Abstract
For oral, oropharyngeal and oesophageal cancer, the early detection of tumours and of residual tumour after surgery are prognostic factors of recurrence rates and patient survival. Here, we report the validation, in animal models and a human, of the use of a previously described fluorescently labelled small-molecule inhibitor of the DNA-repair enzyme poly(ADP-ribose) polymerase 1 (PARP1) for the detection of cancers of the oral cavity, pharynx and oesophagus. We show that the fluorescent contrast agent can be used to quantify the expression levels of PARP1, and to detect oral, oropharyngeal and oesophageal tumours in mice, pigs and fresh human biospecimens when delivered topically or intravenously. The fluorescent PARP1 inhibitor can also detect oral carcinoma in a patient when applied as a mouthwash, and discriminate, between fresh biopsied samples of the oral tumour and the surgical resection margin with larger than 95% sensitivity and specificity. The PARP1 inhibitor could serve as the basis of a fast and sensitive assay for the early detection, and for the surgical-margin assessment, of epithelial cancers of the upper intestinal tract.
Cancers that originate in the epithelium of exposed or accessible body areas, such as oral and oropharyngeal cancers as well as oesophageal cancer, should be easy to screen for by monitoring and identifying visible changes in the mucosal lining. However, cancer registry data reveal that only 29% of oral and oropharyngeal cancers and 19% of oesophageal cancers are detected at early stages. 5-year-survival rates steeply drop with detection at advanced tumour stages from 83.7% (oral and oropharyngeal cancer) and 45.2% (oesophageal cancer) for localized disease to 39.1% and 4.8% for metastatic disease, respectively1,2. Clinical outcome is also negatively affected by the presence of residual tumour at the surgical margin3–5. Intuitively, this suggests that the current clinical practice of visual examination in combination with biopsy-based histopathological diagnosis is insufficient to improve early diagnosis and surgical outcome.
For initial diagnosis, identifying early-stage morphological changes and distinguishing among a variety of inflammatory and benign conditions is difficult. During surgical interventions, success is typically dependent on the absence of positive margins and micro-metastasis. Current clinical practice, e.g., for oral cancer, calls for surgical excision with at least 5 mm margins of normal tissue on histopathologic examination to reduce the risk of residual microscopic disease. Wide margins, however, can lead to additional morbidity and significant, irreversible impairment of phonation, mastication, gustation, and/or swallowing. Intraoperatively, frozen section histopathology is often used to evaluate the presence of tumour cells at the surgical margins. However, this approach leads to significant time delays during surgery and has less accuracy than the gold standard permanent histopathology due to tissue artefacts caused by the freezing process. Furthermore, action is delayed several days until the definitive histopathological diagnosis is available, often leading to secondary surgeries and/or adjuvant treatment with radiation and chemotherapy, which are all associated with compounded morbidity, further cost, and poorer oncologic outcomes.
Enhancing the visibility of malignant lesions in relation to normal tissues contemporaneously—i.e., adding contrast—therefore holds immense potential to address the above-mentioned challenges associated with diagnosis and intraoperative imaging. In pursuit of this goal, a host of methods have been explored as clinical diagnostic adjuncts, including tissue staining with vital dyes (e.g., Toluidine Blue, Methylene Blue, or Lugol’s Iodine), chemiluminescence imaging following an acetic acid wash, and optical-based imaging techniques, which rely on autofluorescence6–10. To date, these methods have shown limited sensitivity and/or specificity and are generally not recommended11.
A new class of emerging diagnostic adjuncts focuses on in vivo microscopic (IVM) techniques. These include optical coherence tomography (OCT), reflectance confocal microscopy (RCM), and high-resolution microendoscopy, which aim to noninvasively visualize the features typically assessed via histopathology during the examination. Currently, IVM techniques for epithelial cancers are in early-stage development and their diagnostic accuracy remains to be determined. Thus, existing approaches possess limitations that have prevented their clinical adoption. Those limitations are i) absence of a tumour-specific molecular target, ii) confinement to either microscopic or macroscopic resolution iii) diagnostic accuracy that is limited by either low sensitivity or low specificity6,7,11–13. In contrast, employing the optically active, molecularly targeted approach presented in this work could not only improve sensitivity and specificity of tumour recognition, but also allow for imaging on both the cellular and macroscopic level, which, if combined, could increase the breadth and depth of the molecular information used for early detection and intraoperative margin delineation.
The molecular target for our optical imaging approach is the DNA repair enzyme Poly(ADP-ribose)Polymerase 1 (PARP1). In addition to its many functions in cell cycle regulation and transcription, high levels of PARP1 expression have been observed in many different tumour types, indicating its potential for diagnostic applications14–20. Higher expression has moreover been linked to worse prognosis21–24. While PARP1 also appears in the nuclei of normal, proliferating cells, expression levels and especially cellularity tend to be lower than in rapidly growing tumours, supporting the rationale that high-contrast imaging can be achieved with PARP-targeted imaging approaches.
