Abstract
Background
The histological grade of pancreatic cancer is an important independent predictor of outcome. However, we lack a method for safely and accurately obtaining the pathological grade before surgery. Radiomics has been used to discriminate between histological grades in tumors. We aimed to develop and validate a radiomics signature for the preoperative prediction of histological grades of pancreatic ductal adenocarcinoma (PDAC) that was based on contrast-enhanced computed tomography (CE-CT).
Methods
This study comprised 301 patients with pathologically confirmed PDAC who were randomly divided into a training (n=151) and test group (n=150). Radiomics features were selected by a support vector machine (SVM) model, and a radiomics signature was generated by the least absolute shrinkage and selection operator (LASSO) model. An additional 100 patients from 2 other medical centers were used for external validation. Receiver operating characteristic (ROC) curve analysis was used to assess the model and to identify the optimal cutoff value.
Results
The radiomics signatures between high-grade and low-grade PDACs in the training and test groups were significantly different (P<0.05). The areas under the curve (AUCs) of the training and test datasets were 0.961 and 0.910, respectively. The optimal cutoff value of the radiomics score was 0.426. In the external validation dataset, the difference between the radiomics signatures of high-grade versus low-grade PDACs was also significant (P<0.05). The radiomics signature for the external validation data had an AUC of 0.770.
Conclusions
The CE-CT-based radiomics signature showed moderate predictive accuracy for differentiating low-grade from high-grade PDAC and should become a new noninvasive method for the preoperative prediction of histological grades of PDAC.
Keywords: Tomography, X-ray computed; carcinoma, pancreatic ductal; receiver operating characteristic curve (ROC curve); support vector machine (SVM)
Introduction
As of 2018, pancreatic cancer is the seventh leading cause of cancer death, having a high mortality rate, with almost as many deaths (n=432,000) as cases (n=459,000) (1). Pancreatic ductal adenocarcinoma (PDAC) is the main histopathological type of pancreatic cancer. Currently, surgery remains the only curative option for PDAC (2) but obtains poor outcomes. The histological grade of pancreatic cancer is an important independent predictor of outcome (3-10). Low-grade PDAC has been associated with long-term postoperative survival (5,8). High-grade PDAC tends to predict shorter survival and increased mortality compared with a low-grade disease (3,7). Even patients with the high-grade disease who undergo surgery did not have a significantly improved prognosis, but they might be affected by surgical complications and have a significantly worse quality of life. To some extent, the histological grade of PDAC might affect the treatment options (11). In order to improve a patient's quality of life, it is critical to avoid unnecessary surgical complications, select the best treatment plan, and prolong survival; furthermore, an accurate preoperative assessment of tumor grade could be essential for individual therapy. To date, the methods for grading pancreatic cancer involve invasive biopsies, which include endoscopic ultrasound-guided fine-needle biopsy (EUS-FNB) and ultrasound/computed tomography (CT)-guided percutaneous biopsy; however, patients who undergo these procedures risk suffering complications such as pancreatitis (12). In addition, because of the heterogeneity of pancreatic tumors, the histopathological evaluation of a specimen obtained by puncture biopsy is not equivalent to the whole tumor (13). A safe and accurate method that can provide a histopathological grade of the tumor before surgery is needed. Traditional imaging examinations are widely used as noninvasive preoperative evaluation methods for patients with PDAC and can provide information, including tumor size, site, and relationship with vessels. We attempted to clarify the differentiation of PDAC from CT images further.
Radiomics (14-17) is a quantitative imaging method that can extract more data from digital medical images to improve the accuracy of clinical diagnosis, prediction, and prognosis, and can support personalized clinical decisions and improve individualized treatment options. Radiomics based on CT and magnetic resonance imaging (MRI) has been used to discern the histological grades of colorectal cancers (18), bladder cancers (19), hepatocellular carcinomas (20), non-small cell lung cancers (21), and gliomas (22). In our study, we attempted to perform radiomics for patients with PDAC by extracting and selecting the features on preoperative contrast-enhanced CT (CE-CT) images that were highly correlated with histological tumor grade, to develop a radiomics signature in the training dataset, and to validate the signature in the test dataset. An external validation dataset was also used for further testing of the accuracy of the model. The aim of our study was to build a predictive model for differentiating low-grade from high-grade PDAC.
Methods
Patients
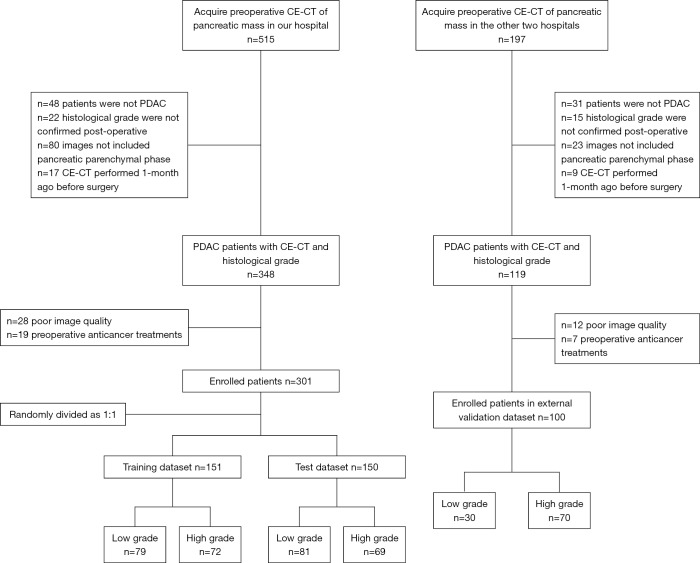
Ethical approval was obtained for this retrospective study, and the requirement for informed consent was waived. The inclusion criteria were as follows: (I) patients were histopathologically confirmed to have PDAC postoperatively; (II) the defined histological grade was on patients’ reports; (III) CE-CT included the pancreatic parenchymal phase; and (IV) CE-CT was performed within 1 month before surgery. The exclusion criteria were as follows: (I) poor image quality, including artifacts (respiratory movements or metallic artifacts associated with a stent) in the tumor or difficulty in identifying the tumor on the CT scans of every phase; or (II) preoperative anticancer treatment. A total of 301 patients admitted to Shengjing Hospital of China Medical University from January 2005 to December 2018, were finally enrolled in this study. They were randomly divided into a training group with 151 patients (89 men, 62 women; age, 60.6±9.4 years) and a test group with 150 patients (94 men, 56 women; age, 60.0±9.6 years). An additional 100 patients (55 men, 45 women; age, 59.3±8.9 years) from The First Affiliated Hospital of China Medical University and Liaoning Cancer Institute and Hospital were recruited as an external validation group from January 2010 to December 2018, with the same criteria (Figure 1). Clinical data such as age, gender, tumor location, and preoperative carbohydrate antigen 19-9 (CA19-9) level were recorded, with CA19-9 level described as normal (0–37 U/mL) or abnormal (>37 U/mL).
