Abstract
Leaf senescence is a programmed developmental process regulated by various endogenous and exogenous factors. Here we report the characterization of the senescence-regulating role of DEAR4 (AT4G36900) from the DREB1/CBF (dehydration-responsive element binding protein 1/C-repeat binding factor) family in Arabidopsis. The expression of DEAR4 is associated with leaf senescence and can be induced by ABA, JA, darkness, drought and salt stress. Transgenic plants over-expressing DEAR4 showed a dramatically enhanced leaf senescence phenotype under normal and dark conditions while the dear4 knock-down mutant displayed delayed senescence. DEAR4 over-expressing plants showed decreased seed germination rate under ABA and salt stress conditions as well as decreased drought tolerance, indicating that DEAR4 was involved in both senescence and stress response processes. Furthermore, we found that DEAR4 protein displayed transcriptional repressor activities in yeast cells. DEAR4 could directly repress the expression of a subset of COLD-REGULATED (COR) and RESPONSIVE TO DEHYDRATION (RD) genes which have been shown to be involved in leaf longevity and stress response. Also we found that DERA4 could induce the production of Reactive oxygen species (ROS), the common signal of senescence and stress responses, which gives us the clue that DEAR4 may play an integrative role in senescence and stress response via regulating ROS production.
Keywords: DEAR4, leaf senescence, stress, ROS, COR, RD, Arabidopsis thaliana
Introduction
Senescence is the last stage of leaf development which is influenced by intrinsic and environmental factors including age, nutrients, hormones, darkness, osmotic stress, extreme temperature and pathogens (Lim et al., 2007; Guo and Gan, 2014). Most of the major plant hormones have been reported to affect leaf senescence process: abscisic acid (ABA), ethylene (ETH), jasmonic acid (JA), salicylic acid (SA) and strigolactones function in promoting senescence, while cytokinins (CK), gibberellic acid (GA) and auxin inhibit senescence (Jibran et al., 2013; Li et al., 2013; Penfold and Buchan-Wollaston, 2014; Mostofa et al., 2018). During the process of leaf senescence, cellular metabolism and structure undergo significant changes, resulting in leaf yellowing. Meanwhile, degradation of macromolecules in senescing leaves functions to remobilize nutrients to support young vegetative organs and reproductive growth. As a mechanism of evolutional fitness, unfavorable environmental conditions can induce precocious senescence leading to reduced yield and quality of crop plants (Wu et al., 2012; Gregersen et al., 2013; Zhang and Zhou, 2013; Yolcu et al., 2017). Senescence execution requires differential expression of a large number of genes a subset of which is called senescence-associated genes (SAGs). A number of SAGs have been identified to play a regulatory role in leaf senescence. These include genes encoding transcription factors of WRKY, NAC, DREB, MYB, and bZIP family (Woo et al., 2001; Yang et al., 2011; Lee et al., 2012; Vainonen et al., 2012).
Dark-induced senescence (DIS) has been widely used as a model system in leaf senescence study (Liebsch and Keech, 2016). Differential expression of a large number of transcription factor genes during dark-induced leaf senescence have been reported (Song, 2014; Song et al., 2014; Yasuhito et al., 2014) and some of them have been studied for their function in regulating senescence. AtWRKY22 is induced by darkness, but suppressed by light. Further study reveals that AtWRKY22 over-expressing plants displayed accelerated senescence, whereas AtWRKY22 loss-of-function plants showed a delay of senescence under dark condition (Zhou et al., 2011b). RD26 loss-of-function plants displayed significantly delayed senescence under normal or dark-induced conditions with a higher chlorophyll level detected. By contrast, over-expression of RD26 led to early leaf senescence (Takasaki et al., 2015; Kamranfar et al., 2018). Over-expression of CBF2 and CBF3, two members of the DREB family in Arabidopsis, significantly delayed the onset of leaf senescence and also delayed leaf senescence induced by hormones and darkness (Sharabi-Schwager et al., 2010a, b). Phytochromes regulate light responses by promoting the degradation of PIFs (Phytochrome-interacting factors). PIF3, 4, and 5 play an important role in natural and dark induced senescence (Yasuhito et al., 2014). Mutations of the PIFs genes resulted in a significant delay of natural and dark induced senescence, whereas over-expression of these genes accelerated senescence. Further study revealed that PIF4 can bind the promoter of NYE1, the chlorophyll degradation regulatory gene, and GLK2, the chloroplast activity maintainer gene, resulting in induction and repression of their expression, respectively (Song et al., 2014; Shi et al., 2017a).
Jasmonic acid is involved in multiple processes including root inhibition, trichome initiation, anthocyanin accumulation, leaf senescence and biotic and abiotic stress responses (Balbi and Devoto, 2008; Wu et al., 2008; Hu et al., 2017; Song et al., 2017; Ono et al., 2019). JA signaling can be initiated by perception of jasmonoyl-L-isoleucine (JA-Ile), which binds to its receptor COI1 (CORONATINE INSENSITIVE1), an F-box domain-containing protein (Balbi and Devoto, 2008; Shan et al., 2011). It has been reported that endogenous JA content increases during leaf senescence and JA biosynthetic genes such as LOX1, LOX3, and LOX4 are up-regulated during leaf senescence (Wasternack, 2007; Seltmann and Berger, 2013). Attached or detached leaves displayed precocious senescence under exogenous application of MeJA (Shan et al., 2011; Hu et al., 2017). Further study revealed that MYC2, 3, and 4 redundantly bind to the SAG29 promoter and activate its expression, leading to activation of JA-induced leaf senescence (Zhu et al., 2015). In contrast, the bHLH family members including bHLH03, bHLH13, bHLH14, and bHLH17 attenuate MYC2/MYC3/MYC4-activated JA-induced leaf senescence by binding to the promoter of SAG29 and repress its expression. It has been suggested that the activators and repressors mediated in JA-induced leaf senescence can enhance the plant survival rate in various environmental conditions (Qi et al., 2015). In addition, the NAC transcriptional factors ANAC019, ANAC055, and RD26 are also direct targets of MYC2 in mediating JA-induced leaf senescence (Zhu et al., 2015). The JA signaling proteins JAZ4 and JAZ8 interact with transcription factor WRKY57 to negatively regulate leaf senescence induced by JA (Jiang et al., 2014). Additionally, the expression of JAZ7 was significantly increased during darkness. jaz7 mutant exhibited precocious senescence induced by darkness, suggesting that JAZ7 plays a negative role in dark-induced leaf senescence (Yu et al., 2016). The Evening Complex (EC), a core component of the circadian oscillator, comprising EARLY FLOWERING3 (ELF3), EARLY FLOWERING4 (ELF4) and LUX ARRHYTHMO (LUX), plays essential roles in the plant circadian clock and negatively regulates leaf senescence in Arabidopsis. It has been reported that EC represses the expression of MYC2 by directly binding to its promoter. myc2myc3myc4 triple mutants abrogate the accelerated leaf senescence induced by JA in EC mutants (Zhang et al., 2018; Thines et al., 2019). Additionally, JA can positively regulate the ICE-CBF signal to enhance cold stress tolerance in Arabidopsis. Interestingly, endogenous JA content was increased under cold stress conditions. Exogenous application of MeJA enhanced cold stress tolerance. Further study revealed that JAZ1 and JAZ4 play negative roles in the ICE-CBF pathway (Hu et al., 2013, 2017).
As sessile organisms, plants have developed sophisticated mechanisms that are activated and integrated by the expression of thousands of genes to cope with variety of environmental stresses (Yeung et al., 2018; Asad et al., 2019). Many transcriptional factors were found to play key roles in stress response and tolerance. As the largest transcription factor family in Arabidopsis, the AP2/ERF family contains 147 members functionally categorized into the development-associated AP2 and RAV subgroups and the stress response-associated DREB and ERF subgroups (Liu et al., 1998; Sakuma et al., 2002). The DREB/CBF proteins are known to directly regulate target genes in response to various stresses including high salinity, drought and cold stress by directly binding the conserved DRE (Dehydration responsive element)/CRT(C-repeat) cis-acting regulatory elements which contains the core sequence CCGAC (Sakuma et al., 2002; Sun et al., 2008). DREB homologs have been identified in a variety of plants species including rice (Dubouzet et al., 2003), cotton (Huang and Liu, 2006; Ma et al., 2014) and soybean (Mizoi et al., 2013). In Arabidopsis, there was 57 DREB transcription factors classified into six groups termed A-1 to A-6 based on the similarities of the AP2/ERF domain (Sazegari et al., 2015). DREB1 including CBF1, CBF2, and CBF3 belongs to the A-1 clade that play an important role in cold stress response and dark induced senescence process (Xu et al., 2010; Wang et al., 2018). Over-expression of CBF1 or CBF3 can also confer plant more capacity of freezing tolerance (Novillo et al., 2004; Zhou et al., 2011a). DREB2 proteins belong to the A-2 clade which is involved in regulation of drought and heat response. Over-expression of DREB2A improved survival rate under drought or heat stress (Schramm et al., 2008). In the A-6 clade, RAP2.4 was induced by salt stress. Over-expression of RAP2.4 can enhance drought tolerance in Arabidopsis. Moreover, RAP2.4A is involved in regulating expression of several chloroplast-targeted antioxidant genes. The expression of RAP2.4B was increased under heat stress conditions. Plants over-expressing RAP2.4B or RAP2.4 were hypersensitive to exogenous ABA at germination. Over-expression of Rap2.4f (At4g28140) caused precocious senescence according to the detection of increasing chlorophyll degradation and up-regulation of many SAGs (Lin et al., 2008; Xu et al., 2010; Rae et al., 2011).
