Abstract
Pathogenic fibrotic diseases, including Idiopathic Pulmonary Fibrosis (IPF), have some of the worst prognoses and affect millions of people worldwide. With unclear etiology and minimally effective therapies, two-thirds of IPF patients die within 2-5 years from this progressive interstitial lung disease. Transforming Growth Factor Beta (TGFβ) and Insulin-like Growth Factor-1 (IGF-1) are known to promote fibrosis; however, myofibroblast specific upregulation of IGF-1 in the initiation and progression of TGFβ-induced fibrogenesis and IPF have remained unexplored. To address this, the current study (1) documents the upregulation of IGF-1 via TGFβ in myofibroblasts and fibrotic lung tissue, as well as its correlation with decreased pulmonary function in advanced IPF; (2) identifies IGF-1’s C1 promoter as mediating the increase in IGF-1 transcription by TGFβ in pulmonary fibroblasts; (3) determines that SMAD2 and mTOR signaling are required for TGFβ-dependent Igf-1 expression in myofibroblasts; (4) demonstrates IGF-1R activation is essential to support TGFβ-driven profibrotic myofibroblast functions and excessive wound-healing; and (5) establishes the effectiveness of slowing the progression of murine lung fibrosis with the IGF-1R inhibitor OSI-906. These findings expand our knowledge of IGF-1’s role as a novel fibrotic-switch, bringing us one step closer to understanding the complex biological mechanisms responsible for fibrotic diseases and developing effective therapies.
Keywords: Signaling, Transcription, Promoter, Fibroblast
INTRODUCTION
Advanced medical care, healthier living conditions, technological discoveries, and a rise in education have contributed to increased longevity and a rapid expansion of the elderly population worldwide [1]. In fact, life expectancy globally has increased nearly 25 years since the 1950’s and the number of people over the age of 60 is projected to double to over 2 billion by 2050 [2, 3]. This enhanced life expectancy has consequentially given rise to a new epidemic of age associated chronic illnesses resulting in an estimated 100,000 deaths each day and a growing economic burden on healthcare [4, 5]. Pulmonary diseases have a disproportionately higher incidence and prevalence in the elderly population, and due to limited and relatively ineffective treatment options they account for 14% of all deaths worldwide [6, 7].
Interstitial Lung Diseases (ILDs) encompass a collection of over 200 lung disorders that result from infection, genetic, and/or environmental elements (e.g., medications, occupational, chemicals or pollutant exposure) that progressively worsen over time [8]. Most notably, Idiopathic Pulmonary Fibrosis (IPF) accounts for 20–30% of all ILDs, affects over 5 million people worldwide, and is more lethal than most cancers [9-11]. IPF is a debilitating, chronic, and irreversible ILD with unclear etiology, characterized by progressive scarring of the lungs in adults between 50 and 85 years old, leading to respiratory failure and death 2–5 years after diagnosis [11, 12]. Prior research has substantiated that repetitive damage to the alveolar epithelium disrupts the balance between injury and repair resulting in exacerbated inflammatory and profibrotic signaling [13]. The subsequent chronic wound-healing response is distinguished from normal physiological repair by uncontrolled tissue remodeling via activated myofibroblasts and excessive accumulation of extracellular matrix (ECM) proteins which destroy healthy lung architecture and diminish pulmonary function [14].
Although antifibrotic medications, Pirfenidone and Nintedanib, were approved for the treatment of IPF, they have only demonstrated a modest delay of pulmonary function decline, are associated with a number of significant side effects, and patients still inevitably progress to end-stage respiratory failure [4]. IPF has recently become the leading indication for lung transplant, which remains the only intervention to improve survival and quality of life [15]. Thus, even though increasing evidence delineating the mechanisms of IPF pathogenesis has led to previously non-existent therapies and given hope to IPF patients, it is all the more urgent to unravel the molecular mechanisms and pathophysiological processes driving IPF to identify novel therapeutic targets and discover effective treatment strategies to improve patient outcomes.
It is widely accepted that while Transforming Growth Factor Beta (TGFβ) is crucial for normal organismal development and tissue homeostasis, it is extensively involved in IPF and even serves as a master regulator of most, if not all, fibrotic diseases [16]. TGFβ is a ubiquitously expressed cytokine whose pleiotropic effects are time, tissue and cell type specific, governed by extracellular conditions, and mediated in collaboration with other growth factors and signaling pathways [17]. Prolonged overexpression of TGFβ suppresses inflammation and promotes fibrogenesis by enhancing migration and proliferation of fibroblasts, activating fibroblasts to differentiate into myofibroblasts, and stimulating myofibroblast synthesis and deposition of ECM proteins [18]. Hyper-activation of the TGFβ receptor complex triggers this excessive fibroproliferative response through a complex network of downstream canonical (SMAD2 & 3) [19] and non-canonical (PI3K → AKT, MEK→ ERK, mTOR, etc.) [20] signaling cascades which regulate the transcription of profibrotic mediators (fatty acid synthase, FASN; glucose transporter-1, GLUT1), additional growth factors (platelet derived growth factor, PDGF; epidermal growth factor, EGF), microRNAs, and ECM proteins [16, 21-23].
In light of TGFβ’s significant role in fibrotic diseases it has recently been explored as an attractive therapeutic target, although few TGFβ-centric approaches have improved outcomes for fibrosis patients [24]. Likely, TGFβ’s ubiquitous and diverse effects on tissue homeostasis and the immune response limit its feasibility as a drug or biologic target due to adverse side effects and lack of tissue specificity [25]. Therefore, the future of effective clinical antifibrotic treatment may require inhibition of several downstream pathways that synergistically drive pathologic tissue remodeling using combination therapy. For this reason, it is imperative to expand our knowledge of the spectrum of profibrotic mediators and their functions in order to specifically treat fibroproliferative diseases, especially IPF. Therefore, we explored the role of Insulin-like Growth Factor-1 (IGF-1) in cooperatively enhancing the chronic wound-healing response with TGFβ.
IGF-1 was initially thought to only mediate the biological effects of growth hormone on endocrine signaling; however, it has since been substantiated that IGF-1 is also responsible for activating autocrine/paracrine responses in peripheral tissues [26]. Beyond IGF-1’s contribution to normal human growth and development, it has a multiplicity of biological effects on cell growth, proliferation, survival, and differentiation of many cell types and tissues through mediating transcription, metabolism, and cytoskeletal reorganization [27]. Increased levels of IGF-1 and/or respective activation of the IGF-1 receptor (IGF-1R) have been implicated in a number of pathologic conditions including cancer, diabetes, inflammation, and fibrosis [28]. Of particular relevance, elevated levels of IGF-1 have been reported in Bleomycin (BLM)-induced murine pulmonary fibrosis [29] and Idiopathic Pulmonary Fibrosis [30, 31] lung tissue. Cumulatively, IGF-1’s stimulatory effects on differentiation and proliferation in addition to its increased presence in IPF tissue suggest IGF-1 influences uncontrolled tissue repair; although IGF-1’s role, regulation, and physiological implications in pulmonary fibroblasts as well as IPF have remained elusive [32].
In the present study, we examined these critical issues and identified that: (1) TGFβ is required for induction of IGF-1 in myofibroblasts; (2) increased IGF-1 is present in IPF tissue and correlated with decreased pulmonary function seen with disease progression; (3) of the two alternative promoters differentially regulating IGF-1 transcription, TGFβ increased expression of the Class-1 promoter; (4) SMAD2 and mTOR signaling are required for TGFβ-dependent Igf-1 expression in myofibroblasts; (5) IGF-1R activation is essential to support TGFβ stimulated profibrotic myofibroblast processes and aberrant wound-healing; and (6) pharmacological inhibition of IGF-1R kinase activity with OSI-906 demonstrated effectiveness in delaying murine lung fibrosis progression and decreased mortality. Collectively, these findings expand our knowledge of the complex biological mechanisms responsible for driving fibrotic diseases and provide preclinical evidence for targeting IGF-1 signaling to effectively treat IPF.
MATERIALS AND METHODS
Cell Culture
All cells were kept under continued incubation at 37°C in 5% CO2. Murine embryonic fibroblast cell lines (AKR-2B, BALB/c-3T3, SWISS-3T3) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, Carlsbad, CA) containing 10% Fetal Bovine Serum (FBS; HyClone Laboratories, Logan, UT) and 1% Penicillin-Streptomycin (P/S; Life Technologies, Carlsbad, CA). Human fetal lung fibroblast cell lines (MRC5 and IMR-90) were maintained in Eagle’s Minimum Essential Medium (EMEM; ATCC, Manassas, VA, USA) containing 20% FBS and 1% P/S. Deidentified patient primary cells including normal human lung fibroblasts (NHLF) and human Idiopathic Pulmonary Fibrosis (IPF) fibroblasts characterized in two previously published studies [33, 34] and obtained from Dr. Nathan Sandbo from the University of Wisconsin-Madison Department of Medicine (TSB BioBank IRB #2011–0521) and Dr. Carol Feghali-Bostwick from the Medical University of South Carolina (University of Pittsburgh IRB #970946) were cultured in DMEM medium containing 10% FBS and 1% P/S. Unless stated otherwise, murine and human fibroblast cells were seeded at 2 × 106 cells/100 mm plate, serum starved for 18–24 hrs in DMEM supplemented with 0.1% FBS and 1% P/S (murine fibroblasts) or EMEM supplemented with no FBS and 1% P/S (human fibroblasts) prior to 2-hour pre-treatment with inhibitor and/or followed by growth factor stimulation for the indicated time. shRNA knockdowns of SMAD2, SMAD3, and mTOR in AKR-2B cells were described previously [35, 36]. A summary of pharmacological inhibitors and growth factor treatments found to be optimal in fibroblasts is provided in Table 1, unless otherwise noted in a specific method.
Table 1.
Summary of cell culture treatments.
| Pharmacological Inhibitors | |||||
|---|---|---|---|---|---|
| Inhibitor | Target | Catalogue # | Source | Working Concentration(s) |
Vehicle |
| SB431542 | TGFβ Type 1 Receptor | S1067 | Selleck Chemicals (Houston, TX, USA) |
10 μM | 0.1% DMSO |
| LY-294,002 | PI3K | S1105 | Selleck Chemicals (Houston, TX, USA) |
20 μM | 0.1% DMSO |
| MK2206 | AKT | S1078 | Selleck Chemicals (Houston, TX, USA) |
300 nM | 0.1% DMSO |
| U0127 | MEK | V1121 | Promega (Madison, WI, USA) |
10 μM | 0.1% DMSO |
| Rapamycin | mTORC1/2 | S1039 | Selleck Chemicals (Houston, TX, USA) |
10 nM | 0.1% DMSO |
| OSI-906 | IGF-1 Receptor | S1091 | Selleck Chemicals (Houston, TX, USA) |
1 nM, 1 μM, 10 μM |
0.1% DMSO |
| Growth Factor Treatments | |||||
| Growth Factor | Catalogue # | Source | Working Concentration(s) |
Vehicle | |
| TGFβ | 240-B | R&D Systems (Minneapolis, MN, USA) |
10 ng/ml | 4 mM HCl, 0.1% BSA |
|
| IGF-1 (mouse) | 9897 | Cell Signaling (Danvers, MA, USA) |
0.2 ng/ml | 20mM citrate, pH 3.0 |
|
| IGF-1 (human) | ab9573 | Abcam (Cambridge, MA, USA) |
2 ng/ml | dH2O | |
Gene Silencing
For siRNA knockdown of IGF-1R in AKR-2B murine fibroblasts or MRC5 human lung fibroblasts, cells were transiently transfected with 40 nM of small interfering RNA (siRNA) to IGF-IR (sc-35638, mouse; sc-29358, human; Santa Cruz Biotechnology Santa Cruz, CA, USA) or non-targeting control (NT, sc-37007; Santa Cruz) according to the manufacturer’s protocol. Each siRNA contains a pool of 3 target-specific siRNAs for murine or human cells. In short, 7.5×104 cells were transfected using Lipofectamine 2000 (Invitrogen) and incubated in Opti-Mem (Invitrogen) for 6 hours. Cells were then grown under normal conditions for 18 hours to recover prior to addition of Vehicle or TGFβ under serum starvation conditions prior to collection. shRNA knockdowns of SMAD2, SMAD3, and mTOR in AKR-2B cells were described previously [35, 36] and shRNA knockdown cells were subsequently cultured in DMEM medium supplemented with 10% FBS, 1% P/S, and 3–5 μg/ml Puromycin (Millipore-Sigma, St. Louis, MO, USA).
RNA-seq and Bioinformatic Analysis of Soft Agar Colonies
To identify differentially expressed genes during TGFβ-stimulated AIG, the soft agar colony formation assay was performed essentially as described in the main text except 3 × 104 AKR-2B cells/well were grown in 6-well plates containing CytoSelect Cell Transformation Assay reagents (CBA-135; Cell Biolabs Inc., San Diego, CA, USA) in the presence or absence of TGFβ for 2, 4, and 8 days before recovering colonies from agarose with Cel Biolab’s matrix solubilization solution. RNA was extracted with Trizol, purified using Qiagen’s RNeasy Kit (Qiagen, Redwood City, CA, USA), and assessed for purity as described for qRT-PCR.
Library preparation (1 μg total RNA) and RNA-sequencing (RNA-seq) was completed by Mayo Clinic’s Genome Analysis Core (Rochester, MN, USA), using previously published methods [37]. Mayo Clinic Division of Biomedical Statistics and Informatics used a comprehensive bioinformatics pipeline, MAP-RSeq [38], for mapping, gene expression counts, alignment to the reference genome for Mus musculus (mm10), and quality control. Differentially expressed genes between TGFβ and vehicle were identified by empirical analysis of RNA-seq gene expression profiles in R, using Bioconductor’s bioinformatics package edgeR (v3.9) and reported as Log2 Fold Change. mRNA with p-values <0.05 (adjusted for false discovery rate of 5%) were considered differentially expressed. The NGS transcriptomics data discussed in this publication have been made publically available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus genomics data repository [39, 40] (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE131253.
