Abstract
The WW domain is a modular protein structure that recognizes the proline-rich Pro-Pro-x-Tyr (PPxY) motif contained in specific target proteins. The compact modular nature of the WW domain makes it ideal for mediating interactions between proteins in complex networks and signaling pathways of the cell (e.g. the Hippo pathway). As a result, WW domains play key roles in a plethora of both normal and disease processes. Intriguingly, RNA and DNA viruses have evolved strategies to hijack cellular WW domain–containing proteins and thereby exploit the modular functions of these host proteins for various steps of the virus life cycle, including entry, replication, and egress. In this review, we summarize key findings in this rapidly expanding field, in which new virus-host interactions continue to be identified. Further unraveling of the molecular aspects of these crucial virus-host interactions will continue to enhance our fundamental understanding of the biology and pathogenesis of these viruses. We anticipate that additional insights into these interactions will help support strategies to develop a new class of small-molecule inhibitors of viral PPxY-host WW-domain interactions that could be used as antiviral therapeutics.
Keywords: virus, microbiology, antiviral agent, E3 ubiquitin ligase, Ebola virus, budding, L domain, PPxY motif, virus-host interaction, WW domain, virus-like particle (VLP), virus budding, modular domains, host-oriented inhibitors, antiviral therapeutic, viral life cycle
Introduction
The modular, compact nature of the WW domain contributes to its status as a prominent mediator of a wide array of protein-protein interactions in the cell. WW-domain ligands include short proline-rich motifs with the proline-proline-x-tyrosine (PPxY)2 motif serving as the consensus ligand (1–3). In mammalian cells, interactions between WW-domain and PPxY-containing proteins regulate pathways that are involved in processes such as protein ubiquitination, signaling, sorting, migration, and degradation (4–27). For example, key WW domain/PPxY interactions are critical for mediating the formation of protein complexes in the Hippo signaling pathway, which regulates cell proliferation, migration, and apoptosis (18, 24, 26, 28–30). In addition, all members of the HECT (homologous to the E6AP C terminus) family of E3 ubiquitin ligases contain multiple WW domains involved in binding to specific host PPxY-containing substrates, leading to modulation of their stability, trafficking/localization, and/or signaling functions (14, 31–37).
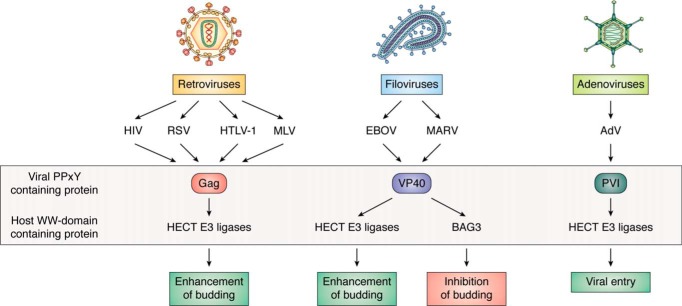
Intriguingly, a growing body of literature has demonstrated that modular WW domain/PPxY interactions not only are critical for regulating cellular pathways, but also play key roles in regulating one or more steps in the life cycles of many viral pathogens (38–46). Not surprisingly, select DNA and RNA viruses have evolved the ability to mimic these protein-protein interactions by using virally encoded PPxY-containing proteins to compete with cellular counterparts for binding to specific host proteins bearing one or more of these small modular WW domains, thus, in essence, hijacking cellular pathways and networks that ultimately impact both the virus and the host (see Figs. 1 and 2 and Table 1). For example, the initial discovery that viruses encode PPxY motifs to hijack host proteins to facilitate virus egress had a tremendous impact on the field because it demonstrated that viruses usurp host proteins and machinery to complete the late stage of virus-cell separation most efficiently. This discovery opened up an entirely new field of research on virus-host interactions that regulate egress. For example, the PPxY motif contained within matrix proteins encoded by RNA viruses, such as retroviruses and filoviruses, was termed a late (L) domain due to its role in facilitating the late step in virus-cell separation or pinching off (budding). In addition to the PPxY motif, PT/SAP and YxxL were also identified as functional L-domain motifs in specific RNA viruses. Intriguingly, the RNA viral L domains were found to be interchangeable and movable while retaining their function in recruiting host proteins to promote virus egress (47–50). In contrast, DNA-containing viruses, such as adenoviruses and herpesviruses, express PPxY-containing proteins that interact with specific host WW domain–bearing proteins to facilitate virus entry/internalization and affect virus replication and cancer progression, respectively.
Figure 1.
Key modular PPxY/WW-domain interactions and outcomes for the retroviruses, filoviruses, and adenoviruses.
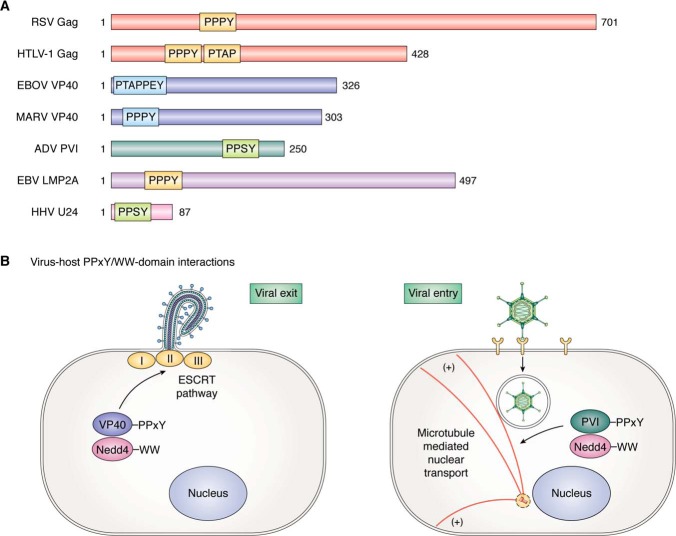
Figure 2.
A, position and sequence of PPxY motifs in key viral proteins. B, representative examples of virus-host PPxY/WW-domain interaction functioning during virus exit (left; EBOV VP40) and virus entry (right; AdV PVI).
Table 1.
Summary of virus-host interactions and effects on RNA and DNA viruses
| Virus family | Virus | Late domain–containing viral proteina,b | WW domain–bearing host interactor | Effect on viral life cycle stage |
|---|---|---|---|---|
| Retroviruses | HIV-1 | Gag | Nedd4, Nedd4.2/Nedd4L, Rsp5 | Enhancement of budding |
| RSV | Gag | LDI-1, LDI-2, Nedd4 | Enhancement of budding | |
| HTLV-1 | Gag | Nedd4, ITCH/AIP4, WWP1, HECW2 | Enhancement of budding | |
| MLV | Gag | Nedd4, WWP1, WWP2, ITCH/AIP4, ART proteins | Enhancement of budding | |
| Filoviruses | EBOV | VP40 | Nedd4, Rsp5, ITCH, WWP1 | Enhancement of budding |
| BAG3 | Inhibition of budding | |||
| MARV | VP40 | Nedd4 | Enhancement of budding | |
| BAG3 | Inhibition of budding | |||
| Herpesviruses | EBV | LMP2A | Nedd4, Nedd4.2/Nedd4L WWP2, ITCH/AIP4 | Viral latency |
| EBV | LMP2A | WOX1 | Cancer progression | |
| HHV (Roseolovirus) | U24 | Nedd4.1, Nedd4.2/Nedd4L, SMURF-2 | Neurologic disease progression | |
| Adenoviruses | AdV | PVI | WWP1, WWP2, AIP4/ITCH, Nedd4.1, Nedd4.2/Nedd4L | Viral entry |
| Tombusviruses | TBSV | p92pol, p33b | Rsp5p | Viral replication |
| Orthomyxoviruses | Influenza | IFITM3 (cellular) | Nedd4 | Immune modulation |
a All proteins listed are viral proteins except for IFITM3.
b p92pol and p33 do not contain canonical late domain motifs.
The viruses chosen for discussion here represent the best-characterized examples of viruses that encode PPxY motifs that mediate physical and functional interactions with host WW domain–bearing proteins to influence specific stages of the virus life cycle. Part of the excitement about this discovery is that viruses that are very different from one another in terms of their genetic material, morphology, life cycle, and disease all have evolved to encode one or more PPxY motifs that interact with specific host proteins to impact their life cycles. We will discuss key findings that highlight the physical and functional nature of a wide array of virus-host PPxY/WW-domain interactions, describe how usurping of host proteins affects the biology and pathogenesis of these viruses, and finally describe recent progress to identify and develop antiviral therapeutics that target and block these modular virus-host interactions as part of a novel host-oriented strategy to combat and treat viral infections.