The optical PARP1-targeted imaging agent PARPi-FL is a fluorescently labelled analogue of the FDA-approved PARP inhibitor olaparib with high affinity and specificity for PARP125. PARPi-FL is a cell penetrating imaging agent that rapidly clears in the absence of target binding. Importantly, compared to other nuclear staining vital dyes, PARPi-FL does not intercalate into the DNA, but instead reversibly binds to PARP1, which locates to DNA strand breaks, and is not mutagenic, enabling in vivo applications in addition to ex vivo applications. The efficacy of high-contrast xenograft imaging with PARPi-FL after intravenous application has been shown on the whole body as well as at the cellular level25,26. Importantly, the tissue penetrating nature of PARPi-FL enables its use as both a systemic and topical agent. Topical application, as an alternative to intravenous injection, enables the application of microdoses of PARPi-FL and allows for almost immediate imaging27. However, this concept has neither been tested systematically nor was it tested in humans. The versatile features of PARPi-FL make it a tool for use in early detection as well as surgical margin assignment applications, both of which could strongly benefit from a diagnostic adjunct.
In this study, we aimed to characterize the potential of PARP1 expression as a quantitative biomarker for early detection and intraoperative delineation in human biospecimens and to subsequently establish PARPi-FL-based imaging strategies using human tissues and preclinical models. Due to differences in anatomical accessibility of oesophageal, oropharyngeal, and oral cancer, we did not seek to create a one-size-fits-all optical imaging approach, but rather pursued individually adapted PARPi-FL imaging approaches to better inform clinical development.
In oesophageal cancer, we validated PARPi-FL as a quantitative marker for PARP1 expression levels and showed that it could be applied topically to the oesophagus. In oropharyngeal cancer, we tested whether combining brush biopsy (a less invasive approach to obtaining diagnostic tissue) with PARPi-FL staining is a feasible approach to identifying tumour cells in a complex tissue sample. In oral cancer, we developed a staining and imaging method for freshly excised biopsies that allows for the identification of positive margins within minutes while preserving the fresh tissue for other downstream applications. We