Abstract
Purpose
To review and critique the current state of liquid biopsy in pHGG.
Materials and Methods
Published literature was reviewed for articles related to liquid biopsy in pediatric glioma and adult glioma with a focus on high-grade gliomas.
Results
This review discusses the current state of liquid biomarkers of pHGG and their potential applications for liquid biopsy development.
Conclusions
While nascent, the progress toward identifying circulating analytes of pHGG primes the field of neuro-oncoogy for liquid biopsy development.
Introduction
Aggressive brain tumors, such as pediatric high-grade gliomas (pHGGs), are the number one cause of cancer-related deaths during childhood (defined as age 0-14 years in the United States).1 Despite decades of clinical trials with aggressive multimodal therapy, disease recurrence is near inevitable, and patient outcomes remain dismal with a combined 5-year survival rate of 17%.2 Clinical advances in central nervous system (CNS) tumors are hampered by inaccurate or incomplete measures of disease owing to sensitive neuroanatomical location, risk of serial biopsy, and limitations in imaging technologies. In addition, a definitive noninvasive biomarker for disease measurement is lacking. The development of liquid biopsies is critical to advance the clinical management of these tumors, with the hope of directing long-term disease control.
Although rare, pHGGs comprise 8% to 12 % of childhood brain tumors with an annual incidence of 0.85 per 100,000 in the United States.1 These tumors are responsible for almost 13,000 years of potential life lost annually.3 Children typically present with headaches, seizures, vision changes, vomiting, or other neurologic deficits related to specific tumor location. Standard evaluation includes magnetic resonance imaging (MRI) and, in most cases, a neurosurgical intervention (i.e., biopsy or resection) for a histologic and molecular diagnosis.
Although pHGGs are distinct diseases from adult high-grade gliomas,4 treatment rationales in clinical trials are often shaped by the literature on adult gliomas.5 Childhood cancer consortiums, such as the Children’s Oncology Group, have conducted numerous clinic trials (e.g., ACNS0126, ACNS0423)6,7 to improve outcomes for this population. However, current management consists of maximal surgical resection followed by radiation therapy with concurrent and adjuvant alkylator therapy. A notable exception is diffuse intrinsic pontine glioma (DIPG), which is an unresectable tumor with no known curative treatment. Radiation therapy is used to extend the symptom-free period, but offers minimal survival advantage.8
Regardless of location and subgroup, patients with pHGGs are commonly monitored with regular clinical examinations and surveillance imaging. Despite the best available therapy, recurrence is near inevitable, and most cases recur locally within the radiation field and diffuse spread occurs in up to a third of patients.9 Clinical advances for this patient population are hampered by the rarity of the disease, sensitive tumor location, risks10 of serial biopsy, disease heterogeneity, limitations of drug delivery (blood-brain barrier), and inaccurate or incomplete measures of disease status. Over the past 50 years, despite hundreds of clinical trials, the dismal outcomes for children with pHGG have not significantly changed.
Although clinical advances are stagnant, remarkable advances have been made in the past decade in the understanding of the bimolecular basis of pHGG. Large cooperative working groups (eg, International Society of Pediatric Oncology Europe),11 patient registries, tissue banking and sharing, and open access data sets (eg, whole genome sequencing, whole exosome sequencing, methylation analysis12) have yielded robust opportunities to study these rare childhood tumors. Interrogations of well-annotated large patient cohorts have revealed the diversity of pHGG with key genetic and epigenetic events associated with distinct age of onset, neuroanatomic locations, and prognosis, allowing for biologically and clinically relevant subgrouping.13
The discovery of novel driver mutations in genes encoding histone 3 K27M (H3K27M) and G34V/R (H3G34R/V) in pHGG implicate chromatin remodeling, developmental signaling pathways, and gene expression mechanisms in disease pathogenesis.14, 15, 16 These unique recurrent mutations underscore fundamental differences between pediatric and adult high-grade gliomas. The World Health Organization classification of CNS tumors now includes some molecularly defined subsets (eg, H3K27M and isocitrate dehydrogenase 1 [IDH1] mutations, which are mutually exclusive and prognostically relevant).17,18 H3K27M-mutant pHGGs confer a worse prognosis compared with wild type, whereas IDH1 mutants (although rare in the pediatrics setting) carry a better prognosis than IDH wild type.19
These observations have ignited efforts to further molecularly subclassify this heterogeneous group of diseases into biologically and prognostically relevant groups through initiatives from the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy.20
Ongoing efforts to dissect and stratify pHGGs continue to yield new insights into pediatric gliomagenesis and identify novel therapeutic opportunities. Leveraging the distinct molecular biology driving pHGG has led to the first study in human clinical trials in the pediatric setting (eg, NCT02960230). However, more than novel therapeutics are needed to improve patient outcomes (eg, inaccurate measures of disease threaten determination of therapeutic efficacy).
Currently few tools exist for the serial measurement of disease in pHGGs, which are predominately neuroimaging and clinical examination and have limited sensitivity and specificity. There is a critical need for minimally invasive biomarkers in pHGG. A real-time readout of tumor dynamics would fundamentally change the care of children with these aggressive tumors. In this review, we highlight the current efforts toward minimally invasive analytes of pHGG and evaluate their overall importance in improving the outcome for children with pHGGs.
Imaging advances in pediatric high-grade gliomas
Gadolinium-based contrast agent (GBCA)–enhanced MRI remains the standard of care for the diagnosis, operative planning, and assessment of the therapeutic response of pHGGs.21,22 The developing brain and biological differences in tumor biology are notable dissimilarities that could affect the imaging assessment of pHGG compared with adult tumors. Warren et al. recognized the potential limitations of applying adult-based response assessment in neuro-oncology23 criteria to pediatric brain tumors and proposed pediatric-centric imaging response criteria.24 The response assessment in pediatric neuro-oncology working group has identified the need for pediatric-centric imaging criteria owing to the heterogeneous group of pediatric brain tumors, relative paucity of clinical trials, and lack of standard metrics to establish radiographic tumor responses to therapy or survival.
