Abstract
The ubiquitin (Ub) proteolysis pathway uses an E1, E2, and E3 enzyme cascade to label substrate proteins with ubiquitin and target them for degradation. The mechanisms of ubiquitin chain formation remain unclear and include a sequential addition model, in which polyubiquitin chains are built unit by unit on the substrate, or a preassembly model, in which polyubiquitin chains are preformed on the E2 or E3 enzyme and then transferred in one step to the substrate. The E2 conjugating enzyme UBE2K has a 150-residue catalytic core domain and a C-terminal ubiquitin-associated (UBA) domain. Polyubiquitin chains anchored to the catalytic cysteine and free in solution are formed by UBE2K supporting a preassembly model. To study how UBE2K might assemble polyubiquitin chains, we synthesized UBE2K-Ub and UBE2K-Ub2 covalent complexes and analyzed E2 interactions with the covalently attached Ub and Ub2 moieties using NMR spectroscopy. The UBE2K-Ub complex exists in multiple conformations, including the catalytically competent closed state independent of the UBA domain. In contrast, the UBE2K-Ub2 complex takes on a more extended conformation directed by interactions between the classic I44 hydrophobic face of the distal Ub and the conserved MGF hydrophobic patch of the UBA domain. Our results indicate there are distinct differences between the UBE2K-Ub and UBE2K-Ub2 complexes and show how the UBA domain can alter the position of a polyubiquitin chain attached to the UBE2K active site. These observations provide structural insights into the unique Ub chain-building capacity for UBE2K.
Significance
The E2 conjugating enzyme UBE2K has catalytic core and C-terminal ubiquitin-associated (UBA) domains and can synthesize polyubiquitin chains anchored to its catalytic cysteine. To understand how the UBA domain directs polyubiquitin chain assembly, we synthesized UBE2K-Ub and UBE2K-Ub2 covalent complexes to mimic a growing ubiquitin chain and analyzed interactions between the E2 enzyme and its attached ubiquitin moieties. NMR spectroscopy showed the directly attached ubiquitin in UBE2K-Ub exists in multiple positions, including the closed state needed for catalysis. Lengthening the ubiquitin chain leads to a more extended conformation facilitated by the UBA domain. The data provide a structural rationale for previous kinetic results and show how the UBA domain can alter the position of a growing ubiquitin chain.
Introduction
Cellular proteins exist in a dynamic state in which both protein synthesis and protein degradation are regulated (1,2). A major mechanism of protein degradation is the ubiquitin proteolysis pathway, which is responsible for the removal of short-lived, damaged, and misfolded proteins within the cell (3). This pathway consists of three key enzymes, the ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligating enzyme (E3), that respectively activate, transfer, and ligate ubiquitin to a substrate protein. The ubiquitin protein is initially activated with ATP to create a highly reactive covalent thioester bond with the E1 enzyme (E1∼Ub). The ubiquitin is then transferred from the E1 active site cysteine to an E2 active site cysteine through a transthiolation reaction to create an E2∼Ub thioester. After this, the pathway uses one of two major types of E3 enzymes: a really interesting new gene E3 aids in the transfer of ubiquitin directly from the E2 enzyme onto the substrate (4) or a homologous to E6AP carboxyl-terminus E3 that first transfers ubiquitin onto the E3 enzyme and then to a substrate (5). In either case, the E3 aids in the transfer of ubiquitin from the E2∼Ub thioester to the ε-amino group of a lysine residue in a substrate. The repetition of this process leads to the assembly of a polyubiquitin chain on a substrate linked through the C-terminal glycine (G76) of one ubiquitin and one of the seven lysine residues on its neighbor. Generally speaking, the linkage type dictates the fate of the ubiquitinated substrate. For example, proteins labeled with K48-linked polyubiquitin chains, composed of four or more ubiquitin molecules, are recognized and degraded by the 26S proteasome (3,6).
Some of the most complicated and least understood facets of ubiquitination are the mechanisms for polyubiquitin chain formation. Several mechanisms have been proposed (7, 8, 9) to account for the positioning of ubiquitin by either an E2 or an E3 that results in the various ubiquitin linkages. The simplest mechanism proposed for polyubiquitin chain formation is the sequential addition model, in which a monoubiquitinated substrate is extended as ubiquitin molecules are connected to each other one at a time. Alternatively, the E2 enzymes Ubc1 (10), UBE2K (E2-25K, HIP2) (11), UBE2R1 (CDC34) (12,13), UBE2G2 (Ubc7) (14), and UBE2N/UBE2V2 (Ubc13/Mms2) (15) have the ability to build polyubiquitin chains in the absence of an E3 enzyme or substrate. In addition, several studies show that some of these E2 enzymes support a mechanism whereby a polyubiquitin chain can be assembled at the catalytic cysteine site, suggesting an entire polyubiquitin chain might be transferred in one step. For example, UBE2K can form thioester complexes with both Ub and Ub2 with near-identical kinetics (11). Further, in vitro ubiquitination assays show that polyubiquitin chains of different lengths can be liberated from UBE2K after thiol reduction, indicating the presence of a polyubiquitin chain linked to the catalytic cysteine (16,17). Similarly, K48-linked polyubiquitin chains are formed and transferred intramolecularly to a nearby lysine in the Saccharomyces cerevisiae E2 enzyme Ubc1 (10). The E2 enzymes AtUBC4 (18) and UBE2G2 (14) can also form thioester complexes with polyubiquitin chains that can subsequently be transferred to a substrate. These types of en bloc polyubiquitin transfer may be specific to enzymes such as Ubc1 and UBE2K that have been shown to be important for ubiquitin chain elongation rather than initiation of a polyubiquitin chain on a substrate (19,20). Interestingly, Ubc1 and UBE2K are class II E2 enzymes that comprise a canonical α/β catalytic domain fold (∼150 residues) but also contain a three-helix ubiquitin-associated (UBA) domain (21). Multiple studies have shown that acceptance of ubiquitin from an E1 enzyme and formation of an E2∼Ub conjugate is not dependent on the presence of the UBA domain (10,11,19,22, 23, 24). In contrast, polyubiquitin chain formation requires both catalytic and C-terminal UBA domains. The UBA domain has been shown to associate weakly with free ubiquitin (21,23,25) but does not appear to have any interaction with the thioester-linked ubiquitin molecule in the E2∼Ub conjugate (26).
The E2 enzyme UBE2K produces unanchored K48-linked polyubiquitin chains in vitro using only an E1 activating enzyme and ubiquitin molecules as substrates (17,23). The biological significance of unanchored chain building by UBE2K remains unknown, but free unconjugated K48-linked polyubiquitin chains are found in vivo (18). In this work, we examined whether the UBA domain in UBE2K has a role in directing ubiquitination through interaction with a preassembled polyubiquitin chain formed at the catalytic cysteine of the E2 enzyme. We used NMR spectroscopy to examine interactions between UBE2K and a series of possible intermediates, including an E2 Ub conjugate (UBE2K∼Ub) and the diubiquitin species linked to the catalytic cysteine of the E2 enzyme (UBE2K∼Ub2). These interactions were used to provide insights into the preassembly of polyubiquitin chains before transfer to a substrate protein.
