Abstract
Purpose
Deformable image registration (DIR) of longitudinal liver cancer computed tomographic (CT) images can be challenging owing to anatomic changes caused by radiation therapy (RT) or disease progression. We propose a workflow for the DIR of longitudinal contrast-enhanced CT scans of liver cancer based on a biomechanical model of the liver driven by boundary conditions on the liver surface and centerline of an autosegmentation of the vasculature.
Methods and Materials
Pre- and post-RT CT scans acquired with a median gap of 112 (32-217) days for 28 patients who underwent RT for intrahepatic cholangiocarcinoma were retrospectively analyzed. For each patient, 5 corresponding anatomic landmarks in pre- and post-RT scans were identified in the liver by a clinical expert for evaluation of the accuracy of different DIR strategies. The first strategy corresponded to the use of a biomechanical model-based DIR method with boundary conditions specified on the liver surface (BM_DIR). The second strategy corresponded to the use of an expansion of BM_DIR consisting of the auto-segmentation of the liver vasculature to determine additional boundary conditions in the biomechanical model (BM_DIR_VBC). The 2 strategies were also compared with an intensity-based DIR strategy using a Demons algorithms.
Results
The group mean target registration errors were 12.4 ± 7.5, 7.7 ± 3.7 and 4.4 ± 2.5 mm, for the Demons, BM_DIR and BM_DIR_VBC, respectively.
Conclusions
In regard to the large and complex deformation observed in this study and the achieved accuracy of 4.4 mm, the proposed BM_DIR_VBC method might reveal itself as a valuable tool in future studies on the relationship between delivered dose and treatment outcome.
Introduction
The incidence of liver and intrahepatic bile duct cancers in the United States continues to increase with 42,220 estimated new cases in 2018.1 Resection of the tumor remains the best therapeutic option but can be offered to only a minority of patients who presents with favorable tumor localization or size and health condition. In recent years, radiation therapy (RT) has continued to gain interest as a treatment alternative owing to the development of both highly conformal techniques such as intensity modulated RT or stereotactic body RT and image guidance techniques with cone beam CT (CBCT) or CT on rails. The ability to image the liver for proper alignment of the patient to the linear accelerator has also enabled dose accumulation using deformable image registration (DIR) during the course of fractionated treatments.2, 3, 4 Accurate DIR between images acquired during the course of treatment for dose accumulation, but also between images acquired before and posttreatment, should lead to the development of better models of the relationship between delivered dose and treatment outcome.
A very limited number of studies have evaluated the accuracy of DIR methods for registration of longitudinal images of liver cancer treated with radiation.5 Registration of such images is challenging owing to tumor or normal tissue responses to radiation and sometimes chemotherapy, which may create inconsistent content between the images and perturb direct intensity or mutual information-based DIR methods. In this case, methods based on the registration of the liver surface only, allowing to disregard changes in intensity, texture, or complex tumor regression schemes, may prove themselves to be more robust. Among approaches using organ contours, the use of a multiorgan biomechanical model-based DIR method, Morfeus,6 has previously been proposed for registration of liver images acquired in radiation therapy treatment protocols.7 This method has been evaluated for the mapping of the tumor from a planning CT to daily CBCT. A similar approach has been investigated for tumor mapping between CT scans acquired before and after thermal ablation.8 For dose accumulation, Morfeus was also used to register different respiratory phases of planning 4-dimensional CT or to register daily CBCT onto the planning CT.2
Quantification of the longitudinal DIR accuracy in terms of target registration error (TRE) was not feasible in these studies, as the visualization of the tumor and liver vasculature was only possible on the planning CT usually acquired after injection of contrast agent. Fukumitsu et al5 evaluated the use of 2 commercially available DIR solutions for radiation oncology when registering contrast enhanced-CT of the liver acquired before and after proton beam therapy. The quantification of the DIR accuracy was, however, limited for a series of patients to the measure of the misalignment of a single metallic marker located close to the tumor. Recently, an expansion of Morfeus to model the volumetric response to the dose of the liver tissues was proposed.9 The accuracy of the method to register CT scans pre- and post-RT of liver cancer was quantified for 7 patients by measuring the TRE of vessel bifurcation points. Modeling the dose response by adding dose boundary conditions in the biomechanic model allowed to significantly improve the DIR accuracy in comparison to using the standard model based on boundary conditions only on the liver surface. However, the TRE remained relatively large with an average error of 7.7 mm, indicating that in the presence of long-term dose-induced volumetric changes, the displacement of internal tissues may present a limited correlation with the displacements of the liver surface. Another challenge for DIR can come from the complexity of the tumor response, which can combine elastic shrinkage and “erosion” as it has been previously illustrated for lung tumors.10 To address this challenge for the registration of longitudinal lung cancer images, a previous study proposed to drive a biomechanic model of the lungs with boundary conditions specified on the vessels surrounding the tumor.11
The goal of this study was to propose a biomechanic model to improve the accuracy and robustness of DIR between pre- and post-RT of liver cancer images by including in a standard biomechanical DIR workflow an autosegmentation of the liver vasculature and the addition of boundary conditions on its centerline. The evaluation of the accuracy was performed on a data set of contrast-enhanced CTs from 28 patients who received radiation therapy and chemotherapy for cholangiocarcinoma.
