Abstract
Objective
To investigate inflammatory cytokines in patients with motor neuron disease (MND) evaluating the putative contribution of amyotrophic lateral sclerosis (ALS)-causing gene variants.
Methods
This study is a retrospective case series with prospective follow-up (1994–2016) of 248 patients with MND, of whom 164 had ALS who were screened for mutations in the genes for SOD1 and C9orf72. Paired CSF and plasma were collected at the diagnostic evaluation before treatment. A panel of cytokines were measured blindly via digital ELISA on the Simoa platform.
Results
Time from disease onset to death was longer for patients with ALS-causing SOD1 mutations (mSOD1, n = 24) than those with C9orf72 hexanucleotide repeat expansion (C9orf72HRE) ALS (n = 19; q = 0.001) and other ALS (OALS) (n = 119; q = 0.0008). Patients with OALS had higher CSF tumor necrosis factor alpha (TNF-α) compared with those with C9orf72HRE ALS (q = 0.014). Patients with C9orf72HRE ALS had higher CSF interferon alpha compared with those with OALS and mSOD1 ALS (q = 0.042 and q = 0.042). In patients with ALS, the survival was negatively correlated with plasma interleukin (IL) 10 (hazard ratio [HR] 1.17, 95% CI 1.05–1.30). Plasma TNF-α, IL-10, and TNF-related apoptosis-inducing ligand (TRAIL) (HR 1.01 [1.00–1.02], 1.15 [1.02–1.30], and 1.01 [1.00–1.01], respectively) of patients with OALS, plasma IL-1β (HR 5.90 [1.27–27.5]) of patients with C9orf72HRE ALS, and CSF TRAIL (10.5 [1.12–98.6]) of patients with mSOD1 ALS all correlated negatively with survival.
Conclusions
Differences in survival times in ALS subtypes were correlated with cytokine levels, suggesting specific immune responses related to ALS genetic variants.
Amyotrophic lateral sclerosis (ALS) is an etiologically and prognostically heterogeneous, progressive neurodegenerative syndrome with a median survival of 2–3 years.1 Several genetic mutations have been associated with ALS, the 2 most prevalent being a hexanucleotide repeat expansion (HRE) in intron 1 of C9orf72 (C9orf72HRE) in 8%–11% and coding mutations in the gene encoding the superoxide-free radical scavenging enzyme Cu,Zn-superoxide dismutase (SOD1) in 4%–7% of patients with ALS in Caucasian populations.2 Type I interferons (IFNs), including IFN alpha (IFN-α), interleukins (ILs), and tumor necrosis factor (TNF), are components of the innate immune system and exert key functions in the inflammatory processes of the CNS.3 Of interest, in a study of mice carrying the human mutated SOD1 (mSOD1) variants, IFN-stimulated genes were upregulated before motor neuron loss and clinical onset, and inhibition of IFN signaling was found to be associated with increased life span.4 Furthermore, elevated levels of IL-6 and IL-1b were observed in CSF and spinal cord tissue from patients with ALS, and activation of the TNF alpha (TNF-α) has been reported in ALS.5,6 However, these previous studies were based on small heterogeneous patient groups not genotyped for ALS-related mutations, which may differentially affect the immune system. The aim of the present study is to analyze a panel of cytokines in patients with different clinical and genetic types of motor neuron disease (MND), including ALS, to gain insights into the role(s) of inflammatory cytokines across subtypes of ALS and to investigate the prognostic potential of these biomarkers.
Methods
Study design
This study is a retrospective case series with a prospective longitudinal follow-up of patients with an MND diagnosis. Clinical information, including certain MND-related mutations, was provided from a clinical registry/database of patients with MND with associated biobank (blood and CSF samples).
Patients
The study cohort consisted of patients referred for possible adult-onset MND to the Department of Neurology, Umeå University Hospital, Sweden, during the period 1994–2016. All patients were evaluated with a uniform diagnostic algorithm from the EFNS consensus criteria.7,8 With a written informed consent, as part of the diagnostic evaluation, an extra 4 mL of uncentrifuged CSF and paired ethylenediamine tetraacetic acid–blood samples (immediately centrifuged into plasma, buffy coat, and erythrocyte fractions) were collected and stored at −80°C. In the initial selection process, patients with a previous medical history of CNS infection, tuberculosis, neuroborreliosis, HIV, syphilis, systemic inflammatory, or neoplastic diseases or an ongoing medication with anti-inflammatory drugs were excluded. The total number of patients selected was 289, but 41 of these were excluded (for incomplete follow-up clinical data, insufficient biological material available, and/or alternative diagnosis). The resulting study population consisted of 248 treatment-naive patients with a clinical phenotype compatible with MND, including probable or definite ALS.7–9
Patients with MND were divided into (1) ALS with adult onset of initial progressive wasting and paresis of spinal-innervated skeletal muscles with definite signs of upper- and lower-motor neuron involvement with preserved or almost-preserved sensory and autonomic functions and (2) other MNDs (OMND; i.e., MND cases that for any reasons did not fulfill the above criteria).7,8
The ALS group was further divided into mSOD1, C9orf72HRE, and cases without mutations in these genes (other ALS [OALS]). An advantage of this study was the prospective clinical follow-up and evaluation of the enrolled patients by the same ALS team at regular intervals until the patient either died or received an alternative non-ALS diagnosis. Furthermore, this procedure facilitated the distinction of patients who had bulbar-onset variant of ALS and later developed generalized ALS from those who had progressive bulbar palsy (PBP) as a manifest condition. None of the included patients later received life-extending invasive ventilation treatment.
