Abstract
Objective
In 2016, Catalonia experienced a pediatric brainstem encephalitis outbreak caused by enterovirus A71 (EV-A71). Conventional testing identified EV in the periphery but rarely in CSF. Metagenomic next-generation sequencing (mNGS) and CSF pan-viral serology (VirScan) were deployed to enhance viral detection and characterization.
Methods
RNA was extracted from the CSF (n = 20), plasma (n = 9), stool (n = 15), and nasopharyngeal samples (n = 16) from 10 children with brainstem encephalitis and 10 children with meningitis or encephalitis. Pathogens were identified using mNGS. Available CSF from cases (n = 12) and pediatric other neurologic disease controls (n = 54) were analyzed with VirScan with a subset (n = 9 and n = 50) validated by ELISA.
Results
mNGS detected EV in all samples positive by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (n = 25). In qRT-PCR-negative samples (n = 35), mNGS found virus in 23% (n = 8, 3 CSF samples). Overall, mNGS enhanced EV detection from 42% (25/60) to 57% (33/60) (p-value = 0.013). VirScan and ELISA increased detection to 92% (11/12) compared with 46% (4/12) for CSF mNGS and qRT-PCR (p-value = 0.023). Phylogenetic analysis confirmed the EV-A71 strain clustered with a neurovirulent German EV-A71. A single amino acid substitution (S241P) in the EVA71 VP1 protein was exclusive to the CNS in one subject.
Conclusion
mNGS with VirScan significantly increased the CNS detection of EVs relative to qRT-PCR, and the latter generated an antigenic profile of the acute EV-A71 immune response. Genomic analysis confirmed the close relation of the outbreak EV-A71 and neuroinvasive German EV-A71. A S241P substitution in VP1 was found exclusively in the CSF.
In early 2016, an outbreak of enterovirus A71 (EV-A71) in Catalonia caused more than 100 pediatric cases of neurologic disease, ranging from aseptic meningitis to brainstem encephalitis with or without myelitis.1 Quantitative reverse transcription polymerase chain reaction (qRT-PCR) for EV with genotyping of peripheral blood, respiratory, and gastrointestinal samples, but not CSF, identified EV-A71 in 40 of 57 subjects. Other EVs were also found in 7 of 57 subjects, including echovirus 30 (E-30), coxsackievirus (CV) B1, and CV A10. Although unable to type the EVs, the BioFire FilmArray Meningitis/Encephalitis panel detected EV in the CSF of 4 of 20 subjects with brainstem encephalitis.2 The EV-A71 strain detected in this study was subtyped as subgenogroup C1, and phylogenetic analyses showed it was closely related to an EV-A71 strain associated with a 2015 German case of brainstem encephalitis.3–5 EV-A71 and other EV-related neurologic disease outbreaks were also reported in France and Denmark in 2016.6,7 Although EV-A71 was detected in peripheral body sites of many of these cases, EV-A71 was only identified in the CSF of 0.02% of subjects in the German study, 3% in the Danish study, and 14% in the French study.
Despite the presence of EV in peripheral samples, questions remained about the Catalonia outbreak. Because pan-EV qRT-PCR was negative in the CSF of subjects with brainstem encephalitis,1,8 concern persisted that neurologic symptoms may have been caused by a parainfectious mechanism or an unidentified coinfection. In addition, standard qRT-PCR assays on peripheral samples recovered small segments of the EV-A71 genome, thereby limiting the ability to assess for viral mutations that may have modulated neurovirulence.
To address these concerns, we deployed metagenomic next-generation sequencing (mNGS) on samples from the 2016 Catalonia outbreak. mNGS is an unbiased assay that can simultaneously identify nucleic acid from viruses, fungi, bacteria, and parasites in subjects with neurologic symptoms. As opposed to traditional PCR assays that amplify limited and usually highly conserved regions of a microbe's genome, the entire genome of a pathogen can often be recovered with mNGS. This makes it possible to identify genomic changes in the virus that may correlate with increased neurovirulence or reveal strain divergence.9 Here, we deployed mNGS on CSF, nasopharyngeal (NP), plasma, and stool samples from children affected by neurologic disease during the 2016 Catalonia outbreak. This allowed us to screen for all EVs and for possible coinfections while simultaneously comparing EV genomes for any differences between the subjects with brainstem encephalitis and other manifestations of neurologic disease (i.e., meningitis with or without encephalitis with self-limited and short-lasting symptoms). We supplemented our investigations with an enhanced version of a previously published assay that comprehensively assesses for antiviral antibodies using phage display (VirScan) and performed orthogonal confirmation with EV ELISA.10–12
Methods
Cohort
The cohort consisted of 20 pediatric cases of EV-related neurologic diseases. Specifically, we selected 10 children diagnosed with brainstem encephalitis or encephalomyelitis and 10 children with aseptic meningitis or uncomplicated encephalitis. Cases were recruited from the Hospital Sant Joan de Deu, University of Barcelona, between April and June 2016. The Hospital Sant Joan de Deu is a 300-bed tertiary care hospital for high-complexity patients across a catchment area with a pediatric population of ∼300,000 and has participated in a Spanish EV molecular surveillance network since 2010. EV-related neurologic disease was defined as the detection of EV by a pan-EV qRT-PCR assay or BioFire FilmArray Meningitis/Encephalitis panel8,13,14 in at least one sample from a subject suffering acute neurologic disease in the absence of another clear cause.1 Case definitions of the World Health Organization's A Guide to Clinical Management and Public Health Response for Hand, Foot and Mouth Disease (HFMD), detailed in table e-1 (links.lww.com/NXI/A216), were followed by a group of trained pediatricians to classify cases.