Materials and Methods
Patient data
Under an institutional review board–approved protocol, 28 patients treated for cholangiocarcinoma with external radiation therapy were retrospectively analyzed. Twenty-one patients received concurrent chemotherapy. For each patient, contrast-enhanced CT scans were acquired with breathhold pre- and posttreatment on average 113 ± 35 days apart. The hepatic phases were selected for image registration. The spatial resolution ranged between 0.66 × 0.66 mm and 0.98 × 0.98 mm in the axial plane. For 22 of the patients, the slice spacing was 2.5 mm for both images, 2.5 and 5 mm for 5 of them, and 5 mm for one patient.
A fully convolutional neural network was used to automatically segment the liver on all images.12 These contours were manually edited for 11 patients. The tumor volume was manually contoured on all images.
For each pair of images, a clinical expert was asked to use the treatment planning system (RayStation version 6.99, RaySearch Laboratories, Stockholm, Sweden) to manually pick 5 corresponding anatomic landmarks spatially distributed in the liver as well as possible. These landmark pairs were used to evaluate the accuracy of the registration strategies.
Rigid alignment
For DIR of 3-dimensional (3D) images, the rigid initialization of the transform defined by translation and rotation parameters is well known to affect the result of standard methods. Three in-house implementations of different initial rigid registration strategies were considered in this study.
The first rigid registration method (Global) corresponded to a global alignment of the patient abdomen based on a classic maximization of the mutual information directly between the 2 abdomen CT scans using a gradient descent.
The second method (COM) corresponded to the translation aligning the center of mass of the liver volumes.
The third method (Chamfer) corresponded to the alignment of the liver surfaces using a Chamfer matching approach allowing rotations.13,14 A distance map was calculated from the pre-RT liver volume while a surface mesh of the post-RT liver was created. Iteratively, the translation and rotation of the mesh were updated after a gradient descent of the sum of the distances given by linear interpolation of the distance map at the locations of all points of the mesh.
Biomechanical model-based DIR of the liver
Standard biomechanical DIR (BM_DIR)
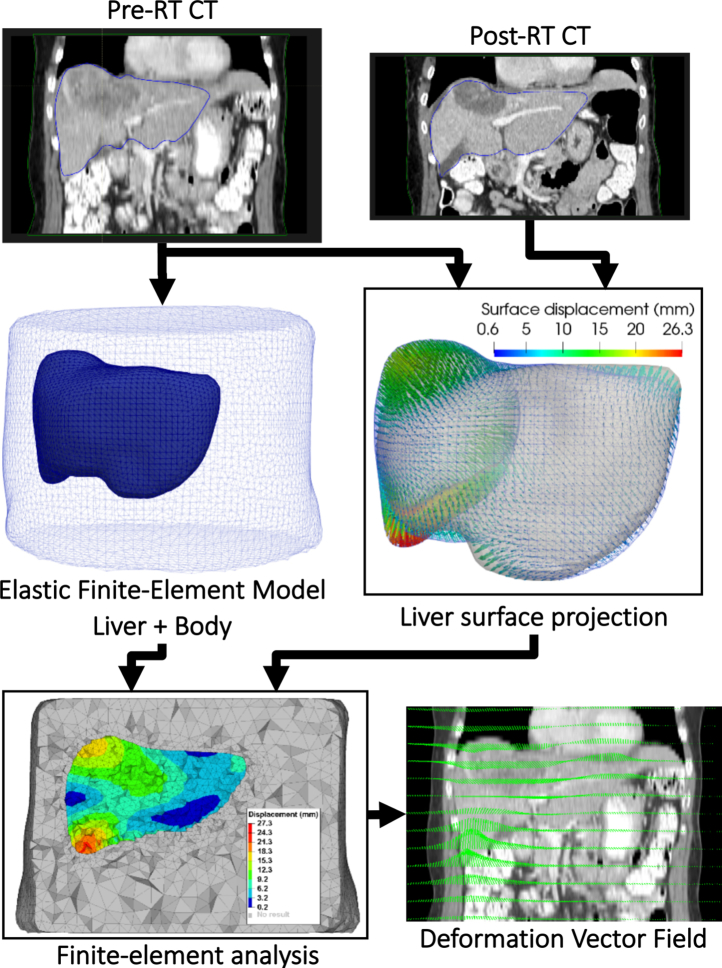
After any of the 3 rigid registration initialization, the workflow used for deformable registration of the liver is depicted Fig 1. First, the auto-segmentations of the liver in the 2 images were converted into smooth surface triangular meshes. The surface mesh corresponding to the pretreatment image was used to generate a solid tetrahedral finite-element model (FEM) of the liver and surrounding anatomy with linear elastic properties. A surface projection algorithm was used to determine the displacement of the surface nodes of the liver FEM. The algorithm considered in this workflow consisted of the computation of signed Euclidean distance maps from the pre- and post-RT liver segmentation and the deformable registration of these maps with a variant of the Demons algorithm.15 For each node of the liver surface in the FEM, a boundary condition as a constrained displacement was obtained by linear interpolation of the displacement vector field (DVF) calculated with the Demons algorithm. A finite-element analysis was then performed using the software Optistruct (Altair, Troy, MI) to solve the displacements of all the nodes of the FEM. Those displacements were finally resampled on the grid of the pre-RT CT to provide a dense DVF.