then tested the diagnostic accuracy of this detection method in human biopsies and evaluated its translational potential. In additional support of the translational potential of PARPi-FL, we report a first-in-human study of in vivo imaging after its topical application (NCT03085147). Finally, we explore the potential to expand PARPi-FL imaging to in situ surgical guidance applications with a large surgical window after intravenous delivery in a large animal. The presented studies are consistent in demonstrating the clinical relevance of PARPi-FL-based optical imaging methods for the early detection and intraoperative delineation of cancers that develop at the epithelial surface. An overview of all conducted studies can be found in Fig. 1.
Fig. 1 |. Study overview.
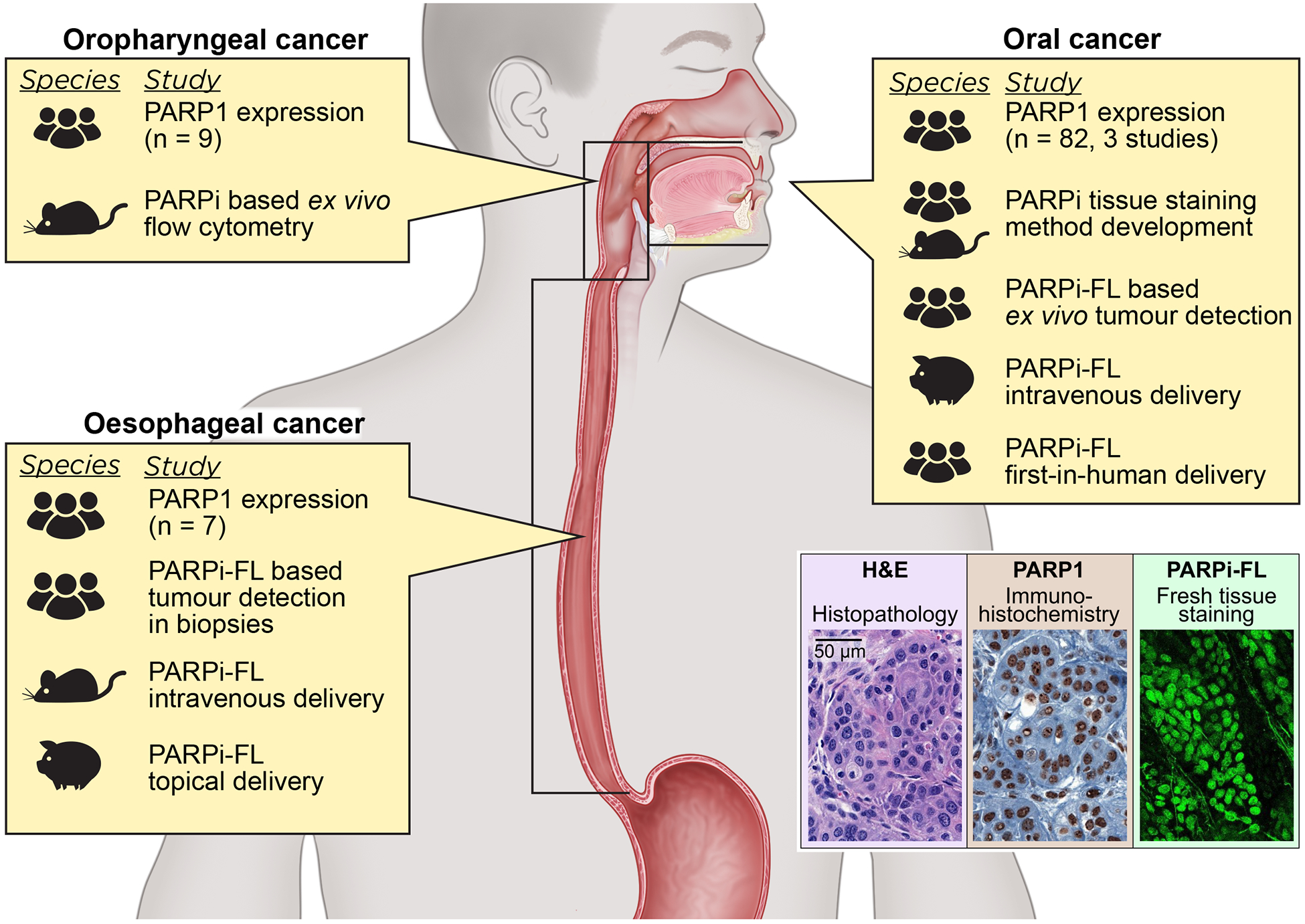
We analysed PARP1 expression via IHC staining in oesophageal, oropharyngeal, and oral cancer samples and carried out different PARP1 imaging studies. The figure illustrates the type of imaging-based study and species used for each tumour type.
Results
PARP1 biomarker validation and imaging in oesophageal cancer.
To explore and confirm the clinical value of a PARP1-targeted imaging approach in oesophageal cancer, we quantified PARP1 expression in human surgical biospecimens (n=7). In all specimens, three regions of interest were analysed: tumour, normal epithelium, and deep margin (comprising submucosa and muscle tissue). We found strongly increased PARP1 expression in tumour compared to deep margin as well as epithelium (Fig. 2a, fig. S1a). Quantification, measured as PARP1-positive area (details in Materials and Methods and fig. S2), revealed a mean value of 26.4% ± 10.8% PARP1-positive area in the tumour, which was significantly higher than the epithelium (5.0% ± 1.9%, p=0.03, Wilcoxon test) and the deep margin (0.9% ± 0.5%, p=0.02, Wilcoxon test) (Fig. 2b and fig. S1, b and c).
Fig. 2 |. PARP1 expression and imaging in oesophageal cancer.
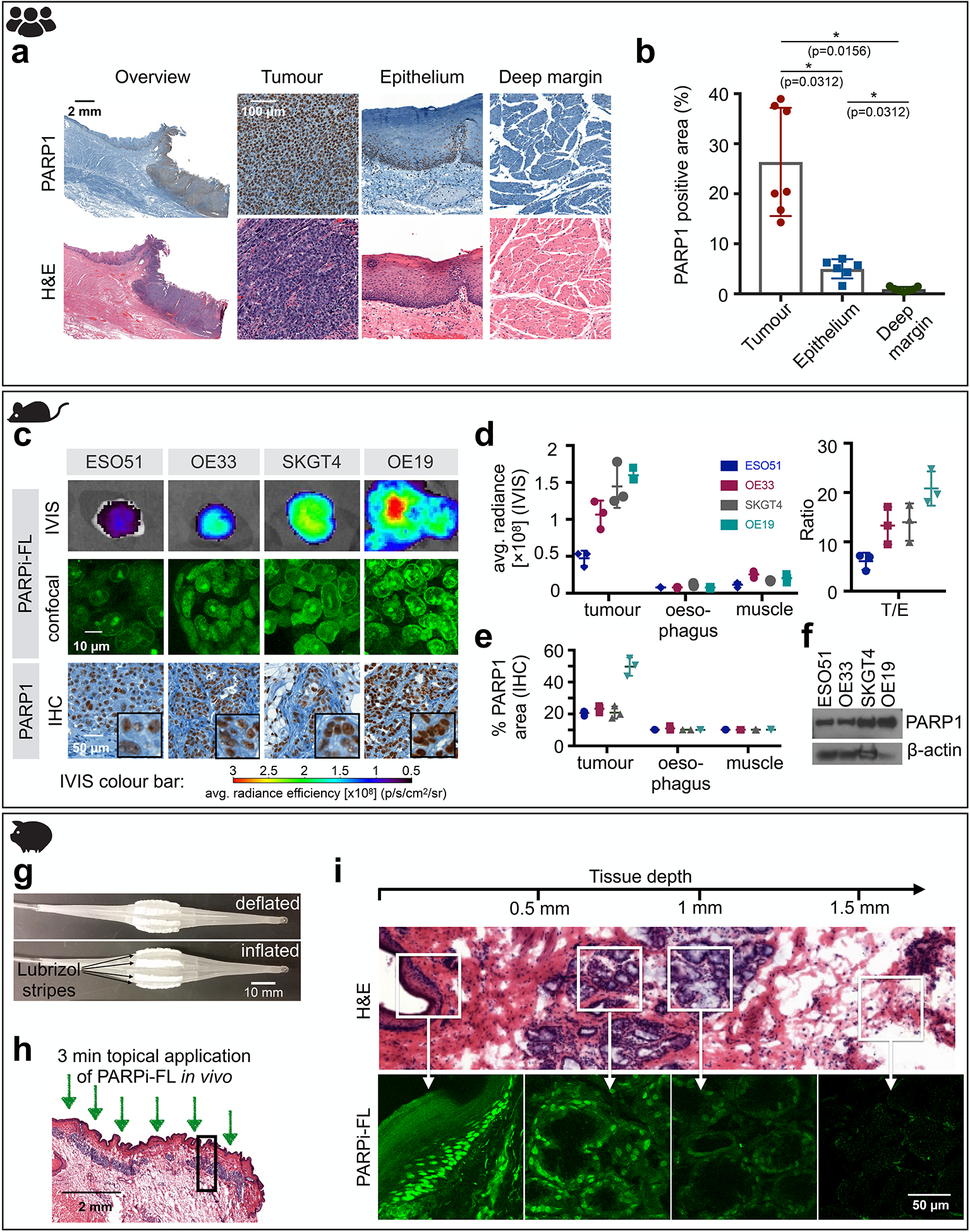
a, Representative PARP1 IHC and H&E histology obtained from human biospecimens (H&E and IHC of all samples (n=7) are included in fig. S1).b, Quantification of PARP1 expression in IHC samples (n=7 patients). Statistical significance was determined using the Wilcoxon matched pairs signed rank test (two-tailed, n=6 pairs). Significance levels: * p<0.05 c, Fluorescence imaging of EAC in xenograft mouse models (n=3 