Abstract
Purpose
As a novel type of non-coding RNAs, circRNAs were found to play important roles in cancer progression. In this study, we aim to investigate genome-wide circRNAs expression and potential functions in glioma to find new therapeutic targets for glioma treatment.
Patients and Methods
A total of 92 pairs of glioma tissue samples and adjacent control samples were enrolled in this study. Microarray and bioinformatics tools were used to identify circRNAs expression in glioma. Relative expression of RNAs was validated by qRT-PCR. Cell viability and migration were measured by Cell Counting Kit-8 and wound-healing assay in U251 and SHG-44 cells. The statistical correlation and overall survival were calculated by Mann–Whitney U-test and Kaplan–Meier method.
Results
A total of 90 differentially expressed circRNAs were identified in glioma tissues compared to control. These circRNAs were mainly back-spliced forms chr1, chr6 and chr12 with a variable number of exons. QRT-PCR showed hsa_circ_0013520 and hsa_circ_0004379 were highly expressed in glioma tissues and cell lines. High expression of hsa_circ_0013520 and hsa_circ_0004379 was significantly correlated with tumor size (P-value < 0.001), Karnofsky Performance Status (KPS, P-value: 0.008 and 0.006) and TNM stage (P-value: 0.004 and 0.005). Moreover, patients with higher expression level of hsa_circ_0013520 and hsa_circ_0004379 had a poorer overall survival (P-value: < 0.01). Besides, loss-of-function experiments revealed that inhibition of hsa_circ_0013520 and hsa_circ_0004379 suppresses proliferation and invasion of glioma cells.
Conclusion
The present study is the first to report that hsa_circ_0013520 and hsa_circ_0004379 were up-regulated in glioma tissues and cell lines, and their expression was positively correlated with poor clinical features of glioma patients, which may serve as novel biomarkers for glioma diagnosis and therapy.
Keywords: circRNAs, glioma, biomarker, survival
Introduction
Glioma is the most common tumor of the central nervous system, with less than 5% survival rate after diagnosis.1 Although radiotherapy and chemotherapy treatments are applied to glioma patients, the clinical effect of the treatment is often poor and unsatisfactory.2,3 Thus, it is therefore of great significance to explore the molecular biology of glioma to find new therapeutic targets for treatment.
Circular RNAs (circRNAs) are emerging as a novel subtype of non-coding RNAs with a special loop structure, which is resistant to RNase R digestion.4,5 The relatively stable structure of circRNAs makes it a potential biomarker for disease diagnosis.6 In recent years, studies have found that circRNAs are abnormally expressed in various cancers, such as papillary thyroid cancer,7 lung cancer,8 and colorectal cancer.9 Previous research has also shown that circRNAs may function as sponges of microRNAs (miRNAs), bind to RNA-binding proteins and modulate transcription and cause interference with splicing.10 However, the expression profile and potential function of circRNAs in glioma remain largely unknown.
In the present study, we use microarray technology combined with bioinformatics tools to identify circRNAs expression in glioma tissues. After differential analysis of circRNAs, we screened out three circRNA candidates with a high expression level in glioma for further study. The high expression of circRNAs were validated in glioma tissues and cell lines by qRT-PCR experiment. Further statistical analysis helped to reveal the potential diagnostic value of circRNAs and their significant correlation with clinical features. Moreover, cellular loss-of-function experiments in glioma cell lines were conducted to investigate the potential function of circRNAs in glioma.
Materials and Methods
Patient Samples and Cell Lines
A total of 92 pairs of glioma tissue samples and adjacent normal samples were obtained from Tongji Hospital Affiliated to Huazhong University of Science and Technology, with an average age of 47.68 ± 2.94 years. None of the patients had other malignancies or had received any treatment before the research. This research was in accordance with the Helsinki Declaration and with the ethical standards of the Tongji Hospital Affiliated to Huazhong University of Science and Technology ethics committee. All patients agreed to participate in the study and signed the written informed consent. Two glioma cell lines (SHG-44 and U251) and human normal brain glial cell line HEB were kindly provided by Wang J Lab (Chinese Academy of Sciences, Beijing, China) and Liu X Lab (Tongji Hospital Affiliated to Huazhong University of Science and Technology, Wuhan, China).