Figure 1.
Standard Biomechanical deformable image registration workflow.
Biomechanic model expansion: addition of vessels boundary conditions (BM_DIR_VBC)
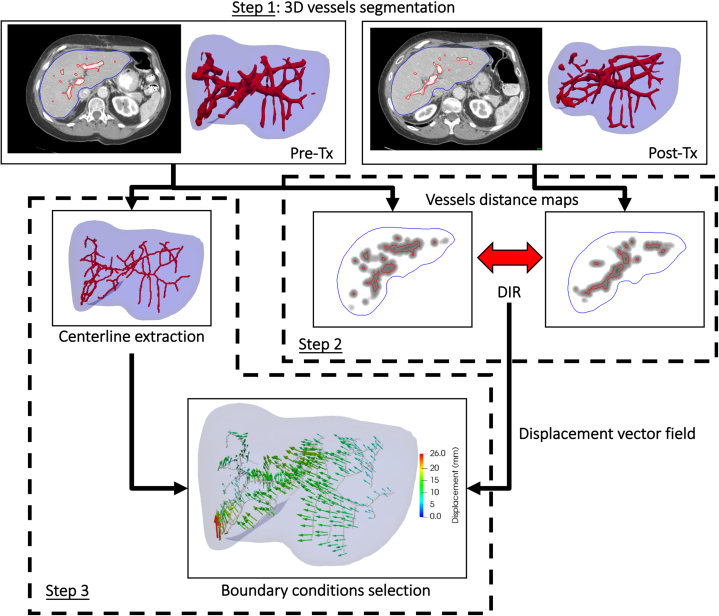
The workflow for the determination of additional boundary on the liver vasculature is shown in Fig 2, with the additional steps detailed below.
Figure 2.
Workflow of the determination of boundary conditions on the liver vasculature.
Step 1 consisted of autosegmenting the vasculature on the 2 CT scans to be registered. Numerous studies have investigated the segmentation of 3D vessels in medical images as reviewed by Lesage et al.16 A popular approach corresponds to the analysis of the eigenvalues of the 3D image Hessian matrix for each voxel. Different functions of the eigenvalues have been proposed to compute a vesselness image representing in each voxel the likelihood for this voxel to belong to a tubular structure.17, 18, 19, 20 To detect tubular structures of various diameters, the vesselness image is usually computed after different scales of a Gaussian smoothing of the original image. In this study, a multiscale vesselness filter based on the formulation by Frangi et al19 and available in the open-source Insight Toolkit (www.itk.org) was applied to the pre- and post-RT images with scales varying by steps of 1 mm from 3 to 7 mm. In a mask of the liver eroded by 5 mm, a first rough segmentation of the vasculature was obtained from the 2 vesselness images by segmenting the liver into 2 classes with an Otsu thresholding.21 Because this segmentation overestimated the volume containing vessels, a second thresholding was applied to it with a Hounsfield Unit lower value set at the mean value minus the standard deviation. A connectivity filter was then applied to keep only segmentation parts with a volume superior to 1000 voxels. Figure 2 shows an example of the segmentation results on 2-dimensional slices and the corresponding surfaces in 3D.
Step 2 consisted of using DIR to establish correspondences between the segmented vasculature volumes. A similar approach as the one described in the Biomechanic model expansion: addition of vessels boundary conditions section for the determination of correspondences on the liver surface was used. Signed squared Euclidean distance maps were calculated from each segmentation of the vasculature and registered using a Demons algorithm. Owing to frequent inconsistencies in the definition of the vasculature obtained for the 2 images, a higher than typical regularization of the DVF in the Demons algorithm was considered by setting the iterative Gaussian smoothing parameter to 2.5 mm.
