Abstract
Osteolineage cell‐derived extracellular vesicles (EVs) play a regulatory role in hematopoiesis and have been shown to promote the ex vivo expansion of human hematopoietic stem and progenitor cells (HSPCs). Here, we demonstrate that EVs from different human osteolineage sources do not have the same HSPC expansion promoting potential. Comparison of stimulatory and non‐stimulatory osteolineage EVs by next‐generation sequencing and mass spectrometry analyses revealed distinct microRNA and protein signatures identifying EV‐derived candidate regulators of ex vivo HSPC expansion. Accordingly, the treatment of umbilical cord blood‐derived CD34+ HSPCs with stimulatory EVs‐altered HSPC transcriptome, including genes with known roles in cell proliferation. An integrative bioinformatics approach, which connects the HSPC gene expression data with the candidate cargo in stimulatory EVs, delineated the potentially targeted biological functions and pathways during hematopoietic cell expansion and development. In conclusion, our study gives novel insights into the complex biological role of EVs in osteolineage cell‐HSPC crosstalk and promotes the utility of EVs and their cargo as therapeutic agents in regenerative medicine.
Keywords: bone, hematopoietic expansion, hematopoietic niche, intercellular communication
Abbreviations
- ANXA2
annexin A2
- EV
extracellular vesicle
- FC
fold change
- GO
gene ontology
- hEV
hFOB‐derived EV
- hOB
hFOB
- HSPC
hematopoietic stem and progenitor cell
- HSC
hematopoietic stem cell
- mEV
hMSC‐TERT‐derived EV
- sEV
SV‐HFO‐derived EV
- sOB
SV‐HFO
- TNC
total number of viable nucleated cells
- UCB
umbilical cord blood
1. INTRODUCTION
Hematopoietic stem cells (HSCs) are multipotent cells that have the capacity to self‐renew and replenish the entire blood system. Accordingly, HSCs are widely used in hematopoietic stem cell transplantations in patients with high‐risk hematological malignancies.1 Umbilical cord (UC) blood is an alternative source of HSCs for both pediatric and adult patients in need for a transplant and lacking an HLA‐matched related or unrelated donor. Interestingly, UC‐derived HSCs are characterized by its rapid availability and less stringent human leucocyte antigen match requirement. However, UC grafts contain a low number of HSCs, resulting in delayed hematopoietic recovery and increased patient morbidity and mortality, particularly in adult patients.2, 3 Development of ex vivo culture systems that support UC‐derived HSC expansion is a promising approach to improve engraftment and consequent post‐transplantation recovery.4, 5
Recent studies reported the success of improved ex vivo HSC expansion by mimicking the bone marrow niche, where HSC fate determination is tightly regulated.6, 7 Osteolineage cells, including primitive mesenchymal cells and bone‐forming immature and mature osteoblasts, represent one of the critical niche components that support self‐renewal and proliferation of HSCs, as well as hematopoietic stem and progenitor cells (HSPCs) in vivo.8, 9, 10, 11, 12, 13 Osteolineage cell‐derived secreted factors and adhesion molecules yield robust in vitro proliferation of HSCs with long‐term repopulation ability stressing the importance of discovering other osteolineage cell components that can support the growth factor‐based expansion culture systems.12, 13, 14, 15, 16, 17 Increasing number of studies report the role of extracellular vesicle (EV)‐mediated intercellular communication within the hematopoietic system.18, 19, 20, 21, 22 EVs are small cellular compartments that regulate the function of their targets by transferring bioactive lipids, proteins, mRNAs, long non‐coding RNAs and small non‐coding RNAs, including microRNAs (miRNAs), between cells.23, 24 We previously demonstrated that human osteoblast‐derived EVs retain HSPC‐supporting capacity ex vivo, as revealed by long‐term cultures and in vivo repopulation assays.25 However, the molecular mechanisms, by which EVs regulate the communication between osteolineage cells and HSPCs, still remains to be elucidated.
In this study, we focused on understanding the key regulatory human osteolineage EV components that modulate the survival, proliferation, and differentiation of human CD34+ UC‐HSPCs. In an effort to generate a candidate EV cargo list, we first investigated the UC‐HSPC supporting capacity of EVs secreted by different human osteolineage cells. Then, we compared the proteomics and miRNA profiles between stimulatory and non‐stimulatory EVs by proteomics analyses and next‐generation sequencing, respectively. Furthermore, we studied the effect of stimulatory EVs on the gene expression of UC‐HSPCs by next‐generation sequencing. Lastly, we employed comprehensive integrative bioinformatics analyses to define and propose biological pathways that are dynamically regulated by stimulatory EV cargo in UC‐HSPC fate regulation.Taken together, our findings demonstrate that fetal calvaria osteoblast EVs prove to be powerful tools to expand UC‐HSPCs ex vivo. This information will likely generate knowledge that is not only relevant for HSC expansion, but also holds the promise of modulating other types of stem cells for regenerative medicine purposes in the future.
2. MATERIALS AND METHODS
2.1. Cell culture
Simian virus 40‐immortalized human osteoblast cells from fetal calvaria, SV‐HFO cells,26 were seeded at a density of 1 × 104 cells/cm2 and cultured in α‐MEM (GIBCO, Paisley, UK) supplemented with 20 mM of HEPES, pH 7.5 (Sigma, St. Louis, MO, USA), streptomycin/penicillin, 1.8 mM CaCl2 (Sigma), 10 mM of β‐glycerophosphate (Sigma) and 2% of FCS (GIBCO) at 37°C in a humidified atmosphere of 5% of CO2 for 12‐14 days. The culture medium was replaced every 2‐3 days. Simian virus 40‐immortalized human osteoblasts from fetal limb tissue (hFOB 1.19 cells, ATCC CRL11372) were seeded at a density of 1.5 × 104 cells/cm2 and cultured in DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 2.5 mM of l‐glutamine, 0.3 mg/mL of G418, streptomycin/ penicillin, and 10% of FCS at 34°C in a humidified atmosphere of 5% of CO2 for 24 hours. hMSC‐TERT cells27 were seeded at a density of 1.5 × 104 cells/cm2 and cultured in MEM (Thermo Fisher Scientific), supplemented with streptomycin/penicillin and 10% of FCS at 37°C in a humidified atmosphere of 5% of CO2 for 24 hours. All cell lines were washed with 1X PBS and refreshed with their respective serum‐free culture medium 24 hours prior to EV isolation. All experiments with human UC were performed in accordance with the Dutch law on Medical Scientific Research with Humans and approved by the Medical Ethical Committee of the Erasmus University Medical Center (MEC‐2009‐410), Rotterdam, the Netherlands and written informed consent from the mothers was obtained prior to UC donation. UC was collected in Erasmus University Medical Center using Stemcare/CB collection blood bag system (Fresenius Kabi Norge AS, Halden, Norway). Within 48 hours after collection, mononuclear cells were isolated using ficoll (Lymphoprep, Fresenius KabiNorge AS). CD34+ cells and viable DAPI‐Lin‐CD34+CD38lowCD45RAlowCD90+ cells were isolated as described previously.28 CD34+ cells and CD34+CD90+ cells were cultured in either serum‐free Cellgenix GMP SCGM (Cellgenix, Freiburg, Germany) or Glycostem Basic Growth Medium (Glycostem, Oss, the Netherlands) supplemented with SCF (50 ng/mL, Cellgenix) and Flt3L (50 ng/mL, Cellgenix), with or without EVs at 37°C in a humidified atmosphere of 5% of CO2. Cells were refreshed every 2‐3 days.
2.2. EV isolation and characterization
EVs were isolated from 20 mL conditioned medium by low speed centrifugation (1500 rpm, 5 minutes; 4500 rpm, 10 minutes) followed by ultracentrifugation (20 000 g, 30 minutes; 100 000 g, 1 hour at 4°C) of the supernatant using the SW32Ti rotor (Beckman Coulter, Fullerton, CA, USA). Transmission electron microscopy (TEM) images were taken by negative staining of the purified EVs. Freshly carbon sputtered and formvar coated copper grids were incubated on EV preparations, washed rapidly in water and contrasted with (3.5%) uranyl acetate. Grids were blotted and dried before the analysis using a Tecnai T12 G2 Biotwin at 120 kV. EV size distribution and concentration was measured with NanoSight LM10 (Nanosight Ltd., Amesbury, UK) equipped with a 405 nm laser. Each sample was tracked for 60 seconds with five repetitions. The data were processed by NTA 2.3 software.
2.3. Immunoblot analysis
Protein samples were prepared by mixing the EVs (in PBS) with 6X reducing sample buffer immediately after isolation. EV proteins (1.8 µg of total protein/sample) were separated by SDS‐PAGE at 200 V and transferred onto a nitrocellulose membrane (Hybond‐ECL, Amersham Biosciences, Buckinghamshire, UK). After blocking non‐specific signal with 5% of BSA in TBS/ 0.1% of Tween‐20, the membrane was incubated with primary antibodies against annexin A2 (ANXA2; rabbit polyclonal. 1:500, Abcam, Cambridge, UK). Membranes were probed with secondary antibody conjugated with IRDye 800CW (1:5000, goat anti‐rabbit, LI‐COR, Lincoln, NE, USA) using Odyssey Infrared Imaging System according to the manufacturer's instructions (LI‐COR).