Modular interactions and virus egress
Retroviruses
HIV-1
Pioneering studies that first identified viral L domains and, subsequently, their ability to interact with modular WW domains of host proteins were undertaken with the Gag proteins encoded by HIV-1 and Rous sarcoma virus (RSV) (50–53). Indeed, the PPPY sequence located in the N-terminal p2b region of the Rous sarcoma virus Gag protein was the first PPxY-type L domain shown to play a key role in facilitating virion egress by interacting with cellular WW domains (51). One of the first WW domain–bearing host proteins implicated in viral PPxY-mediated egress was Nedd4 (neuronal precursor cell–expressed developmentally down-regulated 4), the prototypic member of the HECT family of E3 ubiquitin ligases. Indeed, a plethora of reports soon followed that linked the budding pathways of several RNA viruses to Nedd4, Nedd4 family members, and/or ubiquitin (39, 41, 43–45, 54–67). Although HIV-1 Gag does not possess a PPxY motif, budding of HIV-1 was shown to be positively regulated by ubiquitin and E3 ubiquitin ligases, such as Nedd4, likely via an indirect mechanism. For example, Strack et al. (70) showed that 2–5% of the HIV-1 p6gag found in virions was monoubiquitinated and that chimeric Gag constructs with L domains from RSV and HIV-1 both produced Gag-ubiquitin conjugates in virus-like particles (VLPs). Moreover, inhibition of the proteasome, which reduces the levels of free ubiquitin in the cell, limited Gag VLP formation (69, 70). More recently, Mercenne et al. (71) demonstrated that PPxY-containing host protein angiomotin (Amot) could bind both Nedd4L and HIV Gag and serve as an adaptor protein to promote early stages of HIV-1 assembly and budding. Whereas our understanding of the precise mechanistic role of ubiquitin and E3 ubiquitin ligases in facilitating virus budding still remains unclear, one possibility is that monoubiquitination of viral matrix proteins allows them to engage one or more members of the host endosomal sorting complex required for transport (ESCRT) pathway, which subsequently mediates efficient virus-cell separation and egress of mature virions from the plasma membrane (72, 73). In contrast, other reports suggest that different mechanisms may also be involved. For example, a functional link between Nedd4 family members and HIV-1 budding was revealed by studies demonstrating that overexpression of Nedd4L could rescue budding of an HIV-1 Gag protein lacking an L-domain motif (62, 64, 74). In addition, Nedd4-mediated ubiquitination of ESCRT proteins such as Tsg101, rather than PPxY-deficient viral proteins such as HIV-1 Gag, may serve to activate ESCRT and promote budding of HIV-1 (75). Alternatively, Weiss et al. reported that Nedd4-mediated ubiquitination of HIV-1 Gag was not sufficient to stimulate virus budding, but rather the synthesis of Lys-63–linked ubiquitin chains in the vicinity of the viral bud was critical for E3 ligase-mediated virus budding (65). Indeed, the authors postulate that the Lys-63–linked ubiquitin chains on viral or cellular substrates are sensed or recognized by components of the ESCRT machinery, which is subsequently recruited to facilitate virus egress (65).
RSV
Unlike HIV-1, retroviruses whose Gag proteins do contain PPxY motifs can physically and functionally interact directly with the modular WW domains of either Nedd4 or Nedd4 family members to facilitate their egress and spread. For example, early work by Kikonyogo and colleagues (56), in which they screened a chicken embryo cDNA expression library with a peptide containing the PPxY L-domain sequence from RSV Gag-p2b, identified two WW-domain interactors they named late domain–interacting proteins 1 and 2 (LDI-1 and -2). LDI-1 was found to share sequence homology with Xenopus Nedd4, whereas LDI-2 exhibited homology to other members of the Nedd4 family, specifically human WWP1 and the murine E3 ubiquitin ligase, ITCH (also known as AIP4) (56). The authors also demonstrated that budding of RSV Gag required the PPxY L domain and that overexpression of the isolated WW domains of LDI-1 acted in a dominant-negative manner to inhibit budding (56, 76). In a follow-up study, Vana et al. (45) demonstrated that a catalytically active LDI-1 protein was required to promote RSV Gag egress, as a catalytically inactive LDI-1 mutant blocked Gag release. Whereas they were able to detect both mono- and polyubiquitinated forms of Gag within cells, the monoubiquitinated form of Gag was most prevalent in budded VLPs, and ubiquitination of RSV Gag depended on its PPxY L domain.
Human T-cell leukemia virus 1 (HTLV-1)
The PPPYVEPTAP sequence at the C terminus of the HTLV-1 Gag protein contains a PPxY and PTAP L-domain motif, both of which were shown to play a role in facilitating release of HTLV-1 Gag VLPs; however, the PPxY motif appeared to be more important for efficient egress (39, 43, 59). HTLV-1 Gag was shown to interact with WW domains of E3 ligases Nedd4, BUL1, and HECW2 (HECT, C2, and WW domain–containing E3 ubiquitin protein ligase 2) (39, 43, 59), and results from Blot et al. (59) suggested that the HTLV-1 PPxY motif first recruited host Nedd4, which then allowed the virus to engage the ESCRT complex, specifically Tsg101, to promote efficient assembly and egress of virus particles. Indeed, Tsg101 is a key component of the ESCRT-1 complex that can interact with viral PTAP L-domain motifs and can also bind to ubiquitinated proteins, thus mediating their engagement of the ESCRT machinery. Later studies both confirmed and expanded the repertoire of host WW domain–bearing E3 ligases hijacked by the PPxY motif of HTLV-1 Gag to benefit egress and spread of virus particles. For example, HECT family E3 ubiquitin ligase WWP1, which possesses four WW domains, was shown to ubiquitinate a lysine residue at position 74 of HTLV Gag to promote efficient virus egress (77, 78). In contrast, a lysine 74 mutant of HTLV-1 Gag was not ubiquitinated and was budding-defective. Dorjbal et al. (66) went on to test nine members of the Nedd4 family of E3 ubiquitin ligases (Nedd4, Nedd4L, BUL1, BUL2, WWP1, WWP2, ITCH, SMURF1, and SMURF2) for their ability to promote egress of HTLV-1 and found that ITCH was the main contributor to HTLV-1 budding. Also of interest was their finding that the physical binding efficiencies between the viral PPxY motif and the various host WW domains did not correlate with their functional effect on HTLV-1 budding (66).
Murine leukemia virus (MLV)
Studies by Martin-Serrano and colleagues (79) helped to solidify the functional link between HECT E3 ubiquitin ligases recruited by viral PPxY motifs and the ESCRT pathway during egress of WT or chimeric MLV Gag proteins. Using an array of WW domain–containing E3 ligases and chimeric MLV Gag proteins containing the PPxY motifs and flanking residues from either host amiloride-sensitive sodium channel (ENaC), RSV Gag-p2b, or Ebola virus VP40 proteins, they demonstrated that the E3 ligase WWP1 was recruited to sites of MLV budding in a PPxY-dependent manner and that enhancement of budding required a catalytically active HECT domain (79). They concluded that the HECT ubiquitin ligases likely served as co-factors for viral PPxY-mediated budding and provided a critical link to the ESCRT complex that mediated the final step of virus-cell separation. In a later study by the same group, they demonstrated that host PPxY containing arrestin-related trafficking (ART) proteins could function as adaptors, linking WW domain–containing HECT ubiquitin ligases to the ESCRT machinery (80). Indeed, they showed that the ART proteins could be recruited to sites of PPxY-dependent viral budding and likely served as a platform for ubiquitination during the budding process (80). Thus, it is interesting to speculate that the PPxY-containing viral matrix proteins have evolved to mimic and/or compete with cellular ART-like proteins for binding to modular WW domains to regulate virus egress and spread.
Filoviruses: Ebola (EBOV) and Marburg (MARV) viruses
Filoviruses (EBOV and MARV) are emerging pathogens that can cause severe hemorrhagic disease with high mortality rates in primates and humans (81–83). There has been growing interest in elucidating the molecular mechanisms used by these viruses to bud and spread from cells, as well as in identifying novel virus-host interactions that contribute to the transmission and pathogenesis of these deadly viruses. The filovirus matrix protein, VP40, is the major structural component of filoviruses and plays a key role in directing virion assembly and promoting virion egress during late stages of filovirus replication (54, 84–88). Intriguingly, a number of groups have shown that independent expression of filovirus VP40 leads to the production and egress of filamentous VLPs that mimic the morphology of authentic filoviruses (44, 54, 89–94). Thus, VP40 has been the focus of numerous studies to examine the molecular mechanisms and host interactors that regulate filovirus egress. As described above for the Gag proteins of many retroviruses, L domains are highly conserved in the VP40 matrix proteins of all known EBOV and MARV species. Indeed, both EBOV and MARV VP40 proteins contain a PPxY-type L-domain motif that can compete with cellular counterparts for binding to specific WW domain–bearing host proteins, thus hijacking critical cellular pathways that impact both the virus and the host. It should be mentioned that EBOV VP40 and MARV NP (nucleoprotein) proteins contain a PT/SAP-type L domain as well, and PT/SAP has been proposed to interact with and recruit host Tsg101 to facilitate virion egress (91, 95–98). Recent findings describing key modular interactions between filoviral PPxY motifs and host WW-domain interactors will be discussed below.
Harty et al. (54) were the first to demonstrate a role for the PPxY motif of EBOV VP40 in budding of VLPs and in interacting physically and functionally with host WW domains of E3 ligase Nedd4 and its yeast ortholog Rsp5. A series of reports soon followed that demonstrated that the VP40 PPxY motifs of both EBOV and MARV functioned in a manner similar to that described earlier for both retroviral and rhabdoviral (55, 89, 99–101) PPxY motifs to recruit host interactors and ESCRT to facilitate egress (Fig. 2A) (44, 54, 58, 60, 91, 96, 97, 102). Most of the early work on the filovirus VP40 PPxY motif was centered on its role in hijacking Nedd4 and ultimately the ESCRT complex to facilitate budding. More recent developments were based on utilization of a strategy involving screening of a specially prepared array composed of a complete set of bacterially expressed and purified human WW domains with either WT or PPxY-mutant peptides from EBOV or MARV VP40 to identify novel WW-domain interactors that may regulate filovirus budding (41, 42, 103). Interestingly, the authors found that there was an unprecedented degree of specificity, in that the filoviral WT PPxY motifs bound to only a select number of host WW domains (41, 42, 103). In addition to the expected WW-domain interactors like Nedd4 and Rsp5, the newly identified VP40 hits from this screen included HECT family E3 ubiquitin ligases ITCH (AIP4) and WWP1 (41, 42) previously identified as Gag interactors. In addition, BCL2-associated athanogene 3 (BAG3); a WW domain–bearing member of the BAG family of molecular co-chaperone proteins involved in regulating protein homeostasis and cell survival through chaperone-assisted selective autophagy (CASA), was identified as a novel VP40 interactor (103).
Han et al. (41) confirmed that EBOV VP40 interacted with ITCH in a PPxY/WW domain–dependent manner, as determined by co-immunoprecipitation of full-length proteins, and that the enzymatically active form of ITCH was required to monoubiquitinate EBOV VP40, leading to enhanced egress of VP40 VLPs. In addition, the authors used both siRNA knockdown/rescue assays and ITCH knockout cells to demonstrate further the biological importance of ITCH for VP40 budding in this VLP system and for egress of recombinant VSV engineered to express the PPxY L domain of EBOV VP40 (41, 97). These data demonstrated that EBOV VP40 likely recruits host E3 ligase ITCH via the PPxY/WW-domain modular interaction to promote efficient egress of EBOV VLPs and VSV-EBOV recombinant viruses and, although yet to be proven, imply that a similar modular recruitment of ITCH may occur during authentic EBOV infection.