In step 3, correspondences were selected to be used as boundary conditions in the biomechanical model. For this, the centerline of the vasculature of the pre-RT CT was first obtained using a skeletonization algorithm described by Lee et al.22 For all points of the centerline, the corresponding displacement was obtained by linear interpolation in the DVF calculated by the Demons. When the displaced point fell in the segmentation of the vasculature of the post-RT CT, the correspondence was judged reliable and selected. The displacement was otherwise considered uncertain and discarded. The selected correspondences served to determine boundary conditions to be applied to nodes of the liver tetrahedral mesh. The new boundary conditions were added to the ones determined previously on the liver surface before finite-element analysis.
Intensity based-DIR with the Demons algorithm
To provide an assessment of the difficulty to align the images with a typical intensity-based DIR image algorithm, all the images were also directly first rigidly aligned with the Global method and then registered using the Demons algorithm using 3 resolutions and the iterative Gaussian smoothing parameter set to 1 mm.
Comparison of the registration methods accuracy
For each patient, the Dice score of the liver and TRE were measured after each rigid registration method and after each combination of the rigid and deformable methods. Student 2-tailed paired t tests were performed to determine the statistical significance of the differences between the means obtained for the different methods.
Results
Volume changes
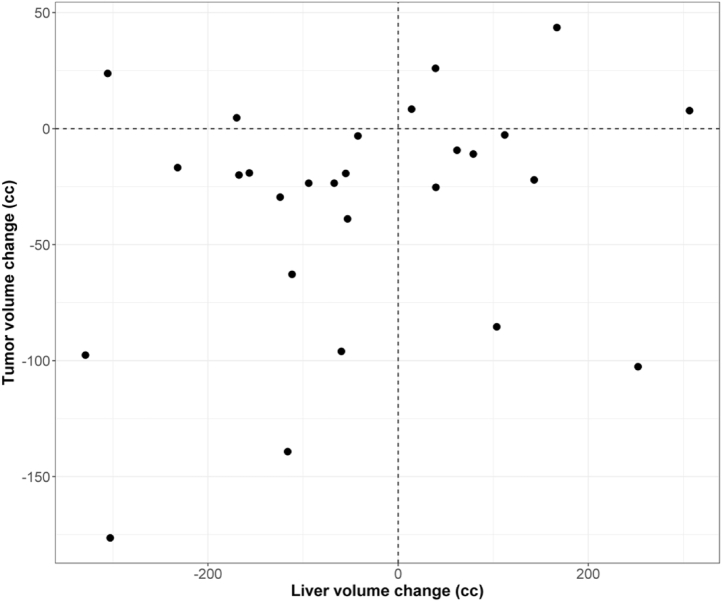
Figure 3 represents the distribution of volume change for the liver and tumor. Volumes changes ranged from –329 to 306 mL and –176 to 44 mL for the total liver and tumor, respectively. No clear relationship could be observed for this population of patients between the variations of the volume of the tumor and total liver, illustrating the existence of various kinds of anatomic changes inside the liver.
Figure 3.
Distribution of volume changes for the liver and tumor of the 28 patients.
Dice scores and target registration errors
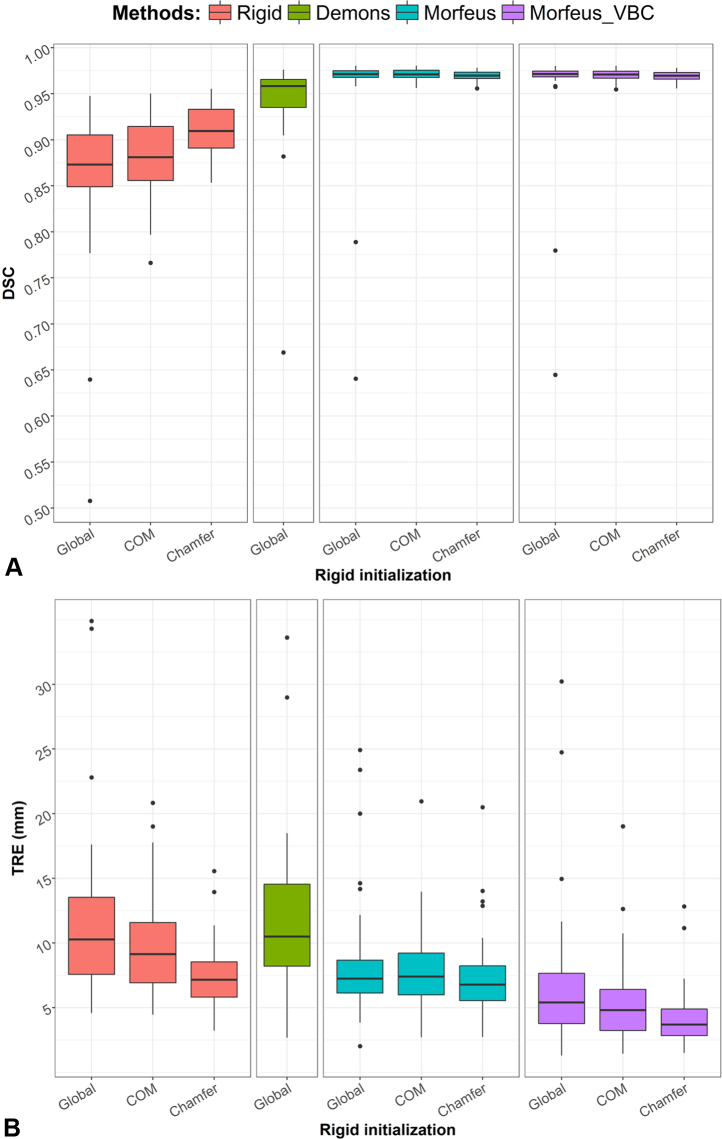
Figure 4 A shows the distributions of the mean Dice scores for the 28 patients after each rigid alignment and DIR method using standard box and whisker plots, which show the median value, interquartile range, minimum and maximum values and outliers. For 2 patients, a very low overlap of the liver was observed after the global rigid alignment of the patient abdomens with Dice scores of 0.51 and 0.64. This could be explained by the large weight loss of these 2 patients. Due to