With a written informed consent, the screening for a panel of ALS-causing genes was performed using DNA extracted from peripheral blood leukocytes as described.10
Protein quantification in CSF and plasma
The assays for measuring TNF-α, IL-1β, IL-6, IL-10, IL-17A, TNF-related apoptosis-inducing ligand (TRAIL), and IFN-α were performed on a Simoa HD-1 Analyzer (Quanterix, Lexington, MA) using the manufacturer's instructions, reagents, and consumables. The instrument has previously been described in detail.11 For each assay run, 2 controls prepared from commercially available control material and 2 pools of >3 anonymized CSF/plasma samples were included as external and in-house quality controls, respectively (table e-1, links.lww.com/NXI/A204). The samples were defined as below lower limit of detection if the signal was below that of (blank + 3 × SD). Cytokines were detectable in >95% of individuals except for CSF IL-17A (39%), plasma IL-1β (69%), and IFN-α in CSF (31%) and plasma (58%). The staff was blinded to the group status of patients when performing the analyses. Analyses were performed in an accredited laboratory at the Department of Biochemistry & Immunology, Lillebaelt Hospital, Vejle, Denmark.
Statistics
The statistical analyses were performed using Stata IC 15 software package (StataCorp, 2017; Stata Statistical Software: Release 15; StataCorp LLC, College Station, TX). All tests were 2 tailed. The Fisher exact test was used to test for differences between distributions of sex or genotype as appropriate and Baptista and Pike' method used to calculate ORs and corresponding 95% CIs. SOD1 and C9orf72 genotypes were used for the substratification of patients. Direct group comparisons were made using the Mann-Whitney or Kruskal-Wallis test, followed by the Dunn post hoc test. Samples below limit of detection were tied at 0.00 pg/mL and included in nonparametric analyses. To address multiple testing, Benjamini-Krieger-Yekutieli correction was applied for serial Mann-Whitney or Dunn testing to obtain a corrected q value. For survival analyses, Cox regression was used to calculate the hazard ratio (HR) for a cytokine with the time frame being the day of biosampling (diagnostic evaluation) until the day of death or, when this information was unavailable, censoring at the latest known follow-up. Adjustment for patient age at the time of collecting of the samples was also completed. To control for any impact of long-term storage at −80°C on detected cytokine levels, bootstrap correlation was performed (500 iterations each). If a significant correlation was found (p < 0.05), robust regression was performed, and the regression coefficient was used as a conservative correction factor to obtain corrected cytokine measurements. Such correction was made for CSF TNF-α, IL-1β, and IL-10 and plasma IL-1β and TRAIL (table e-2, links.lww.com/NXI/A205).
Standard protocol approvals, registrations, and patient consents
Verbal and written informed consent for collecting blood and CSF samples was obtained from all participants. The study was approved by the Swedish Research Ethical Committees (J.no. FEK94-135 with updates, most recently EPN2018-496-32M). Patients were enrolled who qualify according to standard ethical approval and able to give verbal and written informed consent. As the cytokine laboratory work and data analysis were performed in Denmark, approval was also obtained from the Regional Ethics Committee for Southern Denmark (S-20150002 G) and Danish Data Protection Agency. The study was fully performed in accordance with the Declaration of Helsinki (WMA, 1964).
Data availability
Anonymized data of this study will be available from the corresponding author on reasonable request from any qualified investigator, following the EU General Data Protection Regulation.
Results
Demographics and clinical characteristics of patients with ALS and OMND
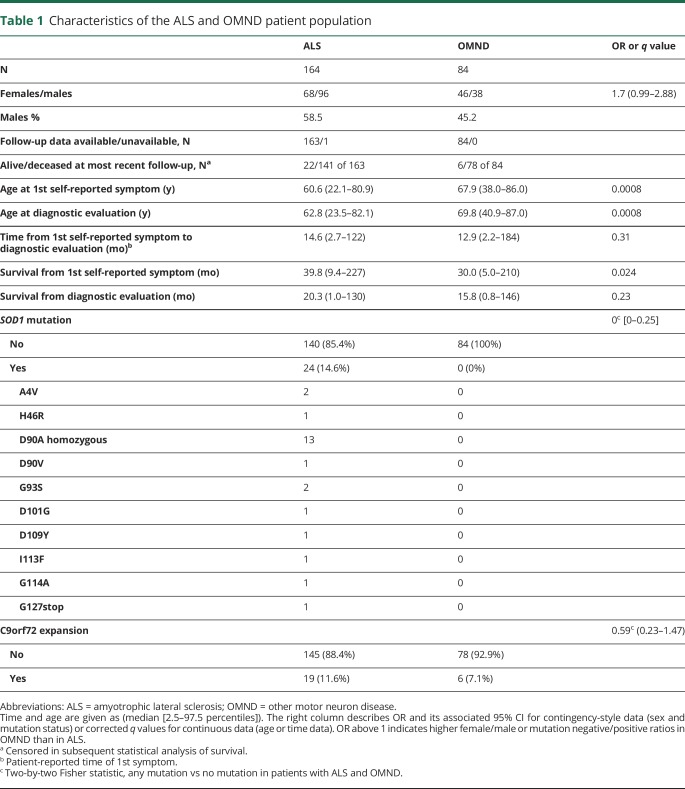
Of the 248 included patients with MND, 164 ended up being diagnosed with classical spinal-onset ALS, 2 had coexisting frontotemporal dementia (FTD), and 1 had Parkinson disease. The remaining 84 patients were categorized as OMND, including PBP (n = 57), progressive muscular atrophy (n = 10), primary lateral sclerosis (n = 2), spinal and bulbar muscular atrophy (DNA verified; n = 3), pseudobulbar palsy (n = 7), combinations of PBP with other neurodegenerative diseases (Parkinson disease, multiple system atrophy, or FTD, with 1 patient each), and atypical motor phenotypes (n = 2). The clinical and demographic data are presented in table 1. No difference was observed between males and females in terms of age at onset of symptoms or in the age at diagnostic evaluation (i.e., at biosampling, q = 0.70 for ALS, q = 0.99 for OMND; data not shown).
Table 1.