There are six DEAR genes named DEAR1 to DEAR6 within the Arabidopsis genome that contain sequences with homology to the DREB domain and EAR motif. Plants over-expressing DEAR1 displayed phenotypes of cell death, increased resistance to pathogen infection and reduced freezing tolerance. Additionally, DEAR1 suppressed the expression of DREB1/CBF family genes induced by cold treatment, which resulted in reduced freezing tolerance (Wang et al., 2008; Tsutsui et al., 2009). DEAR4 was identified as a regulator of cell death in the hypocotyls-root transition zone using the inducible over-expression strategy (Coego et al., 2014). In this study, we characterize the function of DEAR4, a member of DREB/CBF family, in leaf senescence and stress response. The expression of DEAR4 was strongly induced by developmental stage, darkness and multiple stresses. Phenotype analysis revealed that DEAR4 was involved in the senescence process induced by age and darkness. Further study revealed that over-expression of DEAR4 led to reduce expression of COR and RD genes which are involved in senescence and stress regulation. Our findings suggest that DEAR4 is a transcriptional repressor and plays a role in regulating leaf senescence and stress response by repressing the expression of COR and RD genes.
Materials and Methods
Plant Materials, Growth Conditions
Arabidopsis seeds were surface-sterilized by 75% (v/v) ethanol followed by 3 washes with water. Then the seeds were sown on 0.5× Murashige and Skoog medium (MS) and kept at 4°C for 3 day. One-week-old seedlings were transferred into soil. Plants were grown in growth chambers at 22°C under continuously light. The mutant dear4 (Salk_010653c), dear4-1(Salk_045347) and DEAR4 inducible over-expression lines DEAR4-ind-1(cs2102284) and DEAR4-ind-2(cs2102286) used in this study were obtained from the Arabidopsis Biological Resource Center (ABRC).
Detached Leaf Phenotype Investigation
For natural leaf senescence evaluation, the fifth and sixth rosette leaves of 4-week-old plants were detached for measurements of chlorophyll content and ion leakage rate. In the hormone induced leaf senescence assay, the fifth and sixth rosette leaves of 4-week-old plants were detached and floated on 3 mL of treatment buffer (0.5× MS, 3 mM MES, PH: 5.8) supplemented with 50 μM MeJA. Petri dishes were sealed with parafilm tape, wrapped with double-layer aluminum foil. In the dark induced senescence assay, the fully expanded fifth and sixth rosette leaves were detached from 4-week-old plants grown in soil. Then, detached leaves were placed in petri dishes containing two layers of filter paper soaked in 10 mL of treatment buffer (0.5× MS, 3 mM MES adjust PH to 5.8). Then the petri dishes were wrapped in aluminum foil for 5 days. Three biological replicates were performed.
Chlorophyll Content, Ion Leakage Measurement
For chlorophyll content measurement, detached leaf was weighted and soaked in 96% (v/v) ethanol (3–4 mg of tissue in 1 mL of ethanol) overnight at room temperature in the dark condition. Total chlorophyll content was determined by measuring the absorbance at 646.6 and 663.6 nm as described (Zhang and Guo, 2018). For ion leakage measurement, detached leaves were washed three times with deionized water followed by immersion in deionized water, then gentle shaking for 1–2 h at room temperature. Total conductivity was measured as initial readings data, then samples were boiled and cooled down to room temperature and measured again with a bench-top conductivity meter (CON500, CLEAN Instruments) to get the final total conductivity. Total electrolyte leakage is determined by the following formula: Ions leakage (%) = initial / final conductivity ∗100 (Zhang and Guo, 2018). Three biological replicates were performed.
Hormone and Stress Treatments for Gene Expression Analysis
The fifth leaf was detached from 4-week-old plants grown in continuously light condition, then transferred into 0.5× MS liquid culture containing the plant hormones ABA (1 μM), SA (20 μM), IAA (20 μM), MeJA (20 μM), ACC (20 μM) or GR24 (5 μM), incubated for 6 and 8 h, respectively. In environmental condition treatments, the leaves were transferred into 0.5× MS liquid culture containing NaCl (100 mM), mannitol (200 mM) or transferred to 4°C from room temperature for 12 and 24 h.
qRT-PCR
Total RNA was isolated from frozen leaf samples using trizol according to instructions of the manufacturer. The RNA was treated with RNase-free DNaseI to release the genomic DNA. First-strand cDNAs were generated from total RNA by reverse transcription using an AMV reverse transcriptase first-strand cDNA synthesis kit (Life Sciences, Promega). Next, cDNA samples were used for the following qRT-PCR. Triplicate quantitative assays and three biological replicates were performed using the SYBR Green Master mix using an ABI 7500 sequence detection system (Applied Biosystems). The relative quantitation method (ΔΔCT) was used to evaluate quantitative variation among replicates. Actin2 were applied as internal controls to normalize all data. The primers used in this study are as follows: Q_SAG12_F:5′-TCCAATTCTATTCGTCTGGTGTGT-3′; Q_SAG12_R: 5′-CCACTTTCTCCCCATTTTGTTC-3′; Q_SEN4_F: 5′-GACTC TTCTCGTGGCGGCGT-3′;Q_SEN4_R 5′-CCCACGGCCATTC CCCAAGC-3′ Q_ACT2_F: 5′-TGTGCCAATCTACGAGGGTT T-3′; Q_ACT2_R: 5′-TTTCCCGCTCTGCTGTTGT-3′; Q_RBC S3B_F: 5′-AGTAATGGCTTCCTCTATGC-3′; Q_RBCS3B_R: 5′-GTGATGTCCTTGTTGGTCTTG-3′; Q_DEAR4_F:5′-GAGG TCCTTCTGCTCGGCTT-3′; Q_DEAR4_R:5′-CCGCCGACAT ATCTCCACCA-3′.
Stress Response Assays
For seed germination assays in different stress conditions, seeds of DEAR4 over-expression and Col-0 were surface-sterilized and incubated in 70% (v/v) ethanol for 5 min, and then washed five times quickly with water. Then, seeds were distributed on 0.5× MS solid media supplemented with 0 mM, 50 mM, 80 mM NaCl or 0.5 μM, 0.7 μM ABA. Seeds were stratified at 4°C for 3 days then transferred to 22°C under continuously light condition. Germination was monitored every 24 h as percentage of seeds with radicles completely penetrating the seed coat, for up to 5 days. Representative graphs are shown indicating germination up to 5 days. For drought adaption measurement assay, seeds of DEAR4 over-expression and Col-0 were sowed into soil and normally watered for 4 weeks. Then, phenotypes were observed after an additional 10 days without watering. When the Col-0 plants showed lethal phenotypes, watering was resumed, and phenotypes were observed again after an additional 5 days. The survival rate was measured based on three replicates.
Inducible Expression of DEAR4
A total of 3-week-old plants grown in pots were sprayed with 10 μM β-estradiol (EST) once a day for 2 days and incubated for 6 additional days. Phenotype analysis and chlorophyll content measurement were carried out as above described. Three biological replicates were performed.
Determination of H2O2 Accumulation
For NBT staining, the 5th leaves of 4-week old plants were detached, then, soaked in the NBT staining buffer (0.5 mg/mL NBT in 10 mM potassium phosphate buffer, pH 7.6) overnight. Leaf chlorophyll was removed in the fixative solution (ethanol: acetic acid: glycerol, 3:1:1) and then kept in the ethanol: glycerol (4:1) solution at 4°C until photographed. Endogenous hydrogen peroxide content was measured following the instructions of the manufacturer (Nanjing Jiangcheng Company, China). Three biological replicates were performed.
Dual-Luciferase (LUC) Assay
In the dual-LUC reporter assay, CaMV35S:DEAR4 was used as the effector construct. The reporter construct pGreenII 0800-LUC harboring the firefly LUC driven by promoters of COR15a, COR15b, RD29a or RD29b which contains the DRE/CRT element with the length of 481 bp, 361bp, 415 bp, and 429 bp, respectively. Renilla LUC gene driven by the CaMV35S promoter was used as an internal control. The reporter construct was co-transformed with the helper plasmid p19 into Agrobacterium GV3101. The Agrobacterium culture harboring the reporter construct was either incubated alone or as a mixture with the Agrobacterium culture containing the effector construct, and infiltrated manually into the leaves of Nicotiana benthamiana, then the plants were moved into darkness for 3 days. Firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) according to the instruction of the manufacturer. The Firefly/Renilla luciferase ratio indicates transcriptional activity. Three biological replicates were performed.
proDEAR4::GUS Construct and GUS Activity Detection
Firstly, we cloned the 1.4 kb promoter of DEAR4 using primers: proDEAR4-EcoRF:5′-CCGGAATTCattaccgcctcttccct att-3′ and proDEAR4-NcoIR:5′-CATGCCATGGagtggttttc tccggagatttc-3′, then the empty construct pcambia3301 was digested by restriction enzymes Hind III and Nco I, forming the proDEAR4::GUS construct after ligation reaction using T4 ligase (NEB No. M0202). According to the method described by Jefferson et al. (1987), leaves at different developmental stages were detached, then immersed in the histochemical staining buffer (1 mM 5-bromo-4-chloro-3-indolyl-b-glucuronic acid (Gluc) solution in 100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferricyanide and 0.1%Triton X-100), and then incubated at 37°C for 12 h. The leaves were destained in 70% ethanol before photographed.
Transcriptional Repression Assay in Yeast Cells
To detect the transcriptional repression activity of DEAR4, yeast one-hybrid assay was employed in this study. The DEAR4-BD, shDEAR4 (without EAR domain)-BD, DEAR4-BD-VP16, shDEAR4-BD-VP16 constructs were transformed into the yeast strain Y190. To measure the strength of the X-Gal activity, liquid assay was carried out using CPRG as substrate. Three biological replicates were performed.