Figure 1.
Flow chart of the patients enrolled in the training, test, and external validation datasets. PDAC, pancreatic ductal adenocarcinoma; CE-CT, contrast-enhanced computed tomography.
Histological grading
All surgically removed tumors were formalin-fixed, paraffin-embedded, and hematoxylin-eosin stained. According to the WHO classification of tumors of the digestive system (4th edition) (23), the classification of the gland is as follows: well, moderately, poorly differentiated, and undifferentiated. In our study, dichotomous statistics were used to classify well-differentiated (n=56) and moderately differentiated (n=104) versus poorly differentiated (n=133) and undifferentiated (n=8) cancers into low-grade (n=160) versus high-grade groups (n=141), respectively.
Imaging acquisition
The participants in our study underwent CE-CT via multiple CT devices (Brilliance iCT; Philips Healthcare, USA; Aquillion ONE; Toshiba Medical Systems Corporation, Japan; and Somatom Definition AS; Siemens, Germany) at the three participating centers. The imaging conditions satisfied the following requirements: multislice spiral CT scanners; scanning field of view, 35 cm × 35 cm–40 cm × 40 cm; matrix, 512×512; thickness of reconstructed image, 1–3 mm; three-phase scan, preinjection phase, pancreatic parenchymal phase (40–45 s after injection), and portal phase (70–75 s after injection). The images from the pancreatic parenchymal phase were used for feature extraction in our study. The dose of iodine contrast agent was based on the patient’s weight (1 mL/kg), and the flow rate was 2.5–3.5 mL/s.
Tumor segmentation and feature extraction
The segmentation of every tumor on the training, test, and validation datasets was performed by the same junior radiologist with a working experience of 8 years. Imaging Biomarker Explorer (IBEX) software (V1.0β; MD Anderson Cancer Center, Houston, TX, USA) (24) was used. Regions of interest (ROIs) were manually drawn along the borders of the lesions on images of the pancreatic parenchymal phase. Two weeks after the first ROIs were delineated, 30 cases were randomly selected for repeat delineation of the ROIs by the same radiologist, and the extracted features were evaluated by calculating the intraclass correlation coefficient (ICC). Another senior radiologist with a diagnostic experience of 20 years then performed ROI delineation and feature extraction on the same 30 cases. The extracted features from the ROIs of the first less experienced radiologist were compared to those from the ROIs of the experienced radiologist by calculating the ICC. An ICC >0.75 was considered to be excellent for the stability and reproducibility of features. The radiomics features included morphology, intensity-based statistics, intensity histogram, gray level co-occurrence matrix (GLCM), gray level run length matrix, and neighborhood grey tone difference matrix. The parameters of the features are provided in Table S1.
Table S1. Features extracted by IBEX.
| Feature | Definition/formula |
|---|---|
| GLCM (24-28) (direction =0°, 45°, 90°, 135°; offset =1, 4, 7; symmetric =1) | |
| Auto correlation | |
| Cluster prominence | |
| Cluster shade | |
| Cluster tendency | |
| Contrast | |
| Correlation | |
| Difference entropy | |
| Dissimilarity | |
| Energy | |
| Entropy | |
| Homogeneity | |
| Homogeneity 2 | |
| Information measure corr 1 (IMC1) | |
| Information measure corr 2 (IMC2) | |
| Inverse diff moment norm (IDMN) | |
| Inverse diff norm (IDN) | |
| Inverse variance | |
| Max probability | |
| Sum average | |
| Sum entropy | |
| Sum variance | |
| Variance | |
| Gray level run length matrix (24,29) (direction =0°,90°) | |
| Gray level nonuniformity (GLN) | |
| High gray level run empha (HGLRE) | |
| Long run emphasis (LRE) | |
| Long run high gray level empha (LRHGLE) | |
| Long run low gray level empha (LRLGLE) | |
| Low gray level run empha (LGLRE) | |
| Run length nonuniformity (RLN) | |
| Run percentage (RP) | |
| Short run emphasis (SRE) | |
| Short run high gray level empha (SRHGLE) | |
| Short run low gray level empha (SRLGLE) | |
| Intensity direct (24,28) (threshold: 0–8,000) | |
| Energy | |
| Global entropy | The intensity entropy among all the voxels |
| Global max | The intensity maximum among all the voxels |
| Global mean | The intensity mean among all the voxels |
| Global median | The intensity median among all the voxels |
| Global min | The intensity minimum among all the voxels |
| Global std | The intensity standard deviation among all the voxels |
| Global uniformity | The intensity uniformity among all the voxel |
| Inter quartile range | The interquartile range of the intensity values among all the voxels |
| Kurtosis | Measure the peakedness of all the voxels’ intensity |
| Local entropy max | (I) First, at each voxel, compute entropy in its neighborhood region; (II) then, compute the maximum among all the voxel’s entropy calculated from (I) |
| Local entropy mean | (I) First, at each voxel, compute entropy in its neighborhood region; (II) then, compute the mean among all the voxel’s entropy calculated from (I) |
| Local entropy median | (I) First, at each voxel, compute entropy in its neighborhood region; (II) then, compute the median among all the voxel’s entropy calculated from (I) |
| Local entropy min | (I) First, at each voxel, compute entropy in its neighborhood region; (II) then, compute the minimum among all the voxel’s entropy calculated from (I) |
| Local entropy std | (I) First, at each voxel, compute entropy in its neighborhood region; (II) then, compute the standard deviation among all the voxel’s entropy calculated from (I) |
| Local range max | (I) First, at each voxel, compute range value (max value–min value) in its neighborhood region; (II) then, compute the median among all the voxel’s range value calculated from (I) |
| Local range mean | (I) First, at each voxel, compute range value (max value–min value) in its neighborhood region; (II) then, compute the mean among all the voxel’s range value calculated from (I) |
| Local range median | (I) First, at each voxel, compute range value (max value–min value) in its neighborhood region; (II) then, compute the median among all the voxel’s range value calculated from (I) |
| Local range min | (I) First, at each voxel, compute range value (max value–min value) in its neighborhood region; (II) then, compute the minimum among all the voxel’s range value calculated from (I) |
| Local range std | (I) First, at each voxel, compute range value (max value–min value) in its neighborhood region; (II) then, compute the standard deviation among all the voxel’s range value calculated from (I) |
| Local std max | (I) First, at each voxel, compute standard deviation in its neighborhood region; (II) then, compute the maximum among all the voxel’s standard deviation value calculated from (I) |
| Local std mean | (I) First, at each voxel, compute standard deviation in its neighborhood region; (II) then, compute the mean among all the voxel’s standard deviation value calculated from (I) |
