Abstract
Background
In December, 2019, a pneumonia associated with the 2019 novel coronavirus (2019-nCoV) emerged in Wuhan, China. We aimed to further clarify the epidemiological and clinical characteristics of 2019-nCoV pneumonia.
Methods
In this retrospective, single-centre study, we included all confirmed cases of 2019-nCoV in Wuhan Jinyintan Hospital from Jan 1 to Jan 20, 2020. Cases were confirmed by real-time RT-PCR and were analysed for epidemiological, demographic, clinical, and radiological features and laboratory data. Outcomes were followed up until Jan 25, 2020.
Findings
Of the 99 patients with 2019-nCoV pneumonia, 49 (49%) had a history of exposure to the Huanan seafood market. The average age of the patients was 55·5 years (SD 13·1), including 67 men and 32 women. 2019-nCoV was detected in all patients by real-time RT-PCR. 50 (51%) patients had chronic diseases. Patients had clinical manifestations of fever (82 [83%] patients), cough (81 [82%] patients), shortness of breath (31 [31%] patients), muscle ache (11 [11%] patients), confusion (nine [9%] patients), headache (eight [8%] patients), sore throat (five [5%] patients), rhinorrhoea (four [4%] patients), chest pain (two [2%] patients), diarrhoea (two [2%] patients), and nausea and vomiting (one [1%] patient). According to imaging examination, 74 (75%) patients showed bilateral pneumonia, 14 (14%) patients showed multiple mottling and ground-glass opacity, and one (1%) patient had pneumothorax. 17 (17%) patients developed acute respiratory distress syndrome and, among them, 11 (11%) patients worsened in a short period of time and died of multiple organ failure.
Interpretation
The 2019-nCoV infection was of clustering onset, is more likely to affect older males with comorbidities, and can result in severe and even fatal respiratory diseases such as acute respiratory distress syndrome. In general, characteristics of patients who died were in line with the MuLBSTA score, an early warning model for predicting mortality in viral pneumonia. Further investigation is needed to explore the applicability of the MuLBSTA score in predicting the risk of mortality in 2019-nCoV infection.
Funding
National Key R&D Program of China.
Introduction
Since Dec 8, 2019, several cases of pneumonia of unknown aetiology have been reported in Wuhan, Hubei province, China.1, 2, 3 Most patients worked at or lived around the local Huanan seafood wholesale market, where live animals were also on sale. In the early stages of this pneumonia, severe acute respiratory infection symptoms occurred, with some patients rapidly developing acute respiratory distress syndrome (ARDS), acute respiratory failure, and other serious complications. On Jan 7, a novel coronavirus was identified by the Chinese Center for Disease Control and Prevention (CDC) from the throat swab sample of a patient, and was subsequently named 2019-nCoV by WHO.4
Coronaviruses can cause multiple system infections in various animals and mainly respiratory tract infections in humans, such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS).5, 6, 7 Most patients have mild symptoms and good prognosis. So far, a few patients with 2019-nCoV have developed severe pneumonia, pulmonary oedema, ARDS, or multiple organ failure and have died. All costs of 2019-nCoV treatment are covered by medical insurance in China.
At present, information regarding the epidemiology and clinical features of pneumonia caused by 2019-nCoV is scarce.1, 2, 3 In this study, we did a comprehensive exploration of the epidemiology and clinical features of 99 patients with confirmed 2019-nCoV pneumonia admitted to Jinyintan Hospital, Wuhan, which admitted the first patients with 2019-nCoV to be reported on.
Methods
Study design and participants
For this retrospective, single-centre study, we recruited patients from Jan 1 to Jan 20, 2020, at Jinyintan Hospital in Wuhan, China. Jinyintan Hospital is a hospital for adults (ie, aged ≥14 years) specialising in infectious diseases. According to the arrangements put in place by the Chinese Government, adult patients were admitted centrally to the hospital from the whole of Wuhan without selectivity. All patients at Jinyintan Hospital who were diagnosed as having 2019-nCoV pneumonia according to WHO interim guidance were enrolled in this study.4 All the data of included cases have been shared with WHO. The study was approved by Jinyintan Hospital Ethics Committee and written informed consent was obtained from patients involved before enrolment when data were collected retrospectively.
Research in context.
