Abstract
Brain circuits comprise vast numbers of intricately interconnected neurons with diverse molecular, anatomical and physiological properties. To allow “user-defined” targeting of individual neurons for structural and functional studies, we created light-inducible site-specific DNA recombinases (SSRs) based on Cre, Dre and Flp (RecVs). RecVs can induce genomic modifications by one-photon or two-photon light induction in vivo. They can produce targeted, sparse and strong labeling of individual neurons by modifying multiple loci within mouse and zebrafish genomes. In combination with other genetic strategies, they allow intersectional targeting of different neuronal classes. In the mouse cortex they enable sparse labeling and whole-brain morphological reconstructions of individual neurons. Furthermore, these enzymes allow single-cell two-photon targeted genetic modifications and can be used in combination with functional optical indicators with minimal interference. In summary, RecVs enable spatiotemporally-precise optogenomic modifications that can facilitate detailed single-cell analysis of neural circuits by linking genetic identity, morphology, connectivity and function.
Keywords: Light-inducible, optogenomic, recombinase, recombination, RecV, Vivid, Cre, Dre, Flp
INTRODUCTION
The mammalian brain is one of the most complex biological systems. It comprises millions to billions of cells1 with diverse characteristics. To understand this extraordinary complexity, it will be essential to define cell types based on properties such as gene expression, morphology and physiology. Furthermore, the unique properties of individual cells need to be related to their connectivity patterns and their activities in a behavioral context. Anatomical information combined with genetic identity and functional properties at the single-cell level will enable better analysis of brain circuitry underlying complex behaviors in health and disease.
One of the most powerful approaches to characterizing cell types and studying their functions relies on mouse genetics2. Using transgenic or viral expression of recombinases allows a highly specific level of genetic modification3–5. Further improvements on spatiotemporal control can achieve even higher resolution manipulations and studies of biological systems.
Currently, finding individual cells/neurons in vivo, characterizing their function, and genetically manipulating them in a targeted manner is difficult to execute efficiently. The state-of-the-art approach for introducing an exogenous gene to a specific neuron is either by the patch clamp technique6 or via single-cell electroporation7,8. These techniques are highly challenging and usually result in low and variable yields. Sparse neuronal labeling or manipulation can be achieved by lowering the dose of inducers (e.g., tamoxifen, in the case of CreER) or by employing ‘inefficient’ recombinase reporters (e.g., MADM9) to control recombination. However, the sparse genetic modification achieved with these methods is random and difficult to direct to specific cells of interest.
Using optical methods to access and genetically modify individual neurons will offer improvement over the current state-of-the-art. Multi-photon interactions with proteins can generate a spatiotemporally-restricted excitation10. Thus, modifying current genomic manipulation enzymes to make them light-inducible can be a superior approach to reach a high spatiotemporal resolution for targeted single-cell manipulations.
Light-inducible protein-based spatiotemporal systems were developed to control protein states, protein localization, transcription and genetic alterations (Supplementary note 1). Several light-inducible SSRs have been previously reported11–15 paving the path for precision single-cell targeting. Magnets, which are fungal photoreceptor Vivid (VVD) variants that heterodimerize, were used to generate light-inducible Cre recombinase systems, and recently a light-inducible Flp recombinase system14. Optimized cryptochrome-based light-inducible Cre recombinases have been generated11. So have wild type phytochrome-based versions with very similar designs15. However, prior to this study optical manipulation of genomes–optogenomics–of individually targeted single cells had yet to be demonstrated in vivo. We developed, and validated novel light-inducible SSR systems named RecVs. In contrast to other light-inducible systems, RecVs induce robust genomic modifications with no or minimally detectable background recombination under uninduced conditions at different genomic locations and in different species. Our work yielded light-inducible versions of the most commonly used SSRs–Cre, Dre and Flp–that allow population level or target-specific single-cell level optogenomic modifications in vivo.
RESULTS
A split Vivid-Cre enables efficient light-inducible site-specific DNA modifications
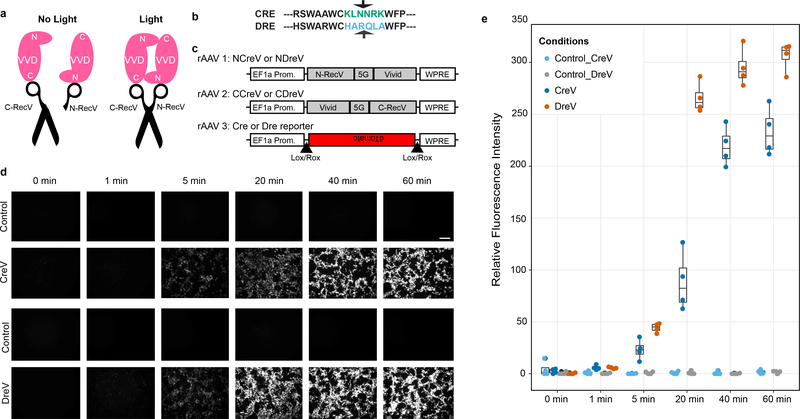
To spatiotemporally regulate site-specific recombination, we generated a light-inducible genetic switch based on a light sensitive fungal protein VVD16 (Supplementary Note 2). We used a split-Cre recombinase17–20 to generate a light-inducible system with VVD dimerizers (Fig. 1a). Guided by the crystal structures21–23 and Cre split location information18,19 (Fig. 1b) our design attempted to bring together the N- and C-portions of the Cre recombinase in the correct orientation upon light exposure. We fused the inactive N-terminal segment of Cre to the N-terminus of one VVD monomer and inactive C-terminal segment of Cre to the C-terminus of another VVD monomer–which we codon diversified. The rAAV expression constructs24 along with fluorescent Cre-reporter constructs (Fig. 1c) were co-transfected into mammalian cells. Recombination with light induction was robust in contrast to no-light conditions (Fig. 1d, e). We named these new proteins CreV, and the general light-inducible Vivid-recombinase system RecV.
Figure 1. Design of the RecV systems.
(a) Based on crystal structures, the N terminus of one VVD protein comes into close proximity to the C terminus of the second in dimer form. Cre is split into two portions: N-Cre and C-Cre. Upon light induced dimerization of the VVDs, the two portions of Cre are brought together and Cre activity is restored (right panel). (b) Alignment of amino-acid sequences of Cre and Dre recombinases with split location noted by arrows. Using this information, we split Dre at the arginine (R) to glutamine (Q) transition marked with the arrow. (c) Schematic of the CreV and DreV rAAV constructs. CreV (NCreV + CCreV) or DreV (NDreV + CDreV) fusion constructs were co-transfected with a Cre- or Dre-dependent red fluorescent reporter. (d) Results of different durations of light induction at 458 nm wavelength of a 1.3 mW/mm2 LED light source (or of darkness for controls) were documented at 48 hours post induction. Scale bar (top right), 200 μm. (e) Quantification of relative fluorescence intensity of reporter constructs shows that light-induced recombination by CreV or DreV depends on duration of light stimulation. Each experiment is represented by 4 replicas. The line across the box represents the median, the lower and upper hinges correspond to the 25th and 75th percentiles, and the upper and lower whiskers extend from the hinge to the largest or smallest values no further than 1.5 * inter-quartile range (IQR) from the hinge.
Vivid-based light-inducible system for Dre recombinase
To broaden the capabilities of the RecV approach we modified another SSR, Dre25, which is homologous to Cre. Dre recombinase recognizes a sequence called Rox26, which is different from LoxP. This creates a possibility to develop Cre/Dre intersectional strategies to further refine cell type-specific genetic manipulation. We reasoned that the sequence homology between Cre and Dre might be used to generate a split Dre (Fig. 1b). Accordingly, we designed and tested DreV constructs using fluorescent Dre reporter plasmids (Fig. 1c and Supp. Fig. 1). Our results indicate that light-induced DreV recombination is efficient using 1P excitation similar to CreV (Fig. 1d, e) and can also be achieved using 2P excitation (Supp. Fig. 2).