| Local std median | (I) First, at each voxel, compute standard deviation in its neighborhood region; (II) then, compute the median among all the voxel’s standard deviation value calculated from (I) |
| Local std min | (I) First, at each voxel, compute standard deviation in its neighborhood region; (II) then, compute the minimum among all the voxel’s standard deviation value calculated from (I) |
| Local std std | (I) First, at each voxel, compute standard deviation in its neighborhood region; (II) then, compute the standard deviation all the voxel’s standard deviation value calculated from (I) |
| Mean absolute deviation | The mean absolute deviation of the intensity values among all the voxels |
| Median absolute deviation | The median absolute deviation of the intensity values among all the voxels |
| Percentile | Percentiles of the intensity values among all the voxels. Percentile =5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95 |
| Quantile | Quantiles of the intensity values among all the voxels. Quantile =0.025, 0.25, 0.5, 0.75, 0.975 |
| Range | The intensity range (max value–min value) among all the voxels |
| Root mean square (RMS) | |
| Skewness | Measure the asymmetry of all the voxels’ intensity |
| Variance | |
| Intensity histogram (24,28) (number of bins =256, range: 0–4,096, range fix =1) | |
| Inter quartile range | The interquartile range of the occurrence probability values in the histogram |
| Kurtosis | Measure the peakedness of the occurrence probability values in the histogram |
| Mean absolute deviation | The mean absolute deviation of the occurrence probability values in the histogram |
| Median absolute deviation | The median absolute deviation of the occurrence probability values in the histogram |
| Percentile | Percentiles of the occurrence probability values in the histogram. Percentile =5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95 |
| Percentile area | Percentiles of values in the accumulative histogram. Percentile =5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95 |
| Quantile | Quantiles of the occurrence probability values in the histogram. Quantile =0.025, 0.25, 0.5, 0.75, 0.975 |
| Range | Measure the range (max value–min value) of the occurrence probability values in the histogram |
| Skewness | Measure the asymmetry of the occurrence probability values in the histogram |
| Intensity histogram gauss fit (24) (number of bins =100, range: 0–4,096, range fix =0, number of gauss =2; number of gauss fix =1) | |
| Gauss amplitude | Amplitude of each gaussian curve |
| Gauss area | Area of each gaussian curve |
| Gauss mean | Mean of each gaussian curve |
| Gauss std | Standard deviation of each gaussian curve |
| Number of gauss | The number of gaussian curve that are used to approximate the curve |
| Neighbor intensity difference (24,30) (the neighborhood matrix size =3, NHoodSym =1, neighborhood matrix size in Y are calculated to best match neighborhood physical length in X dimension, not include edge pixels for analysis, number of bins =256) | |
| Busyness | – |
| Coarseness | – |
| Complexity | – |
| Contrast | – |
| Texture strength | – |
| Shape (24,28,31) | |
| Compactness 1 | |
| Compactness 2 | |
| Convex | (I) First, compute convex value in 2D slice-by-slice. Convex = regionprops (2D mask, ‘solidity’); (II) then, compute the mean of convex value among the slices |
| Convex hull volume | The mean volume of the 2D convex hulls that are the convex envelopes of each slice's binary mask |
| Convex hull volume 3D | 3D volume of the convex hull that is the convex envelope of binary mask |
| Mass | – |
| Max 3D diameter | Max 3D diameter = largest pairwise Euclidean distance between voxels on the surface of the tumor volume |
| Mean breadth | Mean breath = integral of mean curvature |
| Number of objects | – |
| Number of voxel | The number of voxels treating the edge voxels differently |
| Orientation | (I) First, compute orientation value in 2D slice-by-slice. Orientation =regionprops (2D mask, ‘orientation’); (II) then, compute the mean of orientation value among the slices |
| Roundness | – |
| Spherical disproportion | |
| Sphericity | |
| Surface area | |
| Surface area density | |
| Volume | The physical volume treating the edge voxels differently |
| Voxel size | The physical voxel size |
IBEX, Imaging Biomarker Explorer; GLCM, gray level co-occurrence matrix.
Feature selection and development of radiomics signatures
A support vector machine (SVM) method (kernel = rbf, C =0.78, gamma =0.00069) based on recursive feature elimination (RFE) was used to select features for predicting PDAC grade, which trained the classifier using all features, computed the ranking criterion for all features, and removed the features with smallest ranking criterion (15). The procedure was iterated until the number of features was equal to the number of predefined features. All the data of the selected features were normalized with z-score normalization in the training dataset. The test and external validation datasets were also normalized using the mean and standard deviation derived from the features of the training dataset. Redundant features that referred to low discrimination features with variances lower than the threshold were removed. The threshold in our study was 0.5, which was obtained by grid search.
The least absolute shrinkage and selection operator (LASSO) regression model was used to build a predictive classifier machine and was trained by a ten-fold cross-validation method. We calculated a radiomics score (Radscore) for each patient with the following formula:
, where N is the feature number, wi is the weight of the ith feature, xi is the value of the ith selected feature, and b is biased.
Model evaluation
The model that was built from the training dataset was applied to the test dataset and the external validation dataset to assess accuracy. Receiver operating characteristic (ROC) curve analysis and calculations of areas under the curve (AUCs) were performed to determine the discriminative performance of the radiomics signature. The optimal cutoff value of the radiomics score was determined by the Youden index and was determined only in the training dataset, and then directly applied to the testing and external validation dataset. The corresponding sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy were also determined.
Statistical analysis
SPSS software (version 22.0.0.0, IBM Corporation, Armonk, NY, USA) was used for statistical analysis. Continuous variables were expressed as means ± SD or medians and interquartile range (IQR), and categorical variables were expressed as numbers and percentages. Differences between variables of the patient groups were assessed by the independent t-test or Mann-Whitney U test for continuous variables and Fisher’s exact test or chi-squared test for categorical variables. A two-sided P value <0.05 was considered statistically significant.
Multivariate logistic regression analysis was performed to identify independent predictors of PDAC grade and included items such as the Radscore and clinical variables (age, gender, CA19-9 levels, and tumor location).