Institutional Review Board Statement
This study was reviewed and approved by the Ethics Committee of Tongji Hospital Affiliated to Huazhong University of Science and Technology.
circRNAs Microarray
Six pairs of glioma tissues and normal tissues were removed from the liquid nitrogen tank for grinding, and the total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) according to the instruction of the manufacturer. After RNase R digestion, RNAs were supplied to microarray detection using Agilent Human CircRNA Array (V2.0) in CapitalBio company (Capitalbio, Beijing, China).
Bioinformatics and Statistical Analysis
The raw microarray data was processed with Agilent Feature Extraction software and measured by GeneSpring V12.0 software. The differentially expressed circRNAs were identified by EdgeR package from R project with fold change > 2 and P-value < 0.01. The correlation between circRNAs expression and clinical features was evaluated by Mann–Whitney U-test. Overall survival rate was evaluated by Kaplan–Meier method and Log rank test. All statistical analyses were performed using R program, P-value < 0.05 was considered to be statistically significant.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The total RNA was extracted from glioma tissues and normal tissues with Trizol (Invitrogen, CA, USA). The purity and concentration of RNAs were measured by ultraviolet spectrophotometer, and then total RNA of 5 μg was taken for reverse transcription of cDNA using TaqMan Reverse-transcription Kit according to the kit instructions. The PCR experiment was conducted on a Real-Time PCR detection system (Bio-Rad, Hercules, CA) using SYBR Mix. The reaction parameters were as follows: 37°C for 15 minutes, 42°C for 35 minutes and 70°C for 5 minutes. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, F: 5ʹ-GAAGGTGAAGGTC GGAGTC-3ʹ, R: 5ʹ-GAAGATGGTGATGGGATTTC-3ʹ) was used for circRNAs normalization. The 2-ΔΔct method was used to analyze the data.
Cell Culture and Transfection
Two glioma cell lines were cultured in a medium containing 10% PBS DMEM at 37°C and 5% CO2. When the adherent growth and fusion of cells reached 85%, 25% trypsin was added for digestion. Then the cells were placed in the medium for further culture and passage. Cell transfection was performed with Lipofectamine 2000 (Invitrogen, USA) in strict accordance with the corresponding kit instructions. SiRNAs were synthesized by RiboBio biotechnology company (RiboBio, Guangzhou, China).
Cell Counting Kit-8 (CCK-8) Assay and Wound-Healing Assay
After transfection for 48 hours, the glioma cells were inoculated in 96-well plates with about 100 μL cell solution per well and the cell density was adjusted to 3 × 104 cell/mL. Then CCK-8 solution (10 μL) (Dojindo, Japan) was added to each well following the instruction of the manufacturer. After incubation for 24, 48, 72 and 96 hours (at 37°C), the absorbance was measured at 450 nm using a spectrophotometer. For wound-healing assay, the transfected glioma cells were adjusted to 3 x 105 cells/mL and incubated on 6-well plates. A sterile 200 μL pipette tip was used to scratch linear wound. The images were captured by a microscope at 0 and 24 hours, respectively.
Results
Differentially Expressed circRNAs in Glioma
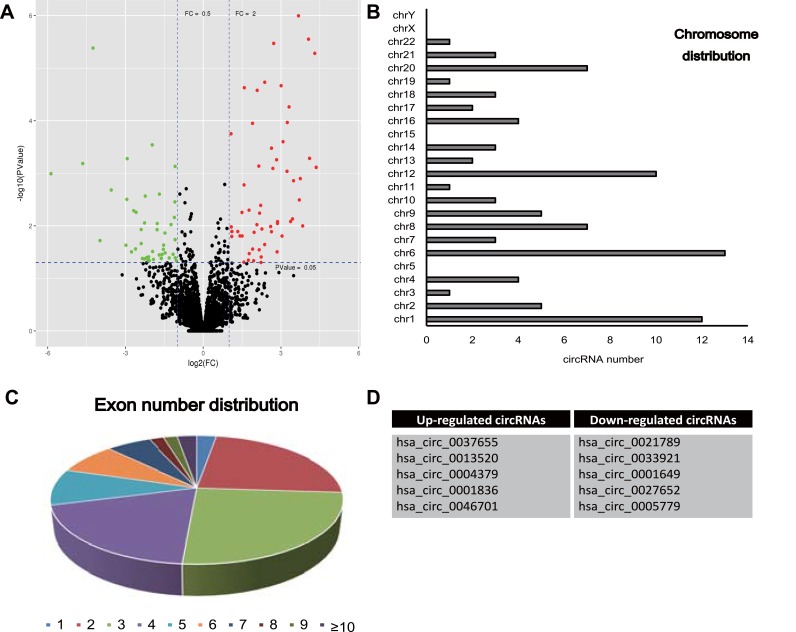
By means of microarray analysis, we identified a total of 90 differentially expressed circRNAs in glioma tissues compared to control, as shown in the the volcano plot in Figure 1A. These differentially expressed circRNAs were mainly back-spliced form chr1, chr6 and chr12 with a variable number of exons (Figure 1B and C). Through comparison of expression alteration between glioma tissues and control, we listed the top 10 differentially expressed circRNAs in Figure 1D, including five up-regulated circRNAs and five down-regulated circRNAs (P-value < 0.01). These abnormally expressed circRNAs may play important roles during pathogenesis of glioma.
