Abstract
Mutations in the Parkinson's disease (PD)-associated protein leucine-rich repeat kinase 2 (LRRK2) commonly lead to a reduction of GTPase activity and increase in kinase activity. Therefore, strategies for drug development have mainly been focusing on the design of LRRK2 kinase inhibitors. We recently showed that the central RocCOR domains (Roc: Ras of complex proteins; COR: C-terminal of Roc) of a bacterial LRRK2 homolog cycle between a dimeric and monomeric form concomitant with GTP binding and hydrolysis. PD-associated mutations can slow down GTP hydrolysis by stabilizing the protein in its dimeric form. Here, we report the identification of two Nanobodies (NbRoco1 and NbRoco2) that bind the bacterial Roco protein (CtRoco) in a conformation-specific way, with a preference for the GTP-bound state. NbRoco1 considerably increases the GTP turnover rate of CtRoco and reverts the decrease in GTPase activity caused by a PD-analogous mutation. We show that NbRoco1 exerts its effect by allosterically interfering with the CtRoco dimer–monomer cycle through the destabilization of the dimeric form. Hence, we provide the first proof of principle that allosteric modulation of the RocCOR dimer–monomer cycle can alter its GTPase activity, which might present a potential novel strategy to overcome the effect of LRRK2 PD mutations.
Keywords: allosteric regulation, GTPases, leucine-rich repeat kinase, Nanobody
Introduction
Roco proteins form a family of large multi-domain proteins, that are characterized by the occurrence of a RocCOR supra-domain consisting of a Ras of complex proteins (Roc) GTPase domain, invariably followed by a C-terminal of Roc (COR) domain [1,2]. The characteristic RocCOR domain module is never found in isolation and is most often preceded by a leucine-rich repeat (LRR) domain, while many representatives of the Roco family also possess other catalytic and protein–protein interaction domains [1,3]. In human, 4 Roco family members are present: MFHAS1 (malignant fibrous histiocytoma amplified sequence 1), LRRK1 and LRRK2 (leucine-rich repeat kinase 1 and 2) and DAPK1 (death-associated protein kinase 1) [4–8]. Mutations in the genes coding for all of these proteins have been linked to human disease, including various types of cancer and neurological disorders [9–11].
The interest in the Roco protein family was especially triggered by the discovery that mutations in the lrrk2 gene are the most common cause of autosomal-dominant Parkinson's disease (PD), while lrrk2 gene variants have also been associated with the idiopathic forms of PD [12–18]. LRRK2 is a very large (2527 amino acids) and complex Roco protein bearing, next to the RocCOR domains and various protein–protein interaction domains, also a Ser/Thr protein kinase domain. Recent results have uncovered several Rab GTPases as the physiological substrates of LRRK2 kinase activity [19–21]. PD-associated mutations in LRRK2 are mainly located in the catalytic RocCOR and kinase domains, and most of them seem to behave as gain of function mutations that lead to an increase in kinase activity and/or a decrease in GTPase activity [22–28]. Considering the increase in kinase activity in the most prevalent PD mutation (G2019S), large efforts have been devoted toward the design of LRRK2 protein kinase inhibitors [29–31]. However, increasingly more studies have shown the importance of the RocCOR domains in LRRK2 functioning, and propose the targeting of these domains as an alternative strategy [32–34].
Although the fine details of the very complex Roco GTPase cycle are not yet completely understood [35], we recently showed, using the Roco protein from the bacterium Chlorobium tepidum (CtRoco), that the RocCOR domain module undergoes a dimer–monomer cycle concomitant with GTP binding and hydrolysis [36,37]. We found that the protein is mainly dimeric in the nucleotide-free state and monomeric in the GTP-bound state, while an intermediate situation occurs in the GDP-bound state. Moreover, an analog of a PD-associated mutation in CtRoco (L487A) decreased the GTPase activity by stabilizing the RocCOR in its dimeric form [36]. While CtRoco shares the central LRR-RocCOR arrangement with LRRK2, but lacks the kinase and some other domains, these results are in line with findings in LRRK2. LRRK2 is mainly purified as a dimeric species, but in vivo studies show that the protein predominantly occurs as a monomeric species with low kinase activity in the cytosol and as a dimeric species with high kinase activity at the membrane [38–42]. Very recently two studies confirmed that also the Roc GTPase domain of human LRRK2 exists in a dynamic dimer–monomer equilibrium, and the important PD mutations R1441G/C/H and N1473H lead to a decrease in GTPase activity by changing this equilibrium [43,44]. Together, these finding illustrate the link between deregulation of the RocCOR GTPase cycle and PD, and thus suggest that modulating the RocCOR dimer–monomer cycle could be a promising approach to overcome the detrimental effect of LRRK2 PD mutations [34,45].
In this study, we report the generation and characterization of Nanobodies (Nbs), the variable domains of camelid heavy chain-only antibodies, that bind the Roco protein from C. tepidum in a conformation-specific way. One Nanobody (NbRoco1) binds the GTP- and GDP-bound states of the RocCOR domain of CtRoco with high affinity, while no binding is observed to the RocCOR in its nucleotide-free state. Another Nanobody (NbRoco2) shows preferential binding to the GTP-bound state over the GDP-bound and nucleotide-free states, while it binds CtRoco via its LRR domain. NbRoco1 increases the turnover rate (kcat) of CtRoco more than four-fold and is able to completely rescue the decrease in GTPase activity caused by the PD-analogous L487A mutation. Analysis by stopped-flow and single-molecule fluorescence resonance energy transfer (FRET) and native mass spectrometry (MS) shows that NbRoco1 exerts its effect by allosterically interfering with the CtRoco dimer–monomer cycle through destabilization of the dimeric form. Together these results show that allosteric modulation of the RocCOR dimer–monomer cycle can alter its GTPase activity.
Experimental procedures
Protein expression and purification
The CtLRR domain (a.a. 1–441) of CtRoco was cloned into the pGEX4T1 plasmid with an N-terminal GST-tag [46]. BL21(DE3) cells were grown at 37°C in Terrific Broth (TB) medium supplemented with 100 µg/ml ampicillin. When an OD600 of 0.7 was reached, expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and expression proceeded overnight at 25°C. The cells were collected by centrifugation and resuspended in buffer A (30 mM Tris/HCl pH 7.5, 150 mM NaCl, 3 mM 2-mercapto-ethanol, 5% glycerol) with 1 µg/ml leupeptin, 0.1 mg/ml AEBSF and 50 µg/ml DNAse. After lysis of the cells with a cell disruptor (Constant Systems), the cell lysate was centrifuged and the supernatant loaded onto glutathione sepharose resin beads equilibrated with buffer A. After extensive washing, another washing step was applied with buffer A supplemented with 300 mM KCl and 1 mM ATP. 1 mg His-tagged TEV protease per 10 mg of protein was added to the beads to cleave off the GST-tag overnight at 4°C. The LRR protein was eluted and was subsequently applied to Ni2+-NTA beads to remove the His-tagged TEV protease. The protein was collected in the flow through. As a final purification step the protein was applied to a Superdex 75 16/90 column (GE Healthcare) equilibrated in buffer consisting of 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2 and 5% glycerol.
The CtRoco (a.a. 1–1102) and CtRocCOR (a.a. 412–946) proteins were produced with an N-terminal His-tag from the pProEX plasmid in Escherichia coli BL21(DE3) cells, as previously described [36,47]. The purification protocol consisted of a Ni2+-NTA immobilized metal affinity chromatography (IMAC) step. Subsequently, the protein was dialyzed against 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM DTT, and after dialysis 1 mM EDTA was added to the protein to remove Mg2+ and disrupt nucleotide binding. Finally, the sample was applied to a Superdex S200 26/60 size exclusion chromatography column (GE Healthcare) equilibrated with the same buffer. After gel filtration 5 mM MgCl2 was added to the protein sample and analytical reversed-phase chromatography was used to confirm the complete removal of nucleotides, as described previously [36]. These nucleotide-free proteins allow subsequent uniform loading with nucleotides of choice and were used in all experiments.
Nanobody generation and purification
Two llamas were immunized with either CtRoco (a.a. 1–1102) or CtRocCOR (a.a. 412–946) in the presence of a large excess (1 mM) of the non-hydrolysable GTP analog 5′-guanylyl imidodiphosphate (GppNHp). A 6-week immunization protocol was followed consisting of weekly immunizations of 200 µg (first 2 weeks) or 100 µg (last 4 weeks) protein in the presence of GERBU adjuvant. All animal vaccinations were performed in strict accordance with good practices and EU animal welfare legislation. Blood was collected 4 days after the last injection. Library construction, Nb selection via phage display and Nb expression and purification were performed as described previously [48], with slight adaptations in order to maximize chances to select Nbs that bind specifically to the GppNHp-bound CtRoco protein. Briefly, the variable domains of the heavy-chain antibody repertoire from each llama were subcloned in a pMESy4 phage display vector, which adds a C-terminal His6-tag and EPEA-tag (= CaptureSelectTM C-tag). This resulted in two independent immune libraries of 5 × 108 transformants and of 6 × 108 transformants from the immunizations with CtRocCOR and CtRoco, respectively. This Nb repertoire was expressed on phage after rescue with the VCSM13 helper phage, and two consecutive rounds of phage display were performed to select for phage expressing Nbs that bind to GppNHp-bound CtRoco/CtRocCOR. Therefore, biotinylated CtRoco/CtRocCOR was captured on neutravidine-coated 96-well plates and all binding and washing steps were performed in a buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.05% Tween20) containing 100 µM GppNHp. After selections, independent clones were chosen for sequence analysis to classify the resulting Nb clones in sequence families.
