Abstract
The skin-colonizing commensal bacterium Staphylococcus epidermidis is a leading cause of hospital-acquired and device-related infections. Its pathogenicity in humans is largely due to its propensity to form biofilms, surface-adherent bacterial accumulations that are remarkably resistant to chemical and physical stresses. Accumulation-associated protein (Aap) from S. epidermidis has been shown to be necessary and sufficient for mature biofilm formation and catheter infection. Aap contains up to 17 tandem B-repeat domains, capable of zinc-dependent assembly into twisted, rope-like intercellular filaments in the biofilm. Using microscopic and biophysical techniques, we show here that Aap B-repeat constructs assemble further into zinc-dependent functional amyloid fibers. We observed such amyloid fibers by confocal microscopy during both early and late stages of S. epidermidis biofilm formation, and we confirmed that extracellular fibrils from these biofilms contain Aap. Unlike what has been observed for amyloidogenic biofilm proteins from other bacteria, which typically use chaperones or initiator proteins to initiate amyloid assembly, our findings indicate that Aap from S. epidermidis requires Zn2+ as a catalyst that drives amyloid fiber formation, similar to many mammalian amyloid-forming proteins that require metals for assembly. This work provides detailed insights into S. epidermidis biofilm formation and architecture that improve our understanding of persistent staphylococcal infections.
Keywords: amyloid, analytical ultracentrifugation, bacterial pathogenesis, biofilm, biophysics, circular dichroism (CD), confocal microscopy, fibril, infection, spectroscopy
Introduction
Staphylococcus epidermidis is a critical component of the normal human flora that helps control the colonization and invasion of potentially dangerous microbial pathogens. However, S. epidermidis has emerged as a leading opportunistic pathogen due to its high prevalence on epithelial surfaces and ability to colonize prosthetic medical devices (1). S. epidermidis specifically is the leading cause of nosocomial infections and device-related infections (2, 3) and is, along with its other coagulase-negative relatives, the leading cause of bacteremia (4, 5). Although S. epidermidis infections are typically nonaggressive, they are extremely resistant to antibiotic therapy (6). Therefore, staphylococcal infections often require invasive treatment methods and frequently lead to chronic morbidity, mortality, and high healthcare costs (5, 7, 8). S. epidermidis pathogenesis and chronic persistence is primarily associated with its ability to form a biofilm (9, 10), a multilayered bacterial aggregation surrounded by an extracellular matrix (11). Biofilm formation occurs in three general stages: primary adherence to a surface by individual staphylococcal cells; accumulation into multicellular colonies through intercellular adhesion events; and formation of a mature biofilm through cycles of remodeling that create characteristic cellular towers separated by channels that allow access to nutrients. The biofilm is typically surrounded by a secreted extracellular matrix composed of macromolecular components that serve to anchor the staphylococcal cells together, including the polysaccharide poly-GlcNAc (PNAG)4 as well as proteins, extracellular DNA, or teichoic acid, depending on the strain and growth conditions (12–14). Biofilm formation can occur through both polysaccharide-dependent and -independent pathways, the latter mediated by protein–protein interactions (15). The protein Aap (accumulation-associated protein) is primarily responsible for protein-dependent intercellular accumulation of S. epidermidis biofilm formation (16), and it is also required for the polysaccharide-dependent mechanism (17). Rohde et al. (18) and Corrigan et al. (19) described a protein-based mechanism for staphylococcal biofilm formation that is independent of PNAG; in several strains of S. epidermidis or Staphylococcus aureus, the protein Aap or its ortholog SasG can mediate biofilm formation in the absence of polysaccharide secretion (18, 19). Indeed, 39% of biofilm-positive clinical isolates are PNAG-negative, whereas 90% are positive for Aap (15, 20, 21). Recent work by Schaeffer et al. (22) demonstrated the critical role of Aap in S. epidermidis infection in vivo. Under fluid shear, Aap-deficient strains formed significantly less biofilm than those expressing normal levels of Aap. A rat catheter model was then used to evaluate the biological implication of these results, which revealed that Aap, but not PNAG, was required for infection (22).
Aap is a multidomain protein consisting of an N-terminal export signal followed by the A-repeat region (11 partially-conserved 16-residue repeats), a lectin domain, the B-repeat region containing 5–17 conserved repeats of a 128-amino acid sequence, a proline/glycine-rich (P/G-rich) region that forms a highly-extended stalk that is resistant to compaction (23), and an LPXTG cell wall anchor motif, as shown in Fig. 1A. The N-terminal portion of Aap containing the A-repeat region and lectin domain (collectively called the A-domain) can be proteolytically cleaved to expose the B-repeat region, which can then initiate bacterial accumulation into microcolonies (18). The staphylococcal metalloprotease SepA is responsible for this proteolytic cleavage event, switching the role of Aap from A-domain–mediated surface adhesion to its namesake role in biofilm accumulation (24). Each B-repeat (Brpt) contains a 78-amino acid G5 domain and a 50-amino acid spacer domain (also called an E domain); the B-repeat sequences in Aap are highly conserved, with 83–100% sequence identity (25). The final repeat in the B-repeat region is composed of a single G5 domain without the spacer motif (Fig. 1A); this C-terminal half-repeat “cap” plays a role in stabilizing the protein (25). We have previously demonstrated that a single B-repeat domain with the half-repeat cap (Brpt1.5) will self-associate in the presence of Zn2+ to form an anti-parallel dimer, leading to a model for Zn2+-mediated protein-dependent intercellular accumulation between staphylococci in a nascent biofilm (25, 26). Subsequent work demonstrated that Brpt1.5 and longer B-repeat constructs can self-assemble in the presence of Zn2+ or Cu2+, whereas other metals (Mn2+, Co2+, and Ni2+) can bind to B-repeats but do not induce assembly (27). Recent work has shown similar Zn2+-dependent self-association behavior for the B-repeat region of SasG, the S. aureus ortholog of Aap (28, 29).
Figure 1.
Characterization of tandem B-repeats from S. epidermidis Aap. A, full-length Aap domain organization, including the A-repeat region and lectin domain that are proteolytically cleaved (dashed line), the B-repeat region (5–17 B-repeats) ending with the conserved half-repeat cap, the proline/glycine-rich region (P/G-rich), and the cell wall anchor motif (LPXTG). The domain boundaries of the Brpt5.5, Brpt3.5, and Brpt1.5 constructs are shown below the cartoon. Each full B-repeat contains a G5 domain and a spacer region, and the half-repeat cap contains only the G5 domain. B, far-UV CD spectra demonstrating similar secondary structure characteristics of the previously characterized Brpt1.5 construct (green circle) and cleaved Brpt3.5 (blue triangle) and Brpt5.5 (red square). The far-UV CD spectrum of Brpt1.5 is adapted from Ref. 25. C, sedimentation coefficient distribution plot c(s) of HMBP–Brpt3.5 (dashed blue line), cleaved Brpt3.5 (solid blue line), and Brpt5.5 (solid red line). All constructs sedimented as monomers in the absence of Zn2+.
Full-length Aap contains 5–17 of these nearly identical B-repeats; Corrigan et al. (19) have demonstrated that at least five tandem B-repeats were required for S. aureus biofilm formation, suggesting that the biological function of SasG and, presumably, Aap relies on longer stretches of at least five consecutive B-repeats. Therefore, the focus of this study is to characterize a biologically-relevant construct of Aap consisting of five consecutive B-repeats and the C-terminal cap (called Brpt5.5; see Fig. 1A) and to determine the role of tandem B-repeats in S. epidermidis biofilm formation. In the presence of Zn2+, tandem B-repeats assembled into a range of oligomeric states, including highly-elongated fibers. We show that Zn2+-induced B-repeat fibers are functional amyloid fibers that assemble in a temperature- and time-dependent fashion. Importantly, fibers formed in vitro by recombinant, tandem B-repeats show at least partial resistance to Zn2+ chelation and acidification, suggesting that these highly stable amyloid fibers may play a critical part in rendering the biofilm resistant to physical or chemical stresses. We apply a newly-developed analytical approach to deconvolute the early-stage assembly of fibril intermediates using analytical ultracentrifugation. Finally, we show that amyloid fibers form during early and late stages of S. epidermidis biofilm growth, and we demonstrate that fibrils isolated from S. epidermidis biofilms are primarily composed of Aap. This is the first report demonstrating that Aap, an essential protein for S. epidermidis infectivity, forms amyloid fibrils; these findings reveal important mechanistic details regarding S. epidermidis infections responsible for extensive morbidity and mortality.
Results
Solution characterization of tandem B-repeats from Aap
To determine the functional relationship between tandem copies of the Aap B-repeat region and S. epidermidis biofilm formation, we have generated a construct containing the C-terminal five intact B-repeats, along with the C-terminal half-repeat (called Brpt5.5) (Fig. 1A). The design of this construct was based on the minimum number of B-repeats previously shown to support biofilm formation by SasG, an Aap ortholog from S. aureus (19). In addition, we utilized a Brpt3.5 construct, both in isolation and as an N-terminally His6-tagged maltose-binding protein (MBP) fusion protein (called HMBP–Brpt3.5). The HMBP–Brpt3.5 construct showed more efficient expression and was more stable in solution, so it was predominantly used for initial experiments.
Far-UV circular dichroism (CD) was used to verify proper folding of the B-repeats in these constructs (Fig. 1B). Based on our previous studies of related Brpt1.5 constructs (25, 26) and the high sequence identity between B-repeats (Fig. S1) (25, 30), it was expected that the Brpt3.5 and Brpt5.5 constructs would have similar secondary structure content. Indeed, these constructs all contain high β-strand and coil content. The increased negativity around 200 nm observed for the Brpt3.5 and Brpt5.5 constructs may be due to greater random coil contribution from the increased proportion of B-repeat spacer motifs compared with G5 domains in Brpt3.5 and Brpt5.5 (i.e. Brpt5.5 contains five spacer motifs versus six G5 domains (∼45% spacer), whereas Brpt1.5 contains one spacer motif versus two G5 domains (∼33% spacer) (see Fig. 1A)). The CD spectrum for uncleaved HMBP–Brpt3.5 (Fig. S2) revealed a combination of α-helical, β-strand, and coil secondary structure, consistent with the α-helical content of MBP (31).
Sedimentation velocity analytical ultracentrifugation (AUC) was used to characterize each construct in solution under native conditions (Fig. 1C). Brpt3.5 and Brpt5.5 sedimented as monomers with very high frictional ratios (2.58 and 3.55, respectively; the frictional ratio f/f0 is the frictional coefficient of the sedimenting species compared with that of an ideal sphere of identical volume). Although these constructs have similar sedimentation coefficients, the difference in frictional ratios is an important indication of the increased mass of Brpt5.5 compared with Brpt3.5. Such high frictional ratios are indicative of highly-elongated global conformations, such as what might be expected based on our prior characterization of Brpt1.5 (25, 26). Furthermore, such an extended conformation for tandem B-repeats, in conjunction with the extended Pro/Gly-rich stalk (23), makes Aap well-suited to project itself from the S. epidermidis surface in order to more easily interact with adjacent cells and surfaces and to avoid steric hindrance from other cell wall–anchored proteins. The HMBP–Brpt3.5 construct also sedimented as a monomer but had a significantly increased sedimentation coefficient due to the additional 42-kDa mass contributed by the more globular HMBP fusion tag (Fig. 1C).
Tandem B-repeats assemble into multiple higher-order species in the presence of Zn2+
We previously reported that shorter Aap B-repeat constructs, Brpt1.5 and Brpt2.5, specifically self-associate to form dimers in the presence of Zn2+ (25–27, 30). These data indicated that tandem B-repeats self-associate in a modular fashion, with each B-repeat capable of forming an adhesive contact with another B-repeat in the presence of approximately two Zn2+ ions, and that longer B-repeats constructs can dimerize at lower free Zn2+ concentrations (25, 32). This phenomenon is known as the chelation effect, in which the binding of Zn2+ and the resulting self-association at the first site in each of two multisite protomers reduce the entropic penalty for neighboring sites to self-assemble in the presence of Zn2+. Thus, the effective free Zn2+ concentration required for assembly of the entire multirepeat protein becomes progressively lower as the number of repeats increases. Prior biophysical work on B-repeat constructs has focused on shorter constructs that were suitable for detailed thermodynamic analyses; these short constructs were limited to monomer–dimer equilibria (25–27, 30). For the longer Brpt3.5 and Brpt5.5 constructs described here, self-association in the presence of Zn2+ could be significantly more complicated than a simple monomer–dimer equilibrium, due to the potential for multiple adhesive interactions within a stretch of three to five intact B-repeats. Indeed, we observed a dramatic change in the sedimentation coefficient distribution for HMBP–Brpt3.5 in the presence of Zn2+ compared with monomeric apo-HMBP–Brpt3.5 (Fig. 2, A and B), consistent with the formation of a wide range of very large oligomeric states (Fig. 2C). In addition to the reaction boundaries visible in the 0–40 s* range, the increasing trend at 40 s* indicates the presence of even larger aggregated species. This aggregation behavior is quite distinct from previously observed oligomerization of B-repeat constructs in the presence of Zn2+. Although the HMBP fusion tag alone is able to assemble in the presence of Zn2+ (Fig. S3), the formation of such enormous aggregates of HMBP–Brpt3.5 in the presence of Zn2+ is unexpected and suggests that a distinct mode of assembly or aggregation is occurring.