Evidence before this study
We searched PubMed on Jan 25, 2020, for articles that describe the epidemiological and clinical characteristics of the 2019 novel coronavirus (2019-nCoV) in Wuhan, China, using the search terms “novel coronavirus” and “pneumonia” with no language or time restrictions. Previously published research discussed the epidemiological and clinical characteristics of severe acute respiratory syndrome coronavirus or Middle East respiratory syndrome coronavirus, and primary study for the evolution of the novel coronavirus from Wuhan. The only report of clinical features of patients infected with 2019-nCoV was published on Jan 24, 2020, with 41 cases included.
Added value of this study
We have obtained data on 99 patients in Wuhan, China, to further explore the epidemiology and clinical features of 2019-nCoV. This study is, to our knowledge, the largest case series to date of 2019-nCoV infections, with 99 patients who were transferred to Jinyintan Hospital from other hospitals all over Wuhan, and provides further information on the demographic, clinical, epidemiological, and laboratory features of patients. It presents the latest status of 2019-nCoV infection in China and is an extended investigation of the previous report, with 58 extra cases and more details on combined bacterial and fungal infections. In all patients admitted with medical comorbidities of 2019-nCoV, a wide range of clinical manifestations can be seen and are associated with substantial outcomes.
Implications of all the available evidence
The 2019-nCoV infection was of clustering onset, is more likely to affect older men with comorbidities, and could result in severe and even fatal respiratory diseases such as acute respiratory distress syndrome. Early identification and timely treatment of critical cases of 2019-nCoV are important. Effective life support and active treatment of complications should be provided to effectively reduce the severity of patients' conditions and prevent the spread of this new coronavirus in China and worldwide.
Procedures
We obtained epidemiological, demographic, clinical, laboratory, management, and outcome data from patients' medical records. Clinical outcomes were followed up to Jan 25, 2020. If data were missing from the records or clarification was needed, we obtained data by direct communication with attending doctors and other health-care providers. All data were checked by two physicians (XD and YQ).
Laboratory confirmation of 2019-nCoV was done in four different institutions: the Chinese CDC, the Chinese Academy of Medical Science, Academy of Military Medical Sciences, and Wuhan Institute of Virology, Chinese Academy of Sciences. Throat-swab specimens from the upper respiratory tract that were obtained from all patients at admission were maintained in viral-transport medium. 2019-nCoV was confirmed by real-time RT-PCR using the same protocol described previously.3 RT-PCR detection reagents were provided by the four institutions. Other respiratory viruses including influenza A virus (H1N1, H3N2, H7N9), influenza B virus, respiratory syncytial virus, parainfluenza virus, adenovirus, SARS coronavirus (SARS-CoV), and MERS coronavirus (MERS-CoV) were also examined with real-time RT-PCR
Sputum or endotracheal aspirates were obtained at admission for identification of possible causative bacteria or fungi. Additionally, all patients were given chest x-rays or chest CT.
Outcomes
We describe epidemiological data (ie, short-term [occasional visits] and long-term [worked at or lived near] exposure to Huanan seafood market); demographics; signs and symptoms on admission; comorbidity; laboratory results; co-infection with other respiratory pathogens; chest radiography and CT findings; treatment received for 2019-nCoV; and clinical outcomes.
Statistical analysis
We present continuous measurements as mean (SD) if they are normally distributed or median (IQR) if they are not, and categorical variables as count (%). For laboratory results, we also assessed whether the measurements were outside the normal range. We used SPSS (version 26.0) for all analyses.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
99 patients with 2019-nCoV were included in this study, two of whom were husband and wife. In total, 49 (49%) patients were clustered and had a history of exposure to the Huanan seafood market. Among them, there were 47 patients with long-term exposure history, most of whom were salesmen or market managers, and two patients with short-term exposure history, who were shoppers. None of the patients were medical staff. Most patients were men, with a mean age of 55·5 years (SD 13·1; table 1 ). 50 (51%) patients had chronic diseases, including cardiovascular and cerebrovascular diseases, endocrine system disease, digestive system disease, respiratory system disease, malignant tumour, and nervous system disease (table 1).
Table 1.
