Summary
Oxidation of formate to CO2 is catalyzed via the donation of electrons from formate dehydrogenase (FDH) to nicotinamide adenine dinucleotide (NAD+), and thus the charge transport characteristics of FDH become essential but remain unexplored. Here, we investigated the charge transport through single-enzyme junctions of FDH using the scanning tunneling microscope break junction technique (STM-BJ). We found that the coupling of NAD+ with FDH boosts the charge transport by ∼2,100%, and the single-enzyme conductance highly correlates with the enzyme activity. The combined flicker noise analysis demonstrated the switching of the coenzyme-mediated charge transport pathway and supported by the significantly reduced HOMO-LUMO gap from calculations. Site-specific mutagenesis analysis demonstrated that FDH-NAD+ stably combined own higher bioactivity and boosts charge transport, and the coupling has been optimized via the natural selection. Our work provides evidence of hydrogen bond coupling in bioactivity but also bridges the charge transport through single-enzyme junctions and enzyme activities.
Subject Areas: Analytical Chemistry, Biotechnology, Molecular Biology
Graphical Abstract
Highlights
-
•
Binding of NAD+ with FDH boosts the charge transport by more than 2,100%
-
•
Single-enzyme conductance highly correlates with the enzyme activity
-
•
Hydrogen bond bridges the charge transport and enzyme activities
-
•
Experiments combined with calculations probe switching of charge transport pathway
Analytical Chemistry; Biotechnology; Molecular Biology
Introduction
The newly registered oxidoreductase on BRENDA accounts for more than 30% of all enzymes, of which about 51% are oxidoreductases using NAD(P)+ or NAD(P)H as coenzyme. Among the bioactive proteins, formate dehydrogenase (FDH) is an important oxidoreductase that catalyzes the dehydrogenation of formate to form carbon dioxide through the electric coupling between FDH and NAD+ (Hirose et al., 2018). FDH also plays an essential role in NADH regeneration of bifunctional enzyme system to produce the intermediates for anti-HIV drugs (L-tert leucine) (Jiang et al., 2017), and many scholars also focused on the modification and design of FDH to catalyze the reverse reaction and applied in the elimination of greenhouse effect (Gleizer et al., 2019, Jayathilake et al., 2019, Nielsen et al., 2019). Thus, the understanding of electron transfer and intramolecular interaction relation to FDH and coenzyme could guide us to clarify the uncharted particulars in the C1 compounds reaction process with carbon dioxide as a substrate, which also benefits the performance improvement of FDH. Although various works have investigated the conformational changes and kinetic behavior of binding using cryo-EM (Sun et al., 2018), DNA origami (Chen et al., 2018), fluorescence microscopy (Bornscheuer, 2016), and force spectroscopy (Pelz et al., 2016), the knowledge of how charge transport through coenzyme coupling correlates to their bioactivities remains missing owing to the technical challenges in the characterization of charge transport through bioactive enzymes (Fried and Boxer, 2017).
The single-molecule measurement technique, originally designed to investigate the charge transport through the small organic compound (Xu and Tao, 2003, Venkataraman et al., 2006) and later applied to the study of biomolecules such as DNA (Huang et al., 2010, Sha et al., 2018), peptides (Sepunaru et al., 2015, Zhao et al., 2014, Aradhya and Venkataraman, 2013), and protein (Ruiz et al., 2017, Fereiro et al., 2018b), may offer new opportunity to solve the long-term challenge for the correlation of bioactivity and conductance at the single protein level. The scanning tunneling microscope break junction (STM-BJ) technique, achieved by forming thousands of single-molecular junctions repeatedly and rapidly, has been widely used in conductance-structure correlation studies to reveal the effect of intramolecular and intermolecular interaction on the single-molecule conductance (Li et al., 2020, Huang et al., 2019, Sha et al., 2018).
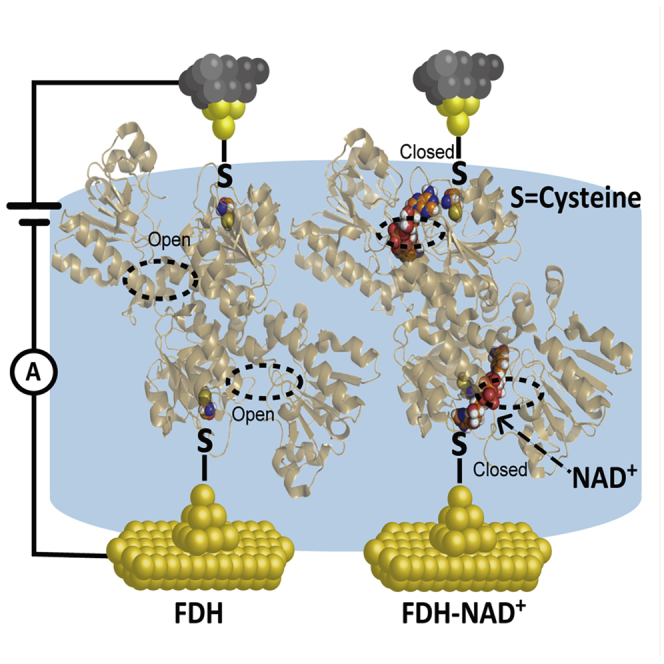
The binding of NAD+ to FDH is through hydrogen bonds between the FDH and NAD+ and provides the activity sites for the reaction (Schirwitz et al., 2010). To investigate the coupling between the FDH and NAD+, we employ the STM-BJ technique to capture the charge transport through single active FDH systems. The active FDH was immobilized between two gold electrodes using the Au-S bond from the L-cysteines (Figure 1A). Two typical coenzymes, NAD+ and nicotinamide adenine dinucleotide phosphate (NADP+) (Figure 1B) were then added into the solution to investigate the correlation between the bioactivities and their charge transport abilities. More importantly, our results provide the first evidence of charge transport and interaction through a single enzyme system and reveal the correlation with the biological functions, which offers a fundamental understanding to design and develop biocatalysis and bioelectronics at the essential molecular level. We also believe that the simple approach developed in this work will be applied to subsequent biophysics studies to understand the electric role of enzymes in various systems.