Case demographics and clinical syndromes are described in table 1. Deidentified samples from encephalomyelitis/brainstem encephalitis (n = 10) and meningitis/encephalitis (n = 10) were transferred to UCSF on dry ice and stored at −80°C. The specimens from cases included CSF, serum, NP, and stool samples (figure 1 and table e-2, links.lww.com/NXI/A216). Among the 60 samples, 35 were EV-negative by clinical pan-EV qRT-PCR (table e-2). The subjects 1–10 were negative by the BioFire FilmArray Meningitis/Encephalitis panel.2 All EV-positive subjects (by clinical PCR) were genotyped at the Enterovirus Unit of the Spanish National Centre for Microbiology according to a previously described procedure.15 For VirScan controls, we used CSF from pediatric subjects with other neurologic diseases (ONDs). VirScan and available clinical data have been previously published on this control cohort.10
Table 1.
Summary of subject group demographics and clinical data
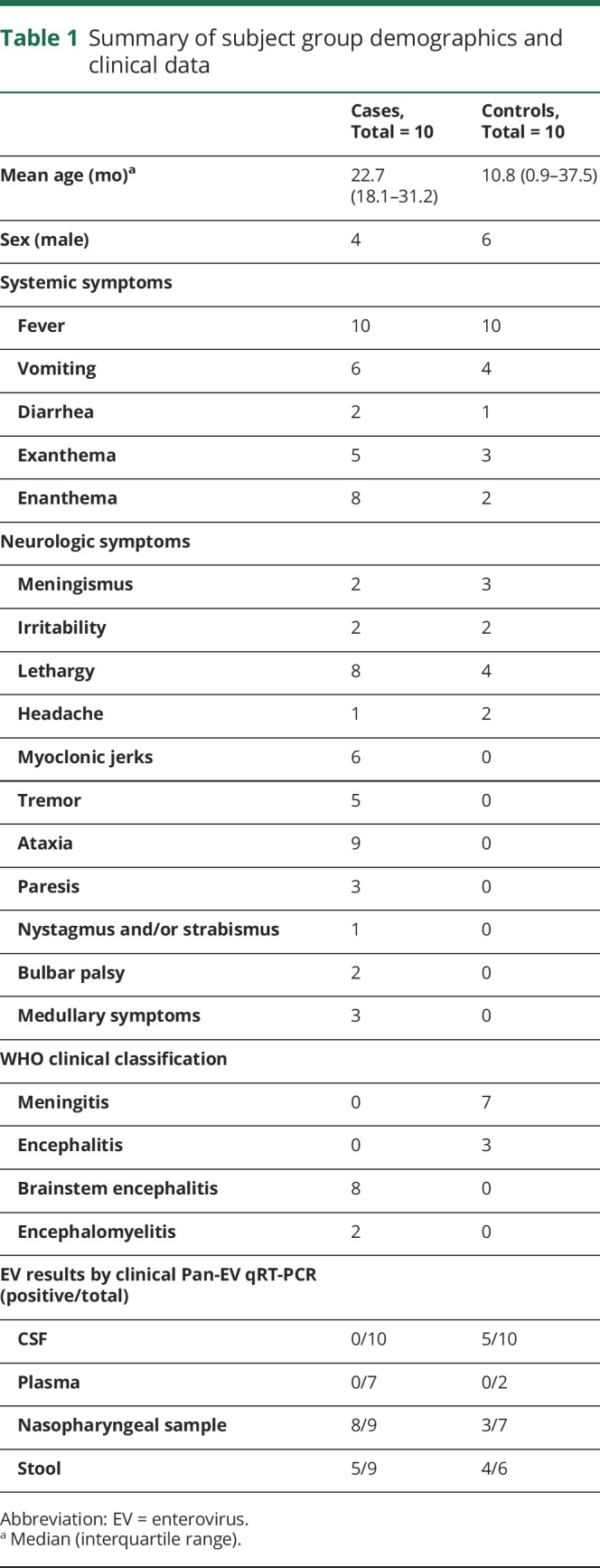
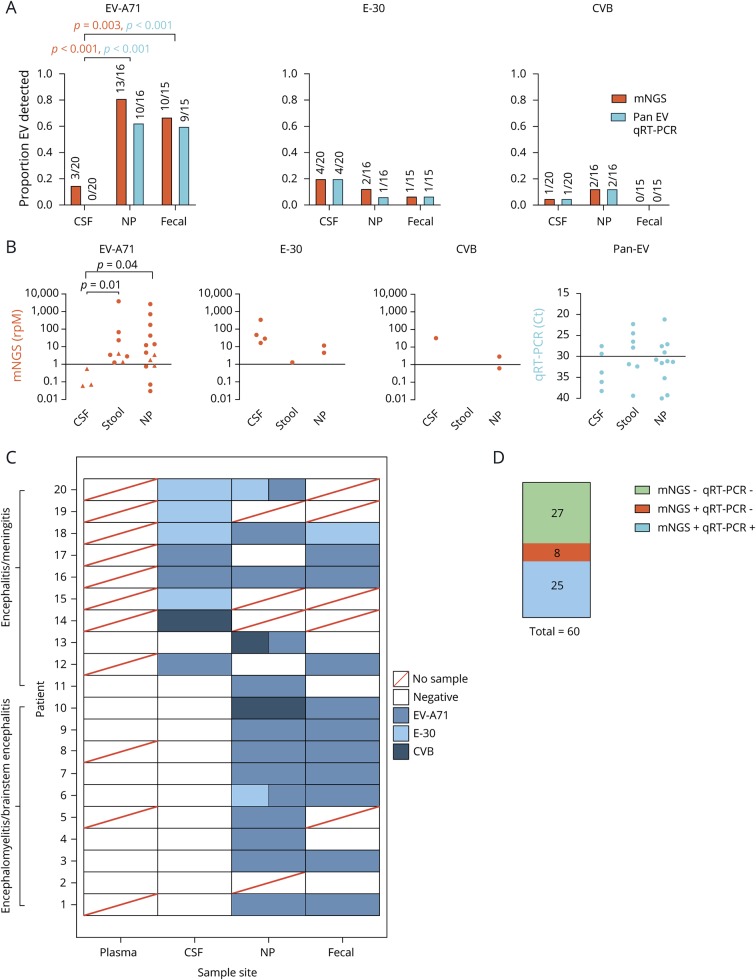
Figure 1. Summary of mNGS diagnostics: improvement over traditional clinical testing.