Bioinformatics using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Qiagen, Redwood City, CA, USA) supported by Ingenuity’s Knowledgebase predicted disease states/biological functions and upstream regulators (Supplemental Fig. 1) associated with in vitro TGFβ-dependent anchorage independent colony formation (pro-fibrotic response) in murine fibroblasts [41]. Activation z-score predicts whether an upstream regulator inhibits or stimulates the downstream effects of TGFβ-dependent AIG. P-values were determined with the Fisher’s Exact Test which represents the likelihood of an association between a set of genes from the experimental data set and a related function.
Real-Time Quantitative PCR (qRT-PCR)
Trizol Reagent (15596018; ThermoFisher Scientific, Waltham, MA) was used to isolate total RNA from cells and tissues. Mouse lung tissues were additionally homogenized using the TissueLyser LT (69980; Qiagen, Redwood City, CA, USA). RNA was purified with Direct-zol RNA MiniPrep Plus (R2072, Zymo Research) according to company specifications, concentration and purity assessed, and a total of 1–3 μg total RNA used for cDNA Synthesis with Maxima Reverse Transcriptase (ThermoFisher Scientific, Waltham, MA).
Quantitative Real-Time PCR (qRT-PCR) reactions were performed with the CFX96 Real-Time PCR system (BioRad) using 25 ng cDNA/16 μl reaction mixture with Platinum SYBR Green SuperMix-UDG (11733046; ThermoFisher Scientific, Waltham, MA) supplemented with MgCl2 (5 mM) and validated primers (200 nM). A summary of primer sets manufactured by Integrated DNA Technologies can be found in Table 2. Each primer set was validated using sequencing of qRT-PCR products. Relative gene expression levels were quantified with the Livak method (2−∆∆CT) [42] by normalizing expression of the gene of interest to the combined expression of two stable housekeeping genes [43] demonstrated to be unaffected by TGFβ (TATA box binding protein, TBP; Beta-2 Microglobulin, B2M), and calculating the relative fold differences between experimental and control samples. Assays were completed in technical triplicate.
Table 2.
Summary of mouse and human primers used for qRT-PCR.
| Gene (species) | Primer Sequence (5’-3’) | Amplicon Size (bp) |
|---|---|---|
| Acta2 (m) | (F) AGCCATCTTTCATTGGGATGG | 117 |
| (R) CCCCTGACAGGACGTTGTTA | ||
| ACTA2 (h) | (F) GACAATGGCTCTGGGCTCTGTAA | 147 |
| (R) CTGTGCTTCGTCACCCACGTA | ||
| β2m (m) | (F) CTCGGTGACCCTGGTCTTTC | 164 |
| (R) GGATTTCAATGTGAGGCGGG | ||
| B2M (h) | (F) GTATGCCTGCCGTGTGAAC | 139 |
| (R) AAAGCAAGCAAGCAGAATTTGG | ||
| Col1α1 (m) | (F) CGATGGATTCCCGTTCGAGT | 197 |
| (R) CGATCTCGTTGGATCCCTGG | ||
| COL1A1 (h) | (F) GAGGGCCAAGACGAAGACATC | 140 |
| (R) CAGATCACGTCATCGCACAAC | ||
| Col5α3 (m) | (F) CTGCAGAACCTGTGGACGTA | 174 |
| (R) AATGCCCATCTGGGAAGAGC | ||
| COL5A3 (h) | (F) AATTCAGCTCTTCTCGAGCGG | 78 |
| (R) ACTTTTGGTTCGTCTGGCCAA | ||
| COL7A1 (h) | (F) TTACGCCGCTGACATTGTGTT | 93 |
| (R) ACCAGCCCTTCGAGAAAGC | ||
| Ctgf (m) | (F) CACAGAGTGGAGCGCCTGTTC | 163 |
| (R) GATGCACTTTTTGCCCTTCTTAATG | ||
| Eda-fn (m) | (F) GAAGGTTTGCAACCCACTGT | 217 |
| (R) CATCCTCAGGGCTCGAGTAG | ||
| EDA-FN (h) | (F) AGTAACCAACATTGATCGCCCTAA | 76 |
| (R) TTCCCAAGCAATTTTGATGGA | ||
| Igf-1 (m) | (F) GATACACATCATGTCGTCTTCACA | 258 |
| (R) CAGTACATCTCCAGTCTCCTCAGA | ||
| Igf-1 C1 Promoter (m) | (F) AGAGCCTGCGCAATGGAATA | 100 |
| (R) GGCAGGGATAATGAGGCGAA | ||
| Igf-1 C2 Promoter (m) | (F) TGCCTGGGTGTCCAAATGTA | 146 |
| (R) ATTTTGGTAGGTCCGGGTCG | ||
| IGF-1 (h) | (F) GAGCTGGTGGATGCTCTTCA | 112 |
| (R) CATCCACGATGCCTGTCTGA | ||
| IGF-1 C1 Promoter (h) | (F) AAGGCAAATGTTCCCCCAGC | 142 |
| (R) CATTGCGCAGGCTCTATCTG | ||
| IGF-1 C2 Promoter (h) | (F) TCACAAACCCCACCCACAAA | 93 |
| (R) TTTACAGCAGGTCAGGGTGG | ||
| Ins1 (m) | (F) TCTTCTACACACCCAAGTCCCG | 101 |
| (R) CTCCAACGCCAAGGTCTGAA | ||
| Ins2 (m) | (F) CTTCTTCTACACACCCATGTCCC | 101 |
| (R) CCAAGGTCTGAAGGTCACCTG | ||
| INS (h) | (F) CTCACACCTGGTGGAAGCTC | 212 |
| (R) AGAGGGAGCAGATGCTGGTA | ||
| Pai-1 (m) | (F) TTCCAACCAGCATCCCAGAC | 92 |
| (R) CCATGAGACCTTTGTGGGGT | ||
| PAI-1 (h) | (F) GGCCATTACTACGACATCCTG | 150 |
| (R) GGTCATGTTGCCTTTCCAGT | ||
| Tbp (m) | (F) GAAGTTCCCTATAAGGCTGGAAG | 117 |
| (R) AGGAGAACAATTCTGGGTTTGA | ||
| TBP (h) | (F) GCCCGAAACGCCGAATATAATC | 136 |
| (R) GTCTGGACTGTTCTTCACTCTTGG |
Mouse (m; Mus musculus, taxid:10090) and human (h; Homo sapiens, taxid: 9606) primer sets: gene name, NCBI accession number, forward (F) and reverse (R) primer sequences, and qRT-PCR amplicon length.
Histology and Immunohistochemistry
Human lung tissue specimens from 7 control and 12 IPF patients undergoing lung surgical biopsy were provided by Mayo Clinic’s Tissue Registry (IRB #15–003622) [21]. All patients had authorized the use of their records and samples for research according to the Minnesota Research Authorization Law. Tissue was histologically reviewed by a Mayo Clinic lung pathologist while patients were clinically diagnosed by a Mayo Clinic pulmonologist as controls or having IPF by clinical history, CT scan, and/or pulmonary function tests. A summary of patients’ demographics and pulmonary function test results is presented in Supplemental Table 1. Murine lung tissue specimens were obtained from the current study with OSI-906 (IACUC #A00003174–17) and a previously published study with Imatinib and Lapatinib (IACUC #A24710) [23].
Human and mouse Formalin Fixed Paraffin Embedded sections (FFPE, 5 μm uncharged) were deparaffinized by incubating for 1 hr at 60°C followed by multiple xylene soaks and rehydrated through a series of ethanol and water washes. Heat Induced Epitope Retrieval (HIER) was done in a steamer for 10 minutes under acidic conditions (CTS014; R&D Systems), followed by 15 minutes of peroxidase blocking solution. Immunohistochemical staining for mouse and human tissues was performed using an Anti-Rabbit HRP-DAB Tissue Staining Kit (CTS005; R&D Systems) according to R&D’s staining procedure. Antigen specific primary antibodies to IGF-1 (4 μg/mL, AB9572; ABCAM, Cambridge, MA, USA) or p-IGF-1R (1:50 dilution, NB100–92555; Novus Biologicals, Centennial, Colorado, USA) were used by overnight 4°C incubation in a hydration container, counterstained with Mayer’s hematoxylin (Gills Formula #2, CS401–1D; ThermoFisher Scientific, Waltham, MA), and dehydrated with ethanol and Clear-Rite 3 (#6901; ThermoFisher Scientific, Waltham, MA) before mounting with non-aqueous VectaMount (H-5000; Vector Laboratories, Burlingame, California, USA). Specificity of IGF-1 and p-IGF-1R staining in both murine and human lung tissues was determined via omitting primary antibody. Masson’s Trichrome, Hematoxylin/Eosin, α Smooth Muscle Actin, Collagen 1, and total Fibronectin staining was performed by the Mayo Pathology Research Core.
Quantitative analyses of the antigen expression intensity (DAB chromogen stain) for IGF-1 or p-IGF-1R was performed by scanning slides with an Aperio Digital Pathology spectral imager at 40x magnification (brightfield scanner; Leica Biosystems Inc, Buffalo Grove, IL) and selecting equal size annotations of fibrotic regions (n ≥ 10/sample; 150 × 150 pixels annotation) in ImageScope prior to measuring positive or strong positive antigen expression using Aperio’s Color Deconvolution Algorithm (Positive Pixel Count macro version 9.1). The percent murine IGF-1 or p-IGF-1R expression was calculated in Aperio (n strong positive + n positive/n total x 100 = % expression) while percent human IGF-1 or p-IGF-1R strong expression was calculated (n strong positive/n total x 100 = % strong expression) based on pixels positive/strongly positive for DAB staining (excluding basal expression).
Microarray Differential Gene Expression Analysis of Human Lung Tissue
Human microarray data from the Lung Genomics Research Consortium (LGRC) publically available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus [39, 40] genomics data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE47460 (ID: 200047460) was used to determine the level of IGF-1 (Probe ID: A_23_P13907) and TGFβ (Probe ID: A_23_P156327) gene expression (Log2 fold change); rank according to expression (fold change), and significance (adj. p-value) from a total of 15,261 differentially expressed genes identified in patients with IPF compared to control lung samples.
Total RNA was isolated from human lung tissue as previously described [44] from 429 deidentified patients undergoing thoracic surgery; provided by the Lung Tissue Research Consortium (LTRC)[45]. From this dataset of 429 samples with multiple subtypes of interstitial lung diseases (ILD), 122 individuals with Idiopathic Pulmonary Fibrosis (IPF) and 91 controls were used in the present study’s analysis. A summary of patients’ demographics and pulmonary function test results is presented in Supplemental Table 2. Differential gene expression of IGF-1 and TGFβ between IPF and controls was identified by comparisons with GEO2R and reported as Log2 normalized expression. The p-value was adjusted (adj. p-value) using the Benjamini & Hochberg method [46] to control the false discovery rate. Additionally, gene expression and the pulmonary function measurement percentage of predicted diffusing capacity for carbon monoxide (% DLCO) were analyzed using linear regression analysis with 95% confidence interval, correlation analysis using Pearson’s r coefficient, and significance associated with correlation (p-value).
RNA-seq Differential Gene Expression Analysis of Human Lung Tissue
FFPE lung tissue specimens (5 μm uncharged section) from 8 control and 8 IPF patients undergoing lung surgical biopsy were procured from the Mayo Clinic Tissue Registry after all patients had authorized the use of their records and samples for research according to the Minnesota Research Authorization Law and approved the Mayo Clinic Institutional Review Board (IRB # 17–006943). Tissue was histologically reviewed by a Mayo Clinic lung pathologist and patients’ clinically diagnosed as having IPF in accordance with ATS/ERS criteria [47]. Demographic features and pulmonary function test results are summarized in Supplemental Table 3.
Differential IGF-1 gene expression and rank according to fold change were identified from a total of 2,560 genes included in HTG’s EdgeSeq Assay’s Oncology Biomarker Panel using next generation sequencing performed by HTG Molecular Diagnostics, Inc. (Tucson, AZ, USA) in a clinically blinded analysis and obtained from Dr. Robert Vassallo (Mayo Clinic Thoracic Disease Research Unit). Differentially expressed genes between IPF and controls were identified by comparison with the DESeq2 package (version 1.14.1), available from the Bioconductor project and reported as Fold Change. The p-value was adjusted (adj. p-value) using the Benjamini & Hochberg method [46].
Soft Agar Colony Formation Assay
Anchorage Independent Growth (AIG) of AKR-2B cells in the presence or absence of 25 ng/mL TGFβ (R&D Systems, Minneapolis, MN, USA) was performed as previously described [35]. Top plugs contained the pharmacologic inhibitor OSI-906 (S1091; Selleck Chemicals, Houston, TX, USA) or vehicle. Colonies ≥ 50 μm in diameter were counted using an optimized CHARM algorithm after 8 days (peak colony formation) and imaged using the Optronix GelCount (Oxford Optronics, Abingdon, United Kingdom). Assays were completed in technical triplicate and images relative to the average shown.
