Abstract
Background
Accurate interpretation of rare genetic variants is a challenge for clinical translation. Updates in recommendations for rare variant classification require the reanalysis and reclassification. We aim to perform an exhaustive re-analysis of rare variants associated with inherited arrhythmogenic syndromes, which were classified ten years ago, to determine whether their classification aligns with current standards and research findings.
Methods
In 2010, the rare variants identified through genetic analysis were classified following recommendations available at that time. Nowadays, the same variants have been reclassified following current American College of Medical Genetics and Genomics recommendations.
Findings
Our cohort included 104 cases diagnosed with inherited arrhythmogenic syndromes and 17 post-mortem cases in which inherited arrhythmogenic syndromes was cause of death. 71.87% of variants change their classification. While 65.62% of variants were classified as likely pathogenic in 2010, after reanalysis, only 17.96% remain as likely pathogenic. In 2010, 18.75% of variants were classified as uncertain role but nowadays 60.15% of variants are classified of unknown significance.
Interpretation
Reclassification occurred in more than 70% of rare variants associated with inherited arrhythmogenic syndromes. Our results support the periodical reclassification and personalized clinical translation of rare variants to improve diagnosis and adjust treatment.
Funding
Obra Social "La Caixa Foundation" (ID 100010434, LCF/PR/GN16/50290001 and LCF/PR/GN19/50320002), Fondo Investigacion Sanitaria (FIS PI16/01203 and FIS, PI17/01690), Sociedad Española de Cardiología, and “Fundacio Privada Daniel Bravo Andreu”.
Keywords: Sudden cardiac death, Arrhythmias, Pathogenicity, Genetics
Research in context.
Evidence before this study
The American College of Medical Genetics and Genomics (ACMG) current recommendations include guidelines for obtaining an accurate assessment of rare variants. However, there is a lack of available data for each rare variant, and many remain of uncertain significance. Therefore, many of the families affected with inherited arrhythmic syndrome remain with an inconclusive genetic diagnosis, which is not helpful in clinical decision-making. Accurate interpretation of rare genetic variants is a challenge for clinical translation. Updates in recommendations for rare variant classification require the reanalysis and reclassification. An exhaustive review of the literature concerning each variant was performed through December 2019. Data was collected from: HGMD (www.hgmd.org), ClinVar (www.ncbi.nlm.nih.gov/clinvar/intro/), the National Center for Biotechnology Information SNP database (www.ncbi.nlm.nih.gov/SNP), Index Copernicus (en.indexcopernicus.com), Google Scholar (scholar.google.es), Springer Link (link.springer.com), Science Direct (www.sciencedirect.com), Excerpta Medica Database (www.elsevier.com/solutions/embase-biomedical-research), and the IEEE Xplore Digital Library (ieeexplore.ieee.org/Xplore/home.jsp). All genetic variants included in our study were compared to data from Exome Variant Server (EVS; evs.gs.washington.edu/EVS), and Genome Aggregation Database (gnomAD gnomad.broadinstitute.org/).
Added value of this study
Establishing a definite pathogenicity of rare genetic variants helps for clinical diagnosis of inherited arrhythmogenic syndromes but also helps to adopt therapeutic measures for prevention of sudden death. We have performed an accurate genetic reinterpretation of variants classified 10 years ago. Reclassification occurred in more than 70% of rare variants associated with inherited arrhythmogenic syndromes. These changes may influence clinical decisions adopted 10 years ago.
Implications of all the available evidence
Currently, classification of a genetic variant follows guidelines published in 2015 by ACMG/AMP. These recommendations are based on available data concerning the variant at the moment of classification. Data available 10 years ago is not the same as now. Therefore, reclassification and reinterpretation of a variant should be updated periodically to improve diagnosis and adjust treatment despite no concrete timeframe for this being established. In the light of our results, we propose that rare variants associated with inherited arrhythmogenic syndromes should be reanalysed within five years if already classified following ACMG recommendations, since this seems to be adequate to manage the rapid obsolescence of genetic data interpretations. In addition, our results support further urgent reanalysis of each IAS rare variant if they were not classified originally following ACMG recommendations.
Alt-text: Unlabelled box
1. Introduction
Advances in gene sequencing technology have made genetic testing in clinical diagnosis more accessible by increasing the number of analysed genes, decreasing costs, and reducing the amount of time required for analysis [1]. For an adequate translation of genetic data to clinical practice, and in order to manage the inherited conditions, it is critical to perform an appropriate interpretation of the genetic variant [2]. Sudden death may be the first manifestation of an inherited arrhythmic syndrome (IAS), thus early identification with genetic technology may help adopt preventive measures and reduce the risk of lethal episodes in family members. Genetic analysis may also be determinant in identifying causality in sudden death deemed inconclusive after a comprehensive autopsy. For these reasons current guidelines recommend genetic analysis in diagnosed patients and relatives who may be at risk, despite remaining asymptomatic [3].
The American College of Medical Genetics and Genomics (ACMG) current recommendations include guidelines for obtaining an accurate assessment of rare variants [4]. However, there is a lack of available data for each rare variant, and many remain of uncertain significance. Therefore, many of the families affected with IAS remain with an inconclusive genetic diagnosis, which is not helpful in clinical decision-making [5]. Rare variants classified as inconclusive are disregarded, and only clinical and family history are referenced in determining risk-assessment and clinical management [4].
Clinical and functional data on rare IAS variants published in the last ten years has helped clarify their roles and improved their classification. Continuous reclassification is recommended to update their roles before clinical translation. Such re-evaluation may serve to improve psychological outcomes and risk stratification while promoting personalized management [6, 7]. Only a few reports have addressed this idea in recent years [8], [9], [10]. In the present study, we describe the reclassification of rare IAS variants reported ten years ago by our laboratory to update their roles following current guidelines.
2. Materials and methods
2.1. Samples
This retrospective study reanalysed rare IAS variants classified in our laboratory ten years ago (during the year 2010). Rare variants were originally classified following recommendations available in 2010 as pathogenic (P), likely pathogenic (LP), variant of unknown significance (VUS), or as likely benign (LB) [11]. Variants classified as Benign in 2010 were not reanalysed due to global frequencies higher than 1%, and already identified ten years ago as common variants. All rare variants were identified in two groups: samples from patients with a definite clinical diagnosis of IAS or post-mortem samples without a conclusive cause of death but with suspected IAS. Genetic analysis was approved by the ethics committee of Hospital Josep Trueta (Girona, Spain) following the Helsinki II declaration. Both clinical and genetic data concerning all patients were kept confidential. Written informed consent was obtained from all patients included in the study. In post-mortem cases, a family member authorized the study or judge/legal authority included molecular autopsy as part of legal process.
2.2. Genetic analysis
Genomic DNA was extracted with Chemagic MSM I from whole blood (Chemagic human blood) or saliva (Chemagic Oragene Saliva). Concentration was determined along with purity using a NanoDrop1000 spectrophotometer (Thermo scientific). Genomic DNA was amplified by polymerase chain reaction (PCR) using intronic primers for each exon of all the genes analysed. The PCR product was purified by ExoSAP-IT (USB Corporation, Cleveland, OH, USA) and directly sequenced by dideoxy chain-termination method in an ABI Prism Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Sequencing was processed in a 3130XL Genetic Analyzer (Applied Biosystems) and analysed by SeqScape Software v2.5 (Life Technologies).