Subsequently, an ELISA screen was performed on crude cell lysates of E. coli cells expressing representatives of each Nb family (and in a later stage on some purified Nbs), in order to confirm binding and to determine the domain and conformational specificity. In a first ELISA experiment CtRoco either in the nucleotide-free state or loaded with 1 mM GDP or GppNHp was coated on the ELISA plate, while in a second ELISA experiment CtRoco, CtRocCOR and CtLRR were coated (in the presence of 1 mM GppNHp). Incubation with the Nb-containing cell extracts and all washing steps were performed in 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.05% Tween20 supplemented with 200 µM of the relevant nucleotide. Binding of the Nbs was detected via their EPEA-tag using a mixture of 1 : 4000 CaptureSelect™ Biotin anti-C-tag conjugate (Thermo Fischer Scientific) and 1 : 1000 Streptavidin Alkaline Phosphatase (Promega). The color was developed by adding 100 µl of a 3 mg/ml disodium 4-nitrophenyl phosphate solution (DNPP, Sigma–Aldrich) and measured at 405 nm.
For Nb production and purification, pMESy4 vectors containing the Nb open reading frames were transformed in non-suppressor E. coli WK6 cells. Cells were grown at 37°C in TB medium and were induced with 1 mM IPTG, after which expression was allowed to proceed overnight at 28°C. Cells were harvested via centrifugation and subjected to an osmotic shock to obtain the periplasmic extract. Subsequently, an affinity purification step on Ni2+-NTA sepharose and a size exclusion chromatography step on a Superdex 75 26/60 column (in buffer 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol) was used to purify the Nbs.
Analytical gel filtration analysis
Prior to mixing with Nb, different CtRoco protein constructs (CtRoco, CtRocCOR, CtLRR) were incubated in the presence of 1 mM GppNHp for 1 h on ice. Subsequently, 50 µM Nb and 75 µM of the different CtRoco protein constructs were mixed in the presence of 1 mM GppNHp in a buffer consisting of 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol in a total volume of 50 µl. Samples were injected on a Shodex KW-803 gel filtration column coupled to a Waters Alliance e2695 HPLC system and run with the same buffer containing 100 µM GppNHp. Runs of the Nbs or the CtRoco constructs alone were used as controls.
Sortase A-mediated site-specific fluorescent labeling of Nbs
To determine the affinity of the Nbs for CtRoco using fluorescence anisotropy titrations, Nbs were site-specifically labeled at their C-terminus with a FITC fluorophore using Sortase A-mediated labeling [49]. Therefore, the Nb-coding open reading frames were recloned to a pHEN29 vector which adds the Sortase-recognition sequence LPETG in front of the C-terminal His6-tag and EPEA-tag. The production and purification of the resulting Nbs was performed as described above. The C-terminal labeling of the Nanobodies with the FITC fluorophore was performed as described previously [49]. In short, 50 µM of Nb produced from the pHEN29 vector was incubated with 150 µM in-house produced Sortase A enzyme and 1.5 mM NH2-GGGK-(FITC)-COOH peptide (GenicBio) in a buffer consisting of 50 mM Tris/HCl pH 7.5, 150 mM NaCl and 10 mM CaCl2 in a total volume of 1.5 ml. The Sortase A-mediated labeling reaction results in an exchange of the His6- and EPEA-tag for the FITC-labeled peptide. After overnight incubation at 37°C, a Ni2+-NTA IMAC purification step was performed to remove the His-tagged Sortase A enzyme and the unreacted (His-tagged) Nbs. The excess of free FITC-labeled peptide was removed by a gel filtration on a Superdex 75 10/30 column (GE Healthcare).
Fluorescence anisotropy titrations
Fluorescence anisotropy titrations with FITC-labeled Nbs and CtRoco were carried out with a LS 55 Fluorescence Spectrometer (PerkinElmer) at an excitation wavelength of 493 nm, an emission wavelength of 516 nm and 25°C. Fifty nanomolars of FITC-labeled Nb with or without 100 µM GppNHp or GDP in 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol was titrated with increasing amounts of CtRoco. The anisotropy signal was measured in function of time using a 5 min interval between each titration step. The anisotropy signal of the last 2 min was averaged and plotted against the corresponding CtRoco concentration. All experiments were performed in triplicate. To obtain Kd values (± standard error), the data were described with the quadratic binding equation using GraphPad Prism 7.
GTP hydrolysis assays
For the multiple turnover steady-state kinetic experiments, 1 µM of CtRoco either in the absence or presence of 100 µM of Nb was incubated with different concentrations of GTP in 20 mM HEPES/NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol at 25°C. After 0, 30, 60, 90 and 120 min, samples of 40 µl were taken and the reaction was stopped by incubation at 95°C for 3 min. After centrifugation, 35 µl of supernatant was mixed with 35 µl of 100 mM KH2PO4 pH 6.4, 10 mM tetrabutylammonium bromide, 7.5% acetonitrile solution. Fifty microliters was injected on a C18 reversed-phase column (Phenomenex, Jupiter 5 µm) using the latter solution as a mobile phase. The area under the 254 nm absorption peak of GDP was converted to a concentration by using a standard curve. All experiments were performed in triplicate and data were fitted on the Michaelis–Menten equation using GraphPad Prism 7.
For the single turnover kinetic experiments, 5 µM of CtRoco L487A mutant or wild type were incubated with 5 µM of GTP, either in the presence or absence of 100 µM of NbRoco1 or in the presence of NbRoco3 as a control. Samples of 100 µl were taken after time points ranging from 0 to 300 min and the samples were analyzed in the same way as for the multiple turnover experiments. All experiments were performed in triplicate and data were fitted with a single exponential using GraphPad Prism 7.
Stopped-flow and single-molecule fluorescence resonance energy transfer (FRET) experiments
A S928C mutation was introduced into a cysteine-free CtRocCOR construct by QuikChange mutagenesis as described previously [36,50]. For (ensemble) stopped-flow FRET experiments, the unique cysteine at position 928 was labeled with a mixture of Sulfo-Cy3 and Sulfo-Cy5 maleimide (Lumiprobe) as previously described [36]. To follow monomerization 0.2 μM Cy3/Cy5-labeled protein was mixed with 100 μM nucleotide (GDP, GppNHp or buffer) in a stopped-flow apparatus (Applied Photophysics), either in the presence or absence of 100 µM of NbRoco1 at 25°C. The Cy3 fluorophore was excited at 548 nm and change in Cy5 emission was monitored using a cut-off filter of 645 nm.
For the single-molecule FRET (smFRET) experiments, the CtRocCOR S928C mutant was labeled with a two-fold molar excess of Alexa555-maleimide (A555, donor) and a five-fold molar excess of Alexa 647-maleimide (A647, acceptor) for 2 h at 25°C in 20 mM HEPES/KOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2 and 5% glycerol in the presence of 1 mM TCEP. Labeling efficiencies were determined from the absorption at 647 nm (A647), 554 nm (A555, corrected for the contribution of A647) and 280 nm (protein, corrected for contributions of A555 and A647). smFRET experiments were performed with 150 pM (donor concentration) of CtRocCOR in the presence of 1 mM DTT, 2 mM nucleotide and 100 µM NbRoco1 at 25°C. Measurements were performed with a Picoquant Microtime 200 confocal microscope equipped with two pulsed diode lasers (LDH-P-FA 530, LDH-D-C-640; pulse duration 100 ps) for pulsed interleaved excitation (PIE), using a 60× water-immersion objective (UPlanAPO NA 1.2; Olympus), a dichroic beam splitter DM505 (Olympus), beam splitters z532/633rpc and 620 dcxr, filters HQ 580/70 (donor) and HQ M690/70 (acceptor) and two SPAD detectors (τ-SPAD). Data were processed with the program SymPhoTime 64 (Picoquant).