Plasmid Construction and Transformation of Arabidopsis
To generate the DEAR4 over-expression constructs, the full length of DEAR4 CDS was amplified by nest PCR method using primers (First round primers: DEAR4-BP-F: 5′-TACAAAAAAGCAGGCTTCATGGAGACGGCGACTGAAGT GG-3′; DEAR4-BP-R:5′-GTACAAGAAAGCTGGGTCATCGT CATCTGAAGTTTCCGG-3′; second round primers: attB-F:5′-GTGGGGACAAGTTTGTACAAAAAAGCAGGCTTC-3′; attB-R:5′-GTGGGGACCACTTTGTACAAGAAAGCTGGGTC-3′). According to the instructions of the invitrogen gateway kit (kit No.11789 (BP Clonase); No.117910 (LR Clonase), the PCR products were cloned into pDnor-207 vector using the BP enzyme. Subsequently, according to Earley et al. (2006), DEAR4 CDS was sub-cloned into pEarleyGate202 using the LR enzyme to form the 35S::DEAR4 construct. Then the constructs was transformed into Agrobacterium tumefaciens strain GV3101. The binary constructs were transformed into Arabidopsis plants via the floral dip method (Clough and Bent, 1998). Transgenic plants were selected by glyphosate resistance.
Results
The Expression Pattern of DEAR4
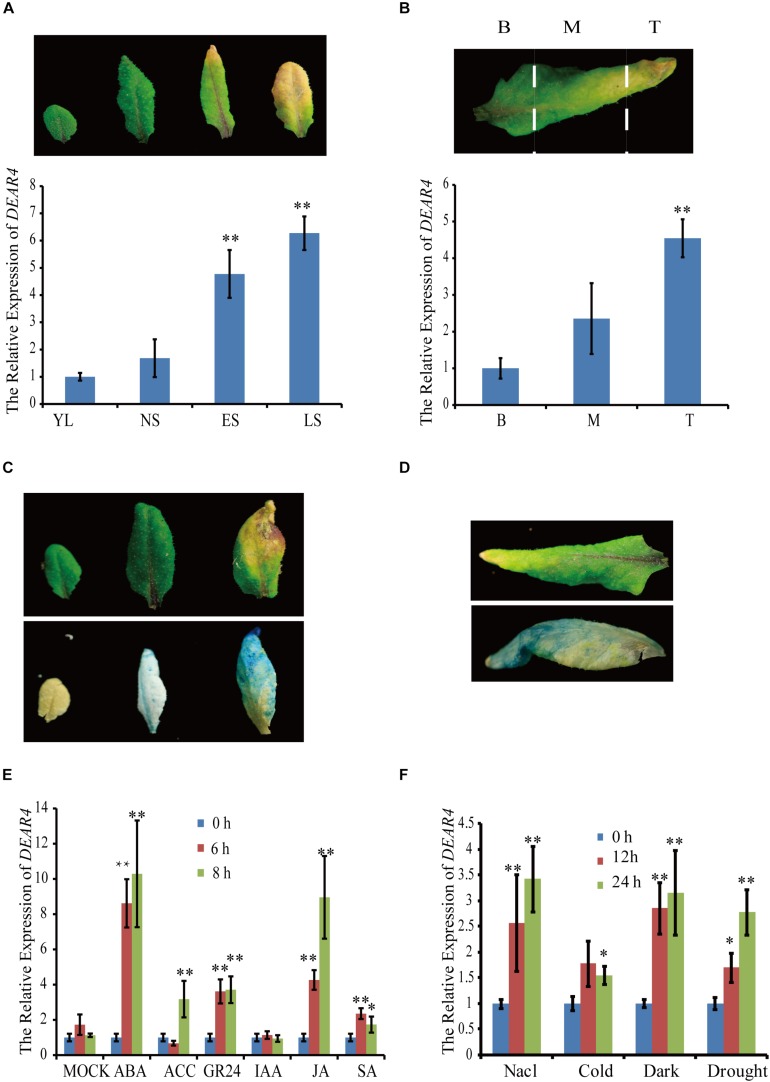
To identify new genes regulated by both senescence and light, the GENEVESTIGATOR database1 was screened and DEAR4 was found to be highly expressed in senescing plant tissues, meanwhile induced by dark conditions. To confirm the in silico data, we performed qRT-PCR to measure the expression of DEAR4. The results showed that expression levels of DEAR4 increased significantly from young leaf to late senescence stage (Figure 1A). In Arabidopsis, senescence proceeds from the tip toward the base of a leaf. When approximately 30% of the leaf area was yellow, the sixth leaves from 4-week-old Arabidopsis plants were detached and dissected into three parts including basal, middle and tip parts (Figure 1B). The expression of DEAR4 in these three parts was determined by qRT-PCR and the results showed that expression of DEAR4 exhibited higher in the tip but lower in the base region of these senescing leaves (Figure 1B).
FIGURE 1.
The expression pattern of DEAR4. (A) The expression pattern of DEAR4 at different developmental stages. YL, young leaves of 2-week old seedlings; NS, fully expanded mature leaves without senescence symptoms; ES, early senescent leaves, with <25% leaf area yellowing; LS, late senescent leaves, with >60% leaf area yellowing. (B) The DEAR4 expression pattern in different parts of a senescing leaf; B, Base; M, Middle; T, Tip. (C) GUS expression detection in different stage rosette leaf of proDEAR4::GUS transgenic plants. Top Leaf, before GUS staining; bottom leaf, after GUS staining. (D) GUS staining results in different parts of a senescing leaf. (E) Effects of plant hormones on DEAR4 expression. (F) Effects of different stress conditions on DEAR4 expression. The bars are standard deviations (SD) of three biological replicates. The one and double asterisks indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
To further examine the expression pattern of DEAR4, we generated proDEAR4::GUS transgenic plants that harbor the 1.4 kb long DEAR4 promoter driving the GUS coding sequence. Analyses of the transgenic plants revealed that strong GUS activity was detected mainly in senescent leaf tissues, which is consistent with the qRT-PCR results (Figures 1C,D). These data indicated that expression of DEAR4 is associated with natural leaf senescence.
The expression changes of DEAR4 after exogenous phytohormones application were also studied. DEAR4 expression was significantly up-regulated to 8 folds higher 6 h after ABA treatments and to 4 folds higher at 6 h after MeJA treatments. But no significant differences in expression of DEAR4 were observed after SA, ACC or IAA treatment (Figure 1E).
To obtain insight into whether DEAR4 was also involved in stress response, the expression pattern of DEAR4 was examined in response to environmental stimuli. As shown in Figure 1F, the transcript levels of DEAR4 increased significantly in response to NaCl, darkness and drought treatments.
DEAR4 Is Involved in Age-Dependent Leaf Senescence
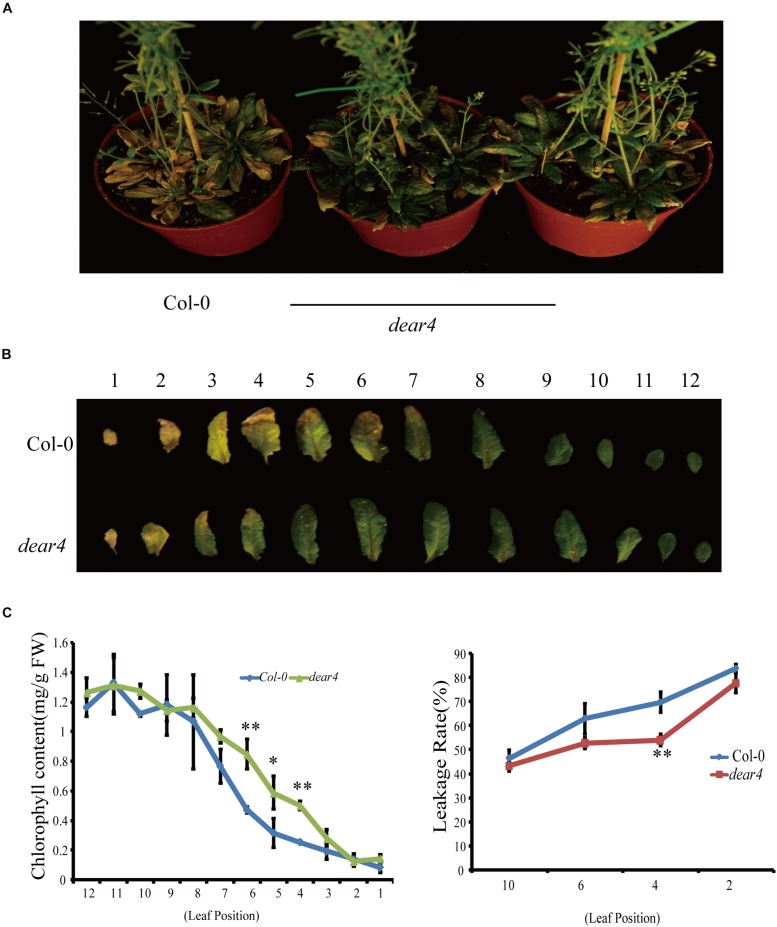
To investigate the function of DEAR4, we obtained two DEAR4 T-DNA insertion mutant lines from ABRC named dear4 (SALK_010653c) and dear4-1 (Salk_045347). The T-DNA insertion in the dear4 mutant is located at the 5′UTR region of DEAR4 (Supplementary Figure 1A) and the qRT-PCR results showed that the transcript of DEAR4 was significantly reduced in the dear4 mutant (Supplementary Figure 1B). Plants of dear4 displayed a delayed senescence phenotype assessed by comparing the degree of leaf yellowing with Col-0 (Figure 2A). In 6-week-old plants, most leaves of Col-0 turned yellow with drying, yet dear4 mutant leaves retained their integrity and displayed only partial yellowing (Figures 2A,B). Consistent with the visual phenotype, the chlorophyll content of Col-0 leaves decline faster in comparison with the counterpart of dear4 mutant plants (Figure 2C upper). Leaf senescence often involves reduction of plasma membrane integrity, as indicated by membrane ion leakage. The delayed senescence symptoms of dear4 can also be evidenced by lower membrane ion leakage of the leaves compared with Col-0 (Figure 2C lower). As seen in Supplementary Figure 2, dear4-1 displayed a similar phenotype with dear4 in delaying leaf senescence. These results demonstrated that DEAR4 plays a potential role in promoting leaf senescence.
