Abstract
Introduction
Type I Interferons (INFαs and INF β) are known to be proinflammatory cytokines that promote atherosclerosis. IFNA21 is a member of alpha Interferon gene cluster on short arm of chromosome 9. We analyzed the potential link between 9p21 coronary artery disease (CAD) risk locus and IFNA21.
Objectives
a) study of association between serum IFNA21 levels and 14 demographic/clinical variables, including age, gender, diabetes, hypertension, and duration of CAD, b) study of association between high serum IFNA21 levels and 30 9p21 SNP genotypes.
Methods
To estimate serum circulating levels of IFNA21, we performed sandwich ELISA in 184 controls and 167 CAD cases. The IFNA21 levels could be classified into two broad classes: a) Low-level group: ≤15.6 pg/ml b) High-level group: >15.6 pg/ml. We also performed SNP genotyping for 30 SNPs at 9p21 locus in all study subjects using Sequenom MassARRAY technology. Statistical software SPSS (Version 21) was used to analyze the data obtained.
Results
Our analysis indicates that there could be an association of high IFNA21 levels with variables – gender, age, and duration of CAD in the study population. SNPs rs10757272 (TT), rs10757274 (GG), rs10757283 (TT), rs1333045 (CC), rs1333048 (CC), rs1333049 (CC), and rs4977574 (GG) showed significant risk association with high-level IFNA21 group.
Conclusions
IFNA21 may be involved in inflammatory processes in an age-dependent manner and in progression of CAD. This IFNA21-mediated mechanism may be more active in females. Several 9p21 SNPs may modulate inflammatory processes mediated by IFNA21 and may, therefore, contribute to pathophysiology of CAD.
Keywords: Coronary artery disease, 9p21.3 CAD risk locus, Interferon alpha 21, Inflammation, Single-nucleotide polymorphism
1. Introduction
Cytokines consist of a large group of peptides or glycoproteins that are secreted by specific types of immune cells. Cytokines constitute a type of signaling molecules that mediate and regulate immunity, inflammation, and hematopoiesis. Interferons (IFNs) belong to the large family of cytokines. IFNs are named after their ability to “interfere” with viral replication within host cells. IFNs are presently classified into three groups: type I, type II, and type III. The type I IFNs include all IFNαs, IFNβ, IFNε, IFNκ, IFNω, and IFNν. Humans have 12 different IFNαs and a single IFNβ. Type I IFN genes are clustered on the human chromosome 9. Each subtype is encoded by its own gene and regulated by its own promoter, and none of them contain introns.
1.1. IFNA21 gene and its association with 9p21.3 CAD risk locus
IFNA21 gene is a member of the alpha interferon gene cluster on the short arm of chromosome 9. The gene is involved in related pathways like: cytokine signaling in immune response and toll-like receptor (TLR) signaling pathway. The encoded protein is a type I interferon and may play a specific role in the antiviral response to rubella virus (Source: GeneCards®- Human Gene Database).
Atherosclerotic Coronary Artery Disease (CAD) is a consequence of a cascade of events that characterize inflammation at the coronary artery vessel wall. Inflammation is mediated by a variety of soluble factors, including cytokines. Inflammatory cytokines are known to be mediators of atherosclerosis. Type I interferons (IFNα and IFNβ) exert either pro- or anti-inflammatory immune functions depending on the context of the disease. However, their actual role in atherogenesis has not been clearly understood. IFNβ has been shown to increase adhesion between macrophage and endothelial cell and promote attraction of leukocyte to atherosclerosis-susceptible sites in mice mediated by chemokines.1,2 Goossens et al demonstrated that cell signaling by type I IFNs is increased in ruptured human atherosclerotic plaques.1 Hence, type I IFNs have been identified as proatherosclerotic cytokines that may serve as additional targets for prevention or treatment of atherosclerotic CAD.
