Abstract
Evolution and selection have shaped diverse immune systems throughout phylogeny, the vast majority of which remain unexplored. Botryllus schlosseri is a colonial tunicate, a sister group to vertebrates, that develops as a chordate, then metamorphoses to an asexually reproductive invertebrate that every week makes the same body plan from budded stem cells. Genetically distinct B. schlosseri colonies can fuse to form a chimera, or reject each other based on allogeneic recognition. In chimeras, circulating germline and somatic stem cells participate in development; stem cells compete in all individuals in the fused colonies, with rejection preventing germline parasitism.
Here we review the isolation and characterization of B. schlosseri hematopoietic stem cells (HSC) and their niches, and the role of the immune effector cells in allorecognition.
Keywords: Comparative Immunology, Allorecognition, Tunicates, HSC, Cytotoxic Cells, Evolution of the Immune System, Hematopoietic System, Innate Immunity
Introduction
Pathogenic microbes and internal pathogenic cells most likely evolved simultaneously with multicellular life forms they could parasitize. The rapid evolution of pathogens was mirrored by the development of innate and anticipatory adaptive immune systems. Throughout evolution the immune system has evolved a heterogeneous and highly diverse repertoire of cells and mechanisms that identify and target pathogens as well as determine the tolerance to harmless antigens like self and symbionts.
While we know a great deal about immunity in a small set of model organisms, the wide array of genes and cells that constitute diverse immune systems in nature are still unknown. The study of immune systems of non-classical model organisms have revealed significant discoveries including the discovery in multicellular organisms phenomena such as phagocytosis (starfish,1), toll-like receptors (flies, 2), VLR novel adaptive immune system (lamprey, 3), somatic immune receptor diversification at a single cell level (urchin,4), resembling somatically recombined Ig and TCR molecules in lymphocytes of most vertebrates, and in single celled organisms restriction endonucleases and the CRISPR–CAS immune system (prokaryotes, 5, 6). Tunicates, like Botryllus schlosseri, the closest living invertebrate relatives of vertebrates, are situated at a critical evolutionary juncture: their larval stage shares developmental and structural characteristics with all chordates, including a notochord, neural tube, segmented musculature and gill slits (7–8). Larvae settle and metamorphose into sessile individuals (Figure 1a), which lose most of their chordate cells by programmed cell death and cell removal. Tunicates reproduce either sexually (solitary species), or sexually and asexually (colonial species like B. schlosseri). These two reproductive modes give rise to nearly identical adult body plans, including digestive and respiratory systems (branchial sac), endostyle, a simple tube-like heart, siphons, a neural complex with larval specific and adult invertebrate specific neural elements, ovary and testis (9; Figure 1b). In B. Schlosseri, the larva develops after fertilization as a chordate, then metamorphoses to an asexually reproductive invertebrate that every week makes the same body plan from budded germline and somatic stem cells. Colonies grow via such cycles of stem cell mediated asexual reproduction, where buds develop and produce genetically identical individuals (zooids; 10). Every week the developed buds in a colony replace the older generation following a synchronized programmed cell death [and macrophage removal] of the parent zooids (11; Figure 1c). A defined vasculature with circulating blood cells including cells with lymphocyte-like and macrophage-like morphology connects all zooids and buds in the colony and extends outward with ampullae into a tunic that covers the entire organism (12–13; Figure 1).
Figure 1: B. schlosseri larva, colony anatomy, and allogeneic reactions.
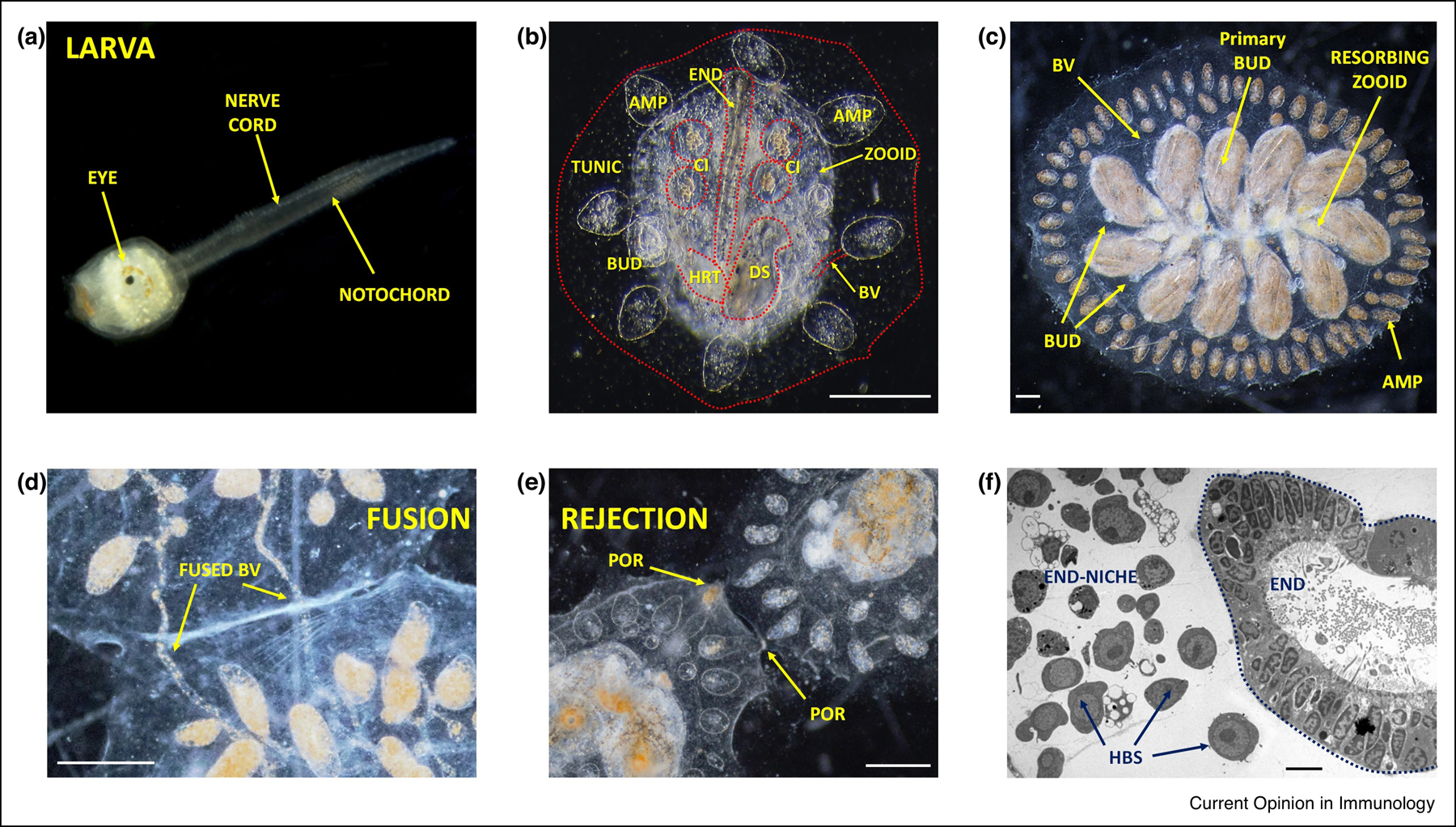