FIGURE 2.
A DEAR4 T-DNA insertion delays leaf senescence process. (A) Leaf phenotype of 6-week-old Col-0 and dear4 mutant plants. (B) Senescence symptoms of detached leaves of Col-0 and dear4 plants. (C) Total chlorophyll content (1th–12th leaf) and ion leakage rate (the leaf position as indicated) in different genotype plants as indicated. The bars are standard deviations (SD) of three biological replicates. The one and double asterisks indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
Over-Expression of DEAR4 Leads to Precocious Senescence
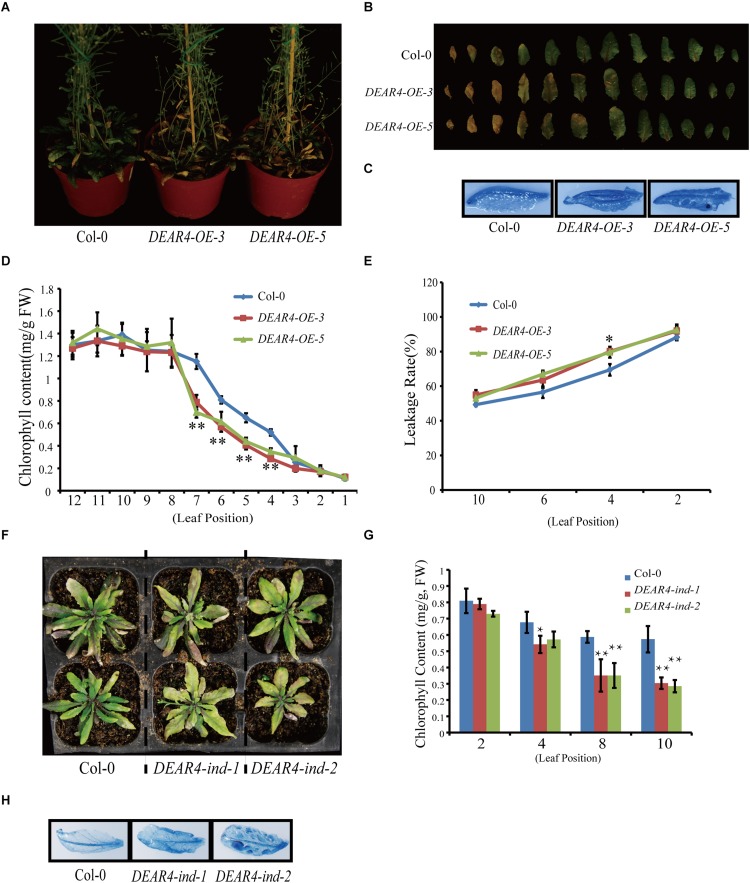
To further explore the biological function of DEAR4, we generate multiple independent DEAR4 over-expression transgenic lines harboring the DEAR4 CDS under control of the CaMV 35S promoter (35S::DEAR4). qRT-PCR analysis showed that DEAR4 transcript levels in DEAR4-OE-3 and DEAR4-OE-5 were 8 to 10 folds higher than that of Col-0 (Supplementary Figure 3A). Phenotypic analysis showed that DEAR4 over-expressing lines displayed precocious leaf senescence judged by the progression of leaf yellowing (Figures 3A,B). Consistent with the visible phenotype, the reduction in chlorophyll contents of leaves from the DEAR4 over-expression lines were greater than in Col-0 (Figure 3D). The precocious senescence of DEAR4 over-expression plants were supported by higher membrane ion leakage of the leaves compared with Col-0 (Figure 3E). These results demonstrated that DEAR4 plays an important role in promoting leaf senescence. In addition, we stained the fully expanded rosette leaves of different genotype plants via Trypan blue staining to assess dead cell rates. As shown in Figure 3C, the staining of dead cells in DEAR4 over-expression lines was higher than that in Col-0. We further examined the expression of senescence marker genes. As showed in Supplementary Figure 4, compare with Col-0, the expression levels of SAG12, SEN4 were dramatically up-regulated in DEAR4 over-expression lines but lower in dear4 mutant, whereas the expression levels of photosynthetic gene such as RBCS were clearly down-regulated in DEAR4 over-expression lines but up-regulated in the dear4 mutant.
FIGURE 3.
Over-expression of DEAR4 accelerates leaf senescence. (A) The phenotypes of DEAR4 over-expression lines. (B) Detached leaf phenotypes of Col-0, DEAR4-OE-3, and DEAR4-OE-5 described in (A). (C) Trypan blue staining of the sixth leaf of 6-week-old Col-0 and DEAR4 over-expression lines. (D) Total chlorophyll content of 1th–12th leaf from 6-week-old Col-0 and DEAR4 over-expression lines. (E) Leakage rate of Col-0 and DEAR4 over-expression lines. (F) Inducible over-expression of DEAR4 causes precocious senescence. (G) Chlorophyll content of different genotype plants that were treated with EST. (H) Trypan blue staining of different plants as indicated. The bars are standard deviations (SD) of three biological replicates. The one and double asterisks indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
To further confirm the DEAR4 gain-of-function phenotype, we investigated the phenotypes of DEAR4 inducible over-expression lines which express DEAR4 under the control of a β-estradiol (EST) inducible promoter (DEAR4-ind-1 and DEAR4-ind-2). The expression of DEAR4 was detected by qRT-PCR (Supplementary Figure 3B). The phenotype analysis revealed that the DEAR4-ind lines displayed precocious senescence after treatment with 10 μM EST compared with Col-0 (Figure 3F). A decline in chlorophyll content was observed in leaves of DEAR4 inducible lines treated with EST (Figure 3G). Moreover, treatment with EST displayed significantly higher cell death ratio in DEAR4 inducible gain-of-function leaves than that of control as revealed by trypan blue staining (Figure 3H). Taken together, these results indicated that DEAR4 functions in accelerating leaf senescence.
DEAR4 Is Involved in Senescence Induced by Darkness and JA
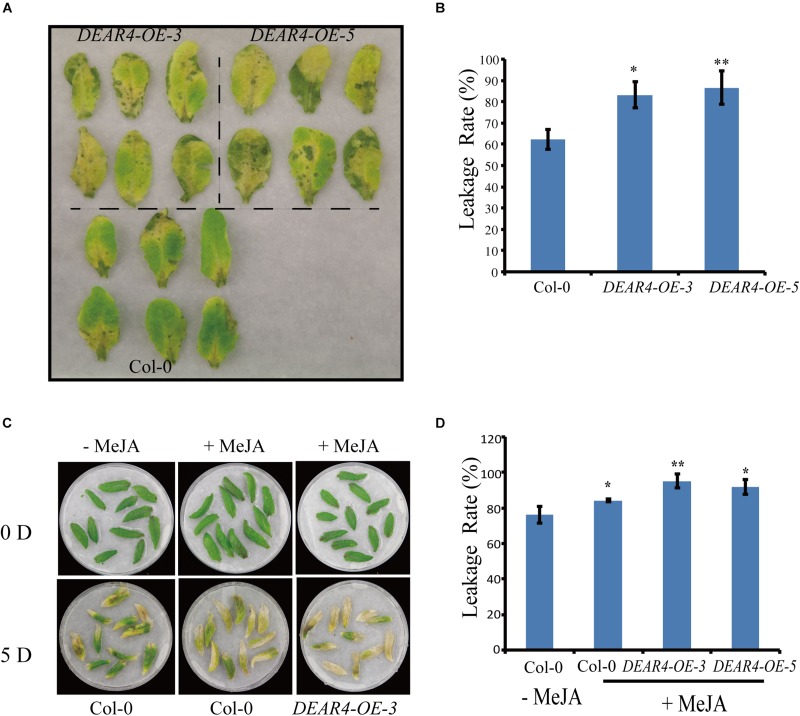
Given that DEAR4 was increased at the transcriptional level under dark condition, we investigated the phenotypes of DEAR4 over-expression plants during dark treatment. Detached leaves of Col-0 and DEAR4 over-expression plants were covered with aluminum foil. Five days after treatment leaves from the two DEAR4 over-expression lines exhibited an accelerated yellowing phenotype compared with those from Col-0 (Figure 4A). Consistent with the visible phenotype, DEAR4 over-expression plants exhibited higher ion leakage rate compared with Col-0 (Figure 4B).
FIGURE 4.
Precocious senescence phenotypes of DEAR4 over-expression lines in dark conditions. (A) Phenotypes of detached leaves in response to dark condition. (B) Ion leakage rate in Col-0, DEAR4-OE-3, and DEAR4-OE-5. (C,D) DEAR4 over-expression plants showed enhanced sensitivity to JA. The bars are standard deviations (SD) of three biological replicates. The one and double asterisks indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
Since expression of DEAR4 could be enhanced by JA treatments (Figure 1E), we sought to explore the relationship between DEAR4 and JA under dark condition. Detached leaves from 4-week-old plants of different genotypes were incubated with MeJA. After 5 days’s MeJA treatment, DEAR4 over-expression leaves displayed serious yellowing compared with Col-0 (Figure 4C). Consistent with the visible precocious senescence, the results of ion leakage rate measurement showed that conductivity in DEAR4 over-expression lines was higher compared with Col-0 after MeJA treatments (Figure 4D). The two independent overexpression lines displayed the similar phenotype.