The genes associated with each IAS subtype were analysed following the prevailing recommendations in 2010 [11]. Genetic analysis for arrhythmogenic cardiomyopathy (ACM) included PKP2, DSP, DSC2, DSG2, DES, JUP, and TGFB3; analysis for Brugada syndrome (BrS) included SCN5A; analysis for catecholaminergic polymorphic ventricular tachycardia (CPVT) included RYR2 and CASQ2; analysis for dilated cardiomyopathy (DCM) included LMNA; analysis for hypertrophic cardiomyopathy (HCM) included MYH7, MYBPC3, TNNT2, and TNNI3; analysis for long QT Syndrome (LQTS) included KCNQ1, KCNH2, and SCN5A; and finally, analysis for sick sinus syndrome (SSS) included SCN5A and HCN4. The genes analysed in post-mortem cases were SCN5A, KCNQ1, KCNH2, KCNE1, KCNE2, KCNE3, RYR2, and CASQ2. All original sequences obtained in 2010 were comprehensively reanalysed with updated software (SeqScape v2.7, Applied Biosystems) to identify any alteration not identified at the time of report.
2.3. Data
An exhaustive review of the literature concerning each variant was performed through December 2019. Data was collected from: HGMD (www.hgmd.org), ClinVar (www.ncbi.nlm.nih.gov/clinvar/intro/), the National Center for Biotechnology Information SNP database (www.ncbi.nlm.nih.gov/SNP), Index Copernicus (en.indexcopernicus.com), Google Scholar (scholar.google.es), Springer Link (link.springer.com), Science Direct (www.sciencedirect.com), Excerpta Medica Database (www.elsevier.com/solutions/embase-biomedical-research), and the IEEE Xplore Digital Library (ieeexplore.ieee.org/Xplore/home.jsp). All genetic variants included in our study were compared to data from Exome Variant Server (EVS; evs.gs.washington.edu/EVS), and Genome Aggregation Database (gnomAD gnomad.broadinstitute.org/).
2.4. Classification
Ten years ago, variants were classified following an in-house algorithm including multiple parameters such as population frequencies, in silico predictions, and published functional data. Algorithms followed recommendations available in 2010 [11]. Part of this in-house algorithm focused on inherited arrhythmogenic disorders was published in 2015 [12]. Nowadays, all variants have been reclassified according to ACMG standards and guidelines for the interpretation of sequence variants as P, LP, VUS, LB, or benign (B) [4]. The PM2 item in the ACMG classification was considered fulfilled if Minor Allele Frequency (MAF) in relevant population databases was ≤0.1% [13]. The vast majority of reported pathogenic variants in IAS are extremely rare (<0.01%) [14]. High degree of pathogenicity (item PVS1) should only be used for rare variants in genes where loss of function is a well-established disease mechanism [15]. Genetic data were independently evaluated and classified by three clinical genetic experts. All investigators discussed and agreed on a final classification of all variants to avoid bias.
3. Results
3.1. Cohort
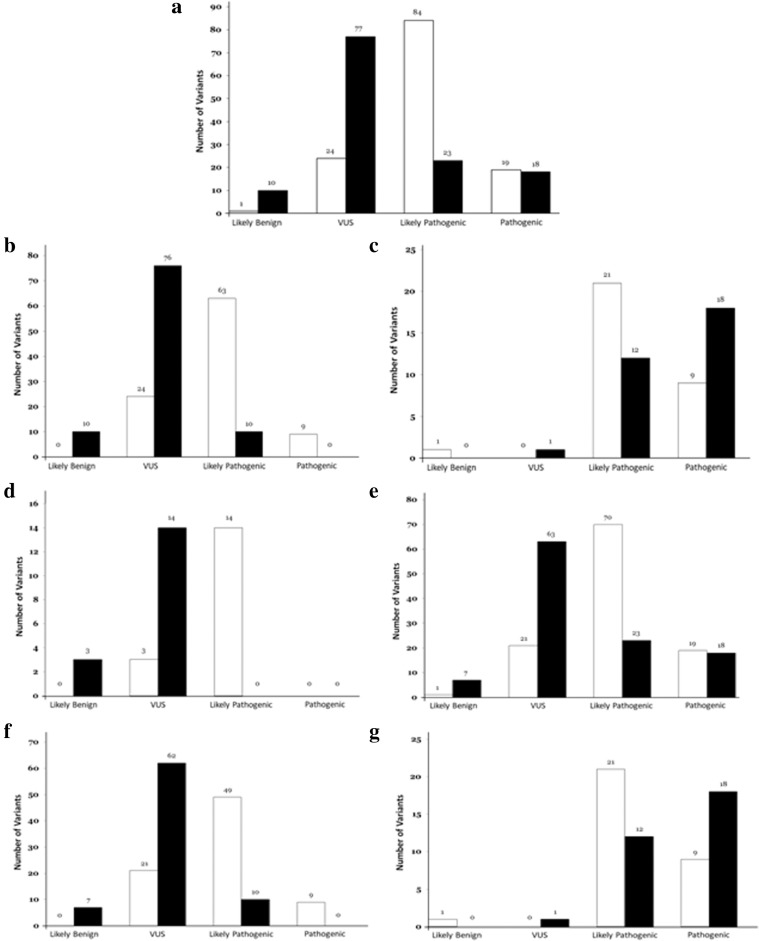
Our retrospective study included 121 cases, all Caucasian, with an even gender distribution (65 men; 53.71% and 56 women; 46.28%). Ages ranged from 19 to 51 years of age (mean age: 39.2 years). The first cohort (named “clinical group” –CL-) included 104 cases (85.95%) with a definite clinical diagnosis of IAS. Definitively, CL included 48 cases of BrS, 34 cases of ACM, 10 cases of LQTS, 7 cases of HCM, 4 cases of CPVT, and only one case diagnosed with DCM (Fig. 1). Suspicious cases with an inconclusive diagnosis were not included to avoid bias in the reclassification of genetic variants. The second cohort (named “post-mortem group” –PM-) comprised 17 post-mortem samples (14.05%) with IAS as the likely cause of death (Fig. 1). Exhaustive complete autopsies were performed in all cases, including toxicological analysis, before final judgment. Review of clinical and forensic data did not change the diagnostic decisions from ten years ago.
Fig. 1.
Distribution of rare variants. White columns represent the original classification (2010). Black columns represent the classification after the reanalysis. (a) Global classification of rare variants. (b) Global classification of rare missense variants. (c) Global classification of rare radical variants. (d) Classification of rare variants in Post-Mortem group. (e) Classification of rare variants in Clinical group. (f) Classification of rare missense variants in Clinical group. (g) Classification of rare radical variants in Clinical group.
3.2. Genetics
One hundred twenty-eight rare variants were localized in 17 genes: 55 in SCN5A, 17 in PKP2, 9 in KCNH2 and RyR2, 8 in DSG2, 6 in KCNQ1, 4 in MYBPC3, MYH7 and DSP, 3 in JUP, 2 in DSC2 and KCNE2, and 1 in KCNE3, DES, LMNA and TGFB3. All identified variants were exonic except one intronic variant (c.3840+1G>A_SCN5A, case 28). Of the 127 exonic variants, 96 were missense and 31 were radical (ten nonsense, fifteen deletions and six insertions). All cases carried at least one rare variant except for seven cases that carried two rare variants (case 26, LQTS; case 28, BrS; case 65, BrS; case 72, LQTS; case 79, ACM; case 107, ACM; and case 113, HCM). Original classification concluded that there were 19 P variants (14.84%), 84 LP variants (65.62%), 24 VUS variants (18.75%), and only one LB variant (0.78%) (Fig. 1) (Tables 1–3).
Table 1.