Native mass spectrometry
To remove traces of glycerol from the buffer, CtRocCOR, the CtRocCOR L487A mutant and NbRoco1 were dialyzed two times for 2 h against 20 mM Tris/HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2. Eleven micromolars of either CtRocCOR or the CtRocCOR L487A mutant was incubated overnight at 4°C with 500 µM of GDP, GppNHp or in the absence of nucleotides, in the presence and absence of 22 µM of NbRoco1. The next day, the buffer was exchanged to 150 mM ammonium acetate pH 7.5, 200 µM MgCl2 and 100 µM of the corresponding nucleotide using Micro Bio-spin columns (Bio-gel P6, Bio-rad). After buffer exchange, the samples were immediately analyzed on a Q-TOF2 instrument (Waters) which is modified for the transmission of macromolecular assemblies [51]. The samples were introduced into the mass spectrometer using nano-ESI with in-house made gold-coated borosilicate capillaries. The capillary voltage ranged between 1.5 and 1.8 kV and the sample and extractor cone were set to 50 V and 10 V, respectively. The collision energy was fixed to 50 V. Gas pressures were 10 and 0.01 mbar for the backing and collision gas, respectively.
Results
Generation and identification of conformation-specific Nanobodies targeting different CtRoco domains
Nanobodies (Nbs) are small (±15 kDa) monomeric single-domain proteins derived from camelid heavy-chain antibodies, that harbor the full antigen-binding capacity of the parental antibody [52,53]. Often, their antigen-recognition sites form a convex surface with a long CDR3 (complementary-determining region) loop that can protrude into cavities of their target epitopes [54]. These unique properties make Nbs exquisite tools for examining dynamic biological systems, and in particular to lock their antigens in a specific conformation [55].
We set out to generate Nbs that specifically recognize the bacterial LRRK2 homolog CtRoco in a GTP-bound conformation. To this end, we immunized two llamas with either the full-length CtRoco protein (CtRoco, amino acids 1–1102) or its RocCOR construct (CtRocCOR, amino acids 412–946) (Supplementary Figure S1). To increase the chances for identifying Nbs specifically recognizing the trinucleotide-bound state of the CtRoco protein, we completely loaded the proteins with GppNHp and added a large excess (1 mM) of the same nucleotide to the protein prior to immunization. The blood lymphocytes from the two immunized llamas were used to construct two independent Nb phage libraries, and two rounds of panning with these two libraries resulted in the identification of a total of 65 Nb families with a unique CDR3 sequence. For the pannings, either biotinylated CtRoco or CtRocCOR were captured on Streptavidin-coated ELISA plates, and an excess of GppNHp was maintained in all binding and washing steps. Subsequently, representatives of the Nb families were expressed in a small scale in the periplasm of E. coli, and the resulting crude cell extracts were tested for binding in two independent ELISA experiments. In a first ELISA, binding of the Nbs was tested on full-length CtRoco either in the nucleotide-free state or bound to (an excess of) GDP or GppNHp. In a second ELISA, binding to different CtRoco constructs was compared (CtRoco, CtRocCOR and CtLRR (amino acids 1–441)). Based on the conformational and domain specificity we decided to characterize the properties of two Nbs in detail: Nb6946 (hereafter called NbRoco1) and Nb8175 (hereafter called NbRoco2) (Supplementary Figure S2). This choice was based on the initial findings in ELISA that NbRoco1 showed a specificity for the GppNHp-bound CtRoco, and that NbRoco2 showed binding to CtRoco, while no or only weak binding to the LRR and RocCOR domains was observed in ELISA (Supplementary Figure S3).
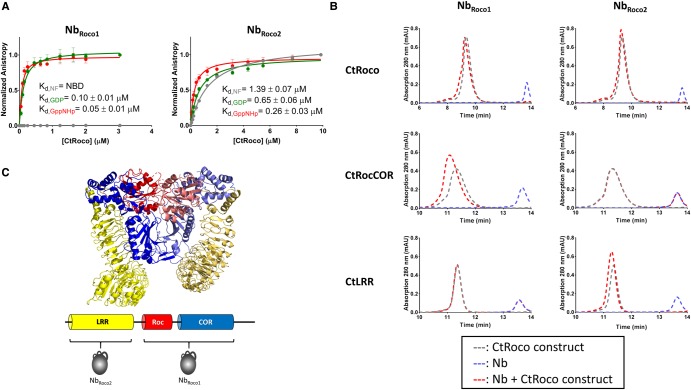
First, we determined the conformational specificity of NbRoco1 and NbRoco2. After purification, both Nbs were C-terminally coupled to a FITC fluorophore using Sortase-mediated labeling, and binding to CtRoco in its different nucleotide-bound states was followed using fluorescence anisotropy. To determine the binding affinities, a fixed amount of FITC-labeled Nb was titrated with increasing concentrations of CtRoco either in the absence of nucleotides, or in the presence of 0.1 mM of GDP or GppNHp (Figure 1A). In agreement with our initial ELISA screening, these data show a clear conformational specificity of NbRoco1. While no binding of NbRoco1 to CtRoco in its nucleotide-free state is observed up to the highest concentrations tested, a high affinity (Kd = 0.05 ± 0.01 µM) is found in the presence of GppNHp. For GDP-bound CtRoco the affinity is intermediate (Kd = 0.10 ± 0.01 µM), showing that NbRoco1 displays preference toward the trinucleotide-state of its target protein. A certain degree of conformational specificity is also observed for NbRoco2, albeit less pronounced (Figure 1A). Also here the highest affinity is observed for the GppNHp-bound state of CtRoco (Kd = 0.26 ± 0.03 µM), with a gradually decreasing affinity for the GDP-bound state (Kd = 0.65 ± 0.06 µM) and the nucleotide-free state (Kd = 1.39 ± 0.07 µM).
Figure 1. NbRoco1 and NbRoco2 are conformation-specific Nbs targeting different domains of CtRoco.
(A) Influence of the nucleotide-bound state of CtRoco on the affinity for NbRoco1 (left) and NbRoco2 (right) assessed by fluorescence anisotropy titrations. The fluorescence anisotropy signal of the FITC-labeled Nbs is monitored upon titration with increasing concentrations of CtRoco in the nucleotide-free state (gray), or bound to GDP (green) or GppNHp (red). The corresponding equilibrium dissociation constants (Kd ± standard error) obtained by fitting with a quadratic binding equation are given (each data point is the average of three independent measurements with the error bars representing the standard deviation; NBD = no binding detectable at the concentrations used). (B) Analysis of the domain specificity of NbRoco1 (left) and NbRoco2 (right) using analytical size exclusion chromatography. NbRoco1 and NbRoco2 were mixed with a small molar excess of the CtRoco protein or either of its constituting domains CtRocCOR or CtLRR, and samples were analyzed on analytical size exclusion chromatography (chromatograms shown as red dotted lines). These elution profiles are compared with the elution profiles of either the Nbs or the CtRoco constructs individually (blue and gray dotted lines, respectively). Binding of the Nbs to CtRoco and to either of its constituting domains, is reflected by a shift of the elution peak corresponding to the CtRoco construct (red vs. gray curve) and a disappearance of the peak corresponding to the Nb elution volume (blue curve). (C) Cartoon representation of the X-ray crystal structure of the homodimer of the LRR-RocCOR construct of CtRoco with the LRR domains shown in yellow, the Roc domains in red and the COR domains in blue. Corresponding domains from adjacent subunits in the homodimer are shown in different shades (PDB code 6HLU) [37]. Below, the domain arrangement of a CtRoco subunit is shown schematically with the binding epitopes of NbRoco1 and NbRoco2 indicated.
Next, using analytical size exclusion chromatography, we established the binding epitopes of NbRoco1 and NbRoco2. Therefore, a 1.5-fold molar excess of CtRoco or its constituting domain constructs, CtRocCOR and CtLRR, were mixed with NbRoco1 or NbRoco2 in the presence of GppNHp and the elution chromatograms were compared with those of the individual proteins (note that the isolated Roc and COR domains of CtRoco cannot be stably expressed individually) (Figure 1B). This analysis shows that NbRoco1 binds to both CtRoco and CtRocCOR, while no binding is observed with CtLRR, thus confirming the results of the initial ELISA. However, in contrast with what was observed in the (crude) initial ELISA (Supplementary Figure S3), NbRoco2 binds to both CtRoco and CtLRR, while no binding is observed to CtRocCOR.
Together, these results show that we were able to generate Nbs that bind to CtRoco with an affinity that depends on the nucleotide state of the protein. Interestingly, while NbRoco1 does this via direct interaction with the RocCOR supra-domain, NbRoco2 binds to the N-terminal LRR domain (Figure 1C). The observation that binding of NbRoco2 to the LRR domain is affected by the nucleotide state of CtRoco indicates some form of nucleotide-induced conformational communication between the LRR and RocCOR domains within CtRoco.