Characteristics of the ALS and OMND patient population
For deceased patients, we calculated the fraction of total disease duration that had passed before the diagnostic evaluation (i.e., [time from onset to diagnostic evaluation]/[total disease duration]). The result ranged from 4% to 98%–99%, and the median was 48% and 43% for ALS and OMND, respectively. Thus, our cohort represented both early disease (survival 3–10 years), intermediate (survival 1–3 years), and terminal stages (less than 12 months of survival). The prevalence and distribution of the investigated ALS-related mutations are presented in table 1. Briefly, pathogenic SOD1 mutations were detected in 24 of 164 investigated patients with ALS (14.6%) compared with none of the 84 patients with OMND (0%; p < 0.0001). The occurrence of C9orf72HRE was observed in 19 patients with ALS and 6 patients with OMND (5 with PBP and 1 with progressive muscular atrophy), not significantly different (11.6% and 7.1%, respectively; p = 0.37). One patient carried a possibly pathogenic VAPBMet170Ile mutation, and this patient was not included in genetic subgrouping. The sex distribution among patients with mutations was comparable to that of OALS (data not shown).
The median time from the first self-reported symptom to the time of diagnostic evaluation was longer in patients with mSOD1 ALS (43.7 months, 2.5th and 97.5th centiles: [2.3–267]) than in patients with OALS (14.2 months [3.6–98.7]; q < 0.0001) and C9orf72HRE ALS (11.9 months [2.5–24.5]; q < 0.0001). The median age at diagnostic evaluation was 54.9 (29.4–72.7) years for patients with mSOD1 ALS, which was significantly younger than that of patients with OALS (62.0 [21.2–81.5]; q = 0.006) but not relative to patients with C9orf72HRE ALS (61.8 years [46.2–75.4]; q = 0.11). In general, patients with ALS were younger at the time of diagnostic evaluation compared with those with OMND (69.8 years [40.9–86.4]; q < 0.0001), and this finding was true even for the oldest ALS patient group (OALS) compared with patients with OMND (q < 0.0001).
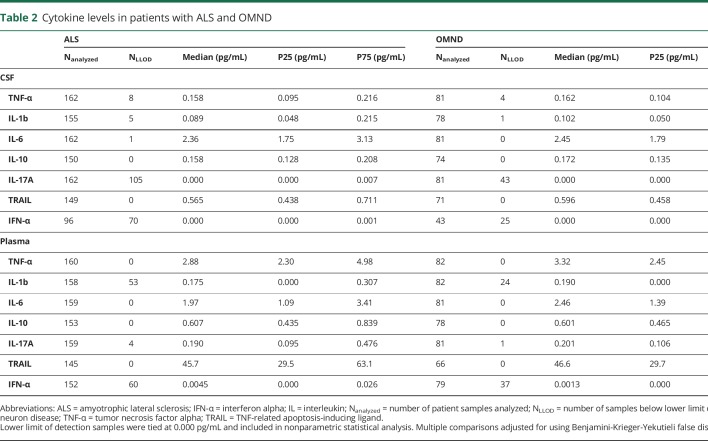
Cytokine levels in patients with ALS and OMND
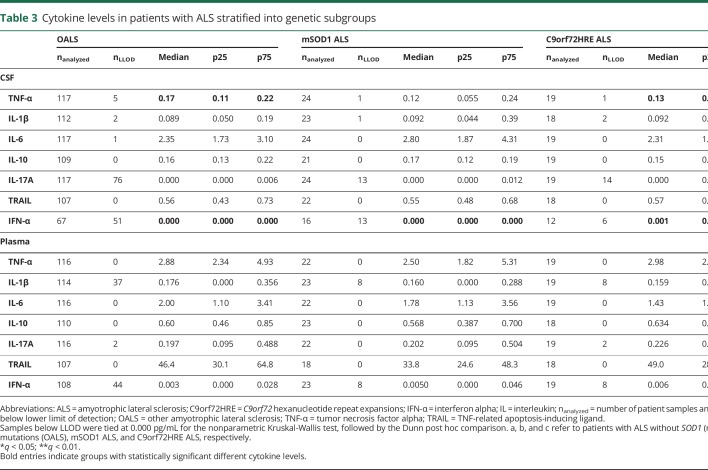
To address the differences in inflammatory processes in patients with ALS and OMND, a panel of cytokines in the plasma and CSF samples were studied. None of the measured cytokine levels in the CSF and plasma samples showed differences between patients with ALS and OMND (table 2). Comparing patients with ALS by their underlying mutations led to the observation that CSF TNF-α was increased in patients with OALS compared with patients with C9orf72HRE (q = 0.014). Finally, patients with C9orf72HRE ALS had higher CSF IFN-α compared with those with OALS and mSOD1 ALS (q = 0.042 and q = 0.042, respectively; table 3).
Table 2.
Cytokine levels in patients with ALS and OMND
Table 3.
Cytokine levels in patients with ALS stratified into genetic subgroups
We determined whether cytokine levels were associated with patients' sex and found no such association (not shown). We also correlated patients' age at the time of diagnostic evaluation with cytokine levels in the CSF and plasma samples. For patients with ALS, we found weak but significant correlations for CSF TNF-α and IL-10 and plasma TNF-α, IL-6, and IL-17A (Spearman rho: 0.16–0.27; not shown). For patients with OMND, a similar correlation test between age at the time of diagnostic evaluation and cytokine levels revealed a correlation for CSF TNF-α and IL-10 and plasma TNF-α, IL-6, and IL-17A (Spearman rho: 0.23–0.39; not shown). Thus, age likely influences CSF and plasma levels of a number of cytokines in patients with ALS and patients with OMND. In addition, the OALS subgroup of patients showed weak correlations for plasma levels of TNF-α, IL-6, IL-10, and IL-17A (table e-3, links.lww.com/NXI/A206). For mSOD1, correlation was found for CSF IL-6 and IL-10 (Spearman rho 0.42, 95% CI [0.054–0.78] and Spearman rho = 0.46 [0.034–0.88], respectively). In patients with C9orf72HRE ALS, CSF TRAIL was negatively correlated with age (Spearman rho = −0.58 [−0.29 to −0.86]). These findings indicate that unique inflammatory signatures may associate with the genetic subtypes of ALS.