2.4. RNA isolation and quantitative real‐time PCR
Total EV‐RNA was isolated using the TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. RNA concentration was determined using Nanodrop (Thermo Fisher Scientific) and size distribution was checked on an Agilent Bioanalyzer RNA 6000 Pico chip (Thermo Fisher Scientific). RNA from CD34+UC‐HSPCs was isolated using NucleoSpin RNA XS kit (Macherey‐Nagel, Duren, Germany) according to the manufacturer's instructions. Quantitative real‐time PCR was performed using the SYBR Green (Eurogentec, Seraing, Belgium) kit, according to the manufacturer's instructions. The following primer sequences were used:
| Gene symbol | Forward sequence (5' → 3') | Reverse sequence (5' → 3') |
|---|---|---|
| CYSLTR2 | GCAGCTGAAAGACAGAGACCT | CCATACCTTGCATGGACCTTCT |
| EGR1 | AGCCCTACGAGCACCTGAC | TGGGTTGGTCATGCTCACTA |
| GAPDH | CCGCATCTTCTTTTGCGTCG | CCCAATACGACCAAATCCGTTG |
| ITGAX | CCTACGGAACCACCATCACC | ACATGTCAGGTGCAGGGAAC |
| MAOA | ATGACACCAAGCCAGATGGG | AAGTCGATCAGCTTTCCGGG |
| NFIB | GTCCAGCCACATCATATCACAG | TTGGCAGGATCATTGTGGCTT |
| S100A9 | GGAATTCAAAGAGCTGGTGCG | AGCTGCTTGTCTGCATTTGTG |
2.5. Next‐generation sequencing and bioinformatic analysis of miRNAs
Sequencing of miRNAs was performed by Illumina MiSeq with samples prepared with the NEBNext Small RNA library preparation kit (New England Biolabs) according to the manufacturer's instructions. Small RNA libraries were quantified on a Bioanalyzer High Sensitivity DNA chip (Agilent) and subsequently normalized and pooled. Single end sequencing was performed on a Miseq (Illumina) single read 50 cycles with 6 cycles for the index read (>1 M reads/sample). Subsequently demultiplexing was done using cassava configure call to FASTQ. Quality metrics on the resulting FASTQ files were generated using fastqc in combination with multiqc. Very sensitive adapter aware alignment was done using novoalign. Alignment rates were measured using the samtools software. Abundance estimation for small RNAs as well as hsa‐miRNA selection was performed using feature counts. Principle component analysis plots were generated using the R software. The sequencing data are available in the Gene Expression Omnibus (GEO) database repository (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143613. Comparative analyses of the EV data were obtained by querying the Vesiclepedia plugin provided by FunRich functional enrichment analysis tool (V3.1.3, 2018). IPA (Ingenuity Systems, http://www.ingenuity.com) was used to classify the miRNA categories and predict target pathways and genes.
2.6. Next‐generation sequencing and bioinformatic analysis of HSPC transcriptome
RNA sequencing and bioinformatic analysis was performed as previously described.29 In brief, sequencing RNA libraries were prepared with the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. The concentration and size distribution of the libraries was determined on an Agilent Bioanalyzer DNA 1000 chip (Thermo Fisher Scientific), and verified with Qubit fluorometry (Thermo Fisher Scientific). Libraries were loaded onto flow cells at concentrations of 8‐10 PM to generate cluster densities of 700 000/mm2 following the standard protocol for the Illumina cBot and cBot Paired‐end cluster kit version 3. Flow cells were sequenced as 51 × 2 paired end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HCS v 2.0.12 data collection software. Base‐calling was performed using Illumina's RTA version 1.17.21.3. The sequencing data were analyzed using CAP‐miRSeq v1.1. Normalization (counts per million mapped reads, CPM; and reads per kilobase pair per million mapped reads, RPKM) analysis was performed using edgeR 2.6.2. The data from the replicates were combined as averages of the normalized read values, and only transcripts with CPM ≥ 1 for all replicates were included in the analysis. The sequencing data are available in the Gene Expression Omnibus (GEO) database repository (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE145178. DAVID Bionformatics Resources v 6.7 was used to categorize the proteins into overrepresented GO annotations using the human expression dataset as a background.30, 31 IPA was used to predict target pathways and analyze the interaction between EV cargo and CD34+ HSPC genes using the path explorer tool in My Pathway analysis.
2.7. Mass spectrometry and bioinformatic analysis of proteins
Tryptic peptides were prepared according to the method described by Kulak et al with some adjustments.32 Briefly, EVs were lysed in 2x concentrated (2%) SDC lysis buffer (Sigma Aldrich, Germany), 20 mM of TCEP (Thermo Scientific), 80 mM of ChloroAcetamide (Sigma Aldrich), and 200 mM of TRIS‐HCl pH 8.0 (Life Technologies, UK), boiled at 95° for 5 minutes and sonicated for 10 cycles of 30 seconds on/off in a Bioruptor (Diagenode, Belgium). After an overnight digestion with 100 ng of Trypsin‐LysC (Promega, USA) at room temperature, sampleswereacidifiedwith 10% trifluoroaceticacid (Thermo Scientific) and loaded on in‐house prepared SDB‐RPS STAGEtips (Empore, USA). The tips werewashedwithethylacetate (Sigma Aldrich) and 0.2% trifluoroaceticacid (Thermo Scientific), and then, the peptides were eluted in three fractions by increasing concentrations (100 and 150 mM) of ammonium formate (VWR Chemicals, Belgium) or 5% (v/v) of ammonium hydroxide (Merck Millipore, Germany) and acetonitrile (40%, 60% and 80% v/v) (BioSolve, France). Sample volume was reduced by SpeedVac and supplemented with 2% of acetonitrile, 0.1% of TFA to a final volume of 10 μL. About 3 μL of each sample was injected for MS analysis. Tryptic peptides were separated by nanoscale C18 reverse phase chromatography coupled on line to an Orbitrap Fusion LumosTribrid mass spectrometer (Thermo Scientific) via a nanoelectrospray ion source (Nanospray Flex Ion Source, Thermo Scientific). All data were acquired with Xcalibur 4.1 software. The raw mass spectrometry files were processed with the MaxQuant computational platform, 1.5.3.30. Proteins and peptides were identified using the Andromeda search engine by querying the human Uniprot database (downloaded March 2017). Standard settings with the additional options match between runs, Label Free Quantification (LFQ), and only unique peptides for quantification were selected. The data were filtered for potential contaminants, reverse hits, and ‘only identified by site’ using Perseus 1.6.5.0.33 The proteins were filtered for three valid values in at least one of the experimental groups. Missing values were imputed by normal distribution (width = 0.3, shift = 1.8), assuming these proteins were close to the detection limit. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE34 partner repository with the dataset identifier PXD017172. A volcano plot from a double‐sided t test (FDR 0.05 and S0 of 2) was used to determine significant differences between the sEV and hEV samples. PANTHER 14.1 classification system and FunRich were used to categorize the proteins into overrepresented processes.
2.8. Flow cytometry
Absolute numbers of viable human CD34+ cells were determined by a single platform flow cytometric assay using anti‐FITC‐CD45, anti‐CD34‐PE, and Stem‐Count Fluorospheres from the Stem‐Kit Reagents kit (Beckman Coulter) and DAPI (Sigma). The frequencies of human phenotypic HSCs were determined using anti‐Lin‐FITC, anti‐CD38‐APC, anti‐CD90‐PE (Thermo Fisher Scientific), anti‐CD34‐PE‐Cy7, anti‐CD45RA‐APC‐H7 (BD Biosciences), and DAPI. All samples were analyzed using BD FACSCanto II (BD Biosciences), and the data were analyzed using FlowJo software (Tree Star, Inc, Ashland, OR, USA).
2.9. Statistics
The results were described as mean ± SD based on at least two independent experiments performed with independent EV isolations and/or different UC donors. Significance was calculated using Student's t test, and P values < .05 were considered significant.
3. RESULTS
3.1. Characterization of EVs secreted by different osteolineage cells
To identify regulatory EV components that support human UC‐HSPC expansion, we used two human fetal osteoblast cell lines of different origin, fetal calvaria‐derived SV‐HFO (sOB), and fetal limb tissue‐derived hFOB 1.19 (hOB) cells, as well as human adult bone marrow‐derived mesenchymal stem cell line (hMSC‐TERT) as EV sources. Throughout this study, the three different EV populations secreted by sOBs, hOBs, and hMSC‐TERTs are referred to as sEVs, hEVs, and mEVs, respectively. EVs were isolated from the conditioned serum‐free cell culture medium by a series of centrifugation steps and verified by morphological and molecular characterization.
TEM analysis showed that all EV populations had spherical vesicular structures in wide ranges of diameters with a surrounding lipid bilayer characteristic of EVs (Figure 1A‐C). According to the nanoparticle tracking analyses (Figure 1A‐C) sEVs were more heterogenous in size with slightly larger mean size of 180 nm compared to the smaller hEVs of 155 nm and mEVs of 140 nm (Supporting Information Table 1). Interestingly, sOBs secreted more EVs than hOBs and hMSC‐TERTs did in the given culture conditions, both in absolute terms as well as on a per cell basis. The majority of the EVs were smaller than 200 nm for all the three EV populations, as expected from the centrifugation protocol. However, the percentage of larger EVs co‐sedimenting at 100 000 g was higher in sEVs (26% compared to 14% in hEVs and 10% in mEVs), in accordance with TEM images (Figure 1D).Western blot analysis in Figure 1E further verified the presence of EVs by testing for the known vesicle marker ANXA2 in all three EV populations.35 Furthermore, representative Bioanalyzer electropherograms showed the typical RNA size distribution profiles for EVs, which were enriched with small RNAs (Figure 1F‐H), and lacked the rRNA peaks characteristic of their parental cells (Figure 1I‐K). These results indicate that osteoblasts and MSCs release EVs matching the characteristics of EVs.
Figure 1.