Demographics, baseline characteristics, and clinical outcomes of 99 patients admitted to Wuhan Jinyintan Hospital (Jan 1–20, 2020) with 2019-nCoV pneumonia
| Patients (n=99) | ||
|---|---|---|
| Age, years | ||
| Mean (SD) | 55·5 (13·1) | |
| Range | 21–82 | |
| ≤39 | 10 (10%) | |
| 40–49 | 22 (22%) | |
| 50–59 | 30 (30%) | |
| 60–69 | 22 (22%) | |
| ≥70 | 15 (15%) | |
| Sex | ||
| Female | 32 (32%) | |
| Male | 67 (68%) | |
| Occupation | ||
| Agricultural worker | 2 (2%) | |
| Self-employed | 63 (64%) | |
| Employee | 15 (15%) | |
| Retired | 19 (19%) | |
| Exposure to Huanan seafood market* | 49 (49%) | |
| Long-term exposure history | 47 (47%) | |
| Short-term exposure history | 2 (2%) | |
| Chronic medical illness | 50 (51%) | |
| Cardiovascular and cerebrovascular diseases | 40 (40%) | |
| Digestive system disease | 11 (11%) | |
| Endocrine system disease† | 13 (13%) | |
| Malignant tumour | 1 (1%) | |
| Nervous system disease | 1 (1%) | |
| Respiratory system disease | 1 (1%) | |
| Admission to intensive care unit | 23 (23%) | |
| Clinical outcome | ||
| Remained in hospital | 57 (58%) | |
| Discharged | 31 (31%) | |
| Died | 11 (11%) | |
Data are n (%) unless specified otherwise. 2019-nCoV=2019 novel coronavirus.
Long-term exposure is having worked at or lived in or around Huanan seafood market, whereas short-term exposure is having been to Huanan seafood market occasionally.
12 were diabetic.
On admission, most patients had fever or cough and a third of patients had shortness of breath (table 2 ). Other symptoms included muscle ache, headache, confusion, chest pain, and diarrhoea (table 2). Many patients presented with organ function damage, including 17 (17%) with ARDS, eight (8%) with acute respiratory injury, three (3%) with acute renal injury, four (4%) with septic shock, and one (1%) with ventilator-associated pneumonia (table 2).
Table 2.
Clinical characteristics and treatment of patients with 2019-nCoV pneumonia
| Patients (n=99) | ||
|---|---|---|
| Signs and symptoms at admission | ||
| Fever | 82 (83%) | |
| Cough | 81 (82%) | |
| Shortness of breath | 31 (31%) | |
| Muscle ache | 11 (11%) | |
| Confusion | 9 (9%) | |
| Headache | 8 (8%) | |
| Sore throat | 5 (5%) | |
| Rhinorrhoea | 4 (4%) | |
| Chest pain | 2 (2%) | |
| Diarrhoea | 2 (2%) | |
| Nausea and vomiting | 1 (1%) | |
| More than one sign or symptom | 89 (90%) | |
| Fever, cough, and shortness of breath | 15 (15%) | |
| Comorbid conditions | ||
| Any | 33 (33%) | |
| ARDS | 17 (17%) | |
| Acute renal injury | 3 (3%) | |
| Acute respiratory injury | 8 (8%) | |
| Septic shock | 4 (4%) | |
| Ventilator-associated pneumonia | 1 (1%) | |
| Chest x-ray and CT findings | ||
| Unilateral pneumonia | 25 (25%) | |
| Bilateral pneumonia | 74 (75%) | |
| Multiple mottling and ground-glass opacity | 14 (14%) | |
| Treatment | ||
| Oxygen therapy | 75 (76%) | |
| Mechanical ventilation | ||
| Non-invasive (ie, face mask) | 13 (13%) | |
| Invasive | 4 (4%) | |
| CRRT | 9 (9%) | |
| ECMO | 3 (3%) | |
| Antibiotic treatment | 70 (71%) | |
| Antifungal treatment | 15 (15%) | |
| Antiviral treatment | 75 (76%) | |
| Glucocorticoids | 19 (19%) | |
| Intravenous immunoglobulin therapy | 27 (27%) | |
2019-nCoV=2019 novel coronavirus. ARDS=acute respiratory distress syndrome. ECMO=extracorporeal membrane oxygenation. CRRT=continuous renal replacement therapy.