a, The larval tadpole phase of the B. schlosseri early lifecycle (adopted from 21). b, An image of the invertebrate stage zooid; its primary buds (BUD), blood vessels (BV) connected to an ampulla (AMP), endostyle (END), cell islands (CI), digestive system (DS), heart (HRT) and the surrounding tunic (TUNIC) are marked. c, A colony during the take-over stage, where the old generation zooids are resorbed and replaced by buds that completed their development. The colony’s individuals are connected to each other by vasculature which extend outward with ampullae (AMP). d-e, Live imaging of colonies undergoing fusion (d) and rejection (e); arrows point to fused vasculature (d; BV), with apparent pigment cells in the fused blood vessels and points of rejection (e; POR). f, Electron microscopy cross section of the endostyle and the sub-endostyle area (endostyle-niche; END). Cells with hemoblast (HBS; candidate HSC) morphology are enriched within the endostyle-niche (adopted from 22). Scale bar b-c, d-e: 0.2 mm; f: 5 μm.
When two colonies touch, they either fuse vasculature and share blood, or undergo a genetically determined immune rejection and permanently separate. (14–15; Figure 1e–f). This self-nonself recognition process is controlled by Botryllus Histocompatability Factor (BHF), a single polymorphic histocompatibility gene (16). For fusion to occur at least one shared BHF allele is required. Thus, B. schlosseri undergoes a natural allorecognition process with features of experimentally produced parabionts, in which successful ‘transplantation’ occurs only with genetically compatible colonies.
Upon fusion, circulating somatic and germline stem cells from both colonies compete for dominance of the somatic and germline organs and contribute to the formation of new buds (10, 17–19). Highly successful germline stem cells can outcompete less successful stem cells in the gonads of their chimeric partner, resulting in heritable dominance of a single germline origin of gonads in the chimera (10, 17–19). In a significant fraction of these chimeras, the budding cycle in all of the individuals of one of the chimeric partners fails to develop or persist (20). It appears that this developmental arrest is an immune cell based rejection of bud cells that operate within a BHF histocompatible pair that involves inflammatory and cytotoxic cells, comparable to mammalian chronic rejections (20). Since fusion and stem cell competition among germ line stem cells is restricted to individuals that share a BHF allele, we hypothesized that allogeneic rejection restricts genotype replacement to histocompatible kin, preventing germline predation from unrelated conspecific colonies.
A comparison of the B. schlosseri genome with those of several invertebrates, solitary tunicates and vertebrates, revealed enrichment for genes essential to the immune system and hematopoiesis that are predicted to have evolved in a common ancestor of B. schlosseri and vertebrates (21). These hematopoietic and immune behaviors and innovations of a colonial tunicate prompted us to characterize the molecular and cellular mechanisms underlying hematopoiesis and immunity in B. schlosseri (22).
Evolution of HSCs and the Hematopoietic Organ
HSCs are multipotent, self-renewing cells that generate all mature blood and immune cell populations in an animal’s life (19). In mammals, the blood-forming organ is the bone marrow, where HSCs reside in specialized niches that support their self-renewal and maintenance of an undifferentiated state. Since the identification of HSCs by prospective isolation (23), several model systems have been studied to elucidate HSC biology (24–27). We found that the mammalian and tunicate candidate HSCs (cHSC) share a transcriptome signature, suggesting that they are homologous and originated from a common ancestor (Figure 2a; 22). Among the genes expressed in B. schlosseri isolated hematopoietic progenitor cell populations, we found high enrichment for gene sets predominantly expressed in human HSC, myeloid populations, and early but not mature lymphoid populations (22). Consistent with previous studies (21, 28–31) this analysis indicates that the evolution of T and B or VLRA and VLRB cell adaptive immunity progressed rapidly in the jawless vertebrates, with much of the genetic repertoire in place by the emergence of jawed vertebrates. However, homologs of human genes with specific expression in HSC and blood progenitor populations, including T and B progenitor cells, appear early in metazoan evolution (21–22, 32–33). It is of interest that the MHC and elements of vertebrate immune receptors appear to be absent in both B. schlosseri and lampreys, and in all other sequenced invertebrates to date; as omnis DNA e DNA, the origin of these elements and how they appeared in jawed vertebrates remains an unsolved mystery.