Variant genetic data.
| Index case | Disease | Gene | Nucleotide | Protein | dbSNP | gnomAD | HGMD (disease) | ClinVar | 2010 classification | 2020 classification (ACMG) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BrS | SCN5A | c.5464_5467delTCTG | p.(Glu1823Hisfs*10) | rs794728924 | 1/249656 (0.0004%) | CD077699 (SSS) | P | P | P |
| 2 | ACM | PKP2 | c.1613G>A | p.(Trp538*) | rs193922672 | 4/251382 (0.001%) | CM061177 (ACM) | LP | LP | P |
| 3 | BrS | SCN5A | c.1717C>T | p.(Gln573*) | NA | NA | CM100660 (BrS) | NA | LP | P |
| 4 | BrS | SCN5A | c.4477A>T | p.(Lys1493*) | NA | NA | CM100735 (BrS) | NA | LP | P |
| 5 | BrS | SCN5A | c.2865_2866delGA | p.(Glu955Aspfs*74) | rs756159737 | 4/248468 (0.001%) | NA | NA | LP | LP |
| 6 | BrS | SCN5A | c.1721delG | p.(Gly574Aspfs*49) | NA | NA | CD100781 (BrS) | NA | LP | LP |
| 7 | ACM | DSG2 | c.146G>A | p.(Arg49His) | rs121913006 | 1/249482 (0.0004%) | CM061700 (ACM) | LP | VUS | VUS |
| 8 | LQTS | KCNQ1 | c.421G>A | p.(Val141Met) | rs199472687 | NA | CM056972 (AF) | LP | P | LP |
| 9 | BrS | SCN5A | c.4562T>A | p.(Ile1521Lys) | rs199473617 | NA | CM100736 (BrS) | NA | LP | VUS |
| 10 | BrS | SCN5A | c.4534C>T | p.(Arg1512Trp) | rs137854602 | 14/251272 (0.005%) | CM994138 (BrS) | VUS | LP | VUS |
| 11 | BrS | SCN5A | c.5272_5274delATC | p.(Ile1758del) | NA | NA | CD1810427 (PCCD) | NA | LB | VUS |
| 12 | BrS | SCN5A | c.707T>G | p.(Leu236Arg) | NA | NA | . | NA | LP | VUS |
| 13 | ACM | DSG2 | c.1381C>T | p.(Gln461*) | rs1212557775 | NA | CM1314709 (ACM) | NA | LP | LP |
| 14 | BrS | SCN5A | c.4978A>G | p.(Ile1660Val) | rs199473625 | 8/251490 (0.003%) | CM057204 (LQTS) | VUS | LP | VUS |
| 15 | BrS | SCN5A | c.2893C>T | p.(Arg965Cys) | rs199473180 | 16/246378 (0.006%) | CM024644 (BrS) | VUS | LP | VUS |
| 16 | ACM | JUP | c.1028G>A | p.(Ser343Asn) | NA | NA | NA | NA | VUS | VUS |
| 17 | BrS | SCN5A | c.4352T>C | p.(Val1451Ala) | NA | NA | NA | NA | LP | VUS |
| 18 | ACM | JUP | c.475G>T | p.(Val159Leu) | rs782702266 | 11/269700 (0.004%) | CM1010258 (ACM) | VUS | VUS | VUS |
| 19 | BrS | SCN5A | c.4493T>C | p.(Met1498Thr) | rs199473263 | NA | CM057203 (LQTS) | VUS | LP | VUS |
| 20 | ACM | DSP | c.2956C>T | p.(Gln986*) | NA | NA | CM1310184 (ACM) | NA | LP | P |
| 21 | ACM | PKP2 | c.2013delC | p.Lys672Argfs*12 | rs764817683 | 2/251350 (0.0007%) | CD061457 (ACM) | P | P | P |
| 22 | BrS | SCN5A | c.2550_2551dupGT | p.Phe851Cysfs*19 | rs397514450 | NA | CI055774 (DCM) | P | P | P |
| 23 | BrS | SCN5A | c.4856delC | p.Pro1619Argfs*12 | NA | NA | CD100798 (BrS) | NA | LP | P |
| 24 | BrS | SCN5A | c.1936del | p.Gln646Argfs*5 | rs727505158 | 1/31374 (0.003%) | CD100782 (BrS) | P | LP | P |
| 25 | BrS | SCN5A | c.5174C>T | p.(Pro1725Leu) | rs199473301 | 5/251170 (0.001%) | CM097849 (LQTS) | VUS | LP | VUS |
| 26 | LQTS | KCNH2 | c.2639G>T | p.(Gly880Val) | NA | NA | CM150041 (LQTS) | NA | P | LP |
| KCNH2 | c.1838C>T | p.(Thr613Met) | rs199473524 | NA | CM990761 (LQTS) | P | LP | VUS | ||
| 27 | ACM | PKP2 | c.2203C>T | p.(Arg735*) | rs121434421 | 1/251356 (0.0003%) | CM043061 (ACM) | P | P | P |
| 28 | BrS | SCN5A | c.3840+1G>A | NA | NA | NA | CS099837 (BrS) | NA | P | LP |
| SCN5A | c.5068G>A | p.(Asp1690Asn) | rs1060499900 | 1/251488 (0.0003%) | CM136071 (BrS) | VUS | LP | VUS | ||
| 29 | BrS | SCN5A | c.2669T>C | p.(Ile890Thr) | NA | NA | CM130365 (BrS) | NA | LP | VUS |
| 30 | BrS | SCN5A | c.1705dupC | p.(Arg569Profs*152) | NA | NA | CI1510495 (BrS) | NA | LP | LP |
| 31 | BrS | SCN5A | c.1872dupA | p.(Glu625Argfs*96) | NA | NA | CI1510496 (BrS) | NA | LP | LP |
| 32 | ACM | PKP2 | c.1912C>T | p.(Gln638*) | rs397517012 | 1/251302 (0.0003%) | CM043056 (ACM) | P | P | P |
| 33 | BrS | SCN5A | c.2729C>T | p.(Ser910Leu) | rs199473175 | 1/250430 (0.0003%) | CM024643 (BrS) | LP | LP | VUS |
| 34 | ACM | PKP2 | c.604dupG | p.(Val202Glyfs*14) | NA | NA | CI146422 (ACM) | NA | LP | LP |
| 35 | ACM | DSG2 | c.137G>A | p.(Arg46Gln) | rs121913008 | 1/280866 (0.00003%) | CM061701 (ACM) | LP | LP | VUS |
| 36 | BrS | SCN5A | c.2962C>T | p.(Arg988Trp) | rs768691853 | 5/238498 (0.002%) | CM137981 (BrS) | VUS | LP | VUS |
| 37 | LQTS | SCN5A | c.5859_5862delTGAG | p.(Ser1953Argfs*84) | rs758317466 | 1/246198 (0.0004%) | NA | NA | LP | LP |
| 38 | BrS | SCN5A | c.4213G>A | p.(Val1405Met) | rs199473239 | NA | CM100715 (BrS) | VUS | P | LP |
| 39 | BrS | SCN5A | c.361C>T | p.(Arg121Trp) | rs199473556 | NA | CM095355 (BrS) | VUS | LP | VUS |
| 40 | BrS | SCN5A | c.1100G>A | p.(Arg367His) | rs28937318 | NA | CM020301 (SUNDS) | LP | P | LP |
| 41 | BrS | SCN5A | c.5177C>G | p.(Pro1726Arg) | NA | NA | NA | NA | LP | VUS |
| 42 | ACM | TGFB3 | c.1230A>C | p.(Lys410Asn) | NA | NA | NA | NA | LP | VUS |