this low initial overlap of the liver after a global rigid initialization, the Demons algorithm and the surface projection method used in the biomechanical DIR workflow, which is also based on a Demons algorithm, failed to match the liver surfaces for these 2 cases. The performance of each method was assessed using the mean and standard deviation and the statistical significance of the differences was evaluated with Student 2-tailed paired t tests. When using the liver contour for the rigid alignment, allowing a rotation of the liver with the Chamfer method significantly improved the mean Dice score in comparison to the COM alignment from 0.88 ± 0.04 to 0.91 ± 0.03 (P < .01). The Demons algorithm significantly improved the Dice score compared with the best rigid alignment (P = .01). Because the biomechanic DIR methods used the liver contours, they all yielded to a high Dice score of on average 0.95 when a global rigid registration was considered and 0.97 for the 2 other rigid initialization methods.
Figure 4.
(A) Box and whisker plots of the 28 mean Dice scores for all rigid and deformable registration methods. The outliers were defined as the values distant from the 75th and 25th percentile by more than 1.5 times the interquartile range. (B) Corresponding box and whisker plots of the 28 mean target registration errors (TRE) for all rigid and deformable registration methods. Abbreviation: VBC = vessels boundary conditions.
Figure 4 B shows the box and whisker plot of the corresponding mean TRE. The same order of performance as suggested by the Dice score results could be observed for the rigid alignment methods with mean TRE of 12.4 ± 7.5, 10.1 ± 4.3, and 7.7 ± 2.8 mm for the Global, COM, and Chamfer methods, respectively. However, the Demons algorithm, which provided significantly better Dice scores yielded to a mean TRE of 11.9 ± 6.9 mm. For 12 out of 28 patients, the TRE in the liver was worse after applying the Demons than after the global rigid alignment alone. Although using the contours in the standard BM_DIR allowed to almost perfectly match the liver surfaces with Dice score greater than 0.95, the corresponding TRE remained high with averages of 9.0 ± 5.6, 8.2 ± 3.7, and 7.7 ± 3.7 mm when considering the Global, COM, or Chamfer initialization, respectively. Considering the Chamfer method, no significant improvement of the mean TRE was found after applying standard BM_DIR (P = .98). Whichever the considered rigid registration, using the additional vessel boundary conditions significantly improved the TRE (P < .01). BM_DIR_VBC achieved TREs of 7.4 ± 6.5, 5.8 ± 3.8, and 4.4 ± 2.5 mm for the Global, COM, and Chamfer initialization methods, respectively.
