Abstract
R-loop structures are a prevalent class of alternative non-B DNA structures that form during transcription upon invasion of the DNA template by the nascent RNA. R-loops form universally in the genomes of organisms ranging from bacteriophages, bacteria, and yeasts to plants and animals, including mammals. A growing body of work has linked these structures to both physiological and pathological processes, in particular to genome instability. The rising interest in R-loops is placing new emphasis on understanding the fundamental physicochemical forces driving their formation and stability. Pioneering work in Escherichia coli revealed that DNA topology, in particular negative DNA superhelicity, plays a key role in driving R-loops. A clear role for DNA sequence was later uncovered. Here, we review and synthesize available evidence on the roles of DNA sequence and DNA topology in controlling R-loop formation and stability. Factoring in recent developments in R-loop modeling and single-molecule profiling, we propose a coherent model accounting for the interplay between DNA sequence and DNA topology in driving R-loop structure formation. This model reveals R-loops in a new light as powerful and reversible topological stress relievers, an insight that significantly expands the repertoire of R-loops' potential biological roles under both normal and aberrant conditions.
Keywords: DNA structure, DNA topology, DNA transcription, DNA topoisomerase, chromosomes, gene transcription, genome stability, R-loop, supercoiling, superhelicity, topological stress
DNA superhelicity: What is it, and why does it matter?
DNA superhelicity, discovered over 50 years ago (1–3), is an essential physical property of the DNA double helix that can be most easily understood for closed circular duplex DNA molecules, such as plasmids. Each strand in a circular duplex DNA is a circle, and these two circles are interlinked due to the helical nature of DNA (4). The number of times either strand crosses through the closed circle formed by the other strand is a fixed integer called the linking number (Lk).3 Lk can only be changed by transiently cutting one or both strands, followed by strand passage or rotation and religation. In the absence of any external stress, the “relaxed” linking number value, denoted here Lk0, will reflect the geometry of the Watson–Crick B form DNA, with one strand crossing every ∼10.5 bp. Molecules with different Lk values (referred to as topoisomers) experience varying degrees of superhelical stress depending on their linking difference, α = Lk − Lk0, also called superhelicity. Superhelicity can be either positive or negative, reflecting an excess or deficit of strand crossings, respectively. The superhelix density, σ = α/Lk0, allows comparisons of the levels of superhelicity in molecules of different lengths.
Superhelicity can also be imposed on noncircular DNA molecules. If a piece of linear DNA is held between semi-rigid attachment points such that the diffusion of superhelical stresses beyond these points is blocked, a topologically constrained domain is formed. Examples include CTCF-anchored topologically associated domains (TADs) and lamin-associated domains in mammalian genomes (5–7). Chromatin templates themselves are also topologically constrained (8, 9). Simple sequence-specific DNA-binding factors, such as the lac repressor, can act as topological domain boundaries (10–13). Prokaryotic circular genomes are partitioned into multiple dynamic, negatively supercoiled domains that contribute to genome architecture (14–16). Thus, a topological domain is any portion of a DNA molecule, linear or circular, on which superhelicity can be imposed. Although the linking difference imposed on a domain can only be changed by transiently cutting one or both of the strands, several processes alter how superhelicity is distributed within a domain (see below).
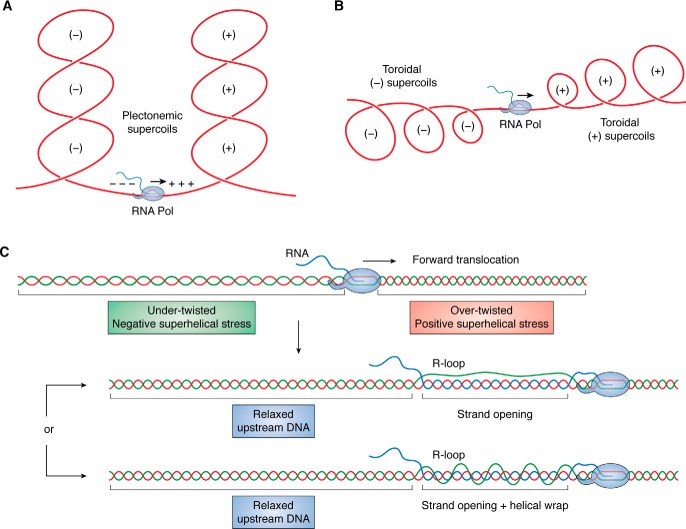
DNA superhelicity is critical to biology for a variety of reasons. First, the three-dimensional (3D) shapes available to a superhelical DNA molecule must satisfy geometrical twisting and writhing constraints, as first elucidated by Vinograd and Lebowitz (1, 17). The strain on a negative superhelical domain can be accommodated either as undertwist of the DNA duplex or by the formation of toroidally or plectonemically writhed structures (18) (Fig. 1). These higher-order structures are biologically relevant because they contribute to genome folding in 3D space, increase local proximity of DNA sequences, and affect local concentrations of DNA- and RNA-binding factors involved in gene expression control and genome dynamics (19). When negative superhelicity is expressed as undertwist, the DNA can undergo strand separation, exposing the two strands in a single-stranded bubble (Fig. 1). Superhelical duplex destabilization can be critical for processes that require strand opening, such as the initiation of transcription and of DNA replication. Promoter and origin regions in prokaryotes have evolved AT-rich DNA sequences that can efficiently transition to a strand-separated state (20–24). Hundreds of Escherichia coli genes respond to superhelicity changes, supporting the notion that topology plays an important role in the control of gene expression (25–27). Negative superhelicity is also thought to facilitate transcription elongation, whereas excessive positive supercoiling impedes it (28).
Figure 1.
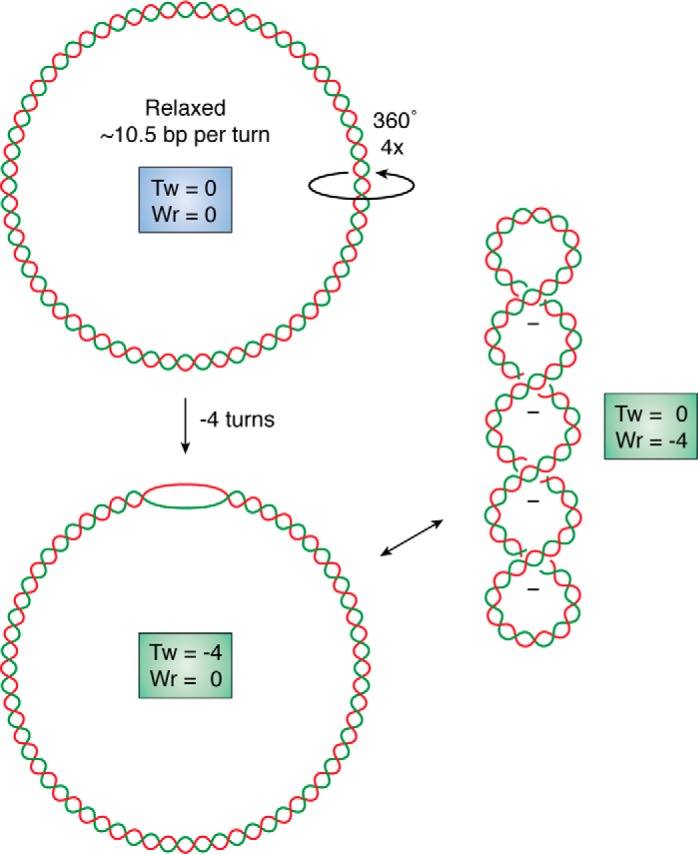
Superhelicity introduces torsional stress and modifies DNA shape. A covalently closed circular DNA molecule is shown at the top in a relaxed state. The introduction of four negative supercoils causes undertwist (Tw) stress in the molecule, which can be expressed as a change in twist shown at the bottom as strand opening or as a change in overall shape shown at the right by the formation of a negatively supercoiled writhed (Wr) structure.