Similar findings were reported by Han et al. (42) for E3 ligase WWP1 and EBOV VP40, again demonstrating that the interaction between WWP1 and VP40 is PPxY/WW domain–dependent and that monoubiquitination of VP40 by enzymatically active WWP1 led to enhanced egress of VP40 VLPs (42). Not surprisingly, both ITCH and WWP1 interact with a plethora of cellular proteins to regulate multiple pathways and networks in mammalian cells (37). For example, ITCH plays a key role in regulating lymphocyte activation and differentiation, key processes in immune regulation and inflammatory signaling (104). Likewise, WWP1 functions in protein signaling and trafficking related to transforming growth factor β and epidermal growth factor signaling, as well as apoptosis and neurological diseases (37). Whereas recruitment or hijacking of E3 ligases, such as Nedd4, ITCH, and WWP1, may facilitate egress and modulate the pathogenesis of infectious EBOV, the potential effects of these modular interactions on normal host protein function remains unclear. In addition, whether the ability of filovirus VP40 and other PPxY-containing viral proteins to interact with a wide array of WW-domain interactors is linked to the expression levels of these ligases in various cell types and reflects the cellular tropism of the virus, or is simply a mechanism of built-in redundancy used by the virus, remains to be determined.
Using the previously mentioned human WW-domain array, Liang et al. (103) identified host BAG3 as a novel WW-domain interactor with the PPxY motifs of both EBOV and MARV VP40. BAG3 is a stress-induced protein that regulates cellular protein homeostasis and cell survival through the CASA pathway (11, 105). The authors confirmed the PPxY/WW domain–dependent interaction between BAG3 and both EBOV and MARV VP40 and, intriguingly, demonstrated that expression of BAG3 negatively regulated budding of both EBOV and MARV VP40 VLPs as well as egress of infectious VSV-EBOV recombinants (103). This finding was novel because all previously identified host WW-domain interactors with VP40 positively regulated VLP and/or virus egress (103). The authors speculated that BAG3 and CASA may function as a host defense mechanism, whereby BAG3 recognizes and interacts with the viral PPxY motifs, thereby dampening virus egress by interfering with the budding function of viral PPxY-containing matrix proteins. Recently, BAG3 and the CASA machinery were reported to regulate both the synthesis and degradation of filamin, a key actin-cross-linking protein at the plasma membrane that modulates cell adhesion and migration (106–110). BAG3 is also involved in regulating the disassembly of the actin-based contractile ring during cytokinesis (cell-cell separation) (111, 112). Cytokinesis is topologically equivalent to a virion budding from the cell surface (virus-cell separation), and both processes involve some of the same host pathways (113–121). One possibility is that BAG3 and the CASA machinery regulate actin-based dynamics, stress, and membrane tension necessary for efficient pinching off of VLPs and/or viruses (122, 123). Liang et al. (103) used confocal microscopy to show that VP40 appeared to be sequestered away from the plasma membrane and accumulated in cytoplasmic aggregates in cells expressing WT BAG3, which correlated with a decreased efficiency of VLP egress. Although more studies are needed, it will also be of interest to investigate the potential interplay and binding competition between WW domain–bearing host proteins that positively regulate virus egress (e.g. ITCH) versus those that negatively regulate virus egress (e.g. BAG3) as a means to better understand the biological impact of these dueling modular interactions.
Modular interactions and virus entry/replication
Adenoviruses
Although most PPxY motifs encoded by RNA viruses hijack HECT-family, WW-domain interactors to promote virion egress, the adenovirus PPxY/host interaction was unique in that it was one of the first discoveries of a viral PPxY motif functioning in virus entry rather than virus egress. Indeed, both the adenoviral membrane lytic internal capsid protein VI (PVI) and the capsid penton base protein possess PPxY motifs that play a role in adenovirus (AdV) entry. AdV is a nonenveloped dsDNA virus, which enters cells by receptor-mediated endocytosis and utilizes the penton base protein for internalization, as well as escape from endosomes. In 2002, Galinier et al. (40) screened a human lung expression library for host proteins that may influence adenoviral entry by interacting with the two PPxY motifs in the N-terminal region of the penton base polypeptide chain. They found that the first PPxY motif was critical for mediating interactions with WW-domain E3 ligases WWP1, WWP2 (also known as AIP2), and ITCH (40), providing the first demonstration that a nonenveloped DNA virus employed a PPxY motif to interact with host HECT-family ligases.
AdV requires exposure of the internal PVI capsid protein to breach the endosomal membrane and escape into the cytosol to complete the internalization step. Wodrich et al. (124) identified a functional PPxY motif within PVI that recruited both Nedd4.1 and Nedd4.2, leading to ubiquitination of PVI. They went on to show that the PPxY motif within PVI was required for microtubule-dependent intracellular trafficking to the nucleus and that PPxY mutants were able to escape the endosome, but were defective in trafficking to the nucleus (Fig. 2B) (124). Later findings by the same group suggested that recruitment of Nedd4 by the PPxY motif of PVI was a strategy used by the virus to divert the host E3 ubiquitin ligase from its physiological function in regulating autophagy, thus allowing the virus to evade the host autophagic response (125, 126). Indeed, the authors observed that the WT PPxY motif of PVI prevented efficient formation of autolysosomes, and they speculated that the WT virus may interfere with elongation of the autophagosomal membrane, thereby preventing autophagosome formation (125, 126). In sum, these novel findings were the first to demonstrate that the modular interaction between the adenoviral PPxY motif and the WW domain(s) of Nedd4 not only promoted efficient viral entry and trafficking, but also represented a unique strategy for the virus to reduce antigenic presentation and evade the host immune response (125, 126).
Herpesviruses
Epstein–Barr virus (EBV; human gammaherpesvirus 4)
Epstein–Barr virus is a human herpesvirus that infects lymphoid and epithelial cells to cause infectious mononucleosis. EBV establishes a lifelong latent state in the absence of replicative gene expression in B-cells, and the viral latent membrane proteins (LMP1, -2A, and -2B) are expressed in latency to antagonize the normal process of B-lymphocyte activation. Indeed, LMP2A plays a key role in enhancing the development of tumors and the transformation potential of cells. LMP2A, which helps maintain EBV latency, contains two highly conserved PPPPY motifs that were first identified by Longnecker and colleagues (127–129) and were shown to interact with WW domains. Two groups went on to show that the PPxY motifs of LMP2A recruited host proteins ITCH (AIP4), WWP2, Nedd4L, and/or Nedd4 via one or more of their modular WW domains (46, 127). Indeed, Winberg et al. (46) found that these E3 ubiquitin ligases formed PPxY/WW domain–dependent complexes with LMP2A in EBV-infected B-cells, which they suggested may lead to ubiquitination of Lyn and Syk tyrosine kinases to ultimately block B-cell signaling. In support, Ikeda et al. (127) demonstrated a reduction in the normal cellular levels of Lyn and LMP2A, suggesting that recruitment of Nedd4-like ubiquitin ligases via the viral PPxY motifs resulted in ubiquitin-mediated degradation of Lyn and LMP2A, thus modulating B-cell signal transduction.
More recently, Lan et al. (130) described an intriguing interaction between the PPxY motifs of EBV LMP2A and a host WW domain–containing oxidoreductase (WOX1) that functions, in part, as a tumor suppressor. They report that WOX1 is important for cancer-promoting effects triggered by LMP2A and, specifically, that the PPxY/WW-domain interactions between LMP2A and WOX1 were important for induction of matrix metalloproteinase 9 (MMP9). These findings suggest that the modular interactions between the PPxY motifs of EBV LMP2A and the WW domain on host WOX1 may be crucial for invasiveness and cancer progression associated with EBV infection (130).
Roseoloviruses (human betaherpesviruses: HHV-6A, HHV-6B, and HHV-7)
Roseoloviruses, also known as human herpesvirus types 6A, 6B, and 7 (HHV-6A, HHV-6B, and HHV-7), have been linked to several neurological diseases, including encephalitis, epilepsy, and multiple sclerosis (MS). The roseoloviral protein U24 is a C-tail–anchored membrane protein shown to affect recycling and endocytosis of cell-surface receptors (131, 132). In addition, U24 function may be linked to MS, as it has been found to mimic the interaction of myelin basic protein's function with its binding partner Fyn (133, 134). The N-terminal region of U24 encoded by HHV-6A and HHV-7 contains a PPxY motif that was shown to interact with HECT-family E3 ligases SMURF2 and Nedd4, albeit more strongly with WW domain 3 of Nedd4 (135). Importantly, because sodium channels have been found to be associated with MS lesions, investigators speculated that U24 may interfere with the endosomal recycling of sodium channels known to be regulated by Nedd4. Although still speculative, results from Sang et al. (133) suggest that the PPxY motif of U24 and its strong interaction with WW domain 3 of Nedd4 may play a role in MS or other related demyelinating diseases.
Tombusviruses: Tomato bushy stunt virus (TBSV)
Tomato bushy stunt virus is an RNA virus with a small positive-stranded RNA genome that infects plants, and the virus encodes a handful of viral proteins that are unable to support viral replication without contributions from host factors. Plants have developed innate and adaptive immune pathways, including cell-intrinsic restriction factors (CIRFs), that limit plant virus replication and disease (136). Indeed, the yeast E3 ubiquitin ligase Rsp5 was identified as a CIRF, as its WW domains interact with the RNA-binding sites on two TBSV replication proteins (p33 and p92pol), thus blocking their ability to function in viral replication (38, 137, 138). Interestingly, the ability to block viral replication was independent of the catalytic HECT domain and was instead dependent on binding of the viral polymerase proteins by the WW domain of Rsp5 itself (38). Although a canonical PPxY motif is not present in either p33 or p92pol, these two viral proteins do share structural similarity that likely renders them capable of interacting with the WW domains of Rsp5. The Rsp5 WW domains not only interfere with the formation and assembly of the viral RNA replicase but also appear to inhibit subversion of additional host proteins required by the vial replicase to complete viral replication (138). In sum, these studies not only have identified host WW-domain interactors that can regulate replication of TBSV and perhaps other related plant viruses, but also have laid the groundwork for future studies to determine whether these modular interactions and WW domains themselves could serve as the basis for new antiviral strategies.