NbRoco1 and NbRoco2 increase the GTPase activity of CtRoco and NbRoco1 reverts the phenotype of the PD-analogous L487A mutation
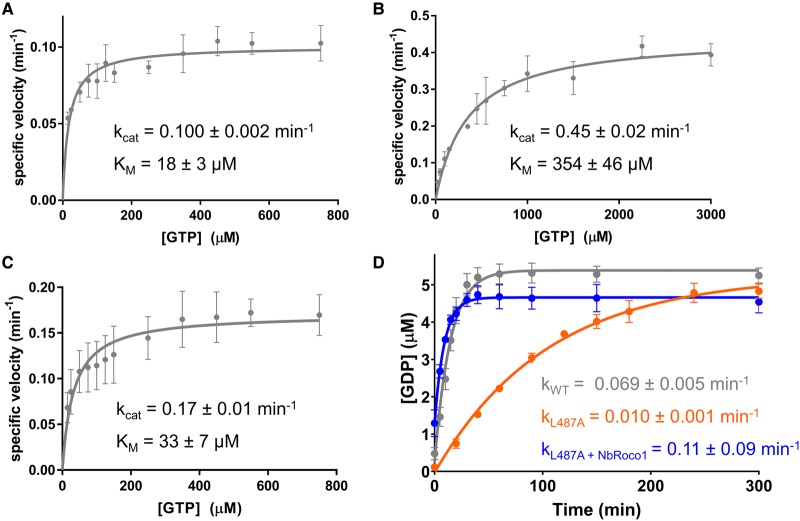
Several studies have indicated that the most common PD mutations in the RocCOR domain of human LRRK2 lead to a decreased GTPase activity, typically by a factor of about two- to four-fold [23,25,43,44,56–58]. Since NbRoco1 and NbRoco2 bind CtRoco in a conformation-specific way, we, therefore, wondered whether these Nbs could influence its GTPase activity. To this end, we determined the steady-state kinetic parameters (kcat and KM) of GTP hydrolysis by CtRoco at 25°C, in either the absence or presence of an excess of NbRoco1 or NbRoco2 (Figure 2A–C). We previously reported that Roco proteins, including CtRoco, follow a complex multi-step GTPase cycle with KM values that are typically much higher than the Kd values for GTP binding, and displaying low GDP affinities caused by high GDP dissociation rates [35]. The steady-state kinetic parameters of CtRoco in the absence of Nb determined here (kcat = 0.100 ± 0.002 min−1; KM = 18 ± 3 µM) are in good agreement with our previous results (Figure 2A) [35]. Remarkably, in the presence of an excess of NbRoco1, the GTP turnover (kcat) of CtRoco is increased more than four-fold, while also the KM value increases (kcat = 0.45 ± 0.02 min−1; KM = 354 ± 46 µM) (Figure 2B). Considering the high cellular GTP concentrations (ranging from 1 to 1.6 mM) [59,60], kcat is most probably the physiologically relevant kinetic parameter, while changes in KM are much harder to interpret for enzymes with complex mechanisms such as CtRoco [61]. In line with NbRoco2 showing a less pronounced conformational specificity compared with NbRoco1, we also observe a less pronounced effect of this Nb on CtRoco GTPase kinetics, with NbRoco2 leading to a less than two-fold increase in the kcat value (kcat = 0.17 ± 0.01 min−1, KM = 33 ± 7 µM) (Figure 2C). As a control, we also measured the effect of Nb9221 (hereafter called NbRoco3) on the GTPase activity of CtRoco. This Nb was selected from the initial ELISA experiment as a non-conformation-specific Nb (Supplementary Figure S4A,B), which was also confirmed using fluorescence anisotropy titrations (Supplementary Figure S4C). In agreement with the lack of conformational specificity, the addition of an excess of NbRoco3 to CtRoco does not considerably influence the GTPase activity (kcat = 0.088 ± 0.004 min−1, KM = 18 ± 4 µM) (Supplementary Figure S4D).
Figure 2. The conformation-specific Nbs increase the GTPase activity of CtRoco and its L487A PD-analogous mutant.
(A–C) Steady-state Michaelis–Menten kinetics of CtRoco alone (A), or in the presence of an excess of NbRoco1 (B) or NbRoco2 (C). The rate of GDP production in function of time was analyzed via reversed-phase HPLC, and specific velocities (initial rate divided by enzyme concentration) were plotted against the GTP concentration. kcat and KM (± standard error) values resulting from fitting with the Michaelis–Menten equation are shown. Each data point is the average of three independent measurements with the error bars representing the standard deviation. (D) Effect of NbRoco1 on the single turnover GTP hydrolysis kinetics of the CtRoco L487A mutant. 5 µM of CtRoco (gray) or the CtRoco L487A mutant either in the absence (orange) or presence (blue) of an excess of NbRoco1 was mixed with 5 µM of GTP, and production of GDP was followed in time. The observed rate constants (k ± standard error) obtained by fitting on a single exponential equation are shown. Each data point is the average of three independent measurements with the error bars representing the standard deviation.
Since NbRoco1 has a profound effect on the CtRoco GTPase activity, we wondered whether this Nb would be able to revert the decrease in GTPase activity of a PD-analogous mutant. Therefore, we turned to the CtRoco L487A mutant, which is analogous to the PD risk variant I1371V in human LRRK2 [37], and measured the CtRoco GTPase activity under single turnover condition by mixing 5 µM CtRoco with 5 µM of GTP and following the production of GDP over time (Figure 2D). Under these conditions, the L487A mutation causes a nearly seven-fold decrease in GTPase activity compared with the wild-type protein (kobs (CtRoco wild-type) = 0.069 ± 0.005 min−1; kobs (CtRoco L487A) = 0.010 ± 0.001 min−1). Interestingly, the addition of an excess of NbRoco1 increases the observed single turnover rate of CtRoco L487A more then 10-fold (kobs = 0.11 ± 0.09 min−1), hence completely nullifying the effect of the PD-analogous mutation and even restoring the GTPase activity beyond that of the wild-type enzyme. In contrast, the non-conformation-specific control Nb, NbRoco3, seems to have only very little effect on the GTPase activity of CtRoco L487A (kobs = 0.014 ± 0.001 min−1; Supplementary Figure S4E). We can thus conclude that our Nbs increase the GTPase activity of CtRoco and have the potential to revert the detrimental effect of PD-analogous mutations. This effect on GTPase activity seems to be coupled directly to the extent of their conformational specificity.
NbRoco1 destabilizes the CtRoco homodimer
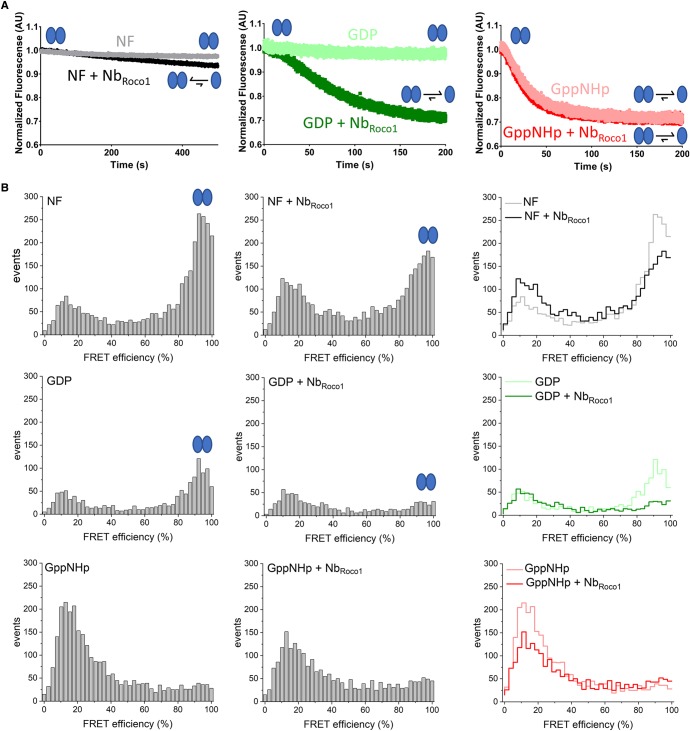
We previously found that CtRoco needs to cycle between a dimeric and monomeric form during GTP binding and hydrolysis, and that the PD-analogous L487A mutation slows down the GTPase activity by stabilizing and blocking the enzyme in its dimeric form [36]. Since we show here that NbRoco1 is able to completely revert the effect of the L487A mutation on GTP hydrolysis, we next investigated whether NbRoco1 influences the CtRoco monomer/dimer equilibrium. To this end, we used both an ensemble and smFRET approach making use of a variant of the CtRocCOR construct containing a single cysteine residue at position 928 of the COR domain (CtRocCOR S928C), which was subsequently site-specifically labeled with a donor/acceptor FRET pair. As previously shown, the decrease in FRET signal of this molecule can be used as a proxy for monomerization [36].
In a first time-resolved ensemble FRET approach, 0.2 µM of a Cy3/Cy5 labeled nucleotide-free CtRocCOR protein was rapidly mixed in the stopped-flow apparatus with either buffer or with 100 µM of GDP or GppNHp, and this in either the presence or absence of an excess of NbRoco1 (Figure 3A). In the absence of NbRoco1 CtRocCOR remains mainly dimeric in the nucleotide-free (NF) and GDP-bound states, while mixing with an excess of GppNHp leads to monomerization, in agreement with our previous results [36]. However, the addition of NbRoco1 clearly induces the monomerization of GDP-bound CtRocCOR. A small decrease in FRET signal is also observed for the nucleotide-free protein in the presence of (an excess of) NbRoco1.
