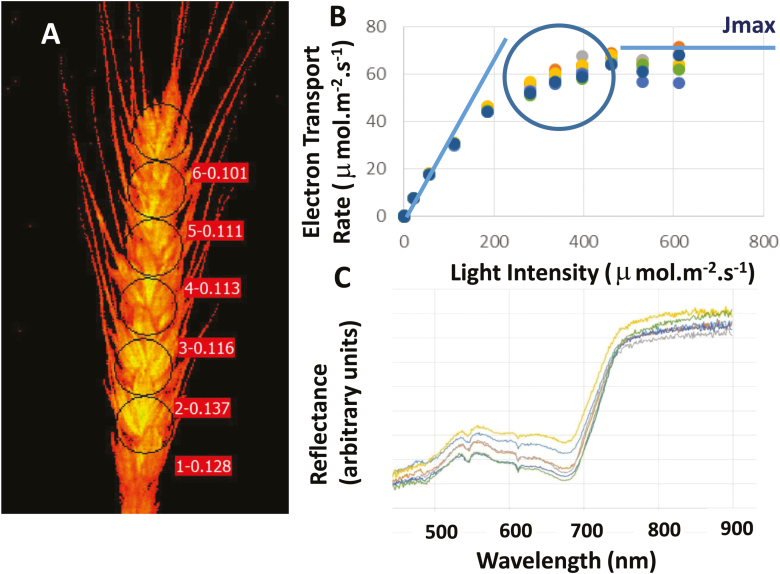
Imaging of chlorophyll fluorescence using PAM of intact or detached ears offers an alternative to estimate photosynthetic capacity of wheat spikes. The contribution of photosynthesis and other energy dissipation processes to ETR (a surrogate for photosynthetic capacity) can be monitored by measuring the quenching of chlorophyll fluorescence over a range of light intensities. A mathematical model (von Caemmerer, 2000) can then be fitted to the ‘light response curve’ of ETR to calculate Φ, the efficiency of light capture; θ, qualitative ‘curvature factor’; and Jmax, maximum potential electron transport rate. Jmax is widely used as a measure of leaf ‘light use capacity’ analogous to the modelled parameter Vcmax as a metric of ‘Rubisco capacity’ (Fig. 6).
Fig. 6.
Hyperspectral reflectance imaging and machine learning for predicting photosynthetic traits in wheat spikes. (A) PAM maximum fluorescence image of a wheat spike showing six regions of interest. The three parameters which can be extracted from a model of the response of ETR to light intensity are: Φ, efficiency of light capture; Jmax, the electron transport ‘capacity’; and θ, a qualitative ‘curvature factor’. ETR is calculated from chlorophyll fluorescence images. (B) Typical light–response curve for individual regions of interest. (C) Average reflectance spectra from hyperspectral imaging for the six regions in (A) and (B).
Fig. 5.
Rapid screening of linear electron flow (LEF) in wheat using chlorophyll fluorescence. Measurements of LEF were taken with three different MultispeQ sensors (Kuhlgert et al., 2016) between 09.00 h and 14.00 h on the same, youngest, fully expanded leaf on a subset (76 lines) of the Vavilov collection and 10 commercial wheat varieties. The box plot shows the spread of LEF values in both sets while the individual points represent the averages for individual lines coloured by the average of the SPAD values simultaneously measured with MultispeQ. In each box plot, the black square and the horizontal bar indicate the mean. The lower and upper edge of each box indicate the interquartile (25–75%) range of the values reported. The whiskers extend to 1.5 times the interquartile range.
However, this technique based on a commercially available PAM fluorometer is not scalable to proximal remote-sensing buggies or to drones, and thus not applicable to canopy-based measurements in the field. Alternatively, hyperspectral imaging sensors mounted on movable platforms would be scalable to field-based application. Machine learning algorithms can deconvolute the hyperspectral signal from wheat leaves to estimate photosynthetic parameters obtained by gas exchange measurements (e.g. Silva-Perez et al., 2018). Similarly, training a statistical model for hyperspectral prediction of wheat spike ETR derived from PAM could be possible. For example, the light response curve of measured ETR with Imaging PAM could be used as a training set and combined with reflectance spectra from hyperspectral imaging of spikes to build predictive models for all key chlorophyll fluorescence parameters (Jmax, θ, and Φ) using statistical approaches such as partial least squares regression, random forest, a support vector machine, or neural networks. The combination of hyperspectral imaging and existing methods to detect spikes using image analyses with deep learning (Hasan et al., 2018) or neural networks (Qiongyan et al., 2017) could be instrumental in identifying genetic diversity in wheat spike photosynthesis.
Phenome to genome: QTLs and the genetic architecture of photosynthesis traits
Establishing allelic variation in genes known to encode important proteins in photosynthesis (genome to phenome) relies upon pre-existing knowledge of these candidates (Fig. 2). Using high-throughput phenomics to generate trait associations with genomic regions can identify completely unknown genes affecting photosynthetic performance. Understanding the genetic architecture of photosynthetic performance can identify quantitative trait loci (QTLs) or markers which are useful breeding tools for marker-assisted selection or, if fine grain genetic maps and re-sequence data are available, single nucleotide polymorphisms (SNPs) can be identified in candidate genes which can be used to understand genetic and biochemical mechanisms and drive gene editing and transgenic approaches.