The SVM model was used to select features, and the LASSO regression model was used to build a radiomics signature. They were all performed by PYTHON (version 3.7.0; www.python.org).
Results
Clinicopathological characteristics
The clinicopathological characteristics of the study patients are shown in Tables 1,2. The differences between age, gender, tumor location, and CA19-9 level of the low-grade versus the high-grade patients were not significant. The P values of age, gender, tumor location, and CA19-9 level were 0.080, 0.420, 0.206, and 0.278 respectively in the training dataset; 0.557, 0.107, 0.674, and 0.271 in the test dataset; and 0.551, 0.511, 0.227, and 0.275 in the external validation dataset. The difference between the histologic grades in the training and external validation datasets was significant (P=0.005). The differences between age, gender, tumor location, and CA19-9 level in the training and external validation datasets were not significant, and P values were 0.301, 0.537, 0.287, and 0.474 respectively.
Table 1. Clinicopathological characteristics and radiomics scores of patients in the training, testing, and external validation datasets.
| Characteristics | Training dataset | Testing dataset | External validation dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low-grade | High-grade | P | Low-grade | High-grade | P | Low-grade | High-grade | P | |||
| Age, mean ± SD, y | 61.823±8.728 | 59.153±9.908 | 0.080 | 60.395±9.230 | 59.464±10.131 | 0.557 | 59.671±8.889 | 58.500±9.176 | 0.551 | ||
| Gender, n (%) | 0.420 | 0.107 | 0.511 | ||||||||
| Male | 49 (62.0) | 40 (55.6) | 46 (56.8) | 48 (69.6) | 40 (57.1) | 15 (50.0) | |||||
| Female | 30 (38.0) | 32 (44.4) | 35 (43.2) | 21 (30.4) | 30 (42.9) | 15 (50.0) | |||||
| Location, n (%) | 0.206 | 0.674 | 0.227 | ||||||||
| Head/neck | 57 (72.2) | 45 (62.5) | 52 (64.2) | 42 (60.9) | 40 (57.1) | 21 (70.0) | |||||
| Body/tail | 22 (27.8) | 27 (37.5) | 29 (35.8) | 27 (39.1) | 30 (42.9) | 9 (30.0) | |||||
| CA19-9, n (%) | 0.278 | 0.271 | 0.275 | ||||||||
| Normal (0–37 U/mL) | 16 (20.3) | 20 (27.8) | 10 (12.3) | 13 (18.8) | 16 (22.9) | 4 (13.3) | |||||
| Abnormal (>37 U/mL) | 63 (79.7) | 52 (72.2) | 71 (87.7) | 56 (81.2) | 54 (77.1) | 26 (86.7) | |||||
| Radscore, median (IQR) | 0.115 (–0.087 to 0.329) | 0.690 (0.516 to 0.908) | <0.001 | 0.304 (0.106 to 0.413) | 0.695 (0.559 to 0.852) | <0.001 | 0.311 (0.147 to 0.427) | 0.535 (0.385 to 0.710) | <0.001 | ||
P<0.05 indicates a statistically significant difference. IQR, interquartile range; CA19-9, carbohydrate antigen 19-9; RADSCORE, radiomics score.
Table 2. Clinicopathological characteristics between the training and external validation datasets.
| Characteristics | Training dataset | External validation dataset | P |
|---|---|---|---|
| Age, mean ± SD, y | 60.550±9.374 | 59.320±8.945 | 0.301 |
| Gender, n (%) | 0.537 | ||
| Male | 89 (58.9) | 55 (55.0) | |
| Female | 62 (41.1) | 45 (45.0) | |
| Location, n (%) | 0.287 | ||
| Head and neck | 102 (67.5) | 61 (61.0) | |
| Body and tail | 49 (32.5) | 39 (39.0) | |
| CA19-9, n (%) | 0.474 | ||
| Normal (0–37 U/mL) | 36 (23.8) | 20 (20.0) | |
| Abnormal (>37 U/mL) | 115 (76.2) | 80 (80.0) | |
| Histopathology grade, n (%) | 0.005 | ||
| Low-grade | 79 (52.3) | 70 (70.0) | |
| High-grade | 72 (47.7) | 30 (30.0) | |
P<0.05 indicates a statistically significant difference. CA19-9, carbohydrate antigen 19-9.
Construction of radiomics signatures
A total of 1,452 features were extracted automatically by IBEX software. The interested reader can find the features in Table S1. Among these features, 614 were considered to have excellent robustness and reproducibility with ICCs >0.75. Finally, 80 important features with correspondingly weighted coefficients were selected from the 614 features and calculated by the model. The top 10 features with the highest weights included homogeneity, entropy, inverse variance, max probability, information measure correlation, and intensity. They are shown in Table S2. The formula for the radiomics score was generated by the LASSO method. The selected features with their corresponding weights and biases are shown in Table S3.
Table S2. The top 10 features with their corresponding weight of the selected 80 features.
| Feature | Weight |
|---|---|
| F2-GLCM 3; 8-4 homogeneity | –0.504 |
| F1-GLCM 25; 90-7 entropy | 0.494 |
| F2-GLCM 3; 1-4 entropy | –0.476 |
| F1-GLCM 25; 90-1 inverse variance | –0.467 |
| F2-GLCM 3; 0-7 entropy | 0.461 |
| F2-GLCM 3; 8-4 homogeneity 2 | 0.288 |
| F2-GLCM 3; 8-7 max probability | 0.237 |
| F2-GLCM 3; –333-4 information measure corr 2 | 0.227 |
| F4-intensity direct; 90 percentile | 0.225 |
| F2-GLCM 3; 5-4 inverse variance | 0.193 |
GLCM, gray level co-occurrence matrix.