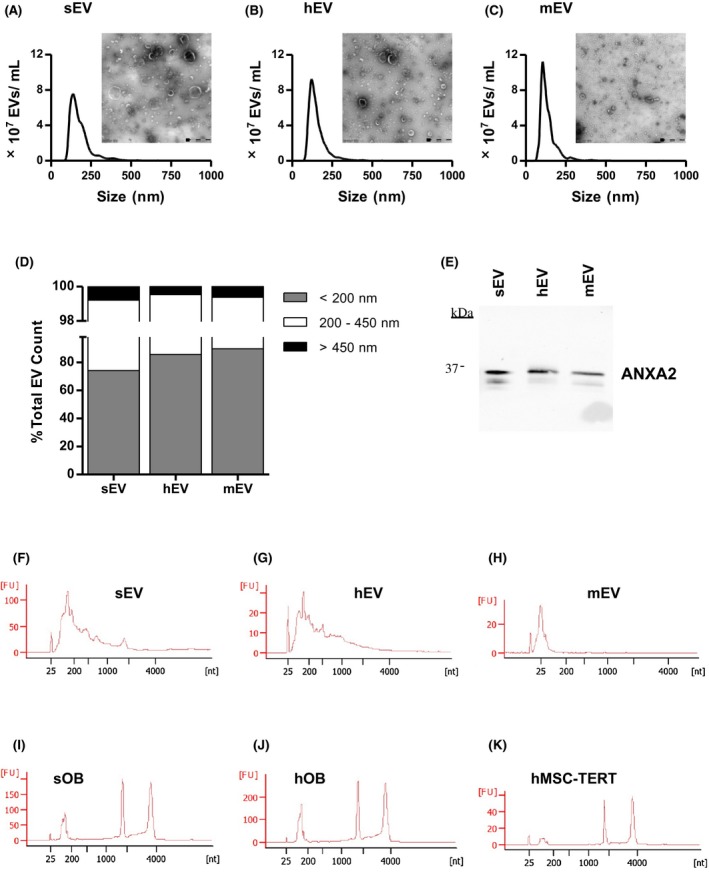
Characterization of EVs secreted by diverse human osteolineage cells. A‐C, Nanoparticle tracking analysis shows the size distribution and concentration of (A) sEVs, (B) hEVs and (C) and mEVs. (N = 3). Representative TEM images (magnification× 49 000) confirm the morphology of EVs. Scale bar: 500 nm. D, The percentage of small (<200 nm), medium (200‐450 nm) and large (>450 nm) EVs varies between the three different EV populations. E) Western blot analysis of EV proteins (1.8 μg/lane) in sEVs, hEVs and mEVs using an antibody against ANXA2, a common EV marker. F‐K, Representative Agilent Bioanalyzer electropherograms show the size distribution of total RNA extracted from (F) sEVs, (G) hEVs, (H) mEVs, (I) sOBs, (J) hOBs and (K) hMSC‐TERTs, and FU, fluorescent units. (N = 3)
Table 1.
The list of sEV‐miRNAs significantly (P < .05) enriched (FC ≥ 2) in sEVs compared to hEVs
| Mature miRNAs | sEVs (Reads) | hEVs (Reads) | Log2 FC | P value |
|---|---|---|---|---|
| miR‐146a‐5p | 14 551.19 | 160.90 | 6.50 | 3.85E−06 |
| miR‐146b‐5p | 1063.74 | 21.49 | 5.63 | 1.62E−04 |
| miR‐32‐5p | 78.91 | 9.62 | 3.04 | 1.29E−02 |
| miR‐641 | 30.59 | 4.39 | 2.80 | 2.53E−02 |
| let‐7i‐5p | 27 918.11 | 5628.69 | 2.31 | 7.04E−04 |
| miR‐101‐3p | 1018.80 | 217.92 | 2.23 | 5.34E−03 |
| miR‐3615 | 40.43 | 9.82 | 2.04 | 3.45E−02 |
| miR‐29b‐3p | 176.45 | 43.11 | 2.03 | 3.00E−03 |
| miR‐30a‐5p | 5788.05 | 1518.24 | 1.93 | 3.13E−03 |
| let‐7b‐5p | 1759.00 | 513.11 | 1.78 | 5.18E−04 |
| miR‐31‐5p | 225.66 | 67.76 | 1.74 | 3.93E−04 |
| miR‐210 | 65.93 | 19.92 | 1.73 | 5.52E−03 |
| miR‐186‐5p | 365.18 | 119.47 | 1.61 | 1.02E−03 |
| miR‐331‐3p | 44.69 | 14.65 | 1.61 | 4.70E−03 |
| miR‐140‐3p | 1082.70 | 354.99 | 1.61 | 1.66E−03 |
| miR‐148a‐3p | 392.39 | 128.89 | 1.61 | 3.48E−02 |
| miR‐30e‐5p | 259.16 | 94.36 | 1.46 | 1.70E−02 |
| miR‐140‐5p | 177.31 | 65.37 | 1.44 | 5.93E−03 |
| miR‐378c | 57.08 | 21.36 | 1.42 | 3.51E−02 |
| miR‐126‐3p | 234.67 | 88.43 | 1.41 | 7.23E−04 |
| miR‐19b‐3p | 117.37 | 46.36 | 1.34 | 1.30E−02 |
| miR‐3613‐5p | 41.63 | 16.57 | 1.33 | 2.97E−02 |
| miR‐598 | 19.70 | 8.18 | 1.27 | 4.44E−02 |
| let‐7b‐3p | 12.53 | 5.21 | 1.27 | 4.65E−02 |
| let‐7g‐5p | 7340.52 | 3236.38 | 1.18 | 2.36E−03 |
| let‐7f‐5p | 27 129.12 | 11 994.55 | 1.18 | 4.10E−03 |
| miR‐192‐5p | 229.02 | 103.88 | 1.14 | 2.11E−02 |
| miR‐28‐5p | 86.46 | 39.48 | 1.13 | 2.06E−04 |
| miR‐194‐5p | 51.00 | 23.67 | 1.11 | 4.85E−02 |
| miR‐143‐3p | 2697.25 | 1260.08 | 1.10 | 1.84E−02 |
| miR‐107 | 97.10 | 46.49 | 1.06 | 1.82E−02 |
| miR‐377‐3p | 10.34 | 5.02 | 1.04 | 3.93E−02 |
| miR‐502‐3p | 35.68 | 17.32 | 1.04 | 1.69E−02 |
| miR‐16‐2‐3p | 150.76 | 73.31 | 1.04 | 2.76E−02 |
| miR‐29c‐3p | 13.52 | 6.69 | 1.01 | 3.75E−02 |
3.2. EVs from different osteolineage sources do not have same HSPC expansion potential
Next, we investigated the potency of the different osteolineage EVs to promote ex vivo expansion of human UC‐HSPCs. CD34+‐selected UC‐HSPCs were expanded in growth factor (SCF and Flt3L)‐driven serum‐free expansion cultures for 10 days with or without EVs. In the absence of EVs, growth factor‐containing medium alone induced a 6.1‐fold and 3.33‐fold expansion of total number of viable nucleated cells (TNCs) (Figure 2A) and CD34+ cells(Figure 2B), respectively. Fold change (FC) expansions of TNCs and CD34+ cells were not significantly altered by the addition of neither hEVs nor mEVs, at any given EV concentration. However, treatment of cells with the low concentration of sEV (0.2 × 107 EVs/μL) induced a small but significantly enhanced number of TNCs (P < .05). Moreover, a five‐fold increased sEV concentration resulted in 13.44‐fold expansion of TNCs, and 7.77‐fold expansion of CD34+ cells, leading to 2.42‐fold more CD34+ cells after 10 days of culture, as compared to the control cultures(P < .005), showing a concentration dependent effect on cell expansion.We also counted the level of exhaustion of the most immature CD34+ cell subset after 10 days by multicolour flow cytometry using markers for primitive HSPCs (Lin‐ CD34+ CD38low CD45RAlow CD90+), referred to as phenotypic HSCs (Supporting Information Figure S1). Similar to TNCs and CD34+ cells, treatment with hEVs and mEVs did not alter the absolute numbers of phenotypic HSCs compared to the control cultures supplemented only with SCF and Flt3L. On the contrary, sEVs increased the number of phenotypic HSCs compared to the control on day 10. Together, these findings demonstrate the unique and strong potency of fetal calvaria osteoblast‐derived sEVs to promote growth‐factor‐driven expansion of both CD34+ UC‐HSPCs and phenotypic HSCs, and show that EV‐effect is parental cell type‐specific.
Figure 2.
