Abstract
Programmed cell death protein 1 (PD-1) is an inhibitory receptor on T lymphocytes that is critical for modulating adaptive immunity. As such, it has been successfully exploited for cancer immunotherapy. Programmed death ligand 1 (PD-L1) and PD-L2 are ligands for PD-1; the former is ubiquitously expressed in inflamed tissues, whereas the latter is restricted to antigen-presenting cells. PD-L2 binds to PD-1 with 3-fold stronger affinity compared with PD-L1. To date, this affinity discrepancy has been attributed to a tryptophan (W110PD-L2) that is unique to PD-L2 and has been assumed to fit snuggly into a pocket on the PD-1 surface. Contrary to this model, using surface plasmon resonance to monitor real-time binding of recombinantly-expressed and -purified proteins, we found that W110PD-L2 acts as an “elbow” that helps shorten PD-L2 engagement with PD-1 and therefore lower affinity. Furthermore, we identified a “latch” between the C and D β-strands of the binding face as the source of the PD-L2 affinity advantage. We show that the 3-fold affinity advantage of PD-L2 is the consequence of these two opposing features, the W110PD-L2 “elbow” and a C–D region “latch.” Interestingly, using phylogenetic analysis, we found that these features evolved simultaneously upon the emergence of placental mammals, suggesting that PD-L2–affinity tuning was part of the alterations to the adaptive immune system required for placental gestation.
Keywords: protein structure, immunotherapy, T-cell biology, protein evolution, glycoprotein structure, immune checkpoint, immune receptors/ligands, PD-L2, programmed cell death protein 1 (PD-1), programmed death ligand 1 (PD-L1)
Introduction
The programmed cell death protein 1 (PD-1) receptor provides an essential constraint on T-cell activation (1). Engagement of PD-1 with either of its two membrane-bound ligands, PD-1 ligand 1 (PD-L1, B7-H1, CD274) or ligand 2 (PD-L2, B7-DC, CD273), suppresses immune responses and promotes self-tolerance (2–4). Expression of both ligands is induced under inflammatory conditions, most notably as a result of interferon γ signaling. PD-L1 is widely expressed in both hematopoietic and nonhematopoietic cells to discourage reactivity to self-antigens (5). In contrast, PD-L2 is restricted to antigen-presenting cells (APCs),3 including dendritic cells, macrophages, monocytes, and some B cells (6). The restricted expression of PD-L2 suggests that its function is distinct from that of PD-L1 and may heighten the tolerogenic hurdle that an immune response must overcome during the priming phase.
Many malignancies co-opt PD-L1 expression, and in some cases PD-L2 (7), in an attempt to hide neoantigens from immune surveillance (8). This provides a rationale for interfering with PD-1 signaling as a key modality in cancer immunotherapy. Indeed, monoclonal antibodies (mAbs) against PD-1 and PD-L1, referred to as immune checkpoint inhibitors, have already achieved long-term responses in the clinic (9–11). Desirable clinical outcomes are particularly evident in malignancies with high mutational burdens predicted to elaborate neoantigens (12–14). Therapeutic mAbs that block the interaction of PD-1 with PD-L1 represent one mode of inhibiting PD-1 signaling. Development of alternative modalities of interfering with PD-1 signaling will require detailed understanding of PD-1 engagement of its ligands, the dynamics of the receptor and ligands, and signaling downstream of PD-1. A key to the success of such strategies will depend on the determination of structural and functional differences between the two ligands.
PD-ligands are members of the B7 family of type 1 transmembrane proteins that also include CD80 and CD86, which engage co-stimulatory or co-inhibitory receptors (15). CD80 and CD86 are both ligands for the CD28 and CTLA-4 receptors, and differential function is mediated by a 100-fold affinity difference (16). In contrast, PD-L2 binds to PD-1 with only ∼3-fold stronger affinity compared with that of PD-L1 (17–19). Affinity differences between PD-L1 and PD-L2 have been ascribed to the presence of an alanine versus tryptophan residue (PD-L1 position 121 and PD-L2 position 110, respectively) in homologous positions within their binding sites (17–20).
We studied the structural, functional, and evolutionary differences that distinguish the two PD-ligands. We find that, contrary to prior reports (17–20), the tryptophan residue at position 110 (W110PD-L2) weakens the interaction between PD-L2 and PD-1 and that the enhanced affinity is instead mediated by a “latch” present only in PD-L2 that evolved along with the W110PD-L2 insertion upon the emergence of placental mammals.