Table S3. The features with their corresponding weight and bias in radiomics score formula.
| Feature | Weight |
|---|---|
| 45-4 cluster shade | –0.032 |
| 90-4 cluster shade | –0.168 |
| 45-4 difference entropy | –0.036 |
| 0-4 entropy | –0.108 |
| 90-7 entropy | 0.494 |
| –333-4 information measure corr 2 | –0.086 |
| 90-1 inverse variance | –0.467 |
| 45-7 max probability | –0.053 |
| 45-4 sum entropy | –0.189 |
| 0-4 cluster shade | 0.001 |
| 4-4 cluster shade | 0.167 |
| 6-7 contrast | –0.033 |
| 8-4 contrast | 0.014 |
| 8-7 contrast | 0.097 |
| 11-4 contrast | 0.112 |
| 5-4 correlation | 0.030 |
| 5-7 correlation | –0.048 |
| 7-7 correlation | 0.049 |
| 10-4 correlation | –0.117 |
| 10-7 correlation | 0.053 |
| –333-4 difference entropy.1 | –0.145 |
| 4-4 difference entropy | 0.041 |
| 8-4 dissimilarity | 0.086 |
| 10-1 dissimilarity | –0.187 |
| 10-7 dissimilarity | 0.053 |
| 0-7 entropy.1 | 0.461 |
| 1-4 entropy | –0.476 |
| 8-4 homogeneity | –0.504 |
| 8-4 homogeneity 2 | 0.288 |
| 10-7 homogeneity 2 | –0.159 |
| –333-4 information measure corr 1.1 | 0.085 |
| 5-1 information measure corr 1 | 0.043 |
| 5-7 information measure corr 1 | 0.038 |
| 9-4 information measure corr 1 | 0.028 |
| 11-4 information measure corr 1 | 0.053 |
| –333-4 information measure corr 2.1 | 0.227 |
| 5-4 information measure corr 2 | –0.042 |
| 6-4 information measur corr 2 | 0.090 |
| 9-7 information measure corr 2 | 0.113 |
| 10-7 information measure corr 2 | –0.121 |
| 10-1 inverse diff norm | –0.022 |
| 0-1 inverse variance.1 | 0.182 |
| 5-1 inverse variance | 0.051 |
| 5-4 inverse variance | 0.193 |
| 7-1 inverse variance | –0.075 |
| 7-4 inverse variance | –0.057 |
| 7-7 inverse variance | –0.094 |
| 8-1 inverse variance | –0.113 |
| 11-7 inverse variance | –0.004 |
| 12-1 inverse variance | 0.161 |
| 2-7 max probability | 0.127 |
| 4-7 max probability | –0.156 |
| 8-7 max probability | 0.238 |
| 11-7 max probability | –0.135 |
| 4-4 sum entropy | –0.191 |
| –333 run length nonuniformity | 0.134 |
| 0 run length nonuniformity | 0.102 |
| Global min | –0.003 |
| Local std std | –0.059 |
| 90 percentile | 0.225 |
| 0.975 quantile | –0.034 |
| Skewness | 0.023 |
| 90 percentile.1 | 0.025 |
| 0.975 quantile.1 | –0.026 |
| Skewness.1 | 0.005 |
| 7 gauss amplitude | 0.158 |
| 3 gauss area | 0.093 |
| 6 gauss area | 0.061 |
| 1 gauss mean | –0.169 |
| 2 gauss mean | –0.109 |
| 3 gauss mean | 0.083 |
| 7 gauss mean | –0.109 |
| 2 gauss std | –0.002 |
| 3 gauss std | –0.078 |
| 7 gauss std | –0.069 |
| Coarseness | –0.191 |
| Complexity.1 | –0.044 |
| Convex | 0.088 |
| Max 3D diameter | 0.073 |
| Surface area density | –0.063 |
| Bias (b) | 0.430 |
Prediction performance of the radiomics signature
The median radiomics scores of the low-grade PDAC patients in the training and testing groups were 0.115 (–0.087 to 0.329) and 0.304 (0.106 to 0.413), respectively. The median radiomics scores of the high-grade PDAC patients in the training and testing groups were 0.690 (0.516 to 0.908) and 0.695 (0.559 to 0.852), respectively. The differences between the radiomics scores of the low-grade versus the high-grade PDAC patients in both the training and test groups were significant (P<0.001; Table 1, Figure 2).
Figure 2.
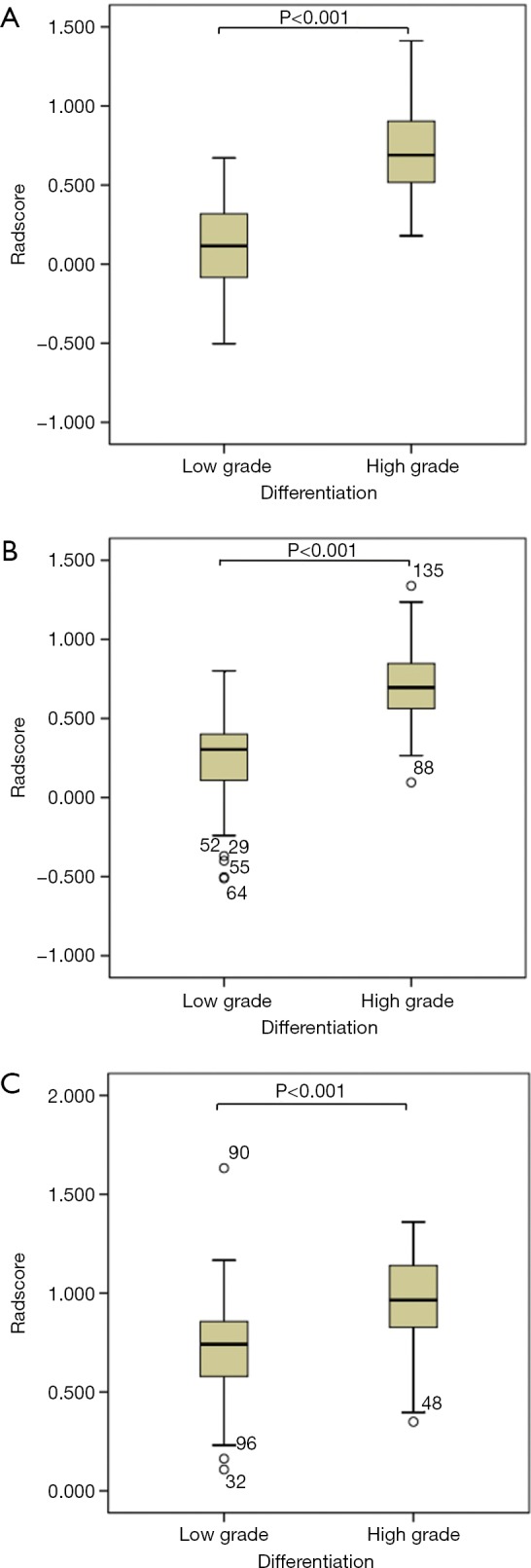
Boxplots of radiomics scores between the patients grouped according to low-grade and high-grade tumors in the training dataset (A), test dataset (B), and external validation dataset (C).
The AUCs of radiomics scores as predictors of tumor differentiation were 0.961 and 0.910 for the training and test groups, respectively (Figure 3). The cutoff value of the radiomics score was 0.426. The sensitivity, specificity, PPV, NPV, and accuracy of the radiomics score are shown in Table 3.