To address this current gap in knowledge, the response assessment in pediatric neuro-oncology working group has set the goal of proposing optimal endpoints and study designs, develop a consensus on the radiologic assessment for clinical trials involving children with brain tumors, and better define response to reflect therapeutic activity in pHGG.24 Jaspan et al. published methodological perspectives on the use of morphologic and physiological MRI response metrics in a randomized phase 2 trial of newly diagnosed pHGG treated with temozolomide and irradiation with or without bevacizumab (HERBY25).
The HERBY study was designed to assess treatment response to inform future clinical trials in pHGG from a biological and imaging perspective. Morphologic imaging response metrics included T2 and GBCA-enhanced T1-weighted sequences. In addition, diffusion- and perfusion-based metrics were assessed and included in the estimation of pHGG response assessments. The rationale behind this was that, compared with morphologic sequences alone, a multimodal MRI approach may provide a more informed biology-based approach to establishing treatment response. Despite initiatives to incorporate physiological sequences into pHGG clinical trial response assessments, GBCA-MRI remains the standard of care.
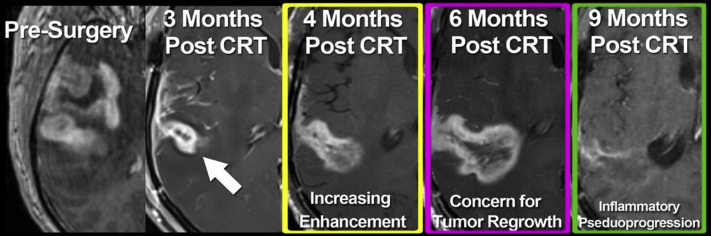
The use of T1-weighted GBCA-MRI as a metric of therapeutic response assessments in pHGGs is problematic. Functionally, GBCA enhancement is a measure of neurovascular unit disruption. As such, GBCA does not specifically localize regions of tumor regrowth. Biologically, tissue enhancement on T1-weighted MRI is a factor of the leakage and pooling of GBCA from the vascular space through the neurovascular unit into the brain tumor interstitial space.25 The limitations of GBCA-MRI in evaluating therapeutic responses in adult gliomas is well established (Fig 1). The nonspecificity of GBCA enhancement for the differentiation of tumor regrowth from treatment-related inflammation necessitates serial follow-up imaging or invasive surgical sampling to establish disease status.
Figure 1.
Dilemma of monitoring pseudoprogression with gadolinium-based contrast agent magnetic resonance imaging. Gadolinium-based contrast agent magnetic resonance imaging after Stupp protocol therapy in isocitrate dehydrogenase-wild type glioblastoma demonstrates the challenge of prospectively diagnosing treatment response. Minimal enhancement after chemoradiation therapy (arrow) increases over time (middle, yellow and purple boxes) but eventually resolves without intervention (right, green box). Retrospectively, this is interpreted as neuroinflammation; however, response assessment in neuro-oncology criteria would not differentiate from disease recurrence necessitating changes in therapy or biopsy.
Furthermore, in certain clinical contexts (e.g., bevacizumab therapy), the absence of GBCA enhancement does not preclude the presence of active tumor growth.26 In either context, this clinical dilemma may delay definitive therapy for disease recurrence and potentially decrease the potential for a response to that therapy. As such, the development of a blood-based biomarker for therapeutic responses to complement imaging would be advantageous in this population.
Progress toward liquid biopsy
Measures of disease status must capture more than anatomic presence, and those capable of assessing pathophysiologic evolution of disease hold the potential to better inform the care of patients with pHGG. The liquid biome, including blood, cerebrospinal fluid (CSF), and urine, provides a rich, noninvasive biosource to interrogate for tumor-associated biomaterials. Biomaterials, such as tumor-associated proteins, nucleic acids, exosomes, and cells, hold the potential to act as analytes for disease diagnosis, prognosis, therapeutic predication, treatment monitoring, and recurrence surveillance.
Robust discovery and exploratory liquid biopsy investigations are ongoing in the adult setting, and emerging work is underway in the pediatric sphere, as reviewed by Bonner et al.27 To advance the care of patients with pHGG, the field must leverage advancing detection technologies to develop sensitive circulating disease analytes that are highly tumor specific and are thereby clinically actionable. Herein the progress toward liquid biopsy in pHGG is critically reviewed and compared with liquid biopsy in adult high-grade glioma.
Protein biomarkers: Powerful peptides
Protein biomarkers are distinct intact peptides that can be differentially regulated in the setting of disease, including alterations in secretion, shedding, and loss from cells. They are detected in blood, urine, cerebral spinal fluid, and generally stable peptides that therefore can provide an objective, quantitative, and noninvasive measure of disease.28 The first biomarker was reported in 1848 with the discovery of the lambda light chain (Bence Jones proteins), which are detected with a urinalysis in 75% of patients with multiple myeloma.29
From this initial discovery, the field of protein biomarkers has grown into the most clinically robust biomaterial for use in cancer monitoring and detection. CA-125, CA19-9, HER2/NEU, and Fibrin/FDP are all blood-based protein biomarkers approved by the US Food and Drug Administration for use in the detection, monitoring, or prognosis of ovarian, pancreatic, breast, and colorectal cancer, respectively.30 Although there are currently no FDA-approved biomarkers for high-grade gliomas, multiple candidate proteins are under exploration.
Protein biomarkers in adult high-grade gliomas
Proteins are the most investigated biomaterial in adult glioma, including studies in serum, plasma, CSF, and urine. Proteins related to angiogenesis, such as vascular endothelial growth factor, have been reported to be more elevated in patients with gliomas compared with controls.31 A variety of proteins related to cell invasion and tissue remodeling, such as matrix metalloproteinases (MMP) and tissue inhibitors of metalloproteinases, have been identified as potential biomarkers. For example, levels of MMP-2, MMP-9, and tissue inhibitor of metalloproteinase-1 could be used to detect the presence of gliomas from healthy controls and distinguish low- from high-grade glioma.32
Many other proteins, including YKL-40, osteopontin, glial fibrillary acid protein, 2-hydroxyglutarte, and growth factors such as hepatocyte placental, insulin binding, and basic fibroblast have been investigated. However, to date no clinically actionable protein biomarkers have emerged.33
Protein biomarkers in pediatric high-grade gliomas
Proteins hold promise as biomarkers in pHGG, but their value remains largely to be realized. Investigations into proteins of interest in adult glioma have not been substantiated potential importance for pHGG, including vascular endothelial and basic fibroblast growth factors, where the levels between control, benign disease, and malignant disease were very similar.34 Tumor-associated proteins have been explored in a handful of pediatric CNS tumors, including cyclophilin A and DDAH1, which were associated with disease presence compared with healthy controls.35
A proteomic analysis of CSF from pediatric CNS tumors, including pHGG, revealed 6 proteins able to discriminate metastatic cases from controls.36 Exploiting the unique drivers of pHGG holds promise for the detection of meaningful biomarkers. Investigations into the posttranslational modification of proteins in the liquid biome may prove a valuable strategy given the known pHGG driver mutations in epigenetic mechanisms, such as chromatin remodeling. Much opportunity exists to discover protein biomarkers that exploit unique features of pHGG and ultimately support the development and utility of a liquid biopsy.