Figure 1.
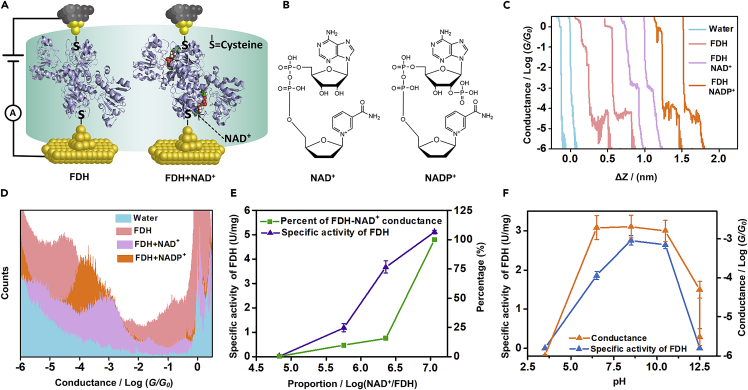
Schematic of STM-BJ and the Molecular Junction and Single-Molecule Conductance Results from STM-BJ Experiments at the Indicated Voltage of 100 mV (Substrate Positive)
(A and B) (A) Schematics of single-molecule FDH and FDH-NAD+ junctions, (B) The structure of NAD+ and NADP+.
(C and D) (C) Typical individual traces for single-molecule conductance measurement of pure solvent (blue), FDH (pink), FDH-NAD+ (purple), and FDH-NADP+ (orange), (D) One-dimensional (1D) conductance histograms of pure solvent (blue), FDH (pink), FDH-NAD+ (purple) and FDH-NADP+ (orange).
(E and F) (E) Plot of conductance in different proportion of FDH and NAD+, the square (green) and triangle (purple) mean the percentage of FDH-NAD+ conductances counted from the total individual traces and the specific activity of FDH in the catalytic reaction in different proportion of FDH to NAD+. (F) Plot of FDH-NAD+ conductance and specific activity in the buffers with different pH. The solid triangle (blue) and solid triangle (orange) mean the specific activity of FDH in the catalytic reaction and FDH-NAD+ conductance in the buffers with different pH, respectively.
See also Figures S1–S4, S6, and S8, Tables S1–S3.
Results and Discussion
Single-Molecule Conductance Measurement of FDH and FDH-NAD+
STM-BJ experiments were carried out in an aqueous solution to investigate the charge transport through single FDH junctions, and a constant bias potential of 100 mV (substrate positive) was applied between the substrate and gold tip covered with an apiezon wax to minimize the leakage current from the solution (STM-BJ setup was shown in Figure S1). The typical individual traces for the conductance of aqueous medium, FDH, FDH-NAD+, and FDH-NADP+ are shown in Figure 1C (see Figure S2 for the control experiment of FDH without the two-terminal L-cysteines to reveal the binding through L-cysteines). During the breaking process, the gold tip moved upward from the substrate until a single gold-gold atomic junction state formed with a plateau at the conductance quantum of G0. After that, a molecular plateau forms, suggesting the formation of FDH (pink) junctions with conductance at ∼10−4 G0. After adding NADP+ and NAD+ to the FDH aqueous solution, we found that the conductance shows a slight increase for NADP+ but increased significantly to ∼10−3 G0 for NAD+ (Figure 1C, orange and purple), suggesting that the coupling between FDH and coenzymes may boost the charge transport process.