DNA superhelicity also affects protein-DNA interactions. A host of DNA-binding proteins sense the topological state of DNA, with negative superhelicity generally facilitating protein-DNA interactions. Nucleosomes, for instance, preferentially form on negatively supercoiled DNA, with each nucleosome-binding event stabilizing one negative superhelical turn (29, 30). The replication initiation proteins of E. coli (DnaA) and of Drosophila (ORC1), along with a number of transcription factors (31–36), also prefer binding to negatively supercoiled DNA. Negatively supercoiled topological domains therefore represent hubs of genome organization and activity in both prokaryotes and eukaryotes (37, 38). Negative superhelicity can also induce the formation of alternative non-B DNA structures (see below), and these structures, in turn, can regulate gene expression (26, 39, 40).
All genomes experience superhelical stresses
All genomes, whether of viral, prokaryotic, archaeal, or eukaryotic origin, experience superhelical stresses as a result of transcription and replication. These processes involve large macromolecular machines that translocate processively along the DNA (41). Impediments to the rotation of these complexes around the DNA axis force the DNA to rotate instead, which imposes large amounts of torque on the fiber (30, 42). This leads to a dynamic repartitioning of superhelicity, whereby a diffusive wave of positive superhelicity is pushed ahead of the advancing replication and transcription forks, and a wave of negative superhelicity of equal magnitude is produced behind. During transcription, the main focus of this review, this repartitioning of positive and negative superhelicity is referred to as the “twin supercoiling domain model” (43, 44) (Fig. 2, A and B).
Figure 2.
Transcription generates superhelical stresses that can be mitigated by R-loop formation. Transcription-driven supercoiling leads to the formation of dual waves of positive (downstream) and negative (upstream) superhelicity depicted here as interlinked plectonemic structures (A) or toroidal structures (B). C, topological disruptions caused by transcription are shown as undertwist (upstream) and overtwist (downstream). As the RNA polymerase (light blue) translocates forward, an R-loop initiates and extends. R-loop formation relaxes the upstream negative superhelical stress by absorbing undertwist within the strand opening that accompanies the formation of long R-loops. In addition, the displaced looped out ssDNA strand may wrap around the RNA:DNA hybrid in a left-handed helical fashion (bottom), further absorbing negative superhelicity.
Transcription-induced supercoiling is a major source of superhelical stress in all genomes. Direct DNA topology measurements in mammalian cells established that transcription creates a ∼1.5-kb negatively supercoiled domain upstream of (i.e. behind) active promoters (45). The superhelix density achieved varied with gene activity, reaching σ = −0.07. Assuming that transcription generates reciprocal superhelical density waves of |σ| = 0.05, then 150 superhelical turns of each sign will be generated during each round of transcription of an average length human gene (30 kb). On a larger scale, Naughton et al. (46) showed that the human genome consists of a series of underwound and overwound domains delineated by both GC/AT sequence transitions and binding sites for the domain-organizing CTCF protein. Underwound domains were transcriptionally active and enriched for open chromatin, and their topological states were dynamically responsive to transcription inhibition. Interestingly, transcription and its ability to form topological domains has been linked to the large-scale folding of chromatin domains (46). Independent modeling experiments have suggested that transcription-induced supercoiling facilitates both cis-interactions between loci in TADs, including promoter-enhancer contacts (47–49), and chromatin loop extrusion (19, 50).
Not surprisingly, mechanisms that manage topological stresses are essential for efficient transcription and replication. DNA topoisomerases, a family of ubiquitous and conserved proteins (for recent reviews, see Refs. 51 and 52) transiently cut, move, and religate DNA strands to relax superhelicity. Type I topoisomerases, which transiently cut one strand of DNA to relax superhelicity, occur in all free-living organisms. Type II topoisomerases make dsDNA cuts and then pass another part of the duplex through the gap before religation, changing ΔLk by 2. Whereas most type II topoisomerases relax DNA, the prokaryotic DNA gyrases introduce negative supercoils to offset positive superhelical stresses (53). As a result, topological domains in prokaryotes are maintained in a negatively superhelical state with an average superhelix density σ = −0.05 (15, 54).
DNA topoisomerases are closely associated with, and necessary for, normal transcription. In mammals, DNA topoisomerase I directly binds to the transcription machinery and becomes catalytically activated upon release into productive elongation (55). This fine-tuned mechanism for DNA topoisomerase I control enables the dynamic removal of positive supercoils ahead of the machinery to facilitate elongation, while preserving promoter-opening negative superhelicity toward the transcription start site. A role for DNA topoisomerase I in favoring elongation was also observed for long genes (56–58). Topo II enzymes, by contrast, have been implicated in the local management of excess supercoils around transcription start sites, particularly for highly expressed genes (45, 59). The Top2B enzyme has been implicated in managing the excess promoter-proximal supercoils generated upon heat shock and serum and hormone induction at the expense of the generation of DNA breaks (60–62).
Superhelicity favors transitions to alternative non-B DNA structures
Superhelicity represents a high energy state for DNA. At equilibrium, the energy associated to DNA superhelicity is quadratic and can be modeled as E(α) = ½Kα2, where α is the linking difference (63, 64). The twisting and writhing of the molecule in three dimensions reflects this energy as the DNA deforms to accommodate the imposed superhelicity. As negative superhelicity increases, transitions to alternative non-B form DNA structures will become favored. Strand opening, for example, absorbs undertwist, thereby allowing the rest of the domain to relax a corresponding amount. Whereas the transition itself costs energy through the formation of junctions between duplex B-DNA and the alternative structure itself, the accompanying relaxation provides an energy return. If this return is larger than the transition cost, then the transition is favored at equilibrium. Negative superhelicity drives transitions to a wide variety of alternative structures in vitro, including strand-separated (i.e. melted) DNA, Z-form DNA, cruciforms, H-form DNA, and R-loops, the focus of this review (39, 40, 65). Which transitions occur in a specific domain depends on base sequence and the level of superhelicity it experiences. Importantly, long genomic sequences will often carry multiple regions susceptible to forming various alternative structures. These structures exist in a competitive equilibrium because the available superhelicity couples together the transition behaviors of all susceptible sites in the domain. This coupling happens because a transition at any one site will absorb superhelicity, thus decreasing the amount remaining to drive transitions elsewhere and thereby lowering their likelihood (66, 67). The existence of many thousands of alternative DNA structures in the genomes of activated B cells in vivo demonstrates the presence of vast stores of negative superhelicity in mammalian genomes (68).
R-loops: Prevalent non-B DNA structures
R-loops are three-stranded nucleic acid structures consisting of an RNA:DNA hybrid and a displaced ssDNA strand (69). R-loops can form in trans as a result of the invasion of an RNA strand into complementary dsDNA. Such invasion typically requires protein-mediated catalysis, either by components of the homology-directed DNA recombination machinery (70, 71) or by CRISPR-Cas systems (72, 73). R-loops also form in cis during transcription upon hybridization of the nascent RNA with the template DNA strand behind the advancing RNA polymerase (RNAP) (74). The formation of a nascent RNA:DNA hybrid leaves the nontemplate DNA strand unpaired and free to wrap around the hybrid duplex (Fig. 2C). Co-transcriptional R-loops are the primary focus of this review.
R-loops are known to form in every genome where they have been looked for, including bacterial plasmids (75, 76), bacterial genomes (77–79), and bacteriophages (80), as well as mitochondrial genomes (81). Genomic profiling studies have confirmed that R-loops are prevalent in the nuclear genomes of eukaryotes, covering 3–5% of the genome in yeasts (82–85), plants (86), and mammals (87–90). In humans, R-loops form over tens of thousands of broadly conserved genic hotspots that are enriched at gene ends (69, 87). Importantly, R-loop formation is a dynamic process that occurs at modest frequencies. In a human cell population at steady state, R-loop frequencies range from 0.5 to 10%, depending on the locus and its sequence, transcription levels, and overall gene length (87, 90). Similar links between R-loop distribution, gene expression, and sequence were observed in yeast and plants (82–84, 86).