Cell-free tumor DNA in circulation and cerebrospinal fluid
Cell-free DNA (cfDNA) are short (approximately 160 base pair fragments of genetic material) and when derived from a tumor are known as circulating tumor DNA (ctDNA), which can be detected in the blood and CSF from patients with cancer.37 These DNA contain both genetic and epigenetic information revealing the tumor site of origin and, importantly, specific DNA alterations (i.e., point mutations, specific methylation patterns).37,38 However, ctDNA has a high turnover rate because of its rapid clearance from the bloodstream, which could make detecting it difficult in cases of low tumor burden.
Beyond diagnostic utility, longitudinal ctDNA has the potential to inform treatment monitoring and, in the era of precision medicine, aid therapeutic predication. Currently, in some diseases, such as melanoma, gastrointestinal, or breast cancers, ctDNA is used as a metric for disease progression, with higher levels correlating to an increased disease burden or recurrence.39
Cell-free DNA in adult high-grade glioma
In adult high-grade glioma, ctDNA has been detected in the blood and CSF. Studies investigating ctDNA have focused on the detection of molecular aberrations from known diagnostic, predictive, prognostic, or monitoring values from the literature on adult tissue biomarkers. These ctDNA targets include IDH1 mutations; EGFRvIII mutation; loss of heterozygosity for 1p, 10q, 19q; and the abnormal methylation of promoters (eg, O-6-methylguanine-DNA methyltransferase) but are currently limited to exploratory investigations and prospective clinical utility testing.40, 41, 42, 43, 44
Tumor hemorrhage, biopsy, and neuroanatomical location may challenge the sensitivity of ctDNA as a diagnostic biomarker. For example, Wang et al. highlight that proximity to CSF reservoirs was associated with increased detection ctDNA in CSF.45 However, as the field of precision medicine grows, the stage may be set for the use of ctDNA as a companion diagnostic and predictive and monitoring biomarker. For example, the detection of EGFRvIII mutation (therapeutic target of interest) can be missed on tissue resection owing to heterogeneous expression but has been detected in the blood as ctDNA.46
Furthermore, the authors report that 2 patients with detectable preresection EGFRvIII ctDNA had undetectable levels after a gross total resection.46 Pathologic variants of EGFR are more common in adult HGG than in pHGG; however, EGFR mutations are present in pHGG and efforts to leverage a molecularly defined lesion not just for therapeutic proposes but also as a minimally invasive biomarker of disease provide a valuable example.
Cell-free DNA in pediatric high-grade glioma
The rapidly advancing molecular subgrouping of pHGG provides a rich landscape of candidate mutations for the development of ctDNA biomarkers, and the detection of ctDNA in the blood and CSF of pediatric patients with HGG has been reported. Multiple reports of H3K27M mutant cfDNA in CSF exist, this prognostic biomarker is known to portend a poor prognosis and to be common in midline gliomas, such as DIPG.47 Diagnostic noninvasive biomarkers for DIPG are highly desirable given that the disease is sensitive to neuroanatomical location and concern for harm with biopsy, and most patients are diagnosed based on imaging features and history. However, a growing number of centers are now safely performing biopsies of these tumors,48 allowing for access to molecularly defined clinical trials.
Recently, the detection of H3K27M has been reported in the plasma of patients with DIPG and when correlated with CSF ctDNA and followed longitudinally, the detectiont correlated with disease progression. The authors suggested that the measurement of H3K27M ctDNA could have diagnostic, prognostic, predictive, and monitoring biomarker potential; however, this has not been prospectively studied. Oncogenic BRAFv600E mutations are seen in pHGG, although more commonly in low-grade gliomas, and hold the potential to be predictive, prognostic, and monitoring biomarkers.
Although not yet reported in the blood of patients with gliomas, the assessment of BRAFv600E cfDNA has been used as a monitoring biomarker in pediatric patients with Langerhans Cell Histocytosis (LCH).49 In this small study, the measurement of BRAFv600E cfDNA correlated with disease stage and treatment response regardless of the treatment agents used.50 The oncogenic neurotrophic tyrosine receptor kinase (NTRK) fusion is present in pHGG and although rare, NTRK is more commonly found in children age <3 years.51 The recent tissue agnostic approval of the pan-tropomyosin receptor kinase inhibitor (TRK), loratrectinib, an inhibitor of the NTRK fusion product, makes the noninvasive detection of NTRK fusions an attractive biomarker.
The literature on adult gliomas measured NTRK cfDNA as a monitoring biomarker of treatment response and predictive biomarker of resistance mechanisms for agents aimed at overcoming the mutational resistance mechanism.52 Pediatric studies, including CNS tumors, with next generation NTRK inhibitors are ongoing (e.g., NCT02650401) and include the measurement of ctDNA as biologic correlates of disease. The continued prospective collection of ctDNA in clinical trials is needed to fully appreciate the utility of ctDNA in pHGG. The clinical development of ctDNA is an exciting tool for pHGG and precision medicine; however, this biomaterial currently falls short of capturing the robust dynamic tumor biology present at the cellular level.