Comparison of single RecV expression constructs with other existing light-inducible recombinase systems
To implement the RecV strategy efficiently and reduce the number of viruses or transgenic mouse lines needed for this system, it is preferable to co-express the two halves of RecVs in a single construct. To achieve this, we tested ways to link the N and C components of CreV and DreV to provide the highest recombination efficiency with least background recombination (Supp. Fig. 3a). We observed highest efficiencies when we used optimized elements from Cre-Magnets (mutant VVDs)27,12. We named the resulting enzymes iCreV and iDreV–i for improved–(Supplementary Note 3).
We next compared iCreV and iDreV with Cre-Magnets. In cell culture Cre-Magnets induced efficient recombination under light conditions however they also induced significant amounts of background recombination without light. iCreV and iDreV exhibited control levels of recombination in the absence of light and retained high levels of inducibility. iCreV elicited ~1.6-fold higher efficiency than split CreV and iDreV elicited ~0.6-fold lower efficiency than split DreV (Supp. Fig. 3b). For both iCreV and Cre-Magnets, we observed efficient recombination after light stimulation in vivo. However, using Cre-Magnets without light induction, background recombination was observed throughout the brain structures–~50 cells per 100 μm thick coronal slice–whereas iCreV induced no significant recombination (Supp. Fig. 4).
We also compared the RecV system with the cryptochrome-based light-inducible Cre recombination system, CRY2 11. The comparison in cell culture revealed that iCreV performs ~fourfold better than CRY2 after 60 minutes of light stimulation. CRY2-based Cre recombination allows only a slight increase (~1.2-fold) over baseline recombination (Supp. Fig. 3c).
Design and screening of light-inducible Flp recombinases
To increase intersectional genomic modification potentials, we turned to a third widely used SSR: Flp28. There is too little protein sequence similarity between Flp, and Cre or Dre to guide a split based on homology. Thus, we resorted to structure-based de novo design and screening for split sites based on the crystal structure of Flp29. Using a more efficient, codon-optimized variant of Flp (FlpO)30; we split the Flp coding sequence at 21 loop locations that correspond to transitions between alpha helices and/or beta sheets to lessen likelihood of altering overall functionality within the dimer form. We modified the iCreV backbone to generate these VVD-fused split iFlpV constructs by replacing the N- and C-termini of Cre with the 21 split Flp variants (Supp. Fig. 5a). iFlpV2, 19 and 20 yielded the most significant light-induced recombination with minimal background activity (Supp. Fig. 5b).
In an effort to further improve the efficiency of light-inducible Flp recombinase activity, we created and tested 62 additional iFlpV variants to scan Flp sites surrounding those of iFlpV2, 19 and 20 (Supp. Fig. 5c). iFlpV2 remained the most efficient.
RecV mediated light-inducible recombination with whole-brain infection in vivo
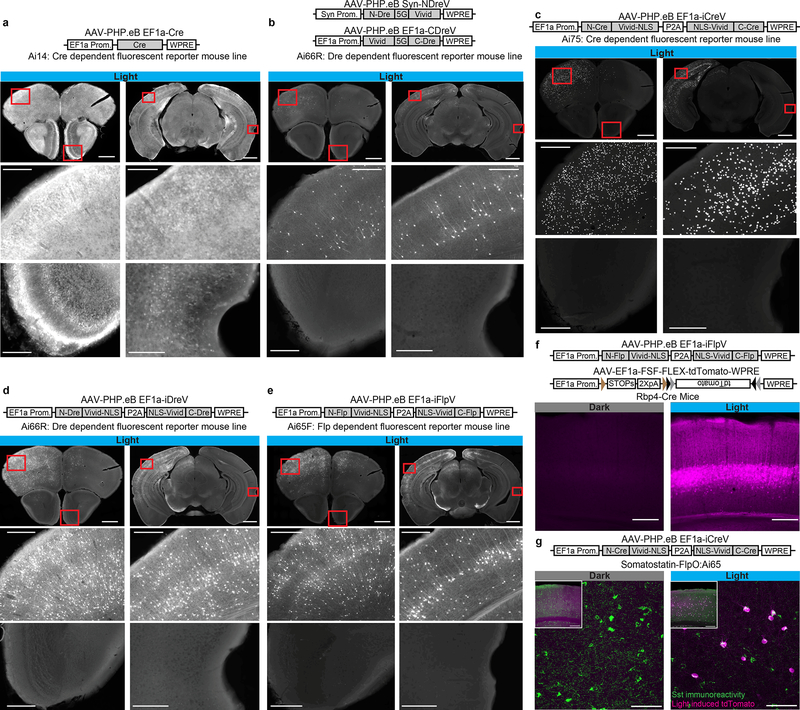
To test RecV constructs in the entire mouse brain we generated RecV- as well as Cre-expressing rAAVs of the PHP.eB serotype31. We found that PHP.eB EF1a-Cre virus, when injected either intracerebroventricularly (ICV) or retro-orbitally (RO) into the fluorescent Cre-reporter mice, efficiently and relatively homogeneously infects the entire brain (Fig. 2a, Supp. Fig. 6a).
Figure 2. Optogenomic modifications by RecV viruses with spatiotemporal and intersectional cell class specific precision in vivo.
Reporter mice (n=2 per case) received right hemisphere intracerebroventricular (ICV) or retro-orbital (RO) injection of PHP.eB rAAVs followed by light stimulation to the left hemisphere two weeks post injection, and imaging two weeks post light stimulation. (a) Injecting Ai14 mice with AAV-PHP.eB EF1a-Cre virus led to widespread recombination throughout the brain (68928 and 182022 cells per section -CPS). (b) Ai66R mice were ICV injected with a 1:1 mixture of AAV-PHP.eB Syn-NDreV and AAV-PHP.eB EF1a-CDreV (204 and 608 CPS). (c) Ai75 mice were ICV injected with AAV-PHP.eB EF1a-iCreV (1323 and 3649 CPS). (d) Ai66R mice were ICV injected with AAV-PHP.eB EF1a-iDreV (1630 and 2670 CPS). (e) Ai65F mice were injected with AAV-PHP.eB EF1a iFlpV. Scale bars a-e: 1 mm for top images, 200 μm for bottom images (1386 and 2471 CPS). (f) L5 pyramidal neuron-specific Rbp4-Cre mice were injected with a mixture of AAV-PHP.eB EF1a-iFlpV and AAV-PHP.eB EF1a-FSF-FLEX-tdTomato. Scale bar: 250 μm (469 of 478 cells in L5). (g) Somatostatin FlpO mouse line, Sst-IRES-FlpO, crossed with a Cre/Flp double-dependent fluorescent reporter mouse line, Ai65, was RO injected with AAV-PHP.eB EF1a-iCreV and light was delivered to the left hemisphere. Specific intersectional light-induced recombination was observed in somatostatin-positive inhibitory interneurons as revealed by immunohistochemistry (100% reporter positive cells (119) were Sst positive (313); 38.1% Sst cells were reporter positive.). Scale bar: 250 μm for inset and 75 μm for zoomed images. All in vivo light activation was applied through the skull on the left hemisphere (opposite ICV injection sites in those cases). For a-e, two coronal planes are shown for each injection (top row) with enlarged views (lower two rows) for areas indicated by red boxes.