Given their prevalence, R-loops represent an important class of non-B DNA structures that is increasingly the subject of investigation. R-loops can be biochemically reconstituted with high efficiency using simple in vitro transcription systems (91–93), and a variety of orthogonal methods have been developed to report their formation in plasmids and chromosomes (88, 94, 95). Studies have linked R-loops to a range of both positive and negative cellular outcomes, suggesting that these structures not only form in genomes but also are biologically relevant. The main purpose of this review is to highlight our understanding of the physiochemical forces that underlie R-loop formation, focusing on the role of DNA topology. We refer readers to recent reviews that cover the possible biological roles of R-loops in health and disease (69, 74, 96–99).
Understanding R-loops from first principles
As with other non-B DNA structures, an R-loop will be favored to form if its energy at equilibrium is lower than that of B-form DNA. As with other alternative DNA structures, the largest energy barrier to R-loop formation is the formation of the two junctions between duplex DNA and the R-loop itself. Junction energies have been measured for B/Z transitions and strand separation in the range of 10–11 kcal/mol/pair of junctions (100–103). It is possible that the value for R-loops is even higher, given that three strands must be accommodated instead of two. A recent energy-based equilibrium energy–modeling approach (104) reveals that R-loops can compensate for this high junctional cost using two complementary paths: DNA sequence and DNA topology.
Making an R-loop involves breaking DNA:DNA base pairs and forming RNA:DNA base pairs. If the energy of the RNA:DNA base pairs is lower than that of the DNA:DNA base pairs with the same sequence, the energy of the structure as a whole will be reduced. When analyzed as dinucleotides, seven of the 16 possible combinations favor the RNA:DNA state over the DNA duplex (105). Most of these sequences are G-rich or purine-rich. This analysis suggests that R-loops should prefer to form in G-rich and G/A-rich regions of transcripts. Experimental evidence indeed shows that R-loops form efficiently from G-rich transcripts (92, 93) and that G clusters constitute strong initiation points for R-loops (106, 107). GC-rich DNA sequences that show strand asymmetry in the distribution of G and C bases (GC skew) are prone to R-loop formation at endogenous loci (88, 94, 108) and in vitro (88, 92–94). Evidence from R-loop mapping studies shows that R-loop hotspots are generally (but not always) enriched for GC-skewed regions and purine-skewed regions (83, 86, 87, 95). These results indicate that R-loop formation in a wide variety of organisms follows at least in part the intrinsic thermodynamic landscape of RNA:DNA versus DNA:DNA base pairing.
DNA topology provides a second way by which R-loops can lower their energy below that of superhelical duplex DNA. A role for negative supercoiling in favoring R-loops has been experimentally established through a strong body of work in E. coli (for a review, see Ref. 109). Strains of E. coli deficient for the DNA relaxing DNA topoisomerase I enzyme were shown to accumulate R-loops (76), highlighting the key role of negative superhelicity in driving R-loops during transcription. DNA gyrase, with its ability to introduce negative supercoiling into DNA (53), was the primary driver of transcription-associated hypernegative supercoiling and of R-loop formation (76). DNA topoisomerase I activity, by contrast, suppressed this phenomenon (76, 91, 110, 111). Strains overexpressing RNase H, an enzyme that specifically degrades RNA in the context of RNA:DNA hybrids, partially rescued the defects arising from DNA topoisomerase I deficiency (112) and abolished the formation of DNA gyrase– and R-loop–dependent hypernegatively supercoiled plasmids (91, 111). This work suggested that R-loop formation is a common occurrence in prokaryotes that is dynamically regulated by both pro-R-loop (negative supercoiling) and anti-R-loop (DNA topoisomerase I, RNase H) factors.
One interpretation of these early findings is that the underwound state of DNA associated with transcription-driven negative supercoiling (43) produces partially melted regions that permit invasion by the nascent RNA, initiating an R-loop. However, analysis of superhelical duplex destabilization shows that sites of strand separation are confined to the AT-richest regions of a domain (24). By contrast, most R-loops occur in G/C-rich locations. Thus, whereas transient supercoiling-induced strand separation or base unstacking cannot be ruled out as contributing to R-loop initiation, this view may not adequately capture the role of negative superhelicity. We suggest instead that, as described for other non-B DNA alternative structures, negative superhelicity constitutes a high-energy, stressed state of the DNA that is efficiently relieved by R-loop formation and the relaxation it produces (104). The structure of an R-loop lends itself well to relaxing negative superhelicity. In an R-loop, the two DNA strands are separated and no longer twist around each other; this allows surrounding undertwist to migrate into the R-loop bubble, relaxing the rest of the domain. Every time an R-loop grows by the helical pitch of DNA (10.5 bp), it absorbs an additional negative superhelical turn. Assuming that the displaced strand is free (which may not necessarily be the case (113, 114)), it can helically wrap around the RNA:DNA hybrid, and, if the wrap is left-handed, this will absorb additional undertwist (Fig. 2C). These two effects together provide significant stress relief to the DNA domain. The ability of R-loops to lower the energy level of the DNA fiber provides a clear alternative explanation for the role of negative superhelicity in their formation and stability. Altogether, if the combined energy saving resulting from favorable base pairing and topological relaxation exceeds the junction energy cost, then R-loop formation will be favored to occur at equilibrium.
R-loops are powerful, reversible, superhelical stress relievers
This model reveals R-loops in a new light as nonenzymatic topological “stress relief valves.” In support of this notion, R-loop formation upon in vitro transcription of supercoiled circular plasmids is well-known to cause significant plasmid relaxation. We showed that a 3.5-kb plasmid carrying ∼18 negative supercoils (assuming a superhelix density of σ = −0.05 for DNA extracted from E. coli) was partially to fully relaxed by an R-loop, indicating that these structures absorb an astounding amount of superhelicity (104). The amount of superhelical stress relief afforded by an R-loop depends primarily on its length. R-loop sizes can be analyzed at the single-molecule level by measuring the single-stranded character of the displaced DNA strand using long-read sequencing (94). Now adapted for PacBio sequencing (115), single-molecule R-loop footprinting (SMRF-Seq) was used to measure the lengths of the in vitro R-loops formed on the plasmid substrate described above. The majority of the structures ranged from 80 to 175 bp, with a median length of 120 bp (104). Such structures are expected to relax ∼8–17 supercoils based on the length of the untwisted region. The helical wrapping of the displaced strand is expected to relax an additional 1–2 supercoils. Thus, R-loops of median length 120 bp relax the large majority of the negative superhelicity present in plasmids nearly 30 times their size, demonstrating the ability of R-loops to act as long-range topological relief valves.
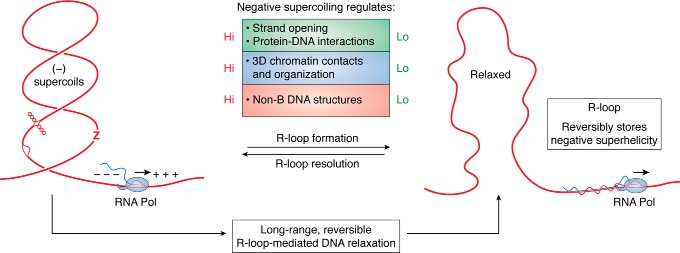
Because the relaxation provided by R-loops is expected to grow roughly linearly with their length (104), it is important to understand the distribution of R-loop sizes in genomic sequences. Using SMRF-Seq, we interrogated a number of R-loop hotspots in the human genome at ultradeep coverage. R-loop lengths typically ranged from 200 to 500 bp, a full order of magnitude larger than other non-B DNA structures (68). Strikingly, kilobase structures, while rarer, were not uncommon (115), consistent with prior mapping data on R-loop–prone murine class switch regions (94, 108, 116, 117). R-loops are giants in the world of non-B DNA structures and are uniquely suited to absorb large amounts of negative DNA superhelicity. A 300-bp-long R-loop (the median length of genomic R-loops (115)) is expected to fully relax a 6–7-kb DNA molecule with a superhelix density σ = −0.05. Rare kilobase-size R-loops are expected to relax vast amounts of negative superhelicity, affecting the topology of the DNA fiber over long distances. Importantly, because R-loops can hold vast stores of negative superhelicity, R-loop resolution can release this superhelicity back into the surrounding DNA domain (see Fig. 4). Indeed, R-loop formation does not involve any change in linking number: the total superhelicity of a given domain is only transiently repartitioned by either R-loop formation or resolution.