(A) Comparison of mNGS and qRT-PCR detection of EV-A71, E-30 and CVB in CSF, NP, and fecal samples. Statistics performed using the Fisher exact test, with orange and blue p-values corresponding with mNGS and qRT-PCR results, respectively. (B) Comparison of detection levels for each different experiment, with rpM representing mNGS and Ct values for qRT-PCR. Triangles denote samples identified by mNGS but not qRT-PCR. Statistics performed using a Mann-Whitney test. (C) Heatmap of each individual subject with each body site represented. Boxes with 2 colors represent codetections of different viral species. (D) Comparison of mNGS detection rates to qRT-PCR. Statistics performed using the McNemar test. CVB = Coxsackievirus B; E-30 = echovirus 30; EV-A71 = enterovirus A71; mNGS = metagenomic next-generation sequencing; NP = nasopharyngeal; qRT-PCR = quantitative reverse transcription polymerase chain reaction.
Standard protocol approvals, registrations, and patient consents
The institutional ethics board approved the study, and informed consent was obtained from parents or caretakers.1
Metagenomic sequencing library preparation
After samples were thawed, RNA was immediately extracted. The plasma, stool, and NP samples were homogenized with Omni-International's 2.8-mm ceramic bead kit and the TissueLyser II (Qiagen) for 5 minutes at 15 Hz. RNA isolation from the CSF samples (250 µL/sample), the plasma samples (250 µL/sample), the NP swab samples, and 2 water controls were performed using the Direct-zol RNA MicroPrep with TRI reagent (Zymo Research) into 20 µL of nuclease-free water (ThermoFisher Scientific). Homogenized stool samples (250 µL/sample) were extracted using the RNeasy PowerMicrobiome Kit (Qiagen) on a Qiacube (Qiagen) into 100 µL of RNase-free water along with a water control. Sequencing libraries were prepared as described previously.16 The libraries were subjected to Depletion of Abundant Sequences by Hybridization, described previously, to remove human mitochondrial cDNA.17 The pooled library was size-selected using AMPure beads (Beckman Coulter), and concentration and quality were determined using a Fragment Analyzer (Advanced Analytical Technologies). The samples were sequenced on an Illumina HiSeq 4000 instrument using 140/140 base pair (bp) paired-end sequencing.
Sequencing libraries produced from the CSF samples were prepared together (including a water control). Sequencing libraries from the plasma, stool, and NP swab samples were prepared together with their respective water controls. The water controls for CSF and NP samples contained no EV reads (i.e., sequences). The stool water control, which had been pooled for sequencing with stool samples containing EV reads, had 0.7 EV reads per million (rpM). To differentiate whether the EV reads present in the stool water control stemmed from sample contamination during library preparation or from sequencer contamination because of barcode hopping,18 we resequenced the same stool water control library independent of the stool samples. When sequenced separately, we found no EV reads in the stool water control. This suggested that the EV reads originally detected in the stool water control resulted from barcode hopping and not from sample contamination during library preparation. To remove any potential that physical cross-contamination had occurred between libraries, we resequenced a subset of the original libraries with pathogen abundance below 0.7 rpM and only considered a case positive if the pathogen was detected in both sequencing runs. If a pathogen was only detected in a sample in one of the 2 sequencing runs, it was considered to have 0 EV reads in that sample.
mNGS bioinformatics
Sample sequences were analyzed for pathogens using a custom pathogen identification pipeline, as previously described.19 Sequences that mapped to the EV genus were assembled de novo using the Geneious version 10.2.3 and St. Petersburg genome assembler (SPAdes) version 3.10.0.20 Phylogenetic trees were created in Geneious using a multiple sequence comparison by log expectation (MUSCLE) or multiple alignment using fast Fourier transform (MAFFT) alignment algorithm, followed by the Geneious Tree Builder tool with Neighbor-Joining build method.3,21 Bootstrapping was performed with 100 replicates. Statistics comparing the degree of concordance between research-based mNGS results and standard clinical diagnostic testing and comparing direct and indirect testing methods results were performed using the Mann-Whitney and McNemar statistical tests.