Results
W110PD-L2 acts as an “elbow” to hinder PD-1 binding
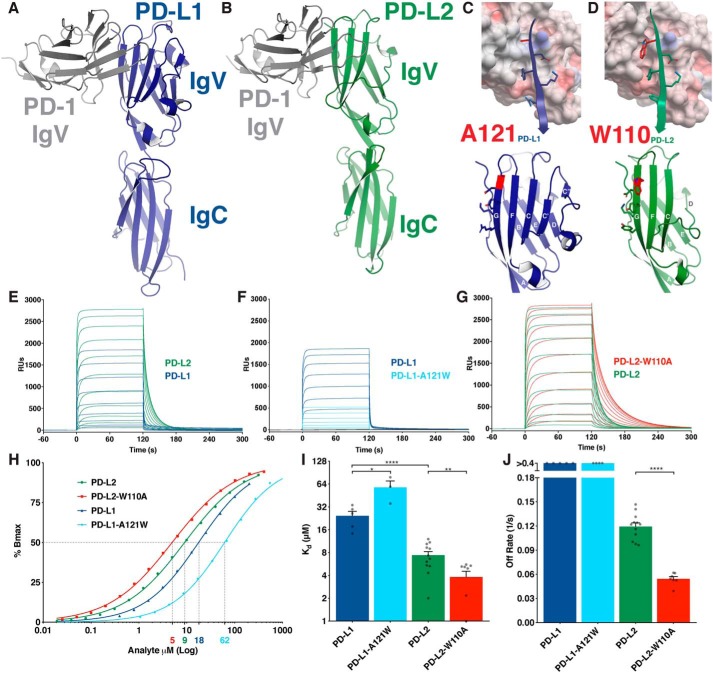
PD-L1 and PD-L2 are B7 protein family members consisting of an N-terminal IgV domain and a membrane-proximal IgC domain (Fig. 1, A and B). Both ligands participate in a front–to–front Ig binding to the PD-1 IgV domain utilizing their front, G-F-C-C′-C″ β-strand IgV face (Fig. 1, A–D). Previous explanations of the ∼3-fold affinity advantage of PD-L2 over PD-L1 identified the conserved alanine at position 121 (A121PD-L1) versus W110PD-L2 on the G-strand of each ligand as the responsible structural feature (Fig. 1, C and D) (17–19). A121PD-L1 sits above a bulge on the PD-1 surface formed by an isoleucine (I126PD-1) (PDB codes 3BIK and 4ZQK, Fig. 1C, and Fig. S1B). This bulge is also evident on apo structures of PD-1 (PDB codes 3RRQ and 1NPU) (Fig. S1A). However, upon PD-L2 binding, I126PD-1 rotates to accommodate W110PD-L2 into a pocket formed on the PD-1 surface (Fig. 1D and Fig. S1C), a feature thought to explain the enhanced affinity of PD-L2.
Figure 1.
W110 of PD-L2 acts as an elbow to hinder PD-1 binding. A and B, ribbon diagrams of the murine PD-1 IgV domain (gray) in complex with the IgV and IgC domains of human PD-L1 (blue) (PDB code 3BIK) (A) and hPD-L2 (green) (hPD-L2 sequence threaded onto PDB code 3BP5) (B). C and D, upper panels show surface electrostatic representation of mPD-1 with the G-strands of PD-L1 (blue) (C) or PD-L2 (green) (D) in ribbon and stick representation. Lower panels show front faces of IgV domains of PD-L1 (blue) (C) and PD-L2 (green) (D) with β-strand lettering and A121PD-L1 and W110PD-L2 highlighted in red. E–G, SPR sensorgrams of the indicated PD-ligand analytes injected over immobilized PD-1. H, representative, normalized binding curves. I, affinity measurements from independent experiments. J, dissociation rates from independent experiments. Dissociation rates exceeding the range of accurate measurement are shown as >0.4 s−1. Unpaired t tests: *, p < 0.05; **, p < 0.01; ****, p < 0.0001; RU, response units.
We tested this assertion by swapping the Trp and Ala residues on the PD-ligands and measuring affinity for PD-1 using surface plasmon resonance (SPR) to monitor real-time binding (Fig. 1, E–G). We observed a 3.3-fold affinity advantage of WT PD-L2 over PD-L1, consistent with previous reports (Fig. 1, E, H, and I) (18, 21). SPR confirmed that the affinity difference between the two WT ligands is a consequence of markedly different dissociation kinetics, with PD-L2 dissociating from the receptor more slowly (Fig. 1, E and J). Contrary to published models (17–20), the A121WPD-L1 substitution decreased affinity as a consequence of very rapid dissociation from the surface, whereas the W110APD-L2 substitution strengthened receptor binding by lengthening dissociation time more than 2-fold (Fig. 1, F–J). We conclude that W110PD-L2 acts as an “elbow” that hinders PD-1 binding by shortening the PD-1/PD-L2 interaction.
W110PD-L2 forces I126PD-1 to rotate away from its position as determined in the crystal structures of PD-1 in both its apo and PD-L1-bound states (Fig. S1, A–C) (PDB codes 3RRQ, 1NPU, 4ZQK, and 3BP5). This rotation is required to form a pocket to accommodate the bulky W110PD-L2 in the PD-L2-bound state. The induced I126PD-1 rotation is energetically unfavorable, and its propensity to return to what is likely a lower energy state may account for the faster off-rate of WT PD-L2 relative to W110APD-L2. Thus, rather than representing an evolutionary change that results in an enhanced affinity, the W110PDL2 elbow weakens the interaction, consistent with the notion that the binding affinity of PD-ligands must be finely tuned to ensure the optimal degree of inhibitory input on T cells.