Modular interactions and innate immunity
Orthomyxoviruses
Influenza A and B viruses
Influenza viruses do not encode any functional PPxY motifs; however, a recent report described the indirect effect of a host PPxY/WW domain modular interaction on the innate immune response and susceptibility to influenza virus infection (139). Interferon-induced transmembrane protein 3 (IFITM3) is a PPxY-containing innate immune protein that restricts influenza virus infection, and levels of IFITM3 are regulated by ubiquitination following binding of IFITM3 to the WW domains of E3 ligase Nedd4 (139). Chesarino et al. (139) demonstrated that endogenous Nedd4 and IFITM3 colocalized with lysosomal markers in cells and that Nedd4-dependent regulation of IFITM3 levels affected the susceptibility of both cells and mice to infection with influenza A and B viruses. For example, knockdown of endogenous Nedd4 resulted in an increase in both IFITM3 levels and in cellular resistance to influenza virus infection, and the PPxY motif of IFITM3 was required for Nedd4-mediated ubiquitination (139). Overall, these findings suggest that short-term pharmacological inhibition of Nedd4's modular interaction with IFITM3 could represent a novel therapeutic strategy to prevent infection by influenza viruses and perhaps other IFITM3-sensitive viruses as well.
Retroviruses and filoviruses
HIV-1 and Ebola virus
Cellular innate immune responses have been identified that target the budding process of invading viral pathogens by affecting specific host proteins associated with virus egress, such as Nedd4 and ESCRT components. For example, interferon stimulated gene 15 (ISG15) was shown to interfere with release of HIV-1 by inhibiting host-mediated ubiquitination of Gag and Tsg101 (140). Indeed, follow-up studies on EBOV VP40-mediated budding demonstrated that ISG15 inhibited the ubiquitin ligase activity of Nedd4, which was required for efficient PPxY-mediated egress of EBOV VP40 VLPs (63, 141, 142). These studies were impactful, in that they identified an indirect mechanism of host inhibition of L domain–mediated virus egress. Subsequent and elegant studies by the Leis group expanded on the ESCRT-associated antiviral role of innate immune protein ISG15 by showing that ISG15 interacted with and/or disrupted the activity of additional ESCRT-related proteins, including CHMP5, LIP5, and Vps4 (143–145). Thus, ISG15 appears to be a key component of the host innate immune response that interferes with the assembly and budding processes of a broad array of viruses at multiple levels (146, 147).
Modular interactions and host-oriented antiviral therapeutics
Filoviruses and other PPxY-containing viruses
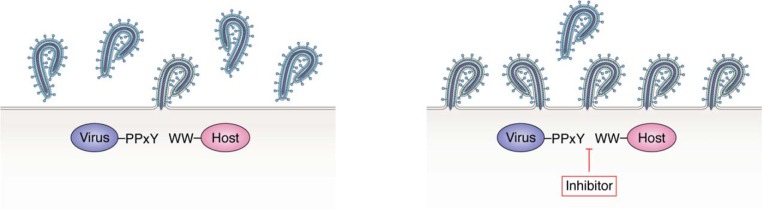
The viral PPxY/host WW-domain interface represents an attractive target for the development of small-molecule or peptide-based inhibitors that are often referred to as “host-oriented” therapeutics because they are designed to target a virus-host interface rather than a viral protein. For some of the viruses discussed above, these therapeutics would be predicted to inhibit efficient virus egress and spread by preventing the viral matrix protein from recruiting WW-domain partners and subsequent egress machinery (Fig. 3). Potential advantages of this host-oriented strategy include the widespread conservation of the viral PPxY as well as other L-domain motifs in a wide array of viral matrix proteins, such that an effective therapeutic may have broad-spectrum activity against numerous viral pathogens. In addition, host-oriented inhibitors may diminish the emergence of drug-resistant viral mutations, particularly for RNA viruses, and may lead to a paradigm shift in the search for better antiviral drugs. Because these PPxY/WW-domain interactions represent a common mechanism for mediating egress for a wide range of RNA viruses, a combination of inhibitors targeting early stages of the virus life cycle along with those targeting this late step in budding may represent a highly effective therapeutic mixture. Considering the small size of functionally folded WW domains (less than 38 amino acids) and the rigid structure of PPxY L domains, which form polyproline type II helices, engineering of their respective polypeptides and peptides into expression vectors may serve as competitors of L domain/E3 ligase interactions. Indeed, a successful interference with Rous sarcoma virus budding from cells by cis overexpression of a WW domain supports this avenue of potential interventions to harness retro- and filoviremias in vivo (76).
Figure 3.
Schematic diagram of filovirus budding in the absence (left) or presence (right) of host-oriented PPxY/WW-domain interaction inhibitors.
Several recent studies support the feasibility of small-molecule inhibition of PPxY/WW-domain interactions as an antiviral strategy for PPxY-containing RNA viruses. First, an in silico strategy was employed by Han et al. (148) to identify existing small-molecule compounds from a ZINC database that could potentially block the interaction between the PPxY L-domain motif of EBOV and the modular WW domain of host protein Nedd4 (149, 150). Functional activity of the best candidate inhibitors was assessed by using filovirus VLP budding assays, as well as infectious virus budding assays employing VSV-WT, VSV recombinants expressing EBOV L-domain motifs, and rabies virus, all of which contain PPxY motifs. The authors showed that the most potent compounds (e.g. compound 5) had broad-spectrum anti-budding activity against PPxY-containing matrix proteins from filoviruses, rhabdoviruses, and arenaviruses with little to no cytotoxicity at the inhibitory concentrations used (148). Importantly, the candidate inhibitors significantly reduced the budding of live infectious VSV and rabies virus in cell culture compared with that observed in the presence of structurally related negative controls.
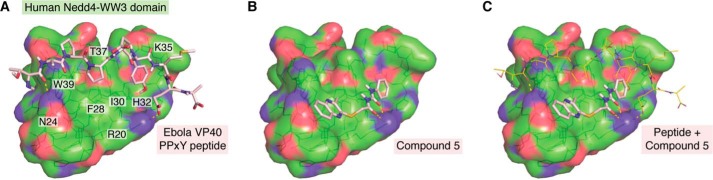
To gain a better understanding of how the viral PPxY motif fits into the WW-domain pocket of Nedd4 and how inhibitors like compound 5 may antagonize this virus-host interaction, modeling and docking analyses were performed using the NMR structure of human Nedd4-WW3 domain and the Ebola VP40-derived PPxY peptide (Protein Data Bank entry 2KQ0), with and without compound 5 (Fig. 4). The peptide-binding site in the WW3 domain contains two subpockets. This study revealed that the second proline residue from the PPxY motif in the EBOV VP40 peptide occupies the first subpocket, surrounded by WW3 residues Asn-24, Phe-28, Thr-37, and Trp-39 (Fig. 4A), and that the tyrosine residue from the EBOV VP40 PPxY motif occupies the second subpocket, surrounded by Arg-20, Ile-30, His-32, and Lys-35 (Fig. 4A). Besides hydrophobic interactions, the peptide also forms one hydrogen bond and one salt bridge with WW3 residues Asn-24 and Arg-20, respectively (Fig. 4A). The docked conformation of compound 5 showed reasonable shape complementarity to this binding site, with its quinoxaline bicyclic moiety occupying the first subpocket, its phenyl ring occupying the second pocket, and its urea moiety forming hydrogen-bonding interactions with Arg-20 in the WW3 domain (Fig. 4B). Thus, compound 5 could directly compete with the EBOV VP40 peptide in binding to the human Nedd4-WW3 domain (Fig. 4C), which would correlate with its anti-budding activity observed by Han et al. (148). Whereas the authors reported little to no cytotoxicity of compound 5 in these in vitro assays, additional experiments will be essential to assess any effects of the inhibitor on the normal cellular functions of Nedd4 and/or other related E3 ligases.
Figure 4.
NMR structure of human Nedd4-WW3 domain in complex with the Ebola VP40 PPxY peptide (A), the docked pose of compound 5 (B), and both the Ebola VP40 PPxY peptide and compound 5 (C).
In a follow-up report by Loughran et al. (151), the authors described the preparation of a series of quinoxaline-2-mercapto-acetyl-urea analogs of compound 5, which were evaluated for their ability to inhibit filoviral VLP and virus egress using assays described above. The new analogs inhibited both VP40 VLP and live recombinant VSV virus budding in the low nanomolar range, with little to no cytotoxicity, good human microsome stability, and no inhibition of P450 3A4 at 33 μm concentration (151). The efficacy of lead candidate inhibitors to block egress of authentic hemorrhagic fever viruses in vitro and in vivo remains to be determined. Identification of inhibitors that block such interactions may be useful for a wide array of PPxY-containing viruses that hijack or recruit WW domain–containing E3 ligases during the late step of budding. Such inhibitors may be particularly useful for the treatment of Ebola virus disease, as EBOV was shown recently to cross the blood-brain barrier and re-emerge months later in the central nervous system and eye, as well as in other immunologically privileged sites that are inaccessible to antibody therapy (152–165).
Whereas progress to date on the identification and development of host-oriented, small-molecule therapeutics that target virus-host interactions is still in the early stages, these studies indicate that this new therapeutic strategy is feasible in vitro and worth pursuing in vivo. However, a number of challenges and hurdles remain, one of which includes the potential for these therapeutics to disrupt key cellular protein WW-domain interactions, leading to possible side effects from these compounds. Indeed, the precise target(s) of these compounds remains to be determined, as does their possible effects on normal host protein functions. Nevertheless, the demonstration that WW domains are potentially druggable (30) and the high specificity of viral L domain interactions with their cognate protein domains support further investigations into this approach to control egress of RNA viruses like EBOV. In support of strategic shifts in drug discovery efforts, recent successes have been achieved with the use of stapled peptides (166–168) and cell-penetrating cyclic peptides (68), which are designed to target specific intracellular protein-protein interactions. Indeed, Zhang et al. (168) demonstrated that stapled peptides targeting dimer formation of the HIV-1 capsid protein showed potent antiviral activity against a broad array of viral isolates, including strains that were resistant to reverse transcriptase and protease inhibitors. In sum, the continued pursuit of fundamental and new insights into the virus-host interface will ultimately lead to the development of novel antiviral targets and strategies.