Pan-viral CSF serologic testing with VirScan
We constructed a T7 bacteriophage display library comprising 481,966 sixty-two amino acid peptides with 14 amino acid overlap tiled across full-length vertebrate, mosquito-borne, and tick-borne viral genomes downloaded from UniProt and RefSeq databases in February 2017, as previously described (VirScan).10,11 VirScan libraries were incubated with 2 µL of CSF overnight, immunoprecipitated with protein A/G beads (ThermoFisher Scientific) for 2 rounds, and sequenced as previously described.10 We normalized individual peptide counts and expressed them as reads per 25,000 reads sequenced (rpqK). A sample was considered EV positive if the total EV rpqK value was greater than the mean signal in the OND controls plus one SD.
Enterovirus ELISA validation
To validate our VirScan results, we generated recombinant EV viral protein 1 (VP1) from EV-A71 and EV-D68, given that complex and cross-reactive serologic responses to EVs are known to occur in subjects and performed 2 independent ELISAs on each sample and considered the higher value, as previously described.10 The signal was measured as the optical density (OD) at 450 nm and reported after background subtraction (background OD = 0.05). A sample was considered positive if the OD was greater than 3 times the background.
Data availability
Assembled viral sequences have been deposited in GenBank (MH484066-MH484076, MN515037), and nonhuman reads have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive BioProject database (PRJNA504776).
Results
Traditional clinical testing vs research mNGS
We obtained an average of 21.8 million (range 4.53–61.1 million) 140 bp paired-end reads per sample (table e-1, links.lww.com/NXI/A216).
As expected, EV-A71 was the most common virus detected across all samples (figure 1A). In the CSF, however, EV-A71 detection remained low relative to NP and fecal testing. By pan-EV qRT-PCR, 0/20 cases were positive in the CSF vs 10/16 in NP and 9/15 in fecal samples (p-value < 0.001 for both comparisons by the Fisher exact test). With mNGS, 3/20 cases were positive in the CSF vs 13/16 in NP and 10/15 in fecal samples (p-value < 0.001 and p = 0.003 by the Fisher exact test). There was no statistically significant difference observed in the number of samples testing positive by NP or fecal testing (p = 1.0 for qRT-PCR and p = 0.43 for mNGS by the Fisher exact test). When EV-A71 was present in the CSF, the number of reads detected was lower than for the other 2 EVs we found, highlighting the difficulty of detecting EV-A71 in CSF (figure 1B). The mean read count for E-30 was greater in the CSF than that for EV-A71 (mean rpM 107.28, range 16.24–337.8 vs 0.23, range 0.06–0.55, p-value = 0.057 by the Mann-Whitney test). We detected CVB in 1 case which, such as E-30, was at a much higher abundance than EV-A71 (32.25 rpM). We did uncover coinfection with more than one EV in 5 subjects (figure 1C). Overall, we tested 60 samples by both mNGS and qRT-PCR. mNGS found EV reads in 100% of qRT-PCR positive samples (n = 25). mNGS detected EV in an additional 22% (n = 8) of samples negative by qRT-PCR, a statistically significant improvement in detection rate for mNGS vs qRT-PCR (Figure 1D, p = 0.01 by the McNemar test).
Phylogenetics
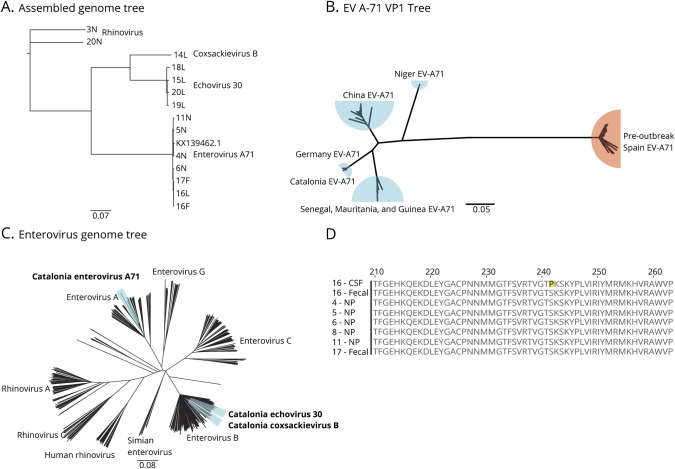
Full-length EV-A71 (n = 7), E-30 (n = 4), and CV-B virus (n = 1) genomes (coverage depth range 22-2,296x) were assembled as described in the Methods. All 7 EV-A71 genomes were essentially identical to the German neuroinvasive EV-A71 strain (Genbank KX139462.1, 99.3%–99.4% nucleotide similarity and 99.7%–99.8% amino acid similarity, figure 2A). In addition, we reproduced a previous but more limited phylogenetic analysis showing greater similarity in the VP1 sequence between the neurovirulent EV-A71 in Catalonia and other neurovirulent EV-A71 strains detected in Germany, China, and West Africa as compared with preoutbreak HFMD EV-A71 in Spain (figure 2B).3,22–24
Figure 2. Phylogenetic and Genomic Analysis of EV-A71.
(A) Phylogenetic tree of full-length viral genomes for enterovirus A71, echovirus 30, coxsackievirus B, and rhinovirus isolated from CSF (L), stool (F), and NP (N) compared with the German neuroinvasive strain (KX139462.1). Rhinovirus obtained from subjects acts as the root. The number refers to the subject. (B) Confirmation of clinical VP1 testing that the Catalonian EV-A71 viral protein 1 (VP1) gene is most closely related to a neuroinfectious German strain. Blue = neuroinvasive EV-A71, Orange = HFMD EV-A71. (C) Phylogenetic tree of 545 EV genomes from every species highlighting the relatedness between the EV strains discovered in this outbreak. (D) Protein alignment of the VP1 gene highlighting the S241P mutation found exclusively in the CSF of subject 16. Scale bars indicate nucleotide substitution rate per position. EV-A71 = enterovirus A71; F = fecal; NP = nasopharyngeal.