Atypical PD-L2 C–D strand region, absent in PD-L1, acts as a latch that enhances binding to PD-1
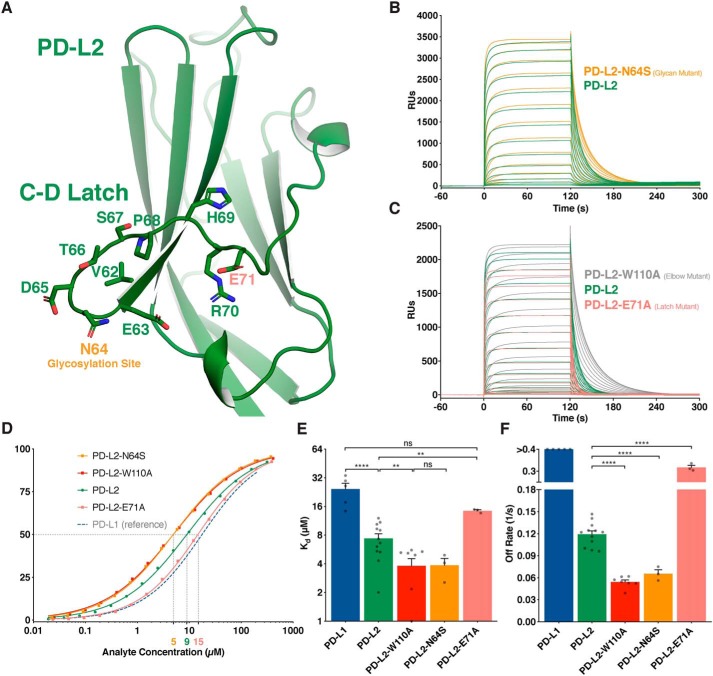
Because W110PD-L2 does not account for the affinity advantage of PD-L2, we re-examined the available PD-L2 crystal structures to find additional structural features that distinguish it from PD-L1. One marked difference between the binding domains of PD-L1 and PD-L2 is evident in their C–D strand regions (Fig. 1, C and D). The PD-L1 IgV domain is conventional, containing both C′ and C″ β-strands (Fig. 1C). In contrast, the PD-L2 IgV domain is atypical because it lacks C′ and C″ β-strands and instead harbors a flexible C–D loop (Fig. 1D). Comparing PD-L2 structures in apo versus PD-1–complexed forms reveals that the C–D loop is dynamic and “latches” up onto PD-1 when in the bound state (Fig. S1, D–F). In fact, the receptor/ligand complex crystal structures demonstrate that PD-L2 makes 40% (∼330 Å2) more contact with PD-1 than does PD-L1 (Fig. S2A). This difference is mostly attributable to the presence of this C–D latch.
The PD-L2 C–D loop region contains a putative N-linked glycosylation site (N64PD-L2), followed by a series of flexible polar residues (Fig. 2A). We confirmed the predicted glycosylation by introducing an N64SPD-L2 mutation and observing the expected molecular weight shift (Fig. S2B). Removing the Asn-64 glycan improved PD-L2 affinity for PD-1 in a manner similar to the W110APD-L2 substitution (Fig. 2, B and D–F). This suggests that the mass, solubility, and/or flexibility of a glycan tree at N64PD-L2 adds to the dynamic nature of the C–D loop region. The enhanced flexibility may contribute to the rate at which the C–D latch opens to permit PD-1/PD-L2 dissociation. Indeed, the N64SPD-L2 glycan mutant exhibited a prolonged dissociation curve relative to WT PD-L2 (Fig. 2, B and F). This finding was substantiated in a recent crystal structure of the human PD-1/PD-L2, in which the three residues around N64PD-L2 were too flexible to resolve (20).
Figure 2.
Atypical C–D region of PD-L2 forms a latch that enhances PD-1 binding. A, ribbon representation of the IgV domain of PD-L2 with the C–D latch residues represented as sticks. B and C, SPR sensorgrams of the indicated PD-ligand analytes injected over immobilized PD-1. D, representative, normalized binding curves. E, affinity measurements from independent experiments. F, dissociation rates from independent experiments. Dissociation rates exceeding the range of accurate measurement are shown as >0.4 s−1. Unpaired t tests: **, p < 0.01; ****, p < 0.0001; ns, not significant; RU, response units.
The most mobile region of the PD-L2 C–D latch is predicted to be around a glutamate at position 71 (E71PD-L2) (Fig. 2A). Indeed, an E71APD-L2 mutation produced a ligand that was intermediate between PD-L2 and PD-L1 in its dissociation kinetics (Fig. 2, C–F). This result supports our model in which the PD-L2 flexible C–D latch is the major structural element responsible for the difference in affinity between the PD-ligands.
Defining structural features of PD-L2 evolved contemporaneously with placental mammal radiation
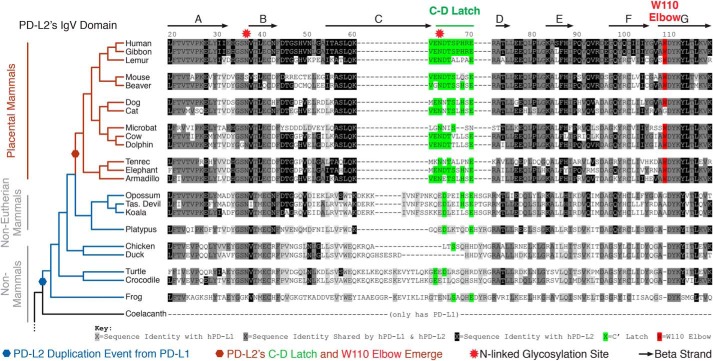
The Trp-110 elbow and C–D latch of PD-L2 are structural features that render it distinct from PD-L1. To explore the evolutionary origins of the two PD-ligands, we compiled a phylogenetic tree (Fig. 3 and Fig. S3) (22). Prior to the divergence of amphibians from fish, vertebrate genomes encoded a single ortholog of PD-ligands, most similar to PD-L1 (Fig. 3 and Fig. S3, A–C). PD-L2 emerged from a gene duplication event after the divergence of coelacanths (an order of lobe-finned fish) and amphibians ∼400 million years ago (23), a conclusion we confirmed by synteny analysis (Fig. S3C). Following this gene duplication event, the PD-L2 primary protein sequence remained largely unchanged and highly similar to PD-L1 through marsupial mammal evolution (Fig. S3, A and B). However, upon placental mammal radiation, the PD-L2 protein sequence significantly and abruptly changed (Fig. 3). Specifically, both the Trp-110 elbow and C–D latch of PD-L2 emerged in placental mammals (Fig. 3). The 110th position in PD-L2 evolved as an amino acid insertion (Gly or Ala) event that occurred between monotreme and marsupial mammal evolution, but it did not become the bulky W110PD-L2 until the emergence of placental mammals (Fig. 3 and Fig. S3D). The PD-L2 C–D latch also became fixed in terms of length in placental mammals (Fig. 3). The N64PD-L2 glycosylation site, which is not present on PD-L1, also emerged only in placental mammals within the PD-L2 C–D latch region (Fig. 3). E71PD-L2, the importance of which is established above (Fig. 2), existed prior to placental mammal evolution, but its position was shifted from a conventional C″ IgV location to the PD-L2 atypical, flexible C–D latch region in placental mammals (Fig. S3, A and B).