Meaningful messengers: MicroRNA in adult high-grade glioma
MicroRNAs (miRNAs) are small, 18 to 22 nucleotide segments of single-stranded noncoding RNA. MiRNAs function as regulators of posttranscriptional and translational processes by targeting specific messenger RNAs.53,54 MiRNAs were first noted in 1993 with the discovery of a novel gene regulation pathway.55 In 2004, Takamizawa et al demonstrated that the let-7 family of miRNAs held prognostic value in lung cancer, with higher expression correlated to a more aggressive disease state.56 In cancer, miRNAs are dysregulated and can act as tumor suppressors or oncogenes depending on their prescribed function. Now, unique miRNA expression profiles have been linked to specific cancers, making miRNAs not only an important effector of disease but also a useful and specific diagnostic tool.53 The presence of miRNAs has been documented in both pediatric and adult HGG. Birks et al. analyzed the miRNA profiles of 24 pediatric CNS tumors and noted a distinct deregulation of miRNA-129, miR-142-5p, and miR-25.57 Although promising, the utility of miRNA in cancer therapeutics and detection needs to be further validated before becoming a common-place cancer detection strategy.
Small but mighty: Exosomes in adult high-grade glioma
Exosomes are small membrane-bound vesicles ranging from 50 to 140 nm and were first discovered in the late 1980s.58 Exosomes have come of age as promising effectors of the tumor microenvironment and liquid biomarkers of disease diagnosis and prognosis. Once thought to be a byproduct underlying a mechanism for transferrin receptor elimination by red blood cells, we now know that many different cell types extrude exosomes in normal physiological conditions. Intravesicular cargo includes proteins, DNA, RNA (including noncoding RNAs), and various lipid species.59, 60, 61 The wide variety of exosome cargo point to the functional relevance of exosomes in intercellular communication. For example, exosomes contain both cytosolic61 and membrane-bound proteins62 (cytoplasmic proteins that support vesicular biogenesis) and membrane-bound proteins (eg, CD963 and CD6364) that imply an immune function. Much of the lipid and protein cargo is thought to reflect the metabolic state of the cell of origin.61
Furthermore, exosomes are thought to mediate intercellular communication within a microenvironment.65 In cancer settings, exosomes are described to mediate angiogenesis,66 establishment of the premetastatic niche, and tumor progression.67 As a liquid biomarker, exosomes remain a promising prognostic or diagnostic analyte, carrying myriad miRNAs, with significant differences seen between healthy controls and various cancers, including glioblastoma,68,69 pancreatic,70,71 colorectal,72 lung,73 and breast74 cancer.
Exosomes function in high-grade glioma
Advances in understanding how exosomes shape HGG are predominantly conducted in adult glioblastoma and shed important insights into the exosome role in tumor progression and its role as a biomarker for disease.68 Currently, studies focus on 3 primary areas: impact of exosomes in shaping the tumor microenvironment, characterization of exosome cargo, and use of exosomes as a liquid biomarker.
Exosomes extruded from irradiated glioblastoma cells alter migratory behaviors of recipient cells in culture, indicating a role for exosomes in shaping the tumor microenvironment.75 These exosomes carry cargo, including connective tissue growth factor mRNA and insulin-like growth factor binding protein, and support the potential to affect tumor cell phenotypes or behaviors. Untreated tumor cells extrude cargo with cell-signaling components, notably farnesylated Ras found in glioblastoma-derived exosomes.76
Furthermore, recent studies from an MD Anderson group led by Dr Heimburger revealed that glioblastoma-derived exosomes affect immunosuppression by targeting tumor-associated monocytes driving differentiation toward an immune-suppressive M2-macrophage fate, specifically elucidating the activation of the STAT3 pathway in this population.77,78 Not only do exosomes harbor effector molecules, they also harbor tumor-associated proteins such as mutant forms of EGFR,79 IDH1,80 and miR21.81 These studies, and other exosome-based biomarkers of glioblastoma, are highlighted in a review by Rennert et al82 focusing on adult glioma. A dearth of studies have extended into pediatric brain tumors, although miRNA that contain exosomes have been documented.83 Exosome detection in CSF and blood84,85 highlight the potential impact of exosomes as biomarkers of disease.
In adult disease, exosomes derived from different subtypes of glioma contain a differential expression of exosome biogenesis mRNA genes,86 which indicates that a potential inequality in exosome extrusion may exist across subtypes. Discrete omics are identified in adult versus pediatric tumors,87 which further calls into question how much of the glioblastoma-derived exosome knowledge from adult disease will translate into pediatric disease. A small handful of studies investigating exosomes in pediatric disease are beginning to provide insights. Recently, miRNA-containing exosomes have been documented in pediatric brain tumors,83 opening the field for potential clinical trials with exosome analytes or the development of exosomes as diagnostic agents for disease progression.61
Circulating cells: Circulating tumor cells, circulating hybrid cells, clusters, and tumor-associated immune cells
Circulating cells with the potential to provide information on tumor state, including its pathology, heterogeneity, mutational burden, and predictive risk for disease progression, remain a holy grail of liquid biopsies. To date, a number of tumor-associated cell populations are detectible in the peripheral blood of patients with cancer, with the most noted being circulating tumor cells (CTCs), which were first detected in 1869 in a patient with metastatic cancer.88 CTCs hold the promise of providing temporal information of the evolving tumor in the form of a noninvasive, liquid biopsy.
The utilization of CTCs have provided prognostic value,89,90 genetic material,91 tumor phenotype (e.g., HER2 for breast cancer92), and a measure of tumor cell heterogeneity.93 Once a barrier, new advances in the isolation of CTCs has led to approval by the U.S. Food and Drug Administration for the use of CTC enumeration for clinical testing.89 Notably, the commercially available CellSearch system94 relies on cytokeratin expression and lack of CD45 expression and magnetic-based separation, microfluidics systems that separate based on size,95 and the Gilupi Cell Collector that extracts CTCs directly from the bloodstream.96
Although promising, the use of CTCs for reliable tumor monitoring has fallen short of expectations, largely owing to the rarity of CTCs in peripheral blood. Reported detectable ranges from 0.1 to 1 cell/mL are dependent on tumor stage and tumor site.97 In addition, the use of common epithelial proteins to identify CTCs may hamper the ability to definitively enumerate tumor-associated cells distinct from circulating epithelial cells.98 Finally, inconsistency among reports on the efficacy and reproducibility of CTCs as a biomarker for treatment response, prognosis, and risk of relapse hamper strong reliance on this biomarker.