animals/group) after intravenous injection of 75 nmol PARPi-FL. Excised tumours were imaged using epifluorescence (IVIS) and confocal microscopy. PARP1 expression was confirmed via IHC staining. d, Quantification of the fluorescence signal 90 min post-injection of PARPi-FL and tumour/oesophagus (T/E) contrast ratios. e, Quantification of PARP1 expression of tumours, oesophagus, and muscle assessed by PARP1 IHC. f, Quantification of PARP1 expression in EAC cell lines via Western blot. Full Blots available in fig. S3b g, Inflatable balloon applicator used to topically apply PARPi-FL to a Yucatan mini swine oesophagus. h, 2 mL of a 1000 nM PARPi-FL solution were loaded into the balloon applicator and topically applied for 3 min onto the oesophagus of the anaesthetized mini swine. i, Penetration of topically applied PARPi-FL was visualized in perpendicular cryosections of the oesophagus using a confocal microscope. H&E staining on adjacent sections was used for anatomical reference. Data in b, d, and e, are presented as mean ± standard deviation.
PARP1-targeted optical imaging in oesophageal cancer.
The ability of PARPi-FL to identify high and low PARP1-expressing tumours was tested in four mouse xenograft models (ESO51, OE33, SKGT3, OE19) of oesophageal adenocarcinoma (EAC). Macroscopic epifluorescence imaging of the excised tumours 90 min after injection revealed increasing fluorescence intensities (ESO51<OE33<SKGT4<OE19). This was corroborated via confocal microscopy, where the nuclear accumulation of PARPi-FL could be clearly seen (Fig. 2c) and increasing PARPi-FL uptake correlated with cellular PARP1 expression (Fig. 2c). Quantification of the fluorescence intensity showed a three-fold increase from the lowest (ESO51) to highest (OE19) PARP1-expressing xenograft model, while fluorescence intensities in normal oesophagus and muscle tissue were low and comparable in all animals (Fig. 2d). Tumour-to-oesophagus ratios were 6.1 ± 1.7 (ESO51) to 20.8 ± 3.5 (OE19), providing adequate contrast to discriminate tumour from normal oesophagus tissue in both high and low PARP1-expressing tumours. PARP1 expression levels were quantified via PARP1 IHC (Fig. 2e) and Western Blot (Fig. 2f). In addition, uptake of the radioactive, fluorine-labelled PARPi [18F]PARPi further corroborated that PARPi-FL uptake was quantitative, showing a similar pattern of in vivo tumour uptake and high tumour-to-muscle ratios (fig. S3).