Figure 3. Binding of NbRoco1 induces (nucleotide-dependent) monomerization of CtRoco.
(A) Influence of NbRoco1 on the stopped-flow FRET traces of labeled CtRocCOR upon mixing with different nucleotides to follow monomerization over time. The FRET signal of Cy3/Cy5-labeled CtRocCOR (S928C) over time is shown after rapid mixing with buffer (left panel), or 100 μM GDP (middle panel) or GppNHp (right panel), either in the absence or presence of an excess of NbRoco1. Monomerization is observed through a decrease in FRET signal. (B) Single-molecule FRET histograms for A555/A647-labeled CtRocCOR in the absence of nucleotide, and in the presence of GDP or GppNHp, and in the absence (left panels) and presence of NbRoco1 (center panels). The right panels show the superposition of the two individual histograms. The population of dimeric CtRocCOR (90% FRET efficiency) decreases in the presence of NbRoco1, indicating monomerization. The GppNHp-bound state of CtRocCOR (lower panel) is predominantly monomeric in the absence and presence of NbRoco1. Signals below 20% FRET efficiency belong to molecules carrying only a donor dye whose signals are not completely suppressed by PIE.
Next, we performed smFRET experiments to look at the effect of NbRoco1 on the CtRocCOR dimer/monomer distribution, using the CtRocCOR S928C mutant labeled with an Alexa555/Alexa647 FRET pair (Figure 3B). In the absence of NbRoco1, the FRET histogram for CtRocCOR in the nucleotide-free (NF) state showed a prominent peak at FRET efficiencies ∼0.9–1.0, again indicative of the presence of a CtRoco dimer. In agreement with previous results [36] and the ensemble stopped-flow FRET measurements, and considering the low protein concentration used in this experiment (150 pM donor concentration), this dimer peak decreases in the presence of GDP and vanishes in the presence of GppNHp (Figure 3B, left column). Like in the stopped-flow FRET experiment, the addition of NbRoco1 leads to a nearly complete dimer to monomer conversion of CtRocCOR in the presence of GDP, while the dimer fraction also decreases in case of the nucleotide-free protein (Figure 3B, middle and right column).
Thus, together these ensemble and smFRET experiments show that NbRoco1 is increasing the GTPase activity of CtRoco by destabilizing the dimeric form of the protein.
NbRoco1 allosterically modulates the CtRoco dimer–monomer cycle
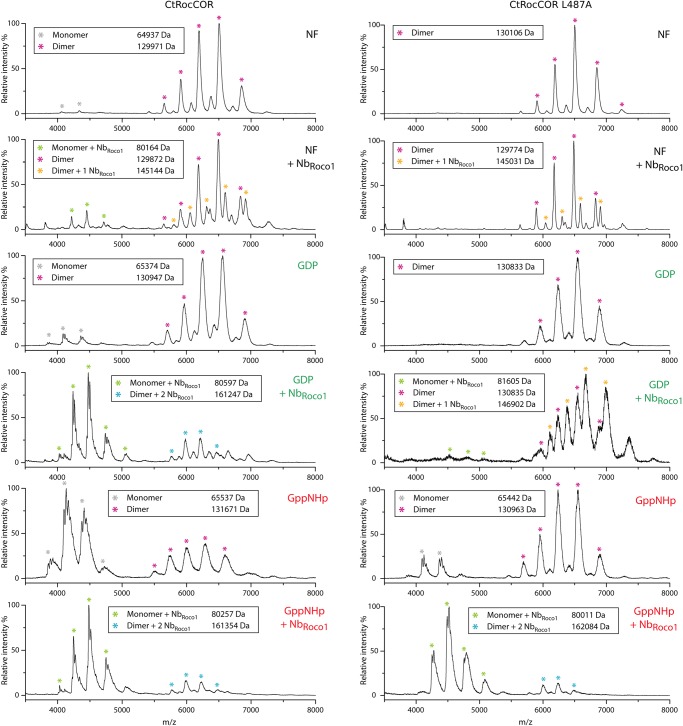
Since our FRET data show that NbRoco1 destabilizes the CtRoco dimer in the GDP and nucleotide-free states, we subsequently used native mass spectrometry (native MS) to directly probe the monomeric and dimeric population of CtRocCOR in the presence and absence of NbRoco1 and in different nucleotide-bound states (nucleotide-free, GDP and GppNHp; Figure 4 and Supplementary Figure S5). The results of these experiments in the absence of NbRoco1 confirm our previous data [36], showing again that CtRocCOR is mainly dimeric in the nucleotide-free state (dimer/monomer ratio ∼97%/3%) and GDP-bound state (dimer/monomer ratio ∼90%/10%), while GppNHp binding leads to a shift toward the monomeric species (dimer/monomer ratio ∼38%/62%). The addition of an excess of NbRoco1 causes an overall shift toward the monomeric species which, in agreement with the FRET data, is most prominent for CtRocCOR in the GDP-bound state (dimer/monomer ratio ∼26%/74%). However, also for CtRocCOR in the nucleotide-free state (dimer/monomer ratio ∼87%/13%) and GppNHp-bound state (dimer/monomer ratio ∼20%/80%) NbRoco1 induces a shift of the dimer–monomer equilibrium toward the monomeric species.
Figure 4. NbRoco1 allosterically modulates the CtRoco dimer/monomer equilibrium.
Native mass spectra of CtRocCOR (left) and CtRocCOR L487A (right) either in the absence or in the presence of a two-fold molar excess of NbRoco1 and for three different nucleotide states: nucleotide free (NF), GDP-bound and GppNHp-bound, as indicated in each mass spectrum. The peaks corresponding to CtRocCOR monomers and dimers either bound to one or two molecules of NbRoco1 are indicated by differently colored * symbols, as indicated, and the corresponding experimental molecular masses are given (Theoretical molecular masses for CtRocCOR: monomer: 64866 Da; dimer: 129 732 Da; monomer + 1 NbRoco1: 80 053 Da; dimer + 1 NbRoco1: 144 919 Da; dimer + 2 NbRoco1: 160 106 Da. Theoretical molecular masses for CtRocCOR L487A: monomer: 64 824 Da; dimer: 129 648 Da; monomer + 1 NbRoco1: 80 011 Da; dimer + 1 NbRoco1: 144 835 Da; dimer + 2 NbRoco1: 160 022 Da). See also Supplementary Figure S5 for the number of charges on the monomer and dimer peaks.
Since NbRoco1 is able to revert the effect of the L487A mutation on GTPase activity (see above), we subsequently looked at the influence of this Nb on the CtRocCOR L487A dimer–monomer equilibrium. In agreement with our previous multi-angle light scattering experiments [36], the native MS data show that the L487A mutation leads to a stabilization of the CtRocCOR dimer for all nucleotide states but most prominently for the GppNHp-bound state, with dimer / monomer ratios ∼100%/0%, 100%/0% and 85%/15%, for nucleotide-free, GDP-bound and GppNHp-bound CtRocCOR, respectively (Figure 4 and Supplementary Figure S5). Correspondingly, the addition of a two-fold molar excess of NbRoco1 is not sufficient to induce a considerable shift of the dimer–monomer equilibrium of CtRocCOR L487A in its nucleotide-free or GDP-bound state, although a small shift toward the monomeric state is observed for the GDP state (dimer/monomer ratio ∼96%/4%). However, in the GppNHp-bound state addition of NbRoco1 causes a very pronounced shift toward the monomeric form (dimer/monomer ratio ∼11%/89%, compared with 85%/15% without NbRoco1), thus completely reverting the dimer-stabilizing effect of the L487A mutation.
Furthermore, the accuracy and resolution provided by native MS allows us to look directly at the binding of NbRoco1 to the monomeric and dimeric fractions of CtRocCOR and its L487A mutant (Figure 4). As expected, the mass of the monomeric species of CtRocCOR (or L487A) in the presence of NbRoco1 corresponds to that of a CtRocCOR (or L487A) monomer with one molecule of NbRoco1 bound, irrespective of the nucleotide present. Interestingly, for the GDP- and GppNHp-bound states of CtRocCOR and for the GppNHp-bound state of CtRocCOR L487A, the mass of the remaining dimeric species correspond to that of the CtRocCOR dimer bound to two molecules of NbRoco1 (Figure 4). This thus means that NbRoco1 is not merely sterically disrupting CtRoco dimerization by binding on the dimer interface, in which case no binding of NbRoco1 to the CtRocCOR (or L487A) dimer would be expected. Rather, these data show that NbRoco1 can bind to both the CtRocCOR (or L487A) dimer and monomer, and is thus shifting the dimer–monomer equilibrium toward the monomer using an allosteric mechanism by favoring the conformation of the monomer. The situation is more complex for CtRocCOR in the nucleotide-free state and CtRocCOR L487A in its nucleotide-free and GDP state. Here, the observed population of dimeric CtRocCOR species in the presence of NbRoco1 correspond to a mixture of unbound CtRocCOR dimers and dimers bound to only one molecule of NbRoco1. This observation indicates the presence of conformational differences between the different nucleotide states of CtRocCOR in the dimeric state and thus prior to monomerization.