Figure 2.
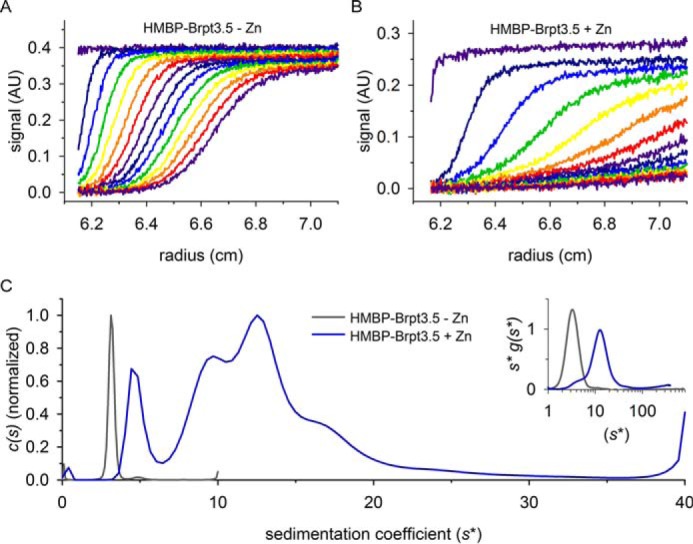
Sedimentation behavior of tandem B-repeats in the presence of Zn2+. A, raw sedimentation velocity data for 5 μm HMBP–Brpt3.5 in the absence of Zn2+, and B, in the presence of 3 mm ZnCl2, both at 36,000 rpm and 20 °C. In both panels, scans 1–100 (5-h elapsed time) were loaded, with every 7th scan plotted. Note the dramatic increase in spacing between scans in B compared with A; the much faster-moving sedimentation boundaries of HMBP–Brpt3.5 in the presence of Zn2+ compared with HMBP–Brpt3.5 alone indicate the sedimentation of very large species. C, sedimentation coefficient distribution of HMBP–Brpt3.5 alone (gray line) and HMBP–Brpt3.5 in the presence of 3 mm ZnCl2 (blue line). The broad peak distribution in the presence of Zn2+ indicates that HMBP–Brpt3.5 sediments as a mixture of assembled oligomers; however, resolution of individual species is difficult due to overlapping sedimentation profiles. The inset of C shows the same data analyzed by Wide Distribution Analysis in SEDANAL; note the x axis is on the natural log scale, and the y scale has been normalized by area. Here, the sample containing 3 mm ZnCl2 shows some material around the 5–10 s* range, with the majority of material sedimenting between 10 and 20 s*, in good agreement with the c(s) distribution calculated by SEDFIT. AU, absorbance units.
2D size–and–shape sedimentation analysis indicates formation of fiber-like species
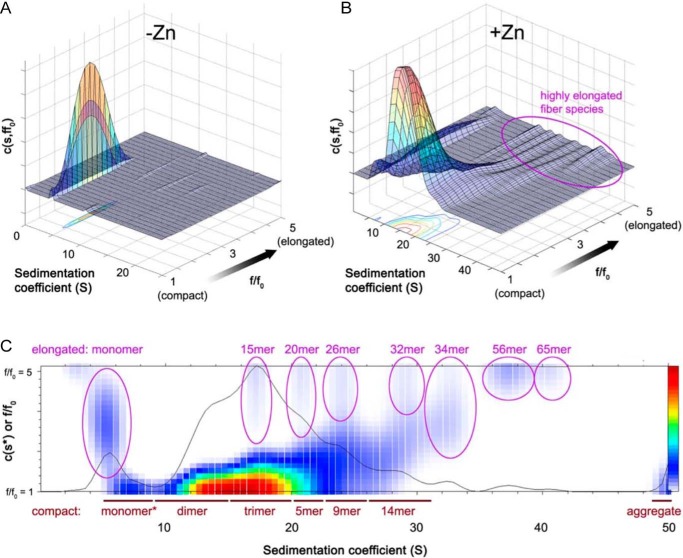
A recently developed AUC analysis method was used to better resolve the multiple sedimentation boundaries observed for HMBP–Brpt3.5 in the presence of Zn2+ and to provide additional information on the size and shape of the sedimenting species. We utilized the 2D size–and–shape c(s,ff0) analysis in SEDFIT that characterizes each sedimenting species in terms of both sedimentation coefficient and frictional ratio (33). This approach is particularly useful when analyzing co-sedimenting species that differ greatly in shape and therefore experience very different degrees of drag (34). For these experiments, we used samples of 5 μm HMBP–Brpt3.5 alone or in the presence of 500 μm or 1 mm ZnCl2. The samples were analyzed at both 25 and 37 °C. To capture the earliest assembly events, the samples were incubated after addition of Zn2+ only for the time required to pull a vacuum (∼30 min). As expected, the HMBP–Brpt3.5 alone sample yielded a single predominant species corresponding to a highly-elongated monomer, as seen in Fig. 3A. Both samples incubated with Zn2+ formed similar oligomeric states, but the higher oligomeric species were more heavily populated for the sample incubated with 1 mm ZnCl2, allowing for easier resolution of the species involved. The c(s,ff0) analysis yields a three-dimensional plot (Fig. 3, A and B) that separates the species based on sedimentation coefficient on the x axis, frictional ratio on the y axis, and peak amplitude on the z axis. We also show a top-down view of the 3D distribution plot (Fig. 3C) superimposed on the standard sedimentation coefficient distribution that has been labeled with the approximate oligomeric states, estimated by Sedfit based on the sedimentation coefficient and apparent frictional ratio values. The data show that HMBP–Brpt3.5 incubated with 1 mm ZnCl2 at 37 °C forms multiple species, including monomeric HMBP–Brpt3.5, several mostly compact oligomeric species (dimer, trimer, and tetramer or pentamer), followed by large oligomers with ever-increasing degrees of elongation. In the presence of Zn2+, peaks for both elongated and compact monomer species are observed (f/f0 values of 3.1 and 1.2, respectively), in contrast to the HMBP–Brpt3.5 alone data that show only elongated monomers (Fig. 3A). The dimer and trimer species are mostly compact, with frictional ratios comparable with those of globular proteins (between 1.2 and 1.4). The putative pentamer species is moderately elongated, with a frictional ratio of 1.7, but all higher oligomers are highly elongated. The putative 9- and 14-mer species show frictional ratios of 2.2 or 2.5, respectively, whereas the 15-mer through 65-mer peaks (circled in magenta ovals in Fig. 3C) gave f/f0 values between 3.9 and 4.7. (As a control, the analysis of the same sedimentation data with an upper limit of 2.5 for f/f0 values resulted in a worse fit to the measured sedimentation boundaries.) These f/f0 values of 3.9 or greater indicate extremely elongated fiber-like morphologies, with approximate axial ratios that range from 72 to 108 (assuming prolate ellipsoids), suggesting that these species may be nascent amyloid fibers. The HMBP–Brpt3.5 sample incubated with 1 mm ZnCl2 analyzed at 25 °C showed a very similar distribution of oligomeric species, although the slower sedimentation rates (due to lower temperature) allowed resolution of a few higher-order oligomeric species, such as putative 75- and 115-mer fibers (Fig. S4).
Figure 3.
AUC c(s,ff0) analysis of early-stage HMBP–Brpt3.5 amyloidogenic intermediates. Sedimentation velocity data (36,000 rpm at 37 °C) were analyzed using the c(s,ff0) analysis model in Sedfit. A, 3D shape and size distribution plot for HMBP–Brpt3.5. Sedimenting species are distinguished based on sedimentation coefficient (plotted uncorrected for buffer conditions) along the x axis and frictional ratio (f/f0) along the y axis. Increasing values of f/f0 correspond to more highly-elongated or nonglobular species. The heat map indicates species concentration, from lowest population density (blue) to highest (red). HMBP–Brpt3.5 alone sediments as a single dominant species of 4.53 S with an elongated frictional ratio (f/f0 = 2.3). B, 3D shape and size distribution plot for HMBP–Brpt3.5 in the presence of 1 mm Zn2+. In the presence of Zn2+, there is a broad distribution of species that vary both in sedimentation coefficient values as well as frictional ratios. Note in particular the series of extremely elongated species (f/f0) values of ∼4 or higher, highlighted by the magenta oval. C, to illustrate the putative species present, the three-dimensional plot from B has been simplified to a two-dimensional distribution of the sedimentation coefficient (x axis) and frictional ratio (y axis), labeled with the putative species present as implied by the given pairs of s and f/f0 values. Elongated species with f/f0 values of ∼3 or greater are highlighted by ovals, and putative species labels are in magenta. Compact species with f/f0 values between 1 and 2.5 are delineated by dark red lines along with putative species labels under the distribution plot. The solid black line depicts a two-dimensional representation of c(s,*), showing the relative total amount of material at any sedimentation coefficient.
Tandem B-repeats form amyloid fibers in the presence of Zn2+
To characterize the nature of these extremely large, fiber-like B-repeat species, samples of HMBP–Brpt3.5 incubated with Zn2+ for 2, 4, or 7 days were visualized by transmission EM (TEM) (Fig. 4A). Negative-stained TEM images revealed large assemblies of protein fibers. One possibility raised by the TEM images is that HMBP–Brpt3.5 can form amyloid fibers in the presence of Zn2+. Amyloid fibers are highly stable protein fibers that have a characteristic β-strand–based fibril architecture (35). Formation of amyloid fibers was initially linked to protein misfolding implicated in disease states; however, a number of “functional” amyloid proteins required for specific cell processes have recently been characterized in organisms ranging from archaea (36) and bacteria to humans (37). Functional amyloid proteins from bacterial species, such as Escherichia coli, Bacillus subtilis, Enterobacteriaceae, and Pseudomonas spp., have recently been implicated in biofilm formation; specifically, these functional amyloid fibers are integral to the overall stability of the biofilm (38–41). To confirm that these HMBP–Brpt3.5 protein fibers were amyloid, samples were incubated with thioflavin T (ThT) and characterized by fluorescence spectroscopy. Thioflavin T is a small-molecule fluorophore that can intercalate within amyloid fibers, which restricts rotation about an internal bond and results in a dramatic increase in quantum yield and characteristic fluorescence emission at 482 nm (42, 43).
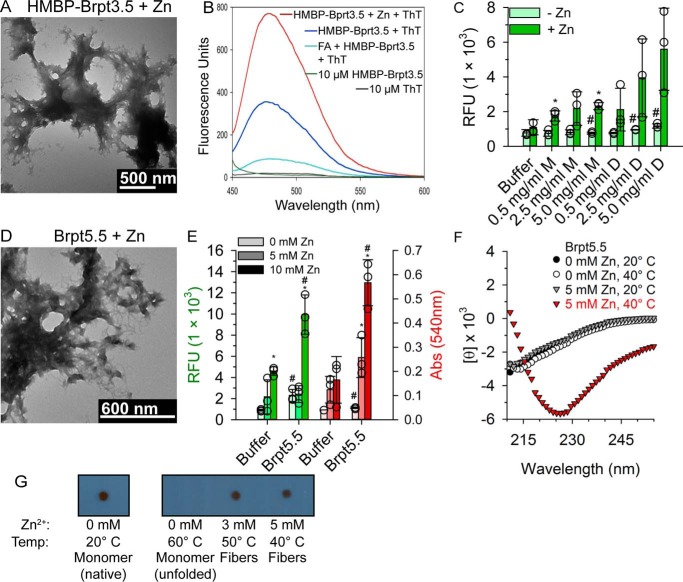
Figure 4.