Single-nucleotide polymorphisms (SNPs) in the chromosomal region 9p21.3 are known to be strongly associated with the risk of CAD.3, 4, 5, 6, 7 However, the mechanisms responsible for this association have not been elucidated clearly. The 9p21 risk locus overlaps exons 13–20 of a large, noncoding, antisense RNA named ANRIL (antisense noncoding RNA in the INK4 locus).8
The ANRIL region is found to have many gene expression enhancers (33 predicted enhancers), and two CAD risk SNPs (rs10811656 and rs10757278) were found to be located in one of these enhancer motifs, consequently disrupting a binding site for the transcription factor STAT 19.
One of the enhancer regions was demonstrated to physically interact with the nearby CDKN2A/B (Cyclin-Dependent Kinase Inhibitor 2A/B) loci, the MTAP (Methylthioadenosine Phosphorylase) gene, and a distant region downstream of IFNA21 gene by using the chromatin conformation capture (3C) method.9
The 9p21 locus may modify the immune responses by regulating expression of IFNA21 or other related type I interferons. Jarinova et al (2009) demonstrated that homozygous carriers of the 9p21 risk allele exhibit higher levels of expression of gene sets involved in cell proliferation in white blood cells.10
The 9p21.3 CAD risk locus and IFNA21, both are loci found only in higher primates.11 There seems to be a potential link between expression of IFNA21 gene and the 9p21.3 CAD risk locus. More studies are needed to elaborate the primate-specific CAD pathogenesis mechanism involving these two loci.
As a step in this direction, we framed the following objectives for this study: a) to study the association between serum IFNA21 levels and 14 demographic/clinical variables, including age, gender, diabetes, hypertension, and duration of CAD and b) to study association between high serum IFNA21 levels and 30 9p21 SNP genotypes.
2. Materials and methods
The study design was case-control association study.
Inclusion criteria for controls: healthy individuals in the age group of 40–85 years.
Exclusion criteria for controls: individuals with a history of CAD, stroke, and Peripheral Artery Disease (PAD).
Inclusion criteria for cases: angiographically documented CAD cases with unstable angina/ST segment elevation Myocardial Infarction (STEMI)/Non-STEMI (NSTEMI), aged 30–85 years.
Exclusion criteria for cases: a) individuals with cardiomyopathies, kidney, liver, gastrointestinal disorders and b) individuals with infectious diseases such as hepatitis, HIV, tuberculosis, and so on.
The study was conducted in the South Indian state of Telangana in the twin cities of Hyderabad and Secunderabad. CAD cases were recruited from Krishna Institute of Medical Sciences (KIMS), Secunderabad, in consultation with the hospital cardiologist. Guidelines of the Ethical Committee of the Hospital and the Helsinki Declaration of 1975 were followed in the sample and data collection from study subjects. Written informed consent was taken from all study subjects. Controls were recruited from the general population. Demographic and health-related data were recorded in a Data Collection Sheet for all study subjects.
About 5 ml of blood sample was collected from each subject, and about 2.5 ml was dispensed in a sterile vacutainer tube coated with clot activator for serum separation. The rest was dispensed in a sterile vacutainer tube coated with Ethylenediaminetetraacetic Acid (EDTA) for genomic DNA extraction from whole blood. For separation of serum, the vacutainer tubes coated with clot activator were kept at room temperature for about 30–60 min and then centrifuged at 3,000 rpm for 5 min. The serum was then separated from the top of the blood clot carefully using a Pasteur pipette and dispensed in 1.5 ml Eppendorf tubes and stored at −80 °C. The serum samples were then used for estimation of IFNA21 by sandwich Enzyme Linked Immuno Sorbent Assay (ELISA) using the kit supplied by Wuhan Fine Biotech Co. Ltd. ELISA was performed for 351 samples, including 184 controls and 167 CAD cases.