Materials and Methods
Plasmid construction
Wild-type UBE2K cDNA in a pET28a-LIC vector was a generous gift of the Structural Genomics Consortium (Toronto, Canada). The QuikChange Site Directed Mutagenesis (Stratagene, San Diego, CA) protocol was used to make C170S (UBE2KC170S) and K97R (UBE2KK97R) substitutions in UBE2K and to convert the protease cleavage site between the N-terminal His6-tag and UBE2K from thrombin to tobacco etch virus. Protein substitutions and cleavage site changes were confirmed by DNA sequencing. The expression plasmids for ubiquitin carrying K48R (UbK48R) or G76C (UbG76C) substitutions were constructed as previously described (10,26).
Protein expression and purification
Ubiquitin variants were expressed as either unlabeled or 15N, 13C-labeled proteins and purified as described previously (26). Uba1 was purified as described previously (27). His6-UBE2KC170S (henceforth His6-UBE2K) was overexpressed in Escherichia coli BL21 CodonPlus(DE3)RIL cells grown at 37°C to an OD600 of 0.6. Uniform 15N or 15N, 13C-labeled UBE2K was expressed using M9 media containing either 1.0 g/L 15NH4Cl or 2.0 g/L 13C-glucose and 1.0 g/L 15NH4Cl. Protein expression was induced with 0.7 mM isopropyl β-D-1-thiogalactopyranoside, and the temperature was lowered to 15°C for 16–20 h. Cells were harvested and homogenized with an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada), and the soluble lysate containing His6-UBE2K was applied to an Ni-NTA column (Qiagen, Toronto, Canada). Fractions containing His6-UBE2K were eluted from the column with 25 mM Tris-HCl, 200 mM NaCl, 250 mM imidazole, and 1 mM TCEP (pH 7.5). The His6-tag was cleaved using tobacco etch virus (1 mg) for 1 h at 22°C, followed by dialysis at 4°C against 25 mM Tris-HCl, 200 mM NaCl, 20 mM imidazole, 1 mM TCEP (pH 7.5). UBE2K was collected as the flow through fractions from application to an Ni-NTA column and further purified by size exclusion chromatography using a Superdex 75 10/300 column (GE Life Sciences, Chicago, IL) eluted with 25 mM Tris-HCl, 200 mM NaCl, 1 mM TCEP, and 1 mM EDTA (pH 7.5). All steps were monitored by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Purified UBE2K was confirmed using mass spectrometry (UBE2K: MWobs 22,534.8 ± 0.3 Da, MWcalc 22,534.7; 15N,13C-labeled UBE2K: MWobs 23,778.9 ± 1.4 Da, MWcalc 23,812.8 Da).
Diubiquitin synthesis
K48-linked diubiquitin chains were constructed using purified UbK48R and UbG76C proteins. In this synthesis, UbG76C can only act as an acceptor molecule, whereas UbK48R acts as the donor molecule. The formation of diubiquitin (UbG76C-UbK48R, Ub2) was catalyzed using the Uba1 activating enzyme UBE2K (28,29) and different combinations of 15N, 13C-labeled and unlabeled ubiquitin proteins to produce species in which only one of the ubiquitin molecules was isotopically labeled.
UBE2K-Ub and UBE2K-Ub2 disulfide complex formation
All covalent complexes were formed using a disulfide linkage between the catalytic cysteine (C92) in UBE2K and the C-terminal cysteine in the UbG76C protein (26). Stock solutions (0.1 mM) of unlabeled or 15N,13C-labeled Ub, Ub2, and UBE2K were freshly reduced using 5 mM TCEP. UBE2K was combined with a twofold excess of UbG76C or UbG76C-UbK48R and dialyzed against several changes of 100 mM Na2HPO4/NaH2PO4, 200 mM NaCl, 10 μM CuCl2 at pH 7.4 and 4°C. The progress of the disulfide complex formation was monitored by nonreducing SDS-PAGE and was considered complete when the reduced UBE2K was exhausted. The protein solution was concentrated and purified by size exclusion chromatography on a Sephadex G-75 column (GE Healthcare, Chicago, IL) with 100 mM Na2HPO4/NaH2PO4, 200 mM NaCl, 3 mM EDTA (pH 7.4). Fractions containing pure UBE2K-UbG76C or UBE2K-UbG76C-UbK48R were pooled and extensively dialyzed against 100 mM Na2HPO4/NaH2PO4, 400 mM NaCl, and 3 mM EDTA buffer (pH 7.4) for NMR experiments. Complex formation was confirmed by mass spectrometry for 15N-labeled UBE2K-UbG76C (MWobs 31,402.9 ± 2.3, MWcalc 31,402.6), UBE2K-UbG76C-(15N,13C) UbK48R (MWobs 40,165.56 ± 4.6, MWcalc 40,184.4), UBE2K-(15N,13C) UbG76C (MWobs 31,602.5 ± 0.8, MWcalc 31,616.6), and UBE2K-(15N,13C) UbG76C-UbK48R (MWobs 40,169.1 ± 2.3, MWcalc 40,183.4).
Ubiquitination assays
Reactions contained 0.2 μM Uba1, 20 μM UBE2K or UBE2KK97R, 80 μM Ub, 20 μM fluorescently labeled ubiquitin (Ub800), 5 mM MgATP, and 50 mM HEPES (pH 7.4) as previously described (30). Ub800 was prepared using an N-terminal cysteine containing ubiquitin protein that was specifically labeled with DyLight800 maleimide (Thermo Fisher Scientific, Waltham, MA). Reactions were quenched after 30 min with 3× SDS sample buffer containing 50 mM dithiothreitol or 0.001% trifluoroacetic acid for reducing or nonreducing conditions, respectively. Protein species were resolved on 15% Bis-Tris (pH 6.4) separating gels using MES running buffer (250 mM MES, 250 mM Tris, 0.5% SDS, 5 mM EDTA (pH 7.4)). Gels were imaged using an Odyssey Imaging system (Li-COR Biosciences, Lincoln, NE) detecting fluorescence at 700 and 800 nm.
NMR spectroscopy
All NMR data were acquired using a Varian INOVA 600 MHz spectrometer (Biomolecular NMR Facility, University of Western Ontario, London, Canada) equipped with a triple resonance cold probe with z gradients. Backbone residue assignments for 15N,13C-labeled UBE2K were completed as previously reported (31). The proteins UbG76C, UbK48R, and UbG76C-UbK48R, as well as the UBE2K-UbG76C and UBE2K-UbG76C-UbK48R disulfide complexes, were dialyzed against 100 mM Na2HPO4/NaH2PO4, 400 mM NaCl, 3 mM EDTA (pH 7.4) in 90% H2O/10% D2O and characterized by 1H-15N HSQC (32), 1H-15N TROSY (33), and HNCA (34) experiments at 30 or 35°C to complete sequential assignments for the backbone residues. Chemical shift perturbation measurements were calculated according to ∑Δδ = [Δδ(1H)2 + ((0.2)Δδ(15N))2]0.5, where Δδ(1H) and Δδ(15N) are the chemical shift differences for each protein component in the complexed and uncomplexed forms (35). All data were processed using NMRPipe and NMRDraw (36) and analyzed with NMRView (37).
Sedimentation equilibrium
Sedimentation equilibrium experiments for UBE2K-Ub2 were performed using a Beckman Optima XL-A analytical ultracentrifuge (Beckman Coulter, Indianapolis, IN) with absorbance optics as previously described (30). Samples were dialyzed into 100 mM Na2HPO4/NaH2PO4, 400 mM NaCl, and 3 mM EDTA (pH 7.33) for 24 h. Experiments were completed in triplicate using 10.9, 8.7, or 5.7 μM protein with matching reference samples containing dialysis buffer.