Figure 1.
(A) Microarray analysis detected differentially expressed circRNAs in glioma tissues compared to control. Red represents the up-regulated circRNAs and green for down-regulated circRNAs. (B, C) Chromosome distribution and exon number distribution of differentially expression circRNAs. (D) List of top 10 differentially expression circRNAs.
Validation of Highly Expressed circRNAs in Glioma
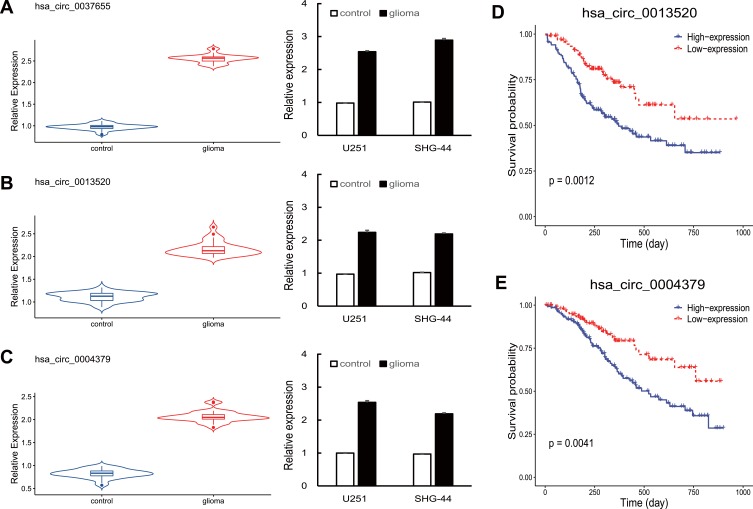
Considering that differentially expressed circRNAs with high expression levels in glioma seem to be more important than those expressed at relatively low levels, we chose three up-regulated circRNAs with higher expression variation in glioma from the microarray data, including hsa_circ_0037655, hsa_circ_0013520 and hsa_circ_0004379. To further validate the expression of these three circRNAs, we performed qRT-PCR experiments in 32 pairs of glioma tissues and controls, as well as two glioma lines (SHG-44 and U251). As shown in Figure 2A–C, the expression of hsa_circ_0037655, hsa_circ_0013520 and hsa_circ_0004379 were significantly up-regulated in glioma tissues and cell lines (P <0.05), consistent with microarray data.
Figure 2.
(A–C) Hsa_circ_0037655, hsa_circ_0013520 and hsa_circ_0004379 were up-regulated in glioma tissues and cell lines, as detected by qRT-PCR. (D, E) Survival analysis of hsa_circ_0013520 and hsa_circ_0004379. P < 0.05.
High Level of hsa_circ_0013520 and hsa_circ_0004379 were Correlated with Poor Clinical Features of Glioma Patients
To investigate the function of hsa_circ_0037655, hsa_circ_0013520 and hsa_circ_0004379 in glioma, we calculated the correlation between clinical features of glioma patients and circRNAs expression. Based on the mean expression level of circRNAs, glioma patients were divided into a low-expression group and a high-expression group for each circRNAs. As shown in Table 1, the expression of hsa_circ_0013520 and hsa_circ_0004379 was significantly correlated with tumor size (P-value < 0.001), Karnofsky Performance Status (KPS, P-value: 0.008 and 0.006) and TNM stage (P-value: 0.004 and 0.005), while hsa_circ_0037655 did not present a significant positive correlation with these clinical features (data not shown). Moreover, Kaplan-Meier survival analysis revealed that glioma patients with higher expression level of hsa_circ_0013520 and hsa_circ_0004379 had a poorer overall survival (P-value: < 0.01, Figure 2D and E), indicating the vital role of hsa_circ_0013520 and hsa_circ_0004379 in glioma progression.