Scratch Assay
The wound-healing response was observed using a scratch assay, as previously described [48]. Briefly, 2 × 105 cells/well were seeded into 6-well plates under normal culture conditions and incubated overnight. Following 18–24 hours serum starvation, a sterile 300 μl pipette tip was used to scrape a straight line across the cell monolayer to create a “wound”. The edge of the scratch was smoothed and cell debris removed by rinsing 3x with PBS. Cells were then refed with low serum media, imaged for baseline measurements, and pre-treated with vehicle or OSI-906 for 2 hours prior to 24 hours stimulation in the presence or absence of 10 ng/ml TGFβ. Additional OSI-906 (or vehicle) was added at hour 12. Images were taken at 10x magnification with an EVOS-XL Core Cell Imaging System (AMEX1000; ThermoFisher Scientific, Waltham, MA) and the distance between leading edges analyzed (wound width at 24 hours/wound width at 0 hours x 100 = % relative wound width). Assays were completed in technical triplicate and images relative to the average shown.
Myofibroblast Morphological Transformation
Morphological transformation from a fibroblast to a myofibroblast phenotype was performed by plating, treating, and imaging confluent cell cultures as described in the scratch assay method above [36]. Assays were done in technical triplicate and images relative to the average shown.
Stress Fiber Immunofluorescence
Capturing myofibroblast stress fiber formation using immunofluorescence was accomplished similar to previous studies [36] by growing 1.75 × 105 cells/well on glass coverslips in 6-well plates and treated as described in the scratch assay method above. Before immunofluorescent staining each well was washed 2x with PBS (5 minutes/wash), fixed with 4% paraformaldehyde in PBS (5 minutes at room temperature), washed again with PBS (2× 5 minutes), permeablized with 0.1% Triton X-100 in PBS (3 minutes at room temperature), and washed a final time in PBS (2× 5 minutes). Cells were then stained for both filamentous actin (1:40 dilution; 1 hour at room temperature) with tetrarhodamine isothiocyanate (TRITC)–labeled phalloidin (SC-301530; Santa Cruz) and nuclei with 4′,6-diamidino-2-phenylindole (1:500 dilution, DAPI; Molecular Probes, Eugene, OR). Coverslips were mounted onto slides using SlowFade Gold Antifade Mountant (S36937; ThermoFisher Scientific, Waltham, MA) and images collected on a Zeiss LSM 780 confocal system (Carl Zeiss, Jena, Germany) with a 40x water immersion objective with the same florescent parameters for all images. Assays were completed in technical triplicate and images relative to the average shown.
Enzyme-Linked Immunosorbent Assay
AKR-2B cells were seeded at 2 × 106 cells/100 mm plate in 10 ml media under normal culture conditions, allowed to adhere overnight, and serum starved in 10 ml media for 18–24 hours. Cells were then treated with vehicle or TGFβ (10 ng/ml) under serum starvation conditions and media harvested after 24 hours. Media was concentrated by centrifugation at 5,000 g for 45 minutes using 3 kDa molecular weight cut off centrifugal filter devices (Amicon Ultra-15, UFC900324; Millipore-Sigma, St. Louis, MO, USA). The level of IGF-1 secreted by murine fibroblasts in 100 μl of concentrated media sample or standard was analyzed using an IGF-1 Enzyme-Linked Immunosorbent Assay (ELISA) kit (ab100695; Abcam, Cambridge, MA, USA) according to the manufacturer’s protocol and absorbance measured using a VersaMax Microplate Reader with SoftMax Pro Software (Molecular Devices LLC, Sunnyvale, CA, USA) at 450 nm. Assays were completed in technical duplicate.
Protein Extraction and Western Blotting
Total proteins were extracted by lysing cells on ice in modified RIPA buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40/IGEPAL, 0.25% sodium deoxycholate (DOC), 1 mM EDTA [pH 8], 1 mM sodium vanadate (NaVO4), 50 mM sodium fluoride (NaF), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1X cOmplete Protease Inhibitor Cocktail (Roche, Indianapolis, IN, USA). Insoluble material was removed by centrifugation (18,000 g for 10 minutes), 2–50 μg protein aliquots were prepared in 2x Laemmli sample buffer, separated by gel electrophoresis on 10–12% SDS-PAGE, and transferred to PVDF membranes. Membranes were blocked for 8 hours with 5% milk in 10 mM Tris (pH 7.4)/0.1 % Tween 20 (TBST) at 4°C, followed by overnight incubation with primary antibodies diluted in blocking solution. After washing the membranes with TBST, they were incubated for 2 hours at room temperature with secondary antibodies in blocking solution, washed with TBST, followed by incubation with Enhanced Chemiluminescence (ECL) reagents (SuperSignal West Pico Chemiluminescent Substrate, 34580; ThermoFisher Scientific, Rockford, IL, USA) and corresponding bands detected with an X-OMAT 200A [36, 48]. A summary of antibodies used for Western blotting and immunoprecipitation can be found in Table 3.
Table 3.
Summary of antibodies used for Western blot and immunoprecipitation.
| Antigen | Primary Antibody | IP/WB | Dilution | Source |
|---|---|---|---|---|
| AKT | 9272 | WB | 1:10,000 | Cell Signaling |
| p-AKT | 9271 | WB | 1:1,000 | Cell Signaling |
| COL 5 | sc-20648 | WB | 1:5,000 | Santa Cruz |
| CTGF | sc-14939 | WB | 1:5,000 | Santa Cruz |
| ERK1/2 | sc-94 | WB | 1:5,000 | Santa Cruz |
| p-ERK1/2 | 9106 | WB | 1:2,000 | Cell Signaling |
| GAPDH | MAB347 | WB | 1:20,000 | Millipore-Sigma |
| IGF-1 | ab9572 | WB | 1:5,000 | Abcam |
| IGF-1R | 3027 | IP | 2 μl antibody, 400 ug protein, 50 μl protein G beads |
Cell Signaling |
| WB | 1:2,000 | |||
| p-IGF-1R | 3024 | IP | 10 μl antibody, 800 ug protein, 50 μl protein G beads |
Cell Signaling |
| WB | 1:1,000 | |||
| p-IRS1 | 44816G | WB | 1:5,000 | ThermoFisher Scientific |
| mTOR | sc-8319 | WB | 1:2,000 | Santa Cruz |
| S6K | 9202 | WB | 1:10,000 | Cell Signaling |
| p-S6K | 9205 | WB | 1:1,000 | Cell Signaling |
| SGK1 | ab43606 | WB | 1:10,000 | Abcam |
| p-SGK1 | sc-16745-R | WB | 1:1,000 | Santa Cruz |
| SMAD2 | 5339 | WB | 1:2,000 | Cell Signaling |
| p-SMAD2 | WB | 1:5,000 | Leof Lab | |
| SMAD3 | ab28379 | WB | 1:10,000 | Abcam |
| p-SMAD3 | WB | 1:5,000 | Leof Lab | |
| αSMA | A5228 | WB | 1:5,000 | Millipore-Sigma |
Immunoprecipitation (IP), Western blot (WB), rabbit anti-p-Smad3 antibody and anti-p-Smad2 antibody were previously generated to the peptide COOH-GSPSIRCSpSVpS and COOH-CGSPSVRCSpSMpS, respectively [48].
BLM-Induced Pulmonary Fibrosis Murine Model
Lung tissue was obtained from paraffin-embedded blocks from two pulmonary fibrosis murine studies whereby the experimental design, procedures, facilities, pharmacologic agents, and humane endpoints were approved by, and performed in accordance with Mayo Clinic’s Institutional Animal Care and Use Committee (IACUC). Immunohistochemical analysis from saline, Bleomycin (BLM), and Bleomycin → Imatinib 50 mg/kg/day + Lapatinib 50 mg/kg/day (BIL) treated mice (Fig. 1B, 2, 5C, and Supplemental Fig. 2E) was derived from tissue preserved from a previously published study [23] (IACUC Protocol A24710).
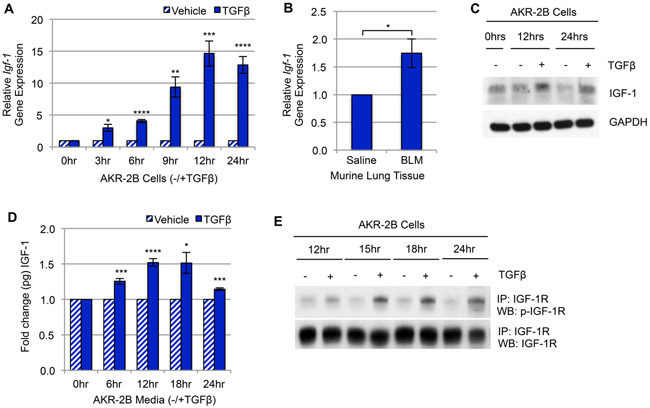
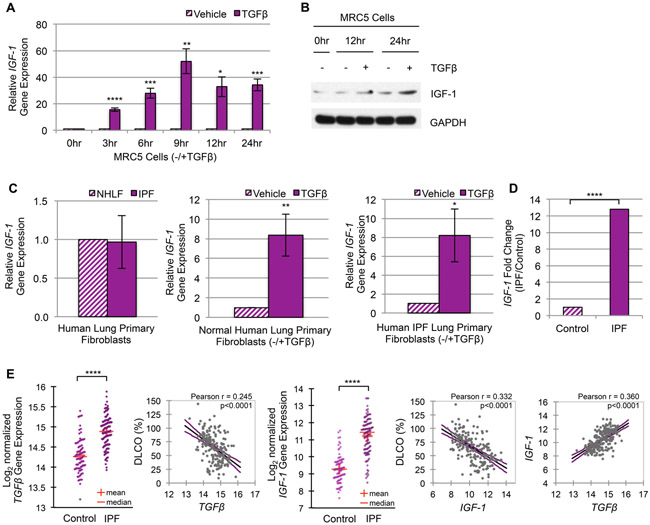
Figure 1. TGFβ upregulates IGF-1 and IGF-1 signaling in murine fibroblasts.
(A) RT-qPCR of Igf-1 gene expression in AKR-2B murine fibroblasts treated with TGFβ compared to vehicle over 24 hours (n=3). (B) RT-qPCR of Igf-1 gene expression in whole lung tissue from intratracheal Bleomycin (BLM, n=5) treated mice compared to Saline (n=5). (C) Western blot and (D) ELISA analysis of IGF-1 ligand expression/secretion following TGFβ or vehicle treatment in AKR-2B fibroblasts over 24 hours (n=3 for both). (E) IGF-1 receptor (IGF-1R) was immunoprecipitated (IP) from AKR-2B murine fibroblasts at the indicated times following treatment with TGFβ (+) or vehicle (−) and Western blotted (WB) for p-IGF-1R (n=3). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that TGFβ stimulates Igf-1 gene expression, ligand production, and IGF-1R activation in murine fibroblasts.
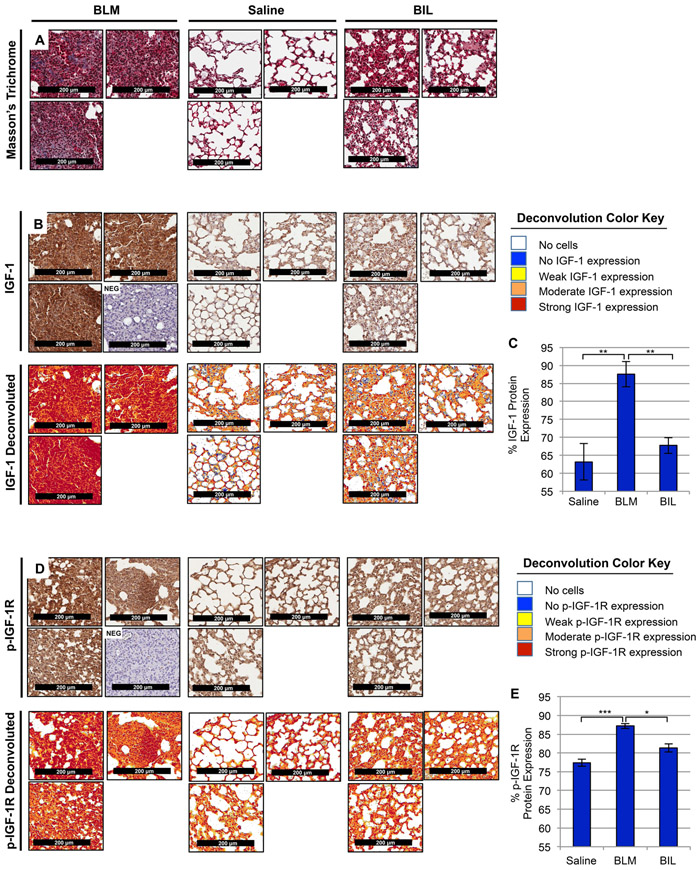
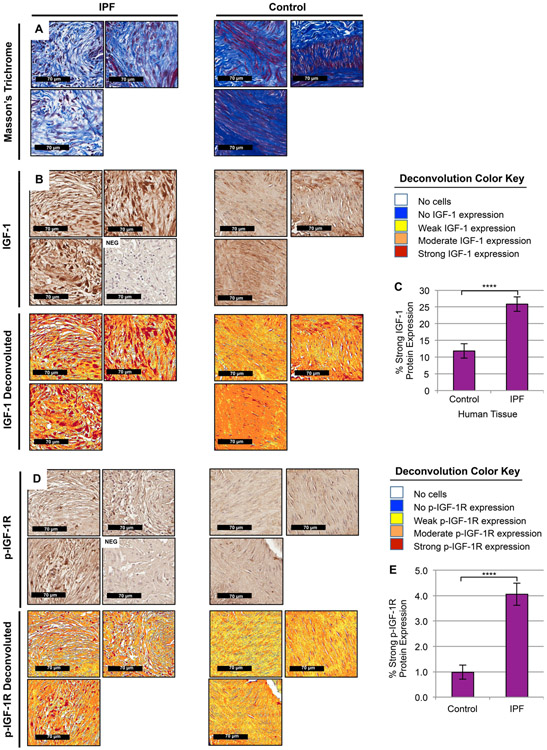
Figure 2. IGF-1 and phosphorylated-IGF-1R are increased in BLM-induced murine lung fibrosis.