DEAR4 Functions in Response to Drought, NaCl and ABA
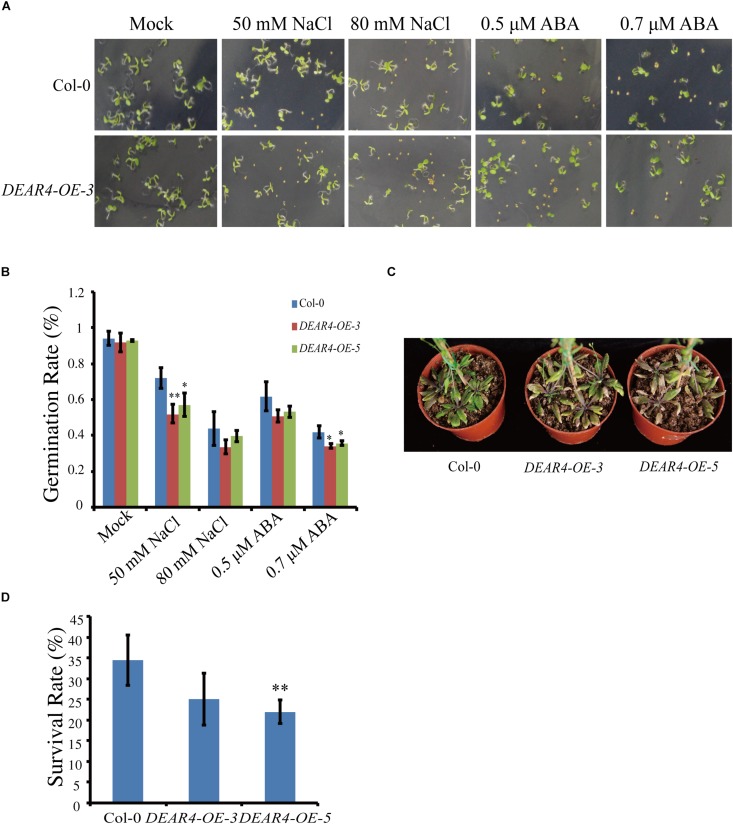
Based on the expression data, DEAR4 expression can be induced by NaCl and ABA (Figures 1C,D). Seeds from DEAR4 over-expression lines and Col-0 were sowed on 0.5× MS plates supplied with different concentrations of NaCl or ABA and the percentages of seed germination were calculated based on the number of seeds showing the radicle emergence. The results showed that the percentage of germination between the Col-0 and DEAR4 over-expression seeds were similar under normal condition. However, in the 50 mM NaCl treatment, the radicle emerged in 71.9 and 93.2% of DEAR4-OE-3 and Col-0 seeds, respectively. Whereas, only 40.6 and 59.9% germination rates were observed in DEAR4 over-expression and Col-0 seeds in the 80 mM NaCl treatment. In the 0.5 μM ABA treatment, 62% of the Col-0 seeds germinated, while this rate was reduced to 53% for DEAR4 over-expression seeds (Figures 5A,B), the two independent overexpression lines displayed the similar results. In the drought treatment, plants of the DEAR4-OE-3, DEAR4-OE-5 lines and Col-0 were grown in soil for 4 weeks under normal condition. Then, drought stress was applied by withholding watering. Survival rates of plants from different genotypes were calculated after15 days of drought stress treatment. Plants of Col-0 exhibited the survival rates of over 34.37%, which was significantly higher than DEAR4-OE-3 and DEAR4-OE-5 transgenic plants with the survival rates of 25 and 21.87%, respectively (Figures 5C,D). Taken together, these results demonstrated that DEAR4 conferred plant more sensitive to drought and salt stress.
FIGURE 5.
Responses of DEAR4 over-expression plants to NaCl, ABA and drought. (A,B) The effect of different concentrations of NaCl and ABA on germination of Col-0 and DEAR4 over-expression plants. (C,D) DEAR4 over-expression plants were sensitive to drought treatment. The bars are standard deviations (SD) of three biological replicates. The one and double asterisks indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
DEAR4 Regulates ROS Production
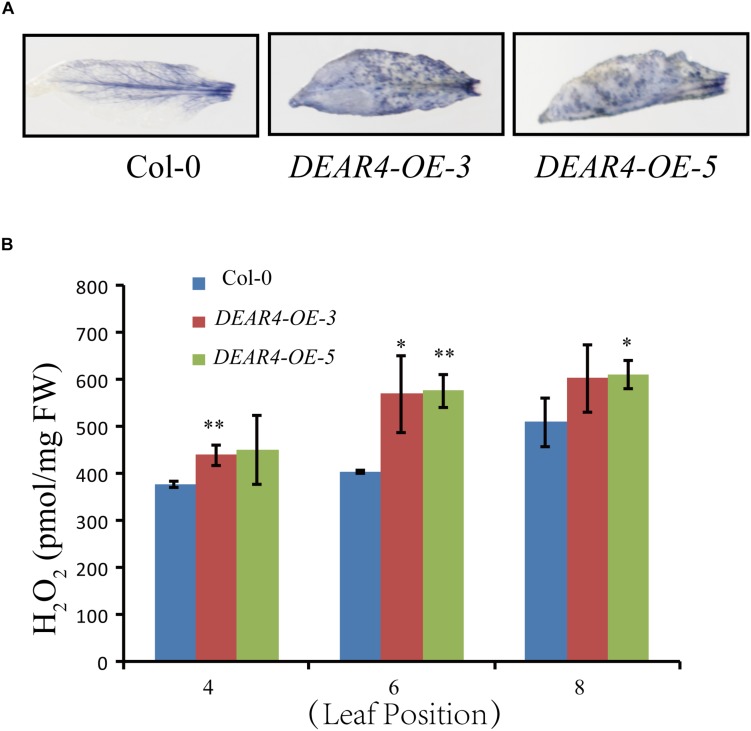
Reactive oxygen species are considered signaling molecules during leaf senescence and stress responses. To further understand the role of DEAR4 in leaf senescence and stress response, we employed NBT staining to visualize the levels of ROS. The fifth leaf from 4-week-old plants of different genotypes including DEAR4 over-expression and Col-0 were detached and analyzed. As shown in Figure 6A, compared with Col-0, leaves of DEAR4-OE-3 and DEAR4-OE-5 were densely stained with dark blue and brown color by NBT staining, suggesting that DEAR4 enhanced ROS production. To further confirm the staining results, quantitative measurement was carried out to determine the endogenous H2O2 levels in plants of different genotypes. The results revealed that DEAR4 over-expression plants accumulated significantly more H2O2 compared with Col-0 (Figure 6B).
FIGURE 6.
DEAR4 promotes ROS production. (A) NBT staining. The 5th leaves of 4-week-old plants were used in NBT staining. (B) Measurement of temporal accumulation of H2O2 in detached leaves of plants with different genotypes. The one and double asterisk indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively. The bars are standard deviations (SD) of three biological replicates.
DEAR4 Is a Transcriptional Repressor
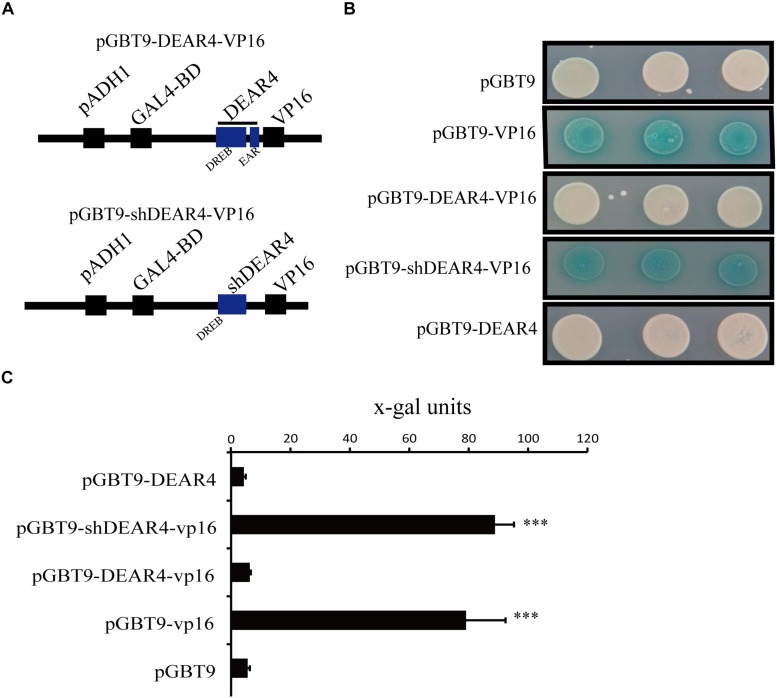
The Arabidopsis DEAR4 protein contains homology to the DREB1/CBF domain and the EAR motif. EAR motifs commonly played as transcriptional repression roles in plants (Yang et al., 2018). Thus, we supposed that DEAR4 has the potential of functioning as a transcriptional repressor. To verify this point, we carried out yeast one-hybrid assay. The full-length or truncated DEAR4 (shDEAR4, without EAR domain, 1-181aa) cDNA was fused to VP16, the activation domain of a potent viral transcriptional activator. The DEAR4 (or shDEAR4)-VP16 component group was then fused downstream of the GAL4 DNA-binding domain (GAL4-BD) in the pGBT9 vector to generate the BD-DEAR4 (or shDEAR4)-VP16 construct (Figure 7A). As a positive control, yeast strain Y190 carrying the reporter genes LacZ transformed with pGBT9-VP16 showed strong blue reaction (Figure 7B). Similar to pGBT9-VP16, Y190 transformed with pGBT9-shDEAR4-VP16 also displayed strong blue reaction. However, Yeast strain harboring pGBT9 empty vector displayed white reaction, similar to yeast transformed with DEAR4 fused with or without VP16 (Figure 7B). These results suggest that the full length DEAR4 protein plays transcriptional repression role which is dependent on the functional EAR domain.
FIGURE 7.
Transcriptional repression activities of DEAR4 in yeast cells. (A) Schematic diagram of pGBT9-DEAR4-VP16 and pGBT9-shDEAR4-VP16. (B) Comparison results of the Y1H assay using full length DEAR4 or shDEAR4 (truncated sequence absenting EAR domain coding region) fused with vp16 and control. (C) Liquid assay to measure x-gal activities in yeast containing different construct using CPRG as substrate. The bars are standard deviations (SD) of three biological replicates. The triple asterisk indicates significant difference to control at P < 0.001 using student’s t-test.