Genomic DNA extraction was carried out from whole blood samples by using the Master Pure™ kit supplied by Epicenter (an Illumina Company). The DNA samples were estimated qualitatively by performing electrophoresis on 0.8% Agarose gels and viewing the ethidium bromide stained bands under a UV transilluminator. The quantitative estimation was performed using NanoDrop spectrophotometer and calculating the ratio of absorbance at 260/280 nm.
The DNA samples were then genotyped for 30 SNPs of the 9p21 region (Table 1). The criteria for selection of SNPs are: a) extensive literature survey and b) Minor Allele Frequency (MAF), taken from the dbSNP site of the National Center for Biotechnology Information (NCBI). SNPs with MAF greater than 0.15 were chosen for the study. SNP genotyping was done by the Sequenom MassARRAY® technology using AGENA protocol.
Table 1.
SNPs studied in the 9p21.3 region.
| S. No. | SNP ID | Chromosomal positiona | Gene view | Functional consequence | Minor allele and global frequency |
|---|---|---|---|---|---|
| 1 | rs1004638 (T/A) | 22115590 | CDKN2B-AS1 | Intron variant | A- 0.31 |
| 2 | rs10116277 (T/G) | 22081398 | CDKN2B-AS1 | Intron variant | G- 0.323 |
| 3 | rs1011970 (G/T) | 22062135 | CDKN2B-AS1 | Intron variant | T- 0.247 |
| 4 | rs1063192 (A/G) | 22003368 | CDKN2B-AS1 | Intron variant, UTR variant 3′ | G- 0.205 |
| 5 | rs10757272 (C/T) | 22088261 | CDKN2B-AS1 | Intron variant | T- 0.45 |
| 6 | rs10757274 (A/G) | 22096056 | CDKN2B-AS1 | Intron variant | G- 0.404 |
| 7 | rs10757278 (A/G) | 22124478 | Near CDKN2B-AS1 | Intron variant | G- 0.408 |
| 8 | rs10757283 (C/T) | 22134173 | Near CDKN2B-AS1 | Intron variant | T- 0.497 |
| 9 | rs10811661 (T/C) | 22134095 | Near CDKN2B-AS1 | Intron variant | C- 0.176 |
| 10 | rs1333040 (T/C) | 22083405 | CDKN2B-AS1 | Intron variant | C- 0.383 |
| 11 | rs1333042 (G/A) | 22103814 | CDKN2B-AS1 | Intron variant | A- 0.321 |
| 12 | rs1333045 (T/C) | 22119196 | CDKN2B-AS1 | Intron variant | C- 0.498 |
| 13 | rs1333048 (A/C) | 22125348 | Near CDKN2B-AS1 | Intron variant | C- 0.442 |
| 14 | rs1333049 (G/C) | 22125504 | Near CDKN2B-AS1 | Intron variant | C- 0.418 |
| 15 | rs16905599 (G/A) | 22069145 | CDKN2B-AS1 | Intron variant | A- 0.190 |
| 16 | rs2383206 (A/G) | 22115027 | CDKN2B-AS1 | Intron variant | G- 0.487 |
| 17 | rs2383207 (G/A) | 22115960 | CDKN2B-AS1 | Intron variant | A- 0.310 |
| 18 | rs2383208 (A/G) | 22132077 | Near CDKN2B-AS1 | Intron variant | G- 0.210 |
| 19 | rs2811712 (A/G) | 21998036 | CDKN2B-AS1 | Intron variant | G- 0.160 |
| 20 | rs2891169 (A/G) | 22131826 | Near CDKN2B-AS1 | Intron variant | G- 0.493 |
| 21 | rs3731239 (A/G) | 21974219 | CDKN2A | Intron variant | G- 0.175 |
| 22 | rs4977574 (A/G) | 22098575 | CDKN2B-AS1 | Intron variant | G- 0.395 |
| 23 | rs4977756 (A/G) | 22068653 | CDKN2B-AS1 | Intron variant | G- 0.288 |
| 24 | rs564398 (T/C) | 22029548 | CDKN2B-AS1 | Intron variant, nc transcript variant | C- 0.184 |
| 25 | rs615552 (T/C) | 22026078 | CDKN2B-AS1 | Intron variant | C- 0.195 |
| 26 | rs6475606 (T/C) | 22081851 | CDKN2B-AS1 | Intron variant | C-0.322 |
| 27 | rs7023329 (A/G) | 21816529 | MTAP | Intron variant | G- 0.449 |
| 28 | rs7865618 (A/G) | 22031006 | CDKN2B-AS1 | Intron variant | G- 0.188 |
| 29 | rs944797 (T/C) | 22115287 | CDKN2B-AS1 | Intron variant | C- 0.487 |
| 30 | rs9632884 (C/G) | 22072302 | CDKN2B-AS1 | Intron variant | G- 0.304 |
Genome build is GRCh38.p12 (taken from dbSNP, NCBI site).