Results
Evidence for polyubiquitin chain formation at the catalytic cysteine of UBE2K
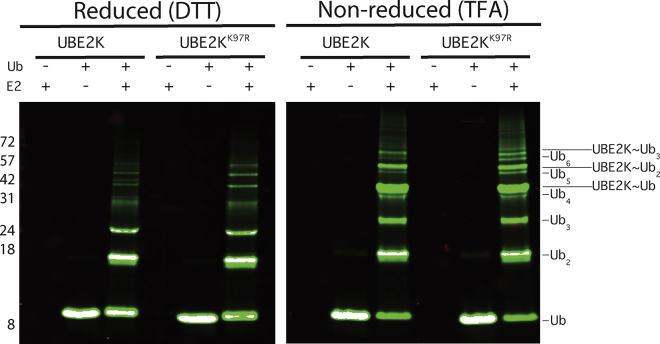
The E2-conjugating enzyme UBE2K (E2-25K) produces unanchored K48-linked polyubiquitin chains in solution using the E1 activating enzyme (Uba1) and free ubiquitin (Ub) proteins (11,17,23). To determine whether UBE2K is able to build polyubiquitin chains on its catalytic cysteine, a series of ubiquitination assays were assessed under reducing and thioester-preserving conditions and monitored using fluorescently labeled ubiquitin (Fig. 1). In these assays, we also utilized a UBE2K protein carrying a substitution at K97 (UBE2KK97R) to distinguish polyubiquitin chains at the catalytic cysteine from those that might have been transferred to K97. It has been noted that transfer of the thioester-linked ubiquitin to this lysine occurs readily in some E2 enzymes, including Ubc1, a relative of UBE2K (10). Under reducing conditions, wild-type UBE2K shows major bands consistent with Ub2, Ub3, and Ubn polyubiquitin chains that are also evident with UBE2KK97R. This indicates the formation of either free polyubiquitin chains or the cleavage of these chains from the E2 enzyme through reduction of the thioester linkage. Above 35 kDa, some minor bands are visible for both UBE2K and UBE2KK97R. Under nonreducing conditions, the intensities of many bands for both UBE2K and UBE2KK97R are significantly different. In particular, a strong band corresponding to the E2∼Ub conjugate is present in both cases. For UBE2K, multiple bands occur above the E2∼Ub conjugate species that are far more intense than similar minor bands under reducing conditions, consistent with the presence of UBE2K∼Ub2, UBE2K∼Ub3, and UBE2K∼Ubn species. These bands are also evident for UBE2KK97R, in which the polyubiquitin chain cannot be transferred to the nearby lysine. Consistent with previous work (16,17), these experiments support the formation of a polyubiquitin chain linked to the catalytic cysteine in UBE2K.
Figure 1.
The catalytic cysteine of UBE2K is an in situ polyubiquitin chain building site. The figure shows UBE2K ubiquitination assays that were conducted for 30 min using Uba1, fluorescently labeled Ub800, and either wild-type UBE2K or UBE2KK97R at 37°C as described in the Materials and Methods. Reactions were visualized by SDS-PAGE, which shows fluorescent ubiquitination products under reducing (left) and nonreducing (right) conditions. Products are identified as isolated polyubiquitin species (Ubn) or polyubiquitin chains linked to the catalytic cysteine in UBE2K (UBE2K∼Ubn) at the right of the gels.
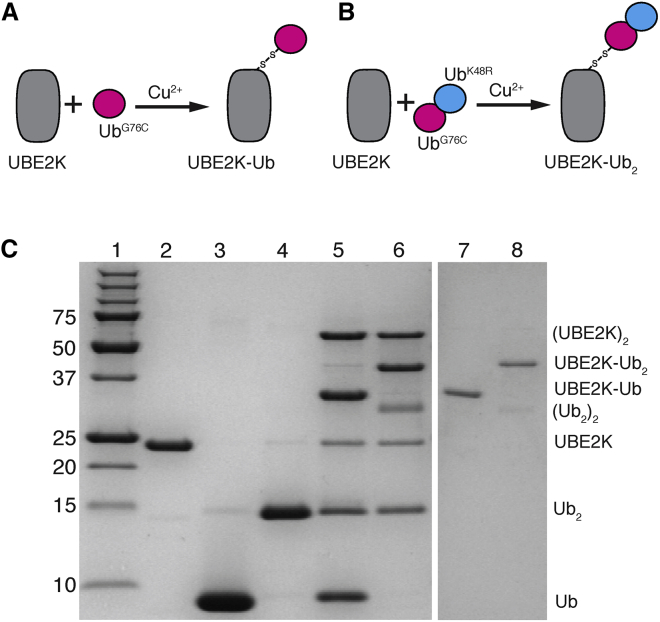
As shown in Fig. 1, thioester-linked ubiquitin or polyubiquitin chains at the catalytic cysteine site are easily reduced or hydrolyzed. To examine the mechanisms of assembly of polyubiquitin chains on the active site cysteine and the nature of a preassembled chain linked to the active site, UBE2K complexes were synthesized carrying either one or two ubiquitin molecules (Fig. 2). To mimic the natural thioester species and limit possible hydrolysis, a disulfide complex was formed between the catalytic cysteine (C92) in UBE2K and the C-terminus of ubiquitin carrying a cysteine substitution (UbG76C). Synthesis of a diubiquitin species (Ub2, UbG76C-UbK48R) was completed using Uba1 and UBE2K enzymes, whereby substitutions at G76 (UbG76C) and K48 (UbK48R) proteins ensured that only one possible K48-linked arrangement of the Ub2 species could be formed. UbG76C or UbG76C-UbK48R proteins were linked to the catalytic cysteine of UBE2K using mild oxidation (Fig. 2). This approach allowed preparative amounts of UBE2K-Ub and UBE2K-Ub2 complexes to be purified with separate isotopic 15N and/or 13C labeling of the individual UBE2K, UbG76C, or UbK48R components for NMR characterization.
Figure 2.
Diagram showing the synthesis of disulfide-linked UBE2K-Ub and UBE2K-Ub2 complexes. Schematic representations showing the formation of the (A) UBE2K-Ub disulfide complex or (B) UBE2K-Ub2 disulfide complex using the E2 ubiquitin-conjugating enzyme UBE2K (gray) and UbG76C-UbK48R (magenta and cyan) components are given. Disulfide formation was accomplished using mild Cu2+ oxidation to link the C-terminal cysteine of UbG76C to the catalytic site (C92) of UBE2K to yield either UBE2K-UbG76C or UBE2K-UbG76C-UbK48R. (C) An SDS-PAGE gel showing the synthesis and purification of UBE2K-UbG76C or UBE2K-UbG76C-UbK48R is given. The gel shows molecular weight standards (lane 1), followed by purified UBE2K (lane 2), UbG76C (lane 3), and UbG76C-UbK48R (lane 4) proteins. The synthesis of UBE2K-UbG76C (lane 5) and UBE2K-UbG76C-UbK48R (lane 6) using Cu2+ oxidation is shown. Side products of these reactions include Ub2 and UBE2K2 as indicated. Size exclusion chromatography was used to obtain purified UBE2K-UbG76C (lane 7) and UBE2K-UbG76C-UbK48R (lane 8), as described in the Materials and Methods.