(A, B, D) Representative images (20x magnification) from histological sections of murine lung tissue with the following treatments: intratracheal Bleomycin (BLM, left, n=4), Saline (center, n=3), and Bleomycin dual-treated with Imatinib and Lapatinib (BIL, right, n=3; previously shown to be effective in treating BLM-induced fibrosis [23]) were stained with Masson’s Trichrome (A, top) to visualize lung tissue architecture (purple, nuclear; pink, cytoplasmic) and collagen composition (blue, collagen fibers), anti-IGF-1 antibodies (B, middle), anti-phosphorylated-IGF-1R antibodies (D, bottom) co-stained with Hematoxylin, or negative controls (NEG) without primary antibody co-stained with Hematoxylin. Color deconvoluted images (B, D) show the intensity of IGF-1 ligand or phosphorylated-IGF-1R present in BLM mice compared to Saline and BIL per the deconvolution color key and quantified using a pixel count algorithm (C, E). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that IGF-1 ligand and IGF-1R activation are increased in fibrotic murine lung tissue and diminished following anti-fibrotic treatment.
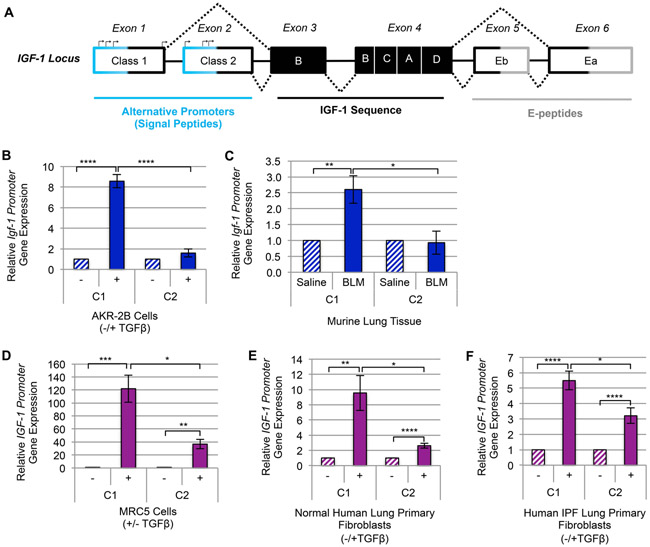
Figure 5. TGFβ upregulates the Class-1 IGF-1 promoter in fibroblasts and pulmonary fibrosis tissue.
(A) A schematic diagram of IGF-1 gene locus [93, 94]. (B) RT-qPCR of IGF-1 gene expression via Class-1 (C1) and Class-2 (C2) alternative promoters following 12 hours (peak IGF-1 induction by TGFβ) treatment with TGFβ (+) or vehicle (−) in AKR-2B (n=3) murine fibroblasts. (C) RT-qPCR gene expression analysis of IGF-1’s C1 and C2 promoters in whole lung tissue from intratracheal Bleomycin (BLM, n=5) treated mice compared to Saline (n=5). (D-F) IGF-1 promoter usage following 24 hours (peak time of TGFβ-dependent stimulation of IGF-1 in human fibroblasts) treatment of (D) MRC5 (n=3), (E) Normal Human Lung Primary Fibroblasts (NHLF, n=5), and (F) Human Idiopathic Pulmonary Fibrosis Primary Fibroblasts (IPF, n=4). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that of the two alternative promoters required for transcription initiation of IGF-1, TGFβ increases transcription of the C1 promoter significantly more than the C2 in both murine and human myofibroblasts.
For the current study (Figure 9 and Supplemental Fig. 7-9; IACUC Protocol A00003174–17), female C57BL/6 mice (9 week old) were purchased from Charles River Laboratories (Wilmington, MA). Upon arrival mice were placed on a breeder chow diet for 2 weeks to increase their body weight to an average of 21.5 grams/mouse and maintained on this diet for the duration. Mice were housed under standard laboratory conditions on a 12 hour light/12 hour dark cycle with ad libitum access to food/water and ongoing veterinary evaluation. Prior to intratracheal aerosolization, mice were weighed and anesthetized with a 200 μl intraperitoneal (IP) injection of Ketamine (90 mg/kg; Ketaset, NADA 043–304; Zoetis Inc., Kalamazoo, MI, USA)/Xylazine (10 mg/kg; AnaSed Injection, NDC 59399–110-20; Akorn Inc., Lake Forest, IL, USA) combination and ophthalmic lubricant (NDC 13985–600-03; Akorn Inc.) applied to their eyes. BLM (2.5 U/kg; NDC 0703–3154-01; Teva Pharmaceuticals Inc., Schaumburg, IL), diluted in 50 μl 0.9% saline (NDC 0338–0048-02; Baxter Healthcare Corporation, Deerfield, IL, USA) or 50 μl 0.9% saline was delivered to the lungs via an intratracheal aerosolizer (Model IA-1C-M; Penn-Century Inc. Wyndmoor, PA, USA) as previously described [22, 48]. While under anesthesia mice were shaved around the collar to prepare for pulse oximetry measurements. Mice were then administered 0.2 mg/mouse Atipamazol (Antisedan, NADA 141–033; Zoetis Inc.) to reverse the sedative and analgesic effects of Xylazine.
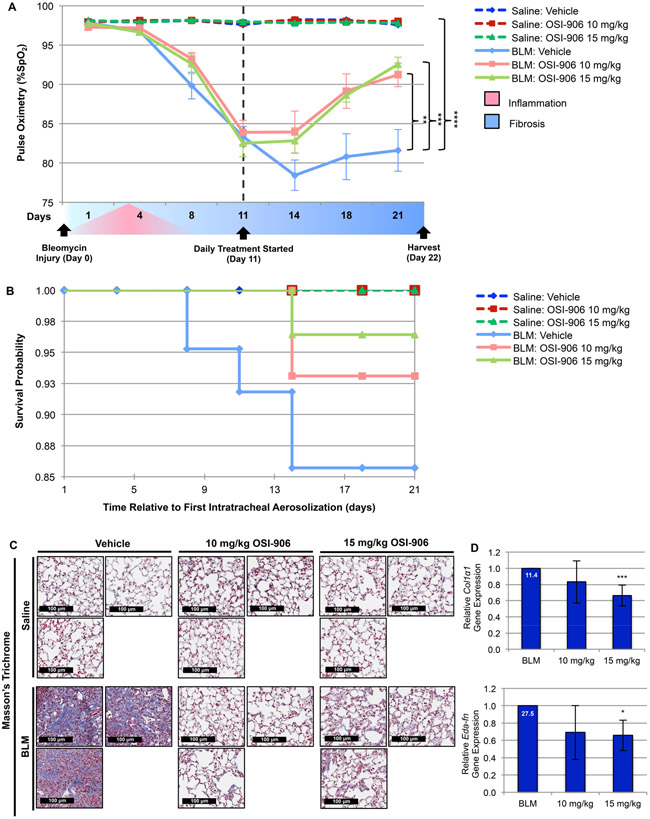
Figure 9. Inhibition of IGF-1R activation with OSI-906 alleviates BLM-induced murine lung fibrosis.
(A) Time-dependent fluctuation of mouse arterial oxygen saturation (%SpO2; determined on room air) in mice intratracheally administered BLM or saline −/+ daily treatment (started 11 days post-intratracheal insult) with vehicle or the indicated concentration (10 or 15 mg/kg) of OSI-906. (B) Kaplan-Meier survival probability estimates animal survival in all treatment groups throughout the duration of the study. (C) Representative lung tissue images stained with Masson’s Trichrome demonstrate lung tissue architecture (purple, nuclear; pink, cytoplasmic; blue, collagen fibers; 20x magnification). (D) RT-qPCR of ECM gene expression (Collagen Type I Alpha 1 Chain, Col1α1, Extra Domain-A Fibronectin, Eda-fn) in lung tissue from BLM mice treated with OSI-906 compared to vehicle. Relative fold gene induction by BLM is indicated at the top of each control bar. (A-D) Saline: Vehicle (n=10); Saline: 10 mg/kg OSI-906 (n=11); Saline 15 mg/kg OSI-906 (n=11); BLM: Vehicle (n=14); BLM: 10 mg/kg OSI-906 (n=9); BLM: 15 mg/kg OSI-906 (n=9). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that OSI-906 exhibits in vivo efficacy in ameliorating pulmonary function, survival, and architectural remodeling of lung tissue in the BLM-induced murine model of pulmonary fibrosis.
Daily gavage treatment (200 μl with 1.5” 22G bulb tipped gastric gavage needles) with vehicle (30% PEG 400 + 0.5% Tween 80 + 5% Propylene glycol) or the indicated concentration (10 or 15 mg/kg) of OSI-906 (IGF-1R kinase inhibitor) was initiated on day 11. Measurement of mouse arterial oxygen saturation (%SpO2; determined on room air) was performed every 3–4 days post intratracheal aerosolization of BLM or saline using a MouseOX Plus pulse oximeter and physiological monitor (Starr Life Science Corp., Oakmont, PA, USA). Animals were monitored for 21 days to analyze the survival of each group using Kaplan Meier Survival Analysis. Those animals that reached the humane endpoint, as specified by Mayo Clinic’s IACUC, were also considered to be dead animals at each time point. On day 22, all mice were euthanized by IP injection of pentobarbital (270 mg/kg, Sodium Pentobarbital-Fatal Plus, NDC 0298–9373-68; Vortech Pharmaceuticals, Ltd., Dearborn, MI, USA), lungs perfused with PBS, the left lung fixed with a 4% Paraformaldehyde solution in PBS (CAS 30525–89-4; Santa Cruz Biotechnology Santa Cruz, CA, USA) for histological analysis, and right lung flash frozen for qPCR analysis.
MTT Cell Viability Assay
Cells were seeded into 96-well plates (2.5 × 103 AKR-2B cells; 4 × 103 MRC5 cells) in 100 μl normal culture media per well. Following overnight incubation cultures were treated with OSI-906 or vehicle for 24 hours under normal (10% FBS, AKR-2B cells; 20% FBS MRC5 cells) or stressful (0.1% FBS, AKR-2B cells; 0% FBS MRC5 cells) growth conditions. Cell viability was determined by cell metabolic activity using the MTT colormetric assay (M5655; 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide, Millipore-Sigma, St. Louis, MO, USA) [22] according to manufacturer’s recommendation and absorbance measured at 570 nm. Assays were completed in technical octuplet.
BrdU Cell Proliferation Assay
Cells were seeded into 96-well plates (2 ×104 AKR-2B cells; 5 ×104 MRC5 cells) and treated with OSI-906 or vehicle as described for MTT Cell Viability Assay. Cell proliferation was measured using the BrdU Cell Proliferation colormetric ELISA Kit (ab126556; Abcam, Cambridge, MA, USA) according to the manufacturer’s protocol and absorbance measured at 450 nm. Assays were completed in technical quadruplicate.
Statistical analyses
Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test. Statistical significance is denoted by the number of asterisks (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Data error bars reflect ± standard error of the mean (SEM). All experiments consisted of ≥ 3 biological repeats as indicated in the figure legend while the number of technical replicates is stated in each method. Statistical analyses were performed using XLSTAT-Biomed statistical software (Addinsoft, Long Island City, NY, USA), unless otherwise noted.
RESULTS
Transcriptional profiling identifies Igf-1 as an initiating event during TGFβ-activated anchorage independent growth (AIG)
While the roles of TGFβ in myofibroblast activation, excessive wound-healing, and the progression of Idiopathic Pulmonary Fibrosis (IPF) are well established [16, 49, 50], the initiating events required for TGFβ-dependent profibrotic responses are undefined. In order to identify dysregulated genes and their profibrotic functions during the onset of fibrogenesis, AKR-2B murine fibroblasts were grown under anchorage independent conditions for 2, 4, and 8 days with 10% FBS −/+ TGFβ and evaluated using RNA-seq. TGFβ increased the expression of 499 genes > 25 fold change (FC) and Igf-1 was the 35th most upregulated gene at day 8 (Supplemental Table 4 and Supplemental Fig. 1A). Bioinformatic analysis with IPA identified upstream regulators that activate (TGFβ, Bleomycin, and IGF-1) or inhibit (SB-431542) the TGFβ-dependent AIG transcriptome (Supplemental Fig. 1B) and associated profibrotic functions (Supplemental Fig. 1C). Together these data suggest IGF-1 may have a critical role during the initiation of TGFβ-dependent profibrotic responses.