Of the 30 9p21 SNPs that were genotyped, four SNPs – rs10757278, rs2891169, rs3731239, and rs944797 – were not in Hardy–Weinberg equilibrium among controls. Hence, these four SNPs were excluded from all analyses.
Statistical analysis: Statistical Package for Social Sciences (SPSS) Windows version 21.0 was used for statistical analysis. The mean and standard deviation values were calculated for quantitative variables, and percentages were calculated for qualitative variables. The chi-square test was used to study the association of demographic variables with IFNA21 levels. Risk estimates were calculated through odds ratio with 95% confidence interval to study the association of SNP genotypes with IFNA21 levels. The level of significance was considered as 0.05.
3. Results
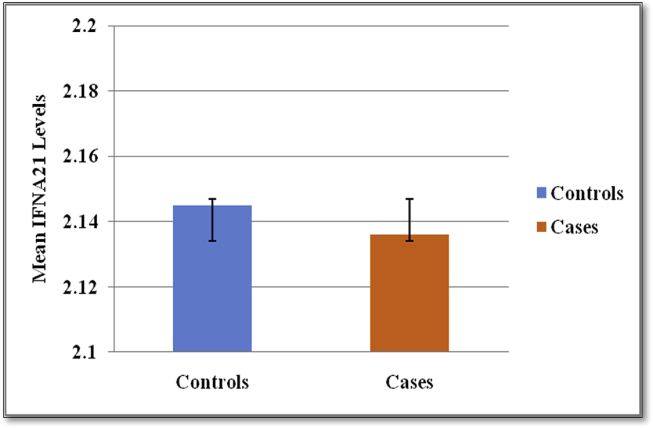
Student's t-test was performed to check the significance of difference in the mean levels of serum circulating IFNA21 between controls and cases, and it was found that the p-value was not significant (p = 0.737) (Table 2, Fig. 1).
Table 2.
IFNA21 descriptives table (log-transformed values).
| Sample size | Mean | Standard deviation | p-value | |
|---|---|---|---|---|
| Controls | n = 184 | 2.145 | 0.268 | 0.737 |
| Cases | n = 167 | 2.136 | 0.227 |
Fig. 1.
Comparison of the mean IFNA21 levels between controls and cases.
The IFNA21 values could be divided mainly into two groups (based on the reference range values provided in the kit manual):
-
•
Values ≤ 15.6 pg/ml (low-level group)
-
•
Values > 15.6 pg/ml (high-level group)
The association of demographic and clinical variables with the two IFNA21 groups was studied using the chi-square test (Table 3). The results indicate that there could be an association of high IFNA21 levels with gender, age, and duration of CAD.
-
•
Gender: Among controls, there is a significantly higher proportion of females in the high-level IFNA21 group than males (p = 0.000). There is a similar trend among cases, but it does not show statistical significance (p = 0.873).