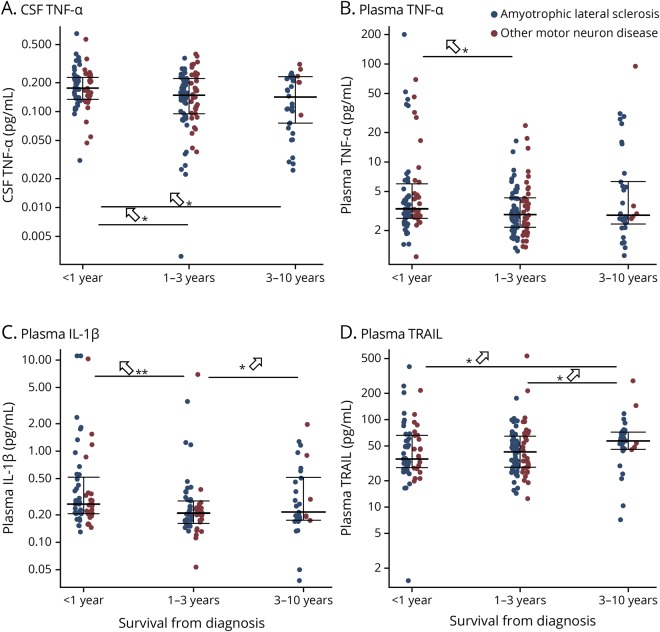
Among the deceased patients (n = 141 for ALS and n = 78 for OMND), cytokine levels were compared in patients who survived less than 12 months from the time of diagnostic evaluation (i.e., late stage disease) to those who had intermediate or early stage diseases (i.e., surviving 1–3 or 3–10 years). In the combined group of patients with ALS and OMND, patients who died within 1 year (n = 74) showed higher levels of 3 cytokines (CSF TNF-α: <1 year vs 1–3 years, q = 0.019; <1 year vs 3–10 years, q = 0.019; and plasma TNF-α: <1 year vs 1–3 years, q = 0.035; IL-1β: <1 year vs 1–3 years, q = 0.0002; 1–3 years vs 3–10 years, q = 0.0122) and reduced plasma TRAIL (<1 year vs 3–10 years, q = 0.020; 1–3 years vs 3–10 years, q = 0.020) compared with those surviving 1–3 years (n = 99) or 3–10 years (n = 46), total (n = 145; figure 1).
Figure 1. Cytokine levels in patients surviving <1 year, 1–3 years, or 3–10 years.
Deceased patients with amyotrophic lateral sclerosis (blue, n = 141), other motor neuron diseases (red, n = 78) were grouped into those surviving <1 year, 1–3 years, or 3–10 years, and cytokine levels compared by the Dunn test. (A) CSF tumor necrosis factor alpha (TNF-α), (B) plasma TNF-α, (C) plasma interleukin (IL)-1β, (D) plasma TNF-related apoptosis-inducing ligand. *p < 0.05. **p < 0.01. Upward arrows indicate which group has the highest relative level. IL = interleukin; TNF-α = tumor necrosis factor alpha; TRAIL = TNF-related apoptosis-inducing ligand.
Disease survival and association with cytokines
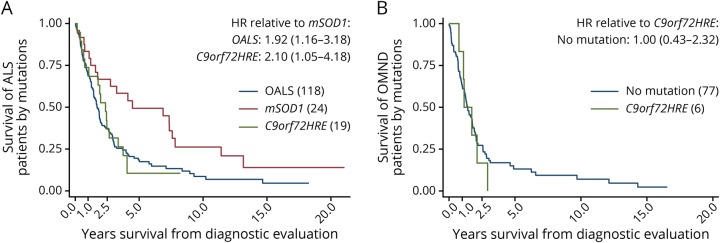
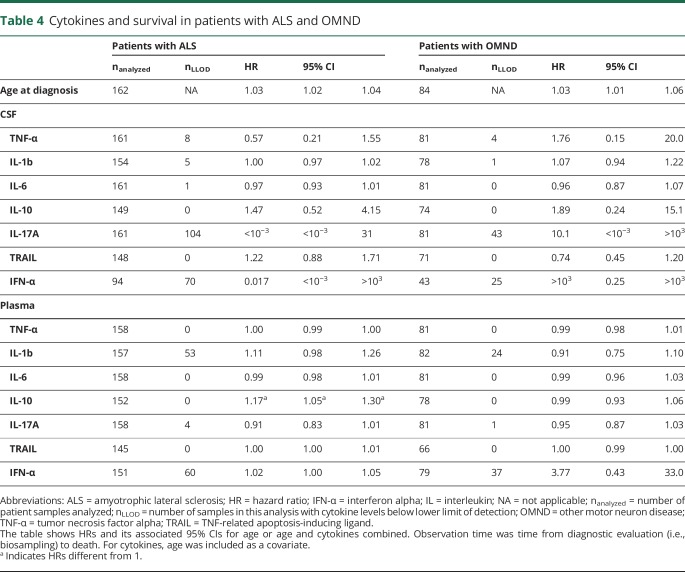
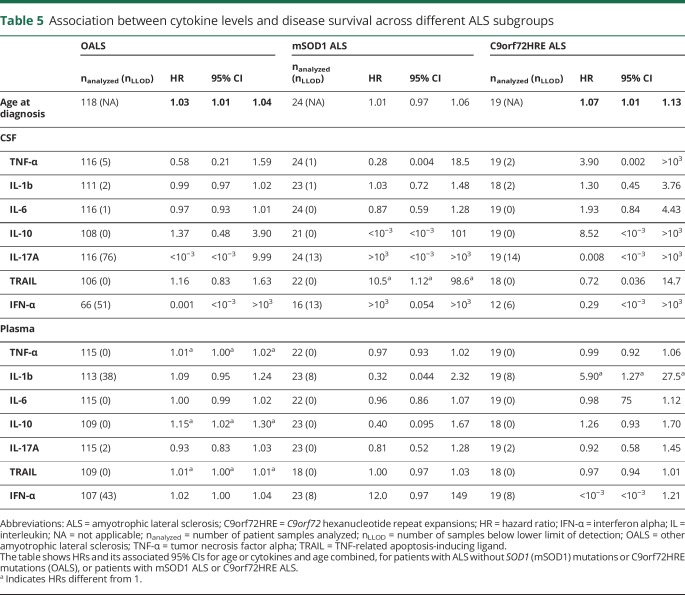
Patients with ALS and OMND showed a similar delay from the first symptom to diagnostic evaluation (table 1). By contrast, survival in ALS was better than in OMND when considering survival time from the first symptom and time of diagnostic evaluation. The median survival time from the first self-reported symptom was higher in patients with ALS (39.8 months, 2.5 and 97.5 percentiles [9.4–227]) than those with OMND (30.0 months [5.0–210]; q = 0.024, table 1). The median survival time from time of diagnostic evaluation was similar between patients with ALS (20.3 months [1.0–130]) and OMND (15.8 months [0.8–146]; q = 0.23, table 1). Within the ALS patient group, survival from the first self-reported symptom was longer in patients with mSOD1 (8.5 years [0.9–26.4]) compared with those with C9orf72HRE ALS and OALS (3.1 years [1.0–5.8] and 3.0 years [0.7–12.6]; p = 0.0020 and p = 0.0008, respectively; not shown). Survival from the time of diagnostic evaluation was shorter in patients with OALS (median survival 18.3 months, 2.5 and 97.5 percentiles [0.7–116] and HR 1.92 [1.16–3.18]) and C9orf72HRE ALS (23.4 months [1.7–48.7] and HR 2.10 [1.05-4.18]) than mSOD1 ALS (38.9 months [1.7–158]; table 1 and figure 2). Survival was particularly heterogeneous between different SOD1 mutations. The D90A homozygous patients, which were also the most frequent carriers in the cohort, were mainly responsible for the wide span in the survival of patients with mSOD1.12 In general, age was a significant factor for survival in ALS and OMND (HR = 1.03; 95% CI [1.02–1.04], and HR = 1.03; 