To test light-inducible recombination using RecVs, first we injected a mixture of PHP.eB NDreV and CDreV viruses into the right ventricle of the Dre-dependent fluorescent reporter mouse, Ai66R. Two weeks later, the left hemisphere was exposed to light. We observed a gradient of recombination from the top of the left hemisphere to deeper structures (Fig. 2b). Sites far away from the light stimulation had no fluorescently labeled cells –except along the ICV needle track, where recombination took place, likely due to infection with multiple viral particles. In addition, we tested the iCreV, iDreV and iFlpV constructs with PHP.eB serotype, in SSR-dependent reporter mouse lines. As in our trials with NDreV and CDreV, we observed substantial recombination in the hemisphere exposed to light (Fig. 2c, 2d, 2e and Supp. Fig. 6b). No significant recombination was observed in iCreV injected no-light control mice after 4 weeks, demonstrating tight optogenomic control (Supp. Fig. 6d). These results show that this technique allows highly specific light-inducible recombination.
Recombinase reporter mouse lines described above were generated via targeted insertions into the Rosa26 locus32. To test a different locus we retro-orbitally injected PHP.eB iCreV rAAVs into the Ai167 ChrimsonR reporter mouse line, which has its reporter insertion within the TIGRE locus33. We again observed a gradient of recombination from the light-stimulated left hemisphere confirming that other genomic loci in mice can be modified by the RecV system (Supp. Fig. 6c).
We also tested the feasibility of light-inducible recombination within a deeper brain area, the striatum. With 1P-light through an optical fiber we induced local recombination using iCreV in the striatum of Ai162 GCaMP6s Cre reporter mice33, and recorded changes in fluorescence due to calcium concentration dynamics before and after light stimulation (Supp. Fig. 7a–d). Due to the localized illumination, GCaMP6s was expressed directly under the fiber, in contrast to the broad expression of red fluorescence from the control virus (Supp. Fig. 7b). Our results demonstrate that optogenomic modifications can be also efficiently spatiotemporally regulated within deep brain targets and thereby provide a tool for restricted reporter expression under the optical device in vivo.
Cell-class specific targeting by intersection of viral RecVs and transgenic recombinases
Versatile and refined cell-type targeting can be achieved by using two or more recombinases with distinct activities33. For example, new RecV tools can be combined with existing transgenic recombinase lines and intersectional reporters.
To test feasibility of this approach, we co-injected PHP.eB iFlpV virus and a PHP.eB Cre/Flp dependent fluorescent reporter virus into the cortex of mice containing the Rbp4-Cre-KL-100 transgene, which drives Cre expression mostly in L5 excitatory cortical cells. After light stimulation we observed L5 specific reporter gene expression (Fig. 2f), suggesting a high degree of intersectional specificity.
To provide further evidence that light-mediated intersectional targeting can be achieved in other neuron types, we used the Sst-IRES-FlpO mouse line–which expresses FlpO recombinase selectively in inhibitory somatostatin (Sst) neurons–in conjunction with iCreV. We crossed this mouse line to a Cre/Flp-double dependent fluorescent reporter mouse line: Ai65. We retro-orbitally injected the resulting mice with PHP.eB iCreV virus and observed reporter gene expression across multiple cortical layers in sparsely distributed neurons close to the light stimulation site (Fig. 2g). To ascertain that the recombination observed within the light induced hemisphere is indeed specific to the Sst-positive neurons, we performed immunohistochemistry and confirmed that all of the fluorescently labeled neurons were also labeled with anti-SST antibody staining. Recombination was not detected in the unstimulated hemisphere (Fig. 2g). These results confirm that reliable intersectional control can be achieved by combining virally delivered iFlpV or iCreV with transgenic cell class-specific Cre or FlpO, respectively.
CreV induces tight light-dependent recombination in zebrafish
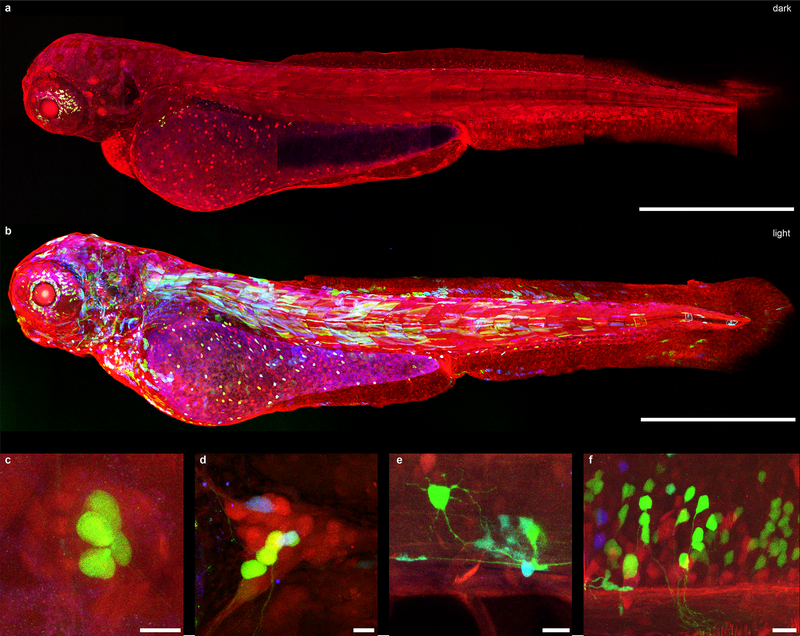
To determine whether the RecV system can effectively induce light-dependent recombination in other loci and in organisms other than mice we tested it in zebrafish. CreV constructs were injected into transgenic zebrabow strain34 embryos which were reared either in light or in darkness. In this line Cre recombination induces YFP and/or CFP expression in place of default state RFP expression. One in 24 viable embryos reared in the dark displayed recombination. Of the 37 viable embryos reared in the light most (32 of 37) had YFP or CFP expression, confirming that CreV-induced recombination was highly light-dependent. We observed recombination in diverse tissues including muscle, skin, heart, spinal cord neurons and in non-neuronal cells, hindbrain, trigeminal ganglion, Rohon-Beard sensory neurons and hair cells of the lateral line (Fig. 3a–f).
Figure 3. CreV allows tight optogenomic modifications in multiple tissues of Danio rerio.
(a) Confocal images of dark reared Zebrabow zebrafish larva (3dpf) co-injected with N-CreV and C-CreV plasmids (see methods section for details). No recombination was observed. In the default state, RFP is expressed in all cells. Apparent green signal is due to autofluorescence. (b) Confocal images of a Zebrabow larva co-injected with N-CreV and C-CreV plasmids and immediately exposed to light. CreV-mediated recombination in multiple tissue types is reflected as expression of YFP and CFP: (c) lateral line hair cells, (d) trigeminal ganglion, (e) spinal neuron and glial cells, and (f) hindbrain neurons. Scale bars: (a-b) 500μm, (c-f) 10μm.
RecV-mediated sparse labeling enables whole-brain reconstructions of single neuron morphologies
Cre-mediated in vivo reporter expression using the Ai139 mouse line results in strong expression of EGFP in multiple cortical layers33. We tested whether light applied to CreV injected Ai139 mice induced sparse yet strong expression at the single cell level, enabling whole-brain reconstruction of single neuron morphologies.