On admission, leucocytes were below the normal range in nine (9%) patients and above the normal range in 24 (24%) patients (table 3 ). 38 (38%) patients had neutrophils above the normal range. Lymphocytes and haemoglobin were below the normal range in many patients (table 3). Platelets were below the normal range in 12 (12%) patients and above the normal range in four (4%). 43 patients had differing degrees of liver function abnormality, with alanine aminotransferase (ALT) or aspartate aminotransferase (AST) above the normal range (table 3); one patient had severe liver function damage (ALT 7590 U/L, AST 1445 U/L). Most patients had abnormal myocardial zymogram, which showed the elevation of creatine kinase in 13 (13%) patients and the elevation of lactate dehydrogenase in 75 (76%) patients, one of whom also showed abnormal creatine kinase (6280 U/L) and lactate dehydrogenase (20 740 U/L). Seven (7%) patients had different degrees of renal function damage, with elevated blood urea nitrogen or serum creatinine. Regarding the infection index, procalcitonin was above the normal range in six (6%) patients. Most patients had serum ferritin above the normal range (table 3). 73 patients were tested for C-reactive protein, most of whom had levels above the normal range (table 3).
Table 3.
Laboratory results of patients with 2019-nCoV pneumonia
| Patients (n=99) | ||
|---|---|---|
| Blood routine | ||
| Leucocytes (× 109 per L; normal range 3·5–9·5) | 7·5 (3·6) | |
| Increased | 24 (24%) | |
| Decreased | 9 (9%) | |
| Neutrophils (× 109 per L; normal range 1·8–6·3) | 5·0 (3·3–8·1) | |
| Increased | 38 (38%) | |
| Lymphocytes (× 109 per L; normal range 1·1–3·2) | 0·9 (0·5) | |
| Decreased | 35 (35%) | |
| Platelets (× 109 per L; normal range 125·0–350·0) | 213·5 (79·1) | |
| Increased | 4 (4%) | |
| Decreased | 12 (12%) | |
| Haemoglobin (g/L; normal range 130·0–175·0) | 129·8 (14·8) | |
| Decreased | 50 (51%) | |
| Coagulation function | ||
| Activated partial thromboplastin time (s; normal range 21·0–37·0) | 27·3 (10·2) | |
| Increased | 6 (6%) | |
| Decreased | 16 (16%) | |
| Prothrombin time (s; normal range 10·5–13·5) | 11·3 (1·9) | |
| Increased | 5 (5%) | |
| Decreased | 30 (30%) | |
| D-dimer (μg/L; normal range 0·0–1·5) | 0·9 (0·5–2·8) | |
| Increased | 36 (36%) | |
| Blood biochemistry | ||
| Albumin (g/L; normal range 40·0–55·0) | 31·6 (4·0) | |
| Decreased | 97 (98%) | |
| Alanine aminotransferase (U/L; normal range 9·0–50·0) | 39·0 (22·0–53·0) | |
| Increased | 28 (28%) | |
| Aspartate aminotransferase (U/L; normal range 15·0–40·0) | 34·0 (26·0–48·0) | |
| Increased | 35 (35%) | |
| Total bilirubin (μmol/L; normal range 0·0–21·0) | 15·1 (7·3) | |
| Increased | 18 (18%) | |
| Blood urea nitrogen (mmol/L; normal range 3·6–9·5) | 5·9 (2·6) | |
| Increased | 6 (6%) | |
| Decreased | 17 (17%) | |
| Serum creatinine (μmol/L; normal range 57·0–111·0) | 75·6 (25·0) | |
| Increased | 3 (3%) | |
| Decreased | 21 (21%) | |
| Creatine kinase (U/L; normal range 50·0–310·0) | 85·0 (51·0–184·0) | |
| Increased | 13 (13%) | |
| Decreased | 23 (23%) | |
| Lactate dehydrogenase (U/L; normal range 120·0–250·0) | 336·0 (260·0–447·0) | |
| Increased | 75 (76%) | |
| Myoglobin (ng/mL; normal range 0·0–146·9) | 49·5 (32·2–99·8) | |
| Increased | 15 (15%) | |
| Glucose (mmol/L; normal range 3·9–6·1) | 7·4 (3·4) | |
| Increased | 51 (52%) | |
| Decreased | 1 (1%) | |
| Infection-related biomarkers | ||
| Procalcitonin (ng/mL; normal range 0·0–5·0) | 0·5 (1·1) | |
| Increased | 6 (6%) | |
| Interleukin-6 (pg/mL; normal range 0·0–7·0) | 7·9 (6·1–10·6) | |
| Increased | 51 (52%) | |
| Erythrocyte sedimentation rate (mm/h; normal range 0·0–15·0) | 49·9 (23·4) | |
| Increased | 84 (85%) | |
| Serum ferritin (ng/mL; normal range 21·0–274·7) | 808·7 (490·7) | |
| Increased | 62 (63%) | |
| C-reactive protein (mg/L; normal range 0·0–5·0)* | 51·4 (41·8) | |
| Increased | 63/73 (86%) | |
| Co-infection | ||
| Other viruses | 0 | |
| Bacteria | 1 (1%) | |
| Fungus | 4 (4%) | |
Data are n (%), n/N (%), mean (SD), and median (IQR). Increased means over the upper limit of the normal range and decreased means below the lower limit of the normal range. 2019-nCoV=2019 novel coronavirus.