Despite promising data on genetic diversity and heritability of photosynthetic traits, there are surprisingly few examples of QTL mapping of these parameters in wheat, given its importance globally. There have, however, been several studies targeted to photosynthesis-related traits such as stress tolerance, canopy temperature, stomatal conductance, and transpiration efficiency (e.g. Mason et al., 2013; Rebetzke et al., 2013). Recently, Molero et al. (2019) explored the genetic basis of biomass accumulation and RUE in wheat by a genome-wide association study (GWAS). A panel of 150 elite spring wheat genotypes including many landrace and synthetically derived lines were examined using more traditional approaches such as measuring yield components and biomass accumulation over time combined with estimated intercepted radiation. Marker–trait association identified 94 SNPs significantly associated with yield, agronomic, and phenology-related traits along with RUE and final biomass (BM_PM) at various growth stages that explained 7–17% of phenotypic variation. Common SNP markers were identified for grain yield, BM_PM, and RUE on chromosomes 5A and 7A. While the density of the genetic map was not sufficient to fine-map and identify single candidate genes, several QTLs encompassed genes involved in processes associated with photosynthesis such as reactive oxygen detoxification and photoprotection of PSII.
In rice, several QTL studies have been carried out mapping measurements of A with QTLs found on chromosomes 3, 4, 5, 6, 8, and 11 (Teng et al., 2004; Adachi et al., 2011, 2019; Gu et al., 2012). Many of these QTLs account for only a small proportion of variation or are dependent on genetic background (Adachi et al., 2019). Consequently, only a small number of these loci have been fine-mapped and causative genic SNPs identified which underpin genetic variation for A in rice. Most notably, GREEN FOR PHOTOSYNTHESIS, originally thought to be associated with Rubisco carboxylation efficiency, has now found to be a determinant of leaf thickness, chlorophyll content, and canopy chlorophyll distribution (Takai et al., 2013; Hirotsu et al., 2017), and Car8, a transcription factor affecting photosynthetic capacity (Adachi et al., 2017), is also involved in control of flowering time and duration. The complex interactions between flowering time, phenology, tillering, N partitioning, and photosynthetic traits add to the difficulty of finding robust trait associations, and sophisticated statistical treatments such as deep learning (Zou et al., 2019) may be necessary to tease apart these confounding factors.
Photosynthetic capacity and leaf nitrogen content
Silva-Pérez et al. (2020), in a study exploring genetic diversity in wheat for photosynthetic traits, introduce the concept of photosynthetic capacity (Pc) and photosynthetic efficiency (Peff, i.e. Vcmax, or J per unit leaf N) to describe the drivers of variation in modelled leaf-level traits. It has been widely reported that in cereal crops Rubisco amount and activity are closely related to N nutrition and leaf N content per unit area (Evans, 1983, 1989; reviewed in Evans and Clarke, 2019). It has also been reported that a reduction in flag leaf size, associated with the introduction of the Rht dwarfing genes to green revolution wheats, increased N and photosynthetic capacity on a leaf area basis in subsequent varieties (Bishop and Bugbee, 1998). Leaf N content is also important in cereals, as a large proportion of grain protein is derived from leaf protein remobilized during senescence (see Evans and Clarke, 2019). There is concern that improvements in photosynthetic capacity may require increased agronomic N fertilizer use over and above that already required to realize the gains of the green revolution genotypes (Parry et al., 2011).
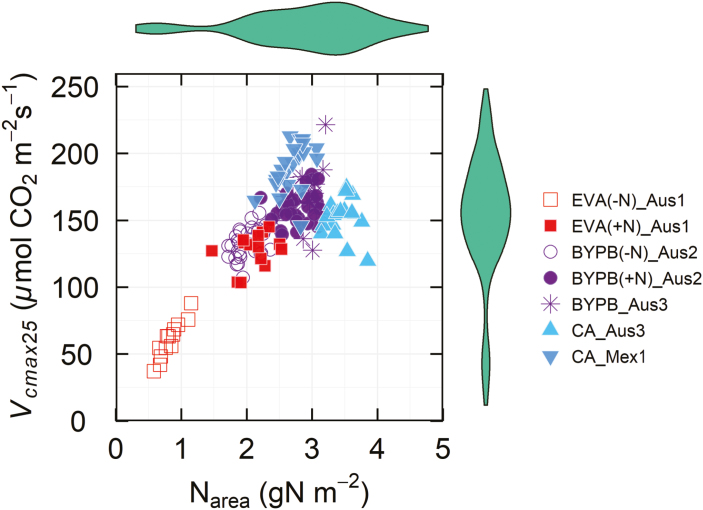
Figure 7 shows the relationship between leaf N and Vcmax on an area basis in diverse wheat genotypes (data from Silva-Pérez et al., 2020). Clearly, the amount and activity of Rubisco are related to leaf N, particularly when N supply is restricted (open squares in Fig. 3), but, at higher leaf N values more relevant to agronomic N application levels, a wide range of Vcmax values were obtained across the genotypes tested. Similar results are also found for J (Silva-Pérez et al., 2020). Given the high heritability of these modelled parameters in wheat (Silva-Pérez et al., 2020 and references therein), either the catalytic efficiency of Rubisco or the electron transport efficiency is superior in some wheat genotypes or the partitioning of leaf N to these protein components in leaves is different between genotypes. These two options cannot be separated with the data available but, clearly, photosynthetic capacity per unit leaf N (Peff) has a strong genetic component and there is considerable variation in wheat. Given the robust models available to predict these parameters from leaf hyperspectral reflectance data (Silva-Perez et al., 2018), it should be possible to screen large diversity sets and mapping populations to understand and exploit the underlying genetic control, and this work is underway in the IWYP consortium (see https://iwyp.org/funded-projects/).