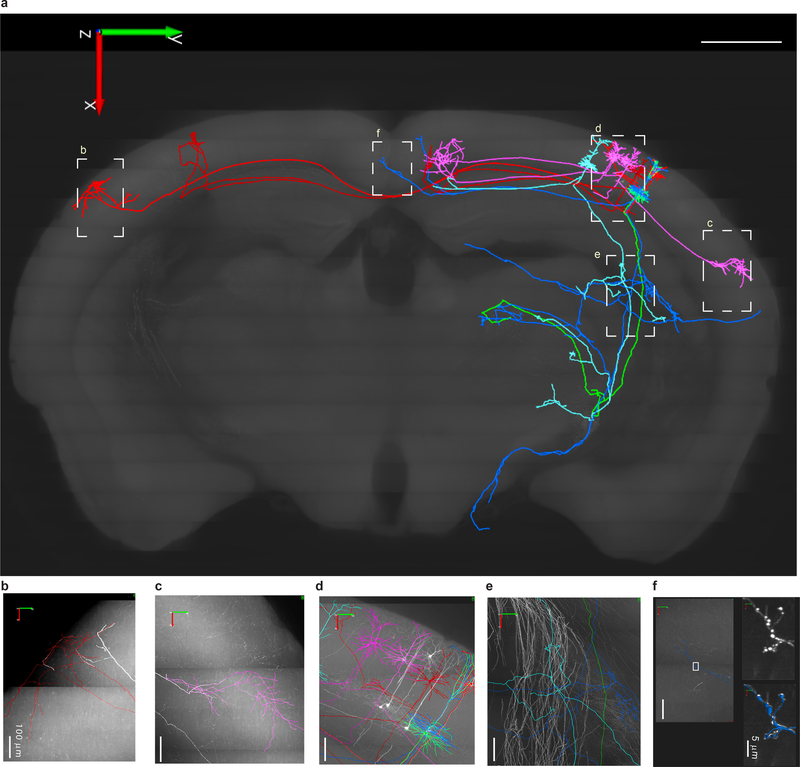
With fluorescence micro-optical sectioning tomography (fMOST)35 we found that low doses of CreV virus and shorter duration light induction yielded sparse and strong labeling of individual neurons in this line (Supp. Fig. 8). This sparse labeling enabled tracing of axons from many individual neurons. In the example brain shown in Figure 4 (also see Supp. Videos 1 and 2), we manually reconstructed eight primary somatosensory cortical neurons. These include three L2/3 pyramidal cells (PCs) with ipsilateral cortico-cortical projections, two L2/3 PCs with contralateral cortico-cortical projections, and three L5 thick-tufted PCs with ipsilateral cortico-subcortical projections, revealing distinct axonal projection patterns.
Figure 4. Cortical pyramidal cells (PCs) labeled with CreV and reconstructed at the whole-brain level.
(a) Eight reconstructed PCs in a mouse somatosensory cortex include three layer 2/3 PCs (in pink) with ipsilateral cortico-cortical projections, two layer 2/3 PCs (in red) with contralateral cortico-cortical projections, and three L5 thick tufted PCs (one green, one blue, one light blue) with ipsilateral cortico-subcortical projections. Local axonal clusters are incomplete because labeling at the somata region is still too dense in this brain for tracing fine axonal branches. The eight reconstructed PCs are superimposed onto a coronal brain plane located 5201–5400 μm posterior to the olfactory bulb (scale bar: 1 mm). (b-f) Enlarged views of areas outlined by dashed boxes in a, with reconstructions (in colors) superimposed on original images with GFP fluorescence shown as white. In f, the two panels on the right (without reconstruction in white, with reconstruction in blue) are enlarged views of the boxed area in the left panel, showing the high-resolution details of a segment of axon with enlarged boutons clearly visible. The whole brain image stack is composed of 12089 images, resolution of XYZ, 0.3 × 0.3 × 1 μm.
RecVs enable 2P-mediated single-cell-specific targeted optogenomic modifications and combinatorial functional imaging in vivo
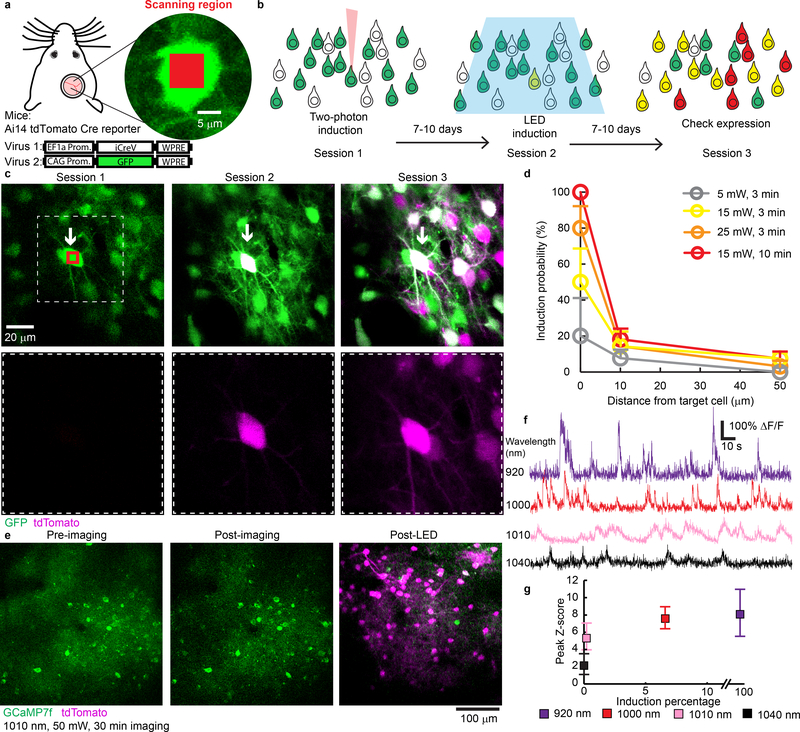
In many cases where restricted genetic access is difficult with conventional methods, it is conceivable to further confine recombination by precise induction through localized illumination. 2P-illumination allows sparse targeting in reporter mice treated with low-concentration iCreV virus (Supp. Fig. 9, Supplementary Note 4). To investigate the feasibility of this approach at the single-cell level in a “user-defined” manner, we tested 2P-illumination in combination with iCreV. In 2P-targeted cortical cells of Ai14 reporter mice we elicited Cre-dependent gene expression with single-cell precision in vivo (Fig. 5a–b). In many cases, we observed reporter expression in the target cell but not in the inducible cells next to it (Fig. 5c and Supp. Fig. 10), demonstrating high spatial accuracy. We tested various 2P-induction protocols and plotted the probability of iCreV activation against the distance to the target cell in each condition (Fig. 5d). Our results suggest that target cell induction rate increases as more laser power and scan time is applied. However, powerful 2P-stimulation in some cases also led to nonspecific induction within a 10 μm radius of the target cell (as high as 18%). Cells within a 10–50 μm radius from the target cell showed a baseline induction rate of 6–7%. The induction rate in no-light conditions was very low (~3% on average, Supp. Fig. 11).
Figure 5. Two-photon guided targeted single-cell optogenomic modifications by iCreV in mouse neocortex.
(a) Diagram of in vivo experiment. Inset shows laser induction area (red square) in target cell (green) within mouse’s craniotomy. Viral vectors and mouse line used are indicated (see methods section for details). (b) Experimental timeline. A month after virus injections, each mouse underwent three imaging sessions with 2P targeted induction (session 1), LED induction (session 2) and a final assessment for expression (session 3). (c) Example 2P images from a targeted induction. Arrows indicate target cell and red box indicates induction scanning region. Bottom row shows magnified regions (dashed box) containing target cell. Similar results were reproduced in five mice. (d) Quantification of in vivo 2P induction rates under different scanning protocols at 0, 10, and 50 μm distance to target cells. N = 4, 5, 3, 5 cells for target cells, 14, 22, 13, 14 cells for 10 μm distance, and 93, 122, 85, 96 cells for 50 μm distance (all in the order of 5 mW 3 min, 15 mW 3 min, 25 mW 3 min and 15 mW 10 min). Data represents mean with standard error of the proportions. (e) Representative 2P images before, and after 30 minutes of calcium imaging with 1000-nm two-photon excitation of jGCaMP7f. Similar results were reproduced in three mice. (f) Example calcium traces from the imaged cortex at different wavelengths. (g) Quantification of in vivo 2P induction rate after 30 minutes of calcium imaging under four excitation wavelengths held at 50 mW, and the quality of calcium signal in each condition. Data shown as median with error bars indicate 25 and 75 percentiles of calcium peak height. N = 100 events for all conditions.