95% CI [1.01–1.06]; table 4). Therefore, in all subsequent Cox regressions, age was used as a covariate. In the general ALS patient group (i.e., independently on underlying mutations), we observed an increased HR for the plasma levels of IL-10 (HR = 1.17; 95% CI: 1.05–1.30; table 4). Next, we identified whether a gene-specific association exists between cytokines and survival. Again, age was a significant factor for the survival of patients with OALS and C9orf72HRE but not for patients with mSOD1 ALS (table 5). When cytokine-specific effects on survival were stratified by gene mutations, IL-10 appeared as a significant factor for patients with OALS (HR = 1.15; 95% CI: 1.02–1.30; table 5) but not for patients with mSOD1 ALS and C9orf72HRE ALS. Thus, the effect of plasma IL-10 found for all patients with ALS irrespective of genotype (table 4) was probably derived from the OALS subgroup. Similar effects were found for plasma TNF-α and TRAIL in patients with OALS (HR = 1.01; 95% CI: 1.00–1.02 and HR = 1.01; 95% CI: 1.00–1.01, respectively). Patients with mSOD1 ALS showed only a strong association with CSF TRAIL (HR = 10.5; 95% CI: 1.12–98.6). In patients with C9orf72HRE, plasma IL-1β appeared related with survival (HR = 5.90; 95% CI: 1.27–27.5), a phenomenon not seen in patients with OALS or mSOD1 ALS. In summary, the data showed significant differences between OALS and the genetic subtypes of ALS in their cytokine reactivity.
Figure 2. Survival of patients with genetic subtypes of ALS and OMND.
(A) The prognosis for patients with different subtypes of ALS is heterogeneous, as survival was shortest in patients carrying C9orf72HRE, followed by cases without mutations in the SOD1 and C9orf72 genes (OALS), and longest for patients with a pathogenic SOD1 mutation (mSOD1). (B) Patients with OMND have similar survival whether carrying a C9orf72 mutation or not. p Values indicate the log-rank test for equality of survivor functions. ALS = amyotrophic lateral sclerosis; HR = hazard ratio; C9orf72HRE = C9orf72 hexanucleotide repeat expansions; OALS = other amyotrophic lateral sclerosis; OMND = other motor neuron disease.
Table 4.
Cytokines and survival in patients with ALS and OMND
Table 5.
Association between cytokine levels and disease survival across different ALS subgroups
Discussion
In the current study, we provide evidence for the effects of age, mutations in ALS-related genes, and cytokine levels on survival. The data indicate that differences in cytokine levels across subtypes of ALS are correlated with survival in a genotype-specific manner, suggesting specific immunopathologic processes related to ALS genetic variants.
Immunopathologic processes related to ALS genetic variants
Previous studies have reported age as a negative prognostic factor for ALS.13 In this study, decreased survival time with increasing age was observed both for patients with OMND and ALS after stratification for gene mutations except in patients with mSOD1 ALS, suggesting that SOD1 mutations as such are a dominating modifying factor. Moreover, the cytokine levels correlated with age. This characteristic may both arise from the pathogenic mechanisms at play in ALS and be a reflection of aging processes, which contribute to cytokine dysregulation.