Amyloid properties of tandem B-repeat constructs in the presence of Zn2+. A, negative-stained TEM image of HMBP–Brpt3.5 protein fibers generated 7 days postincubation with 500 μm ZnCl2. B, fluorescence emission spectra of 10 μm ThT alone (black) or ThT in the presence of HMBP–Brpt3.5 with 500 μm ZnCl2 (red), HMBP–Brpt3.5 alone (dark blue), and HMBP–Brpt3.5 with 10% formic acid (cyan) after excitation at 432 nm. HMBP–Brpt3.5 without ThT (green) did not produce fluorescence. The HMBP–Brpt3.5 alone and HMBP–Brpt3.5 + Zn2+ samples were pretreated with 10% FA to remove aggregates, followed by dialysis back into 50 mm MOPS, pH 7.2, 50 mm NaCl, and addition of ZnCl2. C, ThT fluorescence of cleaved Brpt3.5 tagged with Cys added to the C terminus (Brpt3.5–Cys). At 1 mm ZnCl2, there is increased fluorescence of samples according to overall protein concentration. Furthermore, disulfide-linked Brpt3.5–Cys dimers (0.5–5.0 mg/ml; D) show even higher fluorescence. These results demonstrate the dependence of amyloid formation on local B-repeat concentration. Triplicate samples were examined, with empty circles representing individual measurements and averages shown as filled bars with a standard deviation shown with error bars. Statistically significant increases in the ThT fluorescence of a ZnCl2-containing sample over the sample without ZnCl2 at the same protein concentration is denoted by an asterisk, and a pound sign indicates a significant increase of Brpt3.5–Cys without ZnCl2 over buffer. D, TEM image of native, cleaved Brpt5.5 (20 μm, 1.5 mg/ml) incubated at 37 °C with 5 mm ZnCl2 showing similar fiber morphology as HMBP–Brpt3.5 in A. E shows the ThT fluorescence (green-shaded bars, left) and the Congo Red absorbance (red-shaded bars, right) of Brpt5.5 (20 μm, 1.5 mg/ml) incubated with 0, 5, or 10 mm ZnCl2 (bars shaded from light to dark with increasing ZnCl2 concentration). At 5 and 10 mm ZnCl2, there is increased fluorescence and absorbance of ThT and Congo Red dyes, indicating amyloid-like aggregates in the native, untagged Brpt5.5 construct. Triplicate samples were measured, with individual data points shown in empty circles overlaying bars representing average values (error bars showing one standard deviation). Asterisks signify a statistically significant difference between a zinc-containing condition and the corresponding condition without ZnCl2 (i.e. Brpt5.5 + 10 mm ZnCl2 and Brpt5.5 + 0 mm ZnCl2). The pound sign shows statistical significance of a condition compared with the corresponding buffer condition (i.e. Brpt5.5 + 0 mm ZnCl2 and buffer + 0 mm ZnCl2). F, far-UV CD spectrum of Brpt5.5 (6.5 μm, 0.5 mg/ml) with or without Zn2+, at 20 or 40 °C. Only in the presence of Zn2+ and at higher temperatures does the CD spectrum indicate the rich β-sheet structure observed for amyloid-like aggregate. G, Brpt5.5 samples were tested for recognition by the anti-amyloid antibody OC using a dot-blot assay. Antibody binding was observed for samples incubated at temperatures and Zn2+ concentrations conducive of the CD spectral change to a minimum near 225 nm. Natively-folded Brpt5.5 (without Zn2+) also showed binding, but this was lost when Brpt5.5 was unfolded. Two regions from the same dot-blot assay are shown in G, both after a 5-min exposure. The solid border indicates regions of continuity on the blot. RFU, relative fluorescence units.
To ensure that the ThT fluorescence produced was truly from Zn2+-dependent amyloid fibers, HMBP–Brpt3.5 was first pretreated with 10% formic acid (FA) to depolymerize any fibers that may have spontaneously formed (44). As seen with other amyloid proteins, HMBP–Brpt3.5/Zn2+ produced minimal ThT fluorescence in the presence of FA (Fig. 4B). This effect was not due to FA-induced structural changes in the HMBP–Bprt3.5 monomer, because the secondary structure of the protein was unchanged upon addition of FA (Fig. S2). Upon removal of FA and incubation for 24 h, HMBP–Brpt3.5/Zn2+ protein fibers demonstrated strong ThT fluorescence (Fig. 4B). However, after removal and incubation, HMBP–Brpt3.5 alone also produced a low level of ThT fluorescence, suggesting that some fibers could potentially form in the absence of Zn2+. Based on quantification of monomeric HMBP–Brpt3.5 in the Zn2+-free AUC data, the proportion of protein that forms fibers in the absence of Zn2+ is 3% or less (Figs. 1 and 2). A separate construct was additionally tested to better replicate the natural setting of Aap tethered in dense tufts to the cell wall of S. epidermidis. This construct was Brpt3.5 (cleaved from HMBP) with a C-terminal Cys residue added (Brpt3.5–Cys). Under nonreducing conditions, a disulfide bond would link two Brpt3.5 molecules in a parallel fashion, similar to their orientation on the cell surface, while raising the local B-repeat concentration, a well-known factor in amyloidogenesis. In Fig. 4C, there is an increase in ThT fluorescence corresponding to overall protein concentration. Additionally, there is a dependence on local B-repeat concentration, with disulfide-linked samples (“D” for dimer) yielding higher ThT fluorescence than reduced samples (“M” for monomer).
To ensure that amyloid formation was not due to the presence of the His6–MBP tag (or the C-terminal Cys tag), native Brpt5.5 (tag-free) was incubated with Zn2+ and then visualized by TEM. Brpt5.5 formed fibers (Fig. 4D) primarily of the “branched” morphology seen with HMBP–Brpt3.5 (Fig. 4A). This morphology resembles one often observed with light-chain amyloid (45) but that is also observable with Aβ peptide (46). Analyzing these Brpt5.5 fibers spectroscopically (Fig. 4E, green bars) revealed that incubation with amyloid-binding dyes led to increases in both ThT fluorescence (47) and Congo Red absorbance at 540 nm (Fig. 4E, red bars) (48). Because amyloid fibers have characteristic structures rich in β-strand, we examined the far-UV CD spectrum under conditions that promote these amyloid-like aggregates. It should be noted that based on X-ray crystallography and NMR studies (26, 30, 49), along with CD data presented here and elsewhere (25, 30), natively-folded B-repeats contain β-sheet and random coil. Importantly, however, Fig. 4F demonstrates that when Brpt5.5 is incubated with 5 mm Zn2+ at near-physiological temperature (40 °C), there is a significant change in the CD spectrum, resulting in a strong, broad minimum near 225 nm. This is similar to CD spectra observed with insulin fibrils (50) and glucagon fibrils (51). Together, these spectroscopic results and TEM observations are consistent with amyloid-like fibril formation by native, tandem B-repeats.
A number of antibodies have been raised against amyloid-forming peptides or proteins. Kayed et al. (52) reported an antibody (OC antibody), which could specifically recognize Aβ42 fibers, but not monomers or oligomers. Furthermore, it appeared to recognize generic amyloid fiber conformation(s) irrespective of protein sequence, as it was able to also detect amyloid fibers from islet amyloid polypeptide and α-synuclein (52). Thus, we chose this antibody to test against Brpt5.5 amyloid fibers (Fig. 4G). By dot-blot assays, OC antibody recognized Brpt5.5 that had been incubated with Zn2+ at temperatures that resulted in a significant change in the CD signal in Fig. 4F. Interestingly, OC antibody recognized native, monomeric Brpt5.5 as well. This suggests that conformations present in the amyloid fiber are also present in the natively-folded protein. Incubating Brpt5.5 in the absence of Zn2+ and at a temperature high enough to unfold the protein resulted in loss of OC antibody binding. Collectively, Brpt5.5 and HMBP–Brpt3.5 both share features characteristic of amyloid fibers.
B-repeat fiber assembly is time- and temperature-dependent
Metal ions, namely Zn2+ and Cu2+, play complex roles in the aggregation and amyloidogenesis of β-amyloid peptide important in Alzheimer's disease (53–56). However, functional amyloid-forming proteins from bacteria assemble into amyloid fibers without a known requirement for zinc ions or other triggering molecules. Rather, these amyloid proteins often form as a result of the coordinated action of accessory proteins, including nucleator proteins and chaperones that prevent aberrant polymerization in the cytoplasm (38, 41, 57, 58). In other cases, amyloidogenesis can require proteolytic activity to release the amyloidogenic regions of the protein, such as with Bap (59). Thus, the mechanism of Zn2+-dependent amyloid formation by the B-repeat region of Aap is of significant interest for comparison with other systems. We have therefore used a combination of HPLC, TEM, confocal microscopy, and AUC to analyze the assembly of Aap B-repeat amyloid formation as a function of time, temperature, and solution conditions.
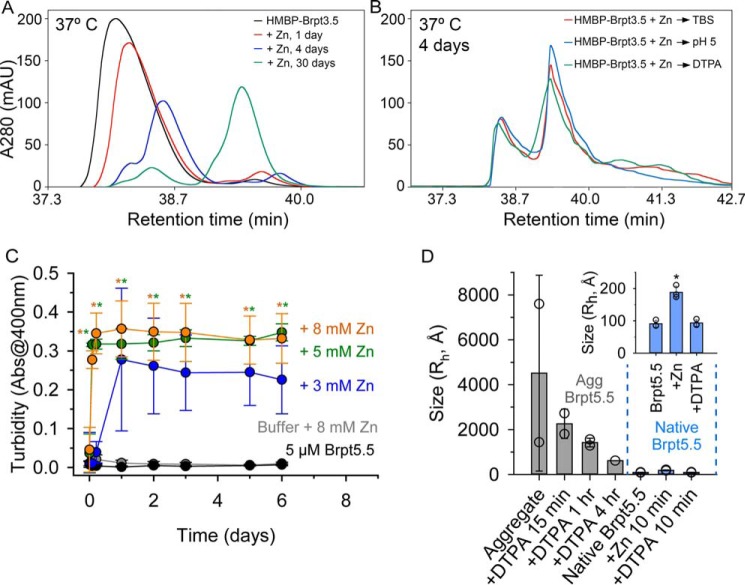
To assess the effect of temperature on the rate and morphology of amyloid fiber formation, we used the HPLC quantification assay previously shown to effectively differentiate between monomer and oligomer or soluble fiber species (60). A relative time course for HMBP–Brpt3.5 fiber formation was established by incubating 10 μm HMBP–Brpt3.5 with 500 μm ZnCl2 for 0, 2, and 6 h and 1, 4, and 30 days at both 20 °C (Fig. S5) and 37 °C (Fig. 5A). Samples were first centrifuged for 1 h at 13,000 rpm to remove any insoluble aggregates prior to separation by a C4 HPLC column; therefore, the elution profile only reports on monomer, oligomer, and/or soluble amyloid fiber species. The HPLC elution profile of HMBP–Brpt3.5 alone (Fig. 5A, black trace) gives a peak eluting at 28.5 ml, which corresponds to a single monomeric species, as shown by its sedimentation coefficient distribution (Fig. 1B). Upon addition of Zn2+ to HMBP–Brpt3.5, samples gradually showed a shift in the distribution of peaks toward higher-elution volumes that represent higher-order oligomer or fiber species. As time progressed, the monomer peak decreased and shifted to the right as the oligomer peak increased. The transition from monomer to putative fiber species was more pronounced after 1, 4, or 30 days for samples incubated at 37 °C (Fig. 5A) compared with the samples incubated at 20 °C (Fig. S5), indicating accelerated HMBP–Brpt3.5 self-assembly at higher temperatures.
Figure 5.
HPLC and turbidity assays to monitor time and temperature dependence of amyloidogenesis. A, C4 reverse-phase HPLC elution profiles of 10 μm HMBP–Brpt3.5 alone (black) or incubated with 500 μm ZnCl2 for 1 (red), 4 (blue), or 30 (green) days at 37 °C. Samples were centrifuged at 13,000 rpm for 1 h prior to HPLC separation to remove insoluble aggregates. B, samples of 10 μm HMBP–Brpt3.5 with 500 μm ZnCl2 were incubated for 4 days at 37 °C prior to addition of Tris-buffered saline (red trace); sufficient diluted HCl to lower the pH to 5 (blue trace); or 2 mm DTPA (green trace). Separation by HPLC revealed that oligomer/soluble fiber species were maintained in the presence of both acid (blue) and DTPA (green) when compared with the buffer-treated control samples (red) at both temperatures. TEM images of these samples are shown in Fig. S5. C, samples of 5 μm Brpt5.5 incubated at 37 °C and followed by absorbance at 400 nm, showing turbidity in the presence of ZnCl2 with maximal turbidity reached by 24 h. Buffer controls for each condition were also monitored. For clarity, only the buffer + 8 mm ZnCl2 data are shown, which had the highest turbidity of all buffer conditions. D, investigation of the DTPA resistance of Brpt5.5 aggregates in 5 mm ZnCl2 from C by dynamic light scattering. Very large particle sizes were observed in the two samples examined. After addition of 5 mm DTPA, measurements were taken at 15 min and 1 h. In each case, particles of very large size were still present. One sample was examined at 4 h, with an average hydrodynamic radius of 620 Å. Natively-folded Brpt5.5 samples (n = 3, blue-shaded bars, also shown in the inset) were measured before and after addition of 5 mm ZnCl2, followed by addition of 5 mm DTPA (measured at 10 min). The average hydrodynamic radius of each sample is shown as a bar with error bars showing one standard deviation. An asterisk shows a significant increase in the Native Brpt5.5 + 5 mm ZnCl2 over both Brpt5.5 without ZnCl2 and after DTPA addition.