Circulating tumor cells in adult high-grade glioma
The use of CTCs for prognosis in glioblastoma is in its infancy. The fact that patients with glioblastoma rarely experience metastatic spread led to the concept that glioblastoma is restricted to the brain. Subsequently, reports of detectable glioblastoma metastatic growth in organ-transplant recipients received from glioblastoma donors challenged this notion.99,100 CTCs were first identified in patients with glioblastoma in 2014.101 Glial fibrillary acid protein-positive CTCs were identified in 20.6% of patients with glioblastoma and found to harbor specific aberrations of the primary tumor, including EGFR gene amplification or chromosome insertions or deletions.
Consistent with CTCs identified in other organ systems, glioblastoma CTCs were rare and only detectible as 1 to 22 CTCs in 2.1 × 106 peripheral blood mononuclear cells. Using a microfluidic approach to deplete blood cells from peripheral blood specimens and thus isolating CTCs, Sullivan et al reported that glioblastoma CTCs have mesenchymal properties. In these studies 13 of 33 adult patients with glioblastoma harbored CTCs.102 Furthermore, Gao et al. enhanced the detection of CTCs using a novel fluorescence in situ hybridization-based approach that allowed for the detection of CTCs in patients with all 7 subtypes of glioblastoma in 77% of patients studied.103
Most recently, in the British Journal of Cancer, temporal analyses of 13 adult patients with glioblastoma undergoing treatment were studied for the presence of CTCs, identifying CTC clusters throughout the disease progression, which suggests that CTCs may not travel the bloodstream as solitary entities.104 This small handful of reports highlights that the study of glioblastoma CTCs lags far behind that of other organ systems. Although glioblastoma CTC is nascent, currently all reported studies of CTCs are in adult cancers. Pediatric glioblastoma provides a needed setting for the development of noninvasive biomarkers for a treatment-resistant disease with unappreciated evolution behaviors and unknown susceptibilities.
Developing the landscape of circulating cellullar biomarkers
Although CTCs are the most well-studied circulating cells in all cancers, including adult glioblastoma, other circulating cells exist and hold value. A novel cancer cell population derived from the fusion between tumor and immune cells, circulating hybrid cells,105 were recently identified in melanoma and pancreatic cancer. Circulating hybrid cells outnumbered conventionally defined CTCs in murine models of cancer and in human patients with pancreatic cancer, which indicates a larger population of tumor burden in peripheral blood than previously appreciated. This revelation provides an opportunity to develop a high-impact liquid biomarker.
In addition, cancer-associated macrophage-like cells106 or tumor epitope detection in monocytes,107 which are immune cells with detectable apoptotic neoplastic cells within their cytoplasm, have been reported in a variety of cancers. Tumor-extrinsic biomarkers, such as circulating immune signatures, may provide an immunomonitoring opportunity that is important to the growing number of immunooncology clinical trials.108 Taken together, the myriad of cells in circulation offer a fertile ground for temporal information of the evolving tumor and its immunogenicity in the form of a noninvasive, liquid biopsy.
Conclusions
Major advances in technology to analyze circulating cancer biomarkers primes the neuro-oncology field to interrogate the liquid biome and develop noninvasive analyses of disease for pHGG. The rarity of pHGG and invasive nature of tumor biopsies hamper major advances in understanding the disease, including disease initiation, progression, and evolution. The development of the liquid biome in pHGG (Table 1) holds promise to improve patient care through the identification of diagnostic, prognostic, predictive, and monitoring biomarkers. The discovery of circulating analytes capable of distinguishing the MRI inaccuracy of pseudo- from true progression can prevent subjecting patients to unnecessary neurosurgical intervention.
Table 1.
Summary of circulating biomaterials with examples
| Biomaterial | Biosources |
Adult examples | Pediatric examples | ||||
|---|---|---|---|---|---|---|---|
| Serum | Plasma | CSF | Urine | ||||
| Protein | x | x | x | x | VEGF30, MMPs/TIMPs31 | VEGF33, bFGF33, CypA34, DDAH134 | |
| Nucleic acids | ctDNA | x | x | x | IDH139, EGFRvIII39, MGMT41, 1p/10q/19q codeletion42 | H3K27M46, BRAFv600E48 | |
| miRNA | x | x | Yet to be reported | miRNA-12956, miR-142-5p56, miR-2556 | |||
| Exosomes | x | x | Ras75, EGFR78, IDH179, miR2180 | Yet to be reported | |||
| Circulating cells | CTCs | x | GFAP100 | Yet to be reported | |||
| CHCs | x | Yet to be reported | Yet to be reported | ||||
Abbreviations: bFGF = basic fibroblast growth factor; CHCs = circulating hybrid cells; CSF = cerebrospinal fluid; CTCs = circulating tumor cells; ctDNA = circulating tumor DNA; CypA = cyclophilin A; DDAH1 = dimethylarginine dimethylaminohydrolase 1; GFAP = glial fibrillary acidic protein; IDH1 = isocitrate dehydrogenase 1; MGMT = O-6-methylguanine-DNA methyltransferase; miRNA = micro RNA; MMP = matrix metalloproteinase; TIMP = tissue inhibitor of metalloproteinase; VEGF = vascular endothelial growth factor.
Biosources, such as serum, plasma, CSF, and urine, can contain multiple biomaterials, including proteins, ctDNA, miRNA, exosomes, CTCs, and CHCs.
Biomarkers capable of delineating pseudo- from true responses will facilitate offering patient’s investigational agents through clinical trials, perhaps earlier in the disease progression course. Although many agents have failed to show activity in early phase clinical trials, the deep and rapid development of the biomolecular landscape of pHGG is unearthing new potential targets.
Furthermore, cutting-edge technology for nucleic acid isolation and sequencing, including cfDNA, RNA, and microRNAs, makes the monitoring of mutations, cellular behaviors, and evolution during treatment of pHGG feasible and ultimately promises biologically relevant insights in the mechanism of disease progression, treatment resistance, and recurrence, as well as associated therapeutic opportunities. The multidisciplinary care of pHGG provides a robust opportunity for each disciple to leverage access to noninvasive patient biosources to advance biomarker development in childhood CNS tumors.