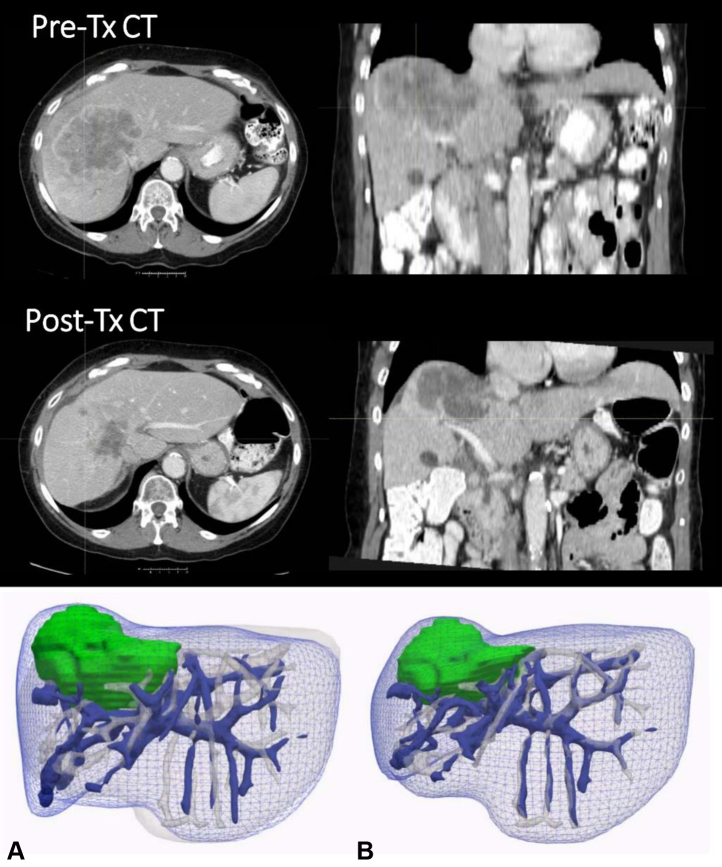
Figure 5 shows an example of complex tumor change, the CT scans presenting a tumor area changing in volume, densities, and textures. The mesh of the liver and the vasculature surfaces are represented after a Chamfer initialization and after applying BM_DIR_VBC. The estimated deformation of the tumor area resulted from the deformation of surrounding vasculature and liver surface only, avoiding to rely on an uncertain definition of the tumor area boundaries.
Figure 5.
Axial and coronal slices of pre- and post-RT computed tomographic scans of a patient showing the tumor area. (A) The blue mesh represents the liver surface of the pretreatment image; the plain blue surface represents the vasculature of the same image; the liver and vasculature corresponding to the posttreatment image are represented in white; the tumor area corresponding to the pretreatment image is represented in green. (B) The surfaces of the tumor in green and of the liver and vasculature in blue represents the result of the deformation estimated with biomechanical model-based deformable image registration with boundary conditions specified on the vessels and liver surface.
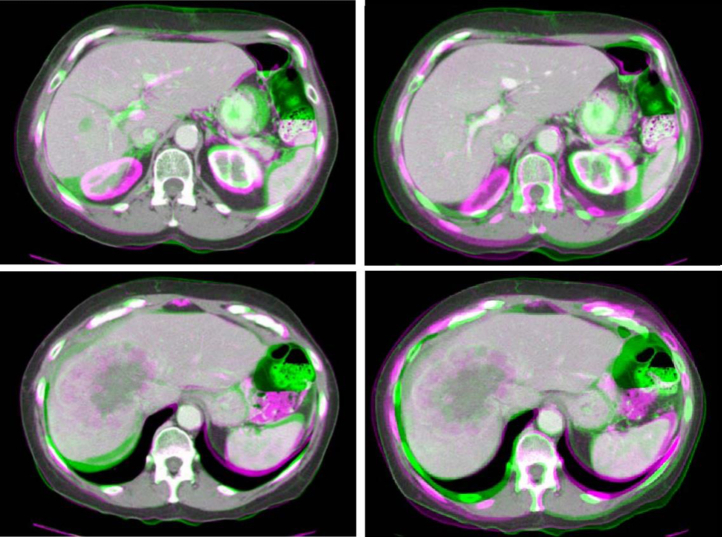
Figure 6 shows overlays of registered image slices for the same patient. The slice representing a normal part of the liver illustrates the ability of the method to align both the liver surface and internal vasculature. The slice representing the tumor area illustrates the ability of the method to provide an apparent reasonable alignment of that region by ignoring image intensity or texture changes, which are challenging to interpret.
Figure 6.
Colored overlays of 2 slices of the pre- and post-RT computed tomographic scans of a patient after a rigid Chamfer alignment (left) and after deformable image registration with boundary conditions specified on the vessels and liver surface (right). Top: slice showing normal tissues in the liver; bottom: slice showing the tumor area.
Discussion
A deformable image registration method has been proposed for the registration of pre- and postradiation therapy contrast-enhanced CT of cholangiocarcinoma cases. Based on a biomechanical model of the liver driven by boundary conditions on the surface and vasculature, the method demonstrated significantly higher accuracy than a previously proposed approach based on boundary conditions on the liver surface only. The previous approach actually failed to provide significantly better accuracy than a rigid alignment based on a chamfer matching of the liver surface. DIR approaches guided by the information of liver surface only should be reserved to the registration of images presenting physiologic-based anatomic variations inside the liver, such as 4-dimensional CT and image guidance at treatment fractions during a course of radiation therapy. Images presenting long-term radiation-induced changes or large disease progression in the liver appeared much more challenging to align.