We verified that our observations were not an artifact of the His6–MBP tag, using an orthogonal approach. Because the HPLC assay is limited to only oligomers and soluble fiber species, we used a turbidity assay that follows light scattering by large assemblies and aggregates, including amyloid fibers (61). With the Brpt5.5 construct, we observed significant increases in the turbidity of the samples containing Zn2+ when incubated at 37 °C (Fig. 5C). These data demonstrate the Zn2+, time, and temperature dependence of B-repeat fiber assembly using native, untagged B-repeats.
B-repeat fibers are resistant to acid and chelator treatment
Our previous work on Brpt1.5 revealed that this shorter construct formed a reversible Zn2+-dependent dimer that was sensitive to removal of Zn2+ (via chelators such as DTPA) or to even a modest decrease in pH (from 7.4 to 6.0) (25). To assess the stability of mature amyloid fibers, we allowed HMBP–Brpt3.5 fibers to form in the presence of Zn2+ over a period of 24 h or 4 days at 20 or 37 °C. We then treated the samples by addition of a sufficient volume of dilute HCl to lower the pH to 5, addition of 2 mm DTPA, or addition of buffer as a control. The samples were incubated for 2 h before loading onto a C4 reverse-phase HPLC column, without spinning out the fibrous aggregates. The elution profile for the 20 °C sample at 4 days resembled the 24-h samples (Fig. S5) in terms of the monomer versus oligomer distribution and showed evidence of limited remodeling upon incubation with DTPA or HCl. In contrast, the elution profile for the 37 °C sample (Fig. 5B) showed that the oligomer/fiber peak predominated and that it was more resistant to the action of DTPA or HCl. TEM was performed on each sample to confirm that fiber structure was maintained (Fig. S6). Furthermore, although some conformational rearrangement of the fibers may occur at lower temperatures and early time points (62), mature fibers at 37 °C are highly resistant to these environmental conditions.
The initial Zn2+-dependent assembly of untagged Brpt5.5 was reversible with the addition of DTPA or HCl, as shown by sedimentation velocity AUC data in Fig. S7. These results coincide with the reversible assembly of Brp1.5 (25), suggesting tandem B-repeats still undergo an initial phase of reversible assembly. Brpt5.5 aggregates from Fig. 5C were examined by dynamic light scattering to determine their resistance against DTPA. Although a decrease in the average hydrodynamic radius of all particles present was observed within 15 min, the average particle size was still about 10 times that of initial, reversible Zn2+-induced complexes (Fig. 5D, inset). Even at 1 and 4 h, the average particle size of the aggregates exceeded that of natively-folded Brpt5.5. This result suggests that whereas some higher-order oligomers or nonamyloid aggregate may have been reversed or solubilized by DTPA, there is a fraction of material that remained in an aggregate form, as indicated by the remaining turbidity. It is possible the DTPA-sensitive material had not yet developed into mature amyloid fibers or that a higher local concentration of the tandem B-repeats (e.g. as seen with HMBP–Brpt3.5 or with native cell wall-anchored Aap) is required for complete DTPA resistance. DTPA resistance is an important feature of Aap functional amyloid, in that we previously reported the addition of Zn2+ chelator could prevent biofilm formation but could not disrupt mature biofilms (25), consistent with the presence of highly-stable Aap-derived amyloid fibers in the biofilm.
Amyloid fibers are structural components in S. epidermidis biofilms
Although it has been well-established that Aap is critical for S. epidermidis biofilm formation (15, 16, 22, 25), the mechanism by which Aap promotes stable, long-term intercellular adhesion is not well-understood. Others have shown that bundles of Aap fibrils extend outward from the cell wall on planktonic S. epidermidis cells (63, 64), but these were presumably free-standing proteins attached to the cell wall rather than amyloid fibrils. We sought to explore the significance of our findings in the context of S. epidermidis biofilms. S. epidermidis strain RP62A biofilms were grown in tryptic soy broth (TSB) supplemented without additional Zn2+ (Fig. 6A) or with additional Zn2+ (Fig. 6B) and visualized by TEM. Large networks of extracellular fibrils were observed around clusters of cells in the biofilm. This morphology closely resembles the fiber morphology observed by native Brpt5.5 when examined at the same magnification (see Fig. 6G for Brpt5.5 incubated at 37 °C with 5 mm ZnCl2, compared with Fig. 6H for RP62A biofilm + 20 μm ZnCl2). These extracellular fibers were observed in biofilms grown in TSB with or without addition of Zn2+, although the presence of Zn2+ increased the prevalence of the fibers. TSB contains Zn2+, which allows for Aap-based, Zn2+-dependent accumulation and biofilm formation (25, 26, 65). Addition of the Zn2+ chelator, DTPA, at the start of growth inhibits biofilm formation (25); by TEM, these S. epidermidis cells showed no evidence of the extracellular fibrous networks seen in the mature biofilms (Fig. 6C). The addition of DTPA at the start of growth would prevent the initial reversible self-assembly of Aap B-repeat regions (Fig. S7), presumably preventing subsequent nucleation of amyloid. However, once a mature biofilm is formed, DTPA can be added with no resulting effect (Fig. 6D) (25), similar to the observation that adding DTPA to mature amyloid fibers formed in vitro does not result in depolymerization (Fig. 5B). The highly-resistant nature of these amyloid fibers would contribute to the strength and stability of the mature biofilm, as shown previously for other systems, including TasA from B. subtilis (40).
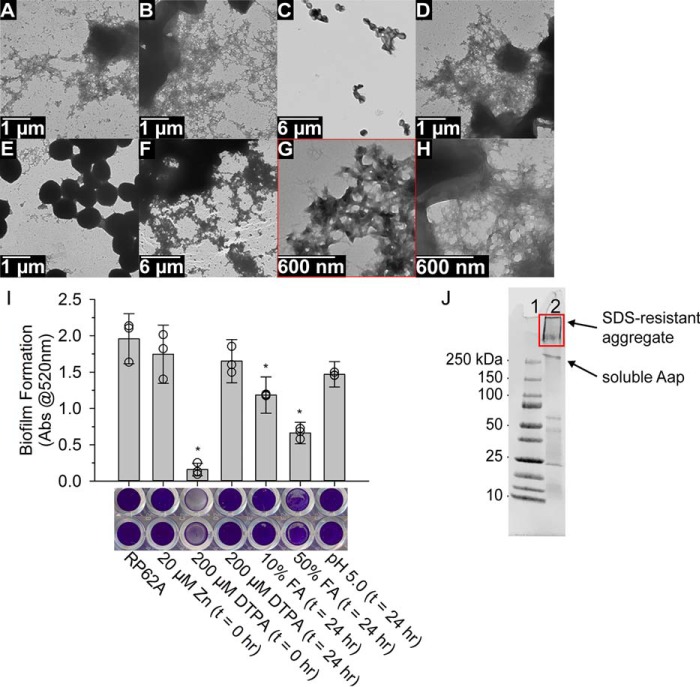
Figure 6.
Amyloid fibers composed of Aap are important structural components in S. epidermidis biofilms. S. epidermidis biofilms were examined by TEM when grown on dialysis membrane on TSA (A), + 20 μm ZnCl2 (B), 100 μm DTPA (C) added at the start of growth (t = 0 h), or 100 μm DTPA (D) at the end of growth and incubated for 1 h (t = 24 h), 50% FA (t = 24 h) (E), and pH lowered to pH 5 (t = 24 h) (F). This series of TEM images displays the resistance of fibers to DTPA and acidification in the setting of a mature biofilm. G, and H, compare the fibers observed by Brpt5.5 incubated at 37 °C with 5 mm ZnCl2 (from Fig. 4) (G) and RP62A biofilms + 20 μm ZnCl2 (H) at similar magnification demonstrating similar morphologies. I examines the ability of S. epidermidis to form biofilms under various conditions. Biofilm formation can be inhibited by addition of DTPA (t = 0 h), but not after biofilm formation has already occurred (t = 24 h), whereas addition of FA can significantly disrupt mature biofilms. Acidification to pH 5, like adding DTPA at t = 24 h, had no significant effect compared with RP62A without treatment. The asterisk denotes statistically significant difference (p < 0.05) to RP62A biofilm formation without treatment, as determined by a two-tailed Student's t test. Three separate experiments were performed for data in C and D, each with four replicate wells. The averages from the three separate experiments are plotted as empty circles, with the average of those plotted as gray bars with error bars showing ± S.D. Images of the crystal violet stained wells (after washing and drying) of two representative samples are also shown. J, bacteria from biofilms shown in B were digested with lysostaphin to remove cell wall-anchored proteins. The supernatant from this mixture was then examined by SDS-PAGE, which revealed a large amount of material unable to migrate beyond the stacking gel. This material (red rectangle) was identified primarily as Aap by nanoLC-MS/MS.
In Fig. 4B, we demonstrated that B-repeat fibers formed in vitro are sensitive to FA. Based on this result, Aap fibers in biofilms should also show sensitivity to FA. Indeed, addition of FA to mature biofilms demonstrated the ability to disrupt the majority of the extracellular fibers in the biofilm (Fig. 6E). Furthermore, we examined the effect of pH on biofilm stability. As with DTPA, we previously observed that mildly-acidic pH disrupts the reversible assembly of B-repeat proteins (25, 32), but low pH has no effect on pre-formed B-repeat amyloid fibrils (Fig. 5B). Consistent with these in vitro results, we observed that lowering the pH of an established biofilm to 5.0 had no effect on the extracellular amyloid fiber network (Fig. 6F).
A biofilm formation assay (Fig. 6I) was performed to complement the TEM observations and evaluate the role of Aap functional amyloid fibers in biofilm formation in a more quantitative way. We once again observed the ability of DTPA to inhibit biofilm formation, but only when present before biofilm formation; when DTPA or pH 5.0 buffer was added after biofilm maturation (t = 24 h), neither was able to disrupt the biofilm, likely due to the resistance of the amyloid fibers (Fig. 6, D, F, and I). Furthermore, addition of FA to mature biofilm was able to significantly disrupt the biofilm, as expected due to its ability to depolymerize the functional amyloid (Fig. 6E). The correlation between depolymerization of amyloid fibrils and weakening of the biofilm structure is a key observation that supports the idea of functional amyloid fibrils contributing strength and stability to the biofilms.
S. epidermidis amyloid fibers are composed of processed Aap
As a final characterization of the extracellular amyloid fibers we observed in S. epidermidis biofilms, we determined the composition of these fibers by a combination of SDS-PAGE and MS. Biofilms were collected as they were for TEM characterization (Fig. 6B). The biofilm mixtures were then centrifuged to separate the bacteria from extracellular material and media. After removing the supernatant, we used lysostaphin to digest the polyglycine cross-links of the cell wall peptidoglycan. The soluble proteins released from the cell wall were examined by SDS-PAGE. We observed SDS-insoluble aggregate trapped in the well and stacking gel (Fig. 6J). This aggregated material contained primarily Aap when examined by nanoLC-MS/MS after in-gel tryptic digestion (Tables S1 and S2). In addition to Aap, peptides from several cytoplasmic proteins were identified, but with sparse coverage, suggesting these proteins are present in small quantities and are likely irrelevant to the amyloid fibers. We observed this result in three separate biological samples, one of which was not exposed to lysostaphin and thus contained only extracellular material and not cell wall-anchored material (Fig. S8 and Table S3 and S4).
Peptides were observed from the B-repeats and the lectin region of Aap, with one peptide including Leu-601, which is one of two SepA cleavage sites (the other being Leu-335) (24), suggesting the lectin domain is still attached to Aap. The Aap A-repeats also contain tryptic sites, so the fact that no A-repeat peptides were detected by MS/MS is highly suggestive that SepA cleavage occurred at Leu-335, downstream of the A-repeat region. This observation is in agreement with Rohde et al. (18), who demonstrated that proteolytic processing of the N terminus of Aap is required for biofilm formation, and with Paharik et al. (24), who observed SepA-processed Aap molecules that retained the lectin domain but not the A-repeat region in the context of biofilms. This suggests the lectin does not need to be removed for biofilm formation (accumulation and amyloidogenesis specifically) to occur, but that it is the A-repeat region that inhibits biofilm formation.