Validation of topical administration route for PARPi-FL in the oesophagus.
Anatomically, access to the oesophagus is limited and therefore requires an alternative topical application strategy. Here, we explored the use of a balloon applicator, soaked with 2 mL of a 1 μM PARPi-FL solution with sponge-like Lubrizol stripes (Fig. 2g) in a pig model and measured penetration depth of PARPi-FL in the oesophagus. After insertion into the pig’s oesophagus in a deflated state, the balloon was inflated to expose the Lubrizol stripes and induce release of PARPi-FL onto the oesophagus wall of the anesthetized pig for a duration of 3 min; 10 min after the application, the pig was sacrificed, followed by necropsy and cryopreservation of the oesophagus (Fig. 2h). On fresh cryosections, without further staining, we detected nuclear PARPi-FL staining in the basal layer of the epithelium (100–200 μm depths) and mucus glands in the submucosa, up to 800 μm below the tissue surface (Fig. 2i).
PARP1 expression in oropharyngeal cancer.
Expanding on whether PARP1 could be a relevant general biomarker to tumours of the oral cavity, we investigated PARP1 expression in a clinical data set of oropharyngeal cancer, which arises at the base of the tongue, tonsils, soft palate, or pharynx wall (n=9). All biospecimens contained invasive carcinomas with high levels of PARP1 expression (Fig. 3a and fig. S4a). In the normal epithelium, PARP1 expression was found in the basal layer and at very low levels in the deep margin (Fig. 3a and fig. S4a). Quantification of the PARP1-positive area confirmed significantly higher expression levels in the tumour (46.5% ± 16.7% PARP1-positive area) than the epithelium (9.4% ± 4.9%; p=0.02, Wilcoxon test) and deep margin (2.0% ± 1.1%; p<0.01, Wilcoxon test) (Fig. 3b and fig. S4b).
Fig. 3 |. PARP1 expression and PARP cytometry in oropharyngeal cancer.
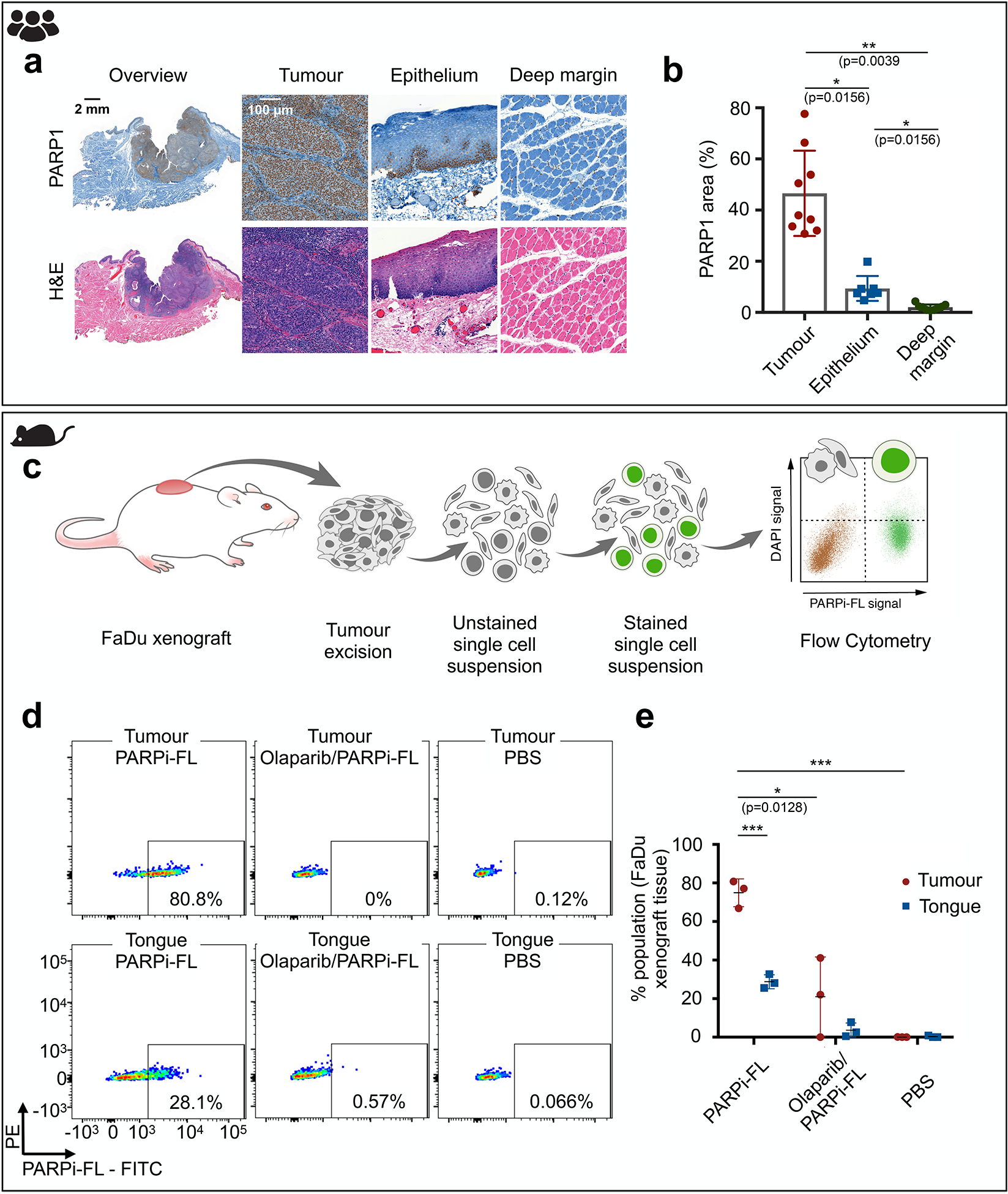
a, Representative PARP1 IHC from human biospecimens of oropharyngeal cancer, displaying PARP1 expression in the epithelium, deep margin, and tumour, with corresponding H&E images. (H&E and IHC of all samples (n=9) are included in fig. S1) b, Quantification of PARP1 expression in IHC samples (n=9 patients). Statistical significance was determined using the Wilcoxon matched pairs signed rank test (two-tailed, n=7 pairs). * p<0.05, ** p<0.01 c, Workflow of PARP cytometry. Tissue samples (FaDu xenograft, tongue) were dissociated into single