TGFβ upregulates IGF-1 in fibroblasts, pulmonary fibrosis tissue, and correlates with decreased lung function
Based on the TGFβ-dependent AIG transcriptome results (Supplemental Fig. 1), we sought to examine whether TGFβ-stimulated Igf-1 gene expression, ligand production, and/or IGF-1-Receptor (IGF-1R) activation in murine fibroblasts as well as in BLM-induced murine pulmonary fibrosis. First, TGFβ increased Igf-1 gene expression in AKR-2B murine fibroblasts in a time-dependent manner (Fig. 1A) in addition to two other murine fibroblast lines, SWISS-3T3 and BALB/c-3T3 (Supplemental Fig. 2A), as well as in BLM-induced murine fibrotic lung tissue (Fig. 1B). Although insulin and IGF-1 ligands [51] and their receptors [52] have high homology, insulin was not upregulated by TGFβ in murine fibroblasts (Supplemental Fig. 2B-D) or fibrotic murine lung tissue (Supplemental Fig. 2E). Second, both intracellular (Fig. 1C) and secreted (Fig. 1D) IGF-1 ligand, and IGF-1R-phsophorylation (Fig. 1E) were increased by TGFβ with peak levels occurring 12–24 hours post stimulation. Finally, the levels of IGF-1 and p-IGF-1R were evaluated using immunohistochemistry in pulmonary tissue from mice intratracheally administered saline or BLM followed by treatment for two weeks with saline or Imatinib (PDGFR and cAbl inhibitor) + Lapatinib (EGFR inhibitor), which was previously shown to be effective in ameliorating murine lung fibrosis [23]. Cell and ECM accumulation (Fig. 2A) as well as levels of IGF-1 (Fig. 2B, C) and p-IGF-1R (Fig. 2D,E) were greater in the BLM model where TGFβ serves as an activator of fibrosis [12, 53], compared to animals treated with saline or antifibrotic agents (e.g., Imatinib +Lapatinib; BIL).
Having established that TGFβ upregulates IGF-1 in murine fibroblasts and BLM-induced murine pulmonary fibrosis (Fig. 1, 2), we next evaluated if IGF-1 was regulated by TGFβ in human pulmonary fibroblasts and IPF. Results with human fibroblasts were consistent with the findings observed in murine fibroblasts as TGFβ increased IGF-1 gene expression in MRC5 (Fig. 3A) and IMR-90 (Supplemental Fig. 2F) human pulmonary fibroblast cells. Moreover, intracellular IGF-1 protein levels increased with TGFβ in MRC5 cells (Fig. 3B). Although primary IPF cells were not observed to have an increased basal level of IGF-1 compared to normal human lung fibroblasts (NHLF), TGFβ increased IGF-1 gene expression in both (Fig. 3C). Insulin was similarly not upregulated (Supplemental Fig. 2G-J) in either cell line or primary fibroblasts.
Figure 3. TGFβ upregulates IGF-1 in human fibroblasts and Idiopathic Pulmonary Fibrosis.
(A) RT-qPCR of IGF-1 gene expression in MRC5 human lung fibroblasts treated with TGFβ or vehicle for the indicated times (n=3). (B) Western blot of cellular IGF-1 protein synthesis in MRC5 cells subsequent to TGFβ (+) or vehicle (−) treatment (n=3). (C) RT-qPCR of IGF-1 gene expression in Normal Human Lung Primary Fibroblasts (NHLF, n=5) compared to Human Idiopathic Pulmonary Fibrosis Primary Fibroblasts (IPF, n=4) after 24 hours growth (left panel) or following 24 hours stimulation with TGFβ or vehicle (middle and right panels). (D) Differential IGF-1 gene expression analysis of RNA-seq data from RNA isolated from whole lung tissue of control (n=8) and IPF (n=8) patients. IGF-1 ranked the 6th most upregulated gene of 2,560 total genes in this small cohort of Mayo Clinic patients. (E) Differential TGFβ (panel 1) and IGF-1 (panel 3) gene expression from microarray data of RNA isolated from whole lung tissue of control (n=91) and IPF (n=122) patients from a published study by the Lung Genomics Research Consortium (LGRC) under accession number GSE47460. IGF-1 ranked the 29th most upregulated gene of 15,261 total genes. Linear regression plots with 95% confidence interval and Pearson’s correlation coefficient of the LGRC patient cohort illustrate the correlation between TGFβ and %DLCO (panel 2), IGF-1 and %DLCO (panel 4), or IGF-1 and TGFβ gene expression (panel 5). The level of fibrosis is determined by decrease in pulmonary function (measured by the percentage of predicted diffusing capacity for carbon monoxide, % DLCO). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that (1) TGFβ promotes IGF-1 gene expression and/or ligand production in human lung fibroblast cell lines and primary human lung fibroblasts; and (2) IGF-1 levels correlate with decreased pulmonary function and increased TGFβ expression.
To investigate whether IGF-1 gene expression was dysregulated in IPF patients, RNA-sequencing analysis of 2,560 genes in a small cohort of Mayo Clinic patients identified IGF-1 as the 6th most upregulated gene with over 12 fold increase compared to control (Fig. 3D). To validate these results, we analyzed a microarray gene expression dataset of 15,261 total genes and level of fibrosis as determined by decreased pulmonary function (measured by the percentage of predicted diffusing capacity for carbon monoxide, % DLCO) of a large patient cohort from the Lung Genomics Research Consortium (LGRC)[45]. From this dataset, we confirmed both TGFβ and IGF-1 were upregulated in IPF patients and correlated with decreased pulmonary function/increased severity of fibrosis (Fig. 3E). IGF-1 was ranked the 29th most upregulated gene and positively correlated with TGFβ expression (Fig. 3E). To explore whether these transcriptional signatures were reciprocated at the protein level, we evaluated another patient cohort from the Mayo Clinic using immunohistochemistry and found cell and ECM accumulation (Fig. 4A) as well as IGF-1 (Fig. 4B, C) and p-IGF-1R (Fig. 4D, E) to be increased in IPF fibroblastic foci compared to regions enriched with fibroblasts in normal human lung. These findings demonstrate that (1) TGFβ upregulates IGF-1 in pulmonary fibroblasts; (2) increased levels of IGF-1 are present in idiopathic pulmonary fibrosis; and (3) an increase in TGFβ and IGF-1 correlates with reduced pulmonary function, as well as each other.
Figure 4. IGF-1 and phosphorylated-IGF-1R are upregulated in IPF Fibroblastic Foci.
(A, B, D) Representative images (30x magnification) from histological sections of control (right, n=7, areas of normal human lung enriched with fibroblasts) and fibrotic (left, n=12, IPF fibroblastic foci) patient tissue were stained with Masson’s Trichrome (A, top) to visualize lung tissue architecture (purple, nuclear; pink, cytoplasmic) and collagen composition (blue, collagen fibers), anti-IGF-1 antibodies (B, middle), anti- phosphorylated-IGF-1R antibodies (D, bottom) co-stained with Hematoxylin, or negative controls without primary antibody co-stained with Hematoxylin (NEG). Color deconvoluted images (B, D) show the area and intensity of IGF-1 ligand (B) or phosphorylated-IGF-1R (D) present in regions of human lung enriched with proliferating myofibroblasts in fibroblastic foci compared to fibroblasts in supportive pulmonary structures per the deconvolution color key and quantified using a pixel count algorithm (C, E). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that IGF-1 ligand and IGF-1R activation are increased in IPF fibroblastic foci.
TGFβ promotes transcription of the Class-1 IGF-1 promoter in fibroblasts and pulmonary fibrosis tissue
Initiation of IGF-1 transcription is regulated by two alternative promoters that harbor multiple transcription start sites (Fig. 5A). While the biological implications are unclear, it’s been hypothesized that the Class-1 (C1) supports IGF-1’s mitogenic autocrine/paracrine actions, and the Class-2 (C2) regulates IGF-1’s endocrine response [54]. Since substantiating TGFβ’s role as a required mediator of IGF-1 upregulation in fibroblasts and pulmonary fibrosis (Fig. 1-4 and Supplemental Fig. 1, 2), we next assessed which of the alternative promoters TGFβ increased. While a minimal response was observed from the C2 promoter, the C1 promoter was upregulated by TGFβ in AKR-2B (Fig. 5B), SWISS-3T3, and BALB/c-3T3 (Supplemental Fig. 3A, B) murine fibroblasts as well as fibrotic murine lung tissue (Fig. 5C). Results with murine fibroblasts were consistent with those observed in human fibroblasts, as TGFβ increased IGF-1’s C1 promoter expression in MRC5 (Fig. 5D) and IMR-90 (Supplemental Fig. 3C) pulmonary fibroblasts as well as in NHLF and IPF primary cells (Fig. 5E, F). Thus, TGFβ induces IGF-1 gene expression in activated lung myofibroblasts primarily through initiating transcription of the C1 promoter.
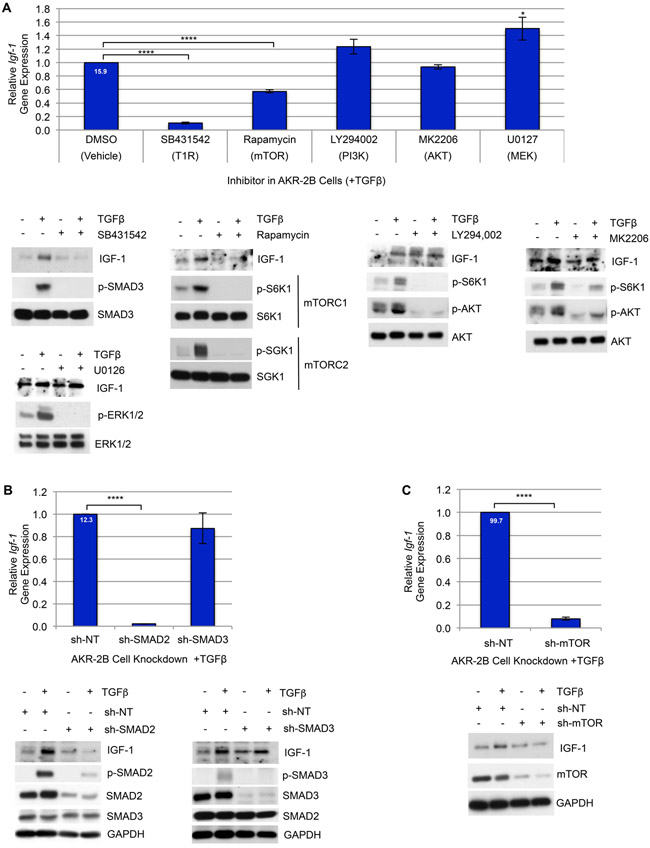
Canonical and non-canonical TGFβ signaling are required for Igf-1 upregulation
Activation of the type 1 TGFβ receptor (T1R) serves as a fibrotic-switch by activating downstream signaling cascades including canonical (SMAD2 & 3) and non-canonical (PI3K → AKT, MEK→ ERK, mTOR, etc.) mediators that govern gene expression [19, 55]. Considering TGFβ’s ability to upregulate Igf-1 transcription at the C1 promoter (Fig. 5), we proceeded to identify which downstream signaling pathways were required for TGFβ-dependent Igf-1 activation. Taking into account how SMAD signaling [56] is perpetuated early, and other targets such as mTOR [35] are induced later, we utilized pharmacologic inhibitors of both canonical and non-canonical targets and found TGFβ-stimulated Igf-1 to be dependent on both T1R activation and mTOR signaling (Fig. 6A). To determine whether canonical signaling by either SMAD2 and/or SMAD3 was required, stable SMAD2 and SMAD3 knockdown AKR-2B cells established the necessity for SMAD2, but not SMAD3 signaling to facilitate an increase in Igf-1 expression (Fig. 6B). Furthermore, while Rapamycin inhibits mTOR with high specificity, like most inhibitors, its efficacy changes with different cell types, tissues, and over time. Moreover, mTOR’s sensitivity to Rapamycin is further complicated by its function as the catalytic subunit in two distinct multi-protein complexes (mTORC1 and mTORC2), which under our conditions signaling from both as well as IGF-1 expression are inhibited [57-59] (Fig. 6A). Additional evaluation with a stable knockdown of mTOR validated its role inducing Igf-1 (Fig. 6C). Collectively, the data establishes that multiple signaling mechanisms are used by TGFβ in the upregulation of Igf-1.
Figure 6. TGFβ-dependent induction of Igf-1 is mediated by SMAD2 & mTOR signaling.
(A) Top: RT-qPCR of Igf-1 gene expression in AKR-2B cells pre-treated for 2 hours with the indicated TGFβ-pathway inhibitors (10 μM SB431542, TGFβ Type-I Receptor (T1R) inhibitor; 20 μM LY294002, PI3K inhibitor; 300 nM MK2206, AKT inhibitor; 10 μM U0126, MEK inhibitor; 10 nM Rapamycin, mTOR inhibitor) or vehicle (0.1% DMSO), followed by stimulation with TGFβ for 12 hours (n=4). Bottom: Western blot of IGF-1 in AKR-2B cells as treated for qPCR and controls show the functionality of inhibitors on target protein activation following 3 (p-ERK/ERK) or 6 hours (p-SMAD3/SMAD3, p-AKT/AKT, p-S6K/S6K, p-SGK1/SGK1) treatment (n=3). (B) Top: RT-qPCR of Igf-1 expression in AKR-2B cells with shRNA-mediated stable knockdown of SMAD2 (sh-SMAD2), SMAD3 (sh-SMAD3), or nontargeting (sh-NT) after 12 hours TGFβ stimulation (n=3). Bottom: Western blot of IGF-1 in AKR-2B KD cells as treated for qPCR and controls confirming the level of knockdown of SMAD2, SMAD3, and p-SMAD2 or p-SMAD3 at 6 hours with TGFβ (+) or vehicle (−) (n=3). (C) Top: RT-qPCR of Igf-1 gene expression in AKR-2B cells with shRNA-mediated stable knockdown of mTOR (sh-mTOR) or NT (sh-NT) following 12 hours TGFβ stimulation (n=4). Bottom: Western blot of IGF-1 in AKR-2B KD cells as treated for qPCR and controls confirm the level of mTOR KD at 6 hours in the presence or absence of TGFβ (n=3). (A-C) Relative fold gene expression by TGFβ compared to control is indicated at the top of the vehicle or NT bars. Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that TGFβ-induction of Igf-1 transcription and translation requires SMAD2 & mTOR signaling.