Fig. 7.
Diversity of Rubisco content (Vcmax25) per unit of nitrogen per leaf area in 74 spring wheat genotypes measured in different environments and growing stages. EV and BYP are two different collections of wheat genotypes grown in glasshouse conditions with two nitrogen levels (–N, +N). BYP and CA are two different wheat collections of wheat genotypes grown in the field in Australia (Aus3) and in Mexico (Mex1). See Silva-Pérez et al., 2020) for experimental details. In the x and y margins, a violin plot is shown representing the distribution of Narea and Vcmax, respectively. EV, early vigour set; BYP, high yielding set of wheat genotypes in Australia. (C) High yielding set of wheat genotypes from CIMMYT measured after anthesis. A and B designations at the end of the genotype acronyms refer to the plant growing stages at the time of the measurements, A, after anthesis, B, before anthesis.
Source/sink regulation: feedback or feedforward?
It has frequently been pointed out that improvements in photosynthetic performance will only be translated into improved grain yield if ‘partitioning’ between source and sink and associated signalling processes are accounted for (reviewed in Reynolds et al., 2012). Indeed, it has long been a point of conjecture among crop physiologists as to whether cereal yields are source or sink limited (Parry et al., 2011; Reynolds et al., 2012). Experiments with free air CO2 enrichment of a variety of crop species, while often showing increases in yield due to elevated photosynthetic activity, also frequently show sink strength-mediated feedback limitation and limited impact on yield, thought to be due to negative effects of sugar signalling on photosynthetic gene expression, particularly Rubisco and leaf N levels (Ainsworth and Long, 2005). Various reports over the last 20 years have indicated strong feedback links between sink and source capacity, but the mechanisms for this signalling are complex (Wingler, 2018; Paul et al., 2018). While sucrose itself is something of a weak signal for sugar feedback on leaf processes, recent evidence suggests that the signalling metabolite trehalose-6-phosphate (T6P) may perform this role and also affect phloem transport processes via regulation of the SWEET membrane transporters (Paul et al., 2018).
While sugar feedback could be an impediment to realization of increased yield from photosynthetic improvement, there is also evidence of feedforward effects of photosynthetic performance on sink development. In wheat, the final size of the ear and floret number is determined early in development when the inflorescence is still within the sheath (Reynolds et al., 2009). Supply of photoassimilate at this stage and at flowering can strongly influence floret number, floret fertility, and final grain number (Fischer, 1985; Reynolds et al., 2005; Broberg et al., 2019). The coordination between sink and source capacity at this developmental stage may be why it is difficult to separate the contribution of photosynthesis from sink strength when examining the basis for historic yield improvement (see Furbank et al., 2019a). Such coordination may also provide hope that increased photosynthetic capacity and efficiency could actually translate to improved yield by feedforward effects on sink capacity.
Translation of photosynthetic performance to yield and resilience on farm
Current large investments in research for improved crop photosynthetic performance have varying timelines for delivery of new cereal varieties to farmers. Improved tools for phenotyping photosynthesis could identify material for crossing which can then be utilized in breeding programmes almost immediately, but understanding the genetic architecture of photosynthesis, and finding QTLs and causative SNPs in new gene targets requires appropriate diversity sets or biparental populations and then creation of near isogenic lines or gene-edited ‘allele mimics’ to demonstrate causality. This may reasonably be expected to take 3–5 years for the results of field-based activities to be available to crop breeders, assuming genetic material is in place. Deployment of genetically modified traits, once prototyped in model systems, may take even longer after careful validation in multiple environments and in multiple transgenic events. Complex multigene pathway engineering, while becoming rapidly more tractable with synthetic biology and gene editing (for C4 rice, see Ermakova et al., 2019b), would require 10 years plus research and then pre-breeding before delivery to breeders as a trait. While a population of 10 billion people on earth seems some way off, there is definitely no time to waste.