Data available for 73 patients.
All patients were tested for nine respiratory pathogens and the nucleic acid of influenza viruses A and B. Bacteria and fungi culture were done at the same time. We did not find other respiratory viruses in any of the patients. Acinetobacter baumannii, Klebsiella pneumoniae, and Aspergillus flavus were all cultured in one patient. A baumannii turned out to be highly resistant to antibiotics. One case of fungal infection was diagnosed as Candida glabrata and three cases of fungal infection were diagnosed as Candida albicans.
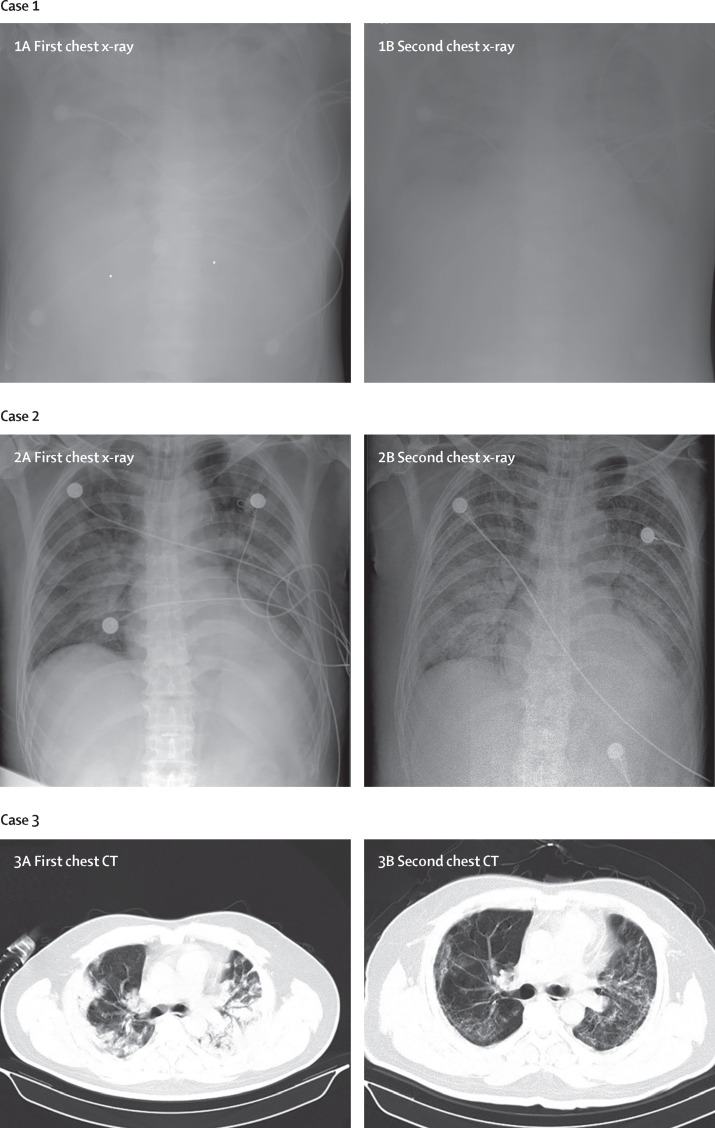
According to chest x-ray and CT, 74 (75%) patients showed bilateral pneumonia (75%) with just 25 (25%) patients showing unilateral pneumonia (table 2). 14 (14%) patients showed multiple mottling and ground-glass opacity (table 2; figure ). Additionally, pneumothorax occurred in one (1%) patient.