Pediatric neuro-oncology as a field is poised to capitalize on the major advances in liquid biopsy. The development of noninvasive disease analytes in pHGG cannot be sidelined owing to the highly resistance, recurrent, and lethal course of these tumors. However, given that the disease course is abbreviated and treatment responses are poor, coordinated efforts across institutions must be engaged and consortiums must maintain focus in collecting the necessary biomaterial to facilitate advancing the treatment and management of pHGG. This type of concerted effort, bringing advances in biomarker knowledge in multianalytes, has the best chance to develop new measures of disease that can affect disease progression and change pHGG from a death sentence to a managed disease.
Footnotes
Sources of support: Grant support includes M.S.D.: NCATS5TL1TR002371; M.H.W.: CA172334, CA2111134, Knight Cancer Institute (P30 CA069533).
Disclosures: No author has any conflicts of interest. No statistics were performed for this series. Authors had full access to all data in this study and take complete responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Ostrom Q.T., de Blank P.M., Kruchko C. Alex's Lemonade Stand Foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 2015;16:x1–x36. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M., Thakkar J.P., Garcia C.R. National Cancer Database analysis of outcomes in pediatric glioblastoma. Cancer Med. 2018;7:1151–1159. doi: 10.1002/cam4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Blank P.M., Ostrom Q.T., Rouse C. Years of life lived with disease and years of potential life lost in children who die of cancer in the United States, 2009. Cancer Med. 2015;4:608–619. doi: 10.1002/cam4.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones C., Perryman L., Hargrave D. Paediatric and adult malignant glioma: Close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9:400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R., Mason W.P., van den Bent M.J. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Cohen K.J., Pollack I.F., Zhou T. Temozolomide in the treatment of high-grade gliomas in children: A report from the Children's Oncology Group. Neuro Oncol. 2011;13:317–323. doi: 10.1093/neuonc/noq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakacki R.I., Cohen K.J., Buxton A. Phase 2 study of concurrent radiotherapy and temozolomide followed by temozolomide and lomustine in the treatment of children with high-grade glioma: A report of the Children's Oncology Group ACNS0423 study. Neuro Oncol. 2016;18:1442–1450. doi: 10.1093/neuonc/now038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallitto M., Lazarev S., Wasserman I. Role of radiation therapy in the management of diffuse intrinsic pontine glioma: A systematic review. Adv Radiat Oncol. 2019;4:520–531. doi: 10.1016/j.adro.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shabason J.E., Sutton D., Kenton O., Guttmann D.M., Lustig R.A., Hill-Kayser C. Patterns of failure for pediatric glioblastoma multiforme following radiation therapy. Pediatr Blood Cancer. 2016;63:1465–1467. doi: 10.1002/pbc.26031. [DOI] [PubMed] [Google Scholar]
- 10.Broniscer A. Past, present, and future strategies in the treatment of high-grade glioma in children. Cancer Invest. 2006;24:77–81. doi: 10.1080/07357900500449702. [DOI] [PubMed] [Google Scholar]
- 11.Veldhuijzen van Zanten S.E., Baugh J., Chaney B. Development of the SIOPE DIPG network, registry and imaging repository: A collaborative effort to optimize research into a rare and lethal disease. J Neurooncol. 2017;132:255–266. doi: 10.1007/s11060-016-2363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajjar A., Bowers D.C., Karajannis M.A., Leary S., Witt H., Gottardo N.G. Pediatric brain tumors: Innovative genomic information is transforming the diagnostic and clinical landscape. J Clin Oncol. 2015;33:2986–2998. doi: 10.1200/JCO.2014.59.9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay A., Burford A., Carvalho D. Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32:520–537.e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones C., Baker S.J. Unique genetic and epigenetic mechanisms driving pediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzentruber J., Korshunov A., Liu X.Y. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 16.Wu G., Broniscer A., McEachron T.A. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm D., Witt H., Hovestadt V. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 19.Turcan S., Rohle D., Goenka A. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis D.N., Aldape K., Brat D.J. Announcing cIMPACT-NOW: The consortium to inform molecular and practical approaches to CNS tumor taxonomy. Acta Neuropathol. 2017;133:1–3. doi: 10.1007/s00401-016-1646-x. [DOI] [PubMed] [Google Scholar]
- 21.Hargrave D., Chuang N., Bouffet E. Conventional MRI cannot predict survival in childhood diffuse intrinsic pontine glioma. J Neurooncol. 2008;86:313–319. doi: 10.1007/s11060-007-9473-5. [DOI] [PubMed] [Google Scholar]
- 22.Poussaint T.Y., Kocak M., Vajapeyam S. MRI as a central component of clinical trials analysis in brainstem glioma: A report from the Pediatric Brain Tumor Consortium (PBTC) Neuro Oncol. 2011;13:417–427. doi: 10.1093/neuonc/noq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen P.Y., Macdonald D.R., Reardon D.A. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 24.Warren K.E., Poussaint T.Y., Vezina G. Challenges with defining response to antitumor agents in pediatric neuro-oncology: A report from the response assessment in pediatric neuro-oncology (RAPNO) working group. Pediatr Blood Cancer. 2013;60:1397–1401. doi: 10.1002/pbc.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaspan T., Morgan P.S., Warmuth-Metz M. Response assessment in pediatric neuro-oncology: Implementation and expansion of the RANO criteria in a randomized phase II trial of pediatric patients with newly diagnosed high-grade gliomas. AJNR Am J Neuroradiol. 2016;37:1581–1587. doi: 10.3174/ajnr.A4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Ali S., Clarke S. Bevacizumab in recurrent glioma: patterns of treatment failure and implications. Brain Tumor Res Treat. 2017;5:1–9. doi: 10.14791/btrt.2017.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonner E.R., Bornhorst M., Packer R.J., Nazarian J. Liquid biopsy for pediatric central nervous system tumors. NPJ Precis Oncol. 2018;2:29. doi: 10.1038/s41698-018-0072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maes E., Mertens I., Valkenborg D., Pauwels P., Rolfo C., Baggerman G. Proteomics in cancer research: Are we ready for clinical practice? Crit Rev Oncol Hematol. 2015;96:437–448. doi: 10.1016/j.critrevonc.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Stone M.J., Frendel E.P. The clinical spectrum of light chain myeloma. A study of 35 patients with special reference to the occurrence of amyloidosis. Am J Med. 1975;58:601–619. doi: 10.1016/0002-9343(75)90496-9. [DOI] [PubMed] [Google Scholar]