| 43 | ACM | DSG2 | c.2440T>C | p.(Cys814Arg) | NA | NA | CM146425 (ACM) | NA | LP | VUS |
| 44 | ACM | PKP2 | c.275T>A | p.(Leu92*) | rs763639737 | 2/251424 (0.0007%) | CM102825 (ACM) | P | P | P |
| 45 | ACM | DES | c.407T>A | p.(Leu136His) | rs397516695 | 15/213206 (0.007%) | CM159728 (DCM) | VUS | VUS | VUS |
| 46 | ACM | DSP | c.6208G>A | p.(Asp2070Asn) | rs41302885 | 1118/282114 (0.39%) | CM198079 (BrS) | LB | VUS | LB |
| 47 | LQTS | KCNQ1 | c.898G>A | p.(Ala300Thr) | rs120074187 | 12/249914 (0.004%) | CM983511 (LQTS) | VUS | P | LP |
| 48 | BrS | SCN5A | c.2548G>A | p.(Val850Met) | rs911293694 | 2/251416 (0.0007%) | NA | NA | LP | VUS |
| 49 | BrS | SCN5A | c.5380T>A | p.(Phe1794Ile) | NA | NA | NA | NA | LP | VUS |
| 50 | BrS | SCN5A | c.4018G>A | p.(Val1340Ile) | rs199473605 | 13/ 282822 (0.004%) | CM100703 (BrS) | VUS | LP | VUS |
| 51 | HCM | MYH7 | c.5779A>T | p.(Ile1927Phe) | rs767300277 | 11/251320 (0.004%) | CM082963 (HCM) | VUS | VUS | VUS |
| 52 | BrS | SCN5A | c.2168dupT | p.(Thr724Hisfs*21) | NA | NA | NA | NA | LP | LP |
| 53 | BrS | SCN5A | c.2314G>A | p.(Asp772Asn) | rs199473157 | 5/249248 (0.002%) | CM097652 (LQTS) | VUS | LP | VUS |
| 54 | BrS | SCN5A | c.4219dupG | p.(Ala1407Glyfs*13) | NA | NA | NA | NA | LP | P |
| 55 | ACM | DSG2 | c.473T>G | p.(Val158Gly) | rs191143292 | 1537/280564 (0.54%) | CM070921 (ACM) | LB | VUS | LB |
| 56 | BrS | SCN5A | c.1577G>A | p.(Arg526His) | rs45627438 | 14/242632 (0.005%) | CM100657 (BrS) | VUS | LP | VUS |
| 57 | BrS | SCN5A | c.1120T>C | p.(Trp374Arg) | NA | NA | NA | NA | LP | VUS |
| 58 | LQTS | KCNQ1 | c.1016T>C | p.(Phe339Ser) | rs199472759 | NA | CM073160 (LQTS) | LP | P | LP |
| 59 | ACM | PKP2 | c.1162C>T | p.(Arg388Trp) | rs766209297 | 1/251320 (0.0003%) | CM097906 (ACM) | LP | LP | VUS |
| 60 | BrS | SCN5A | c.4345T>C | p.(Tyr1449His) | NA | NA | NA | NA | VUS | VUS |
| 61 | BrS | SCN5A | c.481G>A | p.(Glu161Lys) | rs199473062 | 1/240992 (0.0004%) | CM023671 (BrS) | LP | VUS | VUS |
| 62 | ACM | DSG2 | c.166G>A | p.(Val56Met) | rs121913013 | 518/280886 (0.18%) | CM070918 (ACM) | LB | VUS | LB |
| 63 | ACM | PKP2 | c.1378G>A | p.(Asp460Asn) | rs794729106 | NA | CM1213407 (ACM) | NA | P | LP |
| 64 | ACM | DSP | c.6361G>C | p.(Gly2121Arg) | rs368227724 | 1/251360 (0.0003%) | NA | NA | LP | VUS |
| 65 | BrS | SCN5A | c.1579G>C | p.(Gly527Arg) | rs763550164 | 8/243942 (0.003%) | NA | VUS | LP | VUS |
| SCN5A | c.3929C>G | p.(Pro1310Arg) | NA | NA | NA | NA | LP | VUS | ||
| 66 | BrS | SCN5A | c.2236G>A | p.(Glu746Lys) | rs199473582 | 5/248406 (0.002%) | CM100669 (BrS) | VUS | LP | VUS |
| 67 | CPVT | RYR2 | c.14639T>C | p.(Val4880Ala) | rs1242723821 | NA | HM030023 (CPVT) | NA | LP | VUS |
| 68 | ACM | PKP2 | c.2576delA | p.(Lys859Argfs*72) | NA | NA | CD146431 (ACM) | NA | LP | LP |
| 69 | ACM | PKP2 | c.1643delG | p.(Gly548Valfs*15) | rs794729137 | NA | CD043194 (ACM) | P | P | P |
| 70 | BrS | SCN5A | c.2633G>A | p.(Arg878His) | rs199473587 | NA | CM100676 (BrS) | NA | LP | VUS |
| 71 | ACM | PKP2 | c.2060T>G | p.(Leu687Arg) | rs794729113 | NA | NA | VUS | VUS | VUS |
| 72 | LQTS | KCNH2 | c.712G>C | p.(Gly238Arg) | NA | NA | NA | NA | VUS | VUS |
| KCNQ1 | c.944A>G | p.(Tyr315Cys) | rs74462309 | NA | CM981127 (LQTS) | LP | VUS | LP | ||
| 73 | BrS | SCN5A | c.5859_5862delTGAG | p.(Ser1953Argfs*84) | rs758317466 | 1/246198 (0.0004%) | NA | NA | LP | LP |
| 74 | ACM | PKP2 | c.1759G>A | p.(Val587Ile) | rs146102241 | 616/251180 (0.24%) | NA | LB | VUS | LB |
| 75 | BrS | SCN5A | c.4981G>A | p.(Gly1661Arg) | NA | NA | CM100750 (BrS) | NA | LP | VUS |
| 76 | ACM | DSG2 | c.527C>T | p.(Thr176Ile) | rs536617217 | 4/280698 (0.001%) | NA | VUS | LP | VUS |
| 77 | LQTS | KCNH2 | c.1744C>T | p.(Arg582Cys) | rs121912508 | NA | CM990759 (LQTS) | P | P | LP |
| 78 | BrS | SCN5A | c.5177C>A | p.(Pro1726His) | NA | NA | NA | NA | LP | VUS |
| 79 | ACM | DSC2 | c.835C>T | p.(Arg279Cys) | rs193922708 | 12/251360 (0.004%) | CM146543 (ACM) | VUS | LP | LP |
| PKP2 | c.1882delC | p.(Gln628Argfs*28) | NA | NA | CD146544 (ACM) | NA | LP | VUS | ||
| 80 | BrS | SCN5A | c.4052T>G | p.(Met1351Arg) | rs199473232 | NA | CM100707 (BrS) | NA | LP | VUS |
| 81 | BrS | SCN5A | c.5092G>A | p.(Ala1698Thr) | rs199473295 | 3/251490 (0.001%) | CM100753 (BrS) | VUS | VUS | VUS |
| 82 | SCD | RYR2 | c.2047G>A | p.(Glu683Lys) | NA | NA | NA | NA | LP | VUS |
| 83 | SCD | RYR2 | c.12056T>C | p.(Met4019Thr) | rs886039150 | NA | CM173280 (MI) | VUS | LP | VUS |
| 84 | SCD | KCNE2 | c.29C>T | p.(Thr10Met) | rs199473648 | 66/282722 (0.023%) | CM055291 (LQTS) | VUS | VUS | LB |
| 85 | SCD | KCNH2 | c.2674C>T | p.(Arg892Cys) | rs201627778 | 111/277590 (0.039%) | CM1413446 (SCD) | VUS | LP | VUS |
| 86 | SCD | RYR2 | c.12919C>T | p.(Arg4307Cys) | rs200092869 | 86/248746 (0.03%) | NA | VUS | LP | VUS |