The UBE2K-Ub complex displays open and closed states
Recent crystallographic data show that a covalent UBE2K-Ub complex exists in an open state in which the conjugated Ub molecule is not associated with the UBE2K enzyme other than the covalent linkage site (22). Rather, the covalently bound ubiquitin is associated with the UBA domain of an adjacent UBE2K molecule in the crystal lattice that confers the open position. In solution, multiple studies have shown that in the absence of an E3 enzyme, the covalently attached Ub molecule occupies a range of conformations in an E2∼Ub conjugate but with a preference for closed or open states. One example of this is UBE2L3 (UbcH7), for which NMR experiments show a preference for the closed state (38,39), in which the conjugated Ub interacts with a surface on the E2 enzyme in its free state that converts to the open state in the presence of an E3 enzyme or binding partner. Other E2 enzymes such as UBE2R1 (CDC34) and Ubc1, the S. cerevisiae homolog for UBE2K, also appear to prefer a closed E2∼Ub arrangement (40, 41, 42). The choice of open or closed state is required to allow facile access of a lysine residue on a substrate or another ubiquitin molecule to the thioester linkage in the E2∼Ub conjugate.
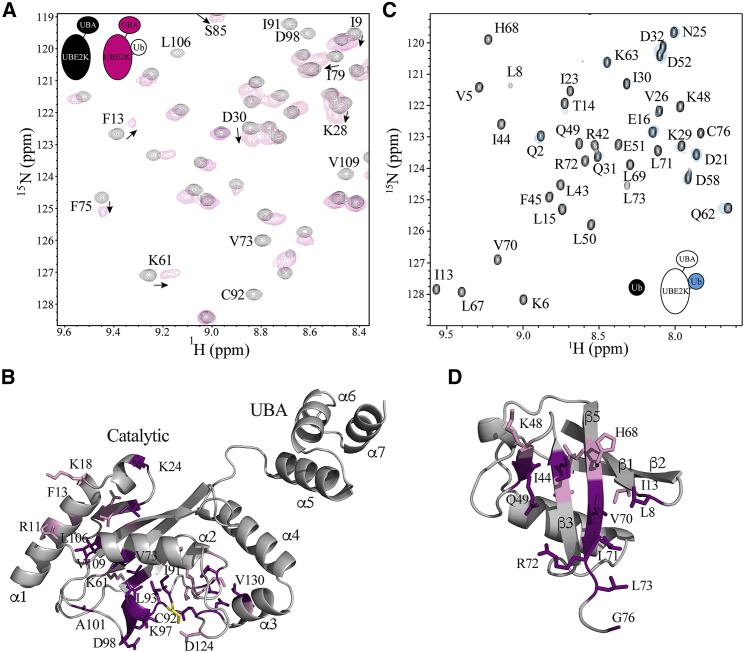
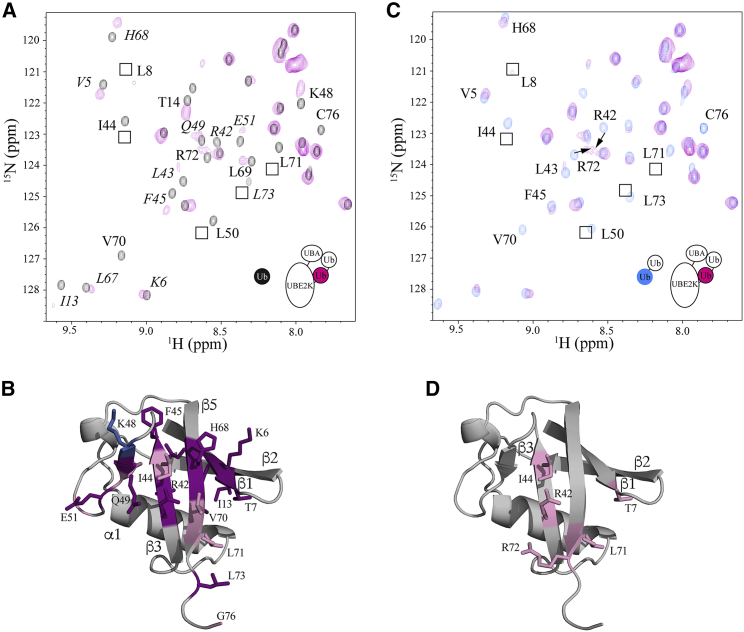
Because UBE2K has the ability to build unaided polyubiquitin chains, we examined whether the UBE2K∼Ub conjugate occupies open or closed states by monitoring NMR spectra of the E2 and Ub components of stable UBE2K-UbG76C conjugates in the absence of an E3 enzyme. To identify the regions in UBE2K most affected by Ub attachment, the positions of each residue in 15N-labeled UBE2K were tracked by comparing 1H-15N TROSY spectra of UBE2K-UbG76C with UBE2K (Fig. 3 A). Overall, two general trends were observed. Firstly, despite the addition of a covalently attached Ub, most visible resonances in UBE2K-UbG76C underwent only minor chemical shift changes compared to the isolated E2 enzyme (Fig. 3 A). Not surprisingly, residues in the loops just before (F75, I79, I84-S86, T88, G89, I91) and across from (D98, L106, V109, D124) the catalytic site had the largest changes. Other changes were noted for residues in α1 (R11, F13, K18), β1 (K28, V29, D30), β2 (G41), β3 (K61), and α3 (V129). However, we observed no significant chemical shift changes beyond K135 in the sequence including the entire UBA domain (α5–α7) in which all signals were visible in UBE2K-UbG76C (Fig. 3 B). Although free ubiquitin has been shown to weakly associate with the UBA domains of both Ubc1 and UBE2K in solution (21,23,25,43), the results here indicate an interaction between the UBA domain and the covalently bound Ub in the UBE2K-Ub conjugate does not occur. Our observations are in agreement with previous studies on Ubc1, the UBE2K yeast homolog (26). Secondly, conjugation of Ub to UBE2K results in a large number of signals that experience significant line broadening (Fig. 3 A). In particular, many residues that straddle the C92 active site where Ub is covalently linked are affected, including those surrounding the active site helix (I91-D98) and across the catalytic cleft (E121, D124, Q126, D127) (Fig. 3 B). We also noted that many residues before (M104-L106) and within helix α2 (V109, L110) are either significantly broadened or absent from the spectrum of UBE2K-UbG76C altogether.
Figure 3.
Identification of the conformation of the UBE2K-Ub complex. (A) Expanded regions for 1H-15N TROSY spectra of UBE2K (black) compared to UBE2K-UbG76C (magenta) are shown. Signals that either shift or are broadened are indicated. (B) A cartoon representation of UBE2K is given, showing signals from (A) that underwent significant chemical shift changes (>average + 1.5 standard deviation (SD)) (pink) or experienced significant line broadening (purple). (C) The expanded region of 1H-15N HSQC of UbG76C (black) and as a component in UBE2K-UbG76C (blue) is shown. Broadened signals are identified based on the chemical shift assignments of UbG76C. (D) A cartoon representation of Ub is given, showing signals from (C) that underwent significant chemical shift changes (>average + 1 SD) (pink) or experienced significant line broadening (purple). The structure of UBE2K (PDB: 1YLA) in this figure is in agreement with small angle X-ray scattering data as described in (30).