Figure 4.
R-loop formation mediates long-range reversible topological relaxation. The consequences of R-loop formation and resolution on the properties of the local chromatin environment are described. When R-loops are resolved, large stores of negative superhelicity are released into the chromatin fiber, driving increased chromatin contacts, increased non-B DNA structure formation (bubble DNA, cruciform, and Z DNA are depicted), and increased protein-DNA interactions. See text for details.
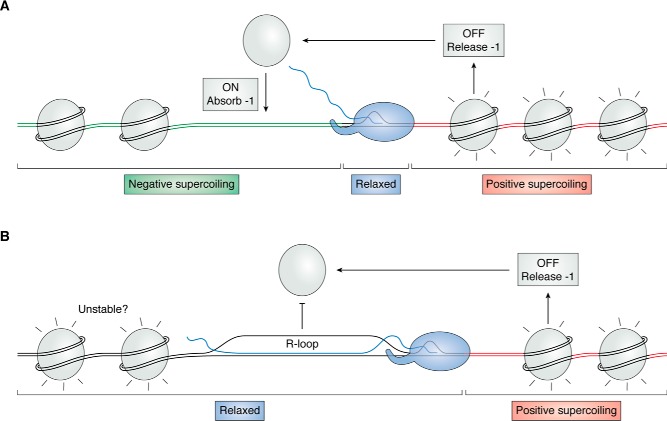
Whereas these considerations hold true for “naked” DNA, eukaryotic chromosomes carry nucleosome arrays. Each nucleosome binds ∼147 bp of DNA and stabilizes one turn of negative superhelicity (29, 118, 119), indicating that the superhelix density of nucleosomal winding is −0.07. The dissociation of nucleosomes downstream (i.e. in front of) the RNA polymerase releases this stored negative superhelicity, counteracting the transcription-driven wave of positive supercoiling (Fig. 3A). Similarly, the reassociation of nucleosome behind the RNAP will stabilize one negative supercoil, lessening the accumulation of negative superhelical stress in the wake of the RNAP complex. The fact that positive supercoiling destabilizes nucleosomes and negative supercoiling facilitates their formation (120–122) nicely agrees with the notion that nucleosomes play important roles in managing supercoiling during transcription (45, 46, 123, 124). Nonetheless, empirical observations show that transcription results in a build-up of upstream negative supercoils (45, 46). In this situation, R-loops can play useful roles in relieving this superhelical stress. This, in turn, is expected to impede or prevent nucleosome redeposition behind the RNAP for two reasons. First, R-loops cannot be wrapped around nucleosomes (125), and second, relaxed DNA is a poor substrate for nucleosome formation (120, 122). This agrees with observations that R-loops are associated with increased chromatin accessibility under normal conditions (87). Because R-loops are capable of absorbing virtually all of the negative superhelicity in large regions, R-loop formation could reduce the strength of nucleosomal binding over neighboring domains (Fig. 3B). Conversely, the release of the topology stored in an R-loop upon its resolution is also expected to favor rapid nucleosome binding. The details of the energetics and kinetics of these important interactions between nucleosomal binding and R-loops largely remain to be elucidated.
Figure 3.
Nucleosomes and R-loops share in topological relief duties. A, positive supercoiling (red) traveling ahead of the translocating RNAP (shown here localized to the linker DNA) destabilizes nucleosomes, favoring their extrusion. Nucleosome release frees one negative supercoil, mitigating the buildup of positive superhelicity. Reassociation of the nucleosome behind the polymerase is favored by the transcription-driven negative supercoiling; sequestration of one negative supercoil upon nucleosome reformation relieves that stress. B, the formation of an R-loop results in two main consequences. First, nucleosome redeposition is inhibited. Second, the accompanying relaxation of the upstream region may weaken surrounding nucleosome-DNA contacts.
The interplay between DNA sequence and DNA topology guides R-loop formation, elongation, and stability
The notion that DNA topology and DNA sequence cooperate to control local R-loop propensity predicts that at one extreme, R-loops could occur over highly favorable sequences without the need for significant superhelical relief. In vitro transcription of highly GC-skewed class switch sequences and CpG island regions results in R-loop formation even on linear templates (104, 126). Measurements of R-loop efficiency as a function of substrate topology nonetheless revealed that negatively supercoiled plasmids favored R-loops by at least one order of magnitude, even in the context of an R-loop prone sequence (104). At the other extreme, very high levels of superhelicity are predicted to support R-loop formation even over unfavorable DNA regions. This was confirmed by bulk in vitro transcription assays (127). Examination of individual in vitro R-loop footprints at deep coverage confirmed that R-loops tend to initiate over unfavorable sequences when plasmids are highly negatively supercoiled (104). Most R-loops observed when negative supercoiling was high were promoter-proximal, likely because these early R-loops formed first, and the relaxation they provide inhibits structure formation over more favorable downstream regions. Thus, increased negative superhelicity can alter both the frequency and the landscape of R-loop formation, at least in vitro.
Under most conditions, R-loop formation is likely to require a balance of contributions from DNA sequence and topology. Calculations suggest that the vast majority of genomic DNA sequences will require some negative superhelicity to permit R-loop formation. This, in turn, implies that the regions observed to form R-loops in vivo are likely experiencing negative supercoiling. Thus, R-loop maps could inform us, albeit indirectly, about local in vivo levels of superhelical stresses. Regions with the most favorable RNA:DNA energetics are expected to permit R-loops even with low superhelicity (104). As such, the conserved class of highly GC-skewed CpG island promoters (128) may represent sensitive R-loop “reporters” that have evolved to transition into R-loop structures at low levels of superhelical stress. By contrast, SMRF-Seq analysis revealed multiple examples of R-loop hotspots whose sequences are not strongly favored to form R-loops (115). This suggests that these regions, which often are located in gene bodies or terminal genic regions, may experience high levels of local superhelicity that allow R-loop formation despite their weaker sequence favorability. However, we cannot rule out the possibility that R-loops might originate via multiple mechanisms, some of which could be less dependent on fundamental nucleic acid thermodynamic properties.
Whereas R-loop sequence signatures can be used to indirectly infer the possible contribution of DNA topology to their formation, direct evidence for its role is also supported in eukaryotes in vivo. In yeast, loss of DNA topoisomerase I leads to increased R-loop levels over the 5′-end of the highly transcribed rDNA region (129). R-loop frequencies were further enhanced upon loss of RNase H activity (129), similar to prior observations in E. coli. In human cells, depletion of Top1 leads to a compensatory accumulation of R-loops in genes with acute topological management needs, namely long, highly transcribed, and physically constrained genes (57). Thus, DNA topology regulates R-loop formation in silico, in vitro, and in vivo, from E. coli to yeast to human cells. Furthermore, the evidence suggests that DNA topoisomerase I and R-loops share topological management duties, with R-loops playing compensatory roles in relieving negative superhelical stress in the absence of Top1.
In addition to regulating R-loop initiation, DNA topology was also predicted to control R-loop extension and therefore length (104). Once initiated, R-loops are likely to extend until the available superhelicity that drives their formation has been (partially or fully) relaxed. Alternatively, R-loops may terminate if the underlying DNA sequence becomes highly unfavorable. Discerning between these two possibilities may be possible by analyzing sequence transitions at the distal edges of individual R-loops. We and others (116) have observed that distal DNA sequence features are often less well-defined than those at proximal edges and that R-loops with the same initiation site can end at multiple downstream locations. Prior observations have shown that, although favorable G stretches are often necessary at the proximal edges of R-loops, these structures can extend through areas that otherwise would not support initiation (106). These results are expected if DNA topology regulates R-loop extension. From this, we can deduce that the length distribution of R-loops may provide information about the level of local superhelical stress that existed in their domains prior to their formation.