Cytokines regulate immune responses and have been linked to the process of neurodegeneration, as drivers of the inflammatory cascade contributing to neuronal death.14 Emerging evidence from experimental and clinical observations suggests that inflammation plays a role in the pathogenesis of ALS.15 It is still uncertain whether the action of cytokines is a primary contributory process or a secondary phenomenon as a consequence of motor neuron death. Various types of neuronal stress and death-inducing mechanisms will trigger different immune pathways in the microenvironment, for instance through activation of various innate receptors.16,17 In mSOD1G37R mice, reducing transgene expression in CD11b+ microglia/macrophages markedly ameliorated late-stage pathology without altering time of clinical onset.18 These data support that neurons are the primary targets during pathology, but that neuron-microglia crosstalk is mechanistically important during progression, and that disease-causing processes in neurons could give rise to certain immunologic fingerprints through downstream induction of different stimuli. In the current study, the survival was negatively associated with plasma levels of IL-10 in patients with ALS. Furthermore, patients who died within 1 year had higher levels of TNF-α in CSF and of TNF-α and IL-1β in plasma, suggesting that these inflammatory markers may serve as negative prognostic markers. Several studies indicate that IL-10 plays a role in inflammatory diseases, such as MS, systemic lupus erythematosus, rheumatoid arthritis, Crohn disease, and psoriasis.19 The survival time in various hematologic malignancies and ovarian cancer has been reported to be negatively correlated with the levels of IL-10.20,21 In ALS, the role of IL-10 has only been studied in a small cohort. One previous retrospective study of 29 patients found that higher plasma IL-10 levels predicted longer disease duration, with a median sampling delay from onset of 17.7 months (range 4.1–189.5 months),22 which is at variance with our findings among 164 patients with ALS. An explanation could be the limited sample size of 29 patients and the subsequent risk of type II error. Furthermore, the previous study defined total disease duration from symptom onset to death,22 whereas we considered from the day of biosampling (initiation of diagnostic process) until death. It could be speculated that the levels of IL-10 change during the disease course as a compensatory mechanism to counteract excess proinflammatory stimuli. The molecular mechanisms regulating IL-10 production by CNS cells, i.e., microglia, astrocytes, and neurons, are sparsely known.23,24 The mechanism of action of IL-10 can include differentiation of regulatory microglia/macrophages that weakens the inflammatory response. Remarkably, in the autopsy materials from the patients with ALS, strong IL-10 immunoreactivity colocalizes with inclusions containing TAR DNA-binding protein (TDP)-43 in motor neurons, supporting the concept that IL-10 is involved in the pathogenesis of ALS.25–27
Specifically, for patients with C9orf72HRE ALS, we observed an association between plasma IL-1β and survival, indicating the involvement of IL-1β in this subset of patients, but not in mSOD1 ALS or OALS. This finding may add to an explanation for the poor prognosis among patients with C9orf72HRE ALS. Of interest, an animal study using C9orf72 knockout mice demonstrated disrupted microglial function, which was linked to neuroinflammation with increased levels of cytokines IL-6 and IL-1β.28 Similar to other neurodegenerative diseases, intracellular protein aggregates are a major characteristic pathologic hallmark in ALS, with TDP-43 protein as a major component.16,29 Of interest, phosphorylated TDP-43 may be identified in the skeletal muscle of patients with ALS as in patients with inclusion body myositis.30 A study reported that exposure of microglial cells to TDP-43 aggregates resulted in the secretion of proinflammatory cytokines including TNF-α and IL-1β, which suggests that TDP-43–mediated signaling contribute to microglial neurotoxic activation in ALS.31 Another recent study of C9orf72HRE knock-in mice highlighted increased innate immune signaling without mentioning specific cytokines/pathways, but IL-1β is central in innate immunity, and produced by all CNS-resident cells downstream of inflammasome signaling.32,33 This could suggest overactivation of inflammasomes as a mechanism behind the observed IL-1β associations. Supporting this, immunopathologic studies of microglia in tissues of patients with C9orf72HRE and non-C9orf72HRE ALS indicate a contribution of C9orf72HRE to the neurodegenerative process, which is paralleled by enhanced inflammation in such patients.34 Recently, it has been suggested that long double-stranded RNA (dsRNA) is a transcription product of the C9orf72HRE gene mutation, and such dsRNA was shown in the neuronal cytoplasm of C9orf72HRE patient brains, coinciding with upregulation of dsRNA sensors that are key in innate immune sensing of viral infections.35 Our finding of increased CSF IFN-α in C9orf72HRE suggests that detrimental antiviral processes are specifically induced by the C9orf72HRE mutation.
In the present study, after stratification for gene mutations, the plasma levels of IL-10, TNF-α, and TRAIL correlated negatively with survival in patients with OALS, suggesting a potential prognostic value of these cytokines for specific subtypes of patients with ALS. Clinical and immunopathologic observations from human and experimental studies support that TNF-α, TRAIL, and their respective receptors are involved in the pathogenesis of ALS and are directly implicated in the motor neuron death via the receptor-mediated induction of apoptosis.6,36,37 However, in previous clinical studies, TNF-α did not correlate with disease duration.38 We also observed higher levels of TNF-α in the end-stage disease of MND, including patients with ALS, paralleled with reduced TRAIL, suggesting that these have a prominent role as prognostic markers. TNF-α and TRAIL belong to the TNF superfamily of cytokines, recognized by their role in controlling survival, proliferation, and cell death.39 This suggests that apoptotic signaling balance is disrupted during late-stage disease, as reflected by an increased TNF-α:TRAIL balance. Collectively, we have identified potential biomarker candidates that are associated with prognosis of ALS and may be useful as therapeutic targets. Clinical trials suggest that IFN-α analogues are not effective in the treatment of ALS,40,41 despite the suggestion that activation of the IFN pathway is linked to disease duration.4 In pilot studies, IV immunoglobulin (IVIg) has been used to treat ALS.42,43 The mechanisms of IVIg therapy include downregulation of cytokine networks; however, no evidence has been provided on any effect on disease progression in patients with ALS. Furthermore, microglial activation and upregulation of TNF-α and IL-6 expression are involved in the proinflammatory mechanisms counteracting ALS pathology and have been suggested as potential therapeutic targets for ALS.37,44,45 Tocilizumab (anti-IL6) was studied in inflammatory gene transcription in peripheral blood mononuclear cells from a limited number of patients with ALS and controls. Infusions of tocilizumab in the patients with ALS with strong inflammatory activation led to downregulation of inflammatory genes including IL1β. TNF-α signaling inhibitors, e.g., thalidomide, may also represent a potential target for intervention. However, solid evidence has not so far appeared for the utility of inflammation-related treatment for ALS, which may reflect multiple and complex pathogenic mechanisms.