Figure 3.
Trp-110 elbow and C–D latch of PD-L2 evolved contemporaneously with placental mammal radiation. Phylogenetic analysis of the PD-L2 IgV domain. PD-L2 emerged from a gene duplication from a primordial PD-L1 between lobe-finned fish and amphibian divergence (blue hexagon). The Trp-110 elbow (red) and C–D latch (green) of PD-L2 emerged exclusively in placental mammals (red hexagon).
Effect of PD-ligand affinity for PD-1 on T-cell inhibition
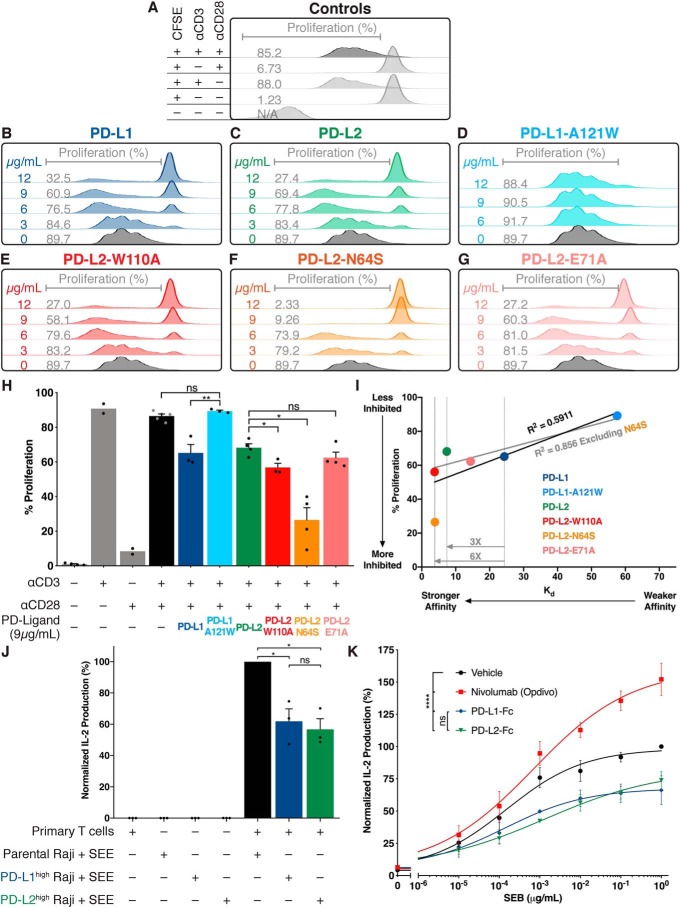
To correlate the structural features described above with function, we tested the ability of WT and mutant PD-ligands to modulate T-cell proliferation (Fig. 4). Primary human CD4+ T-cell blasts were activated in the presence or absence of increasing amounts of PD-ligands adsorbed onto plates, and proliferation was measured by CFSE dilution (Fig. 4, A–I). WT PD-L1 and PD-L2 inhibited T-cell proliferation equally, despite their 3.3-fold affinity differences (Fig. 4, B and C). At similar concentrations, the A121WPD-L1 mutant that bound PD-1 with lower affinity did not inhibit proliferation (Fig. 4D). Conversely, the W110APD-L2 and N64SPD-L2 mutants inhibited proliferation to a greater degree than WT, with the glycan mutant displaying the highest potency (Fig. 4, E and F). Finally, The E71APD-L2 mutant was indistinguishable from WT PD-ligands, consistent with its binding affinity being intermediate between PD-L1 and PD-L2 (Fig. 4, G and H). Thus, while enhancing the affinity difference by introducing W110APD-L2 or N64SPD-L2 translated into increased inhibitory activity (Fig. 4I), the 3.3-fold difference in affinity between WT PD-L1 and PD-L2 was not sufficient to affect inhibitory activity.
Figure 4.
Effect of PD-ligand affinity for PD-1 on T-cell inhibition. A–G, proliferation of primary human CD4+ T-cell blasts in response to anti-CD3 and anti CD-28 antibodies was measured by CFSE dilution without (A) or with the indicated ligand absorbed at the indicated concentration. H, cumulative data from multiple independent experiments with the indicated absorbed concentration. I, correlation between T-cell inhibition and affinity for PD-ligand variants. A–I, data are from a single healthy donor. J, IL-2 release from primary human T cells in response to SEE bound to Raji B cells with or without ectopic and equal expression of PD-ligands (24 h co-culture). Data are from three healthy donors assessed independently. K, effect of 20 μg/ml Nivolumab or the indicated PD-ligand Fc fusion protein on IL-2 secretion from human PBMCs in response to increasing concentrations of SEB superantigen. Data shown represent independent experiments from four healthy donors. Bar graphs: paired t tests: *, p < 0.05; **, p < 0.01. Dose-response curves: two-way analysis of variance; ****, p < 0.0001; ns, not significant.
To verify that the two WT PD-ligands have equivalent inhibitory capabilities when presented on an APC surface, we measured PD-ligand function in the context of a primary T-cell–Raji B-cell co-culture system. Raji B cells were rendered either PD-L1high or PD-L2high via transduction with PD-ligand–mCherry lentiviral constructs. Untransduced Raji cells did not express measurable levels of endogenous PD-ligands (Fig. S4A). Prior to co-culture with human primary T cells, the PD-ligand–expressing Raji cells were sorted to ensure equal PD-ligand expression and subsequently irradiated to block cytokine production and endogenous ligand expression (Fig. S4, B and C). PD-L1 and PD-L2 expressed on Raji cells inhibited T-cell IL-2 production stimulated by engagement of T-cell receptors by superantigen to an equal extent (Fig. 4J).