A reciprocal experiment was completed using 15N,13C-labeled UbG76C to examine the Ub positioning and interaction within the UBE2K-UbG76C complex (Fig. 3 C). Upon formation of the complex, a few residues in UbG76C exhibited minor chemical shift changes accompanied by changes in their linewidths due to the increased size of the UBE2K-UbG76C complex. However, like the UBE2K component of the conjugate, the most prominent observation for UbG76C was the significant line broadening of many resonances such that their position could not be identified in 1H-15N HSQC spectra (Fig. 3 C). This included residues near the β1-β2 loop (L8, I13), β3 (L43-F45), β4 (K48-L50), and β5 (L67-R72) that contribute to the classic I44 hydrophobic patch (Fig. 3 D) recognized by most ubiquitin-binding domains (44,45).
When mapped to the structures of UBE2K and UbG76C, the majority of the residues affected upon UBE2K-Ub formation result from line broadening in NMR spectra. Typically, this type of line broadening occurs when multiple conformations are present that exchange at a rate(s) comparable to the differences in chemical shifts for the individual conformations. Further, the positions of the observed broadened resonances are consistent with an interface between the surfaces of the proteins in the E2-Ub complex (Fig. 3, B and D). Thus, the broadening likely results from conformational exchange between the association and dissociation of UbG76C with residues that surround the catalytic core of UBE2K. This observation is consistent with exchange between the “closed” and “open” states observed for an E2∼Ub conjugate in which Ub associates with the region near helix α2 in the “closed” state or occupies a flexible solvent exposed conformation in the “open” state (46). This conclusion is also supported by the position of Q49 in NMR spectra of UbG76C in the UBE2K-UbG76C conjugate. The position of this signal is a good diagnostic for the open state, in which a limited shift occurs upon conjugation (39), and the closed state, in which a significant change is observed. In our spectra for UBE2K-UbG76C, the signal for Q49 was broadened beyond detection (Fig. 3 C), suggesting exchange between two or more conformations of the E2-Ub conjugate.
Ubiquitin chain extension causes opening of the UBE2K-Ub linkage
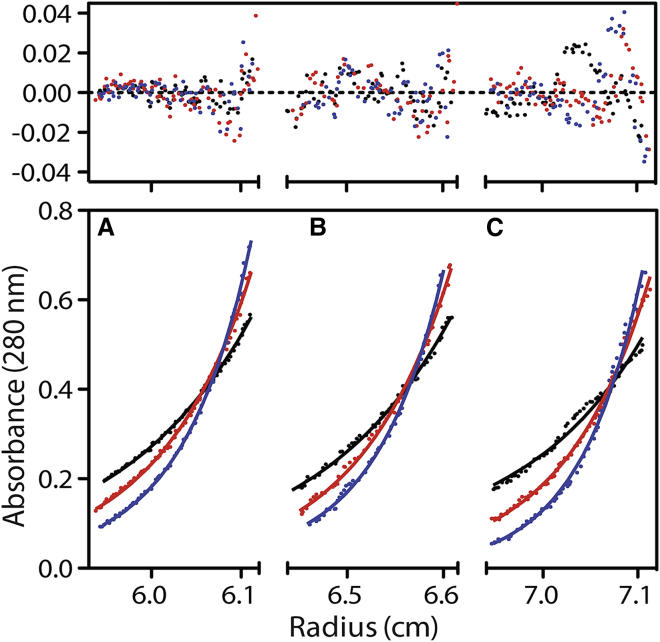
En bloc transfer requires that specifically linked ubiquitin chains be assembled at the catalytic site of an E2 enzyme before transfer to another E2 or E3 enzyme or a substrate. To identify how the position and conformation of the conjugated ubiquitin (UbG76C) might be altered by ubiquitin chain extension, we synthesized several UBE2K-Ub2 complexes in which the UBE2K, proximal Ub (UbG76C), or distal Ub (UbK48R) were isotopically labeled for NMR studies. Sedimentation equilibrium analysis of UBE2K-Ub2 (UBE2K-UbG76C-UbK48R) at multiple concentrations and speeds yielded an average mass of 42,743 ± 427 Da, close to the expected mass of 40,184.37 Da, indicating UBE2K-Ub2 behaves as a homogeneous monomeric species in solution (Fig. 4). Similar behavior has been noted for UBE2K and UBE2K-Ub species (22,30). Our initial focus compared NMR spectra of the proximal 15N-labeled UbG76C in UBE2K-Ub2 to identify how addition of the distal Ub might alter interactions with UBE2K. Fig. 5 shows that conjugation of UbG76C to UBE2K and addition of the distal Ub at K48 leads to surprisingly well-resolved spectra and signal linewidths for a 40 kDa molecule. Despite the increased molecular weight, the 1H-15N HSQC spectrum of 15N-labeled UbG76C within UBE2K-Ub2 (Fig. 5) showed significant improvement over the UbG76C moiety in the UBE2K-UbG76C conjugate (Fig. 3). As expected, chemical shift changes were noted for K48 because of its linkage with G76 in the UbK48R distal protein. However, the most significant observations in the NMR spectrum of UBE2K-Ub2 were the reappearance of many resonances for the proximal UbG76C that were absent in UBE2K-UbG76C because of conformational exchange between open and closed forms of the complex (Fig. 5 A). This included residues in β1 (V5, K6, T7), β2 (I13), β3 (R42, L43, F45), β5 (L67, H68, L69), the C-terminus (L73), and the loop connecting β4 and α2 (Q49, E51). Some of these signals also show minor chemical shift changes from UbG76C alone (Fig. 5 A). In particular, Q49 from UbG76C, which is indicative for open or closed states, is now visible in UBE2K-Ub2 with a chemical shift that is similar to that for unconjugated Ub. These observations indicate that formation of UBE2K-Ub2 through addition of the distal Ub to UBE2K-Ub alters the exchange rate and populations between E2-Ub conformers and promotes a more open state for the UBE2K-Ub2 complex.
Figure 4.
UBE2K-Ub2 is monomeric in solution. The lower panels show triplicate sedimentation equilibrium data sets (A–C) for 10.9 μM UBE2K-Ub2 collected using rotor speeds of 15,000 (solid black circles), 18,000 (solid red circles), and 21,000 (solid blue circles) rotations per minute at 5°C. Fitting of these data yielded a mass of 42,674 ± 564. The upper panel shows the residual errors between the best fit curve and data for each data set. All other details are described in the Materials and Methods.
Figure 5.