cell suspensions, stained with 100 nM PARPi-FL, and subjected to flow cytometry to determine the percentage of PARPi-FL stained cells in the sample. d, Dot plots of the gated samples showing PARPi-FL-positive cells (FITC channel) vs. live/dead stain (DAPI). Representative dot plot for tumour and tongue stained with 100 nM PARPi-FL or controls, which were used to assess specificity of the PARPi-FL binding (Olaparib/PARPi-FL) and specificity of the signal (PBS). e, Quantification of PARPi-FL flow cytometry (n=3 mice processed in 3 separate experiments). Significance was determined using an unpaired, two-tailed t-test, using the Holm-Sidak method to correct for multiple comparisons. Significance levels: * p<0.05, *** p<0.001. Data in b, and e, are presented as mean ± standard deviation.
A brush biopsy approach to PARP1-targeted optical imaging.
Brush biopsy techniques have been investigated as a less invasive alternative to standard punch and scalpel biopsies. We conducted a feasibility study to determine if brush biopsies in combination with PARPi-FL staining could enable rapid detection of tumour cells in cell suspensions derived from solid tissues. In a proof-of-principle approach, we developed an ex vivo staining protocol and flow cytometric analysis of single-cell suspensions derived from pharynx cancer xenografts (FaDu cells) and healthy mouse tongues (Fig. 3c, see fig. S5a for gating procedures). We found a significantly higher number of PARPi-FL-positive cells in tumour-derived suspensions than in tongue-derived suspensions (Fig. 3, d and e; p<0.001, unpaired t-test). Specificity of PARPi-FL staining in cell suspensions was confirmed by blocking PARPi-FL binding sites with the PARP inhibitor olaparib, which reduced the percentage of positive cells from 74.9% ± 7.2% to 21.0% ± 20.0% (p<0.05, unpaired t-test using the Holm-Sidak method without assuming a consistent standard deviation (SD)). A PBS-stained control group showed no PARPi-FL-positive cells. In addition, we were able to confirm these results in oesophageal cancer cells (OE19) (fig. S5b). Thus, this proof-of-principle experiment showed that PARPi-FL staining of a cell suspension derived from a complex tissue can help to distinguish between tumour cell containing and normal samples, encouraging further investigation into combining PARPi-FL staining with brush biopsy approaches.
PARP1 expression during malignant transition and at tumour margins in oral cancer.