| 87 | SCD | KCNE1 | c.253G>A | p.(Asp85Asn) | rs1805128 | 2637/282814 (0.9%) | CM040436 (LQTS, DI) | B | LP | LB |
| 88 | BrS | SCN5A | c.3911C>T | p.(Thr1304Met) | rs199473603 | 46/279030 (0.01%) | CM992663 (LQTS) | VUS | LP | VUS |
| 89 | SCD | SCN5A | c.1440A>C | p.(Lys480Asn) | rs752966781 | 2/249180 (0.0008%) | NA | VUS | LP | VUS |
| 90 | CPVT | RYR2 | c.14667C>G | p.(Phe4889Leu) | NA | NA | NA | NA | LP | VUS |
| 91 | HCM | MYBPC3 | c.2827C>T | p.(Arg943*) | rs387907267 | 3/247124 (0.001%) | CM032959 (HCM) | P | LP | P |
| 92 | SCD | RYR2 | c.8145G>T | p.(Glu2715Asp) | rs200420897 | 14/134624 (0.01%) | NA | VUS | LP | VUS |
| 93 | SCD | KCNH2 | c.865G>A | p.(Glu289Lys) | rs199472880 | 7/35014 (0.01%) | CM097827 (LQTS) | VUS | LP | VUS |
| 94 | SCD | KCNH2 | c.2860C>T | p.(Arg954Cys) | rs141401803 | 8/217960 (0.003%) | CM070176 (SIDS) | VUS | LP | VUS |
| 95 | SCD | SCN5A | c.3530C>G | p.(Pro1177Arg) | NA | NA | NA | NA | LP | VUS |
| 96 | LQTS | KCNQ1 | c.1097G>A | p.(Arg366Gln) | rs199473410 | 1/251240 (0.0003%) | CM002330 (LQTS) | P | P | LP |
| 97 | SCD | SCN5A | c.5054_5055delinsTT | p.(Glu1685Val) | NA | NA | NA | NA | LP | VUS |
| 98 | ACM | JUP | c.2069A>G | p.(Asn690Ser) | rs147628503 | 29/282402 (0.01%) | CM1416877 (Autism) | VUS | VUS | VUS |
| 99 | HCM | MYBPC3 | c.3328delA | p.(Met1110Trpfs*79) | NA | NA | CD1710421 (HCM) | NA | LP | P |
| 100 | SCD | KCNH2 | c.2941A>G | p.(Ser981Gly) | rs76649554 | 75/276264 (0.027%) | NA | VUS | LP | VUS |
| 101 | HCM | MYH7 | c.4377G>C | p.(Lys1459Asn) | rs201307101 | NA | CM042424 (HCM) | NA | VUS | VUS |
| 102 | CPVT | RYR2 | c.217C>G | p.(Leu73Val) | rs777753947 | 1/249224 (0.0004%) | CM1413452 (SCD) | NA | LP | VUS |
| 103 | LQTS | KCNQ1 | c.1861G>A | p.(Gly621Ser) | rs199472820 | 9/177000 (0.005%) | CM1413447 (SCD) | VUS | LP | VUS |
| 104 | ACM | PKP2 | c.1130T>C | p.(Ile377Thr) | rs397516985 | 1/31416 (0.003%) | NA | VUS | LP | VUS |
| 105 | SCD | SCN5A | c.287T>C | p.(Leu96Pro) | NA | NA | NA | NA | LP | VUS |
| 106 | ACM | PKP2 | c.76G>A | p.(Asp26Asn) | rs143004808 | 1427/168710 (0.8%) | CM061172 (ACM) | LB | LP | LB |
| 107 | ACM | DSG2 | c.908C>T | p.(Ser303Phe) | rs757792714 | 2/249400 (0.0008%) | CM1616318 (ACM) | VUS | LP | VUS |
| DSC2 | c.907G>A | p.(Val303Met) | rs145560678 | 282/282824 (0.09%) | CM117222 (DCM) | LB | LP | VUS | ||
| 108 | SCD | KCNE3 | c.248G>A | p.(Arg83His) | rs17215437 | 859/282668 (0.3%) | CM011795 (PP) | LB | VUS | VUS |
| 109 | CPVT | RYR2 | c.12419G>A | p.(Gly4140Glu) | NA | NA | NA | NA | LP | VUS |
| 110 | ACM | PKP2 | c.253_256delGAGT | p.(Glu85Metfs*26) | rs786204388 | 1/251410 (0.0003%) | CD102829 (ACM) | P | P | P |
| 111 | BrS | SCN5A | c.2924G>C | p.(Arg975Pro) | NA | NA | NA | NA | LP | VUS |
| 112 | SCD | RYR2 | c.7201C>T | p.(Arg2401Cys) | rs1321283106 | 2/31398 (0.006%) | NA | NA | LP | VUS |
| 113 | HCM | MYBPC3 | c.1513_1515delAAG | p.(Lys505del) | rs727504287 | 2/249264 (0.0008%) | CD031519 (HCM) | NA | P | LP |
| MYBPC3 | c.565G>A | p.(Val189Ile) | rs11570052 | 637/245140 (0.25%) | CM169151 (HCM) | LB | LP | LB | ||
| 114 | BrS | SCN5A | c.1097T>C | p.(Phe366Ser) | NA | NA | NA | NA | LP | VUS |
| 115 | HCM | MYH7 | c.5117T>C | p.(Leu1706Pro) | rs797044602 | NA | CM042428 (Myopathy) | LP | VUS | VUS |
| 116 | HCM | MYH7 | c.4865T>C | p.(Leu1622Pro) | NA | NA | NA | NA | VUS | VUS |
| 117 | SCD | KCNE2 | c.22A>G | p.(Thr8Ala) | rs2234916 | 1059/282720 (0.37%) | CM003449 (LQTS, DI) | LB | VUS | LB |
| 118 | ACM | DSP | c.88G>A | p.(Val30Met) | rs121912998 | 358/239538 (0.14%) | CM063961 (ACM) | LB | LP | LB |
| 119 | ACM | PKP2 | c.175C>T | p.(Gln59*) | NA | NA | CM1313041 (ACM) | NA | LP | P |
| 120 | DCM | LMNA | c.1056G>T | p.(Met352Ile) | NA | NA | NA | NA | VUS | VUS |
| 121 | LQTS | KCNH2 | c.982C>G | p.(Arg328Gly) | rs199473505 | 161/282246 (0.057%) | NA | VUS | VUS | VUS |
Note – ACM: arrhythmogenic cardiomyopathy, AF: atrial fibrillation, ACMG: American College of Medical Genetics and Genomics, B: benign, BrS: Brugada syndrome, ClinVar: clinical variation, CPVT: catecholaminergic polymorphic ventricular tachycardia, DCM: dilated cardiomyopathy, DM: disease mutation, gnomAD: genome aggregation database, HCM: hypertrophic cardiomyopathy, HGMD: human genome mutation database, LB: likely benign, LP: likely pathogenic, LQTS: long QT syndrome, LQTS-DI: long QT syndrome drug-induced, MI: myocardial infarction, NA: not available data, P: pathogenic, PCCD: progressive cardiac conduction disease, PP: periodic paralysis, SCD: sudden cardiac death, SIDS: sudden infant death syndrome, SSS: sick sinus syndrome, SUNDS: sudden unexpected death syndrome, VUS: variant of uncertain significance.
Table 3.