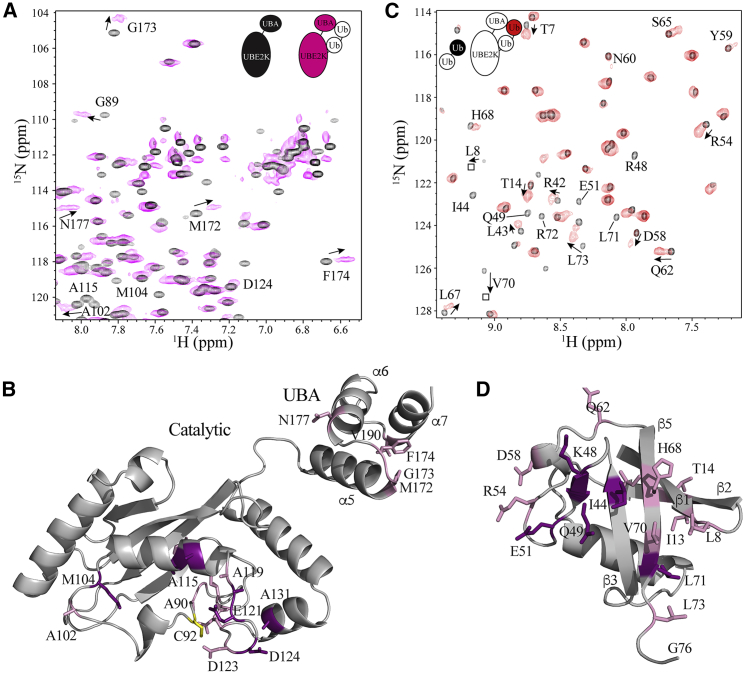
Addition of Ub alters the position of the proximal UbG76C upon chain elongation. (A) Expanded regions for 1H-15N HSQC spectra of UbG76C (black) and labeled UbG76C in the UBE2K-UbG76C-UbK48R conjugate (magenta) are shown. Selected signals that are visible at lower contour levels in the UBE2K-UbG76C-UbK48R conjugate are boxed. Signals that are absent in the UBE2K-UbG76C conjugate but are visible in UBE2K-UbG76C-UbK48R are shown in italics. (B) A cartoon structure is given, showing residues that undergo significant chemical shift changes (average + 1 SD) in UbG76C (pink) and residues that “reappear” in the 1H-15N HSQC spectrum of UBE2K-UbG76C-UbK48R that were absent because of conformational exchange in UBE2K-UbG76C (purple). The site of the K48 linkage in UbG76C with the distal UbK48R in UBE2K-UbG76C-UbK48R is indicated (blue). (C) Expanded regions of 1H-15N HSQC spectra of the proximal UbG76C in UbG76C-UbK48R (blue) and UBE2K-UbG76C-UbK48R (magenta) are shown. Signals with significant shifts (average + 1 SD) are indicated. Boxes indicate the position of a signal that is visible at lower contour levels. (D) A cartoon representation of Ub showing significantly shifted residues from (C) is given.
An extended diubiquitin arrangement exists within UBE2K-Ub2
To identify the conformation of the diubiquitin chain that is conjugated to the catalytic cysteine in UBE2K, we examined 1H-15N HSQC spectra of 15N-labeled UbG76C in UBE2K-Ub2 and in diubiquitin (UbG76C-UbK48R) (Fig. 5 C). First, we examined spectra of UbG76C-UbK48R because the conformational preference for diubiquitin in the absence of E2 conjugation has been extensively characterized. The spectrum of 15N-labeled UbG76C in UbG76C-UbK48R had excellent dispersion and signal intensity (Fig. S1), consistent with previous studies (47,48). The largest chemical shift differences between UbG76C-UbK48R and UbG76C were noted for residues in β1 (T7), β2 (G10, I13), β4 (G47), β5 (H68, V70, R72), and the unstructured C-terminus (L73) (Fig. S1 A). Similarly, comparison of 15N-labeled UbK48R in UbG76C-UbK48R with isolated UbK48R spectra showed chemical shift changes occurred in UbK48R for residues in β1 (T7), β2 (G10, I13), β4 (G47, R48, L50), β5 (H68, V70), the unstructured C-terminus (L73), and the loop between β4 and α2 (G53), as well as significant line broadening for G76, which is linked to K48 of the proximal ubiquitin (Fig. S1 B). These chemical shift changes are similar to those observed by other groups for K48-linked diubiquitin (47,49) and reflect an equilibrium between a “closed” Ub2 conformation, in which the hydrophobic I44 patch is sequestered from solvent (48), and an “open” Ub2 conformation, in which the I44 region is more accessible to solvent (47,50). Our data for UbG76C-UbK48R are consistent with previous studies that show the equilibrium is largely in favor of the open form of diubiquitin at pH 7.0 (47).
Comparison of the 1H-15N HSQC spectra for 15N-labeled UbG76C in UbG76C-UbK48R and UBE2K-UbG76C-UbK48R showed very little difference in the positions of most resonances (Fig. 5 C). In particular, the spectrum of UBE2K-UbG76C-UbK48R is characterized by limited chemical shift changes for residues in the β1 (V5-T9), β2 (K11-I13), and β5 (H68-R72) sheets from UbG76C that are hallmarks of the open state of K48-linked diubiquitin (Fig. 5 D). Although broader in linewidth, several signals including V5, R42, L50, H68, L71, and R72, in UBE2K-UbG76C-UbK48R shift closer to their positions in free UbG76C, showing that the diubiquitin chain linked to UBE2K adopts a more extended conformation. Together with Fig. 3, these data indicate that lengthening of the ubiquitin chain at the catalytic site in UBE2K leads to a more extended arrangement between the E2 and conjugated diubiquitin than in the simpler E2∼Ub conjugate.
The UBA domain in UBE2K recruits the distal ubiquitin in a UBE2K-Ub2 complex
To determine how addition of a distal ubiquitin alters the diubiquitin conformation and interactions with UBE2K, we examined 1H-15N TROSY spectra of 15N-labeled UBE2K in UBE2K-UbG76C and UBE2K-UbG76C-UbK48R (Fig. 6 A). Upon extending the ubiquitin chain, the spectrum of UBE2K in the UBE2K-Ub2 complex displayed broader signals compared to either UBE2K or UBE2K-UbG76C, consistent with its larger size (40 kDa). Uniquely, the spectrum for UBE2K-UbG76C-UbK48R displayed several obvious chemical shift changes in the catalytic domain of UBE2K, even though the added UbK48R is separated from the catalytic site by the directly linked proximal UbG76C. In particular, several chemical shift changes or line broadening were noted near the catalytic site between β4 and α2 (I76, N83, I84, G89, A90, A102, M104) and across the catalytic cleft and the loop between α2 and α3 (A115, A119, D124, Q126) (Fig. 6 A). These results suggest that a distinct region near the active site of UBE2K is altered because of the addition of the distal UbK48R, likely due to a readjustment of the position of the proximal UbG76C upon ubiquitin chain extension.
Figure 6.
The distal UbK48R in UBE2K-Ub2 interacts with the UBE2K UBA domain. (A) Expanded regions of 1H-15N TROSY spectra for 15N-labeled UBE2K alone (black) and in the UBE2K-UbG76C-UbK48R complex (magenta) are shown. Signals with significant shifts are labeled by residue and arrows. (B) A cartoon representation is given, showing residues whose signals underwent a significant chemical shift (>1 SD above the mean) upon addition of the distal UbK48R. (C) Expanded regions of 1H-15N HSQC spectra of 15N-labeled UbK48R in UbG76C-UbK48R (black) and UBE2K-UbG76C-UbK48R (red) are shown. Residues that shift in UbK48R upon formation of the conjugate are labeled. (D) A cartoon representation showing interacting residues on UbK48R is given. Signals that undergo significant chemical shift changes (>average + >1 SD) (pink) and residues whose signals experienced significant line broadening (purple) are indicated.
In contrast to the UBE2K-UbG76C species, the most significant chemical shift changes observed in the UBE2K-UbG76C-UbK48R complex occur in the UBA domain of UBE2K (Fig. 6, A and B). Specifically, several large changes were noted in the loop between helices α5 and α6 (M172, G173, F174), the N-terminus of α6 (N177), and the N-terminus of α7 (V190). This region of the UBA domain comprises the “MGF patch,” which is a well-established ubiquitin-interacting region in other UBA-containing proteins (44,45). Weak noncovalent interactions between ubiquitin and the UBA domains of UBE2K (23,25) and its yeast homolog Ubc1 (21,26) have been shown to occur in trans. However, this work shows that similar interactions between Ub and the UBA domain in an UBE2K-Ub conjugate do not occur (Fig. 3). Conversely, lengthening the ubiquitin chain anchored to the E2 enzyme allows the UBA domain to recruit the distal ubiquitin within a UBE2K-Ub2 complex.