To determine the conductance of single FDH junctions, the one-dimensional (1D) conductance histograms of FDH and FDH-NADP+ are constructed from 3,087 to 2,486 individual traces, respectively. The conductance of single FDH junction at 10−4.28 G0 (Figure 1D, pink) was consistent with that obtained in single-protein measurement through the cysteine-Au junction by using electrochemistry-scanning tunneling microscope (EC-STM) (Ruiz et al., 2017), and similar to the charge transport measurement of oligopeptide (Brisendine et al., 2018) and DNA (Xiang et al., 2017) using STM-BJ, which suggests the enzyme was also detectable in single-molecule scale. However, compared with the technique of EC-STM (Ruiz et al., 2017), and nanowire (Fereiro et al., 2018b) based on the self-assembled monolayer (SAM), we provide the first demonstration of the single-molecule break junction technique for investigating charge transport through a single active enzyme junction. And, it was also found that the charge transport of the protein in the aqueous solution was enhanced than that in the solid state (Mukhopadhyay et al., 2015), which may be from the formation of the intermolecular and intramolecular H-bond (Chen et al., 2016). It was also found that the addition of NAD+ leads to the conductance enhancement to 10−2.96 G0, which is 2,100% higher than single FDH junctions. A similar observation was also found (Amdursky et al., 2012) that the solid-state monolayer was constructed to measure the charge transport performance of protein at low temperature that electron transport across serum albumin was markedly enhanced with the addition of retinoate. As a control experiment, the charge transport through the assembly of FDH and another coenzyme, NADP+, (structure shown in Figure 1B) was also examined. The conductance measurement shows that, after adding NADP+, compared with the conductance of FDH (Figure 1C, 10−4.28 G0), a plateau with most-probable conductance at 10−3.93 G0 formed after the addition of NADP+ to FDH aqueous solutions (Figures 1C and 1D, orange), and the conductance enhancement is much less pronounced, which is almost one order of magnitude lower than that of NAD+. It was reported that the Y194 amino acid of FDH made contact with adenine, D282 and R174 amino acid bound to the nicotinamide group, and the phosphate linker in NAD+, respectively (Schirwitz et al., 2010). The phosphate group combined with the ribose made NADP+ unable to act as the coenzyme of FDH in our work owing to the steric hindrance (Schirwitz et al., 2010) and charge repulsion (Andreadeli et al., 2010), which is also confirmed by the control experiment of dehydrogenation reaction that the reaction could not be catalyzed by FDH when the coenzyme was NADP+ (as shown in Figure S2D). Although NADP+ may also interact with the surface of FDH, the significant lower conductance increase than that of NAD+ suggests that the matching of NAD+ to the active center of FDH provides the better electric coupling between FDH and NAD+ with some specific amino acid residues through the H-bond, which has been proved to be the essential step for the biocatalytic reaction before the binding of the substrate (Guo et al., 2016).
Correlation between the Conductance and Enzyme Activity
To provide further insight into the coupling between FDH and NAD+, the conductance measurement of different proportions of FDH and NAD+ was carried out by changing the concentration of NAD+ (see Figures S3A and S3B for the typical individual traces and 1D conductance histograms). Two-dimensional (2D, Figure S3C) histograms (Liu et al., 2019) demonstrated that there is a conductance intensity cloud for FDH without NAD+ and with 1:67,227 NAD+ (F1). Interestingly, the conductance intensity cloud of FDH-NAD+ (10−2.88 G0) appeared when the proportion changed to 1:571,429 (F2) and 1:2,285,714 (F3). Finally, the plateau of 10−4.12 G0 disappeared when the proportion changed to 1:11,428,571 (F4). We further analyze the percentage of individual conductance-distance traces with low (FDH) and high (FDH-NAD+) conductance features at different FDH-NAD+ ratios. As shown in Figure 1E, as the proportion of NAD+ increased, the percentage of traces with FDH-NAD+ features in total traces increased from 0% at the ratio of 1:67,227 to 99.87% at 1:11,428,571, revealing the complete coupling of FDH with excess NAD+.
To further correlate the charge transport investigations with their bio function, we further examine the enzyme activity through NADH absorbance at 340 nm. More interestingly, it was found that the enzyme activity was also enhanced with an increased proportion of FDH to NAD+, and the trend is highly correlated to the concentration-dependent percentage of high conductance traces in total individual traces during the conductance measurements (Figure 1E). Furthermore, the conductance of FDH-NAD+ in the buffer with different pH was measured and shown in Figure 1F (see Figures S4A and S4B for more details). The conductance of FDH-NAD+ in the buffer with pH of 6.5, 8.5, and 10.5 was around 10−2.7 G0, which is similar to that mentioned above, whereas the conductance decreased when pH rises to 12.5 or declines to 3.5. More importantly, the enzyme activity has the same varying tendency with conductance in these five different pH buffers, suggesting the correlation between the bioactivity and the charge transport characterizations through single active enzyme junctions.