To assess the difficulty in aligning these images, the accuracy obtained with a Demons algorithm was also reported. Although the Dice score improved for all patients with the Demons in comparison to the initial abdomen alignment, the TRE was worse in 43% (12 out of 28) of patients. Other intensity-based DIR methods may prove themselves more accurate, but we believe that all would show poor robustness for the estimation of internal liver deformations.
These results justify the need for defining corresponding landmarks by clinical observers for DIR accuracy evaluation. In this study, the best DIR method provided an accuracy of 4.4 ± 2.5 mm, which has to be relativized considering the slice spacing of 2.5 or 5 mm and the uncertainties in manually picking up landmarks. For the 6 patients who had at least one CT scan with a slice spacing of 5 mm, the mean TRE was 5.0 mm for the best method, which was close to the results for the rest of the patients. This could be explained by the fact that vessel bifurcation points were selected for each patient only when appearing identically in the lower resolution image and other image. For DIR methods relying on the intensity information in the liver, such as the Demons or BM_DIR_VBC in this study, this method of accuracy evaluation remains the most reliable but may be biased and overestimate the accuracy in large areas that are homogeneous in intensity.
Better accuracy than the achieved 4.4 mm may be desirable, for example if the goal of DIR is to map a dose distribution to the posttreatment image for analysis of the dose response or planning a new course of treatment. More advanced segmentation techniques of the vasculature than the method adopted in this study have previously been proposed. However, we believe that this part of the workflow, which could be improved in future work, has little effect on the final achieved TRE. The proposed vessel tree matching method, when combined to a Chamfer matching of the liver surface, seemed very robust to inconsistencies between the segmented vessel trees of the 2 images. Focus will be given to the expansion of the biomechanic model complexity. Future investigations will include the assignment of heterogeneous elastic properties in the liver based on the autosegmentation of the vasculature and the initialization of the deformations based on the simulation of the effect of locally delivered dose on local volume changes as recently proposed.9
Conclusions
A DIR solution has been proposed for the registration of pre- and postradiation therapy CT scans of the liver presenting long-term anatomic changes. The method was based on a biomechanical model of the liver driven by boundary condition on the liver surface and auto-segmented vasculature. For evaluation of the method, corresponding anatomic points were identified between the image pairs of 28 patients. By achieving an average TRE of 4.4 ± 2.5 mm in the presence of the complexity of the anatomic changes owing to treatment response or disease progression, the proposed workflow should prove itself to be a valuable tool for studies of liver cancer dose response.
Footnotes
Sources of support: Dr Cazoulat reports grants from RaySearch Laboratories AB and University of Texas MD Anderson Cancer Center Co-Development and Collaboration Agreement, grants from National Cancer Institute, during the conduct of the study. Dr Brock reports grants from RaySearch Laboratories AB and University of Texas MD Anderson Cancer Center Co-Development and Collaboration Agreement, grants from National Cancer Institute, grants from Helen Black Foundation, during the conduct of the study. Dr Koay reports grants from National Cancer Institute, grants from Stand Up To Cancer, grants from Project Purple, grants from Pancreatic Cancer Action Network, from null, during the conduct of the study; other from Taylor and Francis, LLC, grants from Philips Health care, outside the submitted work. Research reported in this publication was supported in part by RaySearch Laboratories AB and University of Texas MD Anderson Cancer Center through a Co-Development and Collaboration Agreement. Research reported in this publication was supported in part by the Helen Black Image Guided Fund. Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under award number 1R01CA221971-01A1 awarded to Kristy K. Brock. Research reported in this publication was supported in part by resources of the Image Guided Cancer Therapy Research Program from The University of Texas MD Anderson Cancer Center. Research reported in this publication was supported in part by The University of Texas MD Anderson Cancer Center. Dr Eugene Koay was supported by the Andrew Sabin Family Fellowship, Sheikh Ahmed Center for Pancreatic Cancer Research, Khalifa Foundation, equipment support by GE Healthcare and the Center of Advanced Biomedical Imaging, and NIH (U54CA210181.01, U54CA143837 and U01CA196403). The work was also supported by the Cancer Center Support Grant (CA016672) to MD Anderson. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures: Dr Elganainy reports that her husband works in Agios pharmaceuticals. In addition, Dr Brock has a licensing agreement with RaySearch Laboratories.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics, 2018 CA: A cancer. J Clin. 68:7-30. [DOI] [PubMed]