Figure 2: Gene expression analysis of genes whose expression is upregulated in the Botryllus cHSC and endostyle as compared to the expression of their homologues (based on sequence similarities) in the mouse hematopoiesis lineages.
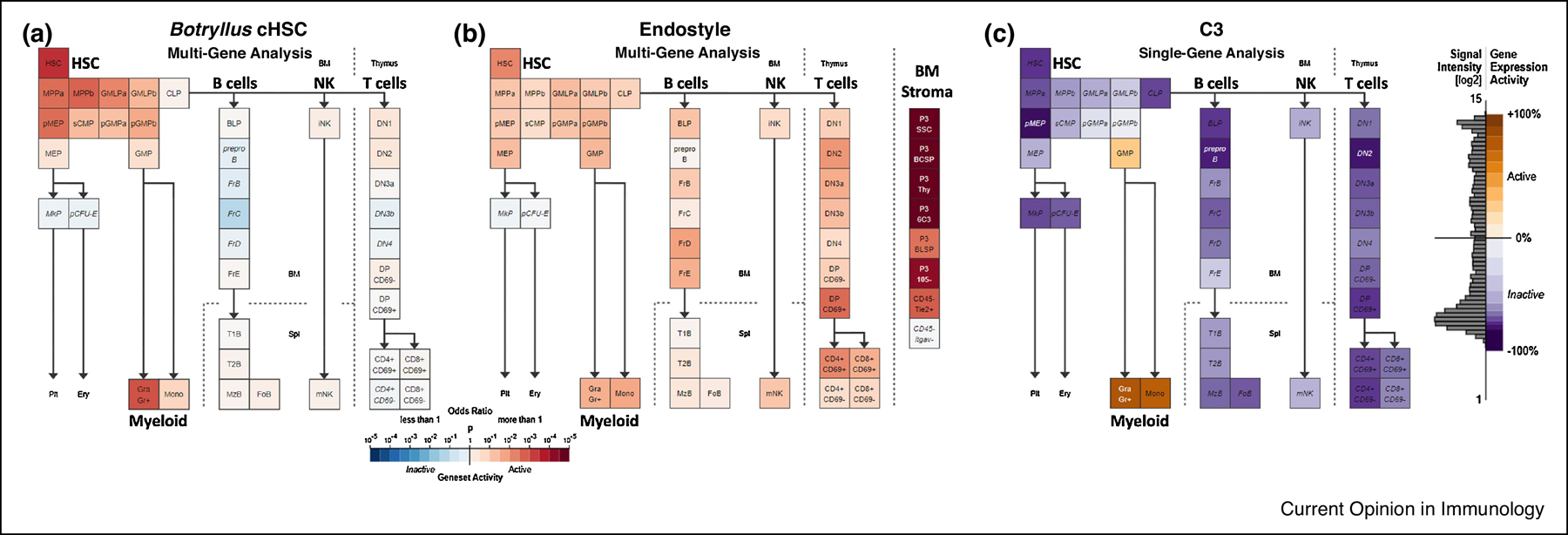
a. Multi-gene analysis (235 genes) using Geneset Activity Analysis of the genes significantly upregulated by the candidate HSCs cell population of B. schlosseri. The enriched populations are HSCs and the myeloid lineage (adopted from Ref. [22]). b. Multi-gene analysis using Geneset Activity Analysis (https://gexc.riken) of top 200 genes significantly upregulated in the B. schlosseri endostyle associated with the blood system, on a mouse hematopoiesis model including the bone marrow (BM) stromal cells. This analysis revealed significant activity between the mouse bone marrow stromal cells and the B. schlosseri endostyle (adopted from Ref. [22]). c. Expression of C3 (complement component 3) gene, as determined by microarray analysis, in 39 different mouse hematopoietic populations. The C3 expresses in the phagocytic myeloid lineage, highest levels of the gene expression activity are detected in the granulocytes (Gra) and monocytes (Mono), and some activity is detected in their common progenitor (GMP).
The hematopoietic stem cell niche is an interactive structural unit organized to facilitate cell-fate decisions in a proper spatiotemporal manner (24, 26, 34). In adult mammals under normal circumstances, the main site of haematopoiesis is the bone marrow (24, 26, 34). The B. schlosseri HSCs are localized in the endostyle; 327 genes that are significantly expressed in the endostyle are also expressed in human hematopoietic bone marrow (Figure 2b; 22, 35), suggesting a common origin for the hematopoietic bone marrow niche and the endostyle. The tunicate endostyle is a long glandular groove extending medially at the ventral face of the zooid branchial sac along its anterior posterior axis consisting of eight distinct anatomical zones and is immersed by blood flow through the large subendostylar sinus and other sinuses (36). The endostyle is connected to the B. schlosseri central nervous system; several zones within it produce mucous and potentially also produce digestive enzymes and hormones (36), indicating a diversity of functions in this organ. The endostyle expresses thyroid transcription factor 1 homologs and has an iodine-concentrating activity, therefore it includes the invertebrate chordate homolog of the vertebrate thyroid gland (36). It exhibits unique expression patterns with site-specific factors that are linked to developmental regulation and stem cell maintenance (37–38). In 2008 we identified a somatic stem cell niche in the anterior subendostylar sinus (37). Recently, we discovered that the specific somatic stem cells that harbor and home to this endostyle niche are hematopoietic stem cells (Figure 1f; 22). In adult mammals and reptiles, the bone marrow is the main center of hematopoiesis, and during embryogenesis the yolk sac, dorsal aorta, placenta, and later fetal liver are centers; but the site of hematopoiesis in other chordates reveals a list of diverse organs and tissues. Hematopoiesis takes place in the head kidney in bony and cartilaginous fish, T cell development in gill tips, and B cell development takes place in the bursa of Fabricius in birds (39). In adult amphibians, hematopoiesis takes place in the bone marrow, but during the larval stage, the head kidney is the hematopoietic tissue (40–41). A bone marrow-like organ, the protovertebral arch, and the kidney are the sites of hematopoiesis in jawless vertebrates (42), and in cephalochordates it is suggested that the aorta-gonads-mesonephros is the site of hematopoiesis (43). Cellular and molecular comparison of these organs and tissues will reveal conserved elements that were maintained throughout evolution essential for hematopoiesis.