Conclusions and outlook
Whereas studies described above highlight the key roles that host WW-domain interactors play in regulating various stages of RNA and DNA virus life cycles, a number of questions remain. For example, are there additional cis- and/or trans-acting factors that influence or regulate the selective binding of the viral PPxY motif to a particular host WW domain during a virus infection? Do host PPxY-bearing proteins that interact with cellular WW domains compete with viral PPxY motif–containing proteins in infected cells? With more than 1,200 PPxY motifs represented in over 1,000 human proteins, and with the number of predicted, functional human WW domains being more than 90, the potential repertoire of WW domain-PPxY complexes in cells seems vast. Therefore, it will be important to understand the spatial, temporal, topological, and structural levels of specificity that make signaling by PPxY L domains and host proteins so discrete and biologically critical for the life cycles of many different viruses. As such, studies of highly conserved PPxY motifs and their shared interactions with modular WW domains of host interactors will likely provide new and valuable insights into the mechanisms of viral budding and transmission, entry, immune evasion, and genome replication for a wide array of viral pathogens.
If indeed antivirals targeting PPxY/WW-domain interactions are to be pursued and developed, it will be critical to assess the potential cytotoxic effects of such antiviral compounds and determine the specificity for these compounds to interfere with virus-host PPxY/WW-domain interactions, rather than with host-host PPxY/WW-domain interactions. Although the treatment regimen for such an inhibitor would likely be brief when used to target acute viral infections, the potential side effects on normal host protein function would still need to be assessed. In addition, the wide array of host WW domains and their potential to be affected by these host-oriented therapeutics must be taken into consideration when developing these compounds. To date, there does appear to be specificity in PPxY binding to select WW domains, and a better understanding of the nature of this specificity will be crucial to ensure that any antiviral inhibitor will exhibit the same specificity for its target.
Acknowledgments
We thank members of the Harty, Fan, and Sudol laboratories for helpful comments and suggestions, Leslie King for critical reading of the manuscript, and Deborah Argento for help in preparing figures. We apologize for any references that were inadvertently omitted from this article.
This work was supported in part by National Institutes of Health Grants AI102104, AI138052, AI138630, and AI139392 and a University Research Foundation (URF) award (to R. N. H.), by internal grants from the National University of Singapore (R-185-000-2710-133 and -733) and the Mechanobiology Institute (R-714-018-006-271) (to M. S.), and by the Biomedical Research Council of A*STAR (to H. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PPxY
- proline-proline-x-tyrosine motif
- AdV
- adenovirus
- ART protein
- arrestin-related trafficking protein
- BAG3
- BCL2-associated athanogene 3
- BUL1 and -2
- binds ubiquitin ligase proteins 1 and 2
- CASA
- chaperone-assisted selective autophagy
- CIRF
- cell-intrinsic restriction factor
- EBV
- Epstein–Barr virus
- EBOV
- Ebola virus
- ESCRT
- endosomal sorting complex required for transport
- HECT
- homologous to the E6AP carboxyl terminus (E3 ligase family)
- HHV
- human herpes virus
- HTLV-1
- human T-cell leukemia virus 1
- IFITM3
- interferon-induced transmembrane protein 3
- ISG15
- interferon stimulated gene 15
- ITCH (AIP4)
- Itchy E3 ubiquitin protein ligase
- LDI-1 and -2
- late domain-interacting proteins 1 and 2
- L domain
- viral late domain
- LMP2A
- latent membrane protein 2A
- MARV
- Marburg virus
- MLV
- murine leukemia virus
- Nedd4
- neuronal precursor cell–expressed developmentally down-regulated 4
- PVI
- preprotein VI (membrane lytic internal capsid protein)
- Rsp5
- yeast reverses SPT-phenotype protein 5 (E3 ubiquitin-protein ligase)
- RSV
- Rous sarcoma virus
- SMURF1 and -2
- Smad ubiquitin-regulating factors 1 and 2
- TBSV
- tomato bushy stunt virus
- Tsg101
- tumor susceptibility gene 101
- VLP
- virus-like particle
- VSV
- vesicular stomatitis virus
- WOX1
- WW domain–containing oxidoreductase
- WWP1 and -2
- WW domain–containing E3 ubiquitin protein ligase 1 and 2
- MS
- multiple sclerosis.
References
- 1. Chen H. I., and Sudol M. (1995) The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 10.1073/pnas.92.17.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kay B. K., Williamson M. P., and Sudol M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 10.1096/fasebj.14.2.231 [DOI] [PubMed] [Google Scholar]
- 3. Sudol M., Cesareni G., Superti-Furga G., and Just W. (2012) Special issue: modular protein domains. FEBS Lett. 586, 2571 10.1016/j.febslet.2012.06.005 [DOI] [PubMed] [Google Scholar]
- 4. Hu H., Columbus J., Zhang Y., Wu D., Lian L., Yang S., Goodwin J., Luczak C., Carter M., Chen L., James M., Davis R., Sudol M., Rodwell J., and Herrero J. J. (2004) A map of WW domain family interactions. Proteomics 4, 643–655 10.1002/pmic.200300632 [DOI] [PubMed] [Google Scholar]
- 5. Sudol M., and Hunter T. (2000) NeW wrinkles for an old domain. Cell 103, 1001–1004 10.1016/S0092-8674(00)00203-8 [DOI] [PubMed] [Google Scholar]
- 6. Einbond A., and Sudol M. (1996) Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 384, 1–8 10.1016/0014-5793(96)00263-3 [DOI] [PubMed] [Google Scholar]
- 7. Ilsley J. L., Sudol M., and Winder S. J. (2002) The WW domain: linking cell signalling to the membrane cytoskeleton. Cell. Signal. 14, 183–189 10.1016/S0898-6568(01)00236-4 [DOI] [PubMed] [Google Scholar]
- 8. Jamous A., and Salah Z. (2018) WW-domain containing protein roles in breast tumorigenesis. Front. Oncol. 8, 580 10.3389/fonc.2018.00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McLoughlin D. M., and Miller C. C. (2008) The FE65 proteins and Alzheimer's disease. J. Neurosci. Res. 86, 744–754 10.1002/jnr.21532 [DOI] [PubMed] [Google Scholar]
- 10. Reuven N., Shanzer M., and Shaul Y. (2015) Tyrosine phosphorylation of WW proteins. Exp. Biol. Med. (Maywood) 240, 375–382 10.1177/1535370214565991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosati A., Graziano V., De Laurenzi V., Pascale M., and Turco M. C. (2011) BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2, e141 10.1038/cddis.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rotin D. (2008) Role of the UPS in Liddle syndrome. BMC Biochem. 9, S5 10.1186/1471-2091-9-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salah Z., Alian A., and Aqeilan R. I. (2012) WW domain-containing proteins: retrospectives and the future. Front. Biosci. (Landmark Ed.) 17, 331–348 10.2741/3930 [DOI] [PubMed] [Google Scholar]
- 14. Staub O., Abriel H., Plant P., Ishikawa T., Kanelis V., Saleki R., Horisberger J. D., Schild L., and Rotin D. (2000) Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int. 57, 809–815 10.1046/j.1523-1755.2000.00919.x [DOI] [PubMed] [Google Scholar]
- 15. Staub O., and Rotin D. (1996) WW domains. Structure 4, 495–499 10.1016/S0969-2126(96)00054-8 [DOI] [PubMed] [Google Scholar]
- 16. Sudol M. (1996) Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65, 113–132 10.1016/S0079-6107(96)00008-9 [DOI] [PubMed] [Google Scholar]
- 17. Sudol M., Chen H. I., Bougeret C., Einbond A., and Bork P. (1995) Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 369, 67–71 10.1016/0014-5793(95)00550-S [DOI] [PubMed] [Google Scholar]
- 18. Sudol M., and Harvey K. F. (2010) Modularity in the Hippo signaling pathway. Trends Biochem. Sci. 35, 627–633 10.1016/j.tibs.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 19. Sudol M., Recinos C. C., Abraczinskas J., Humbert J., and Farooq A. (2005) WW or WoW: the WW domains in a union of bliss. IUBMB Life 57, 773–778 10.1080/15216540500389039 [DOI] [PubMed] [Google Scholar]
- 20. Sudol M., Sliwa K., and Russo T. (2001) Functions of WW domains in the nucleus. FEBS Lett. 490, 190–195 10.1016/S0014-5793(01)02122-6 [DOI] [PubMed] [Google Scholar]
- 21. Zarrinpar A., Bhattacharyya R. P., and Lim W. A. (2003) The structure and function of proline recognition domains. Sci. STKE 2003, RE8 10.1126/stke.2003.179.re8 [DOI] [PubMed] [Google Scholar]
- 22. Chang N. S. (2002) A potential role of p53 and WOX1 in mitochondrial apoptosis (review). Int. J. Mol. Med. 9, 19–24 [PubMed] [Google Scholar]
- 23. Chen S. J., Huang S. S., and Chang N. S. (2013) Role of WWOX and NF-κB in lung cancer progression. Transl. Respir. Med. 1, 15 10.1186/2213-0802-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Y. A., Lu C. Y., Cheng T. Y., Pan S. H., Chen H. F., and Chang N. S. (2019) WW domain-containing proteins YAP and TAZ in the Hippo pathway as key regulators in stemness maintenance, tissue homeostasis, and tumorigenesis. Front. Oncol. 9, 60 10.3389/fonc.2019.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aqeilan R. I., and Croce C. M. (2007) WWOX in biological control and tumorigenesis. J. Cell. Physiol. 212, 307–310 10.1002/jcp.21099 [DOI] [PubMed] [Google Scholar]
- 26. Salah Z., and Aqeilan R. I. (2011) WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2, e172 10.1038/cddis.2011.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanna M., and Aqeilan R. I. (2018) Modeling WWOX loss of function in vivo: what have we learned? Front. Oncol. 8, 420 10.3389/fonc.2018.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hergovich A. (2012) Mammalian Hippo signalling: a kinase network regulated by protein-protein interactions. Biochem. Soc. Trans. 40, 124–128 10.1042/BST20110619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muller R. U., and Schermer B. (2019) Hippo signaling—a central player in cystic kidney disease? Pediatr. Nephrol. 10.1007/s00467-019-04299-3 [DOI] [PubMed] [Google Scholar]
- 30. Sudol M., Shields D. C., and Farooq A. (2012) Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin. Cell Dev. Biol. 23, 827–833 10.1016/j.semcdb.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aki D., Zhang W., and Liu Y. C. (2015) The E3 ligase Itch in immune regulation and beyond. Immunol. Rev. 266, 6–26 10.1111/imr.12301 [DOI] [PubMed] [Google Scholar]