Finally, our work predicts that the relative contributions of DNA sequence and DNA topology to the formation of a particular R-loop will also determine the stability of that structure when faced with a change in the topology of the fiber such as might be expected from DNA topoisomerase action or strand breakage. As expected, R-loops that are primarily driven by DNA topology over less favorable sequences are highly susceptible to spontaneous resolution when that topology is lost (104). By contrast, R-loops that form over sequences with strong RNA:DNA base-pairing potential are much more resistant to topological changes. These findings have implications for our understanding of the instances of RNA:DNA hybrid or R-loop formation observed at sites of DNA double-strand breaks (130–132). Because the loss of topology induced by double-strand breaks is detrimental to both R-loop initiation and stability, we favor models in which two-stranded RNA:DNA hybrids are formed at the break, either through rehybridization of the nascent RNA to a resected ssDNA strand or through de novo loading of RNA polymerase II (130). Overall, the sensitivity of R-loops to DNA topology suggests that superhelicity-relaxing enzymes might provide an attractive mechanism for R-loop resolution. We note that D-loops, which are structurally similar to R-loops and form during recombination, are efficiently resolved through the combined action of the SGS1 helicase and Top3A DNA topoisomerase (133). Interestingly, topoisomerase 3B can cleave R-loops and D-loops (134) and reduce R-loop formation in vitro through its DNA relaxation activity (135). Loss of the Top3B-interacting partner TDRD3 causes slight elevation of R-loop levels and genome instability (135). Whether Top3B directly acts on nuclear R-loops in vivo remains to be determined, given that Top3b also plays an important role as an RNA topoisomerase in the cytoplasm (136, 137).
Rethinking the potential roles of R-loops under normal and pathological conditions
As discussed above, negative superhelicity is an important and often neglected regulator of gene expression and genomic architecture. The observation that R-loops can transiently absorb and release large stores of negative superhelicity expands the repertoire of potential biological roles of these structures (Fig. 4). First, because negative supercoiling favors local strand opening, it may facilitate promoter and/or replication origin firing. An R-loop formed downstream of a promoter region will sequester local superhelicity, which would negatively impact strand opening and hence promoter activity. By contrast, the wave of supercoiling released upon resolution of this R-loop would facilitate promoter firing. The dynamic formation and resolution of R-loops may thus contribute to long-range regulation of gene expression. A similar logic can be applied to replication origins, which are often located near gene ends where R-loops are particularly prevalent (138, 139). Evidence for supercoiling-mediated long-distance communication between RNA polymerases now exists in Bacteria (140), and transcription contributes to the long-range mobility of cis-regulatory elements in mammals (141). Second, given that alternative non-B DNA structures compete for negative superhelicity (39, 40, 65), the sequestration of superhelicity by R-loop formation is expected to dramatically curtail the formation of other non-B DNA structures. Conversely, supercoil release by R-loop resolution will enable the formation of other structures, such as strand-separated bubbles, B/Z transitions, and cruciforms (Fig. 4). Third, negative supercoiling is known to facilitate some protein-DNA interactions, such as nucleosome binding, and to enhance long-range contacts between distant loci, such as promoters and enhancers (48). Transcription-induced supercoiling in particular was proposed to drive the formation of TADs (19, 50). Because the relaxation activity of R-loops is expected to affect large surrounding areas, it is possible that R-loops may exert a dynamic impact on the local structure of chromatin and on its folding in 3D space, affecting long-range contacts between distant loci.
It is interesting to consider these possible roles in light of the fact that dysfunctions of R-loop metabolism have often been invoked as a source of genomic instability. Excess R-loop formation, because it would sequester so much superhelicity, might be expected to cause TAD unfolding and chromatin opening, lower promoter-enhancer contacts, reduce the probability of promoter and origin firing, and lower the frequencies of occurrence of other non-B DNA structures (Fig. 4). This comes in addition to R-loops' documented detrimental effects on transcription elongation (129, 142–146) and on potentiating transcription-replication conflicts (69, 99). By contrast, lower R-loop levels, resulting, for instance, from enhanced R-loop resolution activity, might be expected to lead to higher levels of negative superhelicity throughout the genome. This, in turn, could favor the formation of other alternative non-B DNA structures, such as cruciforms, triplexes, strand-separated bubbles, and B/Z transitions, which might affect normal DNA replication and create sites susceptible to cleavage and mutagenesis (147, 148). Thus, it is possible that altered R-loop homeostasis, whether characterized by increased or decreased R-loop loads, could lead to strong negative consequences for genome function and stability.
Acknowledgments
We thank members of the Chedin laboratory for feedback and useful discussion.
This work was supported by National Institutes of Health Grant R01-GM120607 (to F. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Lk
- linking number
- CTCF
- CCCTC-binding factor
- SMRF-Seq
- single-molecule R-loop footprinting
- TAD
- topologically associated domain
- 3D
- three-dimensional
- RNAP
- RNA polymerase.
References
- 1. Vinograd J., and Lebowitz J. (1966) Physical and topological properties of circular DNA. J. Gen. Physiol. 49, 103–125 10.1085/jgp.49.6.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vinograd J., Lebowitz J., Radloff R., Watson R., and Laipis P. (1965) The twisted circular form of polyoma viral DNA. Proc. Natl. Acad. Sci. U.S.A. 53, 1104–1111 10.1073/pnas.53.5.1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lebowitz J. (1990) Through the looking glass: the discovery of supercoiled DNA. Trends Biochem. Sci. 15, 202–207 10.1016/0968-0004(90)90163-6 [DOI] [PubMed] [Google Scholar]
- 4. Watson J. D., and Crick F. H. (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738 10.1038/171737a0 [DOI] [PubMed] [Google Scholar]
- 5. Dekker J., and Mirny L. (2016) The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rowley M. J., and Corces V. G. (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yáñez-Cuna J. O., and van Steensel B. (2017) Genome-nuclear lamina interactions: from cell populations to single cells. Curr. Opin. Genet. Dev. 43, 67–72 10.1016/j.gde.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 8. Mondal N., and Parvin J. D. (2001) DNA topoisomerase IIα is required for RNA polymerase II transcription on chromatin templates. Nature 413, 435–438 10.1038/35096590 [DOI] [PubMed] [Google Scholar]
- 9. Mondal N., Zhang Y., Jonsson Z., Dhar S. K., Kannapiran M., and Parvin J. D. (2003) Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 31, 5016–5024 10.1093/nar/gkg705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding Y., Manzo C., Fulcrand G., Leng F., Dunlap D., and Finzi L. (2014) DNA supercoiling: a regulatory signal for the λ repressor. Proc. Natl. Acad. Sci. U.S.A. 111, 15402–15407 10.1073/pnas.1320644111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leng F., Chen B., and Dunlap D. D. (2011) Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. U.S.A. 108, 19973–19978 10.1073/pnas.1109854108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan Y., Ding Y., Leng F., Dunlap D., and Finzi L. (2018) Protein-mediated loops in supercoiled DNA create large topological domains. Nucleic Acids Res. 46, 4417–4424 10.1093/nar/gky153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan Y., Leng F., Finzi L., and Dunlap D. (2018) Protein-mediated looping of DNA under tension requires supercoiling. Nucleic Acids Res. 46, 2370–2379 10.1093/nar/gky021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Postow L., Hardy C. D., Arsuaga J., and Cozzarelli N. R. (2004) Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18, 1766–1779 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinden R. R., and Pettijohn D. E. (1981) Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 78, 224–228 10.1073/pnas.78.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le T. B., Imakaev M. V., Mirny L. A., and Laub M. T. (2013) High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vinograd J., Lebowitz J., and Watson R. (1968) Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J. Mol. Biol. 33, 173–197 10.1016/0022-2836(68)90287-8 [DOI] [PubMed] [Google Scholar]