Acknowledgements
This research was funded by the Australian Research Council Centre of Excellence for Translational Photosynthesis (CE140100015), the New South Wales Environmental Trust (2016/RD/0006), the Cotton Research and Development Corporation (CRDC), and the Grains Research and Development Corporation (GRDC)
References
- Adachi S, Tsuru Y, Nito N, Murata K, Yamamoto T, Ebitani T, Ookawa T, Hirasawa T. 2011. Identification and characterization of genomic regions on chromosomes 4 and 8 that control the rate of photosynthesis in rice leaves. Journal of Experimental Botany 62, 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S, Yamamoto T, Nakae T, et al. 2019. Genetic architecture of leaf photosynthesis in rice revealed by different types of reciprocal mapping populations. Journal of Experimental Botany 70, 5131–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S, Yoshikawa K, Yamanouchi U, Tanabata T, Sun J, Ookawa T, Yamamoto T, Sage RF, Hirasawa T, Yonemaru J. 2017. Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Frontiers in Plant Science 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth EA, Long SP. 2005. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytologist 165, 351–371. [DOI] [PubMed] [Google Scholar]
- Ainsworth EA, Serbin SP, Skoneczka JA, Townsend PA. 2014. Using leaf optical properties to detect ozone effects on foliar biochemistry. Photosynthesis Research 119, 65–76. [DOI] [PubMed] [Google Scholar]
- Arneth A, Denton F, Agus F, Elbehri A, Erb K, Osman Elasha B, Rahimi M, Rounsevell M, Spence A, Valentini R. 2019: Framing and context. In: Shukla PR, Skea J, Calvo Buendia E, et al. , eds, Climate change and land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. https://www.ipcc.ch/srccl/ [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. 2010. Photorespiration: players, partners and origin. Trends in Plant Science 15, 330–336. [DOI] [PubMed] [Google Scholar]
- Bishop DL, Bugbee BG. 1998. Photosynthetic capacity and dry mass partitioning in dwarf and semi-dwarf wheat (Triticum aestivum L.). Journal of Plant Physiology 153, 558–565. [DOI] [PubMed] [Google Scholar]
- Boyles RE, Brenton ZW, Kresovich S. 2019. Genetic and genomic resources of sorghum to connect genotype with phenotype in contrasting environments. The Plant Journal 97, 19–39. [DOI] [PubMed] [Google Scholar]
- Broberg MC, Högy P, Feng Z, Pleijel H. 2019. Effects of elevated CO2 on wheat yield: non-linear response and relation to site productivity. Agronomy 9, 243. [Google Scholar]
- Camino C, Gonzalez-Dugoa V, Hernandeza P, Zarco-Tejada PJ. 2019. Radiative transfer Vcmax estimation from hyperspectral imagery and SIF retrievals to assess photosynthetic performance in rainfed and irrigated plant phenotyping trials. Remote Sensing of Environment 231, 111186. [Google Scholar]
- Carmo-Silva E, Andralojc PJ, Scales JC, Driever SM, Mead A, Lawson T, Raines CA, Parry MAJ. 2017. Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. Journal of Experimental Botany 68, 3473–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna S, Demmig-Adams B, Adams WW III. 2010. Exploring photosynthesis and plant stress using inexpensive chlorophyll fluorometers. Journal of Natural Resources and Life Sciences Education 39, 22–30. [Google Scholar]
- Coast O, Shah S, Ivakov A, et al. 2019. Predicting dark respiration rates of wheat leaves from hyperspectral reflectance. Plant, Cell & Environment 42, 2133–2150. [DOI] [PubMed] [Google Scholar]
- Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. 2005. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. [Google Scholar]
- Condon AG. 2020. Drying times: plant traits to improve crop water use efficiency and yield. Journal of Experimental Botany 71, 2239–2252. [DOI] [PubMed] [Google Scholar]
- Condon AG, Richards RA, Rebetzke GJ, Farquhar GD. 2004. Breeding for high water-use efficiency. Journal of Experimental Botany 55, 2447–2460. [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Salvucci ME. 2000. Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proceedings of the National Academy of Sciences, USA 97, 13430–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist E, Mora C, Engelman R. 2017. The interaction of human population, food production, and biodiversity protection. Science 356, 260–264. [DOI] [PubMed] [Google Scholar]
- Dawson TP, Perryman AH, Osborne TM.. 2016. Modelling impacts of climate change on global food security. Climatic Change 134, 429–440. [Google Scholar]
- Driever SM, Lawson T, Andralojc PJ, Raines CA, Parry MA. 2014. Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. Journal of Experimental Botany 65, 4959–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever SM, Simkin AJ, Saqer A, et al. 2017. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philosophical Transactions Royal Society B: Biological Science 372, 20160384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehri A, Challinor A, Verchot L, et al. 2017. https://www.ipcc.ch/publication/fao-ipcc-expert-meeting-on-land-use-climate-change-and-food-security/ FAO–IPCC expert meeting on climate change, land use and food security: final meeting report; January 23–25, 2017 FAO HQ Rome. FAO and IPCC, 2017.