To further explore the lack of a functional difference between the two WT PD-ligands, we stimulated primary human PBMCs with SEB super-antigen in the presence of PD-ligand–Fc fusion proteins as well as nivolumab, a clinically used anti-PD-1 blocking mAb (Fig. 4K). PD-L1–Fc and PD-L2–Fc did not differ in ability to dampen the SEB-induced immune activation (Fig. 4K). Taken together, these results suggest that although evolutionary changes in PD-L2 act to constrain its affinity for PD-1 within tight parameters, this 3.3-fold net change in affinity between the WT proteins alone does not account for biological differences among the PD-ligands.
Discussion
PD-ligands expressed on a wide range of tissues, including APCs, interact with PD-1 on T lymphocytes to inhibit their function (24). As is the case for many immune receptors, the strength of these interactions has been evolutionarily finely tuned to generate a balance between the co-stimulation and co-inhibition of T cells required for adaptive immunity (25). We reveal two of the structural features of PD-L2 that differ from PD-L1 and calibrate the affinities of PD-ligands for PD-1. These are the Trp-110 elbow and the C–D latch. Moreover, we found that these features evolved upon placental mammal radiation, suggesting that PD-L2 had to be retuned at the onset of the maternal–fetal conflict.
The finding that W110PD-L2 acts as an elbow, weakening its affinity for PD-1, was unexpected because inspection of the crystal structure of the mPD-L2/mPD-1 complex (PDB code 3BP5) suggests its snug fit into a pocket on PD-1 might strengthen the interaction (17, 19), recently corroborated by the hPD-L2/hPD-1 complex crystal structure (20). However, our results show that the rotation of I126PD-1 that must occur to accommodate W110PD-L2 is energetically unfavorable, making the evolutionary change in PD-L2 one that weakened the interaction. We also found that the N64PD-L2 glycan that co-evolved with W110PD-L2, and is absent from PD-L1, also serves to weaken the interaction with PD-1. Interestingly, both of these alterations counterbalance the contemporaneous emergence of the PD-L2 C–D latch region, which extends the interaction with PD-1 thereby providing more contact area than exists in the PD-L1/PD-1–bound complex (PDB code 4ZQK) and explaining the 3–4-fold increase in affinity of PD-L2 for PD-1 relative to that of PD-L1 (18).
We theorize that, in the placental mammal common ancestor, the PD-L2 C–D latch evolved to improve binding to PD-1, while contemporaneously evolving the W110PD-L2 elbow and N64PD-L2 glycan to counterbalance its enhanced affinity for PD-1, thus ensuring that the PD-L2 dissociation rate remained within parameters required for optimal engagement of the pathway. A plethora of immunologic alterations likely had to occur during the marsupial to placental mammal evolutionary transition to accommodate long gestation times for an allogeneic fetus. Our phylogenetic and synteny analysis of the PD-ligand sequences clearly demonstrate that PD-L2 evolved as a gene duplication from a primordial PD-L1 (26). We postulate that the function of PD-L2 changed during the marsupial to placental mammal evolutionary transition away from simple redundancy with PD-L1 to facilitate immune tolerance of the placenta. Consistent with this idea, PD-L2 has been reported to be expressed at relatively high levels on placenta (3, 4, 27, 28). A precedent for such immunologic changes during this evolutionary transition has been reported elsewhere (29).
Although the PD-L2 elbow and C–D latch, characterized here, represent the structural difference between the PD-ligands, the 3.3-fold affinity discrepancy alone is insufficient to account for differential function. By exaggerating this affinity difference with mutants such as A121WPD-L1 or W110APD-L2, an affinity/functional correlation can be observed. However, no significant difference between the WT ligands was measured. When viewed in the context of the CD28/CTLA-4 receptor, CD80/CD86 ligand axis in which differential function results from a 100-fold affinity difference (16), it is not surprising that the relatively small PD-ligand affinity difference is insufficient to drive differential function. Furthermore, the markedly greater expression of PD-L1 relative to PD-L2 on APCs (18) renders the small affinity difference unlikely to account for a biological difference. Yet, the restriction of PD-L2 to APCs and the divergence/persistence of PD-L2 through mammalian evolution argues strongly for a nonredundant function of the two ligands.
Given the likely functional differences between PD-ligands, it remains possible that the trans-interaction with PD-1 is not the determining factor. Indeed, recent studies have revealed that PD-L1 interacts with CD80 in cis on the cell surface (30). This interaction has been shown to interrupt the PD-1/PD-L1 trans, CTLA-4/CD80 trans, and CD80 homodimer cis interactions (31, 32). Although it is accepted that PD-L2 does not interact with CD80, we do not know evolutionarily whether PD-L2 lost the capability to bind to CD80 or whether PD-L1 gained the CD80 cis interaction after the PD-L2 gene duplication event. It remains possible that the unique PD-L2 structural features characterized in this study played a role in preventing a cis-CD80 interaction while maintaining canonical PD-1 binding within tight parameters. PD-L2 likely gained a unique functional role exclusively in placental mammals, but the retuning of its affinity indicates an evolutionary pressure to maintain the canonical PD-1 interaction within tight parameters. The precise biological difference between the ligands and the molecular basis for that difference remain to be determined.