- 30.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlsson A., Persson O., Ingvarsson J. Plasma proteome profiling reveals biomarker patterns associated with prognosis and therapy selection in glioblastoma multiforme patients. Proteomics Clin Appl. 2010;4:591–602. doi: 10.1002/prca.200900173. [DOI] [PubMed] [Google Scholar]
- 32.Crocker M., Ashley S., Giddings I. Serum angiogenic profile of patients with glioblastoma identifies distinct tumor subtypes and shows that TIMP-1 is a prognostic factor. Neuro Oncol. 2011;13:99–108. doi: 10.1093/neuonc/noq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Best M.G., Sol N., Zijl S., Reijneveld J.C., Wesseling P., Wurdinger T. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol. 2015;129:849–865. doi: 10.1007/s00401-015-1399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobol-Milejska G., Mizia-Malarz A., Musiol K. Serum levels of vascular endothelial growth factor and basic fibroblast growth factor in children with brain tumors. Adv Clin Exp Med. 2017;26:571–575. doi: 10.17219/acem/62320. [DOI] [PubMed] [Google Scholar]
- 35.Saratsis A.M., Yadavilli S., Magge S. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol. 2012;14:547–560. doi: 10.1093/neuonc/nos067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spreafico F., Bongarzone I., Pizzamiglio S. Proteomic analysis of cerebrospinal fluid from children with central nervous system tumors identifies candidate proteins relating to tumor metastatic spread. Oncotarget. 2017;8:46177–46190. doi: 10.18632/oncotarget.17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donaldson J., Park B.H. Circulating tumor DNA: Measurement and clinical utility. Annu Rev Med. 2018;69:223–234. doi: 10.1146/annurev-med-041316-085721. [DOI] [PubMed] [Google Scholar]
- 38.Neumann M.H.D., Bender S., Krahn T., Schlange T. ctDNA and CTCs in liquid biopsy - Current status and where we need to progress. Comput Struct Biotechnol J. 2018;16:190–195. doi: 10.1016/j.csbj.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bettegowda C., Sausen M., Leary R.J. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kros J.M., Mustafa D.M., Dekker L.J., Sillevis Smitt P.A., Luider T.M., Zheng P.P. Circulating glioma biomarkers. Neuro Oncol. 2015;17:343–360. doi: 10.1093/neuonc/nou207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Ricarte F., Mayor R., Martínez-Sáez E. Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin Cancer Res. 2018;24:2812–2819. doi: 10.1158/1078-0432.CCR-17-3800. [DOI] [PubMed] [Google Scholar]
- 42.Rivera A.L., Pelloski C.E., Gilbert M.R. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12:116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staedtke V., Dzaye O.D.A., Holdhoff M. Actionable molecular biomarkers in primary brain tumors. Trends Cancer. 2016;2:338–349. doi: 10.1016/j.trecan.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thon N., Kreth S., Kreth F.W. Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. Onco Targets Ther. 2013;6:1363–1372. doi: 10.2147/OTT.S50208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Springer S., Zhang M. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salkeni M.A., Zarzour A., Ansay T.Y. Detection of EGFRvIII mutant DNA in the peripheral blood of brain tumor patients. J Neurooncol. 2013;115:27–35. doi: 10.1007/s11060-013-1209-0. [DOI] [PubMed] [Google Scholar]
- 47.Huang T.Y., Piunti A., Lulla R.R. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun. 2017;5:28. doi: 10.1186/s40478-017-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta N., Goumnerova L.C., Manley P. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro Oncol. 2018;20:1547–1555. doi: 10.1093/neuonc/noy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panditharatna E., Kilburn L.B., Aboian M.S. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24:5850–5859. doi: 10.1158/1078-0432.CCR-18-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Héritier S., Hélias-Rodzewicz Z., Lapillonne H. Circulating cell-free BRAF(V600E) as a biomarker in children with Langerhans cell histiocytosis. Br J Haematol. 2017;178:457–467. doi: 10.1111/bjh.14695. [DOI] [PubMed] [Google Scholar]
- 51.Wu G., Diaz A.K., Paugh B.S. The genomic landscape of diffuse intrinsic pontine glioma and pediatric nonbrainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo M., Misale S., Wei G. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6:36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 53.Ono S., Lam S., Nagahara M., Hoon D.S. Circulating microRNA biomarkers as liquid biopsy for cancer patients: Pros and cons of current assays. J Clin Med. 2015;4:1890–1907. doi: 10.3390/jcm4101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 56.Takamizawa J., Konishi H., Yanagisawa K. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 57.Birks D.K., Barton V.N., Donson A.M., Handler M.H., Vibhakar R., Foreman N.K. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56:211–216. doi: 10.1002/pbc.22723. [DOI] [PubMed] [Google Scholar]
- 58.Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 59.Thakur B.K., Zhang H., Becker A. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 61.Rajagopal C., Harikumar K.B. The origin and functions of exosomes in cancer. Front Oncol. 2018;8:66. doi: 10.3389/fonc.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamai K., Tanaka N., Nakano T. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 63.Escola J.M., Kleijmeer M.J., Stoorvogel W., Griffith J.M., Yoshie O., Geuze H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 64.Logozzi M., De Milito A., Lugini L. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005219. e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luga V., Zhang L., Viloria-Petit A.M. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 66.Maji S., Chaudhary P., Akopova I. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L., Zhang S., Yao J. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gourlay J., Morokoff A.P., Luwor R.B., Zhu H.J., Kaye A.H., Stylli S.S. The emergent role of exosomes in glioma. J Clin Neurosci. 2017;35:13–23. doi: 10.1016/j.jocn.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 69.Shao H., Chung J., Lee K. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Armstrong E.A., Beal E.W., Chakedis J. Exosomes in pancreatic cancer: From early detection to treatment. J Gastrointest Surg. 2018;22:737–750. doi: 10.1007/s11605-018-3693-1. [DOI] [PubMed] [Google Scholar]