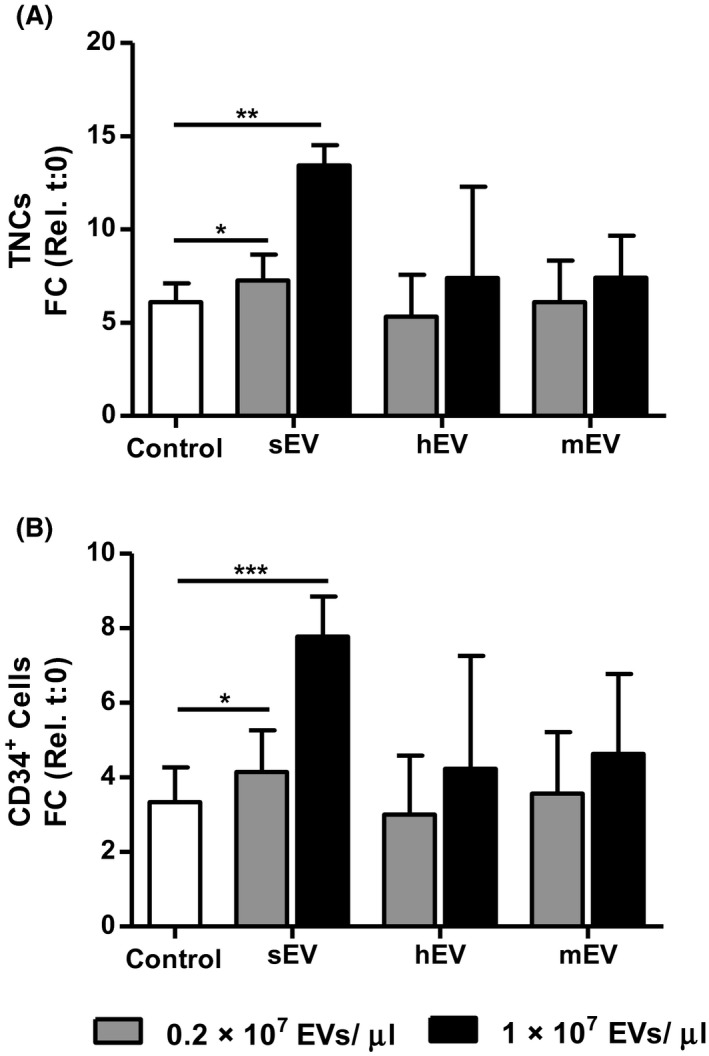
sEVs enhance ex vivo expansion of human CD34+UC‐HSPCs. A and B, sEVs increase the ex vivo expansion of (A) total nucleated cells (TNCs) and (B) CD34+ cells after 10 days of incubation with SCF and Flt3L compared to cells cultured in the absence of EVs (control). hEVs and mEVs do not have a significant effect on expansion. Expansion is shown as fold change (FC) increase (mean ± SD) in total cell number compared to input on day 0. *P < .05, **P < .01, ***P < .005
3.3. sEVs contain an overrepresented set of miRNAs compared to hEVs
MiRNAs are key players in maintaining the fine balance between the number of stem cells and their differentiated progeny.36, 37 To delineate the key EV components that support UC‐HSPC expansion, we compared the miRNA profiles of stimulatory and non‐stimulatory EVs using next‐generation sequencing. In an effort to define a selective list of candidate sEV‐miRNAs responsible for UC‐HSPC support, we focused only on sEVs and hEVs, which derive from extensively characterized immortalized osteoblasts. Interestingly, the majority of the cellular miRNAs were shared between the two osteoblastic parental cells (88.4% of sOB‐ and 83.2% of hOB‐derived miRNAs), and showed similarities in cellular abundance (Supporting Information Figure S2A).In total, we detected 672 mature miRNAs in all replicates for at least one of the samples and 317 miRNAs are detected in all samples (Figure 3A). Smaller portions of cellular miRNAs were represented in their corresponding EVs: only 76.1% (427 out of 561) of sOB‐derived miRNAs and 57% (340 out of 596) of hOB‐derived miRNAs were represented in sEVs and hEVs, respectively. The expression level of vesicular miRNAs was a reflection of the cellular abundance for both EV populations (Supporting Information Figure S2B,C). Nevertheless, 45 miRNAs in sEVs and 43 miRNAs in hEVs (among which 22 are shared) were significantly (P < .05) more abundant (FC ≥ 2) in EVs compared to their parental cells (FC ≥ 2, P < .05), suggesting selectivity of vesicular cargo packaging. Principal component analysis verified the high similarity of the two osteoblastic cell lines on a molecular level, while the clusters of miRNAs found in sEVs and hEVs separated from each other as well as from their parental cells (Figure 3B).These results indicate that molecularly very similar osteoblastic cell lines have different EV miRNA loading mechanisms, which is independent of intracellular miRNA levels.
Figure 3.
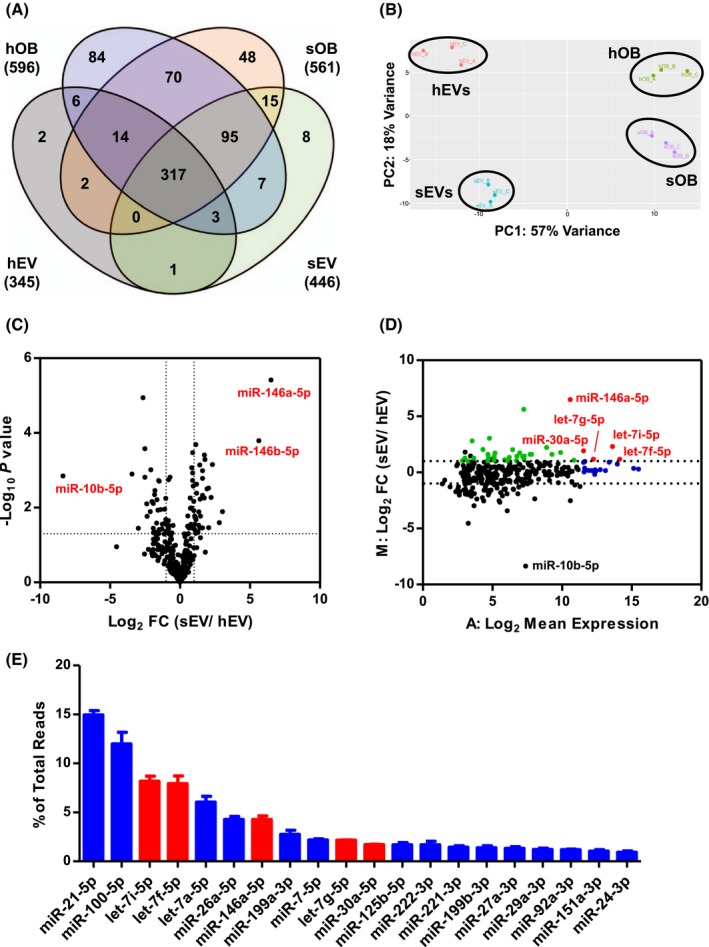
Comparative next‐generation sequencing miRNA profiling of stimulatory and non‐stimulatory EVs. A, Venn diagram shows the number of miRNAs detected in parental cells and their corresponding EVs. Numbers in brackets show the total number of miRNAs detected in a given group. (N = 3). B, Principal component analysis confirms the similarities between the biological/technical replicates and shows the distinct variance between cellular and vesicular miRNA content. C, Volcano plot (significance vs. FC) shows the significantly (P < .05) enriched (FC ≥ 2) miRNAs in sEVs compared to non‐stimulatory hEVs. D, MA plot (FC vs. mean expression) highlights the abundance of the selectively enriched sEV‐miRNAs. The significantly overrepresented sEV‐miRNAs, which are also among the top 20 most abundant sEV‐miRNAs are shown in red. The rest of the significantly overrepresented sEV‐miRNAs are shown in green. The rest of top 20 most abundant sEV‐miRNAs are shown in blue. E, The 20 candidate sEV‐miRNAs are displayed according to the normalized read count proportions (mean ± SD) of the top 20 most abundant sEV‐miRNAs in connection to the color‐coding of the MA plot
Next, we compared the miRNA content of sEVs and hEVs and followed a selection strategy to generate a list of candidate sEV‐miRNAs, which may drive UC‐HSPC expansion. Direct comparison of miRNA profiles of sEVs and hEVs revealed that 35 miRNAs are overrepresented in sEVs (FC ≥ 2, P < .05, Table 1), while 32 miRNAs were significantly depleted in sEVs (Figure 3C).We used Ingenuity Pathway Analysis (IPA) tool to determine the top diseases and biological functions associated with the enriched sEV‐miRNAs. Target genes of the miRNAs enriched in sEVs were mostly annotated(P < .05) to cancer and hematological disease, with most of the molecular and cellular functions annotated to cellular proliferation, including proliferation of lymphoma cells (miR‐101‐3p, miR‐146a‐5p, 19b‐3p, 708‐5p); cell death, including apoptosis of leukemic cells (miR‐29b‐3p);and cell cycle (let‐7a‐5p, miR‐92a‐3p). It is important to note that 30 out of the35 overrepresented sEV‐miRNAs contributed to less than 1% of the total reads, leading to questioning whether they are of biological significance. On the contrary, five overrepresented sEV‐miRNAs, such as miR‐146a‐5p, miR‐30a‐5p, let‐7f‐5p, let‐7g‐5p, and let‐7i‐5p, together made up 24.25% of the total reads (Figure 3D). Moreover, these five overrepresented sEV‐miRNAs were also among the top 20 miRNAs in sEVs with the highest number of reads, making them highly interesting candidates for induction of UC‐HSPC expansion (Figure 3E).
3.4. sEVs and hEVs have distinct protein profiles
Next, we performed label‐free mass spectrometry‐based proteomics analysis to investigate the differential protein expression between stimulatory and non‐stimulatory EVs. We identified a total of 3581 proteins in all replicates for at least one EV group after strict filtering; 2992 proteins were detected in sEVs and 3248 proteins in hEVs. The majority (2659) of the proteins was commonly found in both EVs and had similar abundances based on label‐free quantification (LFQ) values (Supporting Information Figure S3A).The majority (~93%) of the most commonly identified EV markers were detected at high level in both of EV groups.38 Top five most abundant proteins for both groups were the well‐defined EV markers: ANXA1, ANXA2, ANXA5, GAPDH, and HIST1H4A.
Volcano plot in Figure 4A shows the differentially expressed (FDR 0.05, S0 correction of 2—corrected for multiple testing) proteins between sEVs and hEVs. Among the 420 differentially expressed proteins, 152 proteins were overrepresented in sEVs (Table 2), and 268 proteins in hEVs. To gain insights into the potential molecular functions of the overrepresented proteins in sEVs, we conducted several Gene Ontology (GO) enrichment analyses using different tools, such as PANTHER to define the general GO categories (Figure 4B) and FunRich to investigate the detailed functional terms within a category (Figure 4C). The majority of the overrepresented sEV‐proteins were annotated to binding (GO:0005488), including molecular functions such as DNA binding (HMGA1, HIST1H2BM, DDB2, MECP2) and cell adhesion (EPCAM, ICAM1, ITGA7); and structural molecule activity (GO:0005198), such as extracellular matrix structural constituent (LAMA1, THBS2, COL7A1, EMILIN1). The remaining enriched sEV‐proteins were associated with transcription regulator activity (GO:0140110), such as DNA‐directed RNA polymerase activity (POLR3A, POLR1D); and molecular function regulator (GO:0098772), such as peptidase activity (DPP4, PMPCA). On the contrary, hEVs were significantly enriched with proteins mainly involved in transporter activity (GO:0005215), structural molecule activity (GO:0 005 198), and catalytic activity (GO:0003824) (Figure S3B). Figure 4D displays the sEV‐proteins based on the top 20 most abundant proteins among the 152 overrepresented proteins. Interestingly, LAMA1, ABI3BP, and HMGA1, which were the top five most enriched proteins, were also highly abundant, making them interesting candidates to study their significance for induction of cell expansion. Both miRNA and protein profiling analysespropose a list of highly abundant and overrepresented 5sEV‐miRNAs and 3 sEV‐proteins that are associated with the role of fetal calvaria osteoblast‐derived sEVs in supporting UC‐HSPC proliferation.