-
•
Age: Among controls, there is a higher proportion of individuals in the high-level IFNA21 group in both the age categories (<50 years and ≥50 years), with the p-value approaching significance (p = 0.064). A similar trend is observed in cases, but it does not show statistical significance (p = 0.411).
-
•
Duration of CAD: Among cases, individuals affected with CAD for ≥1 year show significantly higher proportion of individuals in the high-level IFNA21 group than individuals affected with CAD for less than 1 year (p = 0.027).
Table 3.
Association of IFNA21 groups with demographic and clinical variables.
| S. No. | Variable | Controls (n = 184) |
Cases (n = 167) |
||||
|---|---|---|---|---|---|---|---|
| IFNA21 ≤ 15.6 pg/ml (%) | IFNA21 > 15.6 pg/ml (%) | p-value | IFNA21 ≤ 15.6 pg/ml (%) | IFNA21 > 15.6 pg/ml (%) | p-value | ||
| 1 | Diabetes | ||||||
| Yes | 43.5 (10) | 56.5 (13) | 0.361 | 41.2 (35) | 58.8 (50) | 0.583 | |
| No | 33.8 (53) | 66.2 (104) | 45.5 (35) | 54.5 (42) | |||
| 2 | Hypertension | ||||||
| Yes | 43.2 (19) | 56.8 (25) | 0.191 | 38.7 (36) | 61.3 (57) | 0.179 | |
| No | 32.4 (44) | 67.6 (92) | 49.3 (34) | 50.7 (35) | |||
| 3 | Family history | ||||||
| Yes | 36.4 (20) | 63.6 (35) | 0.541 | 47.5 (29) | 52.5 (32) | 0.244 | |
| No | 33.0 (38) | 67.0 (77) | 41.8 (41) | 58.2 (57) | |||
| Distant | 50 (5) | 50 (5) | 0 (0) | 100 (3) | |||
| 4 | Gender | ||||||
| Male | 53.3 (32) | 46.7 (28) | 0.000 | 43.5 (57) | 56.5 (74) | 0.873 | |
| Female | 25.8 (31) | 74.2 (89) | 41.9 (13) | 58.1 (18) | |||
| 5 | Marital status | ||||||
| Married | 35 (62) | 65 (115) | 0.951 | 42.8 (68) | 57.2 (91) | 0.408 | |
| Unmarried | 33.3 (1) | 66.7 (2) | 66.7 (2) | 33.3 (1) | |||
| 6 | Hyperlipidemia | ||||||
| Yes | 35.7 (5) | 64.3 (9) | 0.392 | 39.1 (9) | 60.9 (14) | 0.213 | |
| No | 34.5 (57) | 65.5 (108) | 49.4 (43) | 50.6 (44) | |||
| No information | 100 (1) | 0 (0) | 34.6 (18) | 65.4 (34) | |||
| 7 | Food | ||||||
| Vegetarian | 40 (34) | 60 (51) | 0.183 | 35.7 (10) | 64.3 (18) | 0.379 | |
| Mixed | 30.5 (29) | 69.5 (66) | 44.8 (60) | 55.2 (74) | |||
| 8 | Fruits | ||||||
| Daily | 31.9 (15) | 68.1 (32) | 0.900 | 42.2 (27) | 57.8 (37) | 0.592 | |
| Weekly 1–2 times | 40 (14) | 60 (21) | 30 (3) | 70 (7) | |||
| Weekly 3–4 times | 34.5 (19) | 65.5 (36) | 36.4 (8) | 63.6 (14) | |||
| Rarely | 34.9 (15) | 65.1 (28) | 48.5 (32) | 51.5 (34) | |||
| 9 | BMI | ||||||
| <18.5 | 60 (3) | 40 (2) | 0.401 | 100 (2) | 0 (0) | 0.179 | |
| 18.5–25.0 | 38.5 (20) | 61.5 (32) | 45.6 (41) | 54.4 (49) | |||
| ≥25 | 33.1 (40) | 66.9 (81) | 38.6 (27) | 61.4 (43) | |||
| 10 | Age | ||||||
| <50 years | 29.6 (32) | 70.4 (76) | 0.064 | 48 (24) | 52 (26) | 0.411 | |