In oral cancer, we confirmed the gradual increase of PARP1 expression during malignant transition. In healthy epithelial tissues, physiological PARP1 expression was localized predominantly to the basal layer of the epithelium. In benign, dysplastic, and malignant biospecimens (n=60), we found an increase in PARP1 expression, particularly for cases with severe dysplasia (=carcinoma in situ) and in invasive tumours, which showed significantly higher PARP1-positive area compared to benign cases, mild dysplasia, and moderate dysplasia (Fig. 4a, Fig. 4b; p<0.05, Mann-Whitney test). Combining cases that are predominantly treated non-surgically (benign, mild dysplasia) and cases that require surgical resection with adjuvant radiation/chemotherapy (severe dysplasia/tumours), we found a pronounced, statistically significant difference in PARP1 expression (5.6% ± 2.6% vs. 13.1% ± 6.8%, p<0.001, Mann-Whitney test) (Fig. 4C). We determined a sensitivity of 81% and specificity of 83% at the optimal cut-off of 8.1% PARP1-positive area for separation of these two groups, with an area under the curve (AUC) of 0.908 (Fig. 4d). We confirmed these results using a manual scoring method, based on the percentage of PARP1-positive cells and staining intensity, which yielded comparable results (fig. S6a, fig. S6b).
Fig. 4 |. PARP1 expression during malignant transition and at tumour margins in oral cancer.
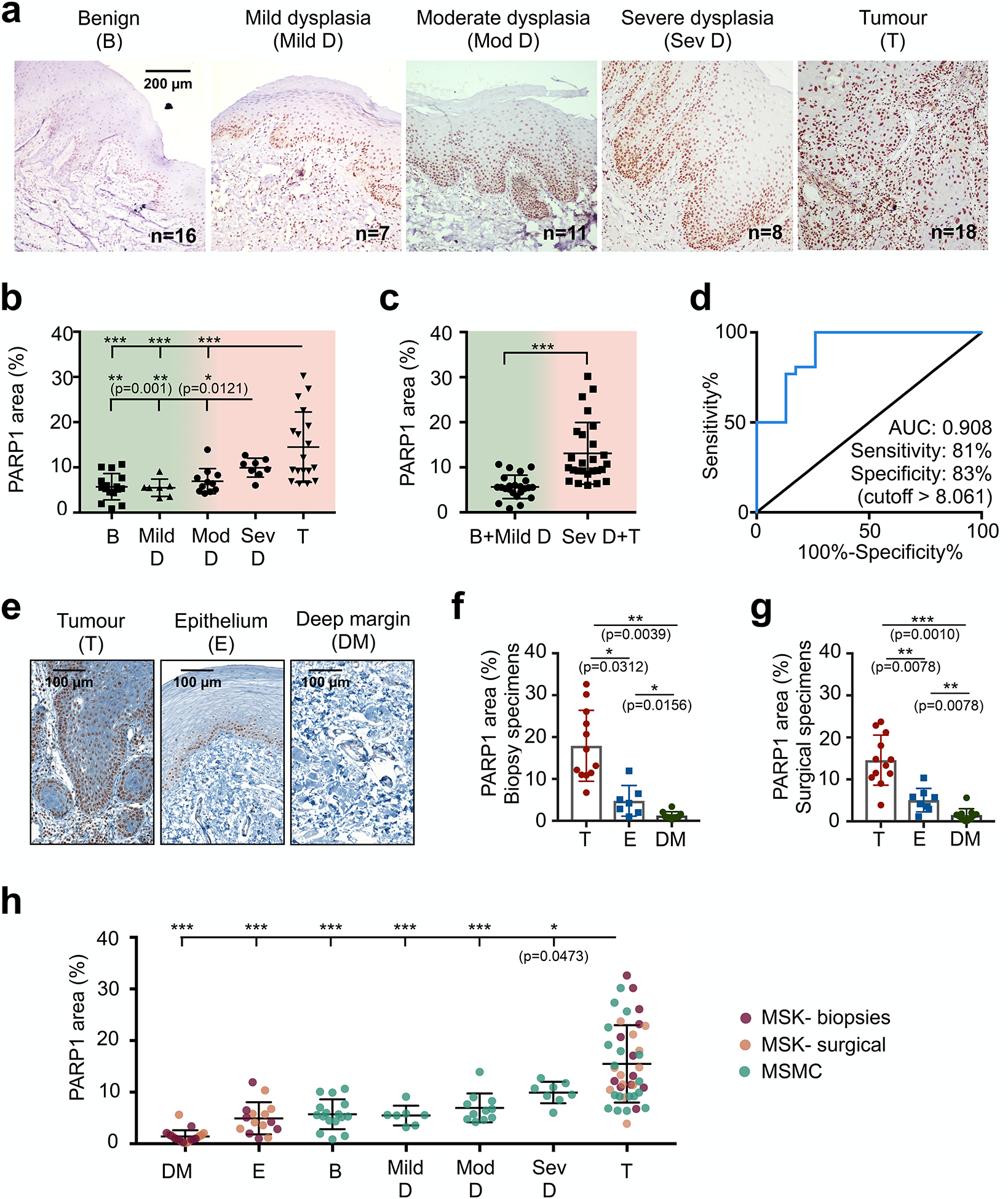
a, Representative PARP1 IHC images in different stages of oral cancer progression (benign (n=16), mild dysplasia (n=7), moderate dysplasia (n=11), severe dysplasia (n=8), malignant(n=18)) of patient biospecimens from Mazumdar Shaw Medical Center (MSMC). b, Quantification of PARP1 expression in IHC samples (n=60 patients). The PARP1-positive area (in relation to the entire tissue area) was quantified in high-resolution images of the epithelium or tumour area (mean of n ≥ 4 images per case). Statistical significance was determined using an unpaired, two-tailed Mann-Whitney rank sum test. c, Comparison of PARP1 expression in low-grade, potentially malignant lesions (benign vs. mild dysplasia) vs. severe dysplasia (carcinoma in situ) and malignant cases. Statistical significance was determined using an unpaired, two-tailed Mann-Whitney rank sum test. ***p<0.001 d, ROC curve for the data represented in c. e, Representative images of PARP1 expression in samples from two populations: presurgical biopsies of tumour and benign tissues (divided in two areas: epithelium and deep margin) and surgical specimens, which included tumour, adjacent benign epithelium, and deep margin on each specimen. f, Quantification of PARP1 expression in IHC samples of biopsy specimens (n=12 patients). Statistical significance was determined using the Wilcoxon matched pairs signed rank test (two-tailed, n=6 pairs). g, Quantification of PARP1 expression in IHC samples of surgical specimens (n=12 patients). Statistical significance was determined using the Wilcoxon matched pairs signed rank test (two-tailed, n=8 pairs). A different analysis of the same dataset has been published previously26. h, Pooling of all three datasets of oral biospecimens investigated for PARP1 expression via IHC. Statistical significance was determined using the unpaired, two-tailed Mann-Whitney rank sum test. Full statistical analysis is reported in Table S1. T-Tumour.. E-Epithelium, DM-deep margin, D-dysplasia, mod-moderate, sev-severe, b-benign. Significance levels: *p<0.05, **p<0.01, ***p<0.001. Data are presented as mean ± standard deviation.