Amyloid fibers form early in biofilm formation and correlate with DTPA resistance of biofilms
To further explore the importance of Aap amyloidogenesis in the context of developing biofilms, we followed biofilm formation by S. epidermidis strain RP62A at distinct time points throughout the first 24 h of biofilm formation. The addition of ThT to the media during the initial inoculation of the bacteria allowed us to follow amyloid formation by CFM (Fig. 7A). In parallel, biofilms were stained with LIVE/DEAD fluorescent dye to ensure ThT was not affecting cell growth or biofilm formation (Fig. 7B). As early as 2 h post-inoculation, punctate ThT fluorescence was visible on planktonic cells. This suggests that fibers start to form to a limited degree prior to cellular accumulation or biofilm assembly (Fig. 7B). At 6 h, the formation of microcolonies, and therefore intercellular accumulation, begins to occur. At this stage, ThT fluorescence is located at the boundaries between associating cells, which is consistent with the role of Aap as the critical factor for intercellular adhesion. ThT fluorescence increases throughout the biofilm over time, with a predominance of fluorescence in the core of the biofilm with the highest cellular densities. Although ThT is regularly used to identify amyloid fibers, there are also other molecules that can interact with ThT (43, 66–69).
Figure 7.
Formation of amyloid fibers is well-correlated to DTPA resistance in S. epidermidis biofilms. A, biofilm formation by S. epidermidis was characterized 2, 6, 12, 18, and 24 h post-inoculation by confocal microscopy, staining with 10 μm ThT (cyan) over a bright field. In the 2-hr panel, a zoomed inset is shown with increased contrast and brightness to highlight punctate ThT fluorescence visible around planktonic S. epidermidis cells. ThT fluorescence is seen primarily at cell–cell junctions at the 6-hour time point. Mature biofilms (24 h) show ThT throughout the biofilm as a major structural component. B, S. epidermidis biofilms at 2, 6, 12, 18, and 24 h post-inoculation were stained in parallel with LIVE/DEAD stain (green/red) as a control (scale bar, 10 μm). Images were generated in Zen 2009 Light Edition. C, biofilm formation assays show DTPA resistance develops within 2–6 h, with full DTPA resistance reached within 24 h. D demonstrates these observations are, in fact, due to addition of DTPA and not mixing. The asterisk denotes statistically significant difference (p < 0.05) to DTPA added at t = 0 h, and the pound sign indicates no significant difference to RP62A without treatment, as determined by a two-tailed Student's t test. Three separate experiments were performed for data in C and D, each with four replicate wells. The averages from the three separate experiments are plotted as empty circles, with the average of those plotted as gray bars with error bars showing ± S.D. Images of the crystal violet–stained wells (after washing and drying) of two representative samples are also shown.
To provide further evidence that the increased ThT fluorescence observed by CFM is indicative of Aap-dependent fibers forming in the developing biofilms, we took advantage of the DTPA resistance of the amyloid fibers observed in vitro and in mature biofilms. Specifically, we tested the ability of DTPA to inhibit biofilm formation when added at distinct time points along a similar time frame. Addition of DTPA during the 1st h of incubation was able to prevent biofilm formation from occurring, and by 2 h, DTPA was significantly less effective. The DTPA resistance of the biofilm correlates with the time frame of amyloid formation in growing biofilms (Fig. 7A). To ensure that the lack of biofilm formation at early time points was not due to mixing of the cultures upon adding DTPA, a duplicate experiment (Fig. 7D) was performed where DTPA was not added but wells were mixed in the same way as in Fig. 7C. Each of the conditions was still able to grow very strong biofilms, similar to the untreated control (“RP62A”), indicating that the mixing does not significantly affect biofilm formation. Therefore, these results support our hypothesis that Aap is important both for intercellular accumulation, by the formation of reversible assemblies and subsequently amyloid fibers between bacteria, and for stabilizing mature biofilms due to the remarkable resistance of amyloid fibers to physical and chemical insults.
Discussion
Our previous work established that the B-repeat region of Aap can undergo Zn2+-mediated self-association to form proteinaceous rope-like filaments between staphylococcal cells in the biofilm and that Zn2+ chelation was able to inhibit biofilm formation by both S. epidermidis and S. aureus (25–27, 30). This initial work was carried out with the short B-repeat constructs Brpt1.5 and Brpt2.5, which showed reversible self-association that could be inhibited upon addition of chelator or moderate reduction in pH. Given that staphylococcal biofilms typically undergo acidification over time, it was unclear how this Zn2+-mediated self-assembly mechanism could be maintained within the biofilm in vivo.
The results presented here using a more biologically relevant construct (Brpt5.5) and a Brpt3.5 fusion protein (HMBP–Brpt3.5) demonstrate that the B-repeat region of Aap is capable of two distinct Zn2+-dependent assembly processes, forming both reversible oligomers and functional amyloid fibers within biofilms. Although the formation of amyloid fibers in biofilms has been established in several bacterial species, the mechanism of Aap amyloid fiber assembly displays some unique features among this group of bacterial biofilm proteins. Unlike the amyloidogenic biofilm proteins curli (E. coli and Salmonella spp.) (57, 70), TasA (B. subtilis) (40, 58), or FapC (Pseudomonas spp.) (41) that use additional chaperones or initiator proteins to help control and initiate amyloid fiber assembly, our data suggest that Aap utilizes Zn2+ as a catalyst to drive amyloid fiber formation. This mechanism for metal-dependent amyloid nucleation is instead reminiscent of several mammalian amyloid-forming proteins, including β-amyloid (54, 71, 72), prions (73, 74), and β2-microglobulin (75, 76). Our proposed mechanism of Aap amyloid assembly shares a few similarities to that reported for S. aureus Bap (59). The N-terminal region of this cell wall–anchored protein is cleaved and released into the local environment, where it can self-assemble into amyloid-like structures in the presence of low Ca2+ concentrations or at acidic pH. Bap, like Aap, can therefore act as both a sensor and a scaffold protein (59). Aap requires cleavage of the N-terminal A-repeat region by staphylococcal proteases or human proteases to unmask the B-repeat region before intercellular contact and amyloidogenesis occur in the presence of Zn2+ (18, 24).
Bap has a similar arrangement of tandemly repeated domains to Aap and SasG (but the domains differ from the A- and B-repeats found in Aap and SasG), and it seems to be critical for infection in a mouse catheter model (59). However, the bap gene has not been found in S. aureus or S. epidermidis human clinical isolates (59). A Bap homolog, Bhp, is present in human isolates of S. epidermidis (77), but no experimental evidence exists regarding its role in biofilm formation or ability to form amyloid fibers, with the exception of a study by Lembre et al. (78), who showed a six-residue peptide from Bhp was able to form amyloid fibers in vitro. The bhp gene was found in less than half of S. epidermidis strains isolated from prosthetic knee joint and hip infections, whereas aap was found in 89% of isolates (15).
A recently identified S. epidermidis protein, Sbp, was shown to form amyloid fibers both in vitro and when expressed in intracellular inclusions in E. coli (79). Wang et al. (79) also showed that an sbp knockout of S. epidermidis strain 1457 did not show thioflavin S (ThS) fluorescence by confocal microscopy nor did it form a biofilm, an observation consistent with the initial study by Decker et al. (80). The WT strain 1457 did show biofilm formation, as expected, along with ThS fluorescence indicative of amyloid-like fibers. However, the authors did not identify Sbp as a component of purified amyloid fibers derived from biofilms, so there is uncertainty whether Sbp directly forms amyloid fibers in the biofilm or whether Sbp plays an indirect role in the nucleation of Aap amyloid fibers. If the latter case were true, then genetic knockout of either aap or sbp would abrograte biofilms as well as eliminate ThT fluorescence. Until there are well-defined mutations identified in either Aap or Sbp that specifically eliminate interactions, it will be difficult to establish the exact role played by Sbp. For this reason, we utilized MS to confirm that the fibers we observed were indeed composed of Aap.
The molecular characteristics of diverse types of amyloid fiber architecture have been well-described in multiple studies using a combination of structural and biophysical techniques (81–89). AUC approaches have also been used to characterize mature amyloid fibers in solution (90, 91). However, there have been few studies about the assembly of early oligomers and proto-fibril intermediates that produce mature amyloid fibers (92–94), particularly in terms of resolving discrete intermediate species. For example, pre-amyloid oligomerization of transthyretin has been characterized by AUC but without resolution of individual species (95). We have applied the c(s,ff0) analysis to sedimentation velocity AUC data to resolve the broad array of species present at early stages of Aap amyloidogenesis, including the oligomeric complexes and nascent fibers. This is one of the first uses of this 2D size–and–shape analysis approach to an amyloid-forming protein, and its ability to separate many species with differing sizes and shapes in solution holds promise for resolving assembly intermediates in other amyloid systems as well. A similar approach, based on the van Holde-Weischet method (96) and implemented in Ultrascan (97) (RRID:SCR_018126), designed to resolve size and shape information from sedimentation velocity data has been applied to mature amyloid fibers formed by Aβ peptides (98, 99), but here we examined amyloidogenesis of a much larger protein. The c(s,ff0) analysis model is capable of distinguishing species of different size and shape that are sedimenting at the same rate (34), which can occur in amyloid assembly systems due to the complicated nature of the assembly process and the number of species to be resolved. This analysis works particularly well in systems such as this one with slow kinetics, which allows resolution of discrete assembly intermediates rather than simply showing broad reaction boundaries representing multiple species in rapid exchange. The c(s,ff0) analysis of Aap presented here clearly discriminates between compact oligomers and fibers that show similar sedimentation coefficients due to the increased drag and resulting slower sedimentation of the highly-elongated fibers. This analytical approach could prove useful in providing mechanistic details for how amyloid nucleation occurs by this or other proteins.
Biofilm formation is the primary characteristic responsible for pathogenicity, and it contributes to antibiotic resistance in chronic infections caused by S. epidermidis. One of the biggest challenges with recurrent infections caused by biofilms is their highly adhesive and cohesive nature and their resistance to chemical and physical insults. Our data indicate that intercellular amyloid fibers appear early during the accumulation phase of nascent biofilms, and they continue to increase until they are ubiquitous throughout the mature biofilm. Using highly-purified tandem B-repeats in solution, we show that mature amyloid fibers are highly resistant to chemical stresses such as Zn2+ chelation or acidic pH, suggesting that the amyloid fibers forming the intercellular network are one reason S. epidermidis biofilms are so resistant to harsh environmental conditions. We have previously shown (25) that addition of Zn2+ chelators to staphylococcal cultures prevents biofilm formation. However, addition of these same chelators to pre-formed mature staphylococcal biofilms does not appreciably disperse the biofilm, which is now explained by the presence of resistant amyloid fibers between cells. Furthermore, we predict that homologous surface proteins containing tandem B-repeats in S. aureus (i.e. SasG and Pls) and other Gram-positive bacteria will form similar Zn2+-dependent amyloid fibers between cells in biofilms. The data presented here provide new insights into the mechanism of S. epidermidis biofilm formation and infection.
Materials and methods
Bacterial strains and media
S. epidermidis strain RP62A (ATCC 35984) was purchased directly from ATCC as a glycerol stock and was cultured in TSB.
Expression construct generation
The Brpt3.5 construct (amino acids 1761–2223) of Aap (NCBI accession no. AAW53239.1 (10)) was PCR-amplified from RP62A genomic DNA and inserted into the expression vector pHisMBP–DEST (kindly provided by Dr. Artem Evdokimov) using Gateway technology, which adds an N-terminal hexahistidine–maltose-binding protein (His–MBP) fusion to the Brpt3.5 construct, with an intervening tobacco etch virus protease site. The Brpt5.5 construct (amino acids 1505–2223) of Aap (NCBI accession no. AAW53239.1 (10)) was synthesized by Life Technologies, Inc., GeneArt®, also containing an intervening tobacco etch virus protease site for removal of the His–MBP tag. The plasmids were then transformed into the E. coli expression cell line BLR(DE3) (Novagen). The Brpt3.5–Cys mutant was synthesized by Life Technologies, Inc., GeneArt®, as well, containing amino acids 1761–2223 of Aap (NCBI accession no. AAW53239.1 (10)), followed by a single cysteine residue inserted before the stop codon.
Protein expression and purification
One-liter cultures were inoculated with 10 ml of His–MBP-Brpt3.5/BLR(DE3) culture at an A600 of 0.6–0.8 and then allowed to incubate overnight with shaking at 37 °C. Protein expression was then induced using 250 μm IPTG for 6 h at 25 °C. The cells were then harvested, resuspended, frozen, and thawed prior to lysis by French press. The cell lysate was centrifuged, and the soluble fraction was decanted onto a nickel-NTA gravity column. The Brpt3.5 protein was eluted by imidazole step gradient at 300 mm imidazole. Fractions confirmed by SDS gel to contain Brpt3.5 were then further purified by anion-exchange using a 5-ml anion Q fast-flow column (GE Healthcare) followed by size-exclusion chromatography using a Superdex 200 column (GE Healthcare). The presence of the His–MBP-fusion tag improved solubility and behavior in solution and was therefore left attached to enhance protein stability, except for initial CD and AUC experiments (Fig. 1).