Figure 4.
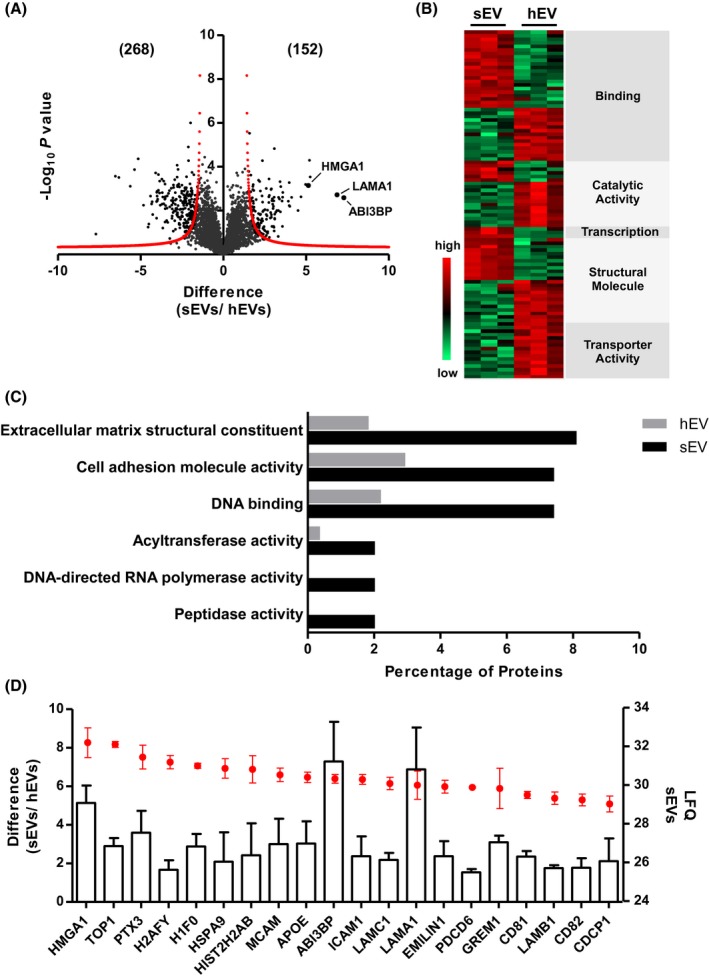
Proteomic profiling of sEVs and hEVs. A, Volcano plot shows the significantly enriched (FDR 0.05, S0 correction of 2, corrected for multiple testing) proteins in sEVs compared to hEVs. Numbers in brackets denote the number of differentially expressed proteins. (N = 3). B, Heat map of significantly enriched sEV‐ and hEV‐proteins categorized within with GO enrichment annotation based on molecular function. C, The percentages of proteins (enrichment factor) annotated to a subset of GO term for sEVs. D, The 20 candidate sEV‐proteins are displayed according to the expression levels (mean ± SD) (red dots, right Y axis) of the top 20 most abundant sEV‐proteins among the overrepresented 152 proteins as well as the enrichment levels (mean ± SD) (columns, left Y axis) of sEV‐proteins compared to hEV‐proteins
Table 2.
The list of significantly enriched (FDR 0.05, S0 correction of 2, corrected for multiple testing) proteins in sEVs compared to hEVs
| Protein Names | LFQ Diffa | P value | Protein Names | LFQ Diff | P value | Protein Names | LFQ Diff | P value |
|---|---|---|---|---|---|---|---|---|
| AAMP | 2.01 | 9.94E‐02 | GAR1 | 1.76 | 1.02E‐02 | NINJ1 | 3.05 | 3.55E‐03 |
| ABHD14B | 2.45 | 1.84E‐01 | GDF15 | 3.98 | 5.92E‐03 | NIT2 | 2.21 | 1.01E‐01 |
| ABI3BP | 7.29 | 2.58E‐03 | GLRX | 1.83 | 5.16E‐02 | NKTR | 2.79 | 2.03E‐03 |
| ACADVL | 3.31 | 1.52E‐01 | GNG10 | 2.64 | 1.74E‐02 | NPEPL1 | 1.70 | 1.55E‐02 |
| ACO2 | 1.94 | 8.91E‐02 | GNPNAT1 | 1.81 | 4.64E‐02 | NTM | 1.81 | 2.11E‐02 |
| ACOT1 | 2.12 | 1.11E‐01 | GREM1 | 3.10 | 1.44E‐02 | NTN1 | 3.17 | 1.17E‐02 |
| ACSF2 | 2.16 | 6.17E‐02 | GSPT1 | 2.62 | 9.36E‐02 | OAT | 2.39 | 6.53E‐02 |
| ADAMTSL1 | 2.61 | 2.46E‐02 | GTF2F1 | 2.76 | 3.59E‐04 | OCLN | 3.34 | 2.49E‐03 |
| AEBP1 | 2.84 | 2.16E‐02 | H1F0 | 2.88 | 1.10E‐03 | OSGEP | 1.62 | 3.18E‐03 |
| AKR1B1 | 1.90 | 7.65E‐02 | H1FX | 2.96 | 2.89E‐03 | PCK2 | 4.52 | 9.84E‐03 |
| AMPD3 | 2.07 | 3.77E‐02 | H2AFY | 1.67 | 3.12E‐03 | PDCD4 | 2.34 | 2.34E‐02 |
| APOC1 | 2.02 | 5.83E‐02 | H2AFY2 | 1.81 | 3.04E‐02 | PDCD6 | 1.54 | 1.64E‐04 |
| APOE | 3.03 | 6.35E‐03 | HAPLN1 | 3.48 | 8.08E‐04 | PLAU | 2.60 | 9.67E‐04 |
| ATE1 | 1.79 | 5.04E‐02 | HDGF | 1.51 | 1.30E‐03 | PMPCA | 2.69 | 9.13E‐02 |
| BCCIP | 1.84 | 1.21E‐03 | HDGFRP3 | 4.97 | 6.38E‐04 | PNPO | 2.15 | 5.05E‐02 |
| BGN | 2.79 | 1.68E‐01 | HIST1H1A | 3.06 | 5.84E‐02 | POLR1D | 2.09 | 3.68E‐02 |
| BRD3 | 1.98 | 5.27E‐02 | HIST1H1D | 1.78 | 7.81E‐04 | POLR3A | 3.40 | 2.97E‐03 |
| C1R | 2.24 | 1.28E‐03 | HIST1H2BM | 4.70 | 2.36E‐03 | POLR3B | 3.09 | 1.49E‐05 |
| C3 | 1.93 | 1.07E‐02 | HIST2H2AB | 2.42 | 1.17E‐01 | PPWD1 | 1.55 | 3.63E‐03 |
| CCNL1 | 1.96 | 4.30E‐02 | HMGA1 | 5.14 | 7.19E‐04 | PTX3 | 3.60 | 1.48E‐03 |
| CD81 | 2.36 | 5.28E‐05 | HMGA2 | 3.58 | 2.45E‐03 | RHBDF2 | 2.75 | 2.36E‐03 |
| CD82 | 1.78 | 1.09E‐03 | HMGN1 | 5.25 | 2.89E‐04 | RPL10 | 1.98 | 4.64E‐02 |
| CDCP1 | 2.12 | 1.37E‐02 | HSPA4L | 1.64 | 3.77E‐03 | RRAD | 2.28 | 3.60E‐02 |
| CDH11 | 1.85 | 6.94E‐03 | HSPA9 | 2.09 | 1.20E‐01 | SFRP1 | 4.02 | 6.37E‐03 |
| CECR5 | 2.42 | 5.76E‐02 | ICAM1 | 2.38 | 1.11E‐02 | SH2D4A | 2.94 | 5.24E‐02 |
| CIAO1 | 2.13 | 4.07E‐02 | ITGA7 | 1.66 | 2.42E‐04 | SHMT2 | 2.51 | 8.20E‐02 |
| CMSS1 | 3.93 | 3.75E‐03 | JAM2 | 2.51 | 8.97E‐03 | SLC29A1 | 1.58 | 2.98E‐06 |
| COL7A1 | 2.08 | 1.04E‐01 | KATNAL2 | 4.99 | 6.46E‐04 | SLC39A14 | 1.81 | 6.45E‐04 |
| COMMD5 | 1.63 | 4.11E‐03 | LAMA1 | 6.88 | 1.91E‐03 | SLIRP | 2.94 | 3.54E‐03 |
| CRIP2 | 2.18 | 4.39E‐02 | LAMB1 | 1.76 | 2.60E‐03 | SNRPC | 2.75 | 4.27E‐03 |
| CS | 2.38 | 2.94E‐02 | LAMB2 | 4.13 | 2.50E‐03 | SNX18 | 2.34 | 2.14E‐03 |
| CXCL1 | 3.55 | 3.68E‐03 | LAMC1 | 2.19 | 4.73E‐04 | SPIN1 | 1.76 | 2.50E‐02 |
| CYR61 | 1.81 | 4.72E‐03 | LHFPL2 | 3.10 | 2.42E‐02 | STC1 | 2.07 | 3.81E‐02 |
| DDB2 | 3.98 | 5.28E‐04 | LONP1 | 2.67 | 1.07E‐01 | SUCLG1 | 3.07 | 8.46E‐02 |
| DDT | 1.58 | 3.73E‐03 | LRIG1 | 2.22 | 1.54E‐02 | SUSD5 | 2.29 | 2.79E‐02 |
| DENR | 2.90 | 1.53E‐02 | LRRC15 | 3.43 | 4.75E‐04 | TEP1 | 3.44 | 1.13E‐02 |
| DNAJC3 | 1.70 | 1.50E‐02 | LUC7L | 3.04 | 5.26E‐02 | TFAM | 2.16 | 8.38E‐02 |
| DPP4 | 2.12 | 2.58E‐02 | LYPLA2 | 2.25 | 1.45E‐02 | TFPI2 | 3.33 | 9.93E‐03 |
| EFNB1 | 2.20 | 9.19E‐02 | MAP1S | 1.80 | 8.95E‐03 | THBS2 | 3.24 | 9.83E‐03 |
| ELP2 | 1.84 | 3.43E‐02 | MCAM | 3.00 | 8.03E‐03 | THY1 | 1.84 | 9.06E‐03 |
| ELTD1 | 2.42 | 1.00E‐02 | MECP2 | 5.20 | 5.03E‐05 | TMEM55B | 2.14 | 1.26E‐01 |
| EMILIN1 | 2.38 | 4.08E‐03 | METTL1 | 1.98 | 3.10E‐03 | TOMM70A | 2.11 | 8.71E‐02 |
| EMILIN2 | 1.81 | 3.70E‐03 | MFI2 | 2.68 | 9.46E‐03 | TOP1 | 2.90 | 1.02E‐03 |
| EPCAM | 2.13 | 4.09E‐04 | MGLL | 2.28 | 1.13E‐01 | TPP2 | 2.59 | 1.52E‐02 |
| F11R | 2.46 | 5.40E‐02 | MMAB | 2.09 | 2.31E‐02 | TRMT1 | 3.08 | 2.46E‐03 |
| FAM45A | 1.66 | 1.37E‐03 | MMP2 | 3.57 | 5.24E‐02 | TRMT11 | 1.99 | 8.35E‐02 |
| FBLN2 | 1.60 | 3.92E‐03 | MRGPRF | 2.91 | 2.02E‐02 | TRMT61A | 2.87 | 1.23E‐02 |
| FDXR | 2.59 | 1.37E‐01 | MYL1 | 2.98 | 5.06E‐03 | TST | 2.46 | 7.43E‐02 |
| FKBP5 | 1.81 | 1.63E‐02 | NADK2 | 2.30 | 1.07E‐01 | USP43 | 1.76 | 6.58E‐03 |
| FLT1 | 2.07 | 2.25E‐02 | NFIC | 2.19 | 4.85E‐02 | WNT5A | 3.11 | 2.27E‐02 |
| FZD2 | 2.14 | 1.14E‐01 | NHP2L1 | 1.72 | 2.84E‐02 |
Diff: LFQ value difference between sEV and hEV
3.5. sEVs alter the gene expression of CD34+UC‐HSPCs