- Ermakova M, Danila FR, Robert T. Furbank RT, Susanne von Caemmerer S. 2019b On the road to C4 rice: advances and perspectives. The Plant Journal doi: 10.1111/tpj.14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova M, Lopez-Calcagno PE, Raines CA, Furbank RT, von Caemmerer S. 2019a Overexpression of the Rieske FeS protein of the Cytochrome b6f complex increases C4 photosynthesis in Setaria viridis. Communications Biology 2, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. 1983. Nitrogen and photosynthesis in the flag leaf of wheat (Triticum aestivum L.). Plant Physiology 72, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. 1989. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78, 9–19. [DOI] [PubMed] [Google Scholar]
- Evans JR. 2019. Where did the carbon go? Science 363, eaat9077. [Google Scholar]
- Evans JR, Clarke VC. 2019. The nitrogen cost of photosynthesis. Journal of Experimental Botany 70, 7–15. [DOI] [PubMed] [Google Scholar]
- Evans LT. 1993. Crop evolution, adaptation and yield. Cambridge: Cambridge University Press. [Google Scholar]
- FAO. 2009. How to feed the world: global agriculture towards 2050; www.fao.org/fileadmin/templates/wsfs/docs/Issues_papers/HLEF2050_Global_Agriculture.pdf
- Farquhar GD, von Caemmerer S, Berry JA. 1980. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90. [DOI] [PubMed] [Google Scholar]
- Feng Z, Calatayud V, Zhu J, Kobayashi K. 2018. Ozone exposure- and flux-based response relationships with photosynthesis of winter wheat under fully open air condition. The Science of the Total Environment 619–620, 1538–1544. [DOI] [PubMed] [Google Scholar]
- Fischer RA. 1985. Number of kernels in wheat crops and the influence of solar radiation and temperature. Journal of Agricultural Science Cambridge 105, 447–461. [Google Scholar]
- Fischer RA, Byerlee D, Edmeades GO. 2010. Can technology deliver on the yield challenge to 2050? Expert Meeting on how to feed the world in 2050. Food and Agriculture Organization of the United Nations Economic and Social Development Department; http://www.fao.org/3/a-ak977e.pdf [Google Scholar]
- Fischer RA, Byerlee D, Edmeades GO. 2014. Crop yields and global food security: will yield increase continue to feed the world? Canberra, Australia: Australian Centre for International Agricultural Research; https://www.aciar.gov.au/node/12101 [Google Scholar]
- Fischer RA, Rees D, Sayre KD, Lu Z-M, Condon AG, Saavedra AL. 1998. Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Science 38, 1467–1475. [Google Scholar]
- Fischer RA, Richards R, Sadras V. 2019. 40% increased growth with genetic engineering. Science 363, eaat9077. [Google Scholar]
- Fortineau A, Bancal P. 2018. An innovative light chamber for measuring photosynthesis by three-dimensional plant organs. Plant Methods 14, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes MJ, Slafer GA, Davies WJ, Berry PM, Sylvester-Bradley R, Martre P, Calderini DF, Griffiths S, Reynolds MP. 2011. Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. Journal of Experimental Botany 62, 469–486. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Chitty JA, Von Caemmerer S, Jenkins C. 1996. Antisense RNA inhibition of RbcS gene expression reduces Rubisco level and photosynthesis in the C4 plant Flaveria bidentis. Plant Physiology 111, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Jimenez-Berni JA, George-Jaeggli B, Potgieter AB, Deery DM. 2019a Field crop phenomics: enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytologist 223, 1714–1727. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Sirault XRR, Stone E. 2019b Plant phenome to genome: a big data challenge. In: Zeigler RS, ed. Sustaining global food security. Melbourne: CSIRO Publishing, 203–223. [Google Scholar]
- Gaju O, DeSilva J, Carvalho P, Hawkesford MJ, Griffiths S, Greenland A, Foulkes MJ. 2016. Leaf photosynthesis and associations with grain yield, biomass and nitrogen-use efficiency in landraces, synthetic-derived lines and cultivars in wheat. Field Crops Research 193, 1–15. [Google Scholar]
- Galmés J, Kapralov MV, Andralojc PJ, Conesa MÀ, Keys AJ, Parry MA, Flexas J. 2014. Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant, Cell & Environment 37, 1989–2001. [DOI] [PubMed] [Google Scholar]
- Gómez R, Carrillo N, Morelli MP, Tula S, Shahinnia F, Hajirezaei MR, Lodeyro AF. 2018. Faster photosynthetic induction in tobacco by expressing cyanobacterial flavodiiron proteins in chloroplasts. Photosynthesis Research 136, 129–138. [DOI] [PubMed] [Google Scholar]
- Groszmann M, Osborn HL, Evans JR. 2017. Carbon dioxide and water transport through plant aquaporins. Plant, Cell & Environment 40, 938–961. [DOI] [PubMed] [Google Scholar]
- Gu J, Yin X, Struik PC, Stomph TJ, Wang H. 2012. Using chromosome introgression lines to map quantitative trait loci for photosynthesis parameters in rice (Oryza sativa L.) leaves under drought and well-watered field conditions. Journal of Experimental Botany 63, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison EP, Olcer H, Lloyd JC, Long SP, Raines CA. 2001. Small decreases in SBPase cause a linear decline in RuBP regeneration rate but do not affect Rubisco carboxylation capacity. Journal of Experimental Botany 52, 1779–1784. [DOI] [PubMed] [Google Scholar]