The Brpt5.5 construct was induced overnight at 20 °C with 200 μm IPTG, lysed by sonication, then run over a homemade 50-ml Ni-NTA column containing 50 ml of IMAC-Sepharose Fast Flow resin (GE Healthcare) charged with Ni2+ and housed inside a XK 16/40 column. After elution by an imidazole linear gradient from 0 to 300 mm imidazole, the fractions containing Brpt5.5 were dialyzed into 20 mm Tris, pH 7.4, 300 mm NaCl and cleaved by TEV for at least 6 h under reducing conditions. Another 50-ml Ni-NTA column was run, this time collecting the flow-through (cleaved Brpt5.5). Finally, a Superdex 200 (GE Healthcare) column was run to separate remaining contaminants and truncations of Brpt5.5.
CD
Brpt3.5 and HMBP–Brpt3.5 samples were dialyzed into 10 mm Tris, pH 7.4, and 150 mm NaF. Far-UV CD spectra were obtained using an Aviv 215 spectrometer (Aviv Biomedical, Inc., Lakewood, NJ). The concentration of cleaved Brpt3.5 (fusion tag removed) was determined using the molar extinction coefficient of 10,430 m−1 cm−1, as calculated using the online server ProtParam, and the concentration of HMBP–Brpt3.5 was determined using the molar extinction coefficient of 78,270 m−1 cm−1 calculated by ProtParam. An additional sample of HMBP–Brpt3.5 was treated with 10% FA to depolymerize amyloid fibers, followed by dialysis of the sample into 10 mm Tris, pH 7.4, and 150 mm NaF. The concentration of Brpt5.5 was based on the molar extinction coefficient of 16,390 m−1 cm−1, and the spectrum was collected in 10 mm Tris, pH 7.4, and 50 mm NaF. Data were analyzed using the CDSSTR program on the online Dichroweb server with reference set 4 (RRID:SCR_018125) (100). Data are plotted as mean residue ellipticity, [θ], which is in units of degrees cm2 dmol−1 residue−1.
Analytical ultracentrifugation
Experiments were performed with a Beckman XL-I analytical ultracentrifuge (Beckman Coulter, Indianapolis, IN) using absorbance optics at 280 nm. Sedimentation velocity experiments were performed at 36,000 rpm at 20 °C with and without 3 mm ZnCl2 (Fig. 2) or at 25 and 37 °C with or without 500 μm or 1 mm ZnCl2 as indicated (Fig. 3). Data were analyzed using Sedfit software (101) and the c(s) (Figs. 1 and 2) or c(s,ff0) models (Fig. 3). The c(s,ff0) model describes the sedimentation behavior of the species in solution as a two-dimensional distribution based on both sedimentation coefficient and frictional ratio, which allows resolution of multiple species of widely-differing size and shape sedimenting at the same sedimentation coefficients (33, 34). Parameters of buffer density and viscosity and the partial specific volume of constructs were calculated using SEDNTERP (102) at all relevant experimental temperatures. The data described in Fig. 3, B and C, were analyzed using several different models in SEDFIT; the c(s,ff0) model yielded fits with the lowest value for the summed square of the residuals (SSR). The standard c(s) model fit gave an SSR value of 0.5140; the best-fit worm-like chain model gave an SSR value of 0.5102; the c(s,ff0) model gave an SSR value of 0.5029 when setting the frictional ratio limits from 1 to 5.
To show reversibility of Brpt5.5 initial assembly (Fig. S7), 6 μm Brpt5.5 was dialyzed overnight into 50 mm MOPS, pH 7.2, 50 mm NaCl, 3.5 mm ZnCl2. A sample was analyzed by sedimentation velocity AUC with the addition of buffer (Brpt5.5 + Zn), 7 mm DTPA (+Zn + DTPA), or enough HCl to reach pH near 6 (+Zn + HCl). As a control, Brpt5.5 was also dialyzed into 20 mm MES, pH 5.0, 50 mm NaCl, 3.5 mm ZnCl2. This sample sedimented as a monomer near 2 S (Fig. S7). The absorbance at 280 nm was used for the c(s) distribution analysis.
Transmission EM
5-μl samples of 10 μm HMBP–Brpt3.5 incubated with 500 μm ZnCl2 were applied to 200 mesh Formvar carbon/copper grids for 2 min, washed with distilled H2O, stained by 1% uranyl acetate dropwise for 30 s, and washed a second time with distilled H2O. Samples were then dried for 1 h prior to viewing on a Hitachi 7600 transmission electron microscope at an accelerating voltage of 80 kV. Images were captured using an AMT 2k CCD camera. Brpt5.5 was incubated at 20 μm with 5 mm Zn2+ and incubated at 37 °C for 3 weeks. Samples were stained using the same protocol as HMBP–Brpt3.5, but with 2% uranyl acetate (Electron Microscopy Sciences). To prepare biofilms for TEM, the protocol for “Harvesting biofilms” was followed up through the step of washing the biofilms off the dialysis tubing (see below). The collected washes were treated with DTPA, FA, or buffer and incubated for 1 h at room temperature while shaking. A 3-μl sample was added to the grids for negative staining as described above. For extracellular biofilm material, washes were centrifuged at 17,000 × g for 5 min before collecting the supernatant.
Harvesting biofilms
This protocol was based on an approach used by Sun et al. (17) to isolate extracellular and cell wall–associated proteins from S. epidermidis biofilms. RP62A was grown overnight on tryptic soy agar (TSA) with sheep blood (Thermo Fisher ScientificTM R01202). Colonies were scraped from the agar plate and suspended in TSB to an absorbance of 0.1. A piece of the 3500 MWCO dialysis tubing was cut down the edge and opened to be arranged over a TSA agar plate. A 2-ml aliquot (with the addition of ZnCl2, DTPA, FA, or 100 mm MES, pH 5.0) was added to the dialysis tubing on top of the TSA agar. After 16 h of growth at 37 °C, the biofilm was washed off of the dialysis tubing by pipetting 1 ml of H2O across the surface of the tubing. For lysostaphin digestion of the cell wall, mixtures were centrifuged for 10 min at 17,000 × g, and the pellet was resuspended in 50 mm Tris, pH 7.4, 150 mm NaCl, 30% raffinose. Samples were then incubated at 37 °C for 1 h with 1 mg/ml of lysostaphin (Sigma L3876).
Thioflavin T protein fluorescence assay
10 μm HMBP–Brpt3.5 and 20 μm Brpt5.5 samples were treated with 10% FA to depolymerize amyloid fibers (44), followed by dialysis into 50 mm MOPS, pH 7.2, 50 mm NaCl. The FA-treated protein was then incubated with or without 500 μm ZnCl2 for 24 h at 20 °C prior to adding thioflavin T (MilliporeSigma) to a final concentration of 10 μm. In parallel, one sample of HMBP–Brpt3.5 was treated with 10% FA and was tested for ThT fluorescence without removing the FA (labeled FA + 3BIKHMBP–Brpt3.5 + ThT in Fig. 4B). Fluorescence was measured using a PerkinElmer Life Sciences LS50B luminescence spectrophotometer or a Biotek Synergy at an excitation of 434 or 440 nm and collecting the complete emission spectrum between 450 and 600 nm. Congo Red absorbance was measured from 400 to 600, and the absorbance at 540 nm was plotted in Fig. 4E. ThT emission was plotted at 482 nm (Fig. 4, C and E).
HPLC assays
For the HPLC fiber/oligomer quantification assay, 10 μm HMBP–Brpt3.5 samples were incubated with or without 500 μm ZnCl2 for 1, 4, or 30 days at 20 or 37 °C. Samples were prepared based on a protocol previously described by O'Nuallain et al. (60). Briefly, samples were centrifuged for 1 h at 13,500 rpm and then resuspended in 250 μl of 5% acetonitrile. Samples were loaded on a Sepax Bio-C4 reverse-phase column (Sepax Technologies, Newark, DE) with a 5-μm particle size and 300 Å pore size and run on an Äkta purifier in a running buffer of 5% acetonitrile and 0.1% trifluoroacetate at a flow rate of 0.75 ml/min. A linear gradient of 0–100% eluent (95% acetonitrile and 0.1% trifluoroacetate) from 13.33 to 66.67 min was used to elute the proteins. Peaks were detected by absorbance at 280 nm and integrated using the Unicorn software, normalizing the peak area to a standardized elution volume for monomer and oligomer peaks. To determine the stability of the Zn2+-induced amyloid fibers, samples of 10 μm HMBP–Brpt3.5 were incubated with 500 μm ZnCl2 for 1 or 4 days at 20 or 37 °C, followed by the addition of an equal volume of the following: 1) buffer (20 mm Tris, pH 7.4, 150 mm NaCl); 2) dilute hydrochloride acid sufficient to lower the pH 5.0; or 3) 2 mm Na5-DTPA. Samples were further incubated for 2 h, mixed with equal volume of 5% acetonitrile, and then separated on a C4 column.
Turbidity assays
5 μm Brpt5.5 in 2 ml of 20 mm MOPS, pH 7.2, 50 mm NaCl was incubated with 3, 5, or 8 mm ZnCl2 at 37 °C in a shaking incubator. The sample was transferred to a cuvette, and the absorbance at 280, 400, and 700 nm was measured in a BioMate 3S (Thermo Fisher Scientific). The reported turbidity value is the absorbance measured at 400 nm (Fig. 5C).
Dynamic light scattering
A Malvern Analytical Zetasizer Nano S was used to perform particle size measurements at 20 °C on 5 μm Brpt5.5 samples incubated with 5 mm ZnCl2 for at least 30 days at 37 °C in a quartz microcuvette. The Z-average of at least three series of measurements was reported. Native Brpt5.5 samples were measured in the same way but were not incubated with ZnCl2 prior to the experiment.
Confocal microscopy
For analysis of Brpt5.5 protein fibers, 10 μm samples were incubated with 500 μm ZnCl2 for 2, 6, 12, 18, or 24 h prior to addition of thioflavin T (10 μm final concentration). Samples were then added to 8-well borosilicate glass slides (Nunc) and viewed using a Zeiss LSM 710 inverted microscope using an Apochromat ×63/1.40 oil differential interference contrast M27 objective for bright field and a laser set to 458 nm with emission filters at 469–580 nm to measure thioflavin T fluorescence. For analysis of amyloid formations in biofilms, overnight cultures of S. epidermidis RP62A were diluted 1:200, and 400-μl aliquots were added to 8-well borosilicate glass slides. The slides were incubated for 18 h at 37 °C without shaking, followed by addition of thioflavin T to 10 μm as indicated. The media were aspirated, and each well was washed three times with distilled H2O. The biofilms were viewed using a Zeiss LSM 510 inverted microscope as described above. Control biofilm samples were alternatively labeled with the LIVE/DEAD stain kit (Invitrogen), using 3 μl of stain diluted in 1 ml of sterile saline buffer.
Biofilm formation assay
To quantify the ability for RP62A to form biofilms under varying conditions, a crystal violet-based biofilm formation assay was utilized (25). An overnight culture grown in TSB was diluted 1:200 into TSB. To wells of a 96-well plate (Corning 351172), 200 μl of diluted culture were added, along with initial treatments of ZnCl2 or DTPA (t = 0 h). DTPA, FA, or 100 mm MES, pH 5.0, was added at the specified time points and incubated for an additional 1 h at 37 °C. The liquid was then removed from the wells, followed by two washes of 200 μl of H2O before being allowed to dry. Next, 100 μl of 0.1% crystal violet was added and allowed to stain the biofilms for 10 min at room temperature. The crystal violet solution was then removed, followed by two more washes with 200 μl of H2O. The plates were allowed to dry before being scanned. Finally, 200 μl of 33% acetic acid was added, and plates were incubated at 4 °C for 30 min to remove crystal violet stain from the adherent biofilms. The solution in the wells was then transferred to new wells and then diluted 1:1 (100 μl of solution to 100 μl of H2O) before scanning at 520 nm. Statistical analysis was performed in Microsoft Excel, using the two-tailed, two-sample unequal variance Student's t test. Significant difference was identified when p < 0.05.
Mass spectrometry
Biofilms were prepared as described under “Harvesting biofilms,” with the exception that no H2O was added to the dialysis membrane during collection. This kept the material in the biofilm at a reasonable concentration to be visualized by SDS-PAGE with Coomassie Brilliant Blue staining. The section outlined in Fig. 6 was reduced with DTT, alkylated with iodoacetamide, and digested with trypsin. The resultant peptides were recovered and dried in a speed vac, before analysis by nanoLC-MS/MS. We provide data from two additional samples in Fig. S8 and Table S3 and S4. These samples were collected in the same way as in Fig. 6, but one sample was not exposed to lysostaphin (details provided in supporting information.