We also carried out liquid assay using CPRG as substrate to quantify the x-gal activities from the above described yeast one-hybrid assays. Consistent with the results from blue-white reactions, the reporter gene activity in the yeast transformed with pGBT9, BD-DEAR4, BD-DEAR4-VP16, and BD-VP16 was 5.57 ± 0.68, 4.24 ± 0.67, 6.29 ± 0.37, and 79.13 ± 13.28, respectively. However, once the EAR domain was deleted, the reporter gene activity in yeast transformed with BD-shDEAR4-VP16 was up to 88.82 ± 6.44 (Figure 7C). Taken together, BD-VP16 protein was able to induce lacZ expression but DEAR4 protein repressed this process. However, DEAR4 protein with EAR domain deleted lost its transcriptional repression ability.
DEAR4 Directly Repress the Expression of COR and RD29
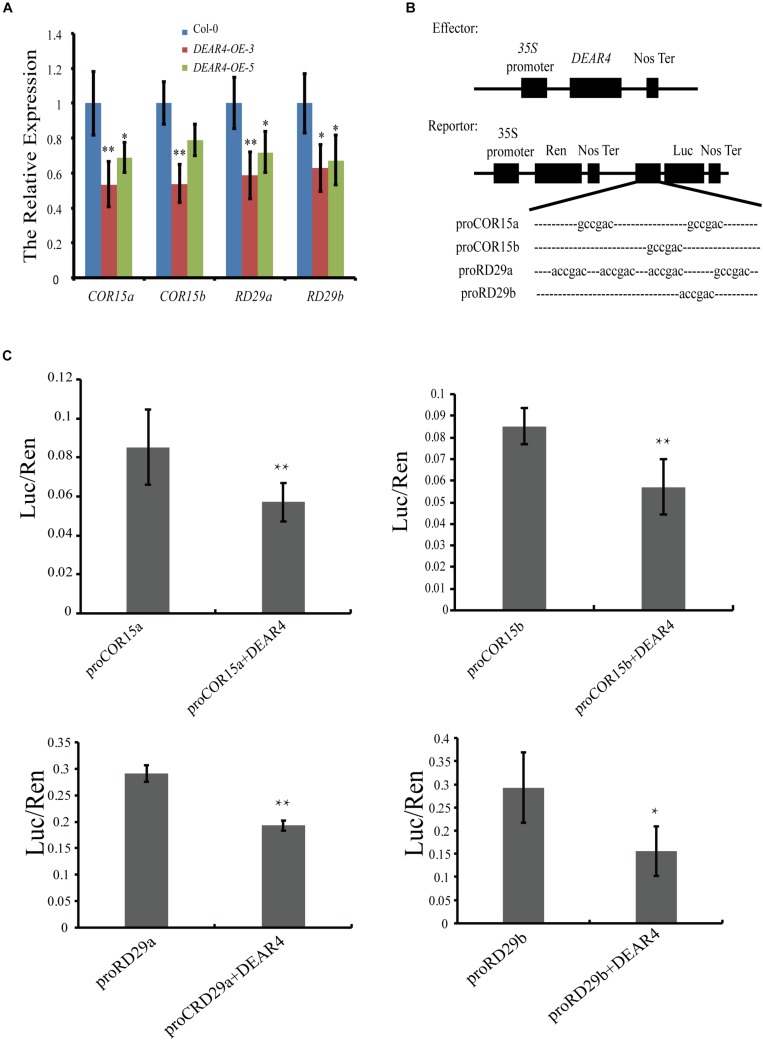
DEAR4 protein contains homology to the DREB1/CBF domain which binds to the DRE/CRT element containing an A/GCCGAC motif within gene promoters to regulate transcription. To further understand the molecular mechanisms underlying DEAR4’s role in leaf senescence and stress responses, we investigated whether DEAR4 could regulate some downstream genes in the DRE/CRT-mediated signaling pathway. There are two putative DRE/CRT elements (−444 to −438 bp, −267 to −261 bp) upstream of the translation start site on the COR15a promoter and one DRE/CRT element at −266 to −260 bp from the translation start site on the promoter of COR15b. Meanwhile, we identified four putative DRE/CRT elements (−353 to −347 bp, −303 to −297 bp, −246 to −240 bp, −209 to −203 bp) upstream of the translation start site on the RD29a promoter and one DRE/CRT element (−321 to −315 bp from the translation start site) on the promoter of RD29b. The expression of the COR and RD genes were significantly decreased in the DEAR4 over-expression transgenic plants (Figure 8A), two independent overexpression lines displayed the similar results, suggesting that DEAR4 is required for reduction of the COR and RD gene expression in leaf senescence regulation.
FIGURE 8.
Transcriptional repression of DEAR4 on COR and RD genes. (A) The expression of COR15a, COR15b, RD29a, and RD29b in Col-0 and DEAR4 over-expression lines. (B) Dual luciferase (LUC) constructs used in this study. Dual LUC constructs contain the double reporter and effector plasmids. In the double reporter construct, Renilla LUC expression was used to normalize the firefly luciferase activities. The effector construct in this study contains DEAR4 driven by the CaMV35S promoter. (C) DEAR4 directly represses the expression of COR15 and RD29. The firefly LUC gene was driven by the COR15 and RD29 promoters, ratio of firefly and RenLuc indicates the relative activities of promoters. The bars are standard deviations (SD) of three biological replicates. The one and double asterisk indicate significant difference to control at the levels of 0.01 < P < 0.05 and P < 0.01 using student’s t-test, respectively.
Next, we tested whether DEAR4 directly regulates the COR and RD genes. We employed the dual luciferase strategy to investigate the direct regulation of COR and RD genes by DEAR4 in Nicotiana benthamiana leaves. Agrobacterium harboring the firefly luciferase-encoding gene driven by the promoter of COR (or RD) was co-transformed with or without 35S::DEAR4. As an internal control, the renilla luciferase gene was driven by the 35S promoter (Figure 8B). The ratio of Firefly/Renilla luciferase activities indicates the transcriptional activity of COR or RD genes. The results showed that the Firefly/Renilla luciferase ratio was significantly lower in leaves co-transformed with DEAR4 compared to that without DEAR4, indicating that DEAR4 repress the expression of COR and RD genes directly (Figure 8C).
Discussion
As the final phase of leaf development, senescence is crucial for plant survival and environmental adaptation. So far, thousands of genes and many signaling pathways have been studied for leaf senescence regulation. Among them, transcription factors are highly effective in engineering stress tolerant plants (Bengoa et al., 2019; Woo et al., 2019). A large number of transcriptional factor genes including NAC, WRKY, MYB, and AP2/EREBP were up- or down-regulated during natural and dark-induced senescence (Zhang and Zhou, 2013). Additionally, transcript profiling studies suggested that many of the genes were affected by senescence and environmental stresses at the same time (Yoshida, 2003; Sharabi-Schwager et al., 2010a; Chen et al., 2017). As the largest transcription factor family in Arabidopsis, the AP2/ERF proteins have roles in the regulation of developmental processes, hormonal signal transduction, and biotic and abiotic stress responses (Smirnoff and Bryant, 1999; Yang et al., 2019b). As one of the AP2/ERF subfamilies, members of the DREB protein family have been extensively studied over the years due to their crucial roles in regulation of abiotic- and biotic-stress responses. It’s well known that DREB protein recognized the dehydration responsive element (DRE)/C-repeat with a core sequence of A/GCCGAC to regulate gene expression (Smirnoff and Bryant, 1999; Sharoni et al., 2011; Kudo et al., 2017). More and more evidence suggested that the DREB proteins play multiple roles during plant development. The CBF genes including CBF1, −2, −3, also known as DREB1b, DREB1c, and DREB1a, respectively, have been identified as key regulators of cold response and drought response (Liu et al., 2004; Novillo et al., 2004). In addition, CBF genes also play important roles in dark induced senescence (Gilmour et al., 2000; Solanke and Sharma, 2008; Zhou et al., 2011a). Here, we investigated the function of DEAR4, which contains the DREB domain but is different from the typical DREB proteins, with an EAR motif at the C terminus. Consistent with in silico data in publically available databases (GENEVESTIGATOR), we found that DEAR4 can be induced by age and dark conditions, as well as multiple abiotic stresses and hormones including ABA, JA, salt, and drought (Figure 1). More and more evidence suggested that plants have the capacity of integrating various signaling pathways to provide a greater regulatory potential for enhancing environment adaption. Our data revealed that DEAR4 was involved in age and dark induced leaf senescence based on phenotype and physiological data (Figures 2–4). At the molecular level, the precocious leaf senescence phenotype is associated with a gradual increase in the transcript levels of SAGs including SAG12, SEN4 and decrease in the expression of the RBCS gene (Supplementary Figure 2). Previous study has reported that DEAR1, one of the DEAR4 homologs, plays roles in mediating crosstalk between biotic and abiotic stresses. DEAR1 expression was enhanced by pathogen infection and cold treatment. Over-expression of DEAR1 caused an activated defense phenotype. Additionally, the induction of DREB1/CBF family genes by cold treatment was suppressed in DEAR1 over-expression lines, leading to a reduction in freezing tolerance (Tsutsui et al., 2009). In addition, DEAR1 is included in the A-5 subgroup of the DREB/family that could form a negative feedback regulation of the DREB1/CBF and DREB2 pathway in response to cold and dehydration (Mizoi et al., 2012). There are five DEAR1 homologs including DEAR4 within the Arabidopsis genome that also contain a DREB domain and an EAR motif. The role of DEAR4 in stress response therefore is not surprising. Interestingly, we found that DEAR4 also plays a role in leaf senescence, suggesting that the DEAR genes may play different roles in plant development and response to environmental stimuli. It will be interesting to find out whether the similar negative feedback mechanism exists in DEAR4’s function in leaf senescence and stress response.