- Hasan MM, Chopin JP, Laga H, Miklavcic SJ. 2018. Detection and analysis of wheat spikes using convolutional neural networks. Plant Methods 14, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann D, Schlüter U, Weber APM. 2017. Machine learning techniques for predicting crop photosynthetic capacity from leaf reflectance spectra. Molecular Plant 10, 878–890. [DOI] [PubMed] [Google Scholar]
- Heureux AMC, Young JN, Whitney SM, Eason-Hubbard MR, Lee RBY, Sharwood RE, Rickaby REM. 2017. The role of Rubisco kinetics and pyrenoid morphology in shaping the CCM of haptophyte microalgae. Journal of Experimental Botany 68, 3959–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu N, Ujiie K, Perera I, Iri A, Kashiwagi T, Ishimaru K. 2017. Partial loss-of-function of NAL1 alters canopy photosynthesis by changing the contribution of upper and lower canopy leaves in rice. Scientific Reports 7, 15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman Z, Gobbett DL, Horan H. 2017. Climate trends account for stalled wheat yields in Australia since 1990. Global Change Biology 23, 2071–2081. [DOI] [PubMed] [Google Scholar]
- Hudson GS, Evans JR, von Caemmerer S, Arvidsson YB, Andrews TJ. 1992. Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase content by antisense RNA reduces photosynthesis in transgenic tobacco plants. Plant Physiology 98, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK, Street NR. 2011. Association genetics of complex traits in plants. New Phytologist 189, 909–922. [DOI] [PubMed] [Google Scholar]
- Jahan E, Amthor JS, Farquhar GD, Trethowan R, Barbour MM. 2014. Variation in mesophyll conductance among Australian wheat genotypes. Functional Plant Biology 41, 568–580. [DOI] [PubMed] [Google Scholar]
- Jahn CE, Mckay JK, Mauleon R, Stephens J, McNally KL, Bush DR, Leung H, Leach JE. 2011. Genetic variation in biomass traits among 20 diverse rice varieties. Plant Physiology 155, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. 1983. Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Archives of Biochemistry and Biophysics 227, 425–433. [DOI] [PubMed] [Google Scholar]
- Keller B, Vass I, Matsubara S, et al. 2019. Maximum fluorescence and electron transport kinetics determined by light-induced fluorescence transients (LIFT) for photosynthesis phenotyping. Photosynthesis Research 140, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP. 2016. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861. [DOI] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. 2008. The biochemistry of Rubisco in Flaveria. Journal of Experimental Botany 59, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Kuhlgert S, Austic G, Zegarac R, et al. 2016. MultispeQ Beta: a tool for large-scale plant phenotyping connected to the open PhotosynQ network. Royal Society Open Science 3, 160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P, Waugh R. 2019. Harnessing the potential of germplasm collections. Nature Genetics 51, 200–201. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Lawson T, Zakhleniuk OV, Lloyd JC, Raines CA, Fryer M. 2005. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiology 138, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu X-G. 2016. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. [DOI] [PubMed] [Google Scholar]
- Mace ES, Tai S, Gilding EK, et al. 2013. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nature Communications 4, 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RE, Hays DB, Mondal S, Ibrahim AMH, Basnet BR. 2013. QTL for yield, yield components and canopy temperature depression in wheat under late sown field conditions. Euphytica 194, 243–259. [Google Scholar]
- Maxwell K, Johnson GN. 2000. Chlorophyll fluorescence—a practical guide. Journal of Experimental Botany 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Meacham-Hensold K, Montes CM, Wu J, et al. 2019. High-throughput field phenotyping using hyperspectral reflectance and partial least squares regression (PLSR) reveals genetic modifications to photosynthetic capacity. Remote Sensing of Environment 231, 111176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner SG, Jost M, Taketa S, et al. 2019. Genebank genomics highlights the diversity of a global barley collection. Nature Genetics 51, 319–326. [DOI] [PubMed] [Google Scholar]
- Molero G, Joynson R, Pinera-Chavez FJ, Gardiner LJ, Rivera-Amado C, Hall A, Reynolds MP. 2019. Elucidating the genetic basis of biomass accumulation and radiation use efficiency in spring wheat and its role in yield potential. Plant Biotechnology Journal 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O. 2017. The diverse AAA+ machines that repair inhibited rubisco active sites. Frontiers in Molecular Biosciences 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Lawson T. 2013. Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. Journal of Experimental Botany 64, 3983–3998. [DOI] [PubMed] [Google Scholar]
- Murthy KK, Singh M. 1979. Photosynthesis, chlorophyll content and ribulose diphosphate carboxylase activity in relation to yield in wheat genotypes. Journal of Agricultural Science 93, 7–11. [Google Scholar]
- Mwale SE, Och-wo-Ssemakula M, Sadik K, Achola E, Okul V, Gibson P, Edema R, Singini W, Rubaihayo P. 2017. Response of Cowpea genotypes to drought stress in Uganda. American Journal of Plant Sciences 8, 720–733. [Google Scholar]