To determine the degree of divergence of neurovirulent EV-A71, E-30, and CV-B from their respective viral species, we built a phylogenetic tree using the Catalonia genomes and 8,841 full-length EV genomes from the NCBI's GenBank database (Accessed February 2018) clustered at 95% similarity. Because we detected more than one EV species in 5 subjects and because EVs are known to recombine, we used this phylogenetic analysis to determine whether any obvious interspecies recombination events had occurred.25,26 However, all 3 viruses clustered within their own species without any major deviation (figure 2C).
Last, an EV-A71 genome assembled from a CSF sample was compared with EV-A71 genomes derived from the same subject's stool and NP samples (subject 16). A single amino acid substitution (S241P) in the VP1 region was found only in the CSF, not the 2 peripheral sites. Indeed, all of the EV-A71 genomes identified in peripheral body sites from this study had a serine in position 241 (figure 2D).
VirScan increases CSF detection of enterovirus
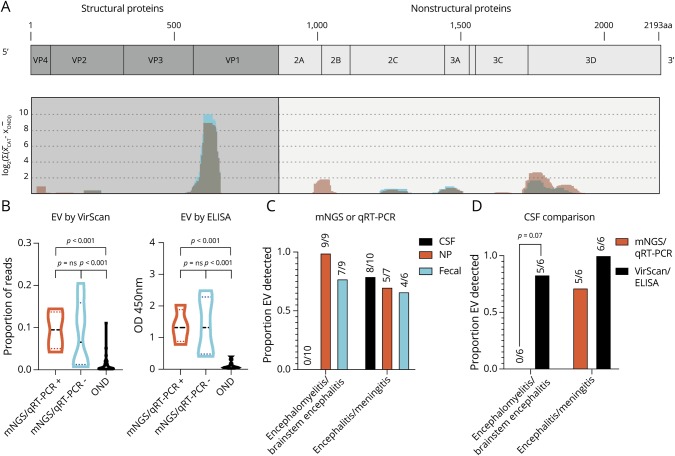
Although mNGS improved the detection of EVs across our 20 subjects compared with traditional qRT-PCR, uncovered EV coinfections in 5 instances, and enabled phylogenetic and mutational analyses, detection of EV-A71 in the CSF was more difficult than in the NP or fecal samples, as described above. Therefore, we deployed VirScan for comprehensive antiviral antibody detection in the CSF. After performing VirScan on 12 cases and 54 pediatric controls with ONDs, the only significantly enriched viral family in our cases was Picornaviridae (figure e-1A, links.lww.com/NXI/A215 and table e-3, links.lww.com/NXI/A216, mean rpqK was 2,366 in cases vs 224 in controls, p-value < 0.001 by the Mann-Whitney with Bonferroni correction). Within Picornaviridae, the genus Enterovirus was the most enriched (figure e1, B and C, and table e-4, median proportion of reads 0.09 in cases vs 0.002 in controls, p-value < 0.001 by the Mann-Whitney with Bonferroni correction). To determine the genomic location of enriched EV antigens in VirScan, we used BLASTp to map all genus Enterovirus peptides against a model EV-A71 genome (Genbank Accession AXK59213.1), recognizing that not all EV subject antibodies are likely to be truly EV-A71 specific. The location and strength of the EV antigen response detected in cases by VirScan is shown normalized against controls, with immunodominant regions visible in VP1 and 3D (figure 3A and table e-5, links.lww.com/NXI/A216). The pattern of EV antigen targeting appears to be highly conserved and similar to a group of previously published pediatric acute flaccid myelitis cases predominantly from the United States (figure 3A, red overlay).10
Figure 3. VirScan Identifies immunodominant enterovirus antigens and improves the detection of CSF enterovirus in encephalomyelitis/brainstem encephalitis.
(A) VirScan identified 136 unique, enriched viral antigens with taxonomies linking them to genus Enterovirus. We mapped 123 of these 136 peptides with BLASTP to a reference EV-A71 genome (Genbank Accession AXK59213.1), as described previously9 (coding genes in light blue, non-coding genes in orange). Mapping revealed the relative locations of the EV antigens identified in the Catalonia cases (graphed blue shading) across the EV genome, which appeared remarkably conserved, as seen previously in pediatric acute flaccid myelitis (graphed light red shading with overlap appearing gray).10 (B) Violin plot revealing enrichment for EV antigens by VirScan and EV VP1 antigen by ELISA in Catalonia cases, regardless of whether an EV had been previously identified by mNGS or qRT-PCR (p = ns for both comparisons). In both groups, EV detection was significantly greater than in the pediatric OND controls (p < 0.001 for all comparisons as indicated). The Mann-Whitney test was used, with Bonferroni correction for VirScan. (C) Differences in virus detection for subjects with encephalomyelitis/brainstem encephalitis or encephalitis/meningitis. The detection of CSF EV by mNGS or qRT-PCR was low (0/10). (D) VirScan or ELISA improved EV detection in encephalomyelitis/brainstem encephalitis (0/6 vs 5/6, p = 0.07 by the McNemar test). CSF = cerebral spinal fluid; EV = enterovirus; mNGS = metagenomic next-generation sequencing; NP = nasopharyngeal; OND = other neurologic disease.