Figure 3.
ROC curves. (A) ROC curve of the training group. The cutoff value of radiomics score was 0.426, and the AUC was 0.961; (B) ROC curve of the test group, with the AUC being 0.910; (C) ROC curve of the external validation group, with AUC being 0.770. ROC, receiver operating characteristic; AUC, area under the curve.
Table 3. Predictive performance of radiomics signatures.
| Groups | AUC (95% CI) | SEN | SPE | PPV | NPV | ACC |
|---|---|---|---|---|---|---|
| Training dataset | 0.961 (0.935–0.987) | 0.886 | 0.917 | 0.921 | 0.880 | 0.901 |
| Testing dataset | 0.910 (0.864–0.956) | 0.778 | 0.899 | 0.900 | 0.775 | 0.833 |
| External validation dataset | 0.770 (0.661–0.878) | 0.757 | 0.667 | 0.841 | 0.541 | 0.730 |
AUC: 0.5–0.7, low accuracy; 0.7–0.9, moderate accuracy; >0.9, high accuracy. AUC, area under the curve; CI, confidence interval; SEN, sensitivity; SPE, specificity; PPV, positive predictive value; NPV, negative predictive value; ACC, accuracy.
External validation of the predictive performance of the radiomics signature
In the external validation group, the median radiomics scores of the low-grade and high-grade patients were 0.311 (0.147 to 0.427) and 0.535 (0.147 to 0.427), respectively, which was significantly different (P<0.001; Table 1, Figure 2). The AUC of the radiomics score for predicting histological grade was 0.770 (Figure 3). The sensitivity, specificity, PPV, NPV, and accuracy of the radiomics score are shown in Table 3.
Independent predictors of PDAC grade
After multivariate logistic regression analysis, the Radscore [odds ratio (OR) =0.003, P<0.001] was included in the predictive model. The P values of the remaining clinicopathological characteristics, including age, gender, CA19-9 level, and tumor site were 0.951, 0.388, 0.066, and 0.709, respectively, and were therefore not included.
Discussion
Radiomics signatures have been widely used in radiomics research. As an important radiomics predictive index, the radiomics signature shows good discrimination performance (32). It combines many features of the image, allowing a more comprehensive analysis that results in a more sensitive and accurate diagnosis of a lesion than a few or single image indicators would (33). In our study, the radiomics signature based on CE-CT showed good performance for predicting the histological grade of PDAC. Our result is consistent with the findings of Huang et al. (18) for colorectal cancers and Chen et al. (21) for non-small cell lung cancers. Our results also validated the conclusions of Choi (34) and Cassinotto (35), who assert that texture analysis is an important component of radiomics that reflects the histological differentiation of PDAC and correlates with patient outcomes.
In our study, the top 10 most important features belonged to the areas of homogeneity, entropy, inverse variance, max probability, information measure correlation, and intensity. Except for intensity, which belongs to the first-order feature that can reflect the gray intensity value of voxels, all the other features were second-order texture features, belonging to the GLCM. The GLCM refers to the spatial correlation characteristics of voxel gray values and is used to reflect texture (26). Homogeneity is a reflection of the uniformity of the distribution of voxels. Entropy refers to the randomness of the distribution of gray values which indicates the complexity of image texture: the more complex the image texture, the larger the entropy value. Inverse variance reflects local changes in image texture. If the image texture was relatively uniform and changed slowly between different regions, the inverse variance was increased. The max probability represents the texture feature that appears most frequently in the image. The features reflected the complexity and heterogeneity of the different aspects of the ROIs in the images and were combined to provide the radiomics signature for predicting the differentiation grade of each tumor.
To test the accuracy of our model further, we performed an external validation study (36-39). In the external validation dataset, the AUC of our model was 0.770, and the accuracy was 0.730, which was decreased in the prediction accuracy. This result might be related to the small sample size in the external validation dataset and the difference in sample distribution. The difference between the histological grades of the training and the external validation groups was significant (P=0.005), and the number of high-grade tumors in the external validation group was significantly lower than the number of low-grade tumors. The differences between the groups might account for the reduced quality of the model evaluation of the external validation group. Furthermore, our data were derived from multiple CT systems used at different medical centers. Differences between acquisition protocols might be the key reason for the reduced predictive power of the model in the external validation group. A multicenter prospective study using standardized acquisition and reconstruction protocols is our goal for the future.
In this study, we classified well-differentiated and moderately differentiated versus poorly differentiated and undifferentiated cancers into low-grade versus high-grade groups, respectively, which was consistent with the classification methods of Wasif and Rochefort et al. (3,4). This dichotomous method has been considered to reflect the biological behavior and outcomes of PDACs (23). Furthermore, there were relatively fewer cases in the well-differentiated group (n=56) and the undifferentiated group (n=8). This can be explained by the combined well and moderately differentiated cases versus the poorly differentiated and undifferentiated cases.
At present, histological grades are mainly obtained from FNB specimens before surgery. However, studies by Larghi et al. (40) have shown that the preoperative grading accuracy, sensitivity, and specificity of EUS-FNB were 56%, 41%, and 78%. This result was mainly attributed to the limited sample size and intra-observer inconsistency. The prediction model we built was based on preoperative CT images, which display the overall characteristics of the lesion, and the model objectively extracts the characteristics of the analyzed image, thereby avoiding the interference associated with subjective assessments and ultimately improving the accuracy of the prediction. Moreover, the risk of complications associated with a biopsy is avoided, and the predictive ability of pathological differentiation of pancreatic adenocarcinoma is improved before surgery.
In order to predict the histological grade of the lesion before surgery, some researchers have attempted to derive increased information from the traditional imaging examinations at the clinics. Elias et al. (41) evaluated the MRIs of 33 patients with pancreatic cancer to determine the degree of tumor differentiation based on the clarity of the margin of the tumor. The results showed that 71.4% of the poorly marginated lesions were well to moderately differentiated, while only 17.6% of focally defined cancers were well to moderately differentiated. In our study, the predictive accuracy of the radiomics signature was 73%, which was based on a larger sample size than the study by Elias et al., and avoided subjective judgment. Some investigators have also used CT perfusion parameters to predict the pathological differentiation of pancreatic cancer (42). The sensitivity and specificity for high-grade pancreatic cancer provided by the assessment of blood flow and blood volume were 79.2% and 82.4%, and 87.5% and 88.2%, respectively. The combination of CT perfusion assessments with radiomics might broaden the application of these techniques and provide improved results.