Next, we investigated the alterations on the transcriptome profiling of CD34+UC‐HSPCs after 24 hours treatment with sEVs. We identified 33 genes, of which 17 genes were upregulated and 16 genes were downregulated more than 1.5‐fold (P < .05) inCD34+ cells by sEVs, as compared to the control treated cells (Figure 5A). We confirmed the differential expression of selected genes by qPCR as shown in Figure 5B. Despite the donor‐related variation, on average gene expression was in the same direction as determined by sequencing for all the tested genes. We then analyzed the patterns of change in the expression levels of the 33 regulated genes between control and sEV‐treated cells (t = 24 hours cell culture) compared to the expression levels at the starting time‐point (t = 0). We identified four different expression profiles: enhanced upregulation (Figure 5C), lowered upregulation (Figure 5D), enhanced downregulation (Figure 5E), and lowered downregulation (Figure 5F) of endogenous (control) gene expression as a response to sEV‐treatment in 24 hours. For the majority of the genes, sEV‐treatment enhanced the endogenous upregulation or downregulation induced by culture conditions (Figure 5C,E). For a small number of genes, however, sEVs altered gene expression by lowering the extent of endogenous upregulation of MAOA and NFIB and TGFB1I1 (Figure 5D), and downregulation of BCL3, DUSP3, PHLDB2, PIM1, and S100A9 (Figure 5F).
Figure 5.
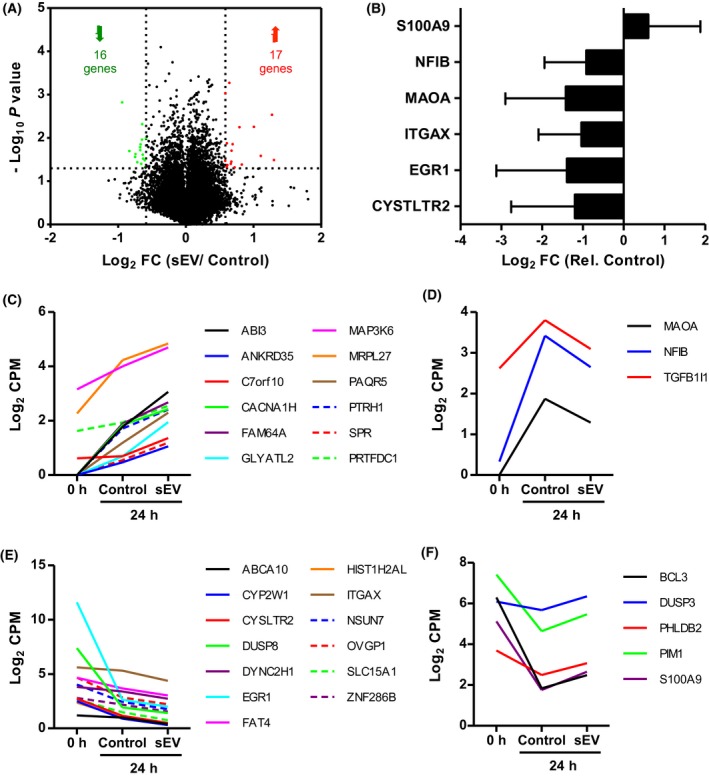
Expression profiles of differentially expressed CD34+UC‐HSPC genes upon sEV treatment. A, Volcano plot shows the CD34+UC‐HSPC genes that are significantly (P < .05) regulated (FC ≥ 1.5) after 24 h of culture in the absence (control) and presence of sEVs (N = 2). Numbers in brackets denote the number of up‐regulated (red, 17 genes) and down‐regulated (green, 16 genes). B, Expression levels (mean ± SD) of selected CD34+UC‐HSPC genes after 24 h of incubation in the presence of sEVs (N = 3). FC is determined compared to control and normalized to average GAPDH expression. The donors used for qPCR analysis are different than the ones used for mRNA sequencing. C‐F, Expression profiles show the temporal pattern of gene expression as a response to sEV treatment defined by normalized read counts. sEVs enhance the (C) up‐regulation and (E) down‐regulation of CD34+UC‐HSPC genes by control alone in 24 h. sEVs lower the control‐induced (D) up‐regulation and (F) down‐regulation of CD34+UC‐HSPC genes by control alone in 24 h
According to GO analysis, the 33 regulated genes were mostly (P < .05) annotated to MAPK signalling pathway (CACNA1H, DUSP3, DUSP8, MAP3K6). Further investigation using IPA indicated that sEVs modulate the expression of CD34+ UC‐HSPC genes mostly annotated (P < .05) to cellular development, cellular growth and proliferation, cell‐to‐cell signaling and interaction, small molecule biochemistry, and cell cycle. A set of genes, including BCL3, EGR1, ITGAX, CYSLTR2, PIM1, and S100A9, were of particular interest as they are mainly related to the expansion of blood cells. These findings show that fetal calvaria osteoblast‐derived sEVs are capable of altering the expression pattern of cellular growth‐related CD34+ UC‐HSPC genes, supporting our previous studies reporting the proliferative effect of sEV cargo.25
3.6. Candidate key molecular players of sEV function
To investigate the molecular basis of sEV‐mediated gene regulation in UC‐HSPCs, we correlated the sEV content to the regulated genes in UC‐HSPCs using an integrative bioinformatics approach. We used IPA to build a network map showing the tentative molecular relationships between the sEV cargo, such as proteins and miRNAs, and the sEV‐regulated CD34+UC‐HSPC genes (Figure 6).Notably, here, we only analyzed the highly abundant miRNAs found in sEVs with normalized read counts above 100, and correlated these miRNAs only to the downregulated genes in treated CD34+UC‐HSPCs. Among the regulated genes, the highest number of EV cargo, including both miRNAs and proteins, targeted EGR1. Furthermore, PHLDB2 and S100A9 were the most common putative targets of the proteins alone. On the contrary, ZNF286B and NFIB were targeted by the highest number of miRNAs, with the former being targeted by miRNAs only. The majority of the mapped proteins were annotated to biological processes, such as cell communication (EPCAM, ICAM1), regulation of nucleic acid metabolism (HMGA1, MECP2, ZNF22), and cell growth and/or maintenance (LAMA1, SPTAN1). Combined analysis of the mapped sEV‐derived protein and miRNAcargo indicated that PI3K/AKT and PTEN signalling were among the top overrepresented canonical pathways that may be targeted in UC‐HSPCs.Remarkably, all three of the candidate sEV‐proteins, speculated to contribute to the sEV‐mediated UC‐HSPC support, were among the mapped proteins. Among these, LAMA1 was mapped to DUSP8, HMGA1 to EGR1, and ABI3BP to ABI3. On the contrary, among the candidate sEV‐miRNAs only miRNAs of the let‐7 family were mapped to an UC‐HSPC gene:ZNF286. Together, these analyses link the fetal calvaria osteoblast‐derived sEV cargo to the regulation of CD34+ UC‐HSPC gene expression, enabling valuable new insights into the molecular complexity of EV‐mediated osteolineage‐cell‐HSPC crosstalk.