| ≥50 years | 43.1 (31) | 56.9 (41) | 41.1 (46) | 58.9 (66) | |||
| 11 | Exercise | ||||||
| Yes | 36.8 (50) | 63.2 (86) | 0.383 | 44 (33) | 56 (42) | 0.850 | |
| No | 29.5 (13) | 70.5 (31) | 42.5 (37) | 57.5 (50) | |||
| 12 | Alcohol | ||||||
| Yes | 47.4 (9) | 52.6 (10) | 0.232 | 46.2 (30) | 53.8 (35) | 0.536 | |
| No | 33.5 (54) | 66.5 (107) | 41.2 (40) | 58.8 (57) | |||
| 13 | Duration of CAD | ||||||
| <1 year | – | – | – | 48.4 (59) | 51.6 (63) | 0.027 | |
| ≥1 year | – | – | 28.2 (11) | 71.8 (28) | |||
Note: Along with percentage, the number has been indicated in parenthesis in all the cells.
These results suggest that IFNA21 may be involved in inflammatory processes in an age-dependent manner and in the progression of CAD. This IFNA21-mediated mechanism may be more active in females.
Statistical analysis was done to determine whether individuals in the high-level IFNA21 group showed association with their 9p21 SNP genotypes using the chi-square test (Table 4).
Table 4.
Association of 9p21 SNP genotypes with the high-level IFNA21 group (>15.6 pg/ml).
| S. No. | SNP ID | Homozygote for normal allele (Reference value 1) | Homozygote for variant allele (OR 95% CI) | p value | Heterozygote (OR 95% CI) | p-value |
|---|---|---|---|---|---|---|
| 1 | rs1004638 | TT (1) | AA-0.546 (0.279–1.068) | 0.077 | AT-0.609 (0.367–1.012) | 0.056 |
| 2 | rs10116277 | TT (1) | GG-0.514 (0.269–0.983) | 0.044 | GT-0.636 (0.389–1.041) | 0.072 |
| 3 | rs1011970 | GG (1) | TT-1.152 (0.516–2.572) | 0.731 | TG-1.027 (0.642–1.643) | 0.913 |
| 4 | rs1063192 | AA (1) | GG-0.708 (0.267–1.880) | 0.489 | GA-0.714 (0.448–1.136) | 0.155 |
| 5 | rs10757272 | CC (1) | TT-2.000 (1.029–3.887) | 0.041 | CT-1.179 (0.620–2.242) | 0.616 |
| 6 | rs10757274 | AA (1) | GG-2.263 (1.212–4.224) | 0.010 | AG-1.093 (0.631–1.893) | 0.752 |
| 7 | rs10757283 | CC (1) | TT-2.217 (1.110–4.427) | 0.024 | CT-0.794 (0.470–1.341) | 0.388 |
| 8 | rs10811661 | TT (1) | CC-0.373 (0.087–1.599) | 0.184 | CT-0.898 (0.543–1.483) | 0.673 |
| 9 | rs1333040 | TT (1) | CC-0.631 (0.316–1.257) | 0.190 | CT-0.768 (0.477–1.236) | 0.276 |
| 10 | rs1333042 | GG (1) | AA-0.694 (0.344–1.404) | 0.310 | AG-0.625 (0.387–1.009) | 0.055 |
| 11 | rs1333045 | TT (1) | CC-2.526 (1.339–4.768) | 0.004 | CT-1.292 (0.741–2.252) | 0.366 |
| 12 | rs1333048 | AA (1) | CC-1.951 (1.022–3.723) | 0.043 | CA-0.951 (0.539–1.679) | 0.862 |
| 13 | rs1333049 | GG (1) | CC-2.186 (1.166–4.098) | 0.015 | CG-1.233 (0.715–2.126) | 0.45 |
| 14 | rs16905599 | GG (1) | AA-1.028(0.438–2.412) | 0.950 | AG-1.115 (0.689–1.805) | 0.656 |
| 15 | rs2383206 | AA (1) | GG-1.423 (0.759–2.668) | 0.271 | AG-0.903 (0.496–1.646) | 0.740 |