Theoretical Investigations of Coenzyme Coupling
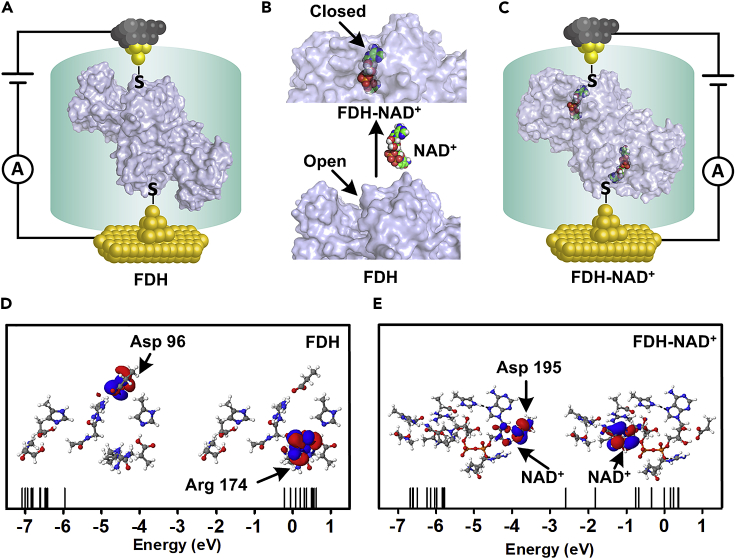
To understand the correlation between the charge transport properties and the bioactivity of FDH-NAD+ assembly, the geometries of FDH and FDH-NAD+ derived from the X-ray crystallographic data (PDB IDs: 5DNA and 5DN9) (Guo et al., 2016) are shown in Figures 2A–2C. It is found that the active center of the FDH shows an open conformation (Figure 2B, FDH) before the entry of NAD+ and is bound to the corresponding amino acid residue through H-bonding. After the entry of NAD+, the active center of FDH was filled with NAD+ and then the active center got closed (Figure 2B, FDH-NAD+). To provide further insight into the electric coupling of this assembly process, DFT calculations of these structures were performed by using the B3LYP to reveal the electron density distribution of the frontier orbitals of FDH and FDH-NAD+ (Ruiz et al., 2017, Fereiro et al., 2018a) (see the Transparent Methods for more details). The key amino acid residues in the active site of the enzyme, which interacts with NAD+ in the self-assembly process, were considered in the quantum calculations (Figures 2D and 2E). In the NAD+-absent FDH model (Figure 2D), the highest occupied molecular orbital (HOMO) in the FDH model was localized mostly on Asp96 and the lowest unoccupied molecular orbital (LUMO) was distributed mostly on Arg174, with a HOMO-LUMO gap of 5.74 eV. After the addition of NAD+, the HOMO was localized on NAD+ and Asp195, which formed obvious H-bonding interactions with the NAD+, and the LUMO was shifted onto NAD+ (Figure 2E). Interestingly, the energy levels of the HOMO in FDH (−5.95 eV) and FDH-NAD+ (−5.78 eV) were quite close to each other, whereas the energy of the LUMO in FDH-NAD+ (−2.59 eV) was much lower than that in the FDH model (−0.22 eV), leading to the reduction of HOMO-LUMO gap to 3.22 eV. All these results support our experimental finding that the addition of NAD+ could significantly enhance conductance.
Figure 2.
Theoretical Calculation
The schematic diagram of geometry structure of (A) FDH, (B) partial schematic of FDH, FDH-NAD+, and (C) FDH-NAD+. The Electronic structure calculations of (D) FDH and (E) FDH-NAD+ and their comparison in HOMO and LUMO; dashed line (black) represents the H-bond.
The Role of the Coenzyme in Charge Transport Pathway
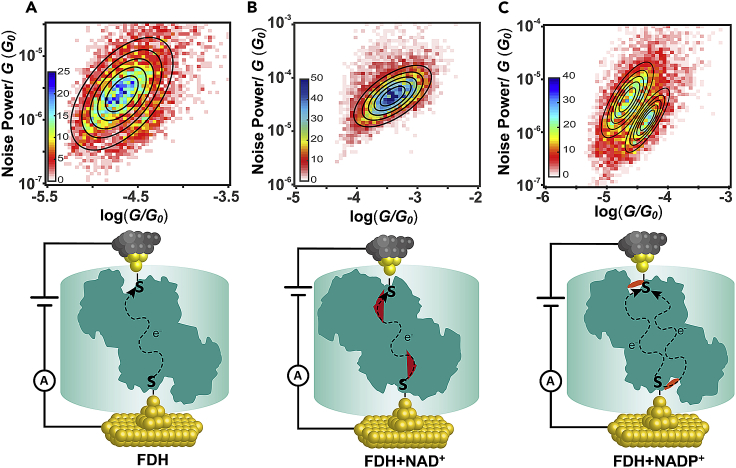
To further reveal the role of coenzymes in the charge transport process, we performed flicker noise analysis to investigate the transmission path of charge transport through FDH and FDH-NAD+ (the experiment and analysis process were described in detail in Transparent Methods). The junction elongation process was paused for 150 ms at the position where the molecule junction is likely to be formed between the gold tip and substrate, and the conductance signals during the period were transformed by the Fourier equation for noise analysis. According to previous results (Tang et al., 2019, Rousseau et al., 2018, Bostick et al., 2018), after integration from 100 to 1,000 Hz, the power spectral density was normalized by the average conductance from the cut-out conductance step. The 2D histograms of normalized flicker noise power versus average conductance are plotted from nearly eight thousand such traces. Then, each 2D histogram fitting was performed by using 2D Gaussian, and the black cycles represent the contour lines of the fitted Gaussian distribution equation. The noise power of the tunneling junction scaled as G2.0, which results from complete through-space transmission and complete through-bond transmission with noise power as G1.0. As shown in Figure 3A and Figure S5B, the noise power of FDH scales as G1.9, suggesting that through-space transmission dominates the charge transport through FDH. Some computational studies (Rousseau et al., 2018, Bostick et al., 2018) reported that through-space was the preferred mode that exists in the charge transport process of the protein itself, which supports the results of this study as well. However, for FDH-NAD+ (Figures 3B and S5C), its noise power was scaled as G1.6 with a bias toward through-bond transmission. The transition of the transmission path from the through-space path to the through-bond path suggests that the inserted NAD+ is involved in the charge transport path through the FDH-NAD+ assembly, which coincides with the DFT calculations that the coupling of NAD+ provides the LUMO and reduces the HOMO-LUMO gap. Interestingly, both through-space and through-bond transmission exists in the charge transport through FDH-NADP+ in the flicker noise analysis process of FDH-NADP+ (Figure 3C). The appearance of both transmission paths revealed that, although NADP+ could not enter the active center of FDH, NADP+ could adsorb to the surface of FDH and form paths that have competitive conductance with initial through-space paths FDH (Figure 3C). The significant different charge transport paths among FDH, FDH-NAD+, and FDH-NADP+ reveal the essential role of the coenzyme in the correlation between the conductance of single active FDH junctions and their bioactivities.