Figure 6.
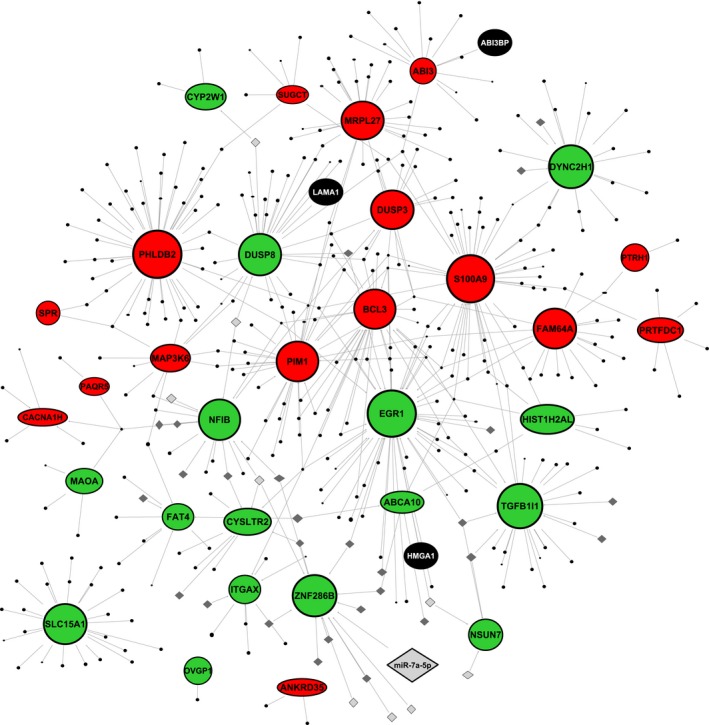
IPA network showing the direct relationship maps between the sEV cargo and sEV‐regulated CD34+UC‐HSPC genes. Green, down‐regulated CD34+UC‐HSPC genes; red, up‐regulated CD34+UC‐HSPC genes; black ovals, sEV‐proteins; grey diamonds, sEV‐miRNAs. The names of the candidate sEV‐proteins and ‐miRNAs, which mapped to the regulated UC‐HSPC genes are displayed on the map
4. DISCUSSION
The discovery of specialized EVs delivering regulatory molecules to target cells has been the hallmark of a recently discovered novel form of intercellular communication. Using comparative proteomics, miRNA sequencing, and bioinformatics analyses, we showed that human fetal calvaria osteoblast‐derived sEVs support human UC‐HSPC expansion and are selectively enriched with a unique set of protein and miRNA cargo. Through an integrative bioinformatics approach, we unified the sEV cargo repositories with the sEV‐regulated CD34+UC‐HSPC transcriptome and identified novel players and molecular mechanisms of EV‐mediated osteolineage cell‐HSPC crosstalk.
Increasing knowledge on key players of the bone marrow niche, where HSC self‐renewal is supported, greatly contributed to the establishment of optimized expansion conditions that support expansion of HSPCs in culture.39, 40, 41, 42 We previously demonstrated that osteoblast‐derived EVs cooperate with hematopoietic growth factors not only to promote ex vivo proliferation, but also to preserve the repopulating activity of CD34+HSPCs.25 However, not all niche cells secrete EVs with the same HSPC supporting potential. In an attempt to avoid interpretation of our results due to the possibility of isolated incidents of the selected cell line, we conducted our experiments using osteolineage cells of different origins. The addition of sEVs deriving from the fetal calvarias OBs significantly increases the number of CD34+UC‐HSPCs in expansion cultures by at least twofold. However, treatments with hEVs deriving from the limb tissue hOBs and immortalized MSC‐derived mEVs do not improve growth factor‐induced UC‐HSPC expansion. It is important to bear in mind that the effect of hEVs and mEVs may be masked by the intrinsic growth rate of the control alone. Moreover, contribution of internalized EVs to CD34 expression levels, as well as the effect of cellular processes, such as circadian rhythm and asymmetric cell division, on CD34 surface marker expression pattern should be further investigated to understand the differences between the EVs. The differential UC‐HSPC growth promoting effects of sEVs, hEVs and mEVs is most likely attributed to the content differences between the EVs.
EV secretion is an evolutionarily conserved form of communication among cells, and therefore, most EVs share biologically relevant EV cargo regardless of the type of parental cells.43, 44 Advancement of systematic and comprehensive studies has helped us identify the biologically active EV components and study their physiological relevance. Proteomic and sequencing analyses demonstrate selective incorporation of a different set of functional proteins and miRNAs inside sEVs and hEVs. Striking examples of candidate cargo include the highly abundant sEV‐miRNAs, such as let‐7i‐5p, let‐7f‐5p, miR‐146a‐5p, let‐7g‐5p, and miR‐30a‐5p, as well as sEV‐proteins, such as LAMA1, ABI3BP, and HMGA1. MiR‐146a, the most overrepresented miRNA in sEVs, plays an important role in hematopoietic stem cell differentiation.45, 46 Starczynowski et al showed that miR‐146a expression is much higher in CD34+ cells compared to the CD34‐ precursor cells, leading to the inhibition of hematopoietic differentiation.47 Interestingly, let‐7 family members are known to be tightly cooperating with miR‐99a/100 and miR‐125b‐1/2, which are also highly abundant in sEVs, to control hematopoietic growth.48 Remarkably, HMGA1 along with HMGA2 is a part of the Lin28‐let7 pathway, which regulates fetal cell‐like stem cell maintenance in adult HSCs.49, 50, 51 The co‐existence of let‐7 and HMGA1 within EVs suggests that the EV cargo consists of a combination of functionally diverse structural and regulatory biomolecules that must work in concert to convey the final message.
The remarkable power of active sorting mechanism renders EVs with unique combinations of EV cargo that may act in concert to modulate the expression levels of their target genes. Understanding the role of EVs in hematopoiesis, therefore, requires unraveling the way EV regulators interact with each other to regulate transcriptional networks. In accordance with HSPC support studies, sEV‐regulated UC‐HSPC genes are mostly annotated to cell growth related processes. Particularly, sEVs alter the expression patterns of key players of HSPC self‐renewal, such as BCL3, EGR1, ITGAX, CYSLTR2, PIM1, and S100A9. 17, 52, 53 Interestingly, some of these genes are among the tentative targets of the sEV cargo. The highest number of sEV cargo, including the candidate EV‐protein and ‐miRNAs, targets EGR1, which encodes for a transcription factor that plays a key role in the regulation of hematopoietic homeostasis. Previous studies showed that EGR1 levels are high in quiescent cells and they are dramatically downregulated upon cell division.54 Accordingly, we demonstrate that EGR1 levels drop after 24 hours as the UC‐HSPCs progress into cell cycle. Interestingly, the downregulation is further enhanced in the presence of sEVs most likely due to the ability of EVs to drive the cell division kinetics of HSPCs.25 S100A9 is another predominantly targeted UC‐HSPC gene. Previous reports showed that S100A9 expression is upregulated during HSC maturation.55 Here, S100A9 levels are downregulated by sEVs, suggestive of an effort for EVs to maintain the cells in a more immature state. Moreover, NFIB is one the targets of the majority of sEV‐miRNAs. The downregulation of NFIB has been shown to be critical during the transition of the immature HSCs to the multipotent progenitors.56 Previously, we demonstrated that osteoblast‐EVs stimulate the expansion of the progenitor cells while maintaining the immature stem cell pool.25 Although it is tempting to speculate a role for NFIB targeting in the expansion of the progenitors, further functional studies are needed to investigate the detailed molecular mechanisms.
In conclusion, the findings described in this study provide a comprehensive human fetal osteoblast‐derived EV cargo repository useful for the functional exploration of EV‐mediated regulation of osteolineage cell‐HSPC crosstalk.UC‐HSPC expansion analyses combined with comparative omics studies identified candidate fetal calvaria osteoblast‐derived sEV cargo, which provided novel insights into the EV‐driven molecular mechanisms that trigger UC‐HSPC expansion. However, it is still challenging to assess the extent to which individual or combinations of cargo contribute to these effects. Further investigation focusing on the generation of modified EVs devoid of particular candidates or engineered EVs loaded with selective cargo is promising to elucidate the biological roles of the EV cargo.
CONFLICT OF INTEREST
The authors declare no competing financial interest.
AUTHOR CONTRIBUTIONS
J. Morhayim, J. van de Peppel, E. Braakman, C. Voermans, J.J. Cornelissen, J.P. van Leeuwen, and S.J. Erkeland conceived the study; J. Morhayim, J. van de Peppel, E. Braakman, C. Voermans, B. van der Eerden and C. Ghebes designed the experiments; J. Morhayim wrote the manuscript; J. Morhayim conducted the experiments and analyzed the data; M. ter Borg performed ex vivo expansion studies; R.M. Hoogenboezem and E.M.J Bindels performed miRNA sequencing and bioinformatic analysis; A.J. van Wijnen performed mRNA sequencing and bioinformatics analysis; F.J. van Alphen performed proteomics studies and bioinformatics analysis; M. Kassem generated the human MSC‐TERT cell line. All authors reviewed the final manuscript.