| 16 | rs2383207 | GG (1) | AA-0.726 (0.351–1.500) | 0.387 | AG-0.575 (0.356–0.928) | 0.023 |
| 17 | rs2383208 | AA (1) | GG-0.306 (0.075–1.255) | 0.100 | GA-0.894 (0.548–1.457) | 0.652 |
| 18 | rs2811712 | AA (1) | GG-0.901 (0.197–4.118) | 0.893 | AG-1.156 (0.692–1.931) | 0.580 |
| 19 | rs4977574 | AA (1) | GG-2.003 (1.067–3.763) | 0.031 | GA-0.963 (0.557–1.666) | 0.893 |
| 20 | rs4977756 | AA (1) | GG-0.587 (0.233–1.479) | 0.258 | GA-0.807 (0.503–1.295) | 0.374 |
| 21 | rs564398 | TT (1) | CC-1.137 (0.460–2.812) | 0.781 | CT-0.765 (0.472–1.239) | 0.276 |
| 22 | rs615552 | TT (1) | CC-0.787 (0.332–1.867) | 0.587 | CT-0.679 (0.424–1.086) | 0.106 |
| 23 | rs6475606 | TT (1) | CC-0.665 (0.339–1.305) | 0.236 | CT-0.679 (0.418–1.103) | 0.118 |
| 24 | rs7023329 | AA (1) | GG-1.250 (0.502–3.114) | 0.632 | GA-1.057 (0.664–1.680) | 0.816 |
| 25 | rs7865618 | AA (1) | GG-0.808 (0.373–1.748) | 0.588 | GA-0.720 (0.443–1.169) | 0.184 |
| 26 | rs9632884 | CC (1) | GG-0.576 (0.269–1.236) | 0.157 | GC-0.770 (0.481–1.233) | 0.277 |
The wild-type homozygote was taken as the normal/standard genotype with reference odds ratio value 1.
SNP IDs marked in bold show significant risk association for the variant allele homozygote.
SNPs that showed significant risk association with the high-level IFNA21 group were: rs10757272 (TT) with OR 2.000 (p = 0.041), rs10757274 (GG) with OR 2.263 (p = 0.010), rs10757283 (TT) with OR 2.217 (p = 0.024), rs1333045 (CC) with OR 2.526 (p = 0.004), rs1333048 (CC) with OR 1.951 (p = 0.043), rs1333049 (CC) with OR 2.186 (p = 0.015), and rs4977574 (GG) with OR 2.003 (p = 0.031).
To analyze association between 9p21.3 SNP genotypes and CAD, we used SNPstat software available online. Of the seven SNPs mentioned above, four SNPs have shown good association results with CAD. SNPstat analysis revealed that the TT and TC genotypes of rs10757272 showed a significant risk association among female CAD patients (OR = 8.15 for TC and 9.27 for TT, p = 0.012 in the codominant model), CC genotype of rs1333045 showed a significant risk association among all CAD patients (OR = 1.46, p = 0.046 in the recessive model), CC genotype of rs1333049 showed a trend toward risk association among premature (age at presentation < 55 years in males and <65 years in females) CAD patients (OR = 1.61, p = 0.061 in the recessive model), and GG genotype of rs4977574 showed a significant risk association among premature CAD patients (OR = 1.77, p = 0.025 in the recessive model).
These results suggest that several 9p21 SNPs may modulate inflammatory processes mediated by the cytokine IFNA21 and may, therefore, contribute to the pathophysiology of CAD.