Classification of rare variants in each gene.
| 2010 |
2020 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Number variants | LB | VUS | LP | P | LB | VUS | LP | P | Modification 2010 vs. 2020 | |
| DES | 1 | . | 1 | . | . | . | 1 | . | . | NO | |
| DSC2 | 2 | . | . | 2 | . | . | 2 | . | . | YES | |
| DSG2 | 8 | . | 3 | 5 | . | 2 | 5 | 1 | . | YES | |
| DSP | 4 | . | 1 | 3 | . | 2 | 1 | . | 1 | YES | |
| JUP | 3 | . | 3 | . | . | . | 3 | . | . | NO | |
| KCNE1 | 1 | . | . | 1 | . | 1 | . | . | . | YES | |
| KCNE2 | 2 | . | 2 | . | . | 2 | . | . | . | YES | |
| KCNE3 | 1 | . | 1 | . | . | . | 1 | . | . | NO | |
| KCNH2 | 9 | . | 2 | 5 | 2 | . | 7 | 2 | . | YES | |
| KCNQ1 | 6 | . | 1 | 1 | 4 | . | 1 | 5 | . | YES | |
| LMNA | 1 | . | 1 | . | . | . | 1 | . | . | NO | |
| MYBPC3 | 4 | . | . | 3 | 1 | 1 | . | 1 | 2 | YES | |
| MYH7 | 4 | . | 4 | . | . | . | 4 | . | . | NO | |
| PKP2 | 17 | . | 2 | 8 | 7 | 2 | 3 | 4 | 8 | YES | |
| RYR2 | 9 | . | . | 9 | . | . | 9 | . | . | YES | |
| SCN5A | 55 | 1 | 3 | 46 | 5 | . | 38 | 10 | 7 | YES | |
| TGFB3 | 1 | . | . | 1 | . | . | 1 | . | . | YES | |
| TOTAL | 17 | 128 | 1 | 24 | 84 | 19 | 10 | 77 | 23 | 18 | |
Note – LB: likely benign, LP: likely pathogenic, P: pathogenic, VUS: variant of uncertain significance.
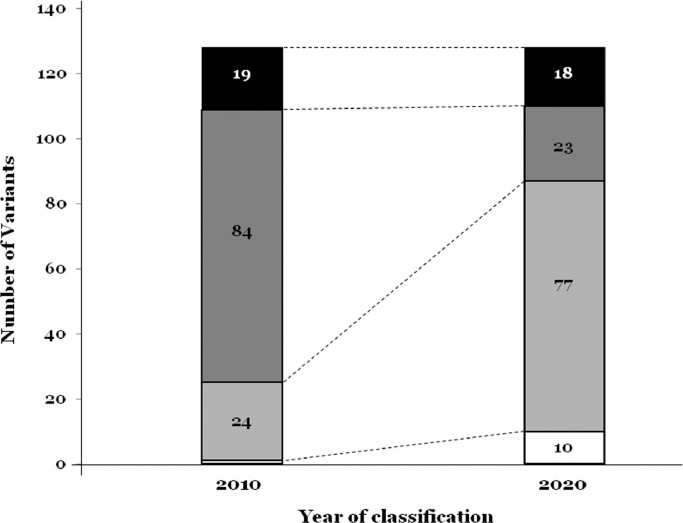
Reanalysis following current ACMG guidelines conferred significant changes in 71.87% (92 of 128) of the rare variants. One variant changed from LB to VUS (0.7%), 6 rare variants changed from VUS to LB (4.6%), and one changed from VUS to LP (0.7%). Four rare variants initially classified as LP in 2010 were changed to LB (3.1%), 59 were changed to VUS (46.09%), and 10 were changed to P (7.8%). Ten rare variants classified as P in 2010 were downgraded to LP in the reclassification (7.8%). In 2010, we classified 24 rare variants as VUS (18.75%) and after reanalysis, there were 77 classified as VUS (60.15%; 17 rare variants had no modification and 60 were reclassified). Originally, the majority were LP variants (84 of 128; 65.62%), but after reanalysis, the predominant classification of the variants was VUS (77 of 128; 60.65%) in contrast to 18.75% (24 of 128), ten years ago. Just twenty-three of 128 (17.96%) were classified as LP. After reanalysis, VUS became the predominant group (77 of 128; 60.65%) in contrast to 18.75% (24 of 128) ten years ago. In 2010, only one variant (0.78%) was classified as LB while after the reanalysis, 7.8% (10 of 128) were classified LB. The percentage of P variants was similar in both classifications (19 of 128; 14.84% in 2010 and 18 of 128; 14.06% after the reanalysis) (Fig. 2).
Fig. 2.
Reclassification of rare variants. White colour represents the number of rare variants classified as LB. Soft grey colour represents the number of rare variants classified as VUS. Dark grey colour represents the number of rare variants classified as LP. Black colour represents the number of rare variants classified as P. Concrete number of variants is included inside each part.
A total of 128 rare variants were analysed from 121 cases of IAS (CL group). In 88 cases (82.72%), the variant classification suffered a modification according to the current ACMG recommendations: 72 of the 104 cases with an IAS diagnosis (69.23%) and 16 of the 17 (94.11%) cases from the post-mortem cohort. Originally, in the post-mortem cohort, 82.23% (14 of 17) of variants were classified as LP, while after the reanalysis the same percentage was classified as VUS (14/17; 82.82%). In the IAS cohort, 63.06% (70 of 111) of the variants were initially classified as LP. However, after the reanalysis, 56.75% (63 of 111) were classified as VUS. Differences on the classification were observed also between missense and radical variants. In the 2010 classification, 49 of the 79 missense variants (62.02%) were classified as LP, while the major classification after the reanalysis was VUS (62 of 70; 78.48%). Radical variants were predominantly classified as LP in 2010 (21 of 31; 67.74%); although after the reanalysis a 58.06% of the radical variants were classified as P. The rare variants located in five genes (DES, JUP, KCNE3, LMNA, and MYH7) did not suffer any difference in final classification (Fig. 1) (Tables 3 and 4).
Table 4.
Classification of rare variants in IAS subtypes.
| All |
Missense |
Radical |
||||||
|---|---|---|---|---|---|---|---|---|
| Disease | Number Variants | 2010 | 2020 | 2010 | 2020 | 2010 | 2020 | |
| BrS | 49 | LB | 1 | . | . | . | 1 | . |
| VUS | 3 | 34 | 3 | 33 | . | 1 | ||
| LP | 41 | 8 | 30 | 2 | 11 | 6 | ||
| P | 4 | 7 | 2 | 0 | 2 | 7 | ||
| LQTS | 12 | LB | . | . | . | . | . | . |
| VUS | 3 | 4 | 3 | 4 | . | . | ||
| LP | 3 | 8 | 2 | 7 | 1 | 1 | ||
| P | 6 | . | 6 | . | . | . | ||
| CPVT | 4 | LB | . | . | . | . | . | . |
| VUS | . | 4 | . | 4 | . | . | ||
| LP | 4 | . | 4 | . | . | . | ||
| P | . | . | . | . | . | . | ||
| ACM | 36 | LB | . | 6 | . | 6 | . | . |
| VUS | 10 | 16 | 10 | 16 | . | . | ||
| LP | 19 | 5 | 12 | 1 | 7 | 4 | ||
| P | 7 | 9 | 1 | . | 6 | 9 | ||
| DCM | 1 | LB | . | . | . | . | . | . |
| VUS | 1 | 1 | 1 | 1 | . | . | ||
| LP | . | . | . | . | . | . | ||
| P | . | . | . | . | . | . | ||
| HCM | 8 | LB | . | 1 | . | 1 | . | . |
| VUS | 4 | 4 | 4 | 4 | . | . | ||
| LP | 3 | 1 | 1 | . | 2 | 1 | ||
| P | 1 | 2 | . | . | 1 | 2 | ||
Note – ACM: arrhythmogenic cardiomyopathy, BrS: Brugada syndrome, CPVT: catecholaminergic polymorphic ventricular tachycardia, DCM: dilated cardiomyopathy, HCM: hypertrophic cardiomyopathy, LB: likely benign, LQTS: long QT syndrome, LP: likely pathogenic, P: pathogenic, VUS: variant of uncertain significance.