Overall, we can conclude from these experiments that NbRoco1 recognizes CtRoco in a conformation-specific way, and allosterically modulates the dimer–monomer cycle finally leading to the observed increase in GTPase activity of wild-type CtRoco and its PD-analogous L487A mutant.
Discussion
Nanobodies (Nbs) are small (15 kDa) and stable single-domain fragments derived from camelid heavy chain-only antibodies that retain the full antigen-binding capacity [52]. Owing to their ease of cloning and production, their small size, and their stability, Nbs have been frequently used in research, but also as potent diagnostic and therapeutic tools [52]. Moreover, because of their prolate shape with a convex antigen-binding region formed by a long CDR3 loop, Nbs have the tendency to bind and insert into clefts or cavities on the surface of the antigen [54]. Correspondingly, Nbs frequently act as enzyme inhibitors by binding to active sites [62–65], while they also form ideal tools to selectively bind and stabilize specific conformational states of proteins [55].
Here, we identified two Nbs (NbRoco1 and NbRoco2) that recognize a bacterial homolog of the PD-associated protein LRRK2 in a conformation-specific way. Both Nbs bind the Roco protein from C. tepidum (CtRoco) preferentially in its GTP (or GppNHp)-bound state, with a gradually lower affinity for the GDP-bound and nucleotide-free states (Figure 1). While this conformational specificity is most pronounced for NbRoco1, it is remarkable that both Nbs achieve their conformational specificity through binding to different domains, with NbRoco1 binding to the RocCOR domain and NbRoco2 binding to the LRR domain. This implies that the conformational changes induced by nucleotide binding to the Roc GTPase domain must be transmitted to the entire CtRoco protein. The latter is in agreement with recent data from negative-stain electron microscopy and from the CtRoco crystal structure in combination with HDX-MS measurements, which suggest that monomerization of CtRoco upon GTP binding is associated with large-scale conformational changes within the subunits including a hinge-like motion of the LRR domain with respect to the RocCOR domain [36,37].
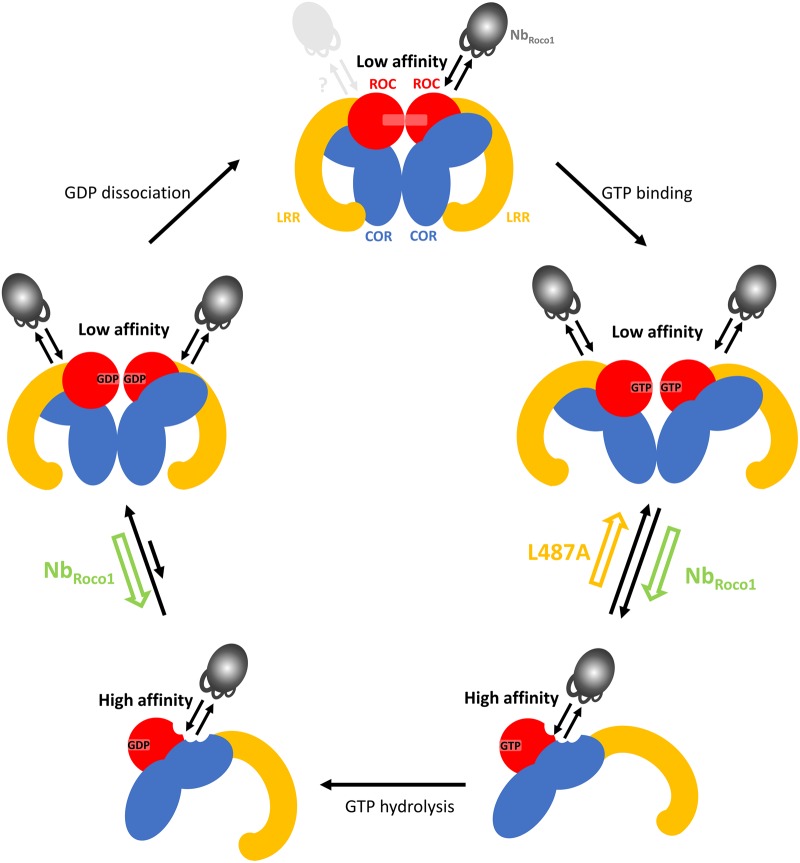
PD mutations in the RocCOR domain of LRRK2 lead to a decrease in GTPase activity [23,25,43,44,56–58], although the affected residues are most often not directly located in the GTP/GDP-binding pocket [37,46]. Using the bacterial CtRoco as a model, we previously showed that the RocCOR domain needs to cycle between a dimeric and monomeric state during the GTPase reaction, where GTP binding induces monomerization while the protein re-dimerizes after GTP hydrolysis [36]. This mechanism thus predicts that either mutations that stabilize the dimer or that block the protein in a monomeric state would impair the GTPase reaction. We concomitantly showed that the L487A mutation (corresponding to the PD-associated mutation I1371V in LRRK2) leads to a decrease in GTP turnover and stabilizes the CtRoco dimer. In agreement with these findings and with our model, it was very recently reported that the R1441G/C/H and N1473H PD mutations decrease the GTPase activity of the Roc domain of LRRK2 by destabilizing or stabilizing the Roc dimer, respectively [43,44]. We now show here that the binding of NbRoco1 increases the CtRoco GTP turnover rate (kcat). NbRoco1 affects the RocCOR dimer–monomer cycle by binding preferentially to the GTP-bound monomeric state (Figures 2 and 3). Interestingly, NbRoco1 is also able to completely revert the effect of the dimer-stabilizing PD-analogous L487A mutation, with the GTP turnover rate of the NbRoco1-bound L487A mutant even surpassing that of the wild-type protein. Our native MS data (Figure 4) demonstrate that NbRoco1 is able to bind to the CtRoco monomer as well as to the dimer, excluding a mode of action where the Nb would disrupt dimerization by simply binding at the RocCOR dimer interface. This thus implies that NbRoco1 shifts the dimer–monomer equilibrium toward the monomeric form using an allosteric mechanism. In the absence of high-resolution structural information the exact binding mode and mechanism of NbRoco1 is not known, but our data suggest that NbRoco1 binds to an epitope that changes conformation between the mainly monomeric GTP-bound protein, the dimeric nucleotide-free protein and the mainly dimeric GDP-bound protein (Figure 5). The law of mass action dictates that by binding with higher affinity to the conformation adopted in the monomeric state than to the dimeric form, NbRoco1 will shift the equilibrium toward the monomer. In our model where Roco proteins need to cycle between a monomeric and a dimeric form, a Nb that completely blocks the protein in a monomeric state by binding on the dimer interface would be expected to induce a decrease in GTPase activity. An allosteric mechanism as found here would modulate the dynamics of interconversion, thereby leading to the shift in equilibrium and the observed increased GTP turnover.
Figure 5. Proposed mechanism of action of NbRoco1.
CtRoco is shown schematically with the Roc domains in red, COR domains in blue and LRR domains in yellow. Upon GTP binding CtRoco undergoes a dimer to monomer transition in addition to intra-subunit conformational changes [36,37]. After GTP hydrolysis CtRoco re-dimerizes. NbRoco1, indicated in gray, binds to an epitope on the RocCOR domain that changes conformation between the dimeric and monomeric states, with the highest affinity for the monomeric state. Through binding, NbRoco1 changes the dimer–monomer equilibrium towards the monomer, thus counteracting the effect of the PD-analogous L487A mutant.
Considering that many prominent mutations in LRRK2 lead to an increase in kinase activity [27,28], drug design for LRRK2-associated PD has so far mainly focused on the development of kinase inhibitors [29–31]. Despite some recent successes with this strategy, the current LRRK2 kinase inhibitors will require further optimization and testing [30,66,67]. However, many LRRK2 PD mutations, and particularly those in the RocCOR domains, also decrease the GTPase activity [23–25]. Therefore, interfering with the GTPase activity of the RocCOR domain has recently come into the picture as a potential alternative therapeutic strategy [32–34]. However, since PD-associated mutations lead to a decreased GTPase activity, classical inhibitor design targeting the GTP/GDP-binding pocket is not a straightforward approach in this case, and might even have a detrimental effect. Here, we provide the proof of principle that the GTPase activity of a bacterial Roco protein can be increased significantly by acting upon the RocCOR dimer–monomer equilibrium, and that this approach can completely abrogate the detrimental effect of a PD-analogous mutation. Moreover, NbRoco1 exerts its effect by changing the dimer–monomer equilibrium via an allosteric mechanism. Such an allosteric mechanism has the clear advantage that it allows to tune the dimer–monomer equilibrium, rather than completely blocking the protein in one or the other state. Additionally, while compounds that target the GTP/GDP-binding pocket of a GTPase might suffer from a lack of specificity and need to compete with very high cellular nucleotide concentrations, allosteric modulators of GTPase activity evade these draw-backs. Potentially, a similar approach could also be amenable to stabilize the dimeric RocCOR state using compounds that specifically bind to a dimer-specific conformation of the protein. In conclusion, this study thus shows that allosteric targeting of the dimer–monomer equilibrium of the RocCOR domain might provide a potentially promising strategy to revert the detrimental effect of (certain) LRRK2 PD mutations.