- 71.Li W., Li C., Zhou T. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145. doi: 10.1186/s12943-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogata-Kawata H., Izumiya M., Kurioka D. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092921. e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rabinowits G., Gerçel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 74.Eichelser C., Stückrath I., Müller V. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arscott W.T., Tandle A.T., Zhao S. Ionizing radiation and glioblastoma exosomes: Implications in tumor biology and cell migration. Transl Oncol. 2013;6:638–648. doi: 10.1593/tlo.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luhtala N., Aslanian A., Yates J.R., 3rd, Hunter T. Secreted glioblastoma nanovesicles contain intracellular signaling proteins and active Ras incorporated in a farnesylation-dependent manner. J Biol Chem. 2017;292:611–628. doi: 10.1074/jbc.M116.747618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Vrij J., Maas S.L., Kwappenberg K.M. Glioblastoma-derived extracellular vesicles modify the phenotype of monocytic cells. Int J Cancer. 2015;137:1630–1642. doi: 10.1002/ijc.29521. [DOI] [PubMed] [Google Scholar]
- 78.Gabrusiewicz K., Li X., Wei J. Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2017.1412909. e1412909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al-Nedawi K., Meehan B., Micallef J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 80.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol Ther Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skog J., Würdinger T., van Rijn S. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rennert R.C., Hochberg F.H., Carter B.S. ExRNA in biofluids as biomarkers for brain tumors. Cell Mol Neurobiol. 2016;36:353–360. doi: 10.1007/s10571-015-0284-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tűzesi Á., Kling T., Wenger A. Pediatric brain tumor cells release exosomes with a miRNA repertoire that differs from exosomes secreted by normal cells. Oncotarget. 2017;8:90164–90175. doi: 10.18632/oncotarget.21621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonda D.D., Akers J.C., Kim R. Neuro-oncologic applications of exosomes, microvesicles, and other nano-sized extracellular particles. Neurosurgery. 2013;72:501–510. doi: 10.1227/NEU.0b013e3182846e63. [DOI] [PubMed] [Google Scholar]
- 85.Shi R., Wang P.Y., Li X.Y. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6:26971–26981. doi: 10.18632/oncotarget.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verhaak R.G., Hoadley K.A., Purdom E. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma X., Liu Y., Liu Y. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Apostolou P., Papadimitriou M., Papapsotiriou I. Stemness gene profiles of circulating tumor cells. J Cancer Ther. 2017;9 [Google Scholar]
- 89.Cristofanilli M., Budd G.T., Ellis M.J. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 90.O'Hara S.M., Moreno J.G., Zweitzig D.R., Gross S., Gomella L.G., Terstappen L.W. Multigene reverse transcription-PCR profiling of circulating tumor cells in hormone-refractory prostate cancer. Clin Chem. 2004;50:826–835. doi: 10.1373/clinchem.2003.028563. [DOI] [PubMed] [Google Scholar]
- 91.Boral D., Vishnoi M., Liu H.N. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun. 2017;8:196. doi: 10.1038/s41467-017-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hayes D.F., Thor A.D. c-erbB-2 in breast cancer: Development of a clinically useful marker. Semin Oncol. 2002;29:231–245. doi: 10.1053/sonc.2002.32899. [DOI] [PubMed] [Google Scholar]
- 93.Brouwer A., De Laere B., Peeters D. Evaluation and consequences of heterogeneity in the circulating tumor cell compartment. Oncotarget. 2016;7:48625–48643. doi: 10.18632/oncotarget.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andreopoulou E., Yang L.Y., Rangel K.M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 95.Stott S.L., Hsu C.H., Tsukrov D.I. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Theil G., Fischer K., Weber E. The use of a new CellCollector to isolate circulating tumor cells from the blood of patients with different stages of prostate cancer and clinical outcomes—A proof-of-concept Study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158354. e0158354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Wit S., van Dalum G., Lenferink A.T. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep. 2015;5:12270. doi: 10.1038/srep12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhan I., Mosesso K., Goyal L. Detection and analysis of circulating epithelial cells in liquid biopsies from patients with liver disease. Gastroenterology. 2018;155:2016–2018.e11. doi: 10.1053/j.gastro.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frank S., Müller J., Bonk C., Haroske G., Schackert H.K., Schackert G. Transmission of glioblastoma multiforme through liver transplantation. Lancet. 1998;352:31. doi: 10.1016/S0140-6736(98)24027-X. [DOI] [PubMed] [Google Scholar]
- 100.Jonas S., Bechstein W.O., Lemmens H.P., Neuhaus R., Thalmann U., Neuhaus P. Liver graft-transmitted glioblastoma multiforme. A case report and experience with 13 multiorgan donors suffering from primary cerebral neoplasia. Transpl Int. 1996;9:426–429. doi: 10.1007/BF00335707. [DOI] [PubMed] [Google Scholar]
- 101.Muller C., Holtschmidt J., Auer M. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 102.Sullivan J.P., Nahed B.V., Madden M.W. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao F., Cui Y., Jiang H. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget. 2016;7:71330–71340. doi: 10.18632/oncotarget.11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krol I., Castro-Giner F., Maurer M. Detection of circulating tumour cell clusters in human glioblastoma. Br J Cancer. 2018;119:487–491. doi: 10.1038/s41416-018-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gast C.E., Silk A.D., Zarour L. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4 doi: 10.1126/sciadv.aat7828. eaat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adams D.L., Martin S.S., Alpaugh R.K. Circulating giant macrophages as a potential biomarker of solid tumors. Proc Natl Acad Sci U S A. 2014;111:3514–3519. doi: 10.1073/pnas.1320198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jansen N., Coy J.F. Diagnostic use of epitope detection in monocytes blood test for early detection of colon cancer metastasis. Future Oncol. 2013;9:605–609. doi: 10.2217/fon.13.8. [DOI] [PubMed] [Google Scholar]
- 108.Müller S., Agnihotri S., Shoger K.E. Peptide vaccine immunotherapy biomarkers and response patterns in pediatric gliomas. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98791. [DOI] [PMC free article] [PubMed] [Google Scholar]