The corresponding surface of interaction for the distal UbK48R within UBE2K-Ub2 was determined by comparing 1H-15N HSQC spectra of UbK48R within UbG76C-UbK48R and UBE2K-UbG76C-UbK48R. In this experiment, significant chemical shift changes and line broadening in UbK48R were observed for residues in β1 (T7), β2 (I13, T14), α2 (D58), β5 (L67, V70), the C-terminus (L73), and the loops between β1 and β2 (L8), β4 and α2 (R54), and α2 and β5 (Q62) (Fig. 6, C and D). Several of these residues showed signals that appeared to be in slow exchange. For example, residues in β2 (T14), β3 (L43), α2 (D58, Y59), β5 (S65), and the loops between β4 and α2 (R54, T55) and α2 and β5 (N60, Q62, K63) showed two broad signals where one was located near that of UbK48R within UbG76C-UbK48R and a second signal was shifted from this position. The most logical interpretation of this observation is that UbK48R occupies both free and bound states with the UBA domain of UBE2K. Significant line broadening in β3 (I44), β4 (R48, Q49, L50), β5 (L69, L71), and the loop between β4 and α2 (E51) was also observed (Fig. 6 C). When combining these results, the affected residues are mostly located on the classic hydrophobic face of UbK48R (Fig. 6 D). The observation that influenced residues are located on the hydrophobic MGF patch of the UBE2K UBA domain and the hydrophobic face of UbK48R within the UBE2K-UbG76C-UbK48R complex indicates these surfaces likely contact each other in the complex. This interaction would require exposure of the hydrophobic face of UbK48R within the UBE2K-Ub2 complex, which would only be possible when Ub2 adopts the “open” or extended conformation, as interpreted from data shown in Fig. 5.
Discussion
The UBE2K enzyme produces unanchored K48-linked polyubiquitin chains, and those chains can also be used to form a UBE2K∼Ub2 thioester with equal kinetics to the UBE2K∼Ub thioester (11). To identify the characteristics of a preassembled ubiquitin chain on the catalytic cysteine and how this might favor chain elongation, we investigated the structures and interactions of UBE2K-Ub and UBE2K-Ub2 complexes.
Upon formation of UBE2K-UbG76C, residues in both UBE2K and ubiquitin experienced chemical shift perturbations and significant broadening of resonances, especially near the active site C92 of UBE2K and the classic hydrophobic face of Ub. Peak broadening is characteristic of intermediate exchange between at least one bound and one unbound position of ubiquitin in association with UBE2K. Many of the residues that experience broadening are consistent with a closed conformation of the E2∼Ub conjugate in which the covalently attached Ub interacts with helix α2 of UBE2K (41,42). In the absence of an E3, the closed E2∼Ub structure is preferred by Ubc1 (42) and UBE2L3 (38,39). Alternatively, multiple orientations of E2∼Ub conjugates have been observed for a variety of E2 enzymes, including hUBE2N-Ub (51), UBE2L6-Ub (52), and UBE2D2-Ub (46,53). Rather than a strict divide between open and closed conformations, a given E2∼Ub conjugate likely exhibits a labile equilibrium in which preference for these conformations is dependent on the E2 enzyme (46) and likely the presence of a substrate and/or E3 ligase enzyme. For UBE2K, even though it is in equilibrium with other conformations, the closed state is necessary for catalysis to form a Ub2 species because substitutions near and along helix α2 inhibit polyubiquitin formation or ubiquitin unloading in the absence of an E3 enzyme (19,22,54). Our data support the idea that this closed state is populated a portion of the time in the UBE2K∼Ub conjugate but that other conformations are also occupied because of the line broadening observed in the NMR spectra. This is not dependent on the UBA domain because our data show this region does not contact the conjugated ubiquitin protein and so has little impact on its positioning. These observations are consistent with kinetic measurements that show the UBA domain does not alter the catalytic rate of Ub2 formation or K48 chain specificity (19,20). Therefore, it is likely that a signal external to the catalytic domain in UBE2K serves as a stimulus to influence the open or closed state equilibrium.
Our data and previous work (11,17) show that polyubiquitin chains can be conjugated to the catalytic cysteine of UBE2K in the absence of an E3 enzyme. Liberation of the chain could be achieved by transfer of the intact chain to a preubiquitinated substrate or free ubiquitin in the cell, with the latter accounting for the formation of free polyubiquitin chains observed for UBE2K and Ubc1 (10,11,18). Presumably, both of these ubiquitin transfer mechanisms would require the closed E2∼Ub conformation. Our data indicate that the arrangement between UBE2K and the proximal donor ubiquitin linked to C92 is altered upon UBE2K-Ub2 formation. Specifically, intermolecular interactions between the UBA domain of UBE2K and the distal Ub in UBE2K-Ub2 draw the ubiquitin chain toward a more open, extended conformation. This open state has been observed in a crystal structure of the UBE2K-Ub conjugate in which the C92-conjugated ubiquitin is held in an open state through interaction with the UBA domain of an adjacent UBE2K molecule in the crystal lattice (22). Current and previous work (22,30) shows that UBE2K, UBE2K-Ub, and UBE2K-Ub2 exist primarily as monomeric proteins, indicating that intermolecular contacts likely contribute minimally to an open state in solution. Nevertheless, the residues and interactions we observe between the distal Ub and the UBA domain in UBE2K-Ub2 and those in the crystal structure (Protein Data Bank, PDB: 5DFL) are very similar, which suggests the crystal structure faithfully recapitulates the intramolecular Ub-UBA interaction observed in UBE2K-Ub2. This extended positioning of a conjugated Ub2 is contradictory to the closed form of UBE2K∼Ub needed to facilitate ubiquitin transfer and suggests the UBE2K∼Ub2 species would have significantly different kinetic properties for this reaction. Indeed, Chen and Pickart (11) showed that UBE2K∼Ub2 is more labile to water or dithiothreitol hydrolysis than UBE2K∼Ub. This would be expected in the extended UBE2K∼Ub2 arrangement observed here, likely because of the accessibility of the thioester linkage that is not as protected as in closed structures. Conversely, reactivity with lysine (aminolysis) or ubiquitin is markedly slower for UBE2K∼Ub2 compared to a UBE2K∼Ub conjugate. This observation is consistent with the more extended conformation observed for UBE2K∼Ub2 that is driven by the UBA interaction with the distal Ub in the conjugated chain. This conformation does not favor nucleophilic attack from a lysine in ubiquitin that is promoted in the closed conformation.