In 48 cases of BrS, most rare variants were originally classified as LP (41 of 49; 83.67%) but after reanalysis, most were classified as VUS (34 of 49; 69.38%). In case number 28, diagnosed with BrS, an intronic variant was originally classified as P but after reanalysis, was classified as LP. In ten cases diagnosed with LQTS, 50% (6 of 12 cases) were classified as P but almost all rare variants have been reclassified as LP (8 of 12; 66.66%). In four cases diagnosed with CPVT, all rare variants (4 of 4; 100%) were originally classified as LP but after reanalysis, the same four rare variants were classified as VUS. A total of 34 cases diagnosed with ACM were originally classified as LP (52.77%; 19 of 36), but after reanalysis, most were classified as VUS (16 of 36; 44.44%). In the one case diagnosed with DCM, the rare variant originally classified as VUS remained at the same level of pathogenicity. In seven cases diagnosed with HCM, 50% of rare variants were originally classified as VUS (4 of 8) and after reanalysis, the same percentage of VUS was maintained (Fig. 1) (Tables 1, 2 and 4).
Table 2.
Modifications in classification of rare variant.
| Total | Total of Changes | PM | Changes in PM | CL | Changes in CL | |
|---|---|---|---|---|---|---|
| Rare Variants | 128 | 92 (71,87%) | 17 | 16 (94,11%) | 111 | 76 (68,46%) |
| Cases | 121 | 88 (72,72%) | 17 | 16 (94,11%) | 104 | 72 (69,28%) |
Note – CL: clinical group, PM: post-mortem group.
4. Discussion
Genetic testing in patients diagnosed with IAS is highly recommended both in clinical and medico-legal settings, since death is often the first manifestation of disease [3]. However, misinterpretation of rare variant designations may lead to inaccurate genetic diagnoses and/or the adoption of unnecessary and/or inappropriate therapeutic approaches resulting in an increased morbidity and mortality. Therefore, one of the main current challenges in genetic analysis is determining the pathogenic role of rare variants. Identifying a genetic cause of IAS allows for accurate clinical diagnosis, risk stratification, adoption of personalized therapeutic measures, and early identification of relatives at risk, while also determining no genetic carriers [16]. Obtaining reliable and accountable interpretations of variant significance is as important as improving molecular diagnostic techniques, and for these reason adequate guidelines, and also a periodic update of the criteria used for interpretation and revision of the variant significances, are fundamental.
In our retrospective study, we have applied current ACMG recommendations for variant classification in a cohort of patients diagnosed with IAS and post-mortem cases with suspected IAS [4]. We determined that over 70% of variants required reclassification after ten years under the updated guidelines. Specifically, after reanalysis of the variants, 69.23% of diagnosed IAS cases required a change in variant classification. In the post-mortem cohort, 94.11% required a change in variant classification. This data reinforces the need for clinical data in genetic diagnoses; a complete clinical history contributes to variant classification and helps to clarify the role of a variant in each patient. Recently, a study focused on the reclassification of VUS in IAS concluded that disease-specific phenotypes significantly increase the accuracy of classification [10]. Interestingly, we determined that many missense variants changed their classification from LP to VUS after reanalysis; this modification is mainly due to the increase of items used in ACMG recommendations. As mentioned above, increase in items implies more accuracy in classification but also stringency. In contrast, many radical variants changed from LP to P, accordingly to Harrison et al. [17] This fact suggests that variants resulting in a premature truncation of proteins and/or frameshifts should be considered highly damaging and therefore should be carefully analysed. Missense variants should be comprehensively analysed in each patient, considering all available data to perform a proper variant prioritization in a personalized clinical context [18]. Family segregation is the most robust tool to corroborate the pathogenic role of a particular variant. Unfortunately, a complete segregation for most rare variants currently associated with IAS is not available. In addition, incomplete penetrance/variable expressivity are hallmarks of IAS, so a segregation analysis of at least three generations should be recommended to obtain helpful data [19, 20]. Thus, the disease manifestation observed at the basal assessment and the clinical evolution on the follow-up, not only in the index cases but in the entire families, may also be highly useful in the understanding of the pathogenic role of the initially identified variants. Despite all the previously mentioned considerations, the frequency of rare variants in the global population is the first tool used to help to discern a potential damaging variant from other rare variants with no potential deleterious role. Nowadays, free and quick access to on-line databases focused on variant population frequencies makes this an easy routine approach.
Classification of a variant as VUS or downgrading a variant status from LP to VUS does not mean that there is less pathogenic risk of IAS for any patient who carries the rare variant; ambiguous significance implies that current evidence does not back a conclusive deleterious role in IAS. Therefore, clinical translation of VUS should be performed with caution and VUS should not be discarded, at least until additional data becomes available focused on clarifying their clinical role. Current recommendations for the interpretation of rare variants [4] include more items to be considered than ten years ago [11]. This increase in items implies more accuracy in classification but also increased stringency; thus, a lack of data for some of these items leads to ambiguous classification [5]. As a consequence, a low percentage of variants classified currently as VUS confer a real of risk in IAS and most are LB [21]. To discriminate a true risk-carrying variant from a non-deleterious variant is a challenge without accurate family segregation and functional studies [7]. Expected frequencies of each IAS variant and constant update of minor allele frequencies in large global population studies should be used to help identify the genes, regions of genes, and/or types of variants strongly associated with IAS which may help to determine the roles of variants in clinical settings, particularly if they are classified as VUS [22].
At the present time, while the ACMG recommend how to classify variants, there is currently no consensus for when and how often variants should be reclassified. Therefore, reinterpretation of genetic variants occurs mainly due to a clinician's request, identification of a previously classified variant in a new patient or new data available concerning the rare variant [23]. These expectations should be explicitly delineated as part of the informed consent process before the sample is obtained and reviewed again when disclosing initial results [24]. Concerning IAS, Smith et al. reported a reclassification after one year of 3% of rare variants [25]. In 2018, a reclassification of rare variants previously considered deleterious in Brugada Syndrome was performed; only 37% were classified as P or LP following current ACMG recommendations [8]. A recent study identified a modification in 52% of rare variants classified as VUS seven years ago [10]. Therefore, the evidence supports the periodic reclassification of the rare variants in IAS despite lack of data concerning the time of re-evaluation. In our report, more than 70% of rare variants were reclassified after ten years, also supporting the necessity of re-evaluation. In the light of all this evidence, we propose that rare variants associated with IAS should be reanalysed within five years if already classified following ACMG recommendations since it seems to be adequate to manage the rapid obsolescence of genetic data interpretations. In addition, our results support further urgent reanalysis of each IAS rare variant if they were not classified originally following ACMG recommendations.
The next step should be clinical translation of the re-evaluation and assessment of the implications in families because the confirmation of a variant as P, or removal of a P designation, may have a significant impact on patients and relatives. Therapeutic management can be modified but emotional and psychological impacts may have lasting effects [26]. New genetic information can lend itself to misinterpretation [5], so we recommended discussions with an expert cardiologist in genetics to explain what reclassification entails for each patient, accordingly to recent American Society of Human Genetics recommendations (ASHG) [27]. One key point is that a change in classification does not necessarily change the fact that a case has an IAS. Finally, it is important to remark that our re-information approach to families follows the ethical premise that definitive consideration for any clinical or research guideline should be improving patient medical care.
We can highlight some major limitations to our study. Variant interpretation is subject to inherent intra- and inter-laboratory differences in data interpretation [28]. In the current study, three of the authors performed independent classification following ACMG recommendations and all authors came to a consensus regarding the final classification decision. All rare variants were identified after a limited analysis of genes; we cannot be sure that patients do not carry other rare variants in genes currently unassociated with IAS. Only genes currently associated with IAS were analysed. The number of cases analysed was small, so comprehensive reassessment should be performed in large cohorts of IAS samples to corroborate a periodic reclassification. Finally, lack of data for some of the rare variants, mainly concerning functional studies as well as familial segregation, impedes comprehensive interpretation of our results and definite classification.