One important strength of our analysis was that laboratory investigations were performed blindly using automated techniques to strengthen data reliability. The quantification of CSF cytokines has until recently presented a technical limitation. However, the ultra-sensitive Simoa technology has led to a substantially improved capability to detect cytokines in the etiology of nonautoimmune diseases, such as ALS.46 Furthermore, this study was performed in a large treatment-naive patient cohort with associated information on clinical and genetic mutations and a longitudinal prospective follow-up. A limitation was the cross-sectional design of our study instead of a longitudinal design with multiple samples; however, our cohort represented all disease stages. A large multipopulation study designed to validate these observations would be an obvious continuation. In conclusion, we found evidence that cytokine levels associate with ALS disease duration in a genotype-specific manner, indicating that specific immunopathologic processes may be at play in the genetic subtypes of ALS and contributing to neuronal death. An aim for future study could be to investigate whether neuronal-glial signaling influences disease progression at different stages of the disease process.
Acknowledgment
The authors are indebted to all the patients who made this study possible. They thank Eva Bern, Helena Alstermark, Ann-Charloth Nilsson, and Eva Jonsson for technical assistance. They also thank Carsten Bisgaard, MD, for inspiring the study design and for creating facilities for patient investigation. Sara Egsgaard, Camilla Davidsen, and Dorte Aalund Olsen at Biochemistry & Immunology, Lillebaelt Hospital, Vejle, and the Autoimmunity Lab at the Department of Clinical Immunology, Odense University Hospital, are acknowledged for their technical assistance.
Glossary
- ALS
amyotrophic lateral sclerosis
- dsRNA
double-stranded RNA
- FTD
frontotemporal dementia
- HR
hazard ratio
- HRE
hexanucleotide repeat expansion
- IFN
interferon
- IFN-α
interferon alpha
- IL
interleukin
- IVIg
IV immunoglobulin
- MND
motor neuron disease
- OALS
other amyotrophic lateral sclerosis
- OMND
other motor neuron diseases
- PBP
progressive bulbar palsy
- TNF
tumor necrosis factor
- TNF-α
tumor necrosis factor alpha
- TRAIL
TNF-related apoptosis-inducing ligand
Appendix. Authors
Study funding
The University of Southern Denmark, Region of Southern Denmark, Merchant L. F. Foght's Foundation, the Swedish Research Council (grants. 2012-0262, 2012-0305, 2013-0279, and 2016-0303), the Swedish Science Council (grants 2012-3167 and 2017-03100), the Knut and Alice Wallenberg Foundation (grants 2012.0091 and 2014.0305), the Bertil Hållsten Foundation, the Ulla-Carin Lindquist Foundation, the Kempe foundation, the Neuroförbundet Association, the Torsten and Ragnar Söderberg Foundation, Umeå University Insamlingsstiftelsen (223-2808-12, 223-1881-13, 2.1.12-1605-14), and the Västerbotten County Council (grants nr 56103-7002829).
Disclosure
The authors report no disclosures. Go to Neurology.org/NN for full disclosures.
References
- 1.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol 2009;65(suppl 1):S3–S9. [DOI] [PubMed] [Google Scholar]
- 2.Zou ZY, Zhou ZR, Che CH, Liu CY, He RL, Huang HP. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 2017;88:540–549. [DOI] [PubMed] [Google Scholar]
- 3.Owens T, Khorooshi R, Wlodarczyk A, Asgari N. Interferons in the central nervous system: a few instruments play many tunes. Glia 2014;62:339–355. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Yang B, Zhang D. Activation of interferon signaling pathways in spinal cord astrocytes from an ALS mouse model. Glia 2011;59:946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Ona VO, Guégan C, et al. Functional role of caspase-1 and caspase-3 in an ALS transgenic mouse model. Science 2000;288:335–339. [DOI] [PubMed] [Google Scholar]
- 6.Tortarolo M, Lo Coco D, Veglianese P, et al. Amyotrophic lateral sclerosis, a multisystem pathology: insights into the role of TNFalpha. Mediators Inflamm 2017;2017:2985051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks BR, Miller RG, Swash M, Munsat TL; World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 8.Andersen PM, Abrahams S, Borasio GD, et al. EFNS guidelines on the clinical management of amyotrophic lateral sclerosis (MALS)—revised report of an EFNS task force. Eur J Neurol 2012;19:360–375. [DOI] [PubMed] [Google Scholar]
- 9.Ludolph A, Drory V, Hardiman O, et al. A revision of the El Escorial criteria—2015. Amyotroph Lateral Scler Frontotemporal Degener 2015;16:291–292. [DOI] [PubMed] [Google Scholar]
- 10.Keskin I, Birve A, Berdynski M, et al. Comprehensive analysis to explain reduced or increased SOD1 enzymatic activity in ALS patients and their relatives. Amyotroph Lateral Scler Frontotemporal Degener 2017;18:457–463. [DOI] [PubMed] [Google Scholar]
- 11.Wilson DH, Rissin DM, Kan CW, et al. The Simoa HD-1 analyzer: a novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J Lab Autom 2016;21:533–547. [DOI] [PubMed] [Google Scholar]
- 12.Andersen PM, Forsgren L, Binzer M, et al. Autosomal recessive adult-onset amyotrophic lateral sclerosis associated with homozygosity for Asp90Ala CuZn-superoxide dismutase mutation: a clinical and genealogical study of 36 patients. Brain 1996;119:1153–1172. [DOI] [PubMed] [Google Scholar]
- 13.Chio A, Logroscino G, Hardiman O, et al. Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler 2009;10:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L, Pei L, Yao S, Wu Y, Shang Y. NLRP3 inflammasome in neurological diseases, from functions to therapies. Front Cell Neurosci 2017;11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sta M, Sylva-Steenland RMR, Casula M, et al. Innate and adaptive immunity in amyotrophic lateral sclerosis: evidence of complement activation. Neurobiol Dis 2011;42:211–220. [DOI] [PubMed] [Google Scholar]
- 16.Parakh S, Atkin JD. Protein folding alterations in amyotrophic lateral sclerosis. Brain Res 2016;1648:633–649. [DOI] [PubMed] [Google Scholar]