- 18. Boles T. C., White J. H., and Cozzarelli N. R. (1990) Structure of plectonemically supercoiled DNA. J. Mol. Biol. 213, 931–951 10.1016/S0022-2836(05)80272-4 [DOI] [PubMed] [Google Scholar]
- 19. Racko D., Benedetti F., Dorier J., and Stasiak A. (2019) Are TADs supercoiled? Nucleic Acids Res. 47, 521–532 10.1093/nar/gky1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., and Benham C. J. (2008) Superhelical destabilization in regulatory regions of stress response genes. PLoS Comput. Biol. 4, e17 10.1371/journal.pcbi.0040017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H., Kaloper M., and Benham C. J. (2006) SIDDBASE: a database containing the stress-induced DNA duplex destabilization (SIDD) profiles of complete microbial genomes. Nucleic Acids Res. 34, D373–D378 10.1093/nar/gkj007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H., and Benham C. J. (2006) Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics 7, 248 10.1186/1471-2105-7-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benham C. J. (1979) Torsional stress and local denaturation in supercoiled DNA. Proc. Natl. Acad. Sci. U.S.A. 76, 3870–3874 10.1073/pnas.76.8.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benham C. J. (1996) Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J. Mol. Biol. 255, 425–434 10.1006/jmbi.1996.0035 [DOI] [PubMed] [Google Scholar]
- 25. Peter B. J., Arsuaga J., Breier A. M., Khodursky A. B., Brown P. O., and Cozzarelli N. R. (2004) Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 5, R87 10.1186/gb-2004-5-11-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatfield G. W., and Benham C. J. (2002) DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 36, 175–203 10.1146/annurev.genet.36.032902.111815 [DOI] [PubMed] [Google Scholar]
- 27. Pruss G. J., and Drlica K. (1989) DNA supercoiling and prokaryotic transcription. Cell 56, 521–523 10.1016/0092-8674(89)90574-6 [DOI] [PubMed] [Google Scholar]
- 28. Gilbert N., and Allan J. (2014) Supercoiling in DNA and chromatin. Curr. Opin. Genet. Dev. 25, 15–21 10.1016/j.gde.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 30. Forth S., Sheinin M. Y., Inman J., and Wang M. D. (2013) Torque measurement at the single-molecule level. Annu. Rev. Biophys. 42, 583–604 10.1146/annurev-biophys-083012-130412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Remus D., Beall E. L., and Botchan M. R. (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23, 897–907 10.1038/sj.emboj.7600077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Donczew R., Mielke T., Jaworski P., Zakrzewska-Czerwińska J., and Zawilak-Pawlik A. (2014) Assembly of Helicobacter pylori initiation complex is determined by sequence-specific and topology-sensitive DnaA-oriC interactions. J. Mol. Biol. 426, 2769–2782 10.1016/j.jmb.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 33. Kasho K., Tanaka H., Sakai R., and Katayama T. (2017) Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA-ATP hydrolysis. J. Biol. Chem. 292, 1251–1266 10.1074/jbc.M116.762815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizutani M., Ohta T., Watanabe H., Handa H., and Hirose S. (1991) Negative supercoiling of DNA facilitates an interaction between transcription factor IID and the fibroin gene promoter. Proc. Natl. Acad. Sci. U.S.A. 88, 718–722 10.1073/pnas.88.3.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobryn K., Lavoie B. D., and Chaconas G. (1999) Supercoiling-dependent site-specific binding of HU to naked Mu DNA. J. Mol. Biol. 289, 777–784 10.1006/jmbi.1999.2805 [DOI] [PubMed] [Google Scholar]
- 36. Fuller R. S., and Kornberg A. (1983) Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. U.S.A. 80, 5817–5821 10.1073/pnas.80.19.5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dekker J., and Heard E. (2015) Structural and functional diversity of topologically associating domains. FEBS Lett. 589, 2877–2884 10.1016/j.febslet.2015.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pope B. D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D. L., Wang Y., Hansen R. S., Canfield T. K., Thurman R. E., Cheng Y., Gülsoy G., Dennis J. H., Snyder M. P., et al. (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kouzine F., and Levens D. (2007) Supercoil-driven DNA structures regulate genetic transactions. Front. Biosci. 12, 4409–4423 10.2741/2398 [DOI] [PubMed] [Google Scholar]
- 40. Baranello L., Levens D., Gupta A., and Kouzine F. (2012) The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim. Biophys. Acta 1819, 632–638 10.1016/j.bbagrm.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cozzarelli N. R., Cost G. J., Nöllmann M., Viard T., and Stray J. E. (2006) Giant proteins that move DNA: bullies of the genomic playground. Nat. Rev. Mol. Cell Biol. 7, 580–588 10.1038/nrm1982 [DOI] [PubMed] [Google Scholar]
- 42. Ma J., Bai L., and Wang M. D. (2013) Transcription under torsion. Science 340, 1580–1583 10.1126/science.1235441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu L. F., and Wang J. C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 10.1073/pnas.84.20.7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma J., and Wang M. D. (2016) DNA supercoiling during transcription. Biophys. Rev. 8, 75–87 10.1007/s12551-016-0215-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T. M., and Levens D. (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 10.1038/nsmb.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naughton C., Avlonitis N., Corless S., Prendergast J. G., Mati I. K., Eijk P. P., Cockroft S. L., Bradley M., Ylstra B., and Gilbert N. (2013) Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 10.1038/nsmb.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Benedetti F., Dorier J., Burnier Y., and Stasiak A. (2014) Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res. 42, 2848–2855 10.1093/nar/gkt1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benedetti F., Dorier J., and Stasiak A. (2014) Effects of supercoiling on enhancer-promoter contacts. Nucleic Acids Res. 42, 10425–10432 10.1093/nar/gku759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benedetti F., Racko D., Dorier J., Burnier Y., and Stasiak A. (2017) Transcription-induced supercoiling explains formation of self-interacting chromatin domains in S. pombe. Nucleic Acids Res. 45, 9850–9859 10.1093/nar/gkx716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Racko D., Benedetti F., Dorier J., and Stasiak A. (2018) Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 46, 1648–1660 10.1093/nar/gkx1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pommier Y., Sun Y., Huang S. N., and Nitiss J. L. (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 10.1038/nrm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vos S. M., Tretter E. M., Schmidt B. H., and Berger J. M. (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 10.1038/nrm3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cozzarelli N. R. (1980) DNA gyrase and the supercoiling of DNA. Science 207, 953–960 10.1126/science.6243420 [DOI] [PubMed] [Google Scholar]
- 54. Roca J. (2011) The torsional state of DNA within the chromosome. Chromosoma 120, 323–334 10.1007/s00412-011-0324-y [DOI] [PubMed] [Google Scholar]
- 55. Baranello L., Wojtowicz D., Cui K., Devaiah B. N., Chung H. J., Chan-Salis K. Y., Guha R., Wilson K., Zhang X., Zhang H., Piotrowski J., Thomas C. J., Singer D. S., Pugh B. F., Pommier Y., et al. (2016) RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 10.1016/j.cell.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. King I. F., Yandava C. N., Mabb A. M., Hsiao J. S., Huang H. S., Pearson B. L., Calabrese J. M., Starmer J., Parker J. S., Magnuson T., Chamberlain S. J., Philpot B. D., and Zylka M. J. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 10.1038/nature12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manzo S. G., Hartono S. R., Sanz L. A., Marinello J., De Biasi S., Cossarizza A., Capranico G., and Chedin F. (2018) DNA topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 19, 100 10.1186/s13059-018-1478-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Solier S., Ryan M. C., Martin S. E., Varma S., Kohn K. W., Liu H., Zeeberg B. R., and Pommier Y. (2013) Transcription poisoning by topoisomerase I is controlled by gene length, splice sites, and miR-142-3p. Cancer Res. 73, 4830–4839 10.1158/0008-5472.CAN-12-3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teves S. S., and Henikoff S. (2014) DNA torsion as a feedback mediator of transcription and chromatin dynamics. Nucleus 5, 211–218 10.4161/nucl.29086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., and Rosenfeld M. G. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 10.1126/science.1127196 [DOI] [PubMed] [Google Scholar]