Author contributions
A. E. Y., S. L. J., and A. B. H. conceptualization; A. E. Y., S. L. J., P. S., and A. B. H. formal analysis; A. E. Y., S. L. J., and A. B. H. investigation; A. E. Y., S. L. J., P. S., and A. B. H. methodology; A. E. Y., S. L. J., and A. B. H. writing-original draft; A. E. Y., S. L. J., P. S., and A. B. H. writing-review and editing; A. B. H. supervision; A. B. H. funding acquisition; A. B. H. project administration.
Supplementary Material
Acknowledgments
We thank Georgianne Ciraolo and the Cincinnati Children's Hospital Medical Center Department of Pathology for assistance and use of the EM facility; Dr. Dan Hassett and lab members and Dr. Birgit Ehmer for assistance with the confocal microscope; Dr. Andy Deng and members of Dr. Tom Thompson's lab for help with the HPLC experiments and analysis; Dr. Nicolas Nassar for use of their fluorescence spectrophotometer; Dr. Deb Conrady for cloning the Brpt3.5 construct; and the University of Cincinnati Cancer Biology Proteomics Core Facility for MS services. We also thank Dr. Rhett Kovall and Dr. Tom Thompson for comments on the manuscript, and Dr. Deb Conrady, Dr. Catherine Chaton, and Dr. Catherine Shelton for helpful discussions.
This work was supported by National Institutes of Health NIGMS Grant R01 GM094363 (to A. B. H.) and NIAID Grant U19 AI070235 (to A. B. H.), by Cincinnati Children's Hospital Research Foundation (to A. B. H.), and in part by the National Institutes of Health Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering (to P. S.). A. B. H. serves as a Scientific Advisory Board member for Hoth Therapeutics, Inc., holds equity in Hoth Therapeutics and Chelexa BioSciences, LLC, and was a co-inventor on three patents broadly related to the subject matter of this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S8 and Tables S1–S4.
- PNAG
- poly-N-acetylglucosamine
- Aap
- accumulation-associated protein
- AUC
- analytical ultracentrifugation
- TEM
- transmission electron microscopy
- CFM
- confocal microscopy
- SasG
- Staphylococcus aureus surface protein G
- MBP
- maltose-binding protein
- HMBP
- His-tagged MBP
- ThT
- thioflavin T
- FA
- formic acid
- DTPA
- 2-[bis[2-[bis(carboxymethyl)amino]ethyl]amino]acetic acid
- Ni-NTA
- nickel-nitrilotriacetic acid
- TSA
- tryptic soy agar
- ThS
- thioflavin S
- TSB
- tryptic soy broth
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- SSR
- summed square of the residual
- Brpt
- B-repeat.
References
- 1. Uçkay I., Pittet D., Vaudaux P., Sax H., Lew D., and Waldvogel F. (2009) Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41, 109–119 10.1080/07853890802337045 [DOI] [PubMed] [Google Scholar]
- 2. National Nosocomial Infections Surveillance System. (2004) National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32, 470–485 10.1016/j.ajic.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 3. Fey P. D., and Olson M. E. (2010) Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933 10.2217/fmb.10.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., and Edmond M. B. (2004) Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 5. Rogers K. L., Fey P. D., and Rupp M. E. (2009) Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23, 73–98 10.1016/j.idc.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 6. Fey P. D. (2010) Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr. Opin. Microbiol. 13, 610–615 10.1016/j.mib.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Eiff C., Peters G., and Heilmann C. (2002) Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2, 677–685 10.1016/S1473-3099(02)00438-3 [DOI] [PubMed] [Google Scholar]
- 8. Otto M. (2009) Staphylococcus epidermidis–the 'accidental' pathogen. Nat. Rev. Microbiol. 7, 555–567 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kristian S. A., Birkenstock T. A., Sauder U., Mack D., Götz F., and Landmann R. (2008) Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 197, 1028–1035 10.1086/528992 [DOI] [PubMed] [Google Scholar]
- 10. Gill S. R., Fouts D. E., Archer G. L., Mongodin E. F., Deboy R. T., Ravel J., Paulsen I. T., Kolonay J. F., Brinkac L., Beanan M., Dodson R. J., Daugherty S. C., Madupu R., Angiuoli S. V., Durkin A. S., et al. (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costerton J. W., Stewart P. S., and Greenberg E. P. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 12. Sadovskaya I., Vinogradov E., Flahaut S., Kogan G., and Jabbouri S. (2005) Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 73, 3007–3017 10.1128/IAI.73.5.3007-3017.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izano E. A., Amarante M. A., Kher W. B., and Kaplan J. B. (2008) Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476 10.1128/AEM.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mack D., Haeder M., Siemssen N., and Laufs R. (1996) Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J. Infect. Dis. 174, 881–884 10.1093/infdis/174.4.881 [DOI] [PubMed] [Google Scholar]
- 15. Rohde H., Burandt E. C., Siemssen N., Frommelt L., Burdelski C., Wurster S., Scherpe S., Davies A. P., Harris L. G., Horstkotte M. A., Knobloch J. K., Ragunath C., Kaplan J. B., and Mack D. (2007) Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720 10.1016/j.biomaterials.2006.11.046 [DOI] [PubMed] [Google Scholar]
- 16. Hussain M., Herrmann M., von Eiff C., Perdreau-Remington F., and Peters G. (1997) A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65, 519–524 10.1128/IAI.65.2.519-524.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun D., Accavitti M. A., and Bryers J. D. (2005) Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin. Diagn. Lab. Immunol. 12, 93–100 10.1128/CDLI.12.1.93-100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rohde H., Burdelski C., Bartscht K., Hussain M., Buck F., Horstkotte M. A., Knobloch J. K., Heilmann C., Herrmann M., and Mack D. (2005) Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895 10.1111/j.1365-2958.2005.04515.x [DOI] [PubMed] [Google Scholar]
- 19. Corrigan R. M., Rigby D., Handley P., and Foster T. J. (2007) The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446 10.1099/mic.0.2007/006676-0 [DOI] [PubMed] [Google Scholar]
- 20. Kogan G., Sadovskaya I., Chaignon P., Chokr A., and Jabbouri S. (2006) Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol. Lett. 255, 11–16 10.1111/j.1574-6968.2005.00043.x [DOI] [PubMed] [Google Scholar]
- 21. Jabbouri S., and Sadovskaya I. (2010) Characteristics of the biofilm matrix and its role as a possible target for the detection and eradication of Staphylococcus epidermidis associated with medical implant infections. FEMS Immunol. Med. Microbiol. 59, 280–291 10.1111/j.1574-695X.2010.00695.x [DOI] [PubMed] [Google Scholar]
- 22. Schaeffer C. R., Woods K. M., Longo G. M., Kiedrowski M. R., Paharik A. E., Büttner H., Christner M., Boissy R. J., Horswill A. R., Rohde H., and Fey P. D. (2015) Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226 10.1128/IAI.02177-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yarawsky A. E., English L. R., Whitten S. T., and Herr A. B. (2017) The proline/glycine-rich region of the biofilm adhesion protein Aap forms an extended stalk that resists compaction. J. Mol. Biol. 429, 261–279 10.1016/j.jmb.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paharik A. E., Kotasinska M., Both A., Hoang T. N., Büttner H., Roy P., Fey P. D., Horswill A. R., and Rohde H. (2017) The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol. Microbiol. 103, 860–874 10.1111/mmi.13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conrady D. G., Brescia C. C., Horii K., Weiss A. A., Hassett D. J., and Herr A. B. (2008) A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461 10.1073/pnas.0807717105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conrady D. G., Wilson J. J., and Herr A. B. (2013) Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 110, E202–E211 10.1073/pnas.1208134110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaton C. T., and Herr A. B. (2017) Defining the metal specificity of a multifunctional biofilm adhesion protein. Protein Sci. 26, 1964–1973 10.1002/pro.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geoghegan J. A., Corrigan R. M., Gruszka D. T., Speziale P., O'Gara J. P., Potts J. R., and Foster T. J. (2010) Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673 10.1128/JB.00628-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Formosa-Dague C., Speziale P., Foster T. J., Geoghegan J. A., and Dufrêne Y. F. (2016) Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. U.S.A. 113, 410–415 10.1073/pnas.1519265113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shelton C. L., Conrady D. G., and Herr A. B. (2017) Functional consequences of B-repeat sequence variation in the staphylococcal biofilm protein Aap: deciphering the assembly code. Biochem. J. 474, 427–443 10.1042/BCJ20160675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spurlino J. C., Lu G. Y., and Quiocho F. A. (1991) The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 266, 5202–5219 10.2210/pdb1mbp/pdb [DOI] [PubMed] [Google Scholar]
- 32. Herr A. B., and Conrady D. G. (2011) Thermodynamic analysis of metal ion-induced protein assembly. Methods Enzymol. 488, 101–121 10.1016/B978-0-12-381268-1.00005-7 [DOI] [PubMed] [Google Scholar]
- 33. Brown P. H., and Schuck P. (2006) Macromolecular size–and–shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90, 4651–4661 10.1529/biophysj.106.081372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaton C. T., and Herr A. B. (2015) Elucidating complicated assembling systems in biology using size–and–shape analysis of sedimentation velocity data. Methods Enzymol. 562, 187–204 10.1016/bs.mie.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sunde M., and Blake C. (1997) The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein Chem. 50, 123–159 10.1016/S0065-3233(08)60320-4 [DOI] [PubMed] [Google Scholar]
- 36. Dueholm M. S., Larsen P., Finster K., Stenvang M. R., Christiansen G., Vad B. S., Bøggild A., Otzen D. E., and Nielsen P. H. (2015) The tubular sheaths encasing Methanosaeta thermophila filaments are functional amyloids. J. Biol. Chem. 290, 20590–20600 10.1074/jbc.M115.654780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fowler D. M., Koulov A. V., Balch W. E., and Kelly J. W. (2007) Functional amyloid–from bacteria to humans. Trends Biochem. Sci. 32, 217–224 10.1016/j.tibs.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 38. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., and Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 10.1126/science.1067484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., and Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090 10.1111/j.1462-2920.2007.01418.x [DOI] [PubMed] [Google Scholar]
- 40. Romero D., Aguilar C., Losick R., and Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dueholm M. S., Petersen S. V., Sønderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., and Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 42. LeVine H., 3rd. (1993) Thioflavine T interaction with synthetic Alzheimer's disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 2, 404–410 10.1002/pro.5560020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolfe L. S., Calabrese M. F., Nath A., Blaho D. V., Miranker A. D., and Xiong Y. (2010) Protein-induced photophysical changes to the amyloid indicator dye thioflavin T. Proc. Natl. Acad. Sci. U.S.A. 107, 16863–16868 10.1073/pnas.1002867107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collinson S. K., Emödy L., Müller K. H., Trust T. J., and Kay W. W. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781 10.1128/JB.173.15.4773-4781.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blancas-Mejía L. M., Hammernik J., Marin-Argany M., and Ramirez-Alvarado M. (2015) Differential effects on light chain amyloid formation depend on mutations and type of glycosaminoglycans. J. Biol. Chem. 290, 4953–4965 10.1074/jbc.M114.615401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kodali R., Williams A. D., Chemuru S., and Wetzel R. (2010) Aβ(1–40) forms five distinct amyloid structures whose β-sheet contents and fibril stabilities are correlated. J. Mol. Biol. 401, 503–517 10.1016/j.jmb.2010.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Navarro S., Carija A., Muñoz-Torrero D., and Ventura S. (2016) A fast and specific method to screen for intracellular amyloid inhibitors using bacterial model systems. Eur. J. Med. Chem. 121, 785–792 10.1016/j.ejmech.2015.10.044 [DOI] [PubMed] [Google Scholar]
- 48. Howie A. J., and Brewer D. B. (2009) Optical properties of amyloid stained by Congo red: history and mechanisms. Micron 40, 285–301 10.1016/j.micron.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 49. Gruszka D. T., Wojdyla J. A., Bingham R. J., Turkenburg J. P., Manfield I. W., Steward A., Leech A. P., Geoghegan J. A., Foster T. J., Clarke J., and Potts J. R. (2012) Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U.S.A. 109, E1011–E1018 10.1073/pnas.1119456109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pedersen J. S., Dikov D., Flink J. L., Hjuler H. A., Christiansen G., and Otzen D. E. (2006) The changing face of glucagon fibrillation: structural polymorphism and conformational imprinting. J. Mol. Biol. 355, 501–523 10.1016/j.jmb.2005.09.100 [DOI] [PubMed] [Google Scholar]