- 17.Van Damme P, Robberecht W, Van Den Bosch L. Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis Model Mech 2017;10:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boillee S, Yamanaka K, Lobsiger CS, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 2006;312:1389–1392. [DOI] [PubMed] [Google Scholar]
- 19.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol 2004;22:929–979. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Wang L, Chi PD, et al. High level of interleukin-10 in serum predicts poor prognosis in multiple myeloma. Br J Cancer 2016;114:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Liu W, Wang X, Wang X, Sun H. Prognostic value of serum IL-8 and IL-10 in patients with ovarian cancer undergoing chemotherapy. Oncol Lett 2019;17:2365–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su XW, Simmons Z, Mitchell RM, Kong L, Stephens HE, Connor JR. Biomarker-based predictive models for prognosis in amyotrophic lateral sclerosis. JAMA Neurol 2013;70:1505–1511. [DOI] [PubMed] [Google Scholar]
- 23.Lobo-Silva D, Carriche GM, Castro AG, Roque S, Saraiva M. Balancing the immune response in the brain: IL-10 and its regulation. J Neuroinflammation 2016;13:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev 2011;91:461–553. [DOI] [PubMed] [Google Scholar]
- 25.Berjaoui S, Povedano M, Garcia-Esparcia P, Carmona M, Aso E, Ferrer I. Complex inflammation mRNA-related response in ALS is region dependent. Neural Plast 2015;2015:573784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon GS, Shim YM, Lee DY, et al. Pathological modification of TDP-43 in amyotrophic lateral sclerosis with SOD1 mutations. Mol Neurobiol 2019;56:2007–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conlon EG, Fagegaltier D, Agius P, et al. Unexpected similarities between C9ORF72 and sporadic forms of ALS/FTD suggest a common disease mechanism. Elife 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burberry A, Suzuki N, Wang JY, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 2016;8:347ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blokhuis AM, Groen EJN, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 2013;125:777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cykowski MD, Powell SZ, Appel JW, Arumanayagam AS, Rivera AL, Appel SH. Phosphorylated TDP-43 (pTDP-43) aggregates in the axial skeletal muscle of patients with sporadic and familial amyotrophic lateral sclerosis. Acta Neuropathol Commun 2018;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W, Beers DR, Bell S, et al. TDP-43 activates microglia through NF-kappaB and NLRP3 inflammasome. Exp Neurol 2015;273:24–35. [DOI] [PubMed] [Google Scholar]
- 32.Hao Z, Liu L, Tao Z, et al. Motor dysfunction and neurodegeneration in a C9orf72 mouse line expressing poly-PR. Nat Commun 2019;10:2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voet S, Srinivasan S, Lamkanfi M, van Loo G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol Med 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brettschneider J, Toledo JB, Van Deerlin VM, et al. Microglial activation correlates with disease progression and upper motor neuron clinical symptoms in amyotrophic lateral sclerosis. PLoS One 2012;7:e39216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez S, Schrank BR, Sahin A, et al. Genome-encoded cytoplasmic double-stranded RNAs, found in C9ORF72 ALS-FTD brain, provoke propagated neuronal death. bioRxiv 2018:248328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang YM, Yamamoto M, Tanaka F, et al. Gene expressions specifically detected in motor neurons (dynactin 1, early growth response 3, acetyl-CoA transporter, death receptor 5, and cyclin C) differentially correlate to pathologic markers in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 2007;66:617–627. [DOI] [PubMed] [Google Scholar]
- 37.Olmos G, Llado J. Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediators Inflamm 2014;2014:861231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poloni M, Facchetti D, Mai R, et al. Circulating levels of tumour necrosis factor-alpha and its soluble receptors are increased in the blood of patients with amyotrophic lateral sclerosis. Neurosci Lett 2000;287:211–214. [DOI] [PubMed] [Google Scholar]
- 39.Sonar S, Lal G. Role of tumor necrosis factor superfamily in neuroinflammation and autoimmunity. Front Immunol 2015;6:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mora JS, Munsat TL, Kao KP, et al. Intrathecal administration of natural human interferon alpha in amyotrophic lateral sclerosis. Neurology 1986;36:1137–1140. [DOI] [PubMed] [Google Scholar]
- 41.Dalakas MC, Aksamit AJ, Madden DL, Sever JL. Administration of recombinant human leukocyte alpha 2-interferon in patients with amyotrophic lateral sclerosis. Arch Neurol 1986;43:933–935. [DOI] [PubMed] [Google Scholar]
- 42.Dalakas MC, Stein DP, Otero C, Sekul E, Cupler EJ, McCrosky S. Effect of high-dose intravenous immunoglobulin on amyotrophic lateral sclerosis and multifocal motor neuropathy. Arch Neurol 1994;51:861–864. [DOI] [PubMed] [Google Scholar]
- 43.Meucci N, Nobile-Orazio E, Scarlato G. Intravenous immunoglobulin therapy in amyotrophic lateral sclerosis. J Neurol 1996;243:117–120. [DOI] [PubMed] [Google Scholar]
- 44.Stommel EW, Cohen JA, Fadul CE, et al. Efficacy of thalidomide for the treatment of amyotrophic lateral sclerosis: a phase II open label clinical trial. Amyotroph Lateral Scler 2009;10:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam L, Halder RC, Montoya DJ, et al. Anti-inflammatory therapies of amyotrophic lateral sclerosis guided by immune pathways. Am J Neurodegener Dis 2015;4:28–39. [PMC free article] [PubMed] [Google Scholar]
- 46.Kothur K, Wienholt L, Brilot F, Dale RC`. CSF cytokines/chemokines as biomarkers in neuroinflammatory CNS disorders: a systematic review. Cytokine 2016;77:227–237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data of this study will be available from the corresponding author on reasonable request from any qualified investigator, following the EU General Data Protection Regulation.