Figure 3.
Flicker Noise Analysis
The conductance data used for flicker noise analysis were also collected from the STM instrument. The gold tip moved downward and forms Au-molecule-Au junction, the junction was held for 150 ms at the position where the molecule junction is likely to be formed, which was controlled by the program automatically. Two-dimensional histogram of normalized flicker noise power versus average conductance for (A) FDH, (B) FDH-NAD+, and (C) FDH-NADP+. And the schematic diagram of the electron transfer process in FDH, FDH-NAD+, FDH-NADP+ were listed under the two-dimensional histogram accordingly.
See also Figure S5.
Conductance Signifies the Effect Derived from Hydrogen Bonds on Enzyme Activity
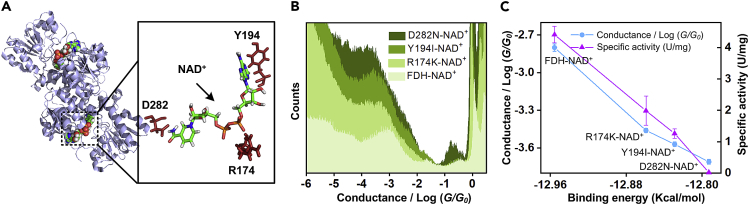
The results above demonstrated the strong correlation between bioactivity and the charge transport and the charge transport enhancement contributed by the coupling of NAD+ to FDH through H-bonds. However, how the H-bonds exert their effects on conductance and enzyme activity was still ambiguous and needed further study. Thus, as shown in the partial schematic diagram (Figure 4A) of NAD+-binding site in FDH from PDB (ID: 5DN9) (Schirwitz et al., 2010, Guo et al., 2016), the three amino acid residues of R174, D282, and Y194 were replaced by K (lysine), N (asparagine), and I (isoleucine) by using site-directed mutagenesis, respectively, to construct R174K-FDH, Y194I-FDH, and D282N-FDH, in which the interaction force of H-bond between amino acid residues in mutant and NAD+ were weakened. As shown in Figure 4B, the conductance of the four FDH-NAD+ (three mutants and a native one) were different, with an order of the contribution from different sites of H-bond, FDH-NAD+ (10−2.8 G0) > R174K-NAD+ (10−3.46 G0) > Y194I-NAD+ (10−3.57 G0) > D282N-NAD+ (10−3.71 G0). Intriguingly, Figure 4C portrays the strong correlation between enzymic activity and conductance in four FDH-NAD+. Nevertheless, the strong correlation between conductance and enzymic activity was not a coincidence but corollary caused by site change of H-bond. As shown in Figure 4C (blue line), the weakening of H-bond in different sites of FDH-NAD+ shows the different effects on the binding energy through calculation (detailed in Transparent Methods). As a result, the reduction of the binding energy leads to a better affinity of FDH to NAD+ and the higher enzymic activity (Liu et al., 2004, Pan et al., 2007). At the same time, it was also consistent with the result reported that weakened or broken H-bond in the catalytic center lowers or even almost completely abrogates catalysis in the enzyme (Dai et al., 2019). Furthermore, as shown in Figure 4C (red line), the correlation between binding energy and conductance was also same with that of enzymic activity-binding energy, which was consistent with the result reported of lower binding energy as higher conductance in single-molecule electron transfer process (Quek et al., 2009, Babanova et al., 2015). We can conclude that the weakening of H-bond in different sites of NAD+ binding center led to changes of binding energy and therefore makes the enzymic activity various at last, which was reflected in the conductance changes due to effect derived from binding energy.
Figure 4.
The Relationship among Conductance, Binding Energy, and Specific Activity
(A) Crystal structure of FDH-NAD+. The inset shows the NAD+ and circumjacent amino acid residue interacted through H-bond.
(B) One-dimensional (1D) conductance histograms of FDH-NAD+ (light green), R174K-NAD+ (jade green), Y194I-NAD+ (green), and D282N-NAD+ (dark green).
(C) Plot of different mutant-NAD+ conductance and enzyme activity in the different binding energy.
See also Figures S7 and S9.