- 61. Madabhushi R., Gao F., Pfenning A. R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T. X., Yamakawa H., Pao P. C., Stott R. T., Gjoneska E., Nott A., Cho S., Kellis M., and Tsai L. H. (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bunch H., Lawney B. P., Lin Y. F., Asaithamby A., Murshid A., Wang Y. E., Chen B. P., and Calderwood S. K. (2015) Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 6, 10191 10.1038/ncomms10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pulleyblank D. E., Shure M., Tang D., Vinograd J., and Vosberg H. P. (1975) Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc. Natl. Acad. Sci. U.S.A. 72, 4280–4284 10.1073/pnas.72.11.4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bauer W., and Vinograd J. (1970) Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J. Mol. Biol. 47, 419–435 10.1016/0022-2836(70)90312-8 [DOI] [PubMed] [Google Scholar]
- 65. Levens D., and Benham C. J. (2011) DNA stress and strain, in silico, in vitro and in vivo. Phys. Biol. 8, 035011 10.1088/1478-3975/8/3/035011 [DOI] [PubMed] [Google Scholar]
- 66. Zhabinskaya D., and Benham C. J. (2012) Theoretical analysis of competing conformational transitions in superhelical DNA. PLoS Comput. Biol. 8, e1002484 10.1371/journal.pcbi.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhabinskaya D., and Benham C. J. (2013) Competitive superhelical transitions involving cruciform extrusion. Nucleic Acids Res. 41, 9610–9621 10.1093/nar/gkt733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kouzine F., Wojtowicz D., Baranello L., Yamane A., Nelson S., Resch W., Kieffer-Kwon K. R., Benham C. J., Casellas R., Przytycka T. M., and Levens D. (2017) Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 4, 344–356.e7 10.1016/j.cels.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santos-Pereira J. M., and Aguilera A. (2015) R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- 70. Zaitsev E. N., and Kowalczykowski S. C. (2000) A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 14, 740–749 [PMC free article] [PubMed] [Google Scholar]
- 71. Kasahara M., Clikeman J. A., Bates D. B., and Kogoma T. (2000) RecA protein-dependent R-loop formation in vitro. Genes Dev. 14, 360–365 [PMC free article] [PubMed] [Google Scholar]
- 72. Doudna J. A., and Charpentier E. (2014) Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 73. Jiang F., Taylor D. W., Chen J. S., Kornfeld J. E., Zhou K., Thompson A. J., Nogales E., and Doudna J. A. (2016) Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chédin F. (2016) Nascent connections: R-loops and chromatin patterning. Trends Genet. 32, 828–838 10.1016/j.tig.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Itoh T., and Tomizawa J. (1980) Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. U.S.A. 77, 2450–2454 10.1073/pnas.77.5.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Drolet M., Bi X., and Liu L. F. (1994) Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 269, 2068–2074 [PubMed] [Google Scholar]
- 77. Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 10.1128/.61.2.212-238.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lang K. S., Hall A. N., Merrikh C. N., Ragheb M., Tabakh H., Pollock A. J., Woodward J. J., Dreifus J. E., and Merrikh H. (2017) Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799.e18 10.1016/j.cell.2017.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Leela J. K., Syeda A. H., Anupama K., and Gowrishankar J. (2013) Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 258–263 10.1073/pnas.1213123110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kreuzer K. N., and Brister J. R. (2010) Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol. J. 7, 358 10.1186/1743-422X-7-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Holt I. J. (2019) The mitochondrial R-loop. Nucleic Acids Res. 47, 5480–5489 10.1093/nar/gkz277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wahba L., Costantino L., Tan F. J., Zimmer A., and Koshland D. (2016) S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hartono S. R., Malapert A., Legros P., Bernard P., Chédin F., and Vanoosthuyse V. (2018) The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J. Mol. Biol. 430, 272–284 10.1016/j.jmb.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. El Hage A., Webb S., Kerr A., and Tollervey D. (2014) Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 10, e1004716 10.1371/journal.pgen.1004716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chan Y. A., Aristizabal M. J., Lu P. Y., Luo Z., Hamza A., Kobor M. S., Stirling P. C., and Hieter P. (2014) Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 10, e1004288 10.1371/journal.pgen.1004288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu W., Xu H., Li K., Fan Y., Liu Y., Yang X., and Sun Q. (2017) The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 3, 704–714 10.1038/s41477-017-0004-x [DOI] [PubMed] [Google Scholar]
- 87. Sanz L. A., Hartono S. R., Lim Y. W., Steyaert S., Rajpurkar A., Ginno P. A., Xu X., and Chédin F. (2016) Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 10.1016/j.molcel.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ginno P. A., Lott P. L., Christensen H. C., Korf I., and Chédin F. (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lim Y. W., Sanz L. A., Xu X., Hartono S. R., and Chedin F. (2015) Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife 4, 10.7554/eLife.08007 10.7554/eLife.08007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stork C. T., Bocek M., Crossley M. P., Sollier J., Sanz L. A., Chedin F., Swigut T., and Cimprich K. A. (2016) Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5, 10.7554/eLife.17548 10.7554/eLife.17548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Phoenix P., Raymond M. A., Massé E., and Drolet M. (1997) Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J. Biol. Chem. 272, 1473–1479 10.1074/jbc.272.3.1473 [DOI] [PubMed] [Google Scholar]
- 92. Daniels G. A., and Lieber M. R. (1995) RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 23, 5006–5011 10.1093/nar/23.24.5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reaban M. E., and Griffin J. A. (1990) Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348, 342–344 10.1038/348342a0 [DOI] [PubMed] [Google Scholar]
- 94. Yu K., Chedin F., Hsieh C. L., Wilson T. E., and Lieber M. R. (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451 10.1038/ni919 [DOI] [PubMed] [Google Scholar]
- 95. Chen L., Chen J. Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H., Zhou Y., Zhang D. E., and Fu X. D. (2017) R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757.e5 10.1016/j.molcel.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Costantino L., and Koshland D. (2015) The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 34, 39–45 10.1016/j.ceb.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Skourti-Stathaki K., and Proudfoot N. J. (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396 10.1101/gad.242990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sollier J., and Cimprich K. A. (2015) Breaking bad: R-loops and genome integrity. Trends Cell Biol. 25, 514–522 10.1016/j.tcb.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Crossley M. P., Bocek M., and Cimprich K. A. (2019) R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 10.1016/j.molcel.2019.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bauer W. R., and Benham C. J. (1993) The free energy, enthalpy and entropy of native and of partially denatured closed circular DNA. J. Mol. Biol. 234, 1184–1196 10.1006/jmbi.1993.1669 [DOI] [PubMed] [Google Scholar]
- 101. Zhabinskaya D., and Benham C. J. (2011) Theoretical analysis of the stress induced B-Z transition in superhelical DNA. PLoS Comput. Biol. 7, e1001051 10.1371/journal.pcbi.1001051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Peck L. J., and Wang J. C. (1983) Energetics of B-to-Z transition in DNA. Proc. Natl. Acad. Sci. U.S.A. 80, 6206–6210 10.1073/pnas.80.20.6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Benham C. J. (1987) Energetics of superhelicity and of B-Z transitions in superhelical DNA. Cell Biophys. 10, 193–204 10.1007/BF02797340 [DOI] [PubMed] [Google Scholar]
- 104. Stolz R., Sulthana S., Hartono S. R., Malig M., Benham C. J., and Chedin F. (2019) Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc. Natl. Acad. Sci. U.S.A. 116, 6260–6269 10.1073/pnas.1819476116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huppert J. L. (2008) Thermodynamic prediction of RNA-DNA duplex-forming regions in the human genome. Mol. Biosyst. 4, 686–691 10.1039/b800354h [DOI] [PubMed] [Google Scholar]