- 32. Flores S. Y., Debonneville C., and Staub O. (2003) The role of Nedd4/Nedd4-like dependent ubiquitylation in epithelial transport processes. Pflugers Arch. 446, 334–338 10.1007/s00424-003-1027-x [DOI] [PubMed] [Google Scholar]
- 33. Ingham R. J., Gish G., and Pawson T. (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–1984 10.1038/sj.onc.1207436 [DOI] [PubMed] [Google Scholar]
- 34. Koganti P., Levy-Cohen G., and Blank M. (2018) Smurfs in protein homeostasis, signaling, and cancer. Front. Oncol. 8, 295 10.3389/fonc.2018.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rotin D., Staub O., and Haguenauer-Tsapis R. (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176, 1–17 10.1007/s00232001079 [DOI] [PubMed] [Google Scholar]
- 36. Yang B., and Kumar S. (2010) Nedd4 and Nedd4–2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17, 68–77 10.1038/cdd.2009.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhi X., and Chen C. (2012) WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell. Mol. Life Sci. 69, 1425–1434 10.1007/s00018-011-0871-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barajas D., Li Z., and Nagy P. D. (2009) The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 83, 11751–11764 10.1128/JVI.00789-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouamr F., Melillo J. A., Wang M. Q., Nagashima K., de Los Santos M., Rein A., and Goff S. P. (2003) PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101 [corrected]. J. Virol. 77, 11882–11895 10.1128/JVI.77.22.11882-11895.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galinier R., Gout E., Lortat-Jacob H., Wood J., and Chroboczek J. (2002) Adenovirus protein involved in virus internalization recruits ubiquitin-protein ligases. Biochemistry 41, 14299–14305 10.1021/bi020125b [DOI] [PubMed] [Google Scholar]
- 41. Han Z., Sagum C. A., Bedford M. T., Sidhu S. S., Sudol M., and Harty R. N. (2016) ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J. Virol. 90, 9163–9171 10.1128/JVI.01078-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Z., Sagum C. A., Takizawa F., Ruthel G., Berry C. T., Kong J., Sunyer J. O., Freedman B. D., Bedford M. T., Sidhu S. S., Sudol M., and Harty R. N. (2017) Ubiquitin ligase WWP1 interacts with Ebola virus VP40 to regulate egress. J. Virol. 91, e00812–17 10.1128/JVI.00812-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakurai A., Yasuda J., Takano H., Tanaka Y., Hatakeyama M., and Shida H. (2004) Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 6, 150–156 10.1016/j.micinf.2003.10.011 [DOI] [PubMed] [Google Scholar]
- 44. Urata S., and Yasuda J. (2010) Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J. Gen. Virol. 91, 228–234 10.1099/vir.0.015495-0 [DOI] [PubMed] [Google Scholar]
- 45. Vana M. L., Tang Y., Chen A., Medina G., Carter C., and Leis J. (2004) Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 78, 13943–13953 10.1128/JVI.78.24.13943-13953.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winberg G., Matskova L., Chen F., Plant P., Rotin D., Gish G., Ingham R., Ernberg I., and Pawson T. (2000) Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell. Biol. 20, 8526–8535 10.1128/MCB.20.22.8526-8535.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Medina G., Zhang Y., Tang Y., Gottwein E., Vana M. L., Bouamr F., Leis J., and Carter C. A. (2005) The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic 6, 880–894 10.1111/j.1600-0854.2005.00323.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ott D. E., Coren L. V., Gagliardi T. D., and Nagashima K. (2005) Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1. J. Virol. 79, 9038–9045 10.1128/JVI.79.14.9038-9045.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yuan B., Campbell S., Bacharach E., Rein A., and Goff S. P. (2000) Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74, 7250–7260 10.1128/JVI.74.16.7250-7260.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parent L. J., Bennett R. P., Craven R. C., Nelle T. D., Krishna N. K., Bowzard J. B., Wilson C. B., Puffer B. A., Montelaro R. C., and Wills J. W. (1995) Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69, 5455–5460 10.1128/JVI.69.9.5455-5460.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garnier L., Wills J. W., Verderame M. F., and Sudol M. (1996) WW domains and retrovirus budding. Nature 381, 744–745 10.1038/381744a0 [DOI] [PubMed] [Google Scholar]
- 52. Göttlinger H. G., Dorfman T., Sodroski J. G., and Haseltine W. A. (1991) Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U.S.A. 88, 3195–3199 10.1073/pnas.88.8.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wills J. W., Cameron C. E., Wilson C. B., Xiang Y., Bennett R. P., and Leis J. (1994) An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68, 6605–6618 10.1128/JVI.68.10.6605-6618.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harty R. N., Brown M. E., Wang G., Huibregtse J., and Hayes F. P. (2000) A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 10.1073/pnas.250277297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Harty R. N., Brown M. E., McGettigan J. P., Wang G., Jayakar H. R., Huibregtse J. M., Whitt M. A., and Schnell M. J. (2001) Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75, 10623–10629 10.1128/JVI.75.22.10623-10629.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kikonyogo A., Bouamr F., Vana M. L., Xiang Y., Aiyar A., Carter C., and Leis J. (2001) Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U.S.A. 98, 11199–11204 10.1073/pnas.201268998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yasuda J., Hunter E., Nakao M., and Shida H. (2002) Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3, 636–640 10.1093/embo-reports/kvf132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yasuda J., Nakao M., Kawaoka Y., and Shida H. (2003) Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77, 9987–9992 10.1128/JVI.77.18.9987-9992.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Blot V., Perugi F., Gay B., Prévost M. C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M. C., and Pique C. (2004) Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117, 2357–2367 10.1242/jcs.01095 [DOI] [PubMed] [Google Scholar]
- 60. Martin-Serrano J., Perez-Caballero D., and Bieniasz P. D. (2004) Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78, 5554–5563 10.1128/JVI.78.11.5554-5563.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Segura-Morales C., Pescia C., Chatellard-Causse C., Sadoul R., Bertrand E., and Basyuk E. (2005) Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280, 27004–27012 10.1074/jbc.M413735200 [DOI] [PubMed] [Google Scholar]
- 62. Chung H. Y., Morita E., von Schwedler U., Muller B., Kräusslich H. G., and Sundquist W. I. (2008) NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82, 4884–4897 10.1128/JVI.02667-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Okumura A., Pitha P. M., and Harty R. N. (2008) ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U.S.A. 105, 3974–3979 10.1073/pnas.0710629105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Usami Y., Popov S., Popova E., and Göttlinger H. G. (2008) Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82, 4898–4907 10.1128/JVI.02675-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weiss E. R., Popova E., Yamanaka H., Kim H. C., Huibregtse J. M., and Göttlinger H. (2010) Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 6, e1001107 10.1371/journal.ppat.1001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dorjbal B., Derse D., Lloyd P., Soheilian F., Nagashima K., and Heidecker G. (2011) The role of ITCH protein in human T-cell leukemia virus type 1 release. J. Biol. Chem. 286, 31092–31104 10.1074/jbc.M111.259945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sette P., Nagashima K., Piper R. C., and Bouamr F. (2013) Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10, 79 10.1186/1742-4690-10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Swedberg J. E., Ghani H. A., Harris J. M., de Veer S. J., and Craik D. J. (2018) Potent, selective, and cell-penetrating inhibitors of kallikrein-related peptidase 4 based on the cyclic peptide MCoTI-II. ACS Med. Chem. Lett. 9, 1258–1262 10.1021/acsmedchemlett.8b00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ott D. E., Coren L. V., Copeland T. D., Kane B. P., Johnson D. G., Sowder R. C. 2nd, Yoshinaka Y., Oroszlan S., Arthur L. O., and Henderson L. E. (1998) Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72, 2962–2968 10.1128/JVI.72.4.2962-2968.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Strack B., Calistri A., Accola M. A., Palu G., and Gottlinger H. G. (2000) A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U.S.A. 97, 13063–13068 10.1073/pnas.97.24.13063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mercenne G., Alam S. L., Arii J., Lalonde M. S., and Sundquist W. I. (2015) Angiomotin functions in HIV-1 assembly and budding. eLife 4 10.7554/eLife.03778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., and Sundquist W. I. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 10.1016/S0092-8674(01)00506-2 [DOI] [PubMed] [Google Scholar]
- 73. Strack B., Calistri A., Craig S., Popova E., and Göttlinger H. G. (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 10.1016/S0092-8674(03)00653-6 [DOI] [PubMed] [Google Scholar]
- 74. Sette P., Jadwin J. A., Dussupt V., Bello N. F., and Bouamr F. (2010) The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 84, 8181–8192 10.1128/JVI.00634-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Usami Y., Popov S., Popova E., Inoue M., Weissenhorn W., and Göttlinger H. G. (2009) The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 37, 181–184 10.1042/BST0370181 [DOI] [PubMed] [Google Scholar]