We next tested the feasibility of combining calcium imaging with the iCreV system. 2P excitation above 1000nm showed minimal iCreV activation and moderately reduced jGCaMP7f36 signals (Fig. 5e–g) (Supplementary note 5). This allows functional imaging of cells with minimal interference to subsequent light-inducible optogenomic modifications.
DISCUSSION
In this study, we used wild-type VVD to create three light-inducible recombinases: Cre, Dre, and Flp. We showed that these enzymes work efficiently and can be used intersectionally in the mouse brain to label specific cell classes. We further demonstrated that RecVs allow effective light-induced optogenomic modifications at multiple loci within the mouse and zebrafish genomes.
We showed that RecV-mediated light-inducible site-specific DNA modifications are possible in the mammalian nervous system at the single cell level. Our data provide a quantitative description of the induction specificity of iCreV at different 2P wavelengths and support the feasibility of combining calcium imaging with the iCreV system in mice in vivo. While we demonstrated the single-cell accuracy of the iCreV system, practical variables (e.g., movement of tissue during induction, ambient light, or high multiplicity of infection) may reduce the accuracy.
Light-inducible Cre recombinases have been reported previously11–13,15. A direct comparison of an improved CRY2 based system11 and the one presented in this study shows that iCreV has higher inducibility in vitro. Magnet-based light-inducible Cre recombination is highly efficient in mammalian cells, but it is also leaky in vitro and in vivo compared to its VVD-based counterpart. In a screen to generate a light-inducible FlpO we tested 82 separate variants and discovered a Flp split site (S27-to-G28) that yielded efficient dimerization-induced recombination. Independent discovery of this site was reported in a recent study14.
The demonstration of the ICV route for whole-brain infections using PHP.eB AAV virus may prove valuable in many scenarios. Brains of embryos might be injected via this route to avoid damage to the eye. In multiple mammalian species, this approach may further promote infection of the nervous system and overcome some obstacles related to intravenous delivery of genes. It may also help avoid immune-response interference with infections. Therefore, it may be useful for gene therapy.
RecV technology may enable loss/gain-of-function studies by switching genes off or on, followed by monitoring effects on development, physiology or behavior. This may provide more refined spatiotemporal specificity than current pharmacologically-gated genetic approaches37(e.g. CreER or tTA system). Spatiotemporally-restricted single-cell-targeted 2P induction combined with prior functional characterization by imaging can provide a powerful means to interrogate cell ensembles. In model systems where germline modification is not available or prohibitively expensive, specificity may be achieved by integrating RecVs into viral vectors equipped with short cell-type-specific promoters or enhancers38,39, or by target-defined retrograde infection40.
Overall, the broad range of potential applications shows that the light-inducible recombinase system presented in this study enables much-improved spatiotemporal precision and multiple combinatorial strategies for the micro- and macro-level analyses of neural circuits, as well as many other biological systems, in a variety of organisms.
ONLINE METHODS
Plasmid and virus construction for RecVs
Sequences of NCre-5G-VVD, VVD-5G-CCre, NCre-5G-VVD-IRES-VVD-5G-CCre, VVD-5G-CCre-IRES-NCre-5G-VVD, NCre-5G-VVD-PQR-VVD-5G-CCre, VVD-5G-CCre-PQR-NCre-5G-VVD, VVD-5G-CDre-IRES-NDre-5G-VVD, NCre-Magnets-NLS-P2A-NLS-Magnets-CCre, NCre-VVD-NLS-P2A-NLS-VVD-CCre, NDre-Magnets-NLS-P2A-NLS-Magnets-CDre, NDre-VVD-NLS-P2A-NLS-VVD-CDre, and all iFlpV versions were chemically synthesized (GenScript, Piscataway, NJ). 19–59 amino acid N terminus and 60–343 C terminus of Cre were used in all CreV cases. To screen poly-cistronic cassettes with the best light-inducible recombinase activity, N and C parts of RecV, as well as IRES-mediated, PQR-mediated and P2A-mediated RecV poly-cistronic cassettes were cloned into pCDNA3.1 with the CMV promoter (Supp. Fig. 1).
To generate recombinant AAV viruses expressing split VVD-Cre (CreV) or VVD-Dre (DreV), the N or C part of CreV and DreV were cloned after the human EF1a promoter, followed by WPRE and hGH-polyA signal (Supp. Fig. 1). The Cre reporters, pAAV-EF1a-Flex-dTomato or EGFP-WPRE-hGHpA, used pairs of double inverted LoxP and Lox2272 sites to flank the reporter dTomato or EGFP sequence. The Dre reporter, pAAV-EF1a-Frex-dTomato-WPRE-hGHpA, was generated by inserting an inverted dTomato sequence flanked with Rox sites after the human EF1a promoter, followed by WPRE and hGH-polyA signal (Supp. Fig. 1).
21 iFlpV variants were generated with custom gene synthesis as follows: iFlpV1: 11 amino acids (aa) N and 412 aa C; iFlpV2: 27 aa N and 396 aa C; iFlpV3: 49 aa N and 374 aa C; iFlpV4: 67 aa N and 356 aa C; iFlpV5 72 aa N and 351 aa C; iFlpV6: 85 aa N and 338 aa C; iFlpV7: 95 aa N and 328 aa C; iFlpV8: 114 aa N and 309 aa C; iFlpV9: 129 aa N and 294 aa C; iFlpV10: 151 aa N and 272 aa C; iFlpV11: 169 aa N and 254 aa C; iFlpV12: 197 aa N and 226 aa C; iFlpV13: 208 aa N and 215 aa C; iFlpV14: 237 aa N and 186 aa C; iFlpV15: 251 aa N and 172 aa C; iFlpV16: 290 aa N and 133 aa C; iFlpV17: 318 aa N and 105 aa C; iFlpV18: 343 aa N and 80 aa C; iFlpV19: 374 aa N and 49 aa C; iFlpV20: 388 aa N and 35 aa C; iFlpV21: 408 aa N and 15 aa C. Additional iFlpV2 variants were generated spanning amino acids 16–39 and 366–405 covering the entire region leading to 61 additional constructs. Construct 62 was generated based on iFlpV2 with an addition of the linker GGSGG -present between the C terminus VVD and FlpV- to also N terminus FlpV and VVD. These constructs were cloned in pcDNA3.1 mammalian expression plasmids.
AAV1, AAV DJ and AAV PHP.eB serotype viruses were produced in house with titers of AAV1-EF1a-NCreV, 1.05 × 1012 genome copies (GC); AAV1-EF1a-CCreV, 5.16 × 1012; AAV1-EF1a-NDreV, 4.20 × 1013; AAV1-EF1a-CDreV, 5.40 × 1013; AAV-DJ-EF1a-Cre, 2.00 × 1013; AAV1-CAG-Flex-EGFP, 1.34 × 1013; AAV-DJ-EF1a-Frex-dTomato, 1.90 × 1012; 7.7 × 1011; 1.6 × 1013; AAV-PHP.eB-EF1a-Cre, 5.8 ×1013; AAV-PHP.eB-Syn-NDreV, 4.2 × 1013; AAV-PHP.eB-EF1a-CDreV, 3.9 × 1013; PHP.eB iCreV, 2.6 × 1013; PHP.eB iDreV, 3.3 × 1013, PHP.eB iFlpV, 2.7 × 1013, AAV-PHP.eB-EF1a eGFP 2.03E+13 and AAV- Cre-Magnets 3.00E+13 per ml. AAV5.CAG.tdTomato (1.0 × 1013 GC/mL) was purchased from UNC Vector core.