Cellular and Molecular Mechanisms of Histocompatibility
The ability to tolerate self and reject a foreign transplant is a critical element of the immune system in all animals. Although this is well characterized in mammals, the cellular and molecular details are not well understood in other groups. Morula cells, cells that contain phenoloxidase and accumulate in rejection points, have been identified as cytotoxic cells that mediate the execution of the rejection reaction in B. schlosseri (44–47). We further identified BHF, a single gene that determines self-recognition in B. schlosseri colonies, and begun characterizing the basic mechanisms of this process (16, 22).
In mammals, the Major Histocompatibility Complex (MHC) encodes the linked genes [haplotype] for tissue-antigens that allows the immune system to recognize and tolerate itself. Histocompatibility is governed by two types of cytotoxic cellular recognition: cytotoxic T-cells and Natural Killer (NK) cells. A subset of T-cells will recognize non-self MHC as foreign, and will be activated to eliminate cells that express this MHC. On the other hand, NK cells are inhibited by self-MHC through Killer Inhibitory Receptors (KIRs), and will eliminate cells that do not express self-MHC. Both the T-cells, which are activated by non-self, and NK cells that are activated by “missing-self” go through an educational process as they mature that allows them to make this distinction (48). Similar to NK inhibitory mechanisms, the allogeneic functional assays with blocking of BHF in B. schlosseri, reveal that the self-BHF recognition is a major inhibitory mechanism of cytotoxicity in allorecognition (22). These assays and the observation that colonies sharing at least one BHF allele fuse (16), demonstrate that the cellular cytotoxicity allorecognition mechanism in B. schlosseri is based on “missing self” and comparable to NK recognition in mammals. Similar to NK mediated cellular recognition through its KIR receptor repertoire we hypothesize that recognition between inhibitory receptors expressed on the effector cells and different BHF alleles is the mechanism that mediates self-recognition in B. schlosseri.
All cell populations sequenced express BHF RNA, however the B. schlosseri cell population that is enriched for the cytotoxic Morula Cells (MC)does not express BHF protein on the membrane (as detected by antibodies), suggesting the existence of an inhibitory receptor on cytotoxic cells that can recognize self-BHF. Interestingly, one of the genes which is differentially expressed by cytotoxic MC is sFuHC. This polymorphic gene was previously suggested to be the Botryllus histocompatibility gene (49), and is located in close proximity to the BHF locus (16). Considering the inhibition mechanism of the BHF and the significantly high expression of sFuHC in the B. schlosseri cytotoxic cells, it is possible that the sFuHC plays the role of an inhibitory recognition receptor for BHF. These observations suggest that there is an educational mechanism in B. schlosseri cytotoxic cells for selection for the inhibitory receptors, similar to NK cell education (50). Further studies will be needed to understand whether the colonial life history of B. schlosseri led to this complex immunity recognition system or if it is shared by all tunicates, both solitary (that do not form chimeras) and colonial. Although the specific molecules are not homologous to mammalian histocompatibility recognition, the features underlying this recognition are similar.
Evolution of the Cellular Immune System
The two best characterized aspects of cellular immunity are cytotoxicity and phagocytosis. B. schlosseri has a myeloid lineage including cells that take part in phagocytosis similar to those found in vertebrates (22). It also contains amoebocytes and large phagocytes with morphologies resembling invertebrate cell types found in Arthropods and Echinoderms (51–57). Jawed vertebrates’ cytotoxic cells belong to the lymphoid lineage and includes NK cells and cytotoxic T cells (58). It has been proposed that the lymphoid lineage VLRA and VLRC In jawless vertebrates also carries cytotoxic activity (32). While B. schlosseri cytotoxic MC share some genes (15%; 22) with the cytotoxic lymphocytes of vertebrates, they mainly express a tunicate specific gene repertoire (85%; 22). Cytotoxic MC have a large granular lymphocyte-like morphology, resembling NK cells, which originally were characterized as large granular lymphocytes (60). Moreover, the activated MC gene expression has resemblance to mouse lymphocytes as analyzed by Geneset Activity Analysis (22). While this activity resemblance is not significant, it raises the question of whether there is a homologous lineage to lymphocytes in colonial chordates. Several studies describe morula-like cells that carry enzymatic activity of phenoloxidase in other invertebrate species (51, 54–56, 61) We hypothesize that the B. schlosseri granular morula cells originate from the same hematopoietic lineage as other invertebrate MC (Figure 3).
Figure 3: Proposed evolution of cellular immune effector lineages.