Overall, we investigated the effect of NAD+ concentration and reveal that the percentage of FDH-NAD+ (10-2.96 G0) and specific activity increased with the increase of NAD+ concentration. The pH experiment also demonstrated the great consistent of conductance and specific activity in diferent pH values. These results suggest that NAD+ contributed to not only the changes but also the correlation between conductance and specific activity. To further verify the correlation between conductance and bioactivity, another experiment was carried out, in which the interaction force of H-bond between amino acid residues in three mutants and NAD+ was weakened by using the site-directed mutagenesis technique. At last, the variation consistency of three mutants in conductance and specific activity (Figure 4C) demonstrated the strong correlation between charge transport and bioactivity.
The site-specific mutagenesis analysis also demonstrates that the bioactivity of FDH has been optimized, as alternative mutations generate enzymes with lower bioactivities. In nature, such optimization has been achieved by the process called natural selection (Futuyma, 2013), which kept mutations with the highest bioactivity while removing any mutations that have lower bioactivities. Furthermore, our study shows that those mutations having lower bioactivities are also with lower conductance and higher binding energy. Therefore, our study demonstrates the pathway of optimization, which is through maximizing the conductance of the charge transport while minimizing the energy of enzyme binding. The optimal bioactivity of FDH has been achieved here, because natural selection is most effective for microbes, which has a large population size and short generation time (Nielsen and Slatkin, 2013). Nevertheless, the more general scenarios would be when natural selection is not so effective and constrained by small population sizes, long generation times, and the linkage structure of genes. Understanding the pathway of optimization provides opportunities to breakdown the constraints on natural selection and evolution and generate innovations in enzyme activity that surpass the evolutionary innovations by nature.
Discussion
In conclusion, we investigated charge transport through single active FDH junctions, and a more than 2,100% increase in conductance was observed owing to the insertion of NAD+ into the active center of FDH. More importantly, the investigation of charge transport of the FDH at different pH and with different coenzymes are highly correlated with their biocatalytic activities in dehydrogenation. To understand the role of coenzyme in the charge transport through FDH junctions, the flicker noise analysis revealed the switching from the through-space pathway in FDH junctions to the through-bond pathway after the binding of NAD+, which supports the density functional theory (DFT) calculation that the NAD+-induced LUMO significantly reduced the HOMO-LUMO gap and thus became the dominant charge transport pathway. We further demonstrate that the charge transport and the bioactivity of the enzyme are bridged by the binding energy of the H-bond, which has been optimized via the natural selection process. The bridge between the charge transport and bio-functions revealed by single enzyme analysis studies in this work offers new insight for the future design and development of bioelectronics and biocatalysis from the molecular level.
Limitations of the Study
We believe that the single-enzyme conductance highly correlates with the enzyme activity. However, as limited by the complicated structure of FDH, the theoretical calculation in this work only considered the active center in this work. Thus, a more complete calculation may be needed to better understand the charge transport process. Further investigation was also essential for guidelines in the futher design and modification of FDH to obtain higher bioactive mutants, which is also challenging at the current stage.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 21978245, 21673195, 21503179, 21490573) and the National Key R&D Program of China (2017YFA0204902). W.H. also acknowledged Dr. Yuan-Y.Z. from College of Environment & Ecology, Xiamen University for the discussion on the mechanism of the evolutions.
Author Contributions
W.H. and B.F. designed the experiments and co-supervised the project. X.Z., A.Z., and S.Q. wrote the manuscript with inputs from all authors. X.Z. and Y.Z were responsible for molecular synthesis and characterization. X.Z. and A.Z. carried out the break junction experiments and analyzed the data. C.T. carried out the flicker noise analysis. S.Z. built the electrical measurement instrument and wrote the software to control the break junction setup. S.Q. and B.W. performed the theoretical modeling. Y.W., Y.Z., and H.L performed the binding affinities calculation. All authors conceived the work and discussed the experiments.
Declaration of Interests
The authors declare no competing financial interests.
Published: April 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101001.
Contributor Information
Baishan Fang, Email: fbs@xmu.edu.cn.
Wenjing Hong, Email: whong@xmu.edu.cn.