Light activation in cultured cells
HEK293T (ATCC) cells were seeded into 6-well plates one day before transfection and reached 80% confluency on the day of transfection. Cells were co-transfected with reporters expressing dTomato –for Cre and Flp- or dTomato -for Dre- for testing Cre, Dre or Flp mediated recombination and various constructs of RecVs. Cells in the control groups were transfected with reporters alone. Each condition contained 4 replicas. Plates were kept in dark immediately after transfection. Twenty-four hours later, experimental cells were exposed to blue light, and were then kept in dark immediately after light exposure. Cells were imaged for fluorescent reporter expression 48 hours after light induction, using a 10x objective on an inverted fluorescence microscope. Reporter expression in each condition was quantified using Image J. The corrected total fluorescence intensity = Integrated Density – (Area of each image X Mean fluorescence of background readings). The mean corrected total fluorescence intensity from 4 replicates was used to represent the relative fluorescence intensity of each condition. Light inducibility of RecV was calculated as the ratio of the relative fluorescence intensity with light to that without light.
Surface/Cortical In vivo 1P optogenomic modifications
All animal experiments were performed in compliance with Allen Institute for Brain Science IACUC guidelines. Stereotaxic injections were performed in adult C57BL/6J (stock no. 00064, The Jackson Laboratory, Bar Harbour, ME) or transgenic reporter mice with a 1:1:1 or 1:1 mixture of three or two different rAAVs. For all experiments, animals were anesthetized with isoflurane (5% induction, 1.5% maintenance) and placed on a stereotaxic frame (model no. 1900, David Kopf Instruments, Tujunga, CA). An incision was made to expose the skull, including bregma, lambda, and the target sites. Stereotaxic coordinates were measured from Bregma and were based on The Mouse Brain in Stereotaxic Coordinates1,2. A burr hole was made above the target by thinning the skull using a small drill bit until only a very thin layer remained. An opening was then made using a microprobe, and the remaining thinned skull was gently pulled away. All animals were injected at each target with 500 nl of virus at a rate of ~150 nl/min using a Nanoject II microinjector (Drummond Scientific, Broomall, PA). Intraventricular injection of AAV-PHP.eB viruses was conducted by injecting 2 μl of virus into the lateral ventricle using a Nanoject II microinjectior. The glass pipettes had inner diameters between 10–20 μm.
Unless noted otherwise two weeks following AAV injection, animals were anesthetized and returned to the stereotaxic frame. An incision was made in the previous location to once again reveal the location of the injection sites. An LED light source (LED-64s, Amscope, Irvine, CA) was mounted to the surgical microscope and positioned 3–4 inches directly above the animal’s skull. The amount of time the animal was exposed to light was varied by experiments. Small amounts of sterile PBS were periodically applied to the scalp and skull to prevent drying.
Two weeks following light exposure, animals were perfused with 4% paraformaldehyde (PFA). Brains were dissected and post-fixed in 4% PFA at room temperature for 3–6 h and then overnight at 4°C. Brains were then rinsed briefly with PBS and stored in PBS with 10% sucrose solution. Brains were then sectioned at a thickness of 100 μm while frozen on a sliding microtome (Leica SM2010 R, Nussloch, Germany). Brain sections were mounted on 1×3 in. Plus slides and coverslipped with Vectashield with DAPI (H-1500, Vector Laboratories, Burlingame, CA). Slides were then imaged using a 10x objective on a Leica TCS SP8 confocal microscope (Leica Microsystems, Buffalo Grove, IL) or using a 10x objective on a VS110-Olympus epifluorescence serial scanning microscope (VS-110 Olympus-America). In certain cases, images were pseudo-colored to grayscale or magenta for presentation purposes.
The Image J trainable WeKa segmentation toolkit was used for image segmentation and cell counting. Two separate classifiers were applied, one for images of reporter mice injected with AAV-PHP.eB EF1a-Cre, which contain dense fluorescent cells, and another for those with sparser fluorescent cells. Six images of 500×500 pixels from Ai14 mice injected with AAV-PHP.eB EF1a-Cre virus, and 5 images of 1000×1000 pixels from Ai75 mice injected with AAV-PHP.eB EF1a-iCreV were used to train these two classifiers. The segmented images were then undergone automatic thresholding, watershed separation, and particle analysis to count the number of cells, followed by manual inspection and correction.
For fMOST imaging, approximately two weeks following light exposure, animals were perfused with 4% paraformaldehyde (PFA). Brains were dissected and post-fixed in 4% PFA at room temperature for 3–6 h and then overnight at 4°C. Brains were then transferred to PBS with 0.1% sodium azide for storage at 4°C until embedding.
Somatostatin immunostaining was conducted using the primary antibody of anti-Somatostatin-28 ( 1:500, T-4546.0400, Peninsula Laboratories, CA, USA), and the secondary antibody of Alexa Fluor 647 donkey anti-mouse IgG (1:500, 711–605-152, Jackson Immunoresearch, PA, USA).
Surface/Cortical in vivo population 2P optogenomic modifications
All animal experiments were performed in compliance with Allen Institute for Brain Science IACUC guidelines. A titanium head plate was attached to the skull of mice to allow positioning and restraint of the mice during imaging. The hole of the head plate was positioned over visual cortical areas, approximately 2.9 mm posterior and 2.7 mm lateral from Bregma. A 5 mm craniotomy was introduced using a dental drill. The dura was removed, and a multilayer glass coverslip was positioned above the craniotomy. The head plate and coverslips were secured using cyanoacrylate glue and metabond. After a period of at least one week, a dental drill was used to remove the cement and metabond holding the coverslip in place, and the coverslip was removed. A Dumont Nanoject II was then used to inject 500 nL of viruses into visual cortex. A new coverslip was placed and adhered. The area above the coverslip was blocked from light using a combination of dental cement and Kwik-cast, both mixed with black acrylic paint powder.
After at least 3 weeks following viral injection, the animal received two-photon laser stimulation. Under dark conditions, the Kwik-cast was removed, and the animal’s head plate was mounted in position. The injection area was identified by the presence of the EGFP labelled cells. Laser output was set to 900 nm to optimally induce recombination. A 600 × 600 μm area was stimulated at three depths (100 μm, 150 μm, and 200 μm) for 15 minutes each. After stimulation, black Kwik-cast was reapplied. Two weeks following stimulation, mice were perfused.
Deep brain in vivo stimulation/imaging experiments
All experiments were performed in compliance with Caltech animal care and use committee and office of laboratory animal resources. For deep brain optogenomic modification and imaging experiments stereotaxic injections were made into the striatum of 11 week old Ai162-GC6s (Stock No. 031562, The Jackson Laboratory, Bar Harbour, ME) Cre dependent GCaMP6s reporter mice with a 1:1 mixture of PHP.eB.iCreV and a control AAV unconditionally expressing red fluorescent protein (AAV5.CAG.tdTomato). For all experiments, animals were anesthetized with isoflurane (5% induction, 1.5% maintenance) and placed on a stereotaxic frame (942, David Kopf Instruments, CA, USA). An incision was made to expose the skull, including bregma, lambda, and the target sites. Stereotaxic coordinates were measured from Bregma and were based on The Mouse Brain Atlas1,2. A burr hole was made above the target. All animals were injected with 400 nl X 2 of virus mixture, at two dorsoventral positions, 300 μm apart, at a rate of ~80 nl/min using UltraMicroPump (UMP3–4, World Precision Instrument, Sarasota, FL). Following virus injection, an optical fiber with cut length of 5 mm and diameter of 400 μm (NA 0.48, Doric lenses, Quebec, QC, Canada) was firmly mounted to a stereotaxic holder. The optical fiber was then inserted to the striatum (AP +1.0 mm, ML ± 1.3 mm, DV −3.5 mm, from either left or right side) through a craniotomy and positioned 300 μm above the deeper viral injection site. A thin layer of metabond was applied on the skull surface to secure the fiber. In addition, a thick layer of black dental cement was applied to secure fiber implant for 1P illumination to allow positioning and restraint of the animal.