Jasmonic acid acts as a crucial signal to modulate multiple plant processes including senescence and stress responses. JA content was much higher in senescent leaves than in non-senescent ones (Shan et al., 2011; Seltmann and Berger, 2013). Additionally, exogenous application of JA enhances plants’ freezing tolerance with or without cold acclimation. Further study revealed that JA positively regulates CBF to up-regulate downstream cold-responsive genes to enhance cold tolerance (Hu et al., 2013, 2017). JAZ proteins were discovered as repressors of JA signaling through the COI1-dependent 26S proteasome pathway for protein degradation. Once JAZ proteins were reduced, various downstream transcription factors including MYC2, MYC3 and MYC4 were activated (Fernandez-Calvo et al., 2011; Hu et al., 2013). In darkness, the mutant of JAZ7 partially liberated MYC2/MYC3/MYC4 from suppression, resulting in the up-regulation of the downstream genes related to indole-glucosinolate biosynthesis, sulfate metabolism, callose deposition, and JA-mediated signaling pathways (Yu et al., 2016). In this study, we found that DEAR4 was induced by exogenous MeJA (Figure 1E), meanwhile, DEAR4 can enhance the role of MeJA in promoting senescence (Figures 4C,D), which provide clues that DEAR4 may play roles in JA regulating senescence and stress responses. The DEAR4 protein contains the EAR motif which plays an important role in ethylene-responsive transcriptional regulation. Interestingly, previous studies have reported the mechanism of crosstalk between JA and other plant hormones including ethylene in plant growth and stress responses. JA and ethylene antagonize or coordinately regulate plant stress response (Zhu, 2014; Yang et al., 2019a). JA and ethylene pathways most likely crosstalk at the levels of JAZ–EIN3 and JAZ–EIL1 (Zhu et al., 2011; Kazan and Manners, 2012; Zhang et al., 2014). The EAR domain proteins could be involved in JA signal pathway. For example, the NINJA (Novel Interactor of JAZ, an EAR motif containing protein) mediated JAZ pathway to block the activity of MYC2, repressing the JA-dependent root growth inhibition and defense processes (Li et al., 2019). Both JA and ethylene were involved in leaf senescence. So far, the crosstalk between JA and ethylene’s functions in leaf senescence is not clear. Based on our data, DEAR4 could potentially play a role in both JA and ethylene pathways in regulating leaf senescence.
Plant ROS include hydrogen peroxide (H2O2), superoxide anion (O2–), hydroxyl radicals (OH) and singlet oxygen (1O2), which can be produced from chloroplast and mitochondrial electron transport chains, and oxidases and peroxidases located in the peroxisomes or in the plasmalemma/apoplast (Wang et al., 2013). ROS are not only as by-products of metabolic pathways but also play signaling roles during normal plant development. Multiple stresses are known to enhance ROS generation (Apel and Hirt, 2004; Mittler et al., 2004). The senescence process also increases accumulation of ROS (Jajic et al., 2015; Li et al., 2016). Thus, ROS production regulation plays key roles in both senescence and stress responses. Given that DEAR4 was involved in age and dark induced senescence, we hypothesized that DEAR4 may be involved in regulating leaf senescence though the proliferation of ROS. Consistent with this hypothesis, over-expression of DEAR4 can induce the production of ROS. DEAR4 over-expression lines displayed substantially induced ROS production based on the NBT staining and H2O2 content measurement data (Figure 6). Leaf senescence is commonly associated with electrolyte leakage which represents loss of membrane integrity. Excessive production of ROS results in membrane lipid peroxidation. Our results demonstrated that DEAR4 was involved in age- and dark induced leaf senescence and multiple stresses responses all of which are associated with membrane damage and accumulation of ROS, suggesting a crucial role of DEAR4 in these processes potentially via ROS regulation.
Leaf senescence is also regulated by environmental stimuli including salinity, drought, low quality light and darkness (Buchanan-Wollaston et al., 2005). Numerous SAGs were influenced by diverse abiotic and biotic stresses (Quirino et al., 1999; Weaver and Amasino, 2001). Additionally, some of the genes that regulate stress responses may also have an important role in regulating leaf senescence (Binyamin et al., 2001). For example, among the 43 transcription factor genes up-regulated during senescence, 28 were also induced by various stresses (Lim et al., 2007). The DREB protein family is known to regulate abiotic stress responses in plants. The DEAR4 protein contains a DREB domain. The expression of DEAR4 is lower in young leaves, but is up-regulated in senescing leaves. Meanwhile, DEAR4 gene transcript was induced by senescence as well as senescence-stress associated hormones including ABA and JA (Figure 1). Detached leaves of DEAR4 gain-of-function plants accelerated the senescence process induced by age, darkness or MeJA, suggesting that DEAR4 possibly integrates the age-dependent leaf senescence with responses to environmental stimuli including darkness and phytohormones. A large amount of evidence suggested that the DREB genes play a major role in cold- and osmotic-stress signal transduction pathways by recognizing the dehydration responsive element (DRE)/C-repeat with a core sequence A/GCCGAC. In this study, we observed that DEAR4 exhibited transcriptional repression activities in yeast cells depending on its EAR motif (Figure 7). As expected, DEAR4 was demonstrated to be able to directly repress the expression of DRE element genes COR and RD genes (Figure 8). Interestingly, it has been reported that COR and RD genes were involved in senescence and stress responses (Lee and Seo, 2015; Bremer et al., 2017; Shi et al., 2017b).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
YG conceived the project. ZZ and YG designed the research, performed the data analysis, and wrote the manuscript. ZZ, WL, MX, and XG performed the experiments.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants from the National Natural Science Foundation of China (No. 31600991) and the Agricultural Science and Technology Innovation Program, Chinese Academy of Agricultural Sciences (ASTIP-TRI02).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00367/full#supplementary-material
References
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Asad M. A. U., Zakari S. A., Zhao Q., Zhou L., Ye Y., Cheng F. (2019). Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: premature leaf senescence in plants. Int. J. Mol. Sci. 20:256. 10.3390/ijms20020256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi V., Devoto A. (2008). Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 177 301–318. 10.1111/j.1469-8137.2007.02292.x [DOI] [PubMed] [Google Scholar]
- Bengoa L. S., Astigueta F. H., Nicosia S., Moschen S., Fernandez P., Heinz R. (2019). Transcription factors associated with leaf senescence in crops. Plants 8:411. 10.3390/plants8100411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binyamin L., Falah M., Portnoy V., Soudry E., Gepstein S. (2001). The early light-induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta 212 591–597. 10.1007/s004250000423 [DOI] [PubMed] [Google Scholar]
- Bremer A., Kent B., Hauss T., Thalhammer A., Yepuri N. R., Darwish T. A., et al. (2017). Intrinsically disordered stress protein COR15A resides at the membrane surface during dehydration. Biophys. J. 113 572–579. 10.1016/j.bpj.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P. O., Nam H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42 567–585. 10.1111/j.1365-313x.2005.02399.x [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang Y., Huang J., Zheng C., Cai C., Wang Q., et al. (2017). Salt and methyl jasmonate aggravate growth inhibition and senescence in Arabidopsis seedlings via the JA signaling pathway. Plant Sci. 261 1–9. 10.1016/j.plantsci.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Coego A., Brizuela E., Castillejo P., Ruiz S., Koncz C., del Pozo J. C., et al. (2014). The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 77 944–953. 10.1111/tpj.12443 [DOI] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. 10.1046/j.1365-313x.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. 10.1111/j.1365-313x.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P., Chini A., Fernandez-Barbero G., Chico J. M., Gimenez-Ibanez S., Geerinck J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23 701–715. 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen P. L., Culetic A., Boschian L., Krupinska K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82 603–622. 10.1007/s11103-013-0013-8 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. S. (2014). Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 65 3901–3913. 10.1093/jxb/eru248 [DOI] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25 2907–2924. 10.1105/tpc.113.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang Y., Han X., Wang H., Pan J., Yu D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68 1361–1369. 10.1093/jxb/erx004 [DOI] [PubMed] [Google Scholar]
- Huang B., Liu J. Y. (2006). Cloning and functional analysis of the novel gene GhDBP3 encoding a DRE-binding transcription factor from Gossypium hirsutum. Biochim. Biophys. Acta 1759 263–269. 10.1016/j.bbaexp.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Jajic I., Sarna T., Strzalka K. (2015). Senescence, stress, and reactive oxygen species. Plants 4 393–411. 10.3390/plants4030393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. 10.1002/j.1460-2075.1987.tb02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26 230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibran R., Hunter A. D., Dijkwel P. P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 82 547–561. 10.1007/s11103-013-0043-2 [DOI] [PubMed] [Google Scholar]
- Kamranfar I., Xue G. P., Tohge T., Sedaghatmehr M., Fernie A. R., Balazadeh S., et al. (2018). Transcription factor RD26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytol. 218 1543–1557. 10.1111/nph.15127 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17 22–31. 10.1016/j.tplants.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Kudo M., Kidokoro S., Yoshida T., Mizoi J., Todaka D., Fernie A. R., et al. (2017). Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 15 458–471. 10.1111/pbi.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. G., Seo P. J. (2015). The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 82 962–977. 10.1111/tpj.12866 [DOI] [PubMed] [Google Scholar]
- Lee S., Seo P. J., Lee H. J., Park C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70 831–844. 10.1111/j.1365-313X.2012.04932.x [DOI] [PubMed] [Google Scholar]
- Li C., Shi L., Wang Y., Li W., Chen B., Zhu L., et al. (2019). Arabidopsis ECAP is a new adaptor protein that connects JAZ repressors with TPR2 co-repressor to suppress jasmonate-responsive anthocyanin accumulation. Mol. Plant 13 246–265. 10.1016/j.molp.2019.10.014 [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang X., Chen J., Gao J., Zhou X., Kuai B. (2016). CCX1, a putative cation/ca2+ exchanger, participates in regulation of reactive oxygen species homeostasis and leaf senescence. Plant Cell Physiol. 57 2611–2619. 10.1093/pcp/pcw175 [DOI] [PubMed] [Google Scholar]