Our NMR experiments and many previous experiments (10,11,17,23,24) have been done in the absence of an E3 ligase enzyme. It has been observed that the UBA domain in Ubc1, the yeast homolog of UBE2K, is required for full processivity to generate longer polyubiquitin chains (19,20) in the presence of an E3 ligase. Further, Ubc1 prefers Ub transfer to pre-existing ubiquitinated substrates rather than as the initial ubiquitin labeling. Interestingly, transfer of a ubiquitin chain from UBE2K∼Ub2 to a Ub2 chain has been shown to be more favorable than transfer to a single ubiquitin (11), suggesting the UBA domain must be directing the transfer in some fashion. Together, these previous observations suggest that the extended UBE2K-Ub2 conformation observed here must be altered by either the E3 ligase or a Ub2 species to position the conjugated Ub or Ub2 in the closed conformation needed to optimize Ub transfer. One possibility is that the UBA domain might preferentially recruit a polyubiquitin chain in a similar manner as proposed here or in other work (22), blocking the intramolecular extended conformation and promoting the conjugated Ub to a closed, more active position. Although further experiments would be needed to show this, our work provides a structural rationale for earlier kinetic studies aimed at understanding the role of the UBA domain on ubiquitin transfer mechanisms by complicated E2 enzymes such as UBE2K.
Author Contributions
B.W.C., R.E.L., and G.S.S. designed experiments. B.W.C. and R.E.L. performed experiments. B.W.C., R.E.L., and G.S.S. analyzed data and wrote the manuscript.
Acknowledgments
The authors thank Kathy Barber and Kishore Basu for preparation of some reagents, Lee-Ann Briere for help with the sedimentation equilibrium analysis, and Anne Rintala-Dempsey for maintenance of the Biomolecular NMR Facility.
This research was supported by a research grant to G.S.S. (05590-17) from the Natural Sciences and Engineering Research Council of Canada.
Editor: Elizabeth Komives.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.02.012.
Supporting Material
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A., Orian A., Schwartz A.L. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Budhidarmo R., Nakatani Y., Day C.L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Weber J., Polo S., Maspero E. HECT E3 ligases: a tale with multiple facets. Front. Physiol. 2019;10:370. doi: 10.3389/fphys.2019.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 7.Wang M., Pickart C.M. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 2005;24:4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 9.Deol K.K., Lorenz S., Strieter E.R. Enzymatic logic of ubiquitin chain assembly. Front. Physiol. 2019;10:835. doi: 10.3389/fphys.2019.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgins R., Gwozd C., Ellison M.J. The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J. Biol. Chem. 1996;271:28766–28771. doi: 10.1074/jbc.271.46.28766. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z., Pickart C.M. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J. Biol. Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 12.Gazdoiu S., Yamoah K., Pan Z.Q. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. USA. 2005;102:15053–15058. doi: 10.1073/pnas.0507646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varelas X., Ptak C., Ellison M.J. Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Mol. Cell. Biol. 2003;23:5388–5400. doi: 10.1128/MCB.23.15.5388-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Tu D., Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 15.McKenna S., Moraes T., Ellison M.J. An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13. The structural basis for lysine 63 chain catalysis. J. Biol. Chem. 2003;278:13151–13158. doi: 10.1074/jbc.M212353200. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.G., Youn H.S., Eom S.H. Crystal structure of the Ube2K/E2-25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis. Biochem. Biophys. Res. Commun. 2018;506:102–107. doi: 10.1016/j.bbrc.2018.10.067. [DOI] [PubMed] [Google Scholar]
- 17.Shin D.Y., Lee H., Yoo Y.J. Assembly of different length of polyubiquitins on the catalytic cysteine of E2 enzymes without E3 ligase; a novel application of non-reduced/reduced 2-dimensional electrophoresis. FEBS Lett. 2011;585:3959–3963. doi: 10.1016/j.febslet.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 18.van Nocker S., Vierstra R.D. Multiubiquitin chains linked through lysine 48 are abundant in vivo and are competent intermediates in the ubiquitin proteolytic pathway. J. Biol. Chem. 1993;268:24766–24773. [PubMed] [Google Scholar]
- 19.Rodrigo-Brenni M.C., Foster S.A., Morgan D.O. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigo-Brenni M.C., Morgan D.O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 21.Merkley N., Shaw G.S. Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J. Biol. Chem. 2004;279:47139–47147. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- 22.Middleton A.J., Day C.L. The molecular basis of lysine 48 ubiquitin chain synthesis by Ube2K. Sci. Rep. 2015;5:16793. doi: 10.1038/srep16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson R.C., Edmondson S.P., Twigg P.D. The E2-25K ubiquitin-associated (UBA) domain aids in polyubiquitin chain synthesis and linkage specificity. Biochem. Biophys. Res. Commun. 2011;405:662–666. doi: 10.1016/j.bbrc.2011.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldeman M.T., Xia G., Pickart C.M. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry. 1997;36:10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- 25.Ko S., Kang G.B., Lee W. Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity. J. Biol. Chem. 2010;285:36070–36080. doi: 10.1074/jbc.M110.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkley N., Barber K.R., Shaw G.S. Ubiquitin manipulation by an E2 conjugating enzyme using a novel covalent intermediate. J. Biol. Chem. 2005;280:31732–31738. doi: 10.1074/jbc.M505205200. [DOI] [PubMed] [Google Scholar]
- 27.Condos T.E., Dunkerley K.M., Shaw G.S. Synergistic recruitment of UbcH7∼Ub and phosphorylated Ubl domain triggers parkin activation. EMBO J. 2018;37:e100014. doi: 10.15252/embj.2018100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thrower J.S., Hoffman L., Pickart C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickart C.M., Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 30.Cook B.W., Barber K.R., Shaw G.S. The HIP2∼ubiquitin conjugate forms a non-compact monomeric thioester during di-ubiquitin synthesis. PLoS One. 2015;10:e0120318. doi: 10.1371/journal.pone.0120318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook B.W., Shaw G.S. Architecture of the catalytic HPN motif is conserved in all E2 conjugating enzymes. Biochem. J. 2012;445:167–174. doi: 10.1042/BJ20120504. [DOI] [PubMed] [Google Scholar]
- 32.Kay L.E., Keifer P., Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 33.Pervushin K., Riek R., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grzesiek S., Anglister J., Bax A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B. 1993;101:114–119. [Google Scholar]
- 35.Shuker S.B., Hajduk P.J., Fesik S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson B.A., Blevins R.A. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Dove K.K., Olszewski J.L., Klevit R.E. Structural studies of HHARI/UbcH7∼Ub reveal unique E2∼Ub conformational restriction by RBR RING1. Structure. 2017;25:890–900.e5. doi: 10.1016/j.str.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dove K.K., Stieglitz B., Klevit R.E. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17:1221–1235. doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams K.M., Qie S., Olsen S.K. Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34. Nat. Commun. 2019;10:3296. doi: 10.1038/s41467-019-11061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceccarelli D.F., Tang X., Sicheri F. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton K.S., Ellison M.J., Shaw G.S. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 43.Raasi S., Varadan R., Pickart C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 44.Mueller T.D., Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 45.Mueller T.D., Kamionka M., Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 46.Pruneda J.N., Stoll K.E., Klevit R.E. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme∼ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano T., Serve O., Kato K. Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution. J. Biol. Chem. 2011;286:37496–37502. doi: 10.1074/jbc.M111.256354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cook W.J., Jeffrey L.C., Pickart C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J. Biol. Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 49.Varadan R., Assfalg M., Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Ryabov Y., Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 51.Eddins M.J., Carlile C.M., Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 52.Serniwka S.A., Shaw G.S. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry. 2009;48:12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- 53.Brzovic P.S., Lissounov A., Klevit R.E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Rout M.K., Lee B.L., Spyracopoulos L. Active site gate dynamics modulate the catalytic activity of the ubiquitination enzyme E2-25K. Sci. Rep. 2018;8:7002. doi: 10.1038/s41598-018-25476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.