Experimental procedures
Construct design and protein expression
Codon-optimized DNAs encoding human PD-1(32–160), PD-L1(19–238), PD-L2(20–220), and CD80(35–242) were synthesized (GenScript) along with a 5′ sequence encoding the signal peptide, DIATMRPTWAWWLFLVLLLALWAPARG. Sequences encoding a thrombin cleavage site, hexahistidine, and AviTagTM were appended sequentially at the 3′ end. These constructs were cloned into the pVRC8400 mammalian expression vector using the XbaI and BamHI restriction sites.
Fc-fusion constructs were also generated in the pVRC8400 expression plasmid between the XbaI and BamHI restriction sites. The PD-ligands, including the signal peptide, were PCR-amplified using the above plasmids as templates with XbaI and Kpn2I sites at the 5′ and 3′ ends, respectively. DNA encoding human IgG1 Fc(99–330) was PCR-amplified from an IgG1 heavy chain cDNA with Kpn2I and BamHI at its 5′ and 3′ ends, respectively. The two PCR fragments generated above were ligated into the pVRC8400 expression plasmid between the XbaI and BamHI sites, thereby introducing a Ser–Gly linker between the PD-ligand ectodomains and the IgG1-Fc, encoded by the Kpn2I nucleotide sequence. Fc-fusion constructs were extended with a hexahistidine tag to aid with purification.
Protein expression was performed using polyethyleneimine lipid-based transfection and 500 μg of plasmid DNA per 5·108 suspension of HEK293F cells in 500 ml of FreeStyleTM 293 Expression Medium (Thermo Fisher Scientific). Cells were incubated for 4–5 days at 37 °C, shaking at 120 rpm, with 5% CO2, and 85% humidity. Proteins were purified from conditioned media diluted 1:2 in 150 mm NaCl and 20 mm Tris, pH 6.8, using a HisTrap FF Crude column (GE Healthcare). Elution was carried out using an imidazole gradient on an FPLC and hexahistidine-tagged proteins eluted from 150 to 450 mm imidazole. Proteins were dialyzed into PBS.
Surface plasmon resonance
Monomeric PD-1 was biotinylated via its C-terminal AviTag codes using the BirA-500 biotin–protein ligase reaction kit (Avidity). 1600–3000 response units of biotinylated PD-1 was immobilized on a streptavidin Biacore sensor chip installed in a Biacore T200. PD-ligand analytes were injected for 120 s at a flow rate of 30 μl/min at concentrations ranging from 400 to 0.01 μm. Dissociation was observed for 180 s followed by 15 s of regeneration with 10 mm glycine, pH 3.0. Steady-state Kd and off-rates were determined by fitting curves with the specific binding with Hill-slope and dissociation one-phase exponential decay curves, respectively, using the Prism software.
CD4+ T-cell proliferation
Primary human CD4+ T cells were isolated from peripheral blood of a healthy volunteer using the CD4 MicroBeads kit (MACS Miltenyi Biotec). CD4+ T-cell blasts were generated by culture for 1 week in complete RPMI media, containing 10% fetal bovine serum, minimal essential medium nonessential amino acids, 1 mm sodium pyruvate, and GlutaMAXTM (Gibco), supplemented with 20–40 units/ml if recombinant human IL-2 (PeproTech). CD4+ T-cell blasts were labeled with 1 μm CFSE for 20 min at 37 °C protected from light. Staining was quenched with complete RPMI media followed by a 5-min 400 × g centrifugation at room temperature. Cells were then subjected to stimulation with plate-bound antibodies as described below for 5 days at 37 °C and 5% CO2. On day 5, the cells were washed once and fixed with 1% paraformaldehyde. Cells were analyzed with a BD FACSCalibur flow cytometer.
Plate bound T-cell activation/inhibition
Plates (48-well) were coated on at 4 °C with 10 μg/ml anti-human CD3 mAb (UCHT1 Ultra-LEAFTM purified, BioLegend) and 1.5 μg/ml PD-ligand monomers or 3 μg/ml PD-ligand–Fc fusion proteins in 200 μl per well. The next day, wells were washed once with media, followed by addition of 5·105 T cells along with 2 μg/ml soluble anti-human CD28 mAb (CD28.2 Ultra-LEAFTM purified, BioLegend) in a 500-μl volume per well.
PBMC superantigen activation
Primary human PBMCs were isolated by density gradient centrifugation (LymphoprepTM). 1·105 PBMCs were mixed with serial dilutions of SEB (Toxin Technologies) along with 20 μg/ml Fc-fusion or mAb constructs in a total volume of 200 μl in a 96-well U-bottom plate. Supernatants were harvested following a 3-day incubation at 37 °C with 5% CO2. IL-2 concentration in the supernatants was measured by enzyme-linked immunosorbent assay (ELISA) (human IL-2 ELISA MAXTM BioLegend).
Raji B-cell–primary T-cell co-culture
Raji B cells were rendered PD-ligandhigh via lentiviral transduction. The PD-L1–mCherry–pHR lentivirus vector was generously provided by Hui et al. (33). The PD-L2–mCherry–pHR vector was created by inserting the PD-L2 cDNA ORF between the MluI and BamHI restriction sites in the pHR plasmid. Lentivirus was generated by transfecting the above pHR plasmids along with psPAX2 and pMD.2G packaging plasmids into 293T cells. Approximately 1 week following transduction, Raji cells were sorted for mCherry expression. Surface expression of the PD-ligand–mCherry constructs was confirmed with surface staining with BV421-conjugated anti-PD-L1 and anti-PD-L2 antibodies (BioLegend). Prior to each primary T-cell co-culture experiment, Raji cells were strictly gated for a narrow range of PD-ligand–mCherry expression. Following sorting, the Raji cells were loaded with SEE (Toxin Technologies), as described previously (33). 200,000 primary CD3+ T cells (isolated using the STEMCELL RosetteSepTM human T-cell enrichment mixture) were added to co-culture with the Raji cells. Supernatants were harvested following a 24-h incubation at 37 °C with 5% CO2. IL-2 concentration in the supernatants was measured by ELISA (human IL-2 ELISA MAXTM BioLegend).