IGF-1R activation is required for myofibroblast differentiation and aberrant wound-healing processes
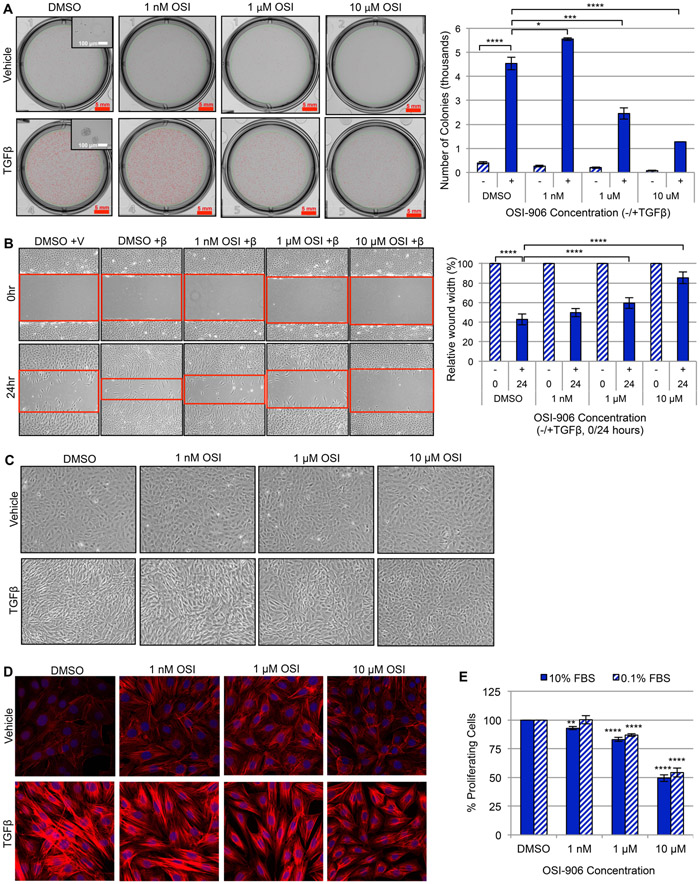
While IGF-1 signaling is required for lung development and homeostasis, stimulation with IGF-1 and IGF-1R activation have also been shown to mediate proliferation, survival, and collagen production in human and murine fibroblasts in vitro [60-65]. Thus, with the previous figures providing evidence for the significance of TGFβ upregulation of IGF-1 in myofibroblasts and pulmonary fibrosis (Fig. 1-6), we next investigated whether inhibiting IGF-1R’s kinase activity with OSI-906 (Linsitinib) [66] would effectively inhibit TGFβ-dependent profibrotic responses. We first examined the dose dependent response of OSI-906 on blocking IGF-1 downstream signaling in murine and human fibroblasts. While OSI-906 inhibited IRS-1 phosphorylation in both cell types (Supplemental Fig. 4A and 5A), this was independent of any major effect on cell viability (Supplemental Fig. 4B and 5B). However, IGF-1R-phosphorylation was required for TGFβ-stimulated anchorage independent growth in soft agar (Fig. 7A), wound-healing (Fig. 7B and Supplemental Fig. 4C), myofibroblast differentiation (Fig. 7C and Supplemental Fig. 4D), actin reorganization (Fig. 7D), proliferation (Fig. 7E and Supplemental Fig. 4E), and extracellular matrix (ECM) gene transcription (Fig. 8A, C) and protein synthesis (Fig. 8B, D). Furthermore, knocking down the IGF-1R (Supplemental Fig. 5C, 6A) validated the requirement for IGF-1 signaling to perpetuate the TGFβ-dependent profibrotic response including wound-healing (Supplemental Fig. 5D, 6B), ECM transcription (Supplemental Fig. 5E, 6C) and protein expression (Supplemental Fig. 5F, 6D) in both murine (Supplemental Fig. 5) and human (Supplemental Fig. 6) fibroblasts. This body of evidence substantiates that (1) IGF-1 signaling is a driver of TGFβ-activated myofibroblast processes; (2) inhibition of IGF-1R activation with OSI-906 is an effective anti-fibrotic approach; and (3) suggest that the IGF-1R may be a potential therapeutic target in the treatment of pulmonary fibrosis.
Figure 7. Inhibition of IGF-1R activation with OSI-906 diminishes TGFβ-induced myofibroblast differentiation and profibrotic responses.
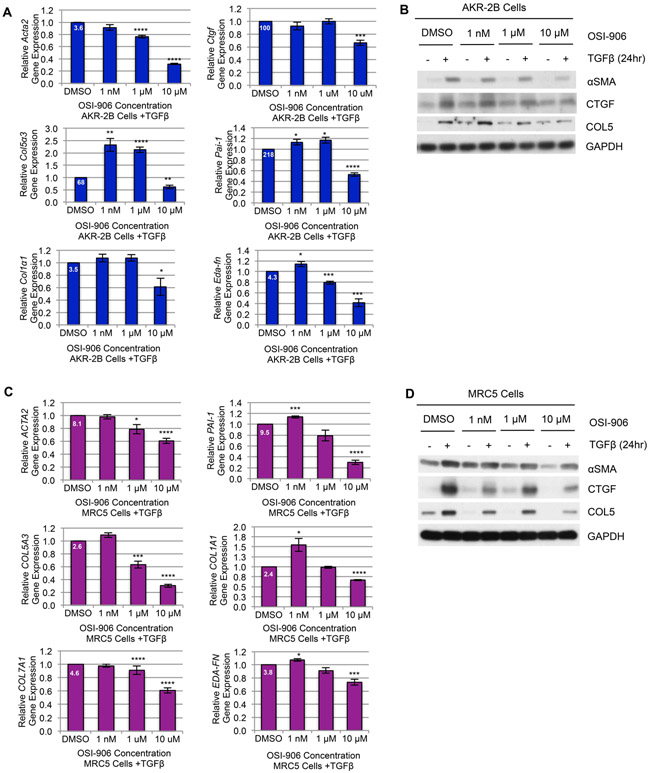
(A) Anchorage Independent Growth (AIG) of AKR-2B murine fibroblast cells stimulated with TGFβ or vehicle in the presence of increasing concentrations of OSI-906 or vehicle (0.1% DMSO) after 8 days. Left: Representative images indicate colonies per treatment condition (red selection marks represent colonies with diameter ≥ 50 μm; scale bars measure 5 mm), and insert depicts a representative colony or individual cell size (scale bars measure 100 μm). Right: AIG was quantified by the number of colonies present under each condition (n=3). (B) Left: Scratch assays were performed with AKR-2B murine fibroblasts treated with vehicle (+V) or TGFβ (+β) and increasing concentrations of OSI-906 or vehicle. Red outlines indicate the leading edges of the wound. Right: The wound-healing response was quantified by the percent relative wound width measured at 0 and 24 hours (n=3). (C) Effect of increasing OSI-906 concentrations or vehicle on TGFβ-stimulated AKR-2B morphological transformation from a fibroblast morphology to differentiated myofibroblast phenotype (10x magnification) at 24 hours (n=3). (D) Stress fiber formation (F-actin polymerization associated with myofibroblast morphology) of confluent AKR-2B fibroblasts grown on glass coverslips and treated as in B (40x magnification). Filamentous actin (F-actin) is stained with phalloidin (red) and nuclei with DAPI (blue) (n=3). (E) Proliferative capacity of AKR-2B cells analyzed under normal (10% FBS) and stressful (0.1% FBS) growth conditions with increasing OSI-906 concentrations using BrdU incorporation (n=3). Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that inhibition of IGF-1R kinase activity with OSI-906 in AKR-2B murine fibroblasts effectively prevents TGFβ-dependent myofibroblast proliferation, differentiation, and profibrotic responses.
Figure 8. Inhibition of IGF-1R activation with OSI-906 reduces TGFβ-stimulated extracellular matrix gene expression and protein synthesis.
(A, C) RT-qPCR or (B, D) Western blot analysis of Extracellular Matrix (ECM) genes/proteins in AKR-2B murine fibroblasts (A, B) or MRC5 human lung fibroblasts (C, D) pre-treated for 2 hours with increasing concentrations of OSI-906 or vehicle (0.1% DMSO) followed by stimulation with TGFβ for 24 hours (n=3). ECM genes/proteins included in the transcriptional/translational profiles regulated by OSI-906 include: Alpha Smooth Muscle Actin, ACTA2/Acta2 (αSMA); Connective Tissue Growth Factor, Ctgf; Collagen Type 5 Alpha 3 Chain, COL5A3/Col5α3; Plasminogen Activator Inhibitor-1, PAI-1/Pai-1; Collagen Type I Alpha 1 Chain, COL1A1/Col1α1; Collagen Type 7 Alpha 1 Chain, COL7A1; Extra Domain-A Fibronectin, EDA-FN/Eda-fn. (A, C) Relative fold gene induction by TGFβ compared to vehicle is indicated at the top of each control bar. Data are presented as means −/+ Standard Error of the Mean (SEM) for the number of biological replicates indicated (n). Statistical significance was determined after computing single factor ANOVA and/or unpaired two-tailed Student’s t-test (p<0.05 (*), p<0.01 (**), p<0.005 (***), p<0.001 (****). Results demonstrate that OSI-906 inhibition of IGF-1R kinase activity is effective in reducing the expression of profibrotic ECM components induced by TGFβ.
Inhibition of IGF-1R activation ameliorates BLM-induced pulmonary fibrosis
Previous evidence using IGF-1R deficient animals [67, 68], anti-IGF-1R antibody [69-71], and IGF-1 overexpression [72-75] in the BLM-induced murine lung fibrosis model have produced opposing evidence as to the involvement of IGF-1 signaling in alveolar injury, inflammation and pulmonary fibrosis. In that, pharmacologic therapies inhibiting IGF-1R activation for the treatment of lung fibrosis have not been explored, and having established OSI-906’s therapeutic potential on TGFβ-dependent profibrotic functions (Fig. 7, 8 and Supplemental Fig. 4, 5), we chose to evaluate the in vivo efficacy of OSI-906 in treating pulmonary fibrosis using the BLM murine model (the most robust experimental model of pulmonary fibrosis [76]). Mice intratracheally administered Saline or BLM began daily treatment with 10 or 15 mg/kg of OSI-906 11 days post-intratracheal insult and were euthanized on day 22. Those animals treated with OSI-906 showed improved pulmonary function per arterial oxygen saturation (%SpO2; Fig. 9A), survival (Fig. 9B), and lung tissue architecture (Fig. 9C and Supplemental Fig. 7, 8A). Additionally, the corresponding lung tissue had decreased accumulation of collagen (Fig. 9C and Supplemental Fig. 7, 8B), Fibronectin (Supplemental Fig. 9A), and α Smooth Muscle Actin (Supplemental Fig. 9B), as well as ECM (Col1α1 and Eda-fn) gene expression (Fig. 9D). Results from this pre-clinical trial demonstrate targeting the IGF-1R with OSI-906 is an effective and novel therapeutic strategy for the treatment of pulmonary fibrosis. A model integrating the fibroproliferative actions of TGFβ with IGF-1 induction/receptor activation is presented in Fig. 10.
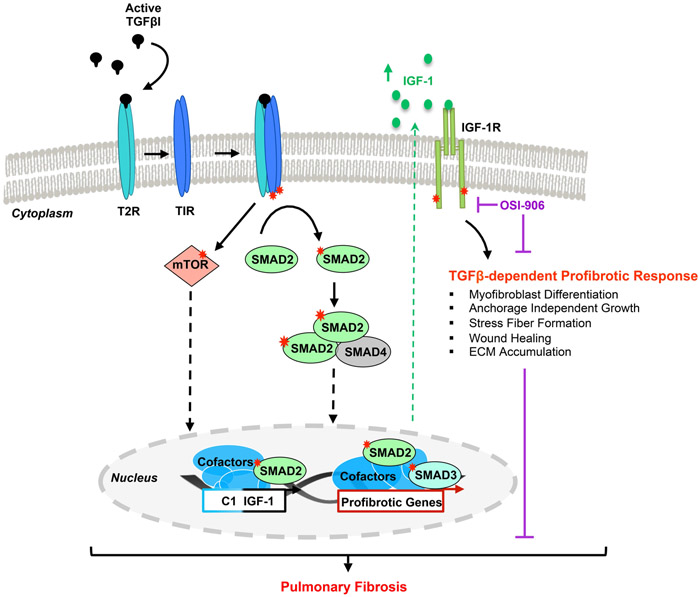
Figure 10. Proposed mechanism for TGFβ-dependent IGF-1 induction regulating the TGFβ-dependent profibrotic response.
A schematic diagram of the molecular mechanisms required for TGFβ-dependent stimulation of IGF-1 and the TGFβ-initiated profibrotic cellular responses mediated by IGF-1R activation.
DISCUSSION
This research study provides insight about the molecular mechanisms and profibrotic processes of myofibroblasts as the principle driver of organ fibrosis, an underrepresented group of deadly diseases [77]. Our results expose IGF-1 as a central mediator of TGFβ-dependent myofibroblast-driven fibrogenesis and the therapeutic effectiveness of targeting IGF-1 signaling in the prevention of IPF progression. These findings not only expand our understanding of IGF-1’s role in disease and identify it as a novel regulator of TGFβ’s profibrotic functions, but most importantly, improve our understanding of fibrosis and advance our capabilities to develop effective therapies for the treatment of IPF as well as other fibrotic diseases.
The heterogeneity of IPF clinically and in the BLM murine model indicates there is likely no single fibrogenic event initiating IPF [78]. In that TGFβ has been shown to be a master regulator of fibrogenesis and a causative element in the early onset of fibrotic diseases [16-18], we transcriptionally profiled the fibroblast response to TGFβ under anchorage independent conditions. These studies not only substantiated this phenotype as a representative model of the fibroproliferative responses associated with pathogenic fibrosis [41], but revealed the upregulation of Igf-1 as a initiating profibrotic event induced by TGFβ (Supplemental Fig. 1).