Supplemental Information
References
- Amdursky N., Pecht I., Sheves M., Cahen D. Doping human serum albumin with retinoate markedly enhances electron transport across the protein. J. Am. Chem. Soc. 2012;134:18221–18224. doi: 10.1021/ja308953q. [DOI] [PubMed] [Google Scholar]
- Andreadeli A., Platis D., Tishkov V., Popov V., Labrou N.E. Structure-guided alteration of coenzyme specificity of formate dehydrogenase by saturation mutagenesis to enable efficient utilization of NADP(+) FEBS J. 2010;275:3859–3869. doi: 10.1111/j.1742-4658.2008.06533.x. [DOI] [PubMed] [Google Scholar]
- Aradhya S.V., Venkataraman L. Single-molecule junctions beyond electronic transport. Nat. Nanotechnol. 2013;8:399–410. doi: 10.1038/nnano.2013.91. [DOI] [PubMed] [Google Scholar]
- Babanova S., Matanovic I., Chavez M.S., Atanassov P. Role of quinones in electron transfer of PQQ-glucose dehydrogenase anodes-mediation or orientation effect. J. Am. Chem. Soc. 2015;137:7754–7762. doi: 10.1021/jacs.5b03053. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U.T. Chemical biology a radical change in enzyme catalysis. Nature. 2016;540:345–346. doi: 10.1038/540345a. [DOI] [PubMed] [Google Scholar]
- Bostick C.D., Mukhopadhyay S., Pecht I., Sheves M., Cahen D., Lederman D. Protein bioelectronics: a review of what we do and do not know. Rep. Prog. Phys. 2018;81:026601. doi: 10.1088/1361-6633/aa85f2. [DOI] [PubMed] [Google Scholar]
- Brisendine J.M., Abramson S.R., Liu Z.F., Cui J., Ng F., Neaton J.B., Koder R.L., Venkataraman L. Probing charge transport through peptide bonds. J. Phys. Chem. Lett. 2018;9:763–767. doi: 10.1021/acs.jpclett.8b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2:e1501240. doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Ke G.L., Ma Y.L., Zhu Z., Liu M.H., Liu Y., Yan H., Yang C.J. A synthetic light-driven substrate channeling system for precise regulation of enzyme cascade activity based on DNA origami. J. Am. Chem. Soc. 2018;140:8990–8996. doi: 10.1021/jacs.8b05429. [DOI] [PubMed] [Google Scholar]
- Dai S.B., Funk L.M., von Pappenheim F.R., Sautner V., Paulikat M., Schröder B., Uranga J., Mata R.A., Tittmann K. Low-barrier hydrogen bonds in enzyme cooperativity. Nature. 2019;573:609–613. doi: 10.1038/s41586-019-1581-9. [DOI] [PubMed] [Google Scholar]
- Fereiro J.A., Yu X., Pecht I., Sheves M., Cahen D. Tunneling explains efficient electron transport via protein junctions. Proc. Natl. Acad. Sci. U S A. 2018;115:4577–4583. doi: 10.1073/pnas.1719867115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereiro J.A., Porat G., Bendikov T., Pecht I., Sheves M., Cahen D. Protein electronics: chemical modulation of contacts control energy level alignment in gold-azurin-gold junctions. J. Am. Chem. Soc. 2018;140:13317–13326. doi: 10.1021/jacs.8b07742. [DOI] [PubMed] [Google Scholar]
- Fried S.D., Boxer S.G. Electric fields and enzyme catalysis. Annu. Rev. Biochem. 2017;86:387–415. doi: 10.1146/annurev-biochem-061516-044432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma D.J. Sinauer Associates Press; 2013. Evolution. [Google Scholar]
- Gleizer S., Ben-Nissan R., Bar-On Y.M., Antonovsky N., Noor E., Zohar Y., Jona G., Krieger E., Shamshoum M., Bar-Even A. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell. 2019;179:1255–1263. doi: 10.1016/j.cell.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Gakhar L., Wickersham K., Francis K., Kilshtain A.V., Major D.T., Cheatum C.M., Kohen A. Structural and kinetic studies of formate dehydrogenase from Candida boidinii. Biochemistry. 2016;55:2760–2771. doi: 10.1021/acs.biochem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose A., Kasai T., Aoki M., Umemura T., Kouzuma A. Electrochemically active bacteria sense electrode potentials for regulating catabolic pathways. Nat. Commun. 2018;9:1083. doi: 10.1038/s41467-018-03416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.Y., Tang C., Li J.Q., Chen L.C., Zheng J.T., Zhang P., Le J.B., Li R.H., Li X.H., Liu J.Y. Electric field-induced selective catalysis of single-molecule reaction. Sci. Adv. 2019;5:eaaw3072. doi: 10.1126/sciadv.aaw3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., He J., Chang S.A., Zhang P.M., Liang F., Li S.Q., Tuchband M., Fuhrmann A., Ros R., Lindsay S. Identifying single bases in a DNA oligomer with electron tunnelling. Nat. Nanotechnol. 2010;5:868–873. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayathilake B.S., Bhattacharya S., Vaidehi N., Narayanan S.R. Efficient and selective electrochemically driven enzyme-catalyzed reduction of carbon dioxide to formate using formate dehydrogenase and an artificial cofactor. Acc. Chem. Res. 2019;52:676–685. doi: 10.1021/acs.accounts.8b00551. [DOI] [PubMed] [Google Scholar]
- Jiang W., Xu C.Z., Jiang S.Z., Zhang T.D., Wang S.Z., Fang B.S. Establishing a mathematical equations and improving the production of l-tert-leucine by uniform design and regression analysis. Appl. Biochem. Biotechnol. 2017;18:1454–1464. doi: 10.1007/s12010-016-2295-1. [DOI] [PubMed] [Google Scholar]