4. Discussion
Our results suggest that SNPs at 9p21.3 CAD risk locus may influence the expression of the IFNA21 gene and may, thus, contribute to the pathogenesis of CAD. INF-γ-induced physical genomic interactions between 9p21.3 enhancers and the gene IFNA21 have been observed by Harismendy et al (2011) using the chromatin conformation capture (3C) method.9 Almontashiri et al (2011) have identified elevated serum IFNA21 as a biomarker for the 9p21.3 CAD risk locus.12
Ours is a preliminary study that has analyzed association between IFNA21 levels and SNP genotypes at 9p21.3 locus and also between IFNA21 levels and demographic/clinical variables. A limitation of this study is the small sample size. The results need to be validated in larger samples. More studies focusing on gene expression profiles in different cell types and tissues such as peripheral blood mononuclear cells (PBMCs) and coronary atherosclerotic plaques (atherectomy samples) are required to unravel the potential link between the expression of type I IFNs, especially IFNA21 and the 9p21.3 CAD risk locus. Also, studies on regulation of type I Interferon signaling in the context of CAD are required to understand its role in the etiopathology of the disease.
5. Conclusions
-
•
SNPs rs10757272, rs10757274, rs10757283, rs133045, rs1333048, rs1333049, and rs4977574 showed a significant risk association with the high-level IFNA21 group. Of these seven SNPs, four SNPs – rs10757274, rs1333045, rs1333049, and rs4977574 – showed a significant risk association with CAD in our study population. Hence, several 9p21.3 SNPs may modulate inflammatory processes mediated by IFNA21 and contribute to the pathophysiology of CAD.
-
•
Demographic variables, gender and duration of CAD, showed significant association with the high-level IFNA21 group in our study population. Hence, inflammatory processes mediated by IFNA21 seem to be more active in females and may be involved in the progression of CAD.
Author contributions
BK has contributed to the design of the work, acquisition of data, and preparation of the final draft of the manuscript.
DKM has contributed to the substantial revision of the manuscript.
NBK has contributed to the statistical analysis and interpretation of the data.
All authors have read and approved the submitted version of the manuscript and have agreed to be personally accountable for their own contribution and all queries related to the study.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
All authors have none to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ihj.2019.10.004.
Contributor Information
Bellary Kalpana, Email: kalpana_bellary@yahoo.com.
Dwarkanath K. Murthy, Email: dwarkanath49@yahoo.co.in.
Nagalla Balakrishna, Email: dr_nbk@yahoo.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Goossens P., Gijbels M.J., Zernecke A. Myeloid type I interferon signaling promotes atherosclerosis by stimulating macrophage recruitment to lesions. Cell Metab. 2010;12:142–153. doi: 10.1016/j.cmet.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Döring Y., Manthey H.D., Drechsler M. Auto-antigenic protein DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–1683. doi: 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 3.McPherson R., Pertsemlidis A., Kavaslar N. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helgadottir A., Thorleifsson G., Manolescu A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 5.Broadbent H.M., Peden J.F., Lorkowski S. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked, SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 6.Schunkert H., Gotz A., Braund P. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schunkert H., König I.R., Kathiresan S. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasmant E., Laurendeau I., Héron D. Characterization of a germ-line deletion including the entire INK4/ARF locus, in a melanoma-neural system tumor family. Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 9.Harismendy O., Notani D., Song X. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarinova O., Stewart A.F., Roberts R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R., Stewart A.F.R. Genes and coronary artery disease: where are we? J Am Coll Cardiol. 2012;60(18):1715–1721. doi: 10.1016/j.jacc.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 12.Almontashiri N.A., Fan M., Chen H.H. Abstract 15730: serum interferon alpha 21 is a biomarker of the 9p21.3 risk locus for coronary artery disease (Abstract) Circulation. 2011;124:A15730. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.