Table 1.
Hsa_circ_0013520/hsa_circ_0004379 Expression and Clinical Features of 92 Glioma Patients
| Clinical Features | hsa_circ_0013520 Expression | P-value | hsa_circ_0004379 Expression | |||
|---|---|---|---|---|---|---|
| Low | High | Low | High | P-value | ||
| Gender | 0.747 | 0.799 | ||||
| Male | 30 | 19 | 27 | 22 | ||
| Female | 22 | 21 | 23 | 20 | ||
| Age (years) | 0.511 | 0.641 | ||||
| <50 | 31 | 24 | 29 | 23 | ||
| ≥50 | 21 | 16 | 23 | 19 | ||
| KPS | 0.008 | 0.006 | ||||
| < 90 | 33 | 37 | 32 | 39 | ||
| ≥ 90 | 19 | 3 | 18 | 3 | ||
| TNM stage | 0.004 | 0.005 | ||||
| I–II | 12 | 2 | 14 | 3 | ||
| >III | 40 | 38 | 36 | 39 | ||
| Tumor size | < 0.001 | <0.001 | ||||
| ≤3 cm | 4 | 1 | 5 | 2 | ||
| ≥3 cm | 48 | 39 | 45 | 40 | ||
Abbreviation: KPS, Karnofsky Performance Status.
Inhibition of hsa_circ_0013520 and hsa_circ_0004379 Suppresses Proliferation and Invasion of Glioma Cells
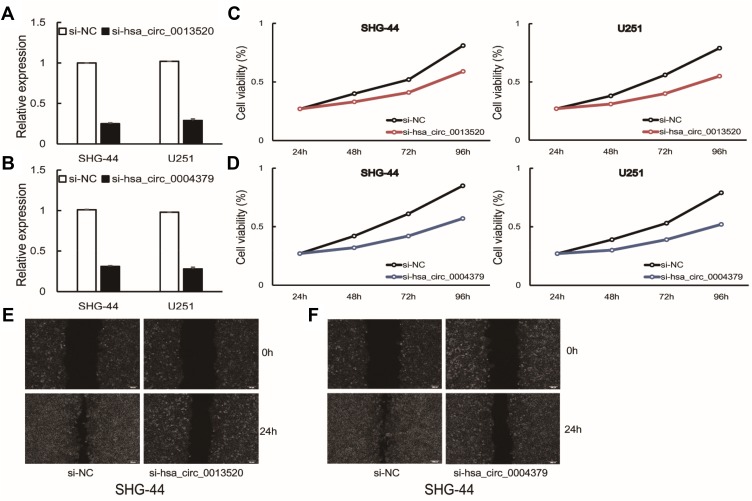
To further explore the function of hsa_circ_0013520 and hsa_circ_0004379 in glioma, cellular experiments were conducted in transfected SHG-44 and U251 cells. As shown in Figure 3A and B, the expression of two circRNAs were significantly reduced in glioma cells after transfection with si-circRNAs. CCK-8 assay revealed that inhibition of hsa_circ_0013520 and hsa_circ_0004379 could significantly suppress proliferation of glioma cells (Figure 3C and D, P < 0.05). In addition, , wound healing assay showed that the migration ability of glioma cells was dramatically suppressed by inhibition of two circRNAs (Figure 3D and E, P < 0.05). The above results indicated the cancer-promoting role of hsa_circ_0013520 and hsa_circ_0004379 in glioma cells.
Figure 3.
(A, B) Glioma cells were transfected with si-hsa_circ_0013520 and si-hsa_circ_0004379. (C–F) Proliferation and migration of transfected cells were detected by CCK-8 and wound-healing assay. P < 0.05.