Evolutionary sequences and synteny analysis
Accession numbers of all the protein and DNA sequences used in this study are listed in Table S1. Blast searches were performed searching the various genome-sequencing projects with the Blast-T program with multiple starting queries using the NCBI and the Ensembl servers as described (34).
Author contributions
E. A. P. and X.-P. K. conceptualization; E. A. P., A. M., and X.-P. K. resources; E. A. P., A. G.-E., A. S. T., and X.-P. K. data curation; E. A. P. and X.-P. K. formal analysis; E. A. P., A. S. T., I. M. A., R. P., A. M., and X.-P. K. supervision; E. A. P., A. M., and X.-P. K. funding acquisition; E. A. P. and X.-P. K. validation; E. A. P., K. R. A., and X.-P. K. investigation; E. A. P. and X.-P. K. visualization; E. A. P., A. S. T., A. M., and X.-P. K. methodology; E. A. P. and X.-P. K. writing-original draft; E. A. P., A. M., and X.-P. K. writing-review and editing.
Supplementary Material
Acknowledgments
We thank the Aaron Diamond AIDS Research Center for access to their Biacore T200, and Vincent Sahi for technical assistance. We thank Brian Lang (GE Healthcare) for technical assistance.
This work was supported in part by National Institutes of Health Grants T32AR069515 (to E. A. P), T32GM007308 (to E. A. P), and R01AI125640 (to A. M.) and Ministerio de Economia y Competitividad FIS Grant PI16/00504 (to A. G.-E.). E. A. P., A. M., and X. P. K. are inventors on a pending patent application pertaining to this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S4 and Table S1.
- APC
- antigen-presenting cell
- SPR
- surface plasmon resonance
- CFSE
- carboxyfluorescein succinimidyl ester
- PBMC
- peripheral blood mononuclear cell,
References
- 1. Boussiotis V. A. (2016) Molecular and biochemical aspects of the PD-1 checkpoint pathway. N. Engl. J. Med. 375, 1767–1778 10.1056/NEJMra1514296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Freeman G. J., Long A. J., Iwai Y., Bourque K., Chernova T., Nishimura H., Fitz L. J., Malenkovich N., Okazaki T., Byrne M. C., Horton H. F., Fouser L., Carter L., Ling V., Bowman M. R., et al. (2000) Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192, 1027–1034 10.1084/jem.192.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Latchman Y., Wood C. R., Chernova T., Chaudhary D., Borde M., Chernova I., Iwai Y., Long A. J., Brown J. A., Nunes R., Greenfield E. A., Bourque K., Boussiotis V. A., Carter L. L., Carreno B. M., et al. (2001) PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2, 261–268 10.1038/85330 [DOI] [PubMed] [Google Scholar]
- 4. Brown J. A., Dorfman D. M., Ma F. R., Sullivan E. L., Munoz O., Wood C. R., Greenfield E. A., and Freeman G. J. (2003) Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J. Immunol. 170, 1257–1266 10.4049/jimmunol.170.3.1257 [DOI] [PubMed] [Google Scholar]
- 5. Liang S. C., Latchman Y. E., Buhlmann J. E., Tomczak M. F., Horwitz B. H., Freeman G. J., and Sharpe A. H. (2003) Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 33, 2706–2716 10.1002/eji.200324228 [DOI] [PubMed] [Google Scholar]
- 6. Keir M. E., Butte M. J., Freeman G. J., and Sharpe A. H. (2008) PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohigashi Y., Sho M., Yamada Y., Tsurui Y., Hamada K., Ikeda N., Mizuno T., Yoriki R., Kashizuka H., Yane K., Tsushima F., Otsuki N., Yagita H., Azuma M., and Nakajima Y. (2005) Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin. Cancer Res. 11, 2947–2953 10.1158/1078-0432.CCR-04-1469 [DOI] [PubMed] [Google Scholar]
- 8. Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., and Minato N. (2002) Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 99, 12293–12297 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Topalian S. L., Hodi F. S., Brahmer J. R., Gettinger S. N., Smith D. C., McDermott D. F., Powderly J. D., Carvajal R. D., Sosman J. A., Atkins M. B., Leming P. D., Spigel D. R., Antonia S. J., Horn L., Drake C. G., et al. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolchok J. D., Kluger H., Callahan M. K., Postow M. A., Rizvi N. A., Lesokhin A. M., Segal N. H., Ariyan C. E., Gordon R. A., Reed K., Burke M. M., Caldwell A., Kronenberg S. A., Agunwamba B. U., Zhang X., et al. (2013) Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133 10.1056/NEJMoa1302369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Topalian S. L., Sznol M., McDermott D. F., Kluger H. M., Carvajal R. D., Sharfman W. H., Brahmer J. R., Lawrence D. P., Atkins M. B., Powderly J. D., Leming P. D., Lipson E. J., Puzanov I., Smith D. C., Taube J. M., et al. (2014) Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol. 32, 1020–1030 10.1200/JCO.2013.53.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rizvi N. A., Hellmann M. D., Snyder A., Kvistborg P., Makarov V., Havel J. J., Lee W., Yuan J., Wong P., Ho T. S., Miller M. L., Rekhtman N., Moreira A. L., Ibrahim F., Bruggeman C., et al. (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman A. M., Kato S., Bazhenova L., Patel S. P., Frampton G. M., Miller V., Stephens P. J., Daniels G. A., and Kurzrock R. (2017) Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riaz N., Havel J. J., Makarov V., Desrichard A., Urba W. J., Sims J. S., Hodi F. S., Martín-Algarra S., Mandal R., Sharfman W. H., Bhatia S., Hwu W. J., Gajewski T. F., Slingluff C. L. Jr., Chowell D., et al. (2017) Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell 171, 934–949.e16 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenwald R. J., Freeman G. J., and Sharpe A. H. (2005) The B7 family revisited. Annu. Rev. Immunol. 23, 515–548 10.1146/annurev.immunol.23.021704.115611 [DOI] [PubMed] [Google Scholar]