- Orr DJ, Alcântara A, Kapralov MV, Andralojc PJ, Carmo-Silva E, Parry MA. 2016. Surveying Rubisco diversity and temperature response to improve crop photosynthetic efficiency. Plant Physiology 172, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Keys AJ, Madgwick PJ, Carmo-Silva AE, Andralojc PJ. 2008. Rubisco regulation: a role for inhibitors. Journal of Experimental Botany 59, 1569–1580. [DOI] [PubMed] [Google Scholar]
- Parry MA, Reynolds M, Salvucci ME, Raines C, Andralojc PJ, Zhu XG, Price GD, Condon AG, Furbank RT. 2011. Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. Journal of Experimental Botany 62, 453–467. [DOI] [PubMed] [Google Scholar]
- Paul MJ, Gonzalez-Uriarte A, Griffiths CA, Hassani-Pak K. 2018. The role of trehalose 6-phosphate in crop yield and resilience. Plant Physiology 177, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, von Caemmerer S, Evans JR, Siebke K, Anderson JM, Badger MR. 1998. Photosynthesis is strongly reduced by antisense suppression of chloroplastic cytochrome bf complex in transgenic tobacco. Australian Journal Plant Physiology 25, 445–452. [Google Scholar]
- Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo-Silva E, Parry MA. 2016. Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. Journal of Experimental Botany 67, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzenberg AE, Víquez-Zamora M, Harbinson J, Lindhout P, van Heusden S. 2018. Chlorophyll fluorescence imaging reveals genetic variation and loci for a photosynthetic trait in diploid potato. Physiologia Plantarum 164, 163–175. [DOI] [PubMed] [Google Scholar]
- Qiongyan L, Cai J, Berger B, Okamoto M, Miklavcic SJ. 2017. Detecting spikes of wheat plants using neural networks with Laws texture energy. Plant Methods 13, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetzke GJ, Rattey AR, Farquhar GD, Richards RA, Condon AG. 2013. Genomic regions for canopy temperature and their genetic association with stomatal conductance and grain yield in wheat. Functional Plant Biology 40, 14–33. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Foulkes J, Furbank R, Griffiths S, King J, Murchie E, Parry M, Slafer G. 2012. Achieving yield gains in wheat. Plant, Cell & Environment 35, 1799–1823. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MA, Snape JW, Angus WJ. 2009. Raising yield potential in wheat. Journal of Experimental Botany 60, 1899–1918. [DOI] [PubMed] [Google Scholar]
- Reynolds MP, Balota M, Delgado MIB, Amani I, Fischer RA. 1994. Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Functional Plant Biology 21, 717–730. [Google Scholar]
- Reynolds MP, Delgado MI, Gutierrez-Rodriguez M, Larque-Saavedra A. 2000. Photosynthesis of wheat in a warm, irrigated environment—I: genetic diversity and crop productivity. Field Crops Research 66, 37–50. [Google Scholar]
- Reynolds MP, Pellegrinesschi A, Skovmand B. 2005. Sink-limitation to yield and biomass: a summary of some investigations in spring wheat. Annals of Applied Biology 146, 39–49. [Google Scholar]
- Riaz A, Hathorn A, Dinglasan E, et al. 2017. Into the vault of the Vavilov wheats: old diversity for new alleles. Genetic Resources and Crop Evolution 64, 531–544. [Google Scholar]
- Romay M. 2018. Rapid, affordable, and scalable genotyping for germplasm exploration in maize. In: Bennetzen J, Flint-Garcia S, Hirsch C, Tuberosa R, eds, The maize genome. Switzerland: Springer Nature, 31–58. [Google Scholar]
- Sadras VO, Lawson C, Montoro A. 2012. Photosynthetic traits in Australian wheat varieties released between 1958 and 2007. Field Crops Research 134, 19–29. [Google Scholar]
- Salesse-Smith CE, Sharwood RE, Busch FA, Kromdijk J, Bardal V, Stern DB. 2018. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nature Plants 4, 802–810. [DOI] [PubMed] [Google Scholar]
- Sanchez-Bragado R, Molero G, Reynolds MP, Araus JL. 2014. Relative contribution of shoot and ear photosynthesis to grain filling in wheat under good agronomical conditions assessed by differential organ δ13C. Journal of Experimental Botany 65, 5401–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbin SP, Dillaway DN, Kruger EL, Townsend PA. 2012. Leaf optical properties reflect variation in photosynthetic metabolism and its sensitivity to temperature. Journal of Experimental Botany 63, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD. 1988. Estimating the rate of photorespiration in leaves. Physiologia Plantarum 73, 147–152. [Google Scholar]
- Sharma DK, Andersen SB, Ottosen CO, Rosenqvist E. 2015. Wheat cultivars selected for high Fv/Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance, transpiration and dry matter. Physiologia Plantarum 153, 284–298. [DOI] [PubMed] [Google Scholar]
- Sharwood RE. 2017. Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytologist 213, 494–510. [DOI] [PubMed] [Google Scholar]
- Sharwood RE, Ghannoum O, Kapralov MV, Gunn LH, Whitney SM. 2016a Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nature Plants 2, 16186. [DOI] [PubMed] [Google Scholar]
- Sharwood RE, Ghannoum O, Whitney SM. 2016bProspects for improving CO2 fixation in C3-crops through understanding C4-Rubisco biogenesis and catalytic diversity. Current Opinion in Plant Biology 31, 135–142. [DOI] [PubMed] [Google Scholar]