Figure.
Chest x-rays and chest CTs of three patients
Case 1: chest x-ray was obtained on Jan 1 (1A). The brightness of both lungs was diffusely decreased, showing a large area of patchy shadow with uneven density. Tracheal intubation was seen in the trachea and the heart shadow outline was not clear. The catheter shadow was seen from the right axilla to the mediastinum. Bilateral diaphragmatic surface and costal diaphragmatic angle were not clear, and chest x-ray on Jan 2 showed worse status (1B). Case 2: chest x-ray obtained on Jan 6 (2A). The brightness of both lungs was decreased and multiple patchy shadows were observed; edges were blurred, and large ground-glass opacity and condensation shadows were mainly on the lower right lobe. Tracheal intubation could be seen in the trachea. Heart shadow roughly presents in the normal range. On the left side, the diaphragmatic surface is not clearly displayed. The right side of the diaphragmatic surface was light and smooth and rib phrenic angle was less sharp. Chest x-ray on Jan 10 showed worse status (2B). Case 3: chest CT obtained on Jan 1 (3A) showed mass shadows of high density in both lungs. Bright bronchogram is seen in the lung tissue area of the lesion, which is also called bronchoinflation sign. Chest CT on Jan 15 showed improved status (3B).
All patients were treated in isolation. 75 (76%) patients received antiviral treatment, including oseltamivir (75 mg every 12 h, orally), ganciclovir (0·25 g every 12 h, intravenously), and lopinavir and ritonavir tablets (500 mg twice daily, orally). The duration of antiviral treatment was 3–14 days (median 3 days [IQR 3–6]).
Most patients were given antibiotic treatment (table 2); 25 (25%) patients were treated with a single antibiotic and 45 (45%) patients were given combination therapy. The antibiotics used generally covered common pathogens and some atypical pathogens; when secondary bacterial infection occurred, medication was administered according to the results of bacterial culture and drug sensitivity. The antibiotics used were cephalosporins, quinolones, carbapenems, tigecycline against methicillin-resistant Staphylococcus aureus, linezolid, and antifungal drugs. The duration of antibiotic treatment was 3–17 days (median 5 days [IQR 3–7]). 19 (19%) patients were also treated with methylprednisolone sodium succinate, methylprednisolone, and dexamethasone for 3–15 days (median 5 [3–7]).
13 patients used non-invasive ventilator mechanical ventilation for 4–22 days (median 9 days [IQR 7–19]). Four patients used an invasive ventilator to assist ventilation for 3–20 days (median 17 [12–19]). The ventilator adopted P-SIMV mode, the inhaled oxygen concentration was 35–100%, and the positive end-expiratory pressure was 6–12 cm H2O. All four patients were still using ventilators at data cutoff. Moreover, nine (9%) patients received continuous blood purification due to renal failure and three (3%) patients were treated with extracorporeal membrane oxygenation (ECMO; table 2).
By the end of Jan 25, 31 (31%) patients had been discharged and 11 (11%) patients had died; all other patients were still in hospital (table 1). The first two deaths were a 61-year-old man (patient 1) and a 69-year-old man (patient 2). They had no previous chronic underlying disease but had a long history of smoking. Patient 1 was transferred to Jinyintan Hospital and diagnosed with severe pneumonia and ARDS. He was immediately admitted to the intensive care unit (ICU) and given an intubated ventilator-assisted breathing therapy. Later, the patient, having developed severe respiratory failure, heart failure, and sepsis, experienced a sudden cardiac arrest on the 11th day of admission and was declared dead. Patient 2 had severe pneumonia and ARDS after admission. The patient was transferred to the ICU and given ventilator-assisted breathing, and received anti-infection and ECMO treatment after admission. The patient's hypoxaemia remained unresolved. On the ninth day of admission, the patient died of severe pneumonia, septic shock, and respiratory failure. The intervals between the onset of symptoms and the use of ventilator-assisted breathing in the two patients were 3 days and 10 days, respectively. The course of the disease and lung lesions progressed rapidly in both patients, with both developing multiple organ failure in a short time. The deaths of these two patients were consistent with the MuLBSTA score, an early warning model for predicting mortality in viral pneumonia.8
Of the remaining nine patients who died, eight patients had lymphopenia, seven had bilateral pneumonia, five were older than 60 years, three had hypertension, and one was a heavy smoker.