We further analysed tumour margins to gain insight into the clinical potential of PARP1-based optical imaging for early detection and intraoperative guidance. In presurgical biopsies (tumour: n=12, benign tissues: n=10 subdivided into epithelium and deep margin from n=12 patients) and surgical specimens (n=12), we quantified the difference in PARP1 expression between the tumour area, epithelium, and deep margin (Fig. 4e). PARP1 staining was significantly higher in the tumour area of both biopsies (17.9% ± 8.5% PARP1-positive area; Fig. 4f) and surgical specimens (14.6% ± 6.0% PARP1-positive area; Fig. 4g), compared to the epithelium (4.8% ± 3.6% and 5.1% ± 2.8%; p=0.03 and p=0.008, respectively, Wilcoxon test) as well as in the deep margin (1.3% ± 0.9% and 1.5% ± 1.5%; p=0.04 and p=0.001, respectively, Wilcoxon test). Importantly, when only looking at paired datasets, PARP1 expression was always higher in the tumour than in the epithelium or deep margin, with no case of overlap, which was also the case in biospecimens of oropharyngeal and oesophageal tumours (fig. S7).
Combining all oral cancer IHC datasets (three datasets, n=84 patients total) confirmed consistently elevated PARP1 expression in tumours and severe dysplasia, whereas a much lower expression was found in the epithelium and deep margin of normal oral tissue and benign and early dysplastic cases (Fig. 4h). Analyses of statistically significant differences in this dataset (Mann-Whitney test, Fig. 4h and Table S1) support PARP1 expression as viable biomarker for both early detection and intraoperative margin delineation of oral cancer. Overall, PARP1 levels increased significantly with disease stage, from a median of 1.0% in deep margin to 13.2% in tumour (Kendall’s tau = 0.67, p<0.0001).
Rapid, tissue-preserving PARPi-FL staining of fresh biopsies.
Motivated by the abrupt drop of PARP1 expression in benign tissues adjacent to the tumour, we wanted to determine whether PARPi-FL uptake can serve as a diagnostic tool in fresh biopsy tissue. We aimed to develop a protocol that does not interfere with tissue integrity or morphology (e.g., by freezing or fixing) and can produce results faster than standard frozen section histopathology. The staining protocol was optimized toward high nuclear staining intensity and low cytosolic and non-specific background staining on freshly excised xenograft tissue (fig. S8). The selected protocol consisted of 5 min PARPi-FL staining followed by 10 min of washing, and was translated to fresh human biopsy samples. A similar protocol can be used for non-fixed cryosections (fig. S9). Staining specificity was confirmed by PARP1 IHC and H&E staining (Fig. 5a).
Fig. 5 |. Rapid PARPi-FL staining on fresh biospecimens.