We next confirmed the EV VirScan results in a subset of cases with remaining CSF using an EV VP1 ELISA. The EV ELISA was confirmatory in all of the cases detected by VirScan (6/6). Thus, we considered the other EV cases detected by VirScan for which there was insufficient CSF to do confirmatory ELISA positive as well (n = 2 cases and n = 1 OND control were positive by VirScan but had insufficient CSF remaining for confirmatory ELISA). In addition, EV ELISA was positive for 3 cases in whom the amount of EV signal by VirScan was below the threshold we set to consider a sample positive. In total, 9/54 (17%) of the pediatric OND controls were positive for CSF EV antibodies. Of note, EV antibody detection by VirScan was not confirmed by EV ELISA in one OND control. This subject had limited clinical information available, and the EV-A71 ELISA had detectable signal above the background but was not above the conservative threshold we used to determine whether a sample was positive.
We found that we were able to detect CSF EV antibodies using VirScan or EV VP1 ELISA, regardless of whether EV had been previously detected in the CSF by mNGS or qRT-PCR (figure 3B, p-value = ns by Mann-Whitney). EV was especially difficult to detect in the CSF of subjects with brainstem encephalitis using mNGS or qRT-PCR (figure 3C). Therefore, we asked whether EV detection in brainstem encephalitis could be improved with EV VirScan or ELISA. Indeed, serologic testing with CSF VirScan or ELISA was able to improve detection of EV in brainstem encephalitis (figure 3D and table e-6, links.lww.com/NXI/A216, and table e-7, links.lww.com/NXI/A216, p-value = 0.07 by the Fisher exact test).
Discussion
The original description of the 2016 pediatric brainstem encephalitis outbreak in Catalonia identified EV-A71 as the likely etiologic agent. However, this conclusion was tempered because (1) there were multiple cocirculating EVs present during the outbreak and (2) EV-A71 was not identified in the CSF of most children with brainstem encephalitis. Identifying EVs in peripheral body sites of children with severe neurologic disease but failing to find it in the CSF mirrors both the recent North American outbreak of acute flaccid myelitis associated with EV-D6821 and outbreaks of EV-A71 neurologic disease.27,28 Here, we used mNGS and serologic testing with VirScan and confirmatory EV ELISA to further investigate samples from the Catalonia outbreak to enhance detection of pathogens, especially in the CSF.
mNGS identified EV-A71 in the CSF of 3 subjects not detected by qRT-PCR. EV-A71 abundance in the CSF was very low (range 0.06–0.55 rpM) compared with other EVs detected in the CSF in this study (i.e., E-30 and CVB). In addition, only 2.4% (4/166) of the EV-A71 reads mapped to the 5′ untranslated region (UTR) targeted by the primers used in the clinical qRT-PCR assay. Because mNGS can identify EV sequences from any part of the viral genome, this may help explain its improved sensitivity over the more targeted clinical qRT-PCR and the BioFire FilmArray Meningitis/Encephalitis panel assays.1,8
In addition, mNGS also identified 5 subjects coinfected by 2 different EVs, and 2 of these subjects had brainstem encephalitis. Owing to the small number of subjects in our study, we are not able to determine whether coinfection contributed to disease severity. These findings highlight that in the midst of an outbreak, unbiased mNGS can detect coinfections, and future studies may uncover ways in which coinfections affect clinical presentations and outcomes.
A limitation of this study is that because of sample availability, we did not perform orthogonal confirmation of the mNGS-only virus identifications. As a result, our evaluation of the performance of the mNGS assay is vulnerable to incorporation bias because the gold standard by which we are evaluating its performance includes the mNGS results.29 However, because we tested low abundance samples on 2 independent sequencing runs, we are confident that even the low levels of EV detected by mNGS in some samples did not result from contamination during sample preparation or from barcode hopping.
Through mNGS, we added significant new knowledge about neurovirulent EV-A71, including 7 full-length genomes. The assembled neurovirulent EV-A71 genomes were related to other neurovirulent EV-A71 strains detected in Germany, West Africa, and China rather than preoutbreak Spanish EV-A71 strains associated with HFMD. Interestingly, we found that the EV-A71 strain circulating in Catalonia appeared to be most closely related to the neurovirulent German strain, followed by the West African and then Chinese strains.22 The German strain reported in 2015 clusters within the C1 subgenogroup when comparing VP1 regions. However, the 5′ UTR clusters more closely with B3 and C2-like strains and the P2 and P3 regions cluster with C4 strains related to strains reported in China.3
In a single subject, we discovered a single amino acid substitution (S241P) that was present only in EV-A71 sequences derived from the CSF and not from viral sequences in the periphery of that same subject. This substitution was not seen in any of the other EV-A71 viruses sequenced from the same outbreak nor was it found in any of the EV-A71 VP1 sequences compared in figure 2B. An amino acid substitution (S241L) at this location has been previously reported to be associated with increased virulence.30 However, those authors also identified coincident, additional mutations that increased the virus's ability to use the P-selectin glycoprotein ligand-1 (PSGL-1) cell receptor.30–33 Here, the S241P mutation we observed occurs without the mutation at position 145 that is required for PSGL-1 binding, suggesting that this variant of unknown significance at position 241 may affect virulence in a PSGL-1 independent manner. Although we were able to obtain EV-A71 sequences in the CSF from subjects 12 and 17, they did not span position 241 of VP1. No CSF remains from these subjects to obtain additional sequence information. Although the clinical and functional significance of the S241P mutation remains uncharacterized, this study highlights the importance of acquiring viral nucleic acid directly from the CNS, as opposed to the periphery alone.