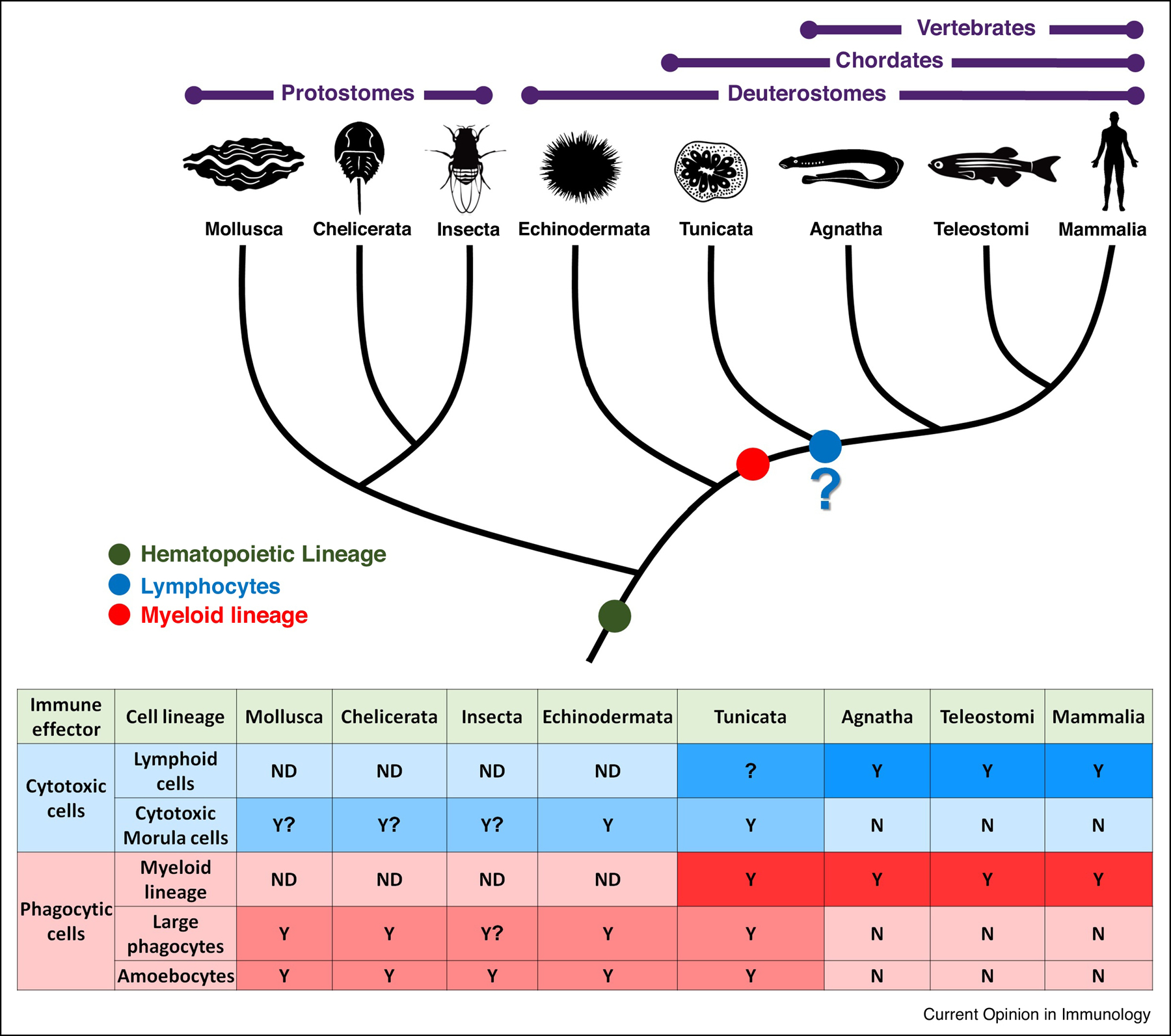
Proposed evolutionary perspective of the cellular immune system, mainly cytotoxic and phagocytic cell lineages. Table at the bottom describes the type of presumed immune associated cells found in each animal group. It appears that myeloid lineage evolved before the branching of the vertebrata from tunicates (red). Amoebocytes and large phagocytes can be found in B. schlosseri and other invertebrate species (light red). While there are some populations and molecular markers resembling lymphoid lineage, whether this lineage evolved in the common ancestor of tunicates and vertebrates is still to be deciphered (blue). On the other hand the cytotoxic morula cells are characterized in tunicates and likely exist in other invertebrates (light blue).Y- Yes, cell lineage present, N- cell lineage Not present, ND- No data, “?”- insufficient data (adopted from 22).
In a recent work on MC and phagocytic cells in B. schlosseri, only the MC expressed the B. schlosseri C3 gene (62), a gene that is exclusively expressed in the mammalian myeloid lineage (Figure 2c). An additional phagocytosis marker, the MFGE8, is also upregulated in the B. schlosseri cytotoxic MC (22). These examples suggest that MC have some commonality with the myeloid lineage (Figure 3). Moreover, in teleosts and later in mammals it was demonstrated that specific cellular immune function is not restricted by cell lineage (63–64). These studies proved that B cells can perform phagocytosis and antigen presentation. Moreover, the same group now suggests a cytotoxic ability of B cells (65). These studies suggest that there might be greater plasticity in function within the different immune lineages. Comprehensive characterization of immune cells from diverse species / clades will be necessary to determine lineage homologies.
In summary, here we limit our discussion of the co-evolution of histocompatibility and migratory stem cells to protection against intra-species ‘pathogens’, primitive germline stem cells that use the bodies of siblings or kin to mature the germline cells that will determine the genotypes for the next reproductive generation. We find parallel blood lineage cells involved in innate immune recognition in a colonial tunicate and mammals, sharing homologous genes restricted to hematopoiesis and immunity. The cytotoxic and phagoptotic cell deaths that prevent germline stem cells from non-kin invasions appear to share common cellular, and some molecular homologues with the vertebrates.
Highlights.
Botryllus schlosseri HSC have been isolated and their niche identified.
Mammalian and B. schlosseri HSCs and their respective niches share many homologous transcripts.
Cytotoxicity is the mechanism of immune allogeneic cellular rejection.
Botryllus Histocompatibility Factor (BHF) determines fusion or rejection of colonies.