Discussion
In recent years, circRNAs have been suggested by some research to play important roles in biological processes, for example cell differentiation, chromatin modification and posttranscriptional regulation.11,12 CircRNAs are characterized by closed loops without a 3′ Poly A tail, this looping structure may enhance the stability of circRNAs and therefore make it a potential molecular biomarker for cancer diagnosis and therapy.6,13
The abnormal circRNAs expression is often associated with the development and progression of cancer, including breast cancer, leukemia, and lung cancer.14 Herein, we investigated the circRNAs expression in glioma by means of microarray technology to identify potential biomarkers for diagnosing glioma patients. A total of 90 differentially expressed circRNAs were identified in six pairs of glioma tissues and control tissues, these abnormally expressed circRNAs may play vital roles during the pathogenesis of glioma. Considering the limits of microarray technology,15,16 we further validated circRNAs expression in glioma tissues using qRT-PCR experiments although the sample size used for microarray is acceptable.17 The top three differentially expressed circRNAs (hsa_circ_0037655, hsa_circ_0013520 and hsa_circ_0004379) with high expression levels in glioma had drawn our attention, and the QRT-PCR results indicated that the three circRNAs were notably up-regulated in glioma tissues and cell lines, consistent with the microarray data.
Previous studies demonstrated that circRNAs may serve as candidate biomarkers and therapeutic targets for cancer,5 for example, Xu et al showed the up-regulation of circRNA_0001178 and circRNA_0000826 contributed to liver metastases in colorectal cancer.18 High expression levels of hsa_circ_0000064 in lung cancer was correlated with clinical characteristics;19 Huang et al found that hsa_circ_0002060 was associated with low bone mineral density in osteoporosis patients.20 However, the biological functions of the three up-regulated circRNAs in glioma were poorly understood. We further confirmed the significant positive correlation between circRNAs expression (hsa_circ_0013520 and hsa_circ_0004379) and clinical features (tumor size, Karnofsky Performance Status and TNM stage) of 92 glioma patients, while no significant correlation was observed in hsa_circ_0037655. Survival analysis revealed that high expression levels of hsa_circ_0013520 and hsa_circ_0004379 led to poorer overall survival. Moreover, the cellular loss-of-function experiments in glioma cell lines demonstrated that silencing of hsa_circ_0013520 and hsa_circ_0004379 suppressed cell proliferation and migration ability. The above findings provide insights into the biological significances and forecast values of hsa_circ_0013520 and hsa_circ_0004379 in glioma.
Conclusion
In conclusion, this study is the first to report that hsa_circ_0013520 and hsa_circ_0004379 were up-regulated in glioma tissues and cell lines, and their expression was positively correlated with poor clinical features of glioma patients. Our study demonstrated that hsa_circ_0013520 and hsa_circ_0004379 may serve as novel biomarkers for glioma diagnosis and therapy.
Author Contributions
Xiaojuan Lyu and Zhen Dong designed the research. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. doi: 10.1016/S0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- 2.Gusyatiner O, Hegi ME. Glioma epigenetics: From subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. doi: 10.1016/j.semcancer.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 3.Barthel FP, Wesseling P, Verhaak RGW. Reconstructing the molecular life history of gliomas. Acta Neuropathol. 2018;135(5):649–670. doi: 10.1007/s00401-018-1842-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi: 10.1186/s12943-018-0812-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 6.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e13. doi: 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi W, Huang J, Nie C, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of beta-catenin pathway. J Exp Clin Cancer Res. 2018;37(1):275. doi: 10.1186/s13046-018-0936-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644. doi: 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]
- 9.Li X-N, Wang Z-J, Ye C-X, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. doi: 10.1186/s13046-018-1006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. [DOI] [PubMed] [Google Scholar]
- 11.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conn SJ, Pillman K, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 13.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi: 10.15252/embj.2018100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Wei H. Designing microarray and RNA-Seq experiments for greater systems biology discovery in modern plant genomics. Mol Plant. 2015;8(2):196–206. doi: 10.1016/j.molp.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 16.Narrandes S, Xu W. Gene expression detection assay for cancer clinical use. J Cancer. 2018;9(13):2249–2265. doi: 10.7150/jca.24744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Wang C, Song H, et al. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. doi: 10.1186/s12943-018-0932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo YH, Zhu X-Z, Huang K-W, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015 [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Xie J, Li E. Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarker for osteoporosis. J Cell Biochem. 2019;120(9):15688–15694. doi: 10.1002/jcb.v120.9 [DOI] [PubMed] [Google Scholar]