One week following AAV injection of the virus mixture baseline signals were recorded with fiberphotometry for 10 minutes in the home cage. Fiberphotometry is a method for measuring population calcium dependent fluorescence from deep brain structures using a single optical fiber for both excitation and emission in freely moving mice. Detailed description of the system can be found elsewhere3. After recordings, mice were connected to a 447nm laser (Opto Engine LLC, UT) using a 200 μm optical fiber, illuminated with 5mW, 100ms pulses, 1Hz for 30 minutes (TTL-controlled by OTPG_4, Doric lenses, Quebec, QC, Canada), in the home-cage. A week following light exposure, fiberphotometry signal was recorded again for 10 minutes. Fiberphotometry peak detection was performed with MATLAB (R2018a), using ‘findpeaks’ function, using a prominence of 2.5. Mice were perfused 4 weeks after illumination.
Animals were perfused with 4% paraformaldehyde (PFA). Brains were dissected and post-fixed in 4% PFA overnight at 4°C. Brains were then rinsed briefly with PBS and then sectioned at a thickness of 100 μm on a vibratome (VT1200 Leica Biosystems, IL, USA).
Fluorescent images from brain tissue were acquired with an LSM 880 confocal microscope (Carl Zeiss, Jena, Germany). We used a 10x Plan Apochromat air objective (NA 0.45), 25x Plan Apochromat water immersion objective (NA 1.2) and three laser wavelengths (488 nm, 561 nm, and 633 nm). Image acquisition was controlled by Zen 2011 software (Zeiss), which also allowed automated tiling, and maximum intensity projection. Images were not further processed. Expression counts were done by summation of the values of the fluorescence within 1mmX1mm below the fiber tip subtracted with the same area at the opposite hemisphere, line by line, and normalized to the maximal value.
Zebrafish experiments
All experiments were performed in compliance with UW institutional animal care and use committee. N-CreV and C-CreV were PCR amplified from pAAV-Ef1a NCreV and pAAV-Ef1a CCreV, and cloned into the pDest-ubi vector (Addgene plasmid # 27323) by Gibson assembly. The pDest-ubi:N-vCre-pDest and pDest-ubi:C-vCre mixture containing equal amounts of each plasmid (25pg each) and tol2 transposase RNA (25 pg) was injected into one-cell stage tg(ubi:Zebrabow-M) zebrafish embryos. Embryos were either light or dark reared for 72 hrs. At 3dpf injected embryos were anesthetized with Mesab, mounted in 2% agarose, and imaged on a Zeiss LSM 880 confocal microscope.
fMOST imaging and reconstructions
All tissue preparation has been described previously4. Following fixation, each intact brain was rinsed three times (6 h for two washes and 12 h for the third wash) at 4°C in a 0.01 M PBS solution (Sigma-Aldrich Inc., St. Louis, US). Then the brain was subsequently dehydrated via immersion in a graded series of ethanol mixtures (50%, 70%, and 95% (vol/vol) ethanol solutions in distilled water) and the absolute ethanol solution three times for 2 h each at 4°C. After dehydration, the whole brain was impregnated with Lowicryl HM20 Resin Kits (Electron Microscopy Sciences, cat.no. 14340) by sequential immersions in 50, 75, 100 and 100% embedding medium in ethanol, 2 h each for the first three solutions and 72 h for the final solution. Finally, each whole brain was embedded in a gelatin capsule that had been filled with HM20 and polymerized at 50°C for 24 h.
The whole brain imaging is performed using a fluorescence microscopic optical sectioning tomography (fMOST) system. The basic structure of the imaging system is the combination of a wide-field upright epifluorescence microscopy with a mechanic sectioning system. This system runs in a wide-field block-face mode but updated with a new principle to get better image contrast and speed and thus enables high throughput imaging of the fluorescence protein labeled sample (manuscript in preparation). Each time we do a block-face fluorescence imaging across the whole coronal plane (X-Y axes), then remove the top layer (Z axis) by a diamond knife, and then expose next layer, and image again. The thickness of each layer is 1.0 micron. In each layer imaging, we used a strip scanning (X axis) model combined with a montage in Y axis to cover the whole coronal plane5. The fluorescence, collected using a microscope objective, passes a bandpass filter and is recorded with a TDI-CCD camera. We repeat these procedures across the whole sample volume to get the required dataset.
The objective used is 40X WI with numerical aperture (NA) 0.8 to provide a designed optical resolution (at 520 nm) of 0.37 μm in XY axes. The imaging gives a sample voxel of 0.30 × 0.30 × 1.0 μm to provide proper resolution to trace the neural process. The voxel size can be varied upon different objectives. Other imaging parameters for GFP include an excitation wavelength of 488 nm, and emission filter with passing band 510–550 nm. 3D reconstructions were performed manually using Neurolucida 360 (NL360).
Cortical targeted in vivo 2P stimulation of single cells
For in vivo targeted single-cell optogenomic modifications as well as simultaneous GCaMP7f imaging experiments Stanford Administrative Panel on Laboratory Animal Care (APLAC) approved all animal procedures. Ai14 mice (The Jackson Laboratory, 07908) of two to four months age were used for experiments. For viral vector injection, mice were anesthetized with isoflurane. iCreV virus AAV2/PHP.eB-EF1a-iCreV, GFP virus AAV2/PHP.eB-CAG-GFP and GCaMP virus AAV2/9-CamkIIa-jGCaMP7f were used in all experiments. Viral vectors were loaded into glass pipettes and injected into cortices with picospritzer (Paker Hannifin). ~500 nL of the virus solution was delivered over 15 minutes and then a 4 mm craniotomy was made with the injection site at the center 30 minutes after the injection. Dura was removed followed by cover glass installation and sealing using UV curable adhesive -Loctite 4305. A custom-made head bar and cover were secured with dental cement on mouse skull. Imaging experiments started one month after the surgery to allow gene expression.
Mouse was mounted on a running wheel with head fixation and was remained awake during the whole experiment. Before imaging, the head mount cover was removed. In order to operate the mouse and the microscope, a red LED light was used for illumination (Wayllshine). Ultrasound gel (Parker, Aquasonic) was put on the cover glass for the water immersion objective lens. The mouse was aligned manually without checking the focal plane with eye piece to avoid iCreV induction during this process.
For induction experiments, a femtosecond Ti:sapphire laser (Spectra-Physics, Mai Tai) was tuned to 920 nm wavelength. The scanning and image acquisition were achieved with a Prairie (Bruker) two-photon microscope, through a 20X 0.95 N.A. water immersion lens (Olympus XLUMPLFLN-W 0.95 NA 20×). For all the imaging sessions, laser power at a specimen was kept at 25 mW and it was monitored (Thorlabs, PM100D and S130C) at an additional output of the optical path before entering the microscope, of which a splitting ratio was calibrated prior the measurements. During the first imaging session, a starting point with unique vessel pattern was identified and recorded. All target cells’ relative coordinates to the starting position were recorded and used for relocation in later sessions. Images were acquired with 4 μs pixel dwell time at 1024 × 1024 pixel frame size (field of view 450 micron x 450 micron), with 3 μm Z-axis step size. After identifying a target cell, an induction scanning was carried out using the ROI function to limit the scanning the middle of the target cell’s somata region. The scanning pixel dwell time is increased to 10 μs and various scanning conditions were used in each mouse tested. After the first induction session, the head cover was re-installed to seal the craniotomy from ambient light. Seven to ten days after the first session, all the cells were re-imaged to check tdTomato expression and at the end underwent a 30 minutes blue LED exposure (5 mW, 470 nm, Thorlabs, M470L3). The third imaging session was carried out seven to ten days after the LED exposure.