- Liebsch D., Keech O. (2016). Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol. 212 563–570. 10.1111/nph.14217 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. [DOI] [PubMed] [Google Scholar]
- Lin R. C., Park H. J., Wang H. Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1 42–57. 10.1093/mp/ssm004 [DOI] [PubMed] [Google Scholar]
- Liu F. X., Tan Z. B., Zhu J. Q., Deng X. J. (2004). [Arabidopsis CBF1 in plant tolerance to low temperature and drought stresses]. Yi Chuan 26 394–398. [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. F., Zhang J. M., Huang G. Q., Li Y., Li X. B., Zheng Y. (2014). Molecular characterization of cotton C-repeat/dehydration-responsive element binding factor genes that are involved in response to cold stress. Mol. Biol. Rep. 41 4369–4379. 10.1007/s11033-014-3308-1 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Mizoi J., Ohori T., Moriwaki T., Kidokoro S., Todaka D., Maruyama K., et al. (2013). GmDREB2A; 2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161 346–361. 10.1104/pp.112.204875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Mostofa M. G., Li W., Nguyen K. H., Fujita M., Tran L. P. (2018). Strigolactones in plant adaptation to abiotic stresses: an emerging avenue of plant research. Plant Cell Environ. 41 2227–2243. 10.1111/pce.13364 [DOI] [PubMed] [Google Scholar]
- Novillo F., Alonso J. M., Ecker J. R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 3985–3990. 10.1073/pnas.0303029101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Kimura M., Matsuura H., Tanaka A., Ito H. (2019). Jasmonate production through chlorophyll a degradation by stay-green in Arabidopsis thaliana. J. Plant Physiol. 238 53–62. 10.1016/j.jplph.2019.05.004 [DOI] [PubMed] [Google Scholar]
- Penfold C. A., Buchan-Wollaston V. (2014). Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 65 3859–3873. 10.1093/jxb/eru054 [DOI] [PubMed] [Google Scholar]
- Qi T., Wang J., Huang H., Liu B., Gao H., Liu Y., Song S., Xie D. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27 1634–1649. 10.1105/tpc.15.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B. F., Normanly J., Amasino R. M. (1999). Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 40 267–278. [DOI] [PubMed] [Google Scholar]
- Rae L., Lao N. T., Kavanagh T. A. (2011). Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234 429–444. 10.1007/s00425-011-1414-z [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Sazegari S., Niazi A., Ahmadi F. S. (2015). A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 11 101–106. 10.6026/97320630011101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., von Koskull-Doring P. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53 264–274. 10.1111/j.1365-313x.2007.03334.x [DOI] [PubMed] [Google Scholar]
- Seltmann M. A., Berger S. (2013). Phenotyping jasmonate regulation of senescence. Methods Mol. Biol. 1011 3–11. 10.1007/978-1-62703-414-2_1 [DOI] [PubMed] [Google Scholar]
- Shan X., Li C., Peng W., Gao B. (2011). New perspective of jasmonate function in leaf senescence. Plant Signal. Behav. 6 575–577. 10.4161/psb.6.4.14899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Lers A., Samach A., Guy C. L., Porat R. (2010a). Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot. 61 261–273. 10.1093/jxb/erp300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Samach A., Porat R. (2010b). Overexpression of the CBF2 transcriptional activator in Arabidopsis counteracts hormone activation of leaf senescence. Plant Signal. Behav. 5 296–299. 10.4161/psb.5.3.10739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni A. M., Nuruzzaman M., Satoh K., Shimizu T., Kondoh H., Sasaya T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52 344–360. 10.1093/pcp/pcq196 [DOI] [PubMed] [Google Scholar]
- Shi Q., Zhang H., Song X., Jiang Y., Liang R., Li G. (2017a). Functional characterization of the maize phytochrome-interacting factors PIF4 and PIF5. Front. Plant Sci. 8:2273. 10.3389/fpls.2017.02273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Huang J., Sun T., Wang X., Zhu C., Ai Y., Gu H. (2017b). The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 59 118–133. 10.1111/jipb.12515 [DOI] [PubMed] [Google Scholar]
- Smirnoff N., Bryant J. A. (1999). DREB takes the stress out of growing up. Nat. Biotechnol. 17 229–230. 10.1038/6968 [DOI] [PubMed] [Google Scholar]
- Solanke A. U., Sharma A. K. (2008). Signal transduction during cold stress in plants. Physiol. Mol. Biol. Plants 14 69–79. 10.1007/s12298-008-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Wang J., Liu B., Qi T., Xie D. (2017). MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses. Plant Cell Physiol. 58 1752–1763. 10.1093/pcp/pcx112 [DOI] [PubMed] [Google Scholar]
- Song Y. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4 and 5. Mol. plant. 7 1776–1787. 10.1093/mp/ssu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yang C., Gao S., Zhang W., Li L., Kuai B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7 1776–1787. 10.1093/mp/ssu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Yu J. P., Chen F., Zhao T. J., Fang X. H., Li Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283 6261–6271. 10.1074/jbc.m706800200 [DOI] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Takahashi F., Fujita M., Yoshida T., Nakashima K., et al. (2015). SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84 1114–1123. 10.1111/tpj.13067 [DOI] [PubMed] [Google Scholar]
- Thines B., Parlan E. V., Fulton E. C. (2019). Circadian network interactions with jasmonate signaling and defense. Plants 8:252. 10.3390/plants8080252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T., Kato W., Asada Y., Sako K., Sato T., Sonoda Y., et al. (2009). DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 122 633–643. 10.1007/s10265-009-0252-6 [DOI] [PubMed] [Google Scholar]
- Vainonen J. P., Jaspers P., Wrzaczek M., Lamminmaki A., Reddy R. A., Vaahtera L., et al. (2012). RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442 573–581. 10.1042/BJ20111739 [DOI] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67 589–602. 10.1007/s11103-008-9340-6 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lin A., Loake G. J., Chu C. (2013). H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. J. Integr. Plant Biol. 55 202–208. 10.1111/jipb.12032 [DOI] [PubMed] [Google Scholar]
- Wang Y., Reiter R. J., Chan Z. (2018). Phytomelatonin: a universal abiotic stress regulator. J. Exp. Bot. 69 963–974. 10.1093/jxb/erx473 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100 681–697. 10.1093/aob/mcm079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. M., Amasino R. M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127 876–886. 10.1104/pp.010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. R., Chung K. M., Park J.-H., Oh S. A., Ahn T., Hong S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790. 10.1105/tpc.13.8.1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. R., Kim H. J., Lim P. O., Nam H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70 347–376. 10.1146/annurev-arplant-050718-095859 [DOI] [PubMed] [Google Scholar]
- Wu K., Zhang L., Zhou C., Yu C. W., Chaikam V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59 225–234. 10.1093/jxb/erm300 [DOI] [PubMed] [Google Scholar]
- Wu X. Y., Kuai B. K., Jia J. Z., Jing H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54 936–952. 10.1111/jipb.12005 [DOI] [PubMed] [Google Scholar]
- Xu H., Wang X., Chen J. (2010). Overexpression of the Rap2.4f transcriptional factor in Arabidopsis promotes leaf senescence. Sci. China Life Sci. 53 1221–1226. 10.1007/s11427-010-4068-3 [DOI] [PubMed] [Google Scholar]
- Yang J., Duan G., Li C., Liu L., Han G., Zhang Y., et al. (2019a). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. 10.3389/fpls.2019.01349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu Y., Yan H., Tian T., You Q., Zhang L., et al. (2018). PlantEAR: functional analysis platform for plant ear motif-containing proteins. Front. Genet. 9:590. 10.3389/fgene.2018.00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. D., Seo P. J., Yoon H. K., Park C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23 2155–2168. 10.1105/tpc.111.084913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Al-Baidhani H. H. J., Harris J., Riboni M., Li Y., Mazonka I., et al. (2019b). DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: impact on plant development, stress tolerance and yield. Plant Biotechnol. J. 18 829–844. 10.1111/pbi.13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhito S., Jinkil J., Min-Young K., Junghyun K., Nam-Chon P., Giltsu C. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- Yeung E., van Veen H., Vashisht D., Sobral Paiva A. L., Hummel M., Rankenberg T., et al. (2018). A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 115 E6085–E6094. 10.1073/pnas.1803841115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolcu S., Li X., Li S., Kim Y. J. (2017). Beyond the genetic code in leaf senescence. J. Exp. Bot. 69 801–810. 10.1093/jxb/erx401 [DOI] [PubMed] [Google Scholar]
- Yoshida S. (2003). Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 6 79–84. 10.1016/s1369526602000092 [DOI] [PubMed] [Google Scholar]
- Yu J., Zhang Y., Di C., Zhang Q., Zhang K., Wang C., et al. (2016). JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 67 751–762. 10.1093/jxb/erv487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou C. (2013). Signal transduction in leaf senescence. Plant Mol. Biol. 82 539–545. 10.1007/s11103-012-9980-4 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhu Z., An F., Hao D., Li P., Song J., et al. (2014). Jasmonate-activated MYC2 represses ethylene insensitive3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26 1105–1117. 10.1105/tpc.113.122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wei H., Li N., Tian W., Chong K., et al. (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11 326–337. 10.1016/j.molp.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Guo Y. (2018). Hormone treatments in studying leaf senescence. Methods Mol. Biol. 1744 125–132. 10.1007/978-1-4939-7672-0_11 [DOI] [PubMed] [Google Scholar]
- Zhou M. Q., Shen C., Wu L. H., Tang K. X., Lin J. (2011a). CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 31 186–192. 10.3109/07388551.2010.505910 [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. (2011b). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31 303–313. 10.1007/s10059-011-0047-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Chen J., Xie Z., Gao J., Ren G., Gao S., et al. (2015). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84 597–610. 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
- Zhu Z. (2014). Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot. 65 5743–5748. 10.1093/jxb/eru349 [DOI] [PubMed] [Google Scholar]
- Zhu Z., An F., Feng Y., Li P., Xue L., Mu A., Jiang Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 12539–12544. 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.