Supporting information
ACKNOWLEDGMENTS
This work was supported by grants from Erasmus MC (Mrace grant 2015) and Sanquin (PPOC‐16‐05). Additional support was provided by US National Institutes of Health grants (R01‐AR049069 to AJvW and F32‐AR066508 to AD). The authors thank M. Kleijer for fruitful scientific discussions, M. van de Biggelaarat Sanquin for mass spectrometry and proteomics analysis, A. Dudakovic, J. Evans and A. Nair at Mayo Clinic for the analysis of high‐throughput mRNA sequencing, N. van der Wel and AE Grootemaat at AMC‐UvA for assisting with electron microscopy, I. Aydogan for help with sample preparation and IJ Robbesom‐van den Berge for cell cultures.
Morhayim J, Ghebes CA, Erkeland SJ, et al. Identification of osteolineage cell‐derived extracellular vesicle cargo implicated in hematopoietic support. The FASEB Journal. 2020;34:5435–5452. 10.1096/fj.201902610R
REFERENCES
- 1. Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759‐806. [DOI] [PubMed] [Google Scholar]
- 2. Brunstein CG, Gutman JA, Weisdorf DJ, et al. Allogeneic hematopoietic cell transplantation for hematologic malignancy: relative risks and benefits of double umbilical cord blood. Blood. 2010;116:4693‐4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker JN, Scaradavou A, Stevens CE. Combined effect of total nucleated cell dose and HLA match on transplantation outcome in 1061 cord blood recipients with hematologic malignancies. Blood. 2010;115:1843‐1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pineault N, Abu‐Khader A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol. 2015;43:498‐513. [DOI] [PubMed] [Google Scholar]
- 5. de Lima M, McNiece I, Robinson SN, et al. Cord‐blood engraftment with ex vivo mesenchymal‐cell coculture. N Engl J Med. 2012;367:2305‐2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su YH, Cai HB, Ye ZY, Tan WS. BMP‐7 improved proliferation and hematopoietic reconstitution potential of ex vivo expanded cord blood‐derived CD34 cells. Hum Cell. 2014;28:14‐21. [DOI] [PubMed] [Google Scholar]
- 7. Krater M, Jacobi A, Otto O, et al. Bone marrow niche‐mimetics modulate HSPC function via integrin signaling. Sci Rep. 2017;7:2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taichman RS, Emerson SG. Human osteoblasts support hematopoiesis through the production of granulocyte colony‐stimulating factor. J Exp Med. 1994;179:1677‐1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836‐841. [DOI] [PubMed] [Google Scholar]
- 10. Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841‐846. [DOI] [PubMed] [Google Scholar]
- 11. Visnjic D, Kalajzic Z, Rowe DW, Katavic V, Lorenzo J, Aguila HL. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258‐3264. [DOI] [PubMed] [Google Scholar]
- 12. Pinho S, Lacombe J, Hanoun M, et al. PDGFRalpha and CD51 mark human nestin+ sphere‐forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med. 2013;210:1351‐1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem‐cell maintenance. Nature. 2013;495:227‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varnum‐Finney B, Brashem‐Stein C, Bernstein ID. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood. 2003;101:1784‐1789. [DOI] [PubMed] [Google Scholar]
- 15. Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin‐1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149‐161. [DOI] [PubMed] [Google Scholar]
- 16. Bhatia M, Bonnet D, Wu D, et al. Bone morphogenetic proteins regulate the developmental program of human hematopoietic stem cells. J Exp Med. 1999;189:1139‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko KH, Holmes T, Palladinetti P, et al. GSK‐3beta inhibition promotes engraftment of ex vivo‐expanded hematopoietic stem cells and modulates gene expression. Stem Cells. 2011;29:108‐118. [DOI] [PubMed] [Google Scholar]
- 18. Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre‐microRNAs. Nucleic Acids Res. 2010;38:215‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nair R, Santos L, Awasthi S, et al. Extracellular vesicles derived from preosteoblasts influence embryonic stem cell differentiation. Stem Cells Dev. 2014;23:1625‐1635. [DOI] [PubMed] [Google Scholar]
- 20. Ekstrom K, Valadi H, Sjostrand M, et al. Characterization of mRNA and microRNA in human mast cell‐derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J Extracell Vesicles. 2012;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goloviznina NA, Verghese SC, Yoon YM, Taratula O, Marks DL, Kurre P. Mesenchymal stromal cell‐derived extracellular vesicles promote myeloid‐biased multipotent hematopoietic progenitor expansion via toll‐like receptor engagement. J Biol Chem. 2016;291:24607‐24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stik G, Crequit S, Petit L, et al. Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells. J Cell Biol. 2017;216:2217‐2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581‐593. [DOI] [PubMed] [Google Scholar]
- 25. Morhayim J, van de Peppel J, Braakman E, et al. Osteoblasts secrete miRNA‐containing extracellular vesicles that enhance expansion of human umbilical cord blood cells. Sci Rep. 2016;6:32034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiba H, Sawada N, Oyamada M, et al. Relationship between the expression of the gap junction protein and osteoblast phenotype in a human osteoblastic cell‐line during cell‐proliferation. Cell Struct Funct. 1993;18:419‐426. [DOI] [PubMed] [Google Scholar]
- 27. Simonsen JL, Rosada C, Serakinci N, et al. Telomerase expression extends the proliferative life‐span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20:592‐596. [DOI] [PubMed] [Google Scholar]
- 28. Duinhouwer LE, Tuysuz N, Rombouts EW, et al. Wnt3a protein reduces growth factor‐driven expansion of human hematopoietic stem and progenitor cells in serum‐free cultures. PLoS ONE. 2015;10:e0119086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dudakovic A, Camilleri E, Riester SM, et al. High‐resolution molecular validation of self‐renewal and spontaneous differentiation in clinical‐grade adipose‐tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115:1816‐1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44‐57. [DOI] [PubMed] [Google Scholar]
- 31. Eijken M, Swagemakers S, Koedam M, et al. The activin A‐follistatin system: potent regulator of human extracellular matrix mineralization. FASEB J. 2007;21:2949‐2960. [DOI] [PubMed] [Google Scholar]
- 32. Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic‐sample processing applied to copy‐number estimation in eukaryotic cells. Nat Methods. 2014;11:319‐324. [DOI] [PubMed] [Google Scholar]
- 33. Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731‐740. [DOI] [PubMed] [Google Scholar]
- 34. Perez‐Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bissels U, Bosio A, Wagner W. MicroRNAs are shaping the hematopoietic landscape. Haematologica. 2012;97:160‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roden C, Lu J. MicroRNAs in control of stem cells in normal and malignant hematopoiesis. Curr Stem Cell Rep. 2016;2:183‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keerthikumar S, Chisanga D, Ariyaratne D, et al. ExoCarta: a web‐based compendium of exosomal cargo. J Mol Biol. 2016;428:688‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fei XM, Wu YJ, Chang Z, et al. Co‐culture of cord blood CD34(+) cells with human BM mesenchymal stromal cells enhances short‐term engraftment of cord blood cells in NOD/SCID mice. Cytotherapy. 2007;9:338‐347. [DOI] [PubMed] [Google Scholar]
- 40. Himburg HA, Muramoto GG, Daher P, et al. Pleiotrophin regulates the expansion and regeneration of hematopoietic stem cells. Nat Med. 2010;16:475‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Renstrom J, Istvanffy R, Gauthier K, et al. Secreted frizzled‐related protein 1 extrinsically regulates cycling activity and maintenance of hematopoietic stem cells. Cell Stem Cell. 2009;5:157‐167. [DOI] [PubMed] [Google Scholar]
- 42. Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF. Angiopoietin‐like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111:3415‐3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yanez‐Mo M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morhayim J, Baroncelli M, van Leeuwen JP. Extracellular vesicles: specialized bone messengers. Arch Biochem Biophys. 2014;561:38‐45. [DOI] [PubMed] [Google Scholar]
- 45. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF‐kappaB‐dependent induction of microRNA miR‐146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481‐12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Labbaye C, Spinello I, Quaranta MT, et al. A three‐step pathway comprising PLZF/miR‐146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788‐801. [DOI] [PubMed] [Google Scholar]
- 47. Starczynowski DT, Kuchenbauer F, Wegrzyn J, et al. MicroRNA‐146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39:167–178.e4. [DOI] [PubMed] [Google Scholar]
- 48. Emmrich S, Rasche M, Schoning J, et al. miR‐99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev. 2014;28:858‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Copley MR, Babovic S, Benz C, et al. The Lin28b‐let‐7‐Hmga2 axis determines the higher self‐renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916‐925. [DOI] [PubMed] [Google Scholar]
- 50. Lee YT, de Vasconcellos JF, Yuan J, et al. LIN28B‐mediated expression of fetal hemoglobin and production of fetal‐like erythrocytes from adult human erythroblasts ex vivo. Blood. 2013;122:1034‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cabezas‐Wallscheid N, Klimmeck D, Hansson J, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507‐522. [DOI] [PubMed] [Google Scholar]
- 52. Walenda T, Bokermann G, Ventura Ferreira MS, et al. Synergistic effects of growth factors and mesenchymal stromal cells for expansion of hematopoietic stem and progenitor cells. Exp Hematol. 2011;39:617‐628. [DOI] [PubMed] [Google Scholar]
- 53. Sauvageau G, Iscove NN, Humphries RK. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene. 2004;23:7223‐7232. [DOI] [PubMed] [Google Scholar]
- 54. Min IM, Pietramaggiori G, Kim FS, Passegue E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380‐391. [DOI] [PubMed] [Google Scholar]
- 55. Kerkhoff C, Hofmann HA, Vormoor J, et al. Binding of two nuclear complexes to a novel regulatory element within the human S100A9 promoter drives the S100A9 gene expression. J Biol Chem. 2002;277:41879‐41887. [DOI] [PubMed] [Google Scholar]
- 56. Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long‐term multilineage engraftment. Science. 2011;333:218‐221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