- 76. Patnaik A., and Wills J. W. (2002) In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76, 2789–2795 10.1128/JVI.76.6.2789-2795.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Heidecker G., Lloyd P. A., Fox K., Nagashima K., and Derse D. (2004) Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78, 6636–6648 10.1128/JVI.78.12.6636-6648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heidecker G., Lloyd P. A., Soheilian F., Nagashima K., and Derse D. (2007) The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J. Virol. 81, 9769–9777 10.1128/JVI.00642-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martin-Serrano J., Eastman S. W., Chung W., and Bieniasz P. D. (2005) HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168, 89–101 10.1083/jcb.200408155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rauch S., and Martin-Serrano J. (2011) Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J. Virol. 85, 3546–3556 10.1128/JVI.02045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rasmussen A. L. (2018) Host factors involved in Ebola virus replication. Curr. Top. Microbiol. Immunol. 419, 113–150 10.1007/82_2017_27 [DOI] [PubMed] [Google Scholar]
- 82. Basler C. F. (2017) Molecular pathogenesis of viral hemorrhagic fever. Semin. Immunopathol. 39, 551–561 10.1007/s00281-017-0637-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baseler L., Chertow D. S., Johnson K. M., Feldmann H., and Morens D. M. (2017) The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 12, 387–418 10.1146/annurev-pathol-052016-100506 [DOI] [PubMed] [Google Scholar]
- 84. Hartlieb B., and Weissenhorn W. (2006) Filovirus assembly and budding. Virology 344, 64–70 10.1016/j.virol.2005.09.018 [DOI] [PubMed] [Google Scholar]
- 85. Jasenosky L. D., and Kawaoka Y. (2004) Filovirus budding. Virus Res. 106, 181–188 10.1016/j.virusres.2004.08.014 [DOI] [PubMed] [Google Scholar]
- 86. Noda T., Sagara H., Suzuki E., Takada A., Kida H., and Kawaoka Y. (2002) Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76, 4855–4865 10.1128/JVI.76.10.4855-4865.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Noda T., Ebihara H., Muramoto Y., Fujii K., Takada A., Sagara H., Kim J. H., Kida H., Feldmann H., and Kawaoka Y. (2006) Assembly and budding of Ebolavirus. PLoS Pathog. 2, e99 10.1371/journal.ppat.0020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Harty R. N. (2018) Hemorrhagic fever virus budding studies. Methods Mol. Biol. 1604, 209–215 10.1007/978-1-4939-6981-4_15 [DOI] [PubMed] [Google Scholar]
- 89. Harty R. N., Paragas J., Sudol M., and Palese P. (1999) A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73, 2921–2929 10.1128/JVI.73.4.2921-2929.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Irie T., Licata J. M., McGettigan J. P., Schnell M. J., and Harty R. N. (2004) Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 78, 2657–2665 10.1128/JVI.78.6.2657-2665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Licata J. M., Simpson-Holley M., Wright N. T., Han Z., Paragas J., and Harty R. N. (2003) Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77, 1812–1819 10.1128/JVI.77.3.1812-1819.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Urata S., and de la Torre J. C. (2011) Arenavirus budding. Adv. Virol. 2011, 180326 10.1155/2011/180326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wirblich C., Tan G. S., Papaneri A., Godlewski P. J., Orenstein J. M., Harty R. N., and Schnell M. J. (2008) PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J. Virol. 82, 9730–9738 10.1128/JVI.00889-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhadina M., and Bieniasz P. D. (2010) Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 6, e1001153 10.1371/journal.ppat.1001153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Martin-Serrano J., Zang T., and Bieniasz P. D. (2001) HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7, 1313–1319 10.1038/nm1201-1313 [DOI] [PubMed] [Google Scholar]
- 96. Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R. W., and Weissenhorn W. (2003) Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326, 493–502 10.1016/S0022-2836(02)01406-7 [DOI] [PubMed] [Google Scholar]
- 97. Irie T., Licata J. M., and Harty R. N. (2005) Functional characterization of Ebola virus L-domains using VSV recombinants. Virology 336, 291–298 10.1016/j.virol.2005.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dolnik O., Kolesnikova L., Stevermann L., and Becker S. (2010) Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J. Virol. 84, 7847–7856 10.1128/JVI.00476-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Craven R. C., Harty R. N., Paragas J., Palese P., and Wills J. W. (1999) Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73, 3359–3365 10.1128/JVI.73.4.3359-3365.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jayakar H. R., Murti K. G., and Whitt M. A. (2000) Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74, 9818–9827 10.1128/JVI.74.21.9818-9827.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jayakar H. R., Jeetendra E., and Whitt M. A. (2004) Rhabdovirus assembly and budding. Virus Res. 106, 117–132 10.1016/j.virusres.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 102. Urata S., Noda T., Kawaoka Y., Morikawa S., Yokosawa H., and Yasuda J. (2007) Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J. Virol. 81, 4895–4899 10.1128/JVI.02829-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liang J., Sagum C. A., Bedford M. T., Sidhu S. S., Sudol M., Han Z., and Harty R. N. (2017) Chaperone-mediated autophagy protein BAG3 negatively regulates Ebola and Marburg VP40-mediated egress. PLoS Pathog. 13, e1006132 10.1371/journal.ppat.1006132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Venuprasad K., Zeng M., Baughan S. L., and Massoumi R. (2015) Multifaceted role of the ubiquitin ligase Itch in immune regulation. Immunol. Cell Biol. 93, 452–460 10.1038/icb.2014.118 [DOI] [PubMed] [Google Scholar]
- 105. Behl C. (2016) Breaking BAG: the co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 37, 672–688 10.1016/j.tips.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 106. Ulbricht A., Gehlert S., Leciejewski B., Schiffer T., Bloch W., and Höhfeld J. (2015) Induction and adaptation of chaperone-assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy 11, 538–546 10.1080/15548627.2015.1017186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ulbricht A., and Höhfeld J. (2013) Tension-induced autophagy: may the chaperone be with you. Autophagy 9, 920–922 10.4161/auto.24213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kathage B., Gehlert S., Ulbricht A., Ludecke L., Tapia V. E., Orfanos Z., Wenzel D., Bloch W., Volkmer R., Fleischmann B. K., Furst D. O., and Hohfeld J. (2017) The cochaperone BAG3 coordinates protein synthesis and autophagy under mechanical strain through spatial regulation of mTORC1. Biochim. Biophys. Acta 1864, 62–75 10.1016/j.bbamcr.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 109. Ulbricht A., Arndt V., and Höhfeld J. (2013) Chaperone-assisted proteostasis is essential for mechanotransduction in mammalian cells. Commun. Integr. Biol. 6, e24925 10.4161/cib.24925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ulbricht A., Eppler F. J., Tapia V. E., van der Ven P. F., Hampe N., Hersch N., Vakeel P., Stadel D., Haas A., Saftig P., Behrends C., Fürst D. O., Volkmer R., Hoffmann B., Kolanus W., and Hohfeld J. (2013) Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430–435 10.1016/j.cub.2013.01.064 [DOI] [PubMed] [Google Scholar]
- 111. Varlet A. A., Fuchs M., Luthold C., Lambert H., Landry J., and Lavoie J. N. (2017) Fine-tuning of actin dynamics by the HSPB8-BAG3 chaperone complex facilitates cytokinesis and contributes to its impact on cell division. Cell Stress Chaperones 22, 553–567 10.1007/s12192-017-0780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fuchs M., Luthold C., Guilbert S. M., Varlet A. A., Lambert H., Jetté A., Elowe S., Landry J., and Lavoie J. N. (2015) A role for the chaperone complex BAG3-HSPB8 in actin dynamics, spindle orientation and proper chromosome segregation during mitosis. PLoS Genet. 11, e1005582 10.1371/journal.pgen.1005582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Scourfield E. J., and Martin-Serrano J. (2017) Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans. 45, 613–634 10.1042/BST20160479 [DOI] [PubMed] [Google Scholar]
- 114. Schöneberg J., Lee I. H., Iwasa J. H., and Hurley J. H. (2017) Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hurley J. H. (2015) ESCRTs are everywhere. EMBO J. 34, 2398–2407 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Votteler J., and Sundquist W. I. (2013) Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 10.1016/j.chom.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. McCullough J., Colf L. A., and Sundquist W. I. (2013) Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 10.1146/annurev-biochem-072909-101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jouvenet N. (2012) Dynamics of ESCRT proteins. Cell. Mol. Life Sci. 69, 4121–4133 10.1007/s00018-012-1035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wollert T., Wunder C., Lippincott-Schwartz J., and Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. McDonald B., and Martin-Serrano J. (2009) No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 122, 2167–2177 10.1242/jcs.028308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lata S., Schoehn G., Solomons J., Pires R., Göttlinger H. G., and Weissenhorn W. (2009) Structure and function of ESCRT-III. Biochem. Soc. Trans. 37, 156–160 10.1042/BST0370156 [DOI] [PubMed] [Google Scholar]
- 122. Lu J., Qu Y., Liu Y., Jambusaria R., Han Z., Ruthel G., Freedman B. D., and Harty R. N. (2013) Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J. Virol. 87, 7777–7780 10.1128/JVI.00470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Han Z., and Harty R. N. (2005) Packaging of actin into Ebola virus VLPs. Virol. J. 2, 92 10.1186/1743-422X-2-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wodrich H., Henaff D., Jammart B., Segura-Morales C., Seelmeir S., Coux O., Ruzsics Z., Wiethoff C. M., and Kremer E. J. (2010) A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog. 6, e1000808 10.1371/journal.ppat.1000808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Montespan C., Marvin S. A., Austin S., Burrage A. M., Roger B., Rayne F., Faure M., Campell E. M., Schneider C., Reimer R., Grünewald K., Wiethoff C. M., and Wodrich H. (2017) Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 13, e1006217 10.1371/journal.ppat.1006217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Montespan C., Wiethoff C. M., and Wodrich H. (2017) A small viral PPxY peptide motif to control antiviral autophagy. J. Virol. 91, e00581–17 10.1128/JVI.00581-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ikeda M., Ikeda A., Longan L. C., and Longnecker R. (2000) The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 268, 178–191 10.1006/viro.1999.0166 [DOI] [PubMed] [Google Scholar]