Allogeneic rejection by cytotoxic cells is inhibited by BHF, similar to NK cells.
Acknowledgments
We would like to thank Mark Kowarsky, Stephen R. Quake, Jun Seita, Katherine J. Ishizuka, Karla J. Palmeri, D. Nathaniel Clarke, Chris Lowe, Lucia Manni, Chiara Anselmi, and Ronnie Voskoboynik for meaningful discussions. This study was supported by NIH grants R56AI089968, R01AG037968 and RO1GM100315 (to I.L.W., S.R.Q., and A.V.), the Chan Zuckerberg investigator program (to I.L.W and A.V), the Virginia and D. K. Ludwig Fund for Cancer Research, a grant from the Siebel Stem Cell Institute and a Stinehart-Reed grant (to I.L.W.). B.R. was supported by ISF grants 1416/19, and HFSP Research Grant, RGY0085/2019.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing to declare.
References
- [1].Mechnikova MO: Life of Elie Metchnikoff, 1921: 1845–1916 (books.google.com). [Google Scholar]
- [2].Anderson KV, Jurgens G, Nusslein-Volhard C: Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell 1985, 42: 779–789. [DOI] [PubMed] [Google Scholar]
- [3].Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD: Pillars Article: Somatic Diversification of Variable Lymphocyte Receptors in the Agnathan Sea Lamprey. Nature. 2004. 430: 174–180. J.Immunol. 2018, 201: 1336–1342. [PubMed] [Google Scholar]
- **[4].Oren M, Rosental B, Hawley TS, Kim GY, Agronin J, Reynolds CR, Grayfer L, Smith LC: Individual Sea Urchin Coelomocytes Undergo Somatic Immune Gene Diversification. Front.Immunol 2019, 10: 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe and characterize a somatic immune gene diversification in an invertebrate species. The authors show on a single cell level of the purple sea urchin coelomocytes, the SpTransformer immune effector gene somatic diversification by gene duplications, deletions, and single nucleotide polymorphisms. This leads to a highly diverse, in frame, SpTransformer gene repertoire in the coelomocytes of the purple sea urchin.
- [5].Meselson M, Yuan R: DNA restriction enzyme from E.coli. Nature 1968, 217: 1110–1114. [DOI] [PubMed] [Google Scholar]
- [6].Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E: Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J.Mol.Evol 2005, 60: 174–182. [DOI] [PubMed] [Google Scholar]
- [7].Darwin C: The origin ofspecies. London: Murray; 1859. [Google Scholar]
- [8].Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. : The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 2002, 298: 2157–2167. [DOI] [PubMed] [Google Scholar]
- **[9].Kowarsky M, Anselmi C, Hotta K, Burighel P, Zaniolo G, Caicci F, Rosental B, Neff NF, Ishizuka KJ, Palmeri KJ, et al. : Molecular and Morphological Signatures of Chordate Development: Two Distinct Pathways, One Tunicate. bioRxiv 2019: 801589. [Google Scholar]; All chordates develop through embryogenesis, but colonial tunicates also develop by asexual reproduction [blastogenesis], whereby stem cells form tissues and organs. These two developmental pathways establish the same body axis, morphogenetic patterning and organ formation. By combining microscopy with transcriptome sequencing, this study revealed that the molecular programs are distinct, but the blastogenic tissue-specific stem cells and embryonic precursor populations share similar molecular profiles. This study establishes a platform for advancing the science of stem cell biology and regulation of development and regeneration.
- [10].Laird DJ, De Tomaso AW, Weissman IL: Stem cells are units of natural selection in a colonial ascidian. Cell 2005, 123: 1351–1360. [DOI] [PubMed] [Google Scholar]
- [11].Lauzon RJ, Ishizuka KJ, Weissman IL: Cyclical generation and degeneration of organs in a colonial urochordate involves crosstalk between old and new: a model for development and regeneration. Dev.Biol 2002, 249: 333–348. [DOI] [PubMed] [Google Scholar]
- [12].Hirose E, Shirae M, Saito Y: Ultrastructures and classification of circulating hemocytes in 9 botryllid ascidians (chordata: ascidiacea). Zoolog Sci. 2003, 20: 647–656. [DOI] [PubMed] [Google Scholar]
- [13].Schlumpberger JM, Weissman IL, Scofield VL: Separation and labeling of specific subpopulations of Botryllus blood cells. J.Exp.Zool 1984, 229: 401–411. [DOI] [PubMed] [Google Scholar]
- [14].Sabbadin A: Le basi geneticha della capacita di fusion fra colonies in B. schlosseri (Ascidiacea). Rend Accad Naz Lincei (Cl Sci FFMMNN) 1962, 32: 1031–1036. [Google Scholar]
- [15].Scofield VL, Schlumpberger JM, West LA, Weissman IL: Protochordate allorecognition is controlled by a MHC-like gene system. Nature 1982, 295: 499–502. [DOI] [PubMed] [Google Scholar]
- [16].Voskoboynik A, Newman AM, Corey DM, Sahoo D, Pushkarev D, Neff NF, Passarelli B, Koh W, Ishizuka KJ, Palmeri KJ, et al. : Identification of a colonial chordate histocompatibility gene. Science 2013, 341: 384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stoner DS, Rinkevich B, Weissman IL: Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc.Natl.Acad.Sci.U.S.A 1999, 96: 9148–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stoner DS, Weissman IL: Somatic and germ cell parasitism in a colonial ascidian: possible role for a highly polymorphic allorecognition system. Proc.Natl.Acad.Sci.U.S.A 1996, 93: 15254–15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weissman IL: In light of evolution: Stem cells are units of natural selection for tissue formation, germline development, and in cancer development. Proc.Natl.Acad.Sci.U.S.A 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Corey DM, Rosental B, Kowarsky M, Sinha R, Ishizuka KJ, Palmeri KJ, Quake SR, Voskoboynik A, Weissman IL: Developmental cell death programs license cytotoxic cells to eliminate histocompatible partners. Proc.Natl.Acad.Sci.U.S.A 2016, 113: 6520–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Voskoboynik A, Neff NF, Sahoo D, Newman AM, Pushkarev D, Koh W, Passarelli B, Fan HC, Mantalas GL, Palmeri KJ, et al. : The genome sequence of the colonial chordate, Botryllus schlosseri. Elife 2013, 2: e00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **[22].Rosental B, Kowarsky M, Seita J, Corey DM, Ishizuka KJ, Palmeri KJ, Chen SY, Sinha R, Okamoto J, Mantalas G, et al. : Complex mammalian-like haematopoietic system found in a colonial chordate. Nature 2018, 564: 425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]; Here for the first time in an invertebrate specie a thorough cellular, molecular, and functional characterization of blood cells was performed. It includes the identification of hematopoietic stem cells (HSCs), myeloid cell lineage progenitors, phagocytic and cytotoxic cells. The hematopoietic organ was also discovered and found to be homologous to human hematopoietic bone marrow. This study revealed significant conservation between the gene expression profiles of the B. schlosseri and mammalian HSC and blood progenitor populations. Immune functional assays revealed that cellular rejection between genetically incompatible colonies is mediated by cytotoxicity that is inhibited by histocompatibility, a mechanism similar to inhibition of NK cell cytotoxicity by MHC in mammals.