Abbreviations
- COR
C-terminal of Roc
- FRET
fluorescence resonance energy transfer
- IMAC
immobilized metal affinity chromatography
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- LRR
leucine-rich repeat
- MS
mass spectrometry
- Nbs
nanobodies
- NF
nucleotide-free
- PD
Parkinson's disease
- Roc
Ras of complex proteins
- smFRET
single-molecule FRET
- TB
Terrific Broth
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Research Foundation Flanders (FWO) (W.V., M.L., J.P.; grant ID G005219N, 11ZH318N and 1S04916N), Strategic Research Program Financing of the VUB (W.V.; grant ID SRP34 and SRP50), The Michael J. Fox Foundation for Parkinson's Research (A.K., W.V.; grant ID 14527), the Hercules Foundation (W.V.), BioStruct-X by the European Community's Seventh Framework Programme (W.V.). We acknowledge the support and the use of resources of Instruct-ERIC, part of the European Strategy Forum on Research Infrastructures (ESFRI) and the Research Foundation Flanders (FWO) for their support to the Nanobody discovery.
Author Contributions
M.L., C.G., E.De., E.Da., L.K. and J.P. performed the experiments. E.P., J.S., A.K., F.S., D.K. and W.V. supervised experiments and aided in interpreting data. E.De. and W.V. designed the study. W.V., M.L. and C.G. wrote the manuscript.
Supplementary Material
References
- 1.Bosgraaf L. and Van Haastert P.J.M. (2003) Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta Mol. Cell Res. 1643, 5–10 10.1016/j.bbamcr.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 2.Wauters L.,Versées W. and Kortholt A. (2019) Roco proteins: GTPases with a baroque structure and mechanism. Int. J. Mol. Sci. 20, 147 10.3390/ijms20010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marín I.,van Egmond W.N. and van Haastert P.J.M. (2008) The Roco protein family: a functional perspective. FASEB J. 22, 3103–3110 10.1096/fj.08-111310 [DOI] [PubMed] [Google Scholar]
- 4.Marín I. (2008) Ancient origin of the Parkinson disease gene LRRK2. J. Mol. Evol. 67, 41–50 10.1007/s00239-008-9122-4 [DOI] [PubMed] [Google Scholar]
- 5.Dihanich S. (2012) MASL1: a neglected ROCO protein. Biochem. Soc. Trans. 40, 1090–1094 10.1042/BST20120127 [DOI] [PubMed] [Google Scholar]
- 6.Civiero L.,Dihanich S.,Lewis P.A. and Greggio E. (2014) Genetic, structural, and molecular insights into the function of RAS of complex proteins domains. Chem. Biol. 21, 809–818 10.1016/j.chembiol.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bialik S. and Kimchi A. (2006) The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75, 189–210 10.1146/annurev.biochem.75.103004.142615 [DOI] [PubMed] [Google Scholar]
- 8.Civiero L. and Bubacco L. (2012) Human leucine-rich repeat kinase 1 and 2: intersecting or unrelated functions? Biochem. Soc. Trans. 40, 1095–1101 10.1042/BST20120123 [DOI] [PubMed] [Google Scholar]
- 9.Tagawa H.,Karnan S.,Kasugai Y.,Tuzuki S.,Suzuki R.,Hosokawa Y. et al. (2004) MASL1, a candidate oncogene found in amplification at 8p23.1, is translocated in immunoblastic B-cell lymphoma cell line OCI-LY8. Oncogene 23, 2576–2581 10.1038/sj.onc.1207352 [DOI] [PubMed] [Google Scholar]
- 10.Raval A.,Tanner S.M.,Byrd J.C.,Angerman E.B.,Perko J.D.,Chen S.S., et al. (2007) Downregulation of Death-Associated Protein Kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell 129, 879–890 10.1016/j.cell.2007.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L.,Girisha K.M.,Iida A.,Hebbar M.,Shukla A.,Shah H., et al. (2017) Identification of a novel LRRK1 mutation in a family with osteosclerotic metaphyseal dysplasia. J. Hum. Genet. 62, 437–441 10.1038/jhg.2016.136 [DOI] [PubMed] [Google Scholar]
- 12.Healy D.G.,Falchi M.,O'Sullivan S.S.,Bonifati V.,Durr A.,Bressman S., et al. (2008) Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 7, 583–590 10.1016/S1474-4422(08)70117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang D.,Nalls M.A.,Hallgrímsdottir I.B.,Hunkapiller J.,Van Der Brug M.,Cai F., et al. (2017) A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 49, 1511–1516 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalls M.A.,Pankratz N.,Lill C.M.,Do C.B.,Hernandez D.G.,Saad M., et al. (2014) Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 46, 989–993 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gass A.,Eisele P.,Rocca M.A.,Agosta F.,Valsasina P.,Filippi M., et al. (2015) MRI monitoring of pathological changes in the spinal cord in patients with multiple sclerosis. Lancet Neurol. 14, 443–454 10.1016/S1474-4422(14)70294-7 [DOI] [PubMed] [Google Scholar]
- 16.Zimprich A.,Biskup S.,Leitner P.,Lichtner P.,Farrer M.,Lincoln S., et al. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 10.1016/j.neuron.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 17.Paisán-Ruíz C.,Jain S.,Evans E.W.,Gilks W.P.,Simón J.,Van Der Brug M., et al. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 44, 595–600 10.1016/j.neuron.2004.10.023 [DOI] [PubMed] [Google Scholar]
- 18.Gilks W.P.,Abou-Sleiman P.M.,Gandhi S.,Jain S.,Singleton A.,Lees A.J., et al. (2005) A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet 365, 415–416 10.1016/S0140-6736(05)17830-1 [DOI] [PubMed] [Google Scholar]
- 19.Steger M.,Tonelli F.,Ito G.,Davies P.,Trost M.,Vetter M., et al. (2016) Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steger M.,Diez F.,Dhekne H.S.,Lis P.,Nirujogi R.S.,Karayel O., et al. (2017) Systematic proteomic analysis of LRRK2-mediated rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6, e31012 10.7554/eLife.31012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z.,Bryant N.,Kumaran R.,Beilina A.,Abeliovich A.,Cookson M.R. et al. (2017) LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 27, 385–395 10.1093/hmg/ddx410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo L.,Gandhi P.N.,Wang W.,Petersen R.B.,Wilson-Delfosse A.L. and Chen S.G. (2007) The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase thatstimulates kinase activity. Exp. Cell Res. 313, 3658–3670 10.1016/j.yexcr.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao J.,Wu C.-X.,Burlak C.,Zhang S.,Sahm H.,Wang M., et al. (2014) Parkinson disease-associated mutation R1441H in LRRK2 prolongs the “active state” of its GTPase domain. Proc. Natl. Acad. Sci. U.S.A. 111, 4055–4060 10.1073/pnas.1323285111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X.,Tan Y.C.,Poulose S.,Olanow C.W.,Huang X.Y. and Yue Z. (2007) Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J. Neurochem. 103, 238–247 10.1111/j.1471-4159.2007.04743.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis P.A.,Greggio E.,Beilina A.,Jain S.,Baker A. and Cookson M.R. (2007) The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357, 668–671 10.1016/j.bbrc.2007.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luzón-Toro B.,de la Torre E.R.,Delgado A.,Pérez-Tur J. and Hilfiker S. (2007) Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum. Mol. Genet. 16, 2031–2039 10.1093/hmg/ddm151 [DOI] [PubMed] [Google Scholar]
- 27.West A.B.,Moore D.J.,Biskup S.,Bugayenko A.,Smith W.W.,Ross C.A. et al. (2005) Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 10.1073/pnas.0507360102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gloeckner C.J.,Kinkl N.,Schumacher A.,Braun R.J.,O'Neill E.,Meitinger T. et al. (2006) The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 15, 223–232 10.1093/hmg/ddi439 [DOI] [PubMed] [Google Scholar]
- 29.Taymans J.-M. and Greggio E. (2016) LRRK2 kinase inhibition as a therapeutic strategy for Parkinson's disease, where do we stand? Curr. Neuropharmacol. 14, 214–225 10.2174/1570159X13666151030102847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott J.D.,DeMong D.E.,Greshock T.J.,Basu K.,Dai X.,Harris J., et al. (2017) Discovery of a 3-(4-pyrimidinyl) indazole (MLi-2), an orally available and selective leucine-rich repeat kinase 2 (LRRK2) inhibitor that reduces brain kinase activity. J. Med. Chem. 60, 2983–2992 10.1021/acs.jmedchem.7b00045 [DOI] [PubMed] [Google Scholar]
- 31.Deng X.,Dzamko N.,Prescott A.,Davies P.,Liu Q.,Yang Q., et al. (2011) Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 7, 203–205 10.1038/nchembio.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T.,Yang D.,Zhong S.,Thomas J.M.,Xue F.,Liu J., et al. (2014) Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson's disease cell and mouse models. Hum. Mol. Genet. 23, 6212–6222 10.1093/hmg/ddu341 [DOI] [PubMed] [Google Scholar]