- 16. Collins A. V., Brodie D. W., Gilbert R. J., Iaboni A., Manso-Sancho R., Walse B., Stuart D. I., van der Merwe P. A., and Davis S. J. (2002) The interaction properties of costimulatory molecules revisited. Immunity 17, 201–210 10.1016/S1074-7613(02)00362-X [DOI] [PubMed] [Google Scholar]
- 17. Lázár-Molnár E., Yan Q., Cao E., Ramagopal U., Nathenson S. G., and Almo S. C. (2008) Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. U.S.A. 105, 10483–10488 10.1073/pnas.0804453105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng X., Veverka V., Radhakrishnan A., Waters L. C., Muskett F. W., Morgan S. H., Huo J., Yu C., Evans E. J., Leslie A. J., Griffiths M., Stubberfield C., Griffin R., Henry A. J., Jansson A., et al. (2013) Structure and interactions of the human programmed cell death 1 receptor. J. Biol. Chem. 288, 11771–11785 10.1074/jbc.M112.448126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Viricel C., Ahmed M., and Barakat K. (2015) Human PD-1 binds differently to its human ligands: a comprehensive modeling study. J. Mol. Graph. Model. 57, 131–142 10.1016/j.jmgm.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 20. Tang S., and Kim P. S. (2019) A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery. Proc. Natl. Acad. Sci. U.S.A. 116, 24500–24506 10.1073/pnas.1916916116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Youngnak P., Kozono Y., Kozono H., Iwai H., Otsuki N., Jin H., Omura K., Yagita H., Pardoll D. M., Chen L., and Azuma M. (2003) Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem. Biophys. Res. Commun. 307, 672–677 10.1016/S0006-291X(03)01257-9 [DOI] [PubMed] [Google Scholar]
- 22. Liu L., Zhang J., Rheindt F. E., Lei F., Qu Y., Wang Y., Zhang Y., Sullivan C., Nie W., Wang J., Yang F., Chen J., Edwards S. V., Meng J., and Wu S. (2017) Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc. Natl. Acad. Sci. U.S.A. 114, E7282–E7290 10.1073/pnas.1616744114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hedges S. B., Kumar S., and Watson J. D. (2009) The Timetree of Life, pp. 309–314, Oxford University Press, New York [Google Scholar]
- 24. Sharpe A. H., Wherry E. J., Ahmed R., and Freeman G. J. (2007) The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 8, 239–245 10.1038/ni1443 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Q., and Vignali D. A. (2016) Co-stimulatory and co-inhibitory pathways in autoimmunity. Immunity 44, 1034–1051 10.1016/j.immuni.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen J. D., Du Pasquier L., Lefranc M. P., Lopez V., Benmansour A., and Boudinot P. (2009) The B7 family of immunoregulatory receptors: a comparative and evolutionary perspective. Mol. Immunol. 46, 457–472 10.1016/j.molimm.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 27. Petroff M. G., Chen L., Phillips T. A., Azzola D., Sedlmayr P., and Hunt J. S. (2003) B7 family molecules are favorably positioned at the human maternal-fetal interface. Biol. Reprod. 68, 1496–1504 10.1095/biolreprod.102.010058 [DOI] [PubMed] [Google Scholar]
- 28. Petroff M. G., and Perchellet A. (2010) B7 family molecules as regulators of the maternal immune system in pregnancy. Am. J. Reprod. Immunol. 63, 506–519 10.1111/j.1600-0897.2010.00841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samstein R. M., Josefowicz S. Z., Arvey A., Treuting P. M., and Rudensky A. Y. (2012) Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150, 29–38 10.1016/j.cell.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaudhri A., Xiao Y., Klee A. N., Wang X., Zhu B., and Freeman G. J. (2018) PD-L1 binds to B7–1 only in cis on the same cell surface. Cancer Immunol. Res. 6, 921–929 10.1158/2326-6066.CIR-17-0316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugiura D., Maruhashi T., Okazaki I. M., Shimizu K., Maeda T. K., Takemoto T., and Okazaki T. (2019) Restriction of PD-1 function by cis-PD-L1/CD80 interactions is required for optimal T cell responses. Science 364, 558–566 10.1126/science.aav7062 [DOI] [PubMed] [Google Scholar]
- 32. Zhao Y., Lee C. K., Lin C. H., Gassen R. B., Xu X., Huang Z., Xiao C., Bonorino C., Lu L. F., Bui J. D., and Hui E. (2019) PD-L1:CD80 cis-heterodimer triggers the co-stimulatory receptor CD28 while repressing the inhibitory PD-1 and CTLA-4 pathways. Immunity 51, 1059–1073.e9 10.1016/j.immuni.2019.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hui E., Cheung J., Zhu J., Su X., Taylor M. J., Wallweber H. A., Sasmal D. K., Huang J., Kim J. M., Mellman I., and Vale R. D. (2017) T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433 10.1126/science.aaf1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desalle R., Chicote J. U., Sun T. T., and Garcia-España A. (2014) Generation of divergent uroplakin tetraspanins and their partners during vertebrate evolution: identification of novel uroplakins. BMC Evol. Biol. 14, 13 10.1186/1471-2148-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.