- Li X.H., Wu Q.Q., Bai J., Hou S.J., Jiang W.L., Tang C., Song H., Huang X.J., Zheng J.T., Yang Y. Structure-independent conductance of thiophene-based single-stacking junctions. Angew. Chem. Int. Ed. 2020 doi: 10.1002/anie.201913344. [DOI] [PubMed] [Google Scholar]
- Liu J.Y., Zhao X.T., Zheng J.T., Huang X.Y., Tang Y.X., Wang F., Li R.H., Pi J.C., Huang C.C., Wang L. Transition from tunneling leakage current to molecular tunneling in single-molecule junctions. Chem. 2019;5:390–401. [Google Scholar]
- Liu Z.J., Dominy B.N., Shakhnovich E.I. Structural mining: self-consistent design on flexible protein-peptide docking and transferable binding affinity potential. J. Am. Chem. Soc. 2004;126:8515–8528. doi: 10.1021/ja032018q. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Dutta S., Pecht I., Sheves M., Cahen D. Conjugated cofactor enables efficient temperature-independent electronic transport across similar to 6 nm long halorhodopsin. J. Am. Chem. Soc. 2015;137:11226–11229. doi: 10.1021/jacs.5b06501. [DOI] [PubMed] [Google Scholar]
- Nielsen C.F., Lange L., Meyer A.S. Classification and enzyme kinetics of formate dehydrogenases for biomanufacturing via CO2 utilization. Biotechnol. Adv. 2019;37:107408. doi: 10.1016/j.biotechadv.2019.06.007. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Slatkin M. Sinauer Associates Press; 2013. An Introduction to Population Genetics: Theory and Applications. [Google Scholar]
- Pan Y.M., Gao D.Q., Yang W.C., Cho H., Zhan C.G. Free energy perturbation (FEP) simulation on the transition states of cocaine hydrolysis catalyzed by human butyrylcholinesterase and its mutants. J. Am. Chem. Soc. 2007;129:13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz B., Zoldak G., Zeller F., Zacharias M., Rief M. Subnanometre enzyme mechanics probed by single-molecule force spectroscopy. Nat. Commun. 2016;7:10848. doi: 10.1038/ncomms10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek S.Y., Kamenetska M., Steigerwald M.L., Choi H.J., Louie S.G., Hybertsen M.S., Neaton J.B., Venkataraman L. Mechanically controlled binary conductance switching of a single-molecule junction. Nat. Nanotechnol. 2009;4:230–234. doi: 10.1038/nnano.2009.10. [DOI] [PubMed] [Google Scholar]
- Rousseau R., Shafei S., Migliore A., Stanley R.J., Beratan D.N. Determinants of photolyase's DNA repair mechanism in mesophiles and extremophiles. J. Am. Chem. Soc. 2018;140:2853–2861. doi: 10.1021/jacs.7b11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M.P., Aragones A.C., Camarero N., Vilhena J.G., Ortega M., Zotti L.A., Pérez R., Cuevas J.C., Gorostiza P., Díez-Pérez I. Bioengineering a single-protein junction. J. Am. Chem. Soc. 2017;139:15337–15346. doi: 10.1021/jacs.7b06130. [DOI] [PubMed] [Google Scholar]
- Schirwitz K., Schmidt A., Lamzin V.S. High-resolution structures of formate dehydrogenase from Candida boidinii. Protein Sci. 2010;16:1146–1156. doi: 10.1110/ps.062741707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepunaru L., Refaely-Abramson S., Lovrincic R., Gavrilov Y., Agrawal P., Levy Y., Kronik L., Pecht I., Sheves M., Cahen D. Electronic transport via homopeptides: the role of side chains and secondary structure. J. Am. Chem. Soc. 2015;137:9617–9626. doi: 10.1021/jacs.5b03933. [DOI] [PubMed] [Google Scholar]
- Sha R.J., Xiang L.M., Liu C.R., Balaeff A., Zhang Y.Q., Zhang P., Li Y.Q., Beratan D.N., Tao N.J., Seeman N.C. Charge splitters and charge transport junctions based on guanine quadruplexes. Nat. Nanotechnol. 2018;13:316–321. doi: 10.1038/s41565-018-0070-x. [DOI] [PubMed] [Google Scholar]
- Sun C., Benlekbir S., Venkatakrishnan P., Wang Y.H., Hong S.J., Hosler J., Tajkhorshid E., Rubinstein J.L., Gennis R.B. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature. 2018;557:123–126. doi: 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Chen L.J., Zhang L.Y., Chen Z.X., Li G.P., Yan Z.W., Lin L.C., Liu J.Y., Huang L.F., Ye Y.L. Multicenter-bond-based quantum interference in charge transport through single-molecule carborane junctions multicenter-bond-based quantum interference in charge transport through single-molecule carborane junctions. Angew. Chem. Int. Ed. 2019;58:10601–10605. doi: 10.1002/anie.201904521. [DOI] [PubMed] [Google Scholar]
- Venkataraman L., Klare J.E., Nuckolls C., Hybertsen M.S., Steigerwald M.L. Dependence of single-molecule junction conductance on molecular conformation. Nature. 2006;442:904–907. doi: 10.1038/nature05037. [DOI] [PubMed] [Google Scholar]
- Xiang L.M., Palma J.L., Li Y.Q., Mujica V., Ratner M.A., Tao N.J. Gate-controlled conductance switching in DNA. Nat. Commun. 2017;8:14471. doi: 10.1038/ncomms14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Tao N.J. Measurement of single-molecule resistance by repeated formation of molecular junctions. Science. 2003;301:1221–1223. doi: 10.1126/science.1087481. [DOI] [PubMed] [Google Scholar]
- Zhao Y.N., Ashcroft B., Zhang P.M., Liu H., Sen S.M., Song W.S., Im J.O., Gyarfas B., Manna S., Biswas S. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol. 2014;9:466–473. doi: 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.