Owing to the limitations of direct detection of viral pathogens using nucleic acid-based methods, we used VirScan to comprehensively profile CSF antiviral antibodies. This technique has previously shown utility in the diagnosis of encephalitis without a clear etiology.12 We show that antibodies directed toward the genus Enterovirus, but not toward other viruses, were present in the CSF of 8/12 cases. When we further examined a subset of these cases with an EV ELISA and combined the results with VirScan, we found evidence for EV antibodies in 11/12 (92%) cases from the Catalonia outbreak vs just 9/54 (17%) pediatric OND controls, with significantly improved detection of brainstem encephalitis due to EV. Although we are able to detect antibodies to EVs broadly, VirScan and ELISA detected antibodies targeting highly conserved segments of EVs and thus did not allow for consistent viral typing. In addition to detecting CSF antibodies to EV, our analysis uncovered immunodominant epitopes at VP1 and 3D, similar to what we found previously in EV-associated acute flaccid myelitis.10,34 However, evaluation of linear peptides from other regions of the EV genome may be more fruitful for subtyping EV infections34 and future work with natively folded EV proteins may also better serologically type EVs.
Although mNGS can detect a variety of pathogens in the CSF,19,35–37 there have been few reports on the actual diagnostic yield of mNGS of CSF compared with traditional assays such as pathogen-specific PCR and the BioFire FilmArray Meningitis/Encephalitis panel. In our study, mNGS was more sensitive in cases with very low EV read abundance.38 Furthermore, identifying an EV-A71 closely related to strains previously associated with neurologic disease in Germany, West Africa, and China in the CSF of 3 additional subjects provides strong evidence that the Catalonia outbreak was because of neuroinvasive EV. Although the experiments described herein used a research-based CSF mNGS assay, a clinically validated CSF mNGS assay with an approximate 7-day turnaround time was recently evaluated in a multicenter prospective study and is now clinically available.38 Thus, the role of mNGS in individual patient care and public health outbreak investigations will likely expand.
Similarly, the detection of CSV EV antibodies in many of these subjects supports our findings of EV nucleic acid in the CSF while also providing more information about the immune response against EV. A limitation of the VirScan method is that we are unable to distinguish between antibodies that have been synthesized intrathecally vs peripheral synthesis, followed by transudation across the blood-brain barrier. Furthermore, it is difficult to definitively say whether these anti-EV IgG antibodies developed in response to this particular outbreak. However, the markedly lower levels of CSF EV antibodies in our pediatric OND control population, many of whom also had blood-brain barrier compromise, support our contention that the high levels of CSF EV antibodies in the cases result from a CNS EV infection and not simply from anti-EV antibodies formed in the periphery in response to the many peripheral EV infections children contract.
Acknowledgment
The authors would like to thank Amy Kistler, PhD, for her intellectual discussions. The authors thank the subjects and their families for their participation in the research study.
Glossary
- CV
coxsackievirus
- mNGS
Metagenomic next-generation sequencing
- NP
nasopharyngeal
- OD
optical density
- OND
other neurologic disease
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- UTR
untranslated region
Appendix. Authors
Study funding
Grants supporting this project include the National Multiple Sclerosis Society and the American Academy of Neurology award FAN-1608-25607 (R.D.S.), Clinical Research Training Scholarship P0534134 (P.S.R.), Sandler and William K. Bowes Jr Foundations (M.R.W., J.L.D., L.M.K., H.A.S., K.C.Z.), Rachleff Family Foundation (M.R.W.), and NINDS of the NIH under award K08NS096117 (M.R.W.) and F31NS113432 (K.E.L.). This study was partially supported by a grant from the Spanish National Health Institute [grant number PI15CIII-00020] and the European Regional Development Fund (FEDER funds). UCSF Biomedical Sciences Program (I.A.H., K.E.L.), UCSF Medical Scientist Training Program (K.E.L.), and the Chan Zuckerberg Biohub (J.E.P., W.W., C.K.C., J.L.D., E.D.C.) also supported this project.
Disclosure
None to report. Go to Neurology.org/NN for full disclosures.