In addition, CA19-9 is an important tumor marker for pancreatic cancer and is closely related to outcomes of patients with pancreatic cancer (43-45). One study found CA19-9 produced in vitro by a pancreatic cancer cell line to be correlated with the histological grade of differentiation in vivo in nude mice (46). CA19-9 as a continuous variable might be more significant than a categorical variable. However, the data in our study came from multiple medical centers that evaluated patients over a long period of time, and the equipment and protocols were inconsistent. Thus, an accurate comparison between the variables of the internal and external validation sets was impossible. Therefore, we were only able to assess them as categorical variables. This issue needs to be resolved in future studies.
Our study has limitations. First, it only explored the relationship between a radiomics signature and the histological differentiation of PDACs but did not investigate the outcomes of the patients, about which patients and surgeons were more concerned, and for which further study is needed. Second, our study only analyzed the images of patients from the pancreatic parenchymal phase. Although PDAC lesions are well depicted in the pancreatic parenchymal phase, we still need to explore whether the noncontrast phase or portal vein phase will provide better results than the phase we investigated.
Conclusions
The CE-CT-based radiomics signature demonstrated moderate predictive accuracy for differentiating low-grade from high-grade PDAC and should become a new noninvasive method for the preoperative prediction of histological grades of PDAC.
Acknowledgments
We gratefully acknowledge the kind cooperation of Lingling Cui (The First Affiliated Hospital of China Medical University) and Yahong Luo (Liaoning Cancer Institute and Hospital) for providing patients datasets from the two other medical centers.
Funding: None.
Ethical Statement: The study protocol was approved by the Institutional Review Board (IRB) of the Shengjing Hospital ethics committee (No. 2019PS186K). A waiver of informed consent was requested and approved by the IRB.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011;378:607-20. 10.1016/S0140-6736(10)62307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasif N, Ko CY, Farrell J, Wainberg Z, Hines OJ, Reber H, Tomlinson JS. Impact of tumor grade on prognosis in pancreatic cancer: should we include grade in AJCC staging? Ann Surg Oncol 2010;17:2312-20. 10.1245/s10434-010-1071-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rochefort MM, Ankeny JS, Kadera BE, Donald GW, Isacoff W, Wainberg ZA, Hines OJ, Donahue TR, Reber HA, Tomlinson JS. Impact of tumor grade on pancreatic cancer prognosis: validation of a novel TNMG staging system. Ann Surg Oncol 2013;20:4322-9. 10.1245/s10434-013-3159-3 [DOI] [PubMed] [Google Scholar]
- 5.Stark AP, Sacks GD, Rochefort MM, Donahue TR, Reber HA, Tomlinson JS, Dawson DW, Eibl G, Hines OJ. Long-term survival in patients with pancreatic ductal adenocarcinoma. Surgery 2016;159:1520-7. 10.1016/j.surg.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellon EA, Jin WH, Frakes JM, Centeno BA, Strom TJ, Springett GM, Malafa MP, Shridhar R, Hodul PJ, Hoffe SE. Predictors and survival for pathologic tumor response grade in borderline resectable and locally advanced pancreatic cancer treated with induction chemotherapy and neoadjuvant stereotactic body radiotherapy. Acta Oncol 2017;56:391-7. 10.1080/0284186X.2016.1256497 [DOI] [PubMed] [Google Scholar]
- 7.Macías N, Sayagués JM, Esteban C, Iglesias M, González LM, Quiñones-Sampedro J, Gutiérrez ML, Corchete LA, Abad MM, Bengoechea O, Muñoz-Bellvis L. Histologic tumor grade and preoperative bilary drainage are the unique independent prognostic factors of survival in pancreatic ductal adenocarcinoma patients after pancreaticoduodenectomy. J Clin Gastroenterol 2018;52:e11-7. 10.1097/MCG.0000000000000793 [DOI] [PubMed] [Google Scholar]
- 8.Golan T, Sella T, Margalit O, Amit U, Halpern N, Aderka D, Shacham-Shmueli E, Urban D, Lawrence YR. Short- and long-term survival in metastatic pancreatic adenocarcinoma, 1993-2013. J Natl Compr Canc Netw 2017;15:1022-7. 10.6004/jnccn.2017.0138 [DOI] [PubMed] [Google Scholar]
- 9.Åkerberg D, Ansari D, Andersson R. Re-evaluation of classical prognostic factors in resectable ductal adenocarcinoma of the pancreas. World J Gastroenterol 2016;22:6424-33. 10.3748/wjg.v22.i28.6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcea G, Dennison AR, Pattenden CJ, Neal CP, Sutton CD, Berry DP. Survival following curative resection for pancreatic ductal adenocarcinoma. A systematic review of the literature. JOP 2008;9:99-132. [PubMed] [Google Scholar]
- 11.Liu L, Xu HX, He M, Wang W, Wang WQ, Wu CT, Wei RQ, Liang Y, Gao HL, Liu C, Xu J, Long J, Ni QX, Shao CH, Wang J, Yu XJ. A novel scoring system predicts postsurgical survival and adjuvant chemotherapeutic benefits in patients with pancreatic adenocarcinoma: implications for AJCC-TNM staging. Surgery 2018;163:1280-94. 10.1016/j.surg.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 12.Eloubeidi MA, Tamhane A, Varadarajulu S, Wilcox CM. Frequency of major complications after EUS-guided FNA of solid pancreatic masses: a prospective evaluation. Gastrointest Endosc 2006;63:622-9. 10.1016/j.gie.2005.05.024 [DOI] [PubMed] [Google Scholar]
- 13.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang TM, Kecman V. Gene extraction for cancer diagnosis by support vector machines--an improvement. Artif Intell Med 2005;35:185-94. 10.1016/j.artmed.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 16.Eilaghi A, Baig S, Zhang Y, Zhang J, Karanicolas P, Gallinger S, Khalvati F, Haider MA. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma - a quantitative analysis. BMC Med Imaging 2017;17:38. 10.1186/s12880-017-0209-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420-8. 10.1037/0033-2909.86.2.420 [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Cheng Z, Huang Y, Liang C, He L, Ma Z, Chen X, Wu X, Li Y, Liang C, Liu Z. CT-based radiomics signature to discriminate high-grade from low-grade colorectal adenocarcinoma. Acad Radiol 2018;25:1285-97. 10.1016/j.acra.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Xu X, Tian Q, Li B, Wu Y, Yang Z, Liang Z, Liu Y, Cui G, Lu H. Radiomics assessment of bladder cancer grade using texture features from diffusion-weighted imaging. J Magn Reson Imaging 2017;46:1281-8. 10.1002/jmri.25669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, Zhu S, Wang M, Dou S, Shi D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol 2019;29:2802-11. 10.1007/s00330-018-5787-2 [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Fang M, Dong D, Wei X, Liu L, Xu X, Jiang X, Tian J, Liu Z. A radiomics signature in preoperative predicting degree of tumor differentiation in patients with non-small cell lung cancer. Acad Radiol 2018;25:1548-55. 10.1016/j.acra.2018.02.019 [DOI] [PubMed] [Google Scholar]