- 33.Nixon-Abell J.,Berwick D.C. and Harvey K. (2016) L'RRK de Triomphe: a solution for LRRK2 GTPase activity? Biochem. Soc. Trans. 44, 1625–1634 10.1042/BST20160240 [DOI] [PubMed] [Google Scholar]
- 34.Civiero L.,Russo I.,Bubacco L. and Greggio E. (2017) Molecular insights and functional implication of LRRK2 dimerization. Adv Neurobiol. 14, 107–121 10.1007/978-3-319-49969-7_6 [DOI] [PubMed] [Google Scholar]
- 35.Wauters L.,Terheyden S.,Gilsbach B.K.,Leemans M.,Athanasopoulos P.S.,Guaitoli G. et al. (2018) Biochemical and kinetic properties of the complex Roco G-protein cycle. Biol. Chem. 399, 1447–1456 10.1515/hsz-2018-0227 [DOI] [PubMed] [Google Scholar]
- 36.Deyaert E.,Wauters L.,Guaitoli G.,Konijnenberg A.,Leemans M.,Terheyden S., et al. (2017) A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover. Nat. Commun. 8, 1008 10.1038/s41467-017-01103-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deyaert E.,Leemans M.,Singh R.K.,Gallardo R.,Steyaert J.,Kortholt A. et al. (2019) Structure and nucleotide-induced conformational dynamics of the Chlorobium tepidum Roco protein. Biochem. J. 476, 51–66 10.1042/BCJ20180803 [DOI] [PubMed] [Google Scholar]
- 38.Civiero L.,Vancraenenbroeck R.,Belluzzi E.,Beilina A.,Lobbestael E.,Reyniers L., et al. (2012) Biochemical characterization of highly purified leucine-rich repeat kinases 1 and 2 demonstrates formation of homodimers. PLoS One 7, e43472 10.1371/journal.pone.0043472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James N.G.,Digman M.A.,Gratton E.,Barylko B.,Ding X.,Albanesi J.P. et al. (2012) Number and brightness analysis of LRRK2 oligomerization in live cells. Biophys. J. 102, L41–L43 10.1016/j.bpj.2012.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen S.,Webber P.J. and West A.B. (2009) Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 284, 36346–36356 10.1074/jbc.M109.025437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger Z.,Smith K.A. and Lavoie M.J. (2010) Membrane localization of LRRK2 is associated with increased formation of the highly active lrrk2 dimer and changes in its phosphorylation. Biochemistry 49, 5511–5523 10.1021/bi100157u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez R.C.,Wawro P.,Lis P.,Alessi D.R. and Pfeffer S.R. (2019) Membrane association but not identity is required for LRRK2 activation and phosphorylation of Rab GTPases. J. Cell Biol. 218, 4157–4170 10.1083/jcb.201902184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C.-X.,Liao J.,Park Y.,Reed X.,Engel V.A.,Hoang N.C., et al. (2019) Parkinson's disease-associated mutations in the GTPase domain of LRRK2 impair its nucleotide-dependent conformational dynamics. J. Biol. Chem. 294, 5907–5913 10.1074/jbc.RA119.007631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X.,Wu C.,Park Y.,Long X.,Hoang Q.Q. and Liao J. (2019) The Parkinson's disease–associated mutation N1437H impairs conformational dynamics in the G domain of LRRK2. FASEB J. 33, 4814–4823 10.1096/fj.201802031R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein C.L.,Rovelli G.,Springer W.,Schall C.,Gasser T. and Kahle P.J. (2009) Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J. Neurochem. 111, 703–715 10.1111/j.1471-4159.2009.06358.x [DOI] [PubMed] [Google Scholar]
- 46.Guaitoli G.,Raimondi F.,Gilsbach B.K.,Gómez-Llorente Y.,Deyaert E.,Renzi F., et al. (2016) Structural model of the dimeric Parkinson's protein LRRK2 reveals a compact architecture involving distant interdomain contacts. Proc. Natl. Acad. Sci. U.S.A. 113, E4357–E4366 10.1073/pnas.1523708113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotthardt K.,Weyand M.,Kortholt A.,Van Haastert P.J.M. and Wittinghofer A. (2008) Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 27, 2352 10.1038/emboj.2008.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardon E.,Laeremans T.,Triest S.,Rasmussen S.G.F.,Wohlkönig A.,Ruf A. et al. (2014) A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 9, 674–693 10.1038/nprot.2014.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massa S.,Vikani N.,Betti C.,Ballet S.,Vanderhaegen S.,Steyaert J., et al. (2016) Sortase A-mediated site-specific labeling of camelid single-domain antibody-fragments: a versatile strategy for multiple molecular imaging modalities. Contrast Media Mol. Imaging 11, 328–339 10.1002/cmmi.1696 [DOI] [PubMed] [Google Scholar]
- 50.Rudi K.,Ho F.Y.,Gilsbach B.K.,Pots H.,Wittinghofer A.,Kortholt A. et al. (2015) Conformational heterogeneity of the Roc domains in C. tepidum Roc-COR and implications for human LRRK2 Parkinson mutations. Biosci. Rep. 35, e00254 10.1042/BSR20150128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sobott F.,Hernández H.,McCammon M.G.,Tito M.A. and Robinson C.V. (2002) A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 10.1021/ac0110552 [DOI] [PubMed] [Google Scholar]
- 52.Muyldermans S. (2013) Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 53.Dmitriev O.Y.,Lutsenko S. and Muyldermans S. (2016) Nanobodies as probes for protein dynamics in vitro and in cells. J. Biol. Chem. 291, 3767–3775 10.1074/jbc.R115.679811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Genst E.,Silence K.,Decanniere K.,Conrath K.,Loris R.,Kinne J. et al. (2006) Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U.S.A. 103, 4586–4591 10.1073/pnas.0505379103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steyaert J. and Kobilka B.K. (2011) Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol. 21, 567–572 10.1016/j.sbi.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danie˜ls V.,Vancraenenbroeck R.,Law B.M.H.,Greggio E.,Lobbestael E.,Gao F., et al. (2011) Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. J. Neurochem. 116, 304–315 10.1111/j.1471-4159.2010.07105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen M.L. and Wu R.M. (2018) LRRK 2 gene mutations in the pathophysiology of the ROCO domain and therapeutic targets for Parkinson's disease: a review. J. Biomed. Sci. 25, 52 10.1186/s12929-018-0454-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong Y.,Coombes C.E.,Kilaru A.,Li X.,Gitler A.D.,Bowers W.J. et al. (2010) GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 6, e1000902 10.1371/journal.pgen.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buckstein M.H.,He J. and Rubin H. (2008) Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 10.1128/JB.01020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bochner B.R. and Ames B.N. (1982) Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257, 9759–9769 PMID: [PubMed] [Google Scholar]
- 61.Cornish-Bowden A. (2012) Fundamentals of Enzyme Kinetics, 4th edn, Wiley-Blackwell, Weinheim, Germany [Google Scholar]
- 62.Lauwereys M.,Arbabi Ghahroudi M.,Desmyter A.,Kinne J.,Holzer W.,De Genst E. et al. (1998) Potent enzyme inhibitors derived from dromedary. Eur. Mol. Biol. Organ. J. 17, 3512–3520 10.1093/emboj/17.13.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conrath K.E.,Lauwereys M.,Galleni M.,Matagne A.,Frère J.M.,Kinne J. et al. (2001) β-Lactamase inhibitors derived from single-domain antibody fragments elicited in the Camelidae. Antimicrob. Agents Chemother. 45, 2807–2812 10.1128/AAC.45.10.2807-2812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oyen D.,Srinivasan V.,Steyaert J. and Barlow J.N. (2011) Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition. J. Mol. Biol. 407, 138–148 10.1016/j.jmb.2011.01.017 [DOI] [PubMed] [Google Scholar]
- 65.Kromann-Hansen T.,Oldenburg E.,Yung K.W.Y.,Ghassabeh G.H.,Muyldermans S.,Declerck P.J. et al. (2016) A camelid-derived antibody fragment targeting the active site of a serine protease balances between inhibitor and substrate behavior. J. Biol. Chem. 291, 15156–15168 10.1074/jbc.M116.732503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuji R.N.,Flagella M.,Baca M.,Baptista M.A.S.,Brodbeck J.,Chan B.K., et al. (2015) Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med. 7, 273ra15 10.1126/scitranslmed.aaa3634 [DOI] [PubMed] [Google Scholar]
- 67.Henderson J.L.,Kormos B.L.,Hayward M.M.,Coffman K.J.,Jasti J.,Kurumbail R.G., et al. (2015) Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor. J. Med. Chem. 58, 419–432 10.1021/jm5014055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.