Discussion
This is an extended descriptive study on the epidemiology and clinical characteristics of the 2019-nCoV, including data on 99 patients who were transferred to Jinyintan Hospital from other hospitals across Wuhan. It presents the latest status of the 2019-nCoV infection in China and adds details on combined bacterial and fungal infections.
Human coronavirus is one of the main pathogens of respiratory infection. The two highly pathogenic viruses, SARS-CoV and MERS-CoV, cause severe respiratory syndrome in humans and four other human coronaviruses (HCoV-OC43, HCoV-229E, HCoV-NL63, HCoV-HKU1) induce mild upper respiratory disease. The major SARS-CoV outbreak involving 8422 patients occurred during 2002–03 and spread to 29 countries globally.9, 10 MERS-CoV emerged in Middle Eastern countries in 2012 but was imported into China.11, 12 The sequence of 2019-nCoV is relatively different from the six other coronavirus subtypes but can be classified as betacoronavirus. SARS-CoV and MERS-CoV can be transmitted directly to humans from civets and dromedary camels, respectively, and both viruses originate in bats, but the origin of 2019-nCoV needs further investigation.13, 14, 15 2019-nCoV also has enveloped virions that measure approximately 50–200 nm in diameter with a single positive-sense RNA genome.16 Club-shaped glycoprotein spikes in the envelope give the virus a crown-like or coronal appearance. Transmission rates are unknown for 2019-nCoV; however, there is evidence of human-to-human transmission. None of the 99 patients we examined were medical staff, but 15 medical workers have been reported with 2019-nCoV infection, 14 of whom are assumed to have been infected by the same patient.17 The mortality of SARS-CoV has been reported as more than 10% and MERS-CoV at more than 35%.5, 18 At data cutoff for this study, mortality of the 99 included patients infected by 2019-nCoV was 11%, resembling that in a previous study.3 However, additional deaths might occur in those still hospitalised.
We observed a greater number of men than women in the 99 cases of 2019-nCoV infection. MERS-CoV and SARS-CoV have also been found to infect more males than females.19, 20 The reduced susceptibility of females to viral infections could be attributed to the protection from X chromosome and sex hormones, which play an important role in innate and adaptive immunity.21 Additionally, about half of patients infected by 2019-nCoV had chronic underlying diseases, mainly cardiovascular and cerebrovascular diseases and diabetes; this is similar to MERS-CoV.19 Our results suggest that 2019-nCoV is more likely to infect older adult males with chronic comorbidities as a result of the weaker immune functions of these patients.19, 20, 21, 22
Some patients, especially severely ill ones, had co-infections of bacteria and fungi. Common bacterial cultures of patients with secondary infections included A baumannii, K pneumoniae, A flavus, C glabrata, and C albicans.8 The high drug resistance rate of A baumannii can cause difficulties with anti-infective treatment, leading to higher possibility of developing septic shock.23 For severe mixed infections, in addition to the virulence factors of pathogens, the host's immune status is also one of the important factors. Old age, obesity, and presence of comorbidity might be associated with increased mortality.24 When populations with low immune function, such as older people, diabetics, people with HIV infection, people with long-term use of immunosuppressive agents, and pregnant women, are infected with 2019-nCoV, prompt administration of antibiotics to prevent infection and strengthening of immune support treatment might reduce complications and mortality.