In summary, reanalysis using current ACMG recommendations showed that 71.87% of rare variants in IAS were given a new classification than originally assigned ten years ago. Many variants, however, remain of ambiguous significance. These findings emphasize the importance of cautious interpretation of variant scoring and comprehensive family segregation, supporting the periodic re-evaluation of rare variants in IAS before clinical translation. It is extremely important that in cases of significance changes after an update the geneticist promptly informs the interested patients.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgments
Funding sources/Acknowledgments
This work was supported by Obra Social "La Caixa Foundation" (ID 100010434, LCF/PR/GN16/50290001 and LCF/PR/GN19/50320002), Fondo Investigacion Sanitaria (FIS PI16/01203 and FIS, PI17/01690) from Instituto Salud Carlos III (ISCIII), Sociedad Española de Cardiología (Proyecto Investigación Básica Cardiología 2015 de los Socios Estratégicos SEC), and “Fundacio Privada Daniel Bravo Andreu”. CIBERCV is an initiative of the ISCIII, Spanish Ministry of Economy and Competitiveness. Funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Author's contributions
Conceptualization_OC, GS, AO, JB, and RB, Data curation_OC, AF, MC, AI, EA, SG, AG, PJ, and RT, Formal analysis_OC, AF, MC, AI, CF, SC, EA, and SG, Funding acquisition_OC, GS, JB, and RB, Investigation_OC, AF, MC, PJ, and RT, Methodology_OC, SC, EA, SG, AG, PJ, and RT, Project administration_OC, GS, AO, JB, and RB, Resources_OC, GS, AO, JB, and RB, Software_CF, SC, Supervision_OC, GS, AO, JB, and RB, Validation_OC, GS, AO, JB, and RB, Visualization_OC, GS EA, JB, and RB, Writing-original draft_OC, GS, AO, JB, and RB, Writing-review & editing_OC, GS, AO, JB, and RB.
Contributor Information
Oscar Campuzano, Email: oscar@brugada.org.
Ramon Brugada, Email: ramon@brugada.org.
References
- 1.Wordsworth S., Doble B., Payne K., Buchanan J., Marshall D.A., McCabe C. Using "Big Data" in the cost-effectiveness analysis of next-generation sequencing technologies: challenges and potential solutions. Value Health: J Int Soc Pharmacoecon Outcomes Res. 2018;21(9):1048–1053. doi: 10.1016/j.jval.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Moreno L., Linossi C., Esteban I., Gadea N., Carrasco E., Bonache S. Germline BRCA testing is moving from cancer risk assessment to a predictive biomarker for targeting cancer therapeutics. Clin Transl Oncol: Official Publ Fed Span Oncol Soc Natl Cancer Inst Mexico. 2016;18(10):981–987. doi: 10.1007/s12094-015-1470-0. [DOI] [PubMed] [Google Scholar]
- 3.Priori S.G., Blomstrom-Lundqvist C. 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs. Eur Heart J. 2015;36(41):2757–2759. doi: 10.1093/eurheartj/ehv445. [DOI] [PubMed] [Google Scholar]
- 4.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackerman M.J. Genetic purgatory and the cardiac channelopathies: exposing the variants of uncertain/unknown significance issue. Heart Rhythm. 2015;12(11):2325–2331. doi: 10.1016/j.hrthm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Macklin S.K., Jackson J.L., Atwal P.S., Hines S.L. Physician interpretation of variants of uncertain significance. Fam Cancer. 2019;18(1):121–126. doi: 10.1007/s10689-018-0086-2. [DOI] [PubMed] [Google Scholar]
- 7.Tsai G.J., Ranola J.M.O., Smith C., Garrett L.T., Bergquist T., Casadei S. Outcomes of 92 patient-driven family studies for reclassification of variants of uncertain significance. Genet Med. 2019;21(6):1435–1442. doi: 10.1038/s41436-018-0335-7. [DOI] [PubMed] [Google Scholar]
- 8.Denham N.C., Pearman C.M., Ding W.Y., Waktare J., Gupta D., Snowdon R. Systematic re-evaluation of SCN5A variants associated with Brugada syndrome. J Cardiovasc Electrophysiol. 2019;30(1):118–127. doi: 10.1111/jce.13740. In press. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.J., Lu T.P., Lin L.Y., Liu Y.B., Ho L.T., Huang H.C. Impact of ancestral differences and reassessment of the classification of previously reported pathogenic variants in patients with Brugada syndrome in the genomic era: a SADS-TW BrS registry. Front genet. 2018;9:680. doi: 10.3389/fgene.2018.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett J.S., Bernhardt M., McBride K.L., Reshmi S.C., Zmuda E., Kertesz N.J. Reclassification of variants of uncertain significance in children with inherited arrhythmia syndromes is predicted by clinical factors. Pediatr Cardiol. 2019;40(8):1679–1687. doi: 10.1007/s00246-019-02203-2. [DOI] [PubMed] [Google Scholar]
- 11.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R. Molecular subcommittee of the ALQAC: ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 12.Campuzano O., Allegue C., Fernandez A., Iglesias A., Brugada R. Determining the pathogenicity of genetic variants associated with cardiac channelopathies. Sci Rep. 2015;5:7953. doi: 10.1038/srep07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi Y., Yang S., Nykamp K., Garcia J., Lincoln S.E., Topper S.E. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9(1):13. doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L. ClinGen sequence variant interpretation working G: recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackerman M.J., Priori S.G., Willems S., Berul C., Brugada R., Calkins H. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13(8):1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 17.Harrison S.M., Rehm H.L. Is 'likely pathogenic' really 90% likely? Reclassification data in ClinVar. Genome Med. 2019;11(1):72. doi: 10.1186/s13073-019-0688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18(10):599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semsarian C., Ingles J. Molecular autopsy in victims of inherited arrhythmias. J Arrhythm. 2016;32(5):359–365. doi: 10.1016/j.joa.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn K.E., Caleshu C., Cirino A.L., Ho C.Y., Ashley E.A. A clinical approach to inherited hypertrophy: the use of family history in diagnosis, risk assessment, and management. Circ Cardiovasc Genet. 2013;6(1):118–131. doi: 10.1161/CIRCGENETICS.110.959387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18(12):883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh R., Thomson K.L., Ware J.S., Funke B.H., Woodley J., McGuire K.J. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Mecky J., Johansson L., Plantinga M., Fenwick A., Lucassen A., Dijkhuizen T. Reinterpretation, reclassification, and its downstream effects: challenges for clinical laboratory geneticists. BMC Med Genom. 2019;12(1):170. doi: 10.1186/s12920-019-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David K.L., Best R.G., Brenman L.M., Bush L., Deignan J.L., Flannery D. Patient re-contact after revision of genomic test results: points to consider—a statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2019;21(4):769–771. doi: 10.1038/s41436-018-0391-z. [DOI] [PubMed] [Google Scholar]
- 25.Smith E.D., Radtke K., Rossi M., Shinde D.N., Darabi S., El-Khechen D. Classification of Genes: standardized clinical validity assessment of gene-disease associations aids diagnostic exome analysis and reclassifications. Hum Mutat. 2017;38(5):600–608. doi: 10.1002/humu.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vears D.F., Niemiec E., Howard H.C., Borry P. Analysis of VUS reporting, variant reinterpretation and recontact policies in clinical genomic sequencing consent forms. Eur J Hum Genet. 2018;26(12):1743–1751. doi: 10.1038/s41431-018-0239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bombard Y., Brothers K.B., Fitzgerald-Butt S., Garrison N.A., Jamal L., James C.A. The responsibility to recontact research participants after reinterpretation of genetic and genomic research results. Am J Hum Genet. 2019;104(4):578–595. doi: 10.1016/j.ajhg.2019.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016;98(6):1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]