- [23].Spangrude GJ, Heimfeld S, Weissman IL: Purification and characterization of mouse hematopoietic stem cells. Science 1988, 241: 58–62. [DOI] [PubMed] [Google Scholar]
- [24].Chen JY, Miyanishi M, Wang SK, Yamazaki S, Sinha R, Kao KS, Seita J, Sahoo D, Nakauchi H, Weissman IL: Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 2016, 530: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Choudhuri A, Fast EM, Zon LI: Using Zebrafish to Study Pathways that Regulate Hematopoietic Stem Cell Self-Renewal and Migration. Stem Cell.Reports 2017, 8: 1465–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Crane GM, Jeffery E, Morrison SJ: Adult haematopoietic stem cell niches. Nat.Rev.Immunol 2017, 17: 573–590. [DOI] [PubMed] [Google Scholar]
- [27].Ghosh S, Singh A, Mandal S, Mandal L: Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev.Cell 2015, 33: 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T: A thymus candidate in lampreys. Nature 2011, 470: 90–94. [DOI] [PubMed] [Google Scholar]
- [29].Dishaw LJ, Leigh B, Cannon JP, Liberti A, Mueller MG, Skapura DP, Karrer CR, Pinto MR, De Santis R, Litman GW: Gut immunity in a protochordate involves a secreted immunoglobulin-type mediator binding host chitin and bacteria. Nat.Commun 2016, 7: 10617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Flajnik MF, Kasahara M: Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat.Rev.Genet 2010, 11: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirano M, Das S, Guo P, Cooper MD: The evolution of adaptive immunity in vertebrates. Adv.Immunol 2011, 109: 125–157. [DOI] [PubMed] [Google Scholar]
- [32].Das S, Li J, Hirano M, Sutoh Y, Herrin BR, Cooper MD: Evolution of two prototypic T cell lineages. Cell.Immunol 2015, 296: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vivier E, van de Pavert SA, Cooper MD, Belz GT: The evolution of innate lymphoid cells. Nat.Immunol 2016, 17: 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Szade K, Gulati GS, Chan CKF, Kao KS, Miyanishi M, Marjon KD, Sinha R, George BM, Chen JY, Weissman IL: Where Hematopoietic Stem Cells Live: The Bone Marrow Niche. Antioxid.Redox Signal 2018, 29: 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, Ehrlich LI, Fathman JW, Dill DL, Weissman IL: Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One 2012, 7: e40321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Burighel P and Cloney RA: Urochordata: Ascidiacea In: Microscopic anatomy of invertebrates, Harrison FW, and Ruppert EE, eds. (Wiley-Liss, Inc., NY: ), 221–347. Edited by Harrison and E.E. Ruppert; 1997: 221–347. [Google Scholar]
- [37].Voskoboynik A, Soen Y, Rinkevich Y, Rosner A, Ueno H, Reshef R, Ishizuka KJ, Palmeri KJ, Moiseeva E, Rinkevich B, Weissman IL: Identification of the endostyle as a stem cell niche in a colonial chordate. Cell.Stem Cell 2008, 3: 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rinkevich Y, Voskoboynik A, Rosner A, Rabinowitz C, Paz G, Oren M, Douek J, Alfassi G, Moiseeva E, Ishizuka KJ, et al. : Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev.Cell 2013, 24: 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].COOPER MD, PETERSON RD, GOOD RA: Delineation of the Thymic and Bursal Lymphoid Systems in the Chicken. Nature 1965, 205: 143–146. [DOI] [PubMed] [Google Scholar]
- [40].Arikan H, Cicek K: Haematology of amphibians and reptiles: a review. North-Western Journal of Zoology. 2014, [Google Scholar]
- [41].Grayfer L, Robert J: Amphibian macrophage development and antiviral defenses. Dev.Comp.Immunol 2016, 58: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Saha NR, Smith J, Amemiya CT: Evolution of adaptive immune recognition in jawless vertebrates. Semin.Immunol 2010, 22: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pascual-Anaya J, Albuixech-Crespo B, Somorjai IM, Carmona R, Oisi Y, Alvarez S, Kuratani S, Munoz-Chapuli R, Garcia-Fernandez J: The evolutionary origins of chordate hematopoiesis and vertebrate endothelia. Dev.Biol 2013, 375: 182–192. [DOI] [PubMed] [Google Scholar]
- [44].Sabbadin A, Zaniolo G, Ballarin L: Genetic and cytological aspects of histocompatibility in ascidians. Ital.J.Zool 1992, 59: 167–173. [Google Scholar]
- [45].Ballarin L, Cima F: Cytochemical properties of Botryllus schlosseri haemocytes: indications for morpho-functional characterisation. Eur.J.Histochem 2005, 49: 255–264. [PubMed] [Google Scholar]