In terms of laboratory tests, the absolute value of lymphocytes in most patients was reduced. This result suggests that 2019-nCoV might mainly act on lymphocytes, especially T lymphocytes, as does SARS-CoV. Virus particles spread through the respiratory mucosa and infect other cells, induce a cytokine storm in the body, generate a series of immune responses, and cause changes in peripheral white blood cells and immune cells such as lymphocytes. Some patients progressed rapidly with ARDS and septic shock, which was eventually followed by multiple organ failure. Therefore, early identification and timely treatment of critical cases is of crucial importance. Use of intravenous immunoglobulin is recommended to enhance the ability of anti-infection for severely ill patients and steroids (methylprednisolone 1–2 mg/kg per day) are recommended for patients with ARDS, for as short a duration of treatment as possible. Some studies suggest that a substantial decrease in the total number of lymphocytes indicates that coronavirus consumes many immune cells and inhibits the body's cellular immune function. Damage to T lymphocytes might be an important factor leading to exacerbations of patients.25 The low absolute value of lymphocytes could be used as a reference index in the diagnosis of new coronavirus infections in the clinic.
In general, the characteristics of patients who died were in line with the early warning model for predicting mortality in viral pneumonia in our previous study: the MuLBSTA score.8 The MuLBSTA score system contains six indexes, which are multilobular infiltration, lymphopenia, bacterial co-infection, smoking history, hypertension, and age. Further investigation is needed to explore the applicability of the MuLBSTA score in predicting the risk of mortality in 2019-nCoV infection.
This study has several limitations. First, only 99 patients with confirmed 2019-nCoV were included; suspected but undiagnosed cases were ruled out in the analyses. It would be better to include as many patients as possible in Wuhan, in other cities in China, and even in other countries to get a more comprehensive understanding of 2019-nCoV. Second, more detailed patient information, particularly regarding clinical outcomes, was unavailable at the time of analysis; however, the data in this study permit an early assessment of the epidemiological and clinical characteristics of 2019-nCoV pneumonia in Wuhan, China.
In conclusion, the infection of 2019-nCoV was of clustering onset, is more likely to infect older men with comorbidities, and can result in severe and even fatal respiratory diseases such as ARDS.
Acknowledgments
Acknowledgments
This study was funded by the National Key R&D Program of China (number 2017YFC1309700). We thank all patients involved in the study.
Contributors
NC, XD, FG, YH, YQ, JW, YL, YW, JX, TY, and LZ collected the epidemiological and clinical data and processed statistical data. NC and MZ drafted the manuscript. JQ and XZ revised the final manuscript. XZ is responsible for summarising all data related to the virus. LZ is responsible for summarising all epidemiological and clinical data.
Declaration of interests
We declare no competing interests.
References
- 1.Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020 doi: 10.1002/jmv.25678. published online Jan 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui DS, Azhar EI, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. published online Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. Jan 11, 2020. https://www.who.int/internal-publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected
- 5.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, Wei D, Zhang X, et al. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B, Zeng LP, Yang XL, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song HD, Tu CC, Zhang GW, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haagmans BL, Al Dhahiry SH, Reusken CB, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 13.Tao Y, Shi M, Chommanard C, et al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. 2017;91:e01953–e02016. doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Fan H, Lan T, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XT, Chen P, Wang JF, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for the risk of human transmission. Sci China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. published online Jan 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinese Academy of Sciences Wuhan coronavirus has strong ability to infect humans. Press release. Jan 21, 2020. https://view.inews.qq.com/w2/20200121A0M08X00?tbkt=F&strategy=&openid=o04IBALMrLyGDxbWNOPoDM1IfG-s&uid=&refer=wx_hot
- 18.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badawi A, Ryoo SG. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int J Infect Dis. 2016;49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Channappanavar R, Fett C, Mack M, Ten Eyck PP, Meyerholz DK, Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaillon S, Berthenet K, Garlanda C. Sexual dimorphism in innate immunity. Clin Rev Allergy Immunol. 2019;56:308–321. doi: 10.1007/s12016-017-8648-x. [DOI] [PubMed] [Google Scholar]
- 22.Dryden M, Baguneid M, Eckmann C, et al. Pathophysiology and burden of infection in patients with diabetes mellitus and peripheral vascular disease: focus on skin and soft-tissue infections. Clin Microbiol Infect. 2015;21(suppl 2):S27–S32. doi: 10.1016/j.cmi.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 23.Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 24.Wang XF, Shi GC, Wan HY, et al. Clinical features of three avian influenza H7N9 virus-infected patients in Shanghai. Clin Respir J. 2014;8:410–416. doi: 10.1111/crj.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu WJ, Zhao M, Liu K, et al. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]