Elevated levels of IGF-1 have been implicated in multiple types of interstitial lung disease [28] and shown to be produced by lung fibroblasts and stimulate mitogenic actions [79-81]; however, there is minimal data as to whether TGFβ is responsible for regulating IGF-1 in fibroblasts or merely acts synergistically in the progression of pulmonary fibrosis [70, 75]. Our results demonstrate that TGFβ is necessary to upregulate IGF-1 gene expression, protein production, and IGF-1R activation in myofibroblasts (Fig. 1, 3). Furthermore, these studies not only confirmed the presence of elevated levels of IGF-1 in BLM-induced murine lung fibrosis (Fig. 2B, C) [29, 69], but showed (1) an increase in IGF-1R-phosphorylation (Fig. 2D, E); (2) that animals treated with Imatinib (PDGFR and cAbl inhibitor) + Lapatinib (EGFR inhibitor), previously shown to ameliorate murine lung fibrosis [23], had decreased IGF-1 expression and IGF-1R activation (Fig. 2); and (3) an analogous increase in IGF-1 ligand and IGF-1R-phosphorylation in IPF lung tissue (Fig. 4). This latter finding was of particular interest as there is conflicting evidence as to whether IGF-1 is upregulated [30, 31] or downregulated [82] in IPF and historic immunohistochemical evidence from IPF tissue implies IGF-1 is localized only to injured alveolar epithelia [72] and inflammatory cells [83]. To further address these issues we, first, determined that IGF-1 gene expression in IPF compared to normal primary fibroblasts is a time sensitive response to TGFβ that is masked by the lack of active TGFβ in cell culture medium compared to that in vivo (Fig. 3C); second, evaluated three different patient cohorts and determined that IGF-1 was increased in all three and positively correlated with TGFβ and decreased pulmonary function associated with increased severity of IPF (Fig. 3C-E); and, third, focused our investigations on the proliferating fibroblasts found in IPF fibroblastic foci which showed an enrichment of IGF-1 and IGF-1R-phosphorylation (Fig. 4). Together these findings recognize TGFβ as an essential upstream regulator of IGF-1 expression and signaling in myofibroblasts which correlates with advanced pulmonary fibrosis. Last, although IGF-1 and insulin as well as their receptors share significant homology, the ability to interact with each other’s receptor, and stimulate growth [51, 84], we additionally found that insulin expression is not increased with TGFβ in fibroblasts and in fibrotic lung tissue; indicating that IGF-1 upregulation and IGF-1R phosphorylation are independent of insulin (Supplemental Fig. 2).
IGF-1 expression is differentially regulated by an alternative promoter system whereby two distinct leader exons composed of mutually exclusive classes of transcription start sites produce several transcript variants, ultimately encoding the same mature IGF-1 ligand [54] (Fig. 5A). Although the biological implications remain unclear, it is hypothesized that the Class-1 promoter (C1) supports IGF-1’s mitogenic autocrine and paracrine actions while the Class-2 (C2) regulates IGF-1’s endocrine response [54]. While the presence of transcriptional IGF-1 isoforms has been investigated in different cell types and disease states [85-88], little consideration has been given to whether IGF-1 promoters are differentially regulated by disease-associated growth factors such as TGFβ. That issue was specifically addressed in Figure 5 and Supplemental Figure 3 where we established that TGFβ upregulated IGF-1’s C1 promoter transcript expression in myofibroblasts and lung fibrosis; supporting the hypothesized role of the C1 promoter to advance mitogenic autocrine/paracrine responses. These findings, together with O’Dwyer and colleagues [89] conclusion that IGF-1 is not enriched in the circulating IPF proteome, further support our determination that localized production of IGF-1 and increased transcription of the autocrine/paracrine C1 promoter by myofibroblasts at the site of epithelial injury perpetuate the uncontrolled wound healing response in the progression of IPF. Moreover, our study further determined that TGFβ’s transcriptional activation of Igf-1 is mediated by canonical SMAD2 signaling independent of SMAD3 (Fig. 6); this is intriguing for two reasons, first, SMAD3 has prevailed in the literature as profibrotic [48] and SMAD2 as antifibrotic transcription factors activated by TGFβ [90], and second, there is no SMAD binding element (SBE) present in either the C1 or C2 Igf-1 promoters [91, 92], suggesting there is a more complex transcriptional network required for SMAD2 activation of Igf-1 transcription. Finally, non-canonical TGFβ-activated mTOR signaling, a known pathway promoting fibrogenesis and myofibroblast differentiation is additionally required for Igf-1 upregulation by TGFβ [35] (Fig. 6). However, it is surprising that inhibition of PI3K or AKT do not abolish TGFβ-dependent upregulation of IGF-1 since PI3K and AKT are well-characterized upstream activators of mTOR signaling; supporting the possibility that there may be additional signaling pathways independent of PI3K/AKT capable of activating mTOR. Collectively, these results give us a glimpse into the intricate web of regulatory mechanisms imperative for TGFβ to enhance Igf-1 activation to promote fibrotic disease and clearly indicate we have more to learn concerning TGFβ signaling and IGF-1 stimulation.
While dysregulated IGF-1 signaling has been implicated in IPF and the BLM lung fibrosis model [29-31], there are conflicting findings as to its participation in the progression, prevention and treatment of pulmonary fibrosis [67-75]. Furthermore, in light of new evidence requiring TGFβ to upregulate IGF-1 signaling in fibroblasts and the lack of research in the use of pharmacologic therapies targeting IGF-1 signaling to treat pulmonary fibrosis, further evaluation of IGF-1R activation as a mediator of fibrogenesis was warranted. To that end, we evaluated the effectiveness of OSI-906 (Linsitinib), an orally bioavailable IGF-1R tyrosine kinase inhibitor; despite the fact that OSI-906 has not had the desired success in treating cancers of epithelial origin, it may be an effective therapy for mesenchymal-derived diseases [66]. First, after determining OSI-906 successfully inhibited downstream IRS-1 phosphorylation in both murine and human fibroblasts without any major cell toxicity, we established that OSI-906 prevented TGFβ-dependent myofibroblast differentiation, proliferation, and profibrotic responses in a dose-dependent manner (Fig. 7, 8 and Supplemental Fig. 4, 5A, B). Profibrotic responses were also diminished in IGF-1R knockdown fibroblast cells (Supplemental Fig. 5C-F, 6), confirming the requirement for IGF-1 signaling in TGFβ-driven aberrant wound healing. Although OSI-906 differentially affected the transcription and translation of specific ECM components (Fig. 8), critical components such as Collagen 1, EDA-Fibronectin, and Smooth Muscle Actin were significantly decreased both in vitro as well as in vivo (Fig. 8, 9 and Supplemental Fig. 7-9). Lastly, we saw an improvement in pulmonary function, survival, and lung tissue architecture in animals treated with low doses (10–15 mg/kg) of OSI-906 following BLM-induced lung fibrosis when compared to BLM-controls (Fig. 9 and Supplemental Fig. 7-9). Results from this preclinical trial and analysis of TGFβ-activated profibrotic myofibroblast actions with OSI-906 demonstrate that inhibition of IGF-1R-phosphorylation is effective in blocking myofibroblast-driven fibrogenesis and effectively delays the progression of lung fibrosis (Fig. 10).
Ultimately, these results contribute to the growing body of biomedical evidence elucidating the unclear etiology, complex molecular mechanisms, myofibroblast-derived profibrotic functions, and heterogeneous pathophysiology of IPF; that may very well translate to other fibroproliferative diseases. Identifying IGF-1’s role and regulation of TGFβ-activated lung myofibroblasts, fibrogenesis, and IPF, as well as addressing previous inconsistent and/or paradoxical evidence of IGF-1 in fibrotic lung diseases, provides new opportunities to accelerate the development of antifibrotic therapies, advance clinical management of IPF, and improve patient prognosis.
Supplementary Material
ACKNOWLEDEMENTS
We would like to thank: Dr. Carol Feghali-Bostwick (Medical University of South Carolina) and Dr. Nathan Sandbo (University of Wisconsin-Madison Department of Medicine, UW-Madison Carbone Cancer Center Biobank, P30 CA014520) for graciously providing primary fibroblasts from healthy and IPF patients; Dr. Robert Vassallo for kindly sharing the NGS profiling dataset; Dr. Eunhee S. Yi and Dr. Anja C. Roden for histological pathology review of patient samples; Kyle J. Schaefbauer for help with animal procedures; Dr. Ruth Lupu for teaching immunohistochemical techniques; and Dr. Douglas Yee, Dr. Scott Kaufmann, and Dr. Dan Billadeau for insightful discussions. This work was supported by grants from the U.S. Department of Health and Human Services National Institute of General Medical Sciences (R01-GM054200, R37-GM055816), the Caerus Foundation (91736058), Hurvis Accelerate the Cure for IPF (91736063), and the Mayo Foundation (to E.B.L.). The authors declare no conflicts of interest.
ABBREVIATIONS
- αSMA/ACTA2
Alpha Smooth Muscle Actin
- BLM
Bleomycin
- B2M
Beta-2 Microglobulin
- CTGF
Connective Tissue Growth Factor
- COL1A1
Collagen Type 1 Alpha 1 Chain
- COL5A3
Collagen Type 5 Alpha 3 Chain
- COL7A1
Collagen Type 7 Alpha 1 Chain
- DMSO
Dimethyl Sulfoxide
- EGFR
Epidermal Growth Factor Receptor
- ERK
Extracellular Signal-Regulated Kinase
- EDA-FN
Extra Domain-A Fibronectin
- FBS
Fetal Bovine Serum
- ILD
Interstitial Lung Diseases
- INS
Insulin
- IGF-1
Insulin-like Growth Factor-1
- IPF
Idiopathic Pulmonary Fibrosis
- mTOR
Mammalian Target of Rapamycin
- mTORC1
Mammalian Target of Rapamycin Complex 1
- mTORC2
Mammalian Target of Rapamycin Complex 2
- MAPK
Mitogen-Activated Protein Kinase
- MEK
Mitogen-Activated Protein Kinase Kinase
- NHLF
Normal Human Lung Fibroblasts
- PI3K
Phosphoinositide 3-Kinase
- PAI-1
Plasminogen Activator Inhibitor-1
- PDGFR
Platelet-Derived Growth Factor Receptor
- AKT
Protein Kinase B
- S6K1
Ribosomal protein S6 kinase beta-1
- SMAD
Sma- and Mad-related Protein
- SBE
SMAD binding element
- SGK1
Serum and glucocorticoid-inducible kinase
- TBP
Tata Binding Protein
- TGFβ
Transforming Growth Factor Beta
- T1R
Type I TGFβ Receptor
REFERENCES
- 1.Brown GC, Living too long: the current focus of medical research on increasing the quantity, rather than the quality, of life is damaging our health and harming the economy. EMBO reports, 2015. 16(2): p. 137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nations, U., World Population Prospects The 2012 Revision, in Highlights and Advance Tables. 2013, Department of Economic and Social Affairs: Population Division: New York. [Google Scholar]
- 3.Nations, U., World Population Prospects 2019, in Highlights. 2019, Department of Econmic and Social Affairs: Population Division: New York. [Google Scholar]
- 4.Hecker L, Mechanisms and consequences of oxidative stress in lung disease: theraputic implications for an aging populace. Am J Physiol Lung Cell Mol Physiol, 2018. 314: p. L642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacNee W, Rabinovich RA, and Choudhury G, Ageing and the border between health and disease. The European respiratory journal, 2014. 44(5): p. 1332–52. [DOI] [PubMed] [Google Scholar]
- 6.Zank DC, et al. , Idiopathic Pulmonary Fibrosis: Aging, Mitochondrial Dysfunction, and Cellular Bioenergetics. Frontiers in medicine, 2018. 5: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divo MJ, Martinez CH, and Mannino DM, Ageing and the epidemiology of multimorbidity. The European respiratory journal, 2014. 44(4): p. 1055–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steele MP and Schwartz DA, Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med, 2013. 64: p. 265–76. [DOI] [PubMed] [Google Scholar]
- 9.Kiaminski N and Gulati M Lung Disease Week at the ATS: Pulmonary Fibrosis. General Information about Pulmonary Fibrosis, 2015. [Google Scholar]
- 10.Allemani C, et al. , Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2). The Lancet, 2015. 385(9972): p. 977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu JH, et al. , Idiopathic Pulmonary Fibrosis: Evolving Concepts. Mayo Clin Proc, 2014. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez IE and Eickelberg O, The impact of TGF-beta on lung fibrosis: from targeting to biomarkers. Proceedings of the American Thoracic Society, 2012. 9(3): p. 111–6. [DOI] [PubMed] [Google Scholar]
- 13.Camelo A, et al. , The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol, 2014. 4: p. 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darby IA, et al. , Fibroblasts and myofibroblasts in wound healing. Clinical, cosmetic and investigational dermatology, 2014. 7: p. 301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George PM, et al. , Lung transplantation for idiopathic pulmonary fibrosis. The Lancet Respiratory Medicine, 2019. 7(3): p. 271–282. [DOI] [PubMed] [Google Scholar]
- 16.Meng XM, Nikolic-Paterson DJ, and Lan HY, TGF-beta: the master regulator of fibrosis. Nature reviews. Nephrology, 2016. 12(6): p. 325–38. [DOI] [PubMed] [Google Scholar]
- 17.Kim KK, Sheppard D, and Chapman HA, TGF-beta1 Signaling and Tissue Fibrosis. Cold Spring Harbor perspectives in biology, 2018. 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prud’homme GJ, Pathobiology of transforming growth factor beta in cancer, fibrosis and immunologic disease, and therapeutic considerations. Laboratory investigation; a journal of technical methods and pathology, 2007. 87(11): p. 1077–91. [DOI] [PubMed] [Google Scholar]