- [46].Franchi N, Ballarin L: Immunity in Protochordates: The Tunicate Perspective. Front.Immunol 2017, 8: 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Oren M, Paz G, Douek J, Rosner A, Amar KO, Rinkevich B: Marine invertebrates cross phyla comparisons reveal highly conserved immune machinery. Immunobiology 2013, 218: 484–495. [DOI] [PubMed] [Google Scholar]
- [48].Ljunggren HG, Karre K: In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol.Today 1990, 11: 237–244. [DOI] [PubMed] [Google Scholar]
- [49].De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL: Isolation and characterization of a protochordate histocompatibility locus. Nature 2005, 438: 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Beziat V, Malmberg KJ, Norman PJ, Guethlein LA, Parham P: Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci.Immunol 2016, 1: 10.1126/sciimmunol.aag1672. Epub 2016 Sep 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kawabata S: Immunocompetent molecules and their response network in horseshoe crabs. Adv.Exp.Med.Biol 2010, 708: 122–136. [DOI] [PubMed] [Google Scholar]
- [52].Smith LC, Ghosh J, Buckley KM, Clow LA, Dheilly NM, Haug T, Henson JH, Li C, Lun CM, Majeske AJ, et al. : Echinoderm immunity. Adv.Exp.Med.Biol 2010, 708: 260–301. [DOI] [PubMed] [Google Scholar]
- [53].Muñoz-Chápuli R, Carmona R, Guadix JA, Macías D, Pérez-Pomares JM: The origin of the endothelial cells: an evo-devo approach for the invertebrate/vertebrate transition of the circulatory system. Evol. Dev 2005, 7: 351–358. [DOI] [PubMed] [Google Scholar]
- [54].Lanot R, Zachary D, Holder F, Meister M: Postembryonic hematopoiesis in Drosophila. Dev.Biol 2001, 230: 243–257. [DOI] [PubMed] [Google Scholar]
- [55].Meister M, Lagueux M: Drosophila blood cells. Cell.Microbiol 2003, 5: 573–580. [DOI] [PubMed] [Google Scholar]
- [56].Nakayama K, Nomoto AM, Nishijima M, Maruyama T: Morphological and Functional Characterization of Hemocytes in the Giant Clam Tridacna crocea. J.Invertebr.Pathol 1997, 69: 105–111. [DOI] [PubMed] [Google Scholar]
- **[57].Vazzana M, Celi M, Chiaramonte M, Inguglia L, Russo D, Ferrantelli V, Battaglia D, Arizza V: Cytotoxic activity of Holothuria tubulosa (Echinodermata) coelomocytes. Fish Shellfish Immunol 2018, 72: 334–341. [DOI] [PubMed] [Google Scholar]; The authors describe and show via functional assays a cytotoxic ability of cells of Echinoderms coelomocytes. By performing in-vitro xenogeneic cytotoxicity assays they demonstrated the ability of the coelomocytes to specifically lyse the target cells. Moreover, they have described that the spherule caulomic cells of a sea cucumber release cytotoxic molecules upon a xenogeneic challenge.
- [58].Sun JC, Lanier LL: Versatility in NK cell memory. Immunol.Cell Biol 2011, 89: 327–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sun JC, Lanier LL: Is There Natural Killer Cell Memory and Can It Be Harnessed by Vaccination? NK Cell Memory and Immunization Strategies against Infectious Diseases and Cancer. Cold Spring Harb Perspect.Biol 2018, 10: 10.1101/cshperspect.a029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Timonen T, Ortaldo JR, Herberman RB: Characteristics of human large granular lymphocytes and relationship to natural killer and K cells. J.Exp.Med 1981, 153: 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ballarin L: Ascidian cytotoxic cells: state of the art and research perspectives. Inv. Surv. J 2012. [Google Scholar]
- **[62].Peronato A, Drago L, Rothbacher U, Macor P, Ballarin L, Franchi N: Complement system and phagocytosis in a colonial protochordate. Dev.Comp.Immunol 2020, 103: 103530. [DOI] [PubMed] [Google Scholar]; The authors characterize the role of Botryllus schlosseri complement C3 (BsC3) in phagocytosis mediation. They show its mediation of phagocytosis in the blastogenetic cycle and suggest its activity increases in allogeneic response. Interestingly, the source of the BsC3 are the cytotoxic morula cells (MC), and not the phagocytic cell populations. This could suggest that phagocytosis in different scenarios such as the blastogenetic cycle, is initiated by the MC.
- [63].Li J, Barreda DR, Zhang YA, Boshra H, Gelman AE, Lapatra S, Tort L, Sunyer JO: B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat.Immunol 2006, 7: 1116–1124. [DOI] [PubMed] [Google Scholar]
- [64].Parra D, Rieger AM, Li J, Zhang YA, Randall LM, Hunter CA, Barreda DR, Sunyer JO: Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+T cells. J.Leukoc.Biol 2012, 91: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sunyer JO, Shibasaki Y, Takizawa F, Gonzales M, Boudinot P,: Discovery of perforin-expressing killer B cells in vertebrates. J. Immunol 2019, 202: 121.17. [Google Scholar]