For calcium imaging experiment, a custom-made two-photon microscope was used with an objective lens Olympus XLUMPLFLN-W 0.95 NA 20×, 8 kHz resonant scanner head (Cambridge Technologies, CRS8K/6215H). Fluorescence light was captured using H11706–40 and H10770PA-40 photomultiplier tubes equipped with low noise amplifiers (Femto, DHPCA-100), with their analog signal subsequently digitized (National Instruments, NI-5732) and formed into images using a field programmable gate array (National Instruments, FPGA NI-7961R) and ScanImage software (Vidrio Technologies)6. GCaMP7f signal was collected at 30 Hz frame repetition rate for 30 minutes in each mouse.
DATA AVAILABILITY STATEMENT
All relevant plasmids will be deposited to Addgene. DNA sequences of the iCreV, iDreV and iFlpV created in this work are curated in National Institute of Health, GenBank. Accession codes are iCreV MN944913, iFlpV MN944914 and iDreV MN944915. All raw data presented in the paper are available from the corresponding author upon reasonable request.
CODE AVAILABILITY STATEMENT
The two-photon microscope was operated using ScanImage v5.3 (Vidrio Technologies, LLC) software and a custom written software written in LabView 2015 (National Instruments). The code is available upon request to authors.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Structured Science teams at the Allen Institute for technical support in stereotaxic injections and mouse colony management. The work was funded by the Allen Institute for Brain Science, NIMH BRAIN Initiative grant RF1MH114106 to A.C., NSFC Science Fund for Creative Research Group of China (Grant No. 61721092) to H.G., Q.L. and S.Z., NIH Brain Initiative RF1MH117069 grant to V.G., Colvin divisional fellowship of Division of Biology and Biological Engineering, California Institute of Technology, to A.K., and NIH Brain Initiative U01NS107610 grant to M.S. The creation of Ai139 mouse line was supported by the NIH grant R01DA036909 to B.T. The authors thank Sevi Durdu, Hilary Bayer, David Schrom, Bilal Kerman and Keisuke Yonehara for critical reading and feedback. The authors wish to thank the Allen Institute founder, Paul G. Allen, for his vision, encouragement, and support.
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Herculano-Houzel S The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci 3, 31, doi: 10.3389/neuro.09.031.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang ZJ & Zeng H Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci 36, 183–215, doi: 10.1146/annurev-neuro-062012-170307 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Nagy A Cre recombinase: the universal reagent for genome tailoring. Genesis 26, 99–109 (2000). [PubMed] [Google Scholar]
- 4.Branda CS & Dymecki SM Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6, 7–28 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Glaser S, Anastassiadis K & Stewart AF Current issues in mouse genome engineering. Nat Genet 37, 1187–1193, doi: 10.1038/ng1668 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Velez-Fort M et al. The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron 83, 1431–1443, doi: 10.1016/j.neuron.2014.08.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshel JH, Mori T, Nielsen KJ & Callaway EM Targeting Single Neuronal Networks for Gene Expression and Cell Labeling In Vivo. Neuron 67, 562–574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rompani SB et al. Different Modes of Visual Integration in the Lateral Geniculate Nucleus Revealed by Single-Cell-Initiated Transsynaptic Tracing. Neuron 93, 767–776 e766, doi: 10.1016/j.neuron.2017.01.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo L Fly MARCM and mouse MADM: genetic methods of labeling and manipulating single neurons. Brain Res Rev 55, 220–227, doi: 10.1016/j.brainresrev.2007.01.012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denk W, Strickler JH & Webb WW Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Taslimi A et al. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol 12, 425–430, doi: 10.1038/nchembio.2063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawano F, Okazaki R, Yazawa M & Sato M A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol 12, 1059–1064, doi: 10.1038/nchembio.2205 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Schindler SE et al. Photo-activatable Cre recombinase regulates gene expression in vivo. Scientific reports 5, 13627, doi: 10.1038/srep13627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung H et al. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat Commun 10, 314, doi: 10.1038/s41467-018-08282-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochrein L, Mitchell LA, Schulz K, Messerschmidt K & Mueller-Roeber B L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast. Nat Commun 9, 1931, doi: 10.1038/s41467-017-02208-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loros JJ & Dunlap JC Genetic and molecular analysis of circadian rhythms in Neurospora. Annual review of physiology 63, 757–794, doi: 10.1146/annurev.physiol.63.1.757 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Chen X & Yang Y Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9, 266–269, doi: 10.1038/nmeth.1892 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Hirrlinger J et al. Split-cre complementation indicates coincident activity of different genes in vivo. Plos One 4, e4286, doi: 10.1371/journal.pone.0004286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jullien N, Sampieri F, Enjalbert A & Herman JP Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res 31, e131 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P et al. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Scientific reports 2, 497, doi: 10.1038/srep00497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaidya AT, Chen CH, Dunlap JC, Loros JJ & Crane BR Structure of a light-activated LOV protein dimer that regulates transcription. Science signaling 4, ra50, doi: 10.1126/scisignal.2001945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoltowski BD et al. Conformational switching in the fungal light sensor Vivid. Science 316, 1054–1057, doi: 10.1126/science.1137128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo F, Gopaul DN & van Duyne GD Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389, 40–46, doi: 10.1038/37925 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Cardin JA et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459, 663–667, doi: 10.1038/nature08002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anastassiadis K et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech 2, 508–515, doi: 10.1242/dmm.003087 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Sauer B & McDermott J DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res 32, 6086–6095, doi: 10.1093/nar/gkh941 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano F, Suzuki H, Furuya A & Sato M Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun 6, 6256, doi: 10.1038/ncomms7256 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Andrews BJ, Proteau GA, Beatty LG & Sadowski PD The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell 40, 795–803 (1985). [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Narendra U, Iype LE, Cox MM & Rice PA Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Molecular cell 6, 885–897 (2000). [PubMed] [Google Scholar]
- 30.Raymond CS & Soriano P High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PloS one 2, e162, doi: 10.1371/journal.pone.0000162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20, 1172–1179, doi: 10.1038/nn.4593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70–71, doi: 10.1038/5007 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Daigle TL et al. A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480 e422, doi: 10.1016/j.cell.2018.06.035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan YA et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846, doi: 10.1242/dev.094631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong H et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat Commun 7, 12142, doi: 10.1038/ncomms12142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dana H et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16, 649–657, doi: 10.1038/s41592-019-0435-6 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Joyner AL & Zervas M Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn 235, 2376–2385, doi: 10.1002/dvdy.20884 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Dimidschstein J et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 19, 1743–1749, doi: 10.1038/nn.4430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu YJ et al. Tracing inputs to inhibitory or excitatory neurons of mouse and cat visual cortex with a targeted rabies virus. Curr Biol 23, 1746–1755, doi: 10.1016/j.cub.2013.07.033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron, doi: 10.1016/j.neuron.2016.09.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 1.Cetin A, Komai S, Eliava M, Seeburg PH & Osten P Stereotaxic gene delivery in the rodent brain. Nature Protocols 1, 3166–3173 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Franklin KBJ, & Paxinos G The mouse brain in stereotaxic coordinates. Vol. 18th Edition (1997). [Google Scholar]
- 3.Cho JR et al. Dorsal Raphe Dopamine Neurons Modulate Arousal and Promote Wakefulness by Salient Stimuli. Neuron 94, 1205–1219 e1208, doi: 10.1016/j.neuron.2017.05.020 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Gang Y et al. Embedding and Chemical Reactivation of Green Fluorescent Protein in the Whole Mouse Brain for Optical Micro-Imaging. Front Neurosci 11, 121, doi: 10.3389/fnins.2017.00121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li A et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408, doi: 10.1126/science.1191776 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Pologruto TA, Sabatini BL & Svoboda K ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online 2, 13, doi: 10.1186/1475-925X-2-13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant plasmids will be deposited to Addgene. DNA sequences of the iCreV, iDreV and iFlpV created in this work are curated in National Institute of Health, GenBank. Accession codes are iCreV MN944913, iFlpV MN944914 and iDreV MN944915. All raw data presented in the paper are available from the corresponding author upon reasonable request.