- 128. Longnecker R., Merchant M., Brown M. E., Fruehling S., Bickford J. O., Ikeda M., and Harty R. N. (2000) WW- and SH3-domain interactions with Epstein-Barr virus LMP2A. Exp. Cell Res. 257, 332–340 10.1006/excr.2000.4900 [DOI] [PubMed] [Google Scholar]
- 129. Cen O., and Longnecker R. (2015) Latent Membrane Protein 2 (LMP2). Curr. Top. Microbiol. Immunol. 391, 151–180 10.1007/978-3-319-22834-1_5 [DOI] [PubMed] [Google Scholar]
- 130. Lan Y. Y., Wu S. Y., Lai H. C., Chang N. S., Chang F. H., Tsai M. H., Su I. J., and Chang Y. (2013) WW domain-containing oxidoreductase is involved in upregulation of matrix metalloproteinase 9 by Epstein-Barr virus latent membrane protein 2A. Biochem. Biophys. Res. Commun. 436, 672–676 10.1016/j.bbrc.2013.06.014 [DOI] [PubMed] [Google Scholar]
- 131. Sullivan B. M., and Coscoy L. (2008) Downregulation of the T-cell receptor complex and impairment of T-cell activation by human herpesvirus 6 u24 protein. J. Virol. 82, 602–608 10.1128/JVI.01571-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sullivan B. M., and Coscoy L. (2010) The U24 protein from human herpesvirus 6 and 7 affects endocytic recycling. J. Virol. 84, 1265–1275 10.1128/JVI.01775-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sang Y., Zhang R., Scott W. R., Creagh A. L., Haynes C. A., and Straus S. K. (2017) U24 from Roseolovirus interacts strongly with Nedd4 WW domains. Sci. Rep. 7, 39776 10.1038/srep39776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tejada-Simon M. V., Zang Y. C., Hong J., Rivera V. M., and Zhang J. Z. (2003) Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Ann. Neurol. 53, 189–197 10.1002/ana.10425 [DOI] [PubMed] [Google Scholar]
- 135. Sang Y., Zhang R., Creagh A. L., Haynes C. A., and Straus S. K. (2017) Interactions of U24 from Roseolovirus with WW domains: canonical vs noncanonical. Biochem. Cell Biol. 95, 350–358 10.1139/bcb-2016-0250 [DOI] [PubMed] [Google Scholar]
- 136. Sasvari Z., Alatriste Gonzalez P., and Nagy P. D. (2014) Tombusvirus-yeast interactions identify conserved cell-intrinsic viral restriction factors. Front. Plant Sci. 5, 383 10.3389/fpls.2014.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Qin J., Barajas D., and Nagy P. D. (2012) An inhibitory function of WW domain-containing host proteins in RNA virus replication. Virology 426, 106–119 10.1016/j.virol.2012.01.020 [DOI] [PubMed] [Google Scholar]
- 138. Barajas D., Kovalev N., Qin J., and Nagy P. D. (2015) Novel mechanism of regulation of tomato bushy stunt virus replication by cellular WW-domain proteins. J. Virol. 89, 2064–2079 10.1128/JVI.02719-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chesarino N. M., McMichael T. M., and Yount J. S. (2015) E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 11, e1005095 10.1371/journal.ppat.1005095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Okumura A., Lu G., Pitha-Rowe I., and Pitha P. M. (2006) Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U.S.A. 103, 1440–1445 10.1073/pnas.0510518103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Harty R. N., Pitha P. M., and Okumura A. (2009) Antiviral activity of innate immune protein ISG15. J. Innate Immun. 1, 397–404 10.1159/000226245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Malakhova O. A., and Zhang D. E. (2008) ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283, 8783–8787 10.1074/jbc.C800030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kuang Z., Seo E. J., and Leis J. (2011) Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J. Virol. 85, 7153–7161 10.1128/JVI.02610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Pincetic A., Kuang Z., Seo E. J., and Leis J. (2010) The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J. Virol. 84, 4725–4736 10.1128/JVI.02478-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Seo E. J., and Leis J. (2012) Budding of enveloped viruses: interferon-induced ISG15-antivirus mechanisms targeting the release process. Adv. Virol. 2012, 532723 10.1155/2012/532723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Morales D. J., and Lenschow D. J. (2013) The antiviral activities of ISG15. J. Mol. Biol. 425, 4995–5008 10.1016/j.jmb.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Villarroya-Beltri C., Guerra S., and Sánchez-Madrid F. (2017) ISGylation - a key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 130, 2961–2969 10.1242/jcs.205468 [DOI] [PubMed] [Google Scholar]
- 148. Han Z., Lu J., Liu Y., Davis B., Lee M. S., Olson M. A., Ruthel G., Freedman B. D., Schnell M. J., Wrobel J. E., Reitz A. B., and Harty R. N. (2014) Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J. Virol. 88, 7294–7306 10.1128/JVI.00591-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sterling T., and Irwin J. J. (2015) ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., and Coleman R. G. (2012) ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Loughran H. M., Han Z., Wrobel J. E., Decker S. E., Ruthel G., Freedman B. D., Harty R. N., and Reitz A. B. (2016) Quinoxaline-based inhibitors of Ebola and Marburg VP40 egress. Bioorg. Med. Chem. Lett. 26, 3429–3435 10.1016/j.bmcl.2016.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Billioux B. J. (2017) Neurological complications and sequelae of Ebola virus disease. Curr. Infect. Dis. Rep. 19, 19 10.1007/s11908-017-0573-x [DOI] [PubMed] [Google Scholar]
- 153. Jacobs M., Rodger A., Bell D. J., Bhagani S., Cropley I., Filipe A., Gifford R. J., Hopkins S., Hughes J., Jabeen F., Johannessen I., Karageorgopoulos D., Lackenby A., Lester R., Liu R. S., et al. (2016) Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 388, 498–503 10.1016/S0140-6736(16)30386-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Steptoe P. J., Scott J. T., Sahr F., Vandy M., Harding S. P., Beare N. A. V., and Semple M. G. (2018) Re: Shantha et al.: Ophthalmic manifestations and causes of vision impairment in Ebola virus disease survivors in Monrovia, Liberia (Ophthalmology. 2017;124:170–177). Ophthalmology 125, e19 10.1016/j.ophtha.2017.09.009 [DOI] [PubMed] [Google Scholar]
- 155. Shantha J. G., Crozier I., and Yeh S. (2017) An update on ocular complications of Ebola virus disease. Curr. Opin. Ophthalmol. 28, 600–606 10.1097/ICU.0000000000000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Hereth-Hebert E., Bah M. O., Etard J. F., Sow M. S., Resnikoff S., Fardeau C., Toure A., Ouendeno A. N., Sagno I. C., March L., Izard S., Lama P. L., Barry M., Delaporte E., and Postebogui Study Group (2017) Ocular complications in survivors of the Ebola outbreak in Guinea. Am. J. Ophthalmol. 175, 114–121 10.1016/j.ajo.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 157. Shantha J. G., Yeh S., and Nguyen Q. D. (2016) Ebola virus disease and the eye. Curr. Opin. Ophthalmol. 27, 538–544 10.1097/ICU.0000000000000313 [DOI] [PubMed] [Google Scholar]
- 158. Jampol L. M., Ferris F. L. 3rd, Bishop R. J. (2015) Ebola and the eye. JAMA Ophthalmol. 133, 1105–1106 10.1001/jamaophthalmol.2015.2400 [DOI] [PubMed] [Google Scholar]
- 159. Moshirfar M., Fenzl C. R., and Li Z. (2014) What we know about ocular manifestations of Ebola. Clin. Ophthalmol. 8, 2355–2357 10.2147/OPTH.S73583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Caswell R. J., and Manavi K. (2017) Emerging sexually transmitted viral infections: 1. Review of Ebola virus disease. Int. J. STD AIDS 28, 1352–1359 10.1177/0956462417703572 [DOI] [PubMed] [Google Scholar]
- 161. Sissoko D., Keïta M., Diallo B., Aliabadi N., Fitter D. L., Dahl B. A., Akoi Bore J., Raymond Koundouno F., Singethan K., Meisel S., Enkirch T., Mazzarelli A., Amburgey V., Faye O., Alpha Sall A., et al. (2017) Ebola virus persistence in breast milk after no reported illness: a likely source of virus transmission from mother to child. Clin. Infect. Dis. 64, 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Crozier I. (2016) Ebola virus RNA in the semen of male survivors of Ebola virus disease: the uncertain gravitas of a privileged persistence. J. Infect. Dis. 214, 1467–1469 10.1093/infdis/jiw079 [DOI] [PubMed] [Google Scholar]
- 163. Uyeki T. M., Erickson B. R., Brown S., McElroy A. K., Cannon D., Gibbons A., Sealy T., Kainulainen M. H., Schuh A. J., Kraft C. S., Mehta A. K., Lyon G. M. 3rd, Varkey J. B., Ribner B. S., Ellison R. T. 3rd, et al. (2016) Ebola virus persistence in semen of male survivors. Clin. Infect. Dis. 62, 1552–1555 10.1093/cid/ciw202 [DOI] [PubMed] [Google Scholar]
- 164. Chughtai A. A., Barnes M., and Macintyre C. R. (2016) Persistence of Ebola virus in various body fluids during convalescence: evidence and implications for disease transmission and control. Epidemiol. Infect. 144, 1652–1660 10.1017/S0950268816000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Rogstad K. E., and Tunbridge A. (2015) Ebola virus as a sexually transmitted infection. Curr. Opin. Infect. Dis. 28, 83–85 10.1097/QCO.0000000000000135 [DOI] [PubMed] [Google Scholar]
- 166. Verdine G. L., and Hilinski G. J. (2012) Stapled peptides for intracellular drug targets. Methods Enzymol. 503, 3–33 10.1016/B978-0-12-396962-0.00001-X [DOI] [PubMed] [Google Scholar]
- 167. Tan B. X., Brown C. J., Ferrer F. J., Yuen T. Y., Quah S. T., Chan B. H., Jansson A. E., Teo H. L., Nordlund P., and Lane D. P. (2015) Assessing the efficacy of Mdm2/Mdm4-inhibiting stapled peptides using cellular thermal shift assays. Sci. Rep. 5, 12116 10.1038/srep12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhang H., Curreli F., Zhang X., Bhattacharya S., Waheed A. A., Cooper A., Cowburn D., Freed E. O., and Debnath A. K. (2011) Antiviral activity of α-helical stapled peptides designed from the HIV-1 capsid dimerization domain. Retrovirology 8, 28 10.1186/1742-4690-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]