- 106. Roy D., and Lieber M. R. (2009) G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 29, 3124–3133 10.1128/MCB.00139-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Roy D., Yu K., and Lieber M. R. (2008) Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 28, 50–60 10.1128/MCB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Huang F. T., Yu K., Hsieh C. L., and Lieber M. R. (2006) Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 103, 5030–5035 10.1073/pnas.0506548103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Drolet M. (2006) Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 59, 723–730 10.1111/j.1365-2958.2005.05006.x [DOI] [PubMed] [Google Scholar]
- 110. Massé E., and Drolet M. (1999) Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 274, 16659–16664 10.1074/jbc.274.23.16659 [DOI] [PubMed] [Google Scholar]
- 111. Massé E., Phoenix P., and Drolet M. (1997) DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J. Biol. Chem. 272, 12816–12823 10.1074/jbc.272.19.12816 [DOI] [PubMed] [Google Scholar]
- 112. Drolet M., Phoenix P., Menzel R., Massé E., Liu L. F., and Crouch R. J. (1995) Overexpression of RNase H partially complements the growth defect of an Escherichia coli δ topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 92, 3526–3530 10.1073/pnas.92.8.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nguyen H. D., Yadav T., Giri S., Saez B., Graubert T. A., and Zou L. (2017) Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847.e4 10.1016/j.molcel.2017.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. De Magis A., Manzo S. G., Russo M., Marinello J., Morigi R., Sordet O., and Capranico G. (2019) DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 816–825 10.1073/pnas.1810409116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Malig M., Hartono S. R., Giafaglione J. M., Sanz L. A., and Chedin F. (2020) Ultra-Deep Coverage Single-Molecule R-loop Footprinting Reveals Principles of R-loop Formation. J. Mol. Biol. 10.1016/j.jmb.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang F. T., Yu K., Balter B. B., Selsing E., Oruc Z., Khamlichi A. A., Hsieh C. L., and Lieber M. R. (2007) Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol. Cell. Biol. 27, 5921–5932 10.1128/MCB.00702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kao Y. P., Hsieh W. C., Hung S. T., Huang C. W., Lieber M. R., and Huang F. T. (2013) Detection and characterization of R-loops at the murine immunoglobulin Sα region. Mol. Immunol. 54, 208–216 10.1016/j.molimm.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 118. Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., and Klug A. (1977) Structure of nucleosome core particles of chromatin. Nature 269, 29–36 10.1038/269029a0 [DOI] [PubMed] [Google Scholar]
- 119. Richmond T. J., and Davey C. A. (2003) The structure of DNA in the nucleosome core. Nature 423, 145–150 10.1038/nature01595 [DOI] [PubMed] [Google Scholar]
- 120. Patterton H. G., and von Holt C. (1993) Negative supercoiling and nucleosome cores. I. The effect of negative supercoiling on the efficiency of nucleosome core formation in vitro. J. Mol. Biol. 229, 623–636 10.1006/jmbi.1993.1068 [DOI] [PubMed] [Google Scholar]
- 121. Pfaffle P., Gerlach V., Bunzel L., and Jackson V. (1990) In vitro evidence that transcription-induced stress causes nucleosome dissolution and regeneration. J. Biol. Chem. 265, 16830–16840 [PubMed] [Google Scholar]
- 122. Pfaffle P., and Jackson V. (1990) Studies on rates of nucleosome formation with DNA under stress. J. Biol. Chem. 265, 16821–16829 [PubMed] [Google Scholar]
- 123. Corless S., and Gilbert N. (2016) Effects of DNA supercoiling on chromatin architecture. Biophys. Rev. 8, 245–258 10.1007/s12551-016-0210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Teves S. S., and Henikoff S. (2014) Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 21, 88–94 10.1038/nsmb.2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dunn K., and Griffith J. D. (1980) The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 8, 555–566 10.1093/nar/8.3.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Roy D., Zhang Z., Lu Z., Hsieh C. L., and Lieber M. R. (2010) Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159 10.1128/MCB.00897-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Baaklini I., Usongo V., Nolent F., Sanscartier P., Hraiky C., Drlica K., and Drolet M. (2008) Hypernegative supercoiling inhibits growth by causing RNA degradation. J. Bacteriol. 190, 7346–7356 10.1128/JB.00680-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hartono S. R., Korf I. F., and Chédin F. (2015) GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Res. 43, 9729–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. El Hage A., French S. L., Beyer A. L., and Tollervey D. (2010) Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Puget N., Miller K. M., and Legube G. (2019) Non-canonical DNA/RNA structures during transcription-coupled double-strand break repair: roadblocks or bona fide repair intermediates? DNA Repair (Amst.) 81, 102661 10.1016/j.dnarep.2019.102661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cohen S., Puget N., Lin Y. L., Clouaire T., Aguirrebengoa M., Rocher V., Pasero P., Canitrot Y., and Legube G. (2018) Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9, 533 10.1038/s41467-018-02894-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Michelini F., Pitchiaya S., Vitelli V., Sharma S., Gioia U., Pessina F., Cabrini M., Wang Y., Capozzo I., Iannelli F., Matti V., Francia S., Shivashankar G. V., Walter N. G., and d'Adda di Fagagna F. (2017) Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 10.1038/ncb3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Oh S. D., Lao J. P., Hwang P. Y., Taylor A. F., Smith G. R., and Hunter N. (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 10.1016/j.cell.2007.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wilson-Sali T., and Hsieh T. S. (2002) Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIβ. Proc. Natl. Acad. Sci. U.S.A. 99, 7974–7979 10.1073/pnas.122007999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yang Y., McBride K. M., Hensley S., Lu Y., Chedin F., and Bedford M. T. (2014) Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 10.1016/j.molcel.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stoll G., Pietiläinen O. P. H., Linder B., Suvisaari J., Brosi C., Hennah W., Leppä V., Torniainen M., Ripatti S., Ala-Mello S., Plöttner O., Rehnström K., Tuulio-Henriksson A., Varilo T., Tallila J., et al. (2013) Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237 10.1038/nn.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J., Fox D. 3rd, Qian Y., Martindale J. L., Piao Y., Machamer J., et al. (2013) Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 16, 1238–1247 10.1038/nn.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Petryk N., Kahli M., d'Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C. L., and Hyrien O. (2016) Replication landscape of the human genome. Nat. Commun. 7, 10208 10.1038/ncomms10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Prioleau M. N. (2009) CpG islands: starting blocks for replication and transcription. PLoS Genet. 5, e1000454 10.1371/journal.pgen.1000454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kim S., Beltran B., Irnov I., and Jacobs-Wagner C. (2019) Long-distance cooperative and antagonistic RNA polymerase dynamics via DNA supercoiling. Cell 179, 106–119.e16 10.1016/j.cell.2019.08.033 [DOI] [PubMed] [Google Scholar]
- 141. Gu B., Swigut T., Spencley A., Bauer M. R., Chung M., Meyer T., and Wysocka J. (2018) Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Belotserkovskii B. P., and Hanawalt P. C. (2011) Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys. J. 100, 675–684 10.1016/j.bpj.2010.12.3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Belotserkovskii B. P., Liu R., Tornaletti S., Krasilnikova M. M., Mirkin S. M., and Hanawalt P. C. (2010) Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 107, 12816–12821 10.1073/pnas.1007580107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Belotserkovskii B. P., Soo Shin J. H., and Hanawalt P. C. (2017) Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 45, 6589–6599 10.1093/nar/gkx403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hraiky C., Raymond M. A., and Drolet M. (2000) RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J. Biol. Chem. 275, 11257–11263 10.1074/jbc.275.15.11257 [DOI] [PubMed] [Google Scholar]
- 146. Baaklini I., Hraiky C., Rallu F., Tse-Dinh Y. C., and Drolet M. (2004) RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 54, 198–211 10.1111/j.1365-2958.2004.04258.x [DOI] [PubMed] [Google Scholar]
- 147. Bacolla A., and Wells R. D. (2004) Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 279, 47411–47414 10.1074/jbc.R400028200 [DOI] [PubMed] [Google Scholar]
- 148. Zhao J., Bacolla A., Wang G., and Vasquez K. M. (2010) Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 67, 43–62 10.1007/s00018-009-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]