- 19.Massague J, TGFbeta signalling in context. Nat Rev Mol Cell Biol, 2012. 13(10): p. 616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YE, Non-Smad pathways in TGF-β signaling. Cell Research, 2009. 19(1): p. 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrianifahanana M, et al. , Profibrotic up-regulation of glucose transporter 1 by TGF-beta involves activation of MEK and mammalian target of rapamycin complex 2 pathways. FASEB J, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung MY, et al. , Fatty acid synthase is required for profibrotic TGF-beta signaling. FASEB J, 2018. 32(7): p. 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrianifahanana M, et al. , Profibrotic TGFbeta responses require the cooperative action of PDGF and ErbB receptor tyrosine kinases. FASEB J, 2013. 27(11): p. 4444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walton KL, Johnson KE, and Harrison CA, Targeting TGF-beta Mediated SMAD Signaling for the Prevention of Fibrosis. Front Pharmacol, 2017. 8: p. 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyorfi AH, Matei AE, and Distler JHW, Targeting TGF-beta signaling for the treatment of fibrosis. Matrix biology : journal of the International Society for Matrix Biology, 2018. 68–69: p. 8–27. [DOI] [PubMed] [Google Scholar]
- 26.Puche JE and Castilla-Cortazar I, Human conditions of insulin-like growth factor-I (IGF-I) deficiency. Journal of Translational Medicine, 2012. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y, et al. , Mechanisms of disease: signaling of the insulin-like growth factor 1 receptor pathway--therapeutic perspectives in cancer. Nature clinical practice. Oncology, 2007. 4(10): p. 591–602. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, et al. , Insulin-Like Growth Factor-1 Signaling in Lung Development and Inflammatory Lung Diseases. BioMed research international, 2018. 2018: p. 6057589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honeyman L, et al. , MicroRNA profiling implicates the insulin-like growth factor pathway in bleomycin-induced pulmonary fibrosis in mice. Fibrogenesis & Tissue Repair, 2013. 6(16): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu E, et al. , Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis & Rheumatism, 2011. 63(3): p. 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selman M, Pardo A, and Kaminski N, Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS medicine, 2008. 5(3): p. e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiles AD and D’Ercole A, The Insulin-like Growth Factors and the Lung. American journal of respiratory cell and molecular biology, 1990. 3. [DOI] [PubMed] [Google Scholar]
- 33.Esnault S, et al. , Endogenous Semaphorin-7A Impedes Human Lung Fibroblast Differentiation. PLoS ONE, 2017. 12(1): p. e0170207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilewski JM, et al. , IGFBP3 and 5 are Overexpressed in IPF and contribute to ECM deposition. American Journal of Pathology, 2005. 166(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahimi RA, et al. , Distinct roles for mammalian target of rapamycin complexes in the fibroblast response to transforming growth factor-beta. Cancer research, 2009. 69(1): p. 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrianifahanana M, et al. , ERBB receptor activation is required for profibrotic responses to transforming growth factor beta. Cancer Res, 2010. 70(19): p. 7421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudakovic A, et al. , High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem, 2014. 115(10): p. 1816–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalari KR, et al. , MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics, 2014. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar R, Domrachev M, and Lash AE, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 2002. 30(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, et al. , NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research, 2013. 41(Database issue): p. D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torry DJ, et al. , Anchorage-independent Colony Growth of Pulmonary Fibroblasts Derived from Fibrotic Human Lung Tissue. J. Clin. Invest, 1994. 93(April): p. 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ and Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 2001. 25(4): p. 402–8. [DOI] [PubMed] [Google Scholar]
- 43.Vandesompele J, et al. , Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 2002. 3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauer Y, et al. , A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. American journal of respiratory cell and molecular biology, 2015. 52(2): p. 217–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartholmai B, et al. , The Lung Tissue Research Consortium: An extensive open database containing histological, clinical, and radiological data to study chronic lung disease Insight Journal, 2006(2006 MICCAI Open Science Workshop; ). [Google Scholar]
- 46.Benjamini Y and Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 1995. 57(1). [Google Scholar]
- 47.Raghu G, et al. , An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med, 2011. 183(6): p. 788–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang JH, et al. , Cell-penetrating peptides selectively targeting SMAD3 inhibit profibrotic TGF-beta signaling. J Clin Invest, 2017. 127(7): p. 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kottmann RM, et al. , Determinants of initiation and progression of idiopathic pulmonary fibrosis. Respirology, 2009. 14(7): p. 917–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kendall RT and Feghali-Bostwick CA, Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol, 2014. 5: p. 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brzozowski AJ, et al. , Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin. Biochemistry, 2002. 41: p. 9389–9397. [DOI] [PubMed] [Google Scholar]
- 52.Cai W, et al. , Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat Commun, 2017. 8: p. 14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sime PJ and O’Reilly KM, Fibrosis of the lung and other tissues: new concepts in pathogenesis and treatment. Clin Immunol, 2001. 99(3): p. 308–19. [DOI] [PubMed] [Google Scholar]
- 54.Oberbauer AM, The Regulation of IGF-1 Gene Transcription and Splicing during Development and Aging. Frontiers in Endocrinology, 2013. 4: p. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahimi RA and Leof EB, TGF-beta signaling: a tale of two responses. J Cell Biochem, 2007. 102(3): p. 593–608. [DOI] [PubMed] [Google Scholar]
- 56.Brown KA, Pietenpol JA, and Moses HL, A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem, 2007. 101(1): p. 9–33. [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay S, et al. , The Enigma of Rapamycin Dosage. Mol Cancer Ther, 2016. 15(3): p. 347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toschi A, et al. , Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol, 2009. 29(6): p. 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Kim SG, and Blenis J, Rapamycin: one drug, many effects. Cell Metab, 2014. 19(3): p. 373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstein RH, et al. , Stimulation of Collagen Formation by Insulin and Insulin-Like Growth Factor I in Cultures of Human Lung Fibroblasts. Endocrinology, 1989. 124(2). [DOI] [PubMed] [Google Scholar]
- 61.Wu Y, et al. , Activation of the Insulin-Like Growth Factor-I Receptor Inhibits Tumor Necrosis Factor-Induced Cell Death. Journal of Cellular Physiology, 1996. 168: p. 499–509. [DOI] [PubMed] [Google Scholar]
- 62.Chetty A, Faber S, and Nielsen HC, Epithelial-mesenchymal interaction and insulin-like growth factors in hyperoxic lung injury. Experimental Lung Research, 1999. 25: p. 701–718. [DOI] [PubMed] [Google Scholar]
- 63.Valentinis B, et al. , Anti-apoptotic signaling of the IGF-1 receptor in fibroblasts following loss of matriz adhesion. Oncogene, 1999. 18: p. 1827–1836. [DOI] [PubMed] [Google Scholar]
- 64.Chetty A, Cao GJ, and Nielsen HC, Insulin-like Growth Factor-I signaling mechanisms, type I collagen and alpha smooth muscle actin in human fetal lung fibroblasts. Pediatric research, 2006. 60(4): p. 389–94. [DOI] [PubMed] [Google Scholar]
- 65.Warnken M, et al. , Characterization of proliferative effects of insulin, insulin analogues and insulin-like growth factor-1 (IGF-1) in himan lung fibroblasts. Naunyn-Schmied Arch Pharmacol, 2010. 382: p. 511–524. [DOI] [PubMed] [Google Scholar]
- 66.Mulvihill MJ, et al. , Discovery of OSI-906: a slective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor, in Future Medical Chemistry. 2009. p. 1153–1171. [DOI] [PubMed] [Google Scholar]
- 67.Ahamed K, et al. , Deficiency in type 1 insulin-like growth factor receptor in mice protects against oxygen-induced lung injury. Respir Res, 2005. 6: p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pineiro-Hermida S, et al. , IGF1R deficiency attenuates acute inflammatory response in a bleomycin-induced lung injury mouse model. Sci Rep, 2017. 7(1): p. 4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi JE, et al. , Insulin-like growth factor-I receptor blockade improves outcome in mouse model of lung injury. Am J Respir Crit Care Med, 2009. 179(3): p. 212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hung CF, et al. , Role of IGF-1 pathway in lung fibroblast activation. Respir Res, 2013. 14: p. 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim TH, et al. , Effect of insulin-like growth factor blockade on hyperoxia-induced lung injury. Am J Respir Cell Mol Biol, 2012. 47(3): p. 372–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kotarkonda LK, Kulshrestha R, and Ravi K, Role of insulin like growth factor axis in the bleomycin induced lung injury in rats. Experimental and molecular pathology, 2017. 102(1): p. 86–96. [DOI] [PubMed] [Google Scholar]
- 73.Frankel SK, et al. , Human insulin-like growth factor-IA expression in transgenic mice promotes adenomatous hyperplasia but not pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol, 2004. 288. [DOI] [PubMed] [Google Scholar]
- 74.Léger C, et al. , Adenovirus-mediated gene transfer of hIGF-IB in mouse lungs induced prolonged inflammation but no fibroproliferation. Am J Physiol Lung Cell Mol Physiol, 2010. 298(4): p. L492–L500. [DOI] [PubMed] [Google Scholar]
- 75.Andonegui G, et al. , Sequential expression of IGF-1B followed by active TGF-b1 induces synergistic pulmonary fibroproliferation in vivo. Am J Physiol Lung Cell Mol Physiol, 2012. 303: p. L788–L798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Della Latta V, et al. , Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacological research, 2015. 97: p. 122–30. [DOI] [PubMed] [Google Scholar]
- 77.Hewlett JC, Kropski JA, and Blackwell TS, Idiopathic pulmonary fibrosis: Epithelial-mesenchymal interactions and emerging therapeutic targets. Matrix biology : journal of the International Society for Matrix Biology, 2018. 71–72: p. 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reiser KM and Last JA, Early cellular events in pulmonary fibrosis. Exp Lung Res, 1986. 10(4): p. 331–55. [DOI] [PubMed] [Google Scholar]
- 79.Stiles AD and Moats-Staats BM, Production and Action of Insulin-like Growth Factor I/ Somatomedin C in Primary Cultures of Fetal Lung Fibroblasts. Am J Respir. Cell Mol. Biol, 1989. 1: p. 21–26. [DOI] [PubMed] [Google Scholar]
- 80.Clemmons DR, Underwood LE, and Van Wyk JJ, Hormonal control of immunoreactive somatomedin production by cultured human fibroblasts. J Clin Invest, 1981. 67(1): p. 10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atkinson PR, et al. , Release of Somatonedin-Like Activity by Cultured WI-38 Human Fibroblasts. Endocrinology, 1980. 106(6). [DOI] [PubMed] [Google Scholar]
- 82.Antoniou KM, et al. , Expression analysis of angiogenic growth factors and biological axis CXCL12/CXCR4 axis in idiopathic pulmonary fibrosis. Connective tissue research, 2010. 51(1): p. 71–80. [DOI] [PubMed] [Google Scholar]
- 83.Pala L, et al. , Direct measurement of IGF-1 and IGFBP-3 in bronchoalveolar lavage fluid from idiopathic pulmonary fibrosis. J. Endocrinol. Invest, 2001. 24: p. 856–864. [DOI] [PubMed] [Google Scholar]
- 84.Laron Z, IGF-1 and insulin as growth hormones. Novartis Found Symp., 2004. 262: p. 56–77. [PubMed] [Google Scholar]
- 85.Bloor CA, et al. , Differential mRNA Expression of Insulin-like Growth Factor-1 Splice Variants in Patients With Idiopathic Pulmonary Fibrosis and Pulmonary Sarcoidosis. Am J Respir Crit Care Med, 2001. 164: p. 265–272. [DOI] [PubMed] [Google Scholar]
- 86.Allen JT, et al. , Expression of growth hormone-releasing factor, growth hormone, insulin-like growth factor-1 and its binding proteins in human lung. Neuropeptides, 2000. 34(2): p. 98–107. [DOI] [PubMed] [Google Scholar]
- 87.Kasprzak A, et al. , Differential expression of IGF-1 mRNA isoforms in colorectal carcinoma and normal colon tissue. International journal of oncology, 2013. 42(1): p. 305–16. [DOI] [PubMed] [Google Scholar]
- 88.Koczorowska MM, Kwasniewska A, and Gozdzicka-Jozefiak A, IGF1 mRNA isoform expression in the cervix of HPV-positive women with pre-cancerous and cancer lesions. Experimental and therapeutic medicine, 2011. 2(1): p. 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Dwyer DN, et al. , The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci Rep, 2017. 7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meng XM, et al. , Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol, 2010. 21(9): p. 1477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jonk LJC, et al. , Identification and Functional Characterization of a Smad Binding Element (SBE) in the JunB Promoter That Acts as a Transforming Growth Factor-β, Activin, and Bone Morphogenetic Protein-inducible Enhancer. Journal of Biological Chemistry, 1998. 273(33): p. 21145–21152. [DOI] [PubMed] [Google Scholar]
- 92.Morikawa M, et al. , Genome-wide mechanisms of Smad binding. Oncogene, 2013. 32(13): p. 1609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brisson BK and Barton ER, New Modulators for IGF-I Activity within IGF-I Processing Products. Front Endocrinol (Lausanne), 2013. 4: p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vassilakos G, et al. , Biological activity of the e domain of the IGF-1Ec as addressed by synthetic peptides. Hormones Review, 2014. 13(2): p. 182–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.