References
- 1.Casas-Alba D, de Sevilla MF, Valero-Rello A, et al. . Outbreak of brainstem encephalitis associated with enterovirus-A71 in Catalonia, Spain (2016): a clinical observational study in a children's reference centre in Catalonia. Clin Microbiol Infect 2017;23:874–881. [DOI] [PubMed] [Google Scholar]
- 2.Leber AL, Everhart K, Balada-Llasat J, et al. . Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol 2016;54:2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher S, Obermeier PE, Neubauer K, Diedrich S. Recombinant enterovirus A71 subgenogroup C1 strains, Germany, 2015. Emerg Infect Dis 2016;22:1843–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Sanz R, Casas-Alba D, Launes C, et al. . Molecular epidemiology of an enterovirus A71 outbreak associated with severe neurological disease, Spain, 2016. Euro Surveill 2019;24:1800089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karrasch M, Fischer E, Scholten M, et al. . A severe pediatric infection with a novel enterovirus A71 strain, Thuringia, Germany. J Clin Virol 2016;84:90–95. [DOI] [PubMed] [Google Scholar]
- 6.Midgley SE, Nielsen AG, Trebbien R, Poulsen MW, Andersen PH, Fischer TK. Co-circulation of multiple subtypes of enterovirus A71 (EV- A71) genotype C, including novel recombinants characterised by use of whole genome sequencing (WGS), Denmark 2016. Euro Surveill 2017;22:30565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antona D, Kossorotoff M, Schuffenecker I, et al. . Severe paediatric conditions linked with EV-A71 and EV-D68, France, May to October 2016. Euro Surveill 2016;21:30402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Launes C, Casas-Alba D, Fortuny C, Valero-Rello A, Cabrerizo M, Muñoz-Almagro C. Utility of FilmArray meningitis/encephalitis panel during outbreak of brainstem encephalitis caused by enterovirus in Catalonia in 2016. J Clin Microbiol 2017;55:336–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudas G, Carvalho LM, Bedford T, et al. . Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature 2017;544:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert RD, Hawes I, Ramachandran PS, et al. . Pan-viral serology implicates enteroviruses in acute flaccid myelitis. Nat Med 2019;25:1748–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu GJ, Kula T, Xu Q, et al. . Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015;348:aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson TP, Larman HB, Lee M, et al. . Chronic dengue virus panencephalitis in a patient with progressive dementia with extrapyramidal features. Ann Neurol 2019;86:695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selva L, Martinez-Planas A, García-García JJ, Casadevall R, Luaces C, Muñoz-Almagro C. Comparison of an in-house real-time RT-PCR assay with a commercial assay for detection of enterovirus RNA in clinical samples. Eur J Clin Microbiol Infect Dis 2012;31:715–719. [DOI] [PubMed] [Google Scholar]
- 14.Verstrepen WA, Kuhn S, Kockx MM, Van De Vyvere ME, Mertens AH. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J Clin Microbiol 2001;39:4093–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabrerizo M, Echevarria JE, González I, de Miguel T, Trallero G. Molecular epidemiological study of HEV-B enteroviruses involved in the increase in meningitis cases occurred in Spain during 2006. J Med Virol 2008;80:1018–1024. [DOI] [PubMed] [Google Scholar]
- 16.Wilson MR, Suan D, Duggins A, et al. . A novel cause of chronic viral meningoencephalitis: cache valley virus. Ann Neurol 2017;82:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu W, Crawford ED, O'Donovan BD, et al. . Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol 2016;17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MR, Fedewa G, Stenglein MD, et al. . Multiplexed metagenomic deep sequencing to analyze the composition of high-priority pathogen reagents. mSystems 2016;1:e00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson MR, O'Donovan BD, Gelfand JM, et al. . Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol 2018;75:947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greninger AL, Naccache SN, Messacar K, et al. . A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis 2015;15:671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Garcia MD, Volle R, Joffret M, et al. . Genetic characterization of enterovirus A71 circulating in Africa. Emerg Infect Dis 2018;24:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrerizo M, Tarragó D, Muñoz-Almagro C, et al. . Molecular epidemiology of enterovirus 71, coxsackievirus A16 and A6 associated with hand, foot and mouth disease in Spain. Clin Microbiol Infect 2014;20:O150–O156. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, Zheng H, Zheng H, et al. . An enterovirus 71 epidemic in Guangdong province of China, 2008: epidemiological, clinical, and virogenic manifestations. Jpn J Infect Dis 2011;64:13–18. [PubMed] [Google Scholar]
- 25.McWilliam Leitch EC, Cabrerizo M, Cardosa J, et al. . The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol 2012;86:2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmonds P, Welch J. Frequency and dynamics of recombination within different species of human enteroviruses. J Virol 2006;80:483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Vélez CM, Anderson MS, Robinson CC, et al. . Outbreak of neurologic enterovirus type 71 disease: a diagnostic challenge. Clin Infect Dis 2007;45:950–957. [DOI] [PubMed] [Google Scholar]
- 28.Messacar K, Spence-Davizon E, Osborne C, et al. . Clinical characteristics of enterovirus A71 neurological disease during an outbreak in children in Colorado, USA, in 2018: an observational cohort study. Lancet Infect Dis 2019;20:P230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohn MA, Carpenter CR, Newman TB. Understanding the direction of bias in studies of diagnostic test accuracy. Acad Emerg Med 2013;20:1194–1206. [DOI] [PubMed] [Google Scholar]
- 30.Chang C, Wu S, Chen Y, et al. . Mutations in VP1 and 5'-UTR affect enterovirus 71 virulence. Sci Rep 2018;8:8744–8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura Y, Lee H, Hafenstein S, et al. . Enterovirus 71 binding to PSGL-1 on leukocytes: VP1-145 acts as a molecular switch to control receptor interaction. PLoS Pathog 2013;9:e1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J, Shih S. Cell and tissue tropism of enterovirus 71 and other enteroviruses infections. J Biomed Sci 2014;21:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura Y, Shimojima M, Tano Y, Miyamura T, Wakita T, Shimizu H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat Med 2009;15:794–797. [DOI] [PubMed] [Google Scholar]
- 34.Mishra N, Ng TFF, Marine RL, et al. . Antibodies to enteroviruses in cerebrospinal fluid of patients with acute flaccid myelitis. MBio 2019;10:e01903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson MR, Naccache SN, Samayoa E, et al. . Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014;370:2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naccache SN, Peggs KS, Mattes FM, et al. . Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 2015;60:919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller S, Naccache SN, Samayoa E, et al. . Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res 2019;29:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson MR, Sample HA, Zorn KC, et al. . Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 2019;380:2327–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Assembled viral sequences have been deposited in GenBank (MH484066-MH484076, MN515037), and nonhuman reads have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive BioProject database (PRJNA504776).