- 51. Macchi F., Eisenkolb M., Kiefer H., and Otzen D. E. (2012) The effect of osmolytes on protein fibrillation. Int. J. Mol. Sci. 13, 3801–3819 10.3390/ijms13033801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., and Glabe C. G. (2007) Fibril specific, conformation-dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18 10.1186/1750-1326-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tõugu V., Karafin A., Zovo K., Chung R. S., Howells C., West A. K., and Palumaa P. (2009) Zn(II)- and Cu(II)-induced non-fibrillar aggregates of amyloid-β(1–42) peptide are transformed to amyloid fibrils, both spontaneously and under the influence of metal chelators. J. Neurochem. 110, 1784–1795 10.1111/j.1471-4159.2009.06269.x [DOI] [PubMed] [Google Scholar]
- 54. Bush A. I., Pettingell W. H., Multhaup G., d Paradis M., Vonsattel J. P., Gusella J. F., Beyreuther K., Masters C. L., and Tanzi R. E. (1994) Rapid induction of Alzheimer Aβ amyloid formation by zinc. Science 265, 1464–1467 10.1126/science.8073293 [DOI] [PubMed] [Google Scholar]
- 55. Adlard P. A., and Bush A. I. (2006) Metals and Alzheimer's disease. J. Alzheimers Dis. 10, 145–163 10.3233/jad-2006-102-303 [DOI] [PubMed] [Google Scholar]
- 56. Faller P. (2009) Copper and zinc binding to amyloid-β: coordination, dynamics, aggregation, reactivity and metal-ion transfer. Chembiochem 10, 2837–2845 10.1002/cbic.200900321 [DOI] [PubMed] [Google Scholar]
- 57. Hammer N. D., Schmidt J. C., and Chapman M. R. (2007) The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U.S.A. 104, 12494–12499 10.1073/pnas.0703310104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Romero D., Vlamakis H., Losick R., and Kolter R. (2011) An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol. Microbiol. 80, 1155–1168 10.1111/j.1365-2958.2011.07653.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taglialegna A., Navarro S., Ventura S., Garnett J. A., Matthews S., Penades J. R., Lasa I., and Valle J. (2016) Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 12, e1005711 10.1371/journal.ppat.1005711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Nuallain B., Thakur A. K., Williams A. D., Bhattacharyya A. M., Chen S., Thiagarajan G., and Wetzel R. (2006) Kinetics and thermodynamics of amyloid assembly using a high-performance liquid chromatography-based sedimentation assay. Methods Enzymol. 413, 34–74 10.1016/S0076-6879(06)13003-7 [DOI] [PubMed] [Google Scholar]
- 61. Zhao R., So M., Maat H., Ray N. J., Arisaka F., Goto Y., Carver J. A., and Hall D. (2016) Measurement of amyloid formation by turbidity assay-seeing through the cloud. Biophys. Rev. 8, 445–471 10.1007/s12551-016-0233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Plakoutsi G., Bemporad F., Calamai M., Taddei N., Dobson C. M., and Chiti F. (2005) Evidence for a mechanism of amyloid formation involving molecular reorganisation within native-like precursor aggregates. J. Mol. Biol. 351, 910–922 10.1016/j.jmb.2005.06.043 [DOI] [PubMed] [Google Scholar]
- 63. Banner M. A., Cunniffe J. G., Macintosh R. L., Foster T. J., Rohde H., Mack D., Hoyes E., Derrick J., Upton M., and Handley P. S. (2007) Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are composed of the accumulation-associated protein. J. Bacteriol. 189, 2793–2804 10.1128/JB.00952-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Macintosh R. L., Brittan J. L., Bhattacharya R., Jenkinson H. F., Derrick J., Upton M., and Handley P. S. (2009) The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 191, 7007–7016 10.1128/JB.00764-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Damo S. M., Kehl-Fie T. E., Sugitani N., Holt M. E., Rathi S., Murphy W. J., Zhang Y., Betz C., Hench L., Fritz G., Skaar E. P., and Chazin W. J. (2013) Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, 3841–3846 10.1073/pnas.1220341110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vassar P. S., and Culling C. F. (1959) Fluorescent stains, with special reference to amyloid and connective tissues. Arch. Pathol. 68, 487–498 [PubMed] [Google Scholar]
- 67. Naiki H., Higuchi K., Hosokawa M., and Takeda T. (1989) Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- 68. Biancalana M., and Koide S. (2010) Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 1804, 1405–1412 10.1016/j.bbapap.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fedunova D., Huba P., Bagelova J., and Antalik M. (2013) Polyanion induced circular dichroism of thioflavin T. Gen. Physiol. Biophys. 32, 215–219 10.4149/gpb_2013020 [DOI] [PubMed] [Google Scholar]
- 70. Nenninger A. A., Robinson L. S., and Hultgren S. J. (2009) Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U.S.A. 106, 900–905 10.1073/pnas.0812143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dong J., Shokes J. E., Scott R. A., and Lynn D. G. (2006) Modulating amyloid self-assembly and fibril morphology with Zn(II). J. Am. Chem. Soc. 128, 3540–3542 10.1021/ja055973j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Talmard C., Leuma Yona R., and Faller P. (2009) Mechanism of zinc(II)-promoted amyloid formation: zinc(II) binding facilitates the transition from the partially α-helical conformer to aggregates of amyloid β protein(1–28). J. Biol. Inorg. Chem. 14, 449–455 10.1007/s00775-008-0461-9 [DOI] [PubMed] [Google Scholar]
- 73. Edgeworth J. A., Gros N., Alden J., Joiner S., Wadsworth J. D., Linehan J., Brandner S., Jackson G. S., Weissmann C., and Collinge J. (2010) Spontaneous generation of mammalian prions. Proc. Natl. Acad. Sci. U.S.A. 107, 14402–14406 10.1073/pnas.1004036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jobling M. F., Huang X., Stewart L. R., Barnham K. J., Curtain C., Volitakis I., Perugini M., White A. R., Cherny R. A., Masters C. L., Barrow C. J., Collins S. J., Bush A. I., and Cappai R. (2001) Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP106-126. Biochemistry 40, 8073–8084 10.1021/bi0029088 [DOI] [PubMed] [Google Scholar]
- 75. Calabrese M. F., Eakin C. M., Wang J. M., and Miranker A. D. (2008) A regulatable switch mediates self-association in an immunoglobulin fold. Nat. Struct. Mol. Biol. 15, 965–971 10.1038/nsmb.1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eakin C. M., Knight J. D., Morgan C. J., Gelfand M. A., and Miranker A. D. (2002) Formation of a copper specific binding site in non-native states of β2-microglobulin. Biochemistry 41, 10646–10656 10.1021/bi025944a [DOI] [PubMed] [Google Scholar]
- 77. Tormo M. Á., Knecht E., Götz F., Lasa I., and Penadés J. R. (2005) Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151, 2465–2475 10.1099/mic.0.27865-0 [DOI] [PubMed] [Google Scholar]
- 78. Lembré P., Vendrely C., and Martino P. D. (2014) Identification of an amyloidogenic peptide from the Bap protein of Staphylococcus epidermidis. Protein Pept. Lett. 21, 75–79 10.2174/09298665113209990072 [DOI] [PubMed] [Google Scholar]
- 79. Wang Y., Jiang J., Gao Y., Sun Y., Dai J., Wu Y., Qu D., Ma G., and Fang X. (2018) Staphylococcus epidermidis small basic protein (Sbp) forms amyloid fibrils, consistent with its function as a scaffolding protein in biofilms. J. Biol. Chem. 293, 14296–14311 10.1074/jbc.RA118.002448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Decker R., Burdelski C., Zobiak M., Büttner H., Franke G., Christner M., Sass K., Zobiak B., Henke H. A., Horswill A. R., Bischoff M., Bur S., Hartmann T., Schaeffer C. R., Fey P. D., and Rohde H. (2015) An 18-kDa scaffold protein is critical for Staphylococcus epidermidis biofilm formation. PLoS Pathog. 11, e1004735 10.1371/journal.ppat.1004735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pedersen J. S., and Otzen D. E. (2008) Amyloid–a state in many guises: survival of the fittest fibril fold. Protein Sci. 17, 2–10 10.1110/ps.073127808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., and Eisenberg D. (2005) Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 10.1038/nature03680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shewmaker F., McGlinchey R. P., Thurber K. R., McPhie P., Dyda F., Tycko R., and Wickner R. B. (2009) The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 284, 25065–25076 10.1074/jbc.M109.007054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Andersen C. B., Hicks M. R., Vetri V., Vandahl B., Rahbek-Nielsen H., Thøgersen H., Thøgersen I. B., Enghild J. J., Serpell L. C., Rischel C., and Otzen D. E. (2010) Glucagon fibril polymorphism reflects differences in protofilament backbone structure. J. Mol. Biol. 397, 932–946 10.1016/j.jmb.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 85. Morris K., and Serpell L. (2010) From natural to designer self-assembling biopolymers, the structural characterisation of fibrous proteins and peptides using fibre diffraction. Chem. Soc. Rev. 39, 3445–3453 10.1039/b919453n [DOI] [PubMed] [Google Scholar]
- 86. Zhang R., Hu X., Khant H., Ludtke S. J., Chiu W., Schmid M. F., Frieden C., and Lee J. M. (2009) Interprotofilament interactions between Alzheimer's Aβ1–42 peptides in amyloid fibrils revealed by cryoEM. Proc. Natl. Acad. Sci. U.S.A. 106, 4653–4658 10.1073/pnas.0901085106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sikorski P., Atkins E. D., and Serpell L. C. (2003) Structure and texture of fibrous crystals formed by Alzheimer's Aβ(11–25) peptide fragment. Structure 11, 915–926 10.1016/S0969-2126(03)00149-7 [DOI] [PubMed] [Google Scholar]
- 88. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., Madsen A. Ø., Riekel C., and Eisenberg D. (2007) Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 10.1038/nature05695 [DOI] [PubMed] [Google Scholar]
- 89. Liu C., Sawaya M. R., Cheng P. N., Zheng J., Nowick J. S., and Eisenberg D. (2011) Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic β-sheet mimics. J. Am. Chem. Soc. 133, 6736–6744 10.1021/ja200222n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. MacRaild C. A., Hatters D. M., Lawrence L. J., and Howlett G. J. (2003) Sedimentation velocity analysis of flexible macromolecules: self-association and tangling of amyloid fibrils. Biophys. J. 84, 2562–2569 10.1016/S0006-3495(03)75061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pham Cle L., Mok Y. F., and Howlett G. J. (2011) Sedimentation velocity analysis of amyloid fibrils. Methods Mol. Biol. 752, 179–196 10.1007/978-1-60327-223-0_12 [DOI] [PubMed] [Google Scholar]
- 92. Smith D. P., Radford S. E., and Ashcroft A. E. (2010) Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry–mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 107, 6794–6798 10.1073/pnas.0913046107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Binger K. J., Pham C. L., Wilson L. M., Bailey M. F., Lawrence L. J., Schuck P., and Howlett G. J. (2008) Apolipoprotein C-II amyloid fibrils assemble via a reversible pathway that includes fibril breaking and rejoining. J. Mol. Biol. 376, 1116–1129 10.1016/j.jmb.2007.12.055 [DOI] [PubMed] [Google Scholar]
- 94. Ryan T. M., Teoh C. L., Griffin M. D., Bailey M. F., Schuck P., and Howlett G. J. (2010) Phospholipids enhance nucleation but not elongation of apolipoprotein C-II amyloid fibrils. J. Mol. Biol. 399, 731–740 10.1016/j.jmb.2010.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lashuel H. A., Lai Z., and Kelly J. W. (1998) Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: implications for wild-type, V30M, and L55P amyloid fibril formation. Biochemistry 37, 17851–17864 10.1021/bi981876+ [DOI] [PubMed] [Google Scholar]
- 96. Van Holde K. E., and Weischet W. O. (1978) Boundary analysis of sedimentation-velocity experiments with monodisperse and paucidisperse solutes. Biopolymers 17, 1387–1403 10.1002/bip.1978.360170602 [DOI] [Google Scholar]
- 97. Demeler B., and van Holde K. E. (2004) Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 335, 279–288 10.1016/j.ab.2004.08.039 [DOI] [PubMed] [Google Scholar]
- 98. Demeler B., Brookes E., and Nagel-Steger L. (2009) Analysis of heterogeneity in molecular weight and shape by analytical ultracentrifugation using parallel distributed computing. Methods Enzymol. 454, 87–113 10.1016/S0076-6879(08)03804-4 [DOI] [PubMed] [Google Scholar]
- 99. Nagel-Steger L., Demeler B., Meyer-Zaika W., Hochdörffer K., Schrader T., and Willbold D. (2010) Modulation of aggregate size- and shape-distributions of the amyloid-β peptide by a designed β-sheet breaker. Eur. Biophys. J. 39, 415–422 10.1007/s00249-009-0416-2 [DOI] [PubMed] [Google Scholar]
- 100. Whitmore L., and Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 10.1093/nar/gkh371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., and Schubert D. (2002) Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82, 1096–1111 10.1016/S0006-3495(02)75469-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Laue T. M., Shah B. D., Ridgeway T. M., and Pelletier S. L. (1992) Computer Aided Interpretation of Analytical Sedimentation Data for Proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science, Royal Society of Chemistry, (Harding S. and Rowe A., eds.) pp. 90–125, Cambridge, UK [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.