- 2.Velec M., Moseley J.L., Craig T., Dawson L.A., Brock K.K. Accumulated dose in liver stereotactic body radiotherapy: positioning, breathing, and deformation effects. Int J Radiat Oncol Biol Phys. 2012;83:1132–1140. doi: 10.1016/j.ijrobp.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaminath A., Massey C., Brierley J.D. Accumulated delivered dose response of stereotactic body radiation therapy for liver metastases. Int J Radiat Oncol Biol Phys. 2015;93:639–648. doi: 10.1016/j.ijrobp.2015.07.2273. [DOI] [PubMed] [Google Scholar]
- 4.Huang P., Yu G., Kapp D.S. Cumulative dose of radiation therapy of hepatocellular carcinoma patients and its deterministic relation to radiation-induced liver disease. Med Dosim. 2018;43:258–266. doi: 10.1016/j.meddos.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Fukumitsu N., Nitta K., Terunuma T. Registration error of the liver CT using deformable image registration of MIM Maestro and Velocity AI. BMC Med Imaging. 2017;17:30. doi: 10.1186/s12880-017-0202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock K., Sharpe M., Dawson L., Kim S., Jaffray D. Accuracy of finite element model-based multi-organ deformable image registration. Med Phys. 2005;32(6Part1):1647–1659. doi: 10.1118/1.1915012. [DOI] [PubMed] [Google Scholar]
- 7.Brock K.K., Hawkins M., Eccles C. Improving image-guided target localization through deformable registration. Acta Oncol. 2008;47:1279–1285. doi: 10.1080/02841860802256491. [DOI] [PubMed] [Google Scholar]
- 8.Anderson BM, Lin EY, Cazoulat G, Gupta S, Odisio BC, Brock KK. Improvement of liver ablation treatment for colorectal liver metastases. Paper presented at: Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling 2018. doi:10.1117/12.2294554
- 9.Polan D.F., Feng M., Lawrence T.S., Haken T.K., Brock K.K. Implementing Radiation Dose-Volume Liver Response in Biomechanical Deformable Image Registration. Int J Radiat Oncol Biol Phys. 2017;99:1004–1012. doi: 10.1016/j.ijrobp.2017.06.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonke J.J., Belderbos J. Adaptive radiotherapy for lung cancer. Semin Radiat Oncol. 2010;20:94–106. doi: 10.1016/j.semradonc.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Cazoulat G., Owen D., Matuszak M.M., Balter J.M., Brock K.K. Biomechanical deformable image registration of longitudinal lung CT images using vessel information. Phys Med Biol. 2016;61:4826. doi: 10.1088/0031-9155/61/13/4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson B.M., Lin E., Cardenas C.E., Koay E.J., Odisio B., Brock K.K. Automated Contouring of Contrast and Non-Contrast CT Liver Images with Fully Convolutional Neural Networks. Int J Radiat Oncol Biol Phys. 2018;102:S55. doi: 10.1016/j.adro.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang H., Holton K.S., Robb R.A. Image registration of multimodality 3-D medical images by chamfer matching. Biomedical Image Processing and Three-Dimensional MicroscopySPIE. 1992;1660:356–373. [Google Scholar]
- 14.van Herk M., Kooy H.M. Automatic three-dimensional correlation of CT-CT, CT-MRI, and CT-SPECT using chamfer matching. Med Phys. 1994;21:1163–1178. doi: 10.1118/1.597344. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Dong L., O'Daniel J. Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy. Phys Med Biol. 2005;50:2887. doi: 10.1088/0031-9155/50/12/011. [DOI] [PubMed] [Google Scholar]
- 16.Lesage D., Angelini E.D., Bloch I., Funka-Lea G. A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes. Med Image Anal. 2009;13:819–845. doi: 10.1016/j.media.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Koller TM, Gerig G, Szekely G, Dettwiler D. 1995. Multiscale detection of curvilinear structures in 2-D and 3-D image data. In Proceedings of the Fifth International Conference on Computer Vision(ICCV '95). Washington, DC: IEEE Computer Society; 1995:864-869.
- 18.Sato Y., Nakajima S., Shiraga N. Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal. 1998;2:143–168. doi: 10.1016/s1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- 19.Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. In: Wells WM, Colchester A, Delp S, eds. Medical Image Computing and Computer-Assisted Intervention—MICCAI’98. MICCAI1998 Lecture Notes in Computer Science, vol 1496. Berlin, Heidelberg: Springer; 1998.
- 20.Li Q., Sone S., Doi K. Selective enhancement filters for nodules, vessels, and airway walls in two-and three-dimensional CT scans. Medical Physics. 2003;30:2040–2051. doi: 10.1118/1.1581411. [DOI] [PubMed] [Google Scholar]
- 21.Otsu N. A Threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 1979;9:62–66. [Google Scholar]
- 22.Lee T.-C., Kashyap R.L., Chu C.-N. Building skeleton models via 3-D medial surface axis thinning algorithms. CVGIP: Graphical Models and Image Processing. 1994;56:462–478. [Google Scholar]