- 22.Hwan-Ho Cho , Hyunjin Park. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. Conf Proc IEEE Eng Med Biol Soc 2017;2017:3081-4. [DOI] [PubMed] [Google Scholar]
- 23.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. World Health Organization, 2010. [Google Scholar]
- 24.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42:1341-53. 10.1118/1.4908210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soh LK, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Transactions on Geoscience and Remote Sensing 1999;37:780-95. 10.1109/36.752194 [DOI] [Google Scholar]
- 26.Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics 1973;SMC-3:610-21. 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 27.Haralick RM, Shapiro LG. Computer and robot vision. Reading: Addison-Wesley Longman Publishing Company, 1992. [Google Scholar]
- 28.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X. Texture information in run-length matrices. IEEE Transactions on Image Processing 1998;7:1602-9. 10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- 30.Amadasun M, King R. Textural Features Corresponding to Texural Properties. IEEE Transactions on Systems, Man and Cybernetics 1989;19:1264-74. 10.1109/21.44046 [DOI] [Google Scholar]
- 31.Basu S. Developing predictive models for lung tumor analysis. 2012. Available online: https://scholarcommons.usf.edu/etd/3963
- 32.Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, Wang G, Liang C, Li ZC. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep 2017;7:14331. 10.1038/s41598-017-14753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1-73. 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 34.Choi MH, Lee YJ, Yoon SB, Choi JI, Jung SE, Rha SE. MRI of pancreatic ductal adenocarcinoma: texture analysis of T2-weighted images for predicting long-term outcome. Abdom Radiol (NY) 2019;44:122-30. 10.1007/s00261-018-1681-2 [DOI] [PubMed] [Google Scholar]
- 35.Cassinotto C, Chong J, Zogopoulos G, Reinhold C, Chiche L, Lafourcade JP, Cuggia A, Terrebonne E, Dohan A, Gallix B. Resectable pancreatic adenocarcinoma: role of CT quantitative imaging biomarkers for predicting pathology and patient outcomes. Eur J Radiol 2017;90:152-8. 10.1016/j.ejrad.2017.02.033 [DOI] [PubMed] [Google Scholar]
- 36.Foley KG, Shi Z, Whybra P, Kalendralis P, Larue R, Berbee M, Sosef MN, Parkinson C, Staffurth J, Crosby TDL, Roberts SA, Dekker A, Wee L, Spezi E. External validation of a prognostic model incorporating quantitative PET image features in oesophageal cancer. Radiother Oncol 2019;133:205-12. 10.1016/j.radonc.2018.10.033 [DOI] [PubMed] [Google Scholar]
- 37.Kang D, Park JE, Kim YH, Kim JH, Oh JY, Kim J, Kim Y, Kim ST, Kim HS. Diffusion radiomics as a diagnostic model for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol 2018;20:1251-61. 10.1093/neuonc/noy021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leijenaar RT, Carvalho S, Hoebers FJ, Aerts HJ, van Elmpt WJ, Huang SH, Chan B, Waldron JN, O'Sullivan B, Lambin P. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 2015;54:1423-9. 10.3109/0284186X.2015.1061214 [DOI] [PubMed] [Google Scholar]
- 39.Lucia F, Visvikis D, Vallieres M, Desseroit MC, Miranda O, Robin P, Bonaffini PA, Alfieri J, Masson I, Mervoyer A, Reinhold C, Pradier O, Hatt M, Schick U. External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging 2019;46:864-77. 10.1007/s00259-018-4231-9 [DOI] [PubMed] [Google Scholar]
- 40.Larghi A, Correale L, Ricci R, Abdulkader I, Monges G, Iglesias-Garcia J, Giovannini M, Attili F, Vitale G, Hassan C, Costamagna G, Rindi G. Interobserver agreement and accuracy of preoperative endoscopic ultrasound-guided biopsy for histological grading of pancreatic cancer. Endoscopy 2015;47:308-14. [DOI] [PubMed] [Google Scholar]
- 41.Elias J, Jr, Semelka RC, Altun E, Tsurusaki M, Pamuklar E, Zapparoli M, Voultsinos V, Armao DM, Rubinas T. Pancreatic cancer: correlation of MR findings, clinical features, and tumor grade. J Magn Reson Imaging 2007;26:1556-63. 10.1002/jmri.21210 [DOI] [PubMed] [Google Scholar]
- 42.Kovač JD, Đurić-Stefanović A, Dugalić V, Lazić L, Stanisavljević D, Galun D, Mašulović D. CT perfusion and diffusion-weighted MR imaging of pancreatic adenocarcinoma: can we predict tumor grade using functional parameters? Acta Radiol 2019;60:1065-73. 10.1177/0284185118812202 [DOI] [PubMed] [Google Scholar]
- 43.Williams JL, Kadera BE, Nguyen AH, Muthusamy VR, Wainberg ZA, Hines OJ, Reber HA, Donahue TR. CA19-9 normalization during pre-operative treatment predicts longer survival for patients with locally progressed pancreatic cancer. J Gastrointest Surg 2016;20:1331-42. 10.1007/s11605-016-3149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Shao Z, Chen W, Xie H, Wu Z, Qin G, Zhao N. A varying-coefficient cox model for the effect of CA19-9 kinetics on overall survival in patients with advanced pancreatic cancer. Oncotarget 2017;8:29925-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni XG, Bai XF, Mao YL, Shao YF, Wu JX, Shan Y, Wang CF, Wang J, Tian YT, Liu Q, Xu DK, Zhao P. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur J Surg Oncol 2005;31:164-9. 10.1016/j.ejso.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 46.Iwamura T, Taniguchi S, Kitamura N, Yamanari H, Kojima A, Hidaka K, Setoguchi T, Katsuki T. Correlation between CA19-9 production in vitro and histological grades of differentiation in vivo in clones isolated from a human pancreatic cancer cell line (SUIT-2). J Gastroenterol Hepatol 1992;7:512-9. 10.1111/j.1440-1746.1992.tb01030.x [DOI] [PubMed] [Google Scholar]