- Shen BR, Wang LM, Lin XL, et al. 2019. Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Molecular Plant 12, 199–214. [DOI] [PubMed] [Google Scholar]
- Silva-Pérez V, De Faveri J, Molero G, Deery DM, Condon AG, Reynolds MP, Evans JR, Furbank RT. 2020. Genetic variation for photosynthetic capacity and efficiency in spring wheat. Journal of Experimental Botany 71, 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Perez V, Molero G, Serbin SP, Condon AG, Reynolds MP, Furbank RT, Evans JR. 2018. Hyperspectral reflectance as a tool to measure biochemical and physiological traits in wheat. Journal of Experimental Botany 69, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, López-Calcagno PE, Raines CA. 2019. Feeding the world: improving photosynthetic efficiency for sustainable crop production. Journal of Experimental Botany 70, 1119–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, McAusland L, Lawson T, Raines CA. 2017. Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield. Plant Physiology 175, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Serbin SP, McNeil BE, Kingdon CC, Townsend PA. 2015. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecological Applications 25, 2180–2197. [DOI] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR. 2019. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinziano JR, Morgan PB, Lynch DJ, Saathoff AJ, McDermitt DK, Hanson DT. 2017. The rapid A–Ci response: photosynthesis in the phenomic era. Plant, Cell & Environment 40, 1256–1262. [DOI] [PubMed] [Google Scholar]
- Takai T, Adachi S, Taguchi-Shiobara F, et al. 2013. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Scientific Reports 3, 2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wu X, Li C, Yang W, Huang M, Ma X, Li S. 2017. Yield, growth, canopy traits and photosynthesis in high-yielding, synthetic hexaploid-derived wheats cultivars compared with non-synthetic wheats. Crop & Pasture Science 68, 115–125. [Google Scholar]
- Teng S, Qian Q, Zeng D, Kunihiro Y, Fujimoto K, Huang D, Zhu L. 2004. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.). Euphytica 135, 1–7. [Google Scholar]
- Traub J, Porch T, Naeem M, Urrea CA, Austic G, Kelly JD, Loescher W. 2018. Screening for heat tolerance in Phaseolus spp. using multiple methods. Crop Science 58, 2459–2469. [Google Scholar]
- von Caemmerer S. 2000. Biochemical models of leaf photosynthesis. Collingwood: CSIRO Publishing. [Google Scholar]
- von Caemmerer S, Hendrickson L, Quinn V, Vella N, Millgate AG, Furbank RT. 2005. Reductions of Rubisco activase by antisense RNA in the C4 plant Flaveria bidentis reduces Rubisco carbamylation and leaf photosynthesis. Plant Physiology 137, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Quick WP, Furbank RT. 2012. The development of C4 rice. Progress and future challenges. Science 336, 1671–1672. [DOI] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, et al. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmés J. 2011. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proceedings of the National Academy of Sciences, USA 108, 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RH, Martin-Avila E, Conlan C, and Whitney SM. 2018. An improved Escherichia coli screen for Rubisco identifies a protein–protein interface that can enhance CO2-fixation kinetics. Journal Biological Chemistry 293, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A. 2018. Transitioning to the next phase: the role of sugar signaling throughout the plant life cycle. Plant Physiology 176, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Hammer GL, Doherty A, von Caemmerer S, Farquhar GD. 2019. Quantifying impacts of enhancing photosynthesis on crop yield. Nature Plants, 54, 380–388. [DOI] [PubMed] [Google Scholar]
- Wu A, Song Y, van Oosterom EJ, Hammer GL. 2016. Connecting biochemical photosynthesis models with crop models to support crop improvement. Frontiers in Plant Science 7, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendrek CR, Tomaz T, Montes CM, Cao Y, Morse AM, Brown PJ, McIntyre LM, Leakey AD, Ainsworth EA. 2017. High-throughput phenotyping of maize leaf physiological and biochemical traits using hyperspectral reflectance. Plant Physiology 173, 614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Struik PC. 2017. Can increased leaf photosynthesis be converted into higher crop mass production? A simulation study for rice using the crop model GECROS. Journal of Experimental Botany 68, 2345–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JN, Heureux AM, Sharwood RE, Rickaby RE, Morel FM, Whitney SM. 2016. Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. Journal of Experimental Botany 67, 3445–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guanter L, Joiner J, Song L, Guan K. 2018. Spatially-explicit monitoring of crop photosynthetic capacity through the use of space-based chlorophyll fluorescence data. Remote Sensing of Environment 210, 362–374. [Google Scholar]
- Zhu XG, de Sturler E, Long SP. 2007. Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: a numerical simulation using an evolutionary algorithm. Plant Physiology 145, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. 2019. A primer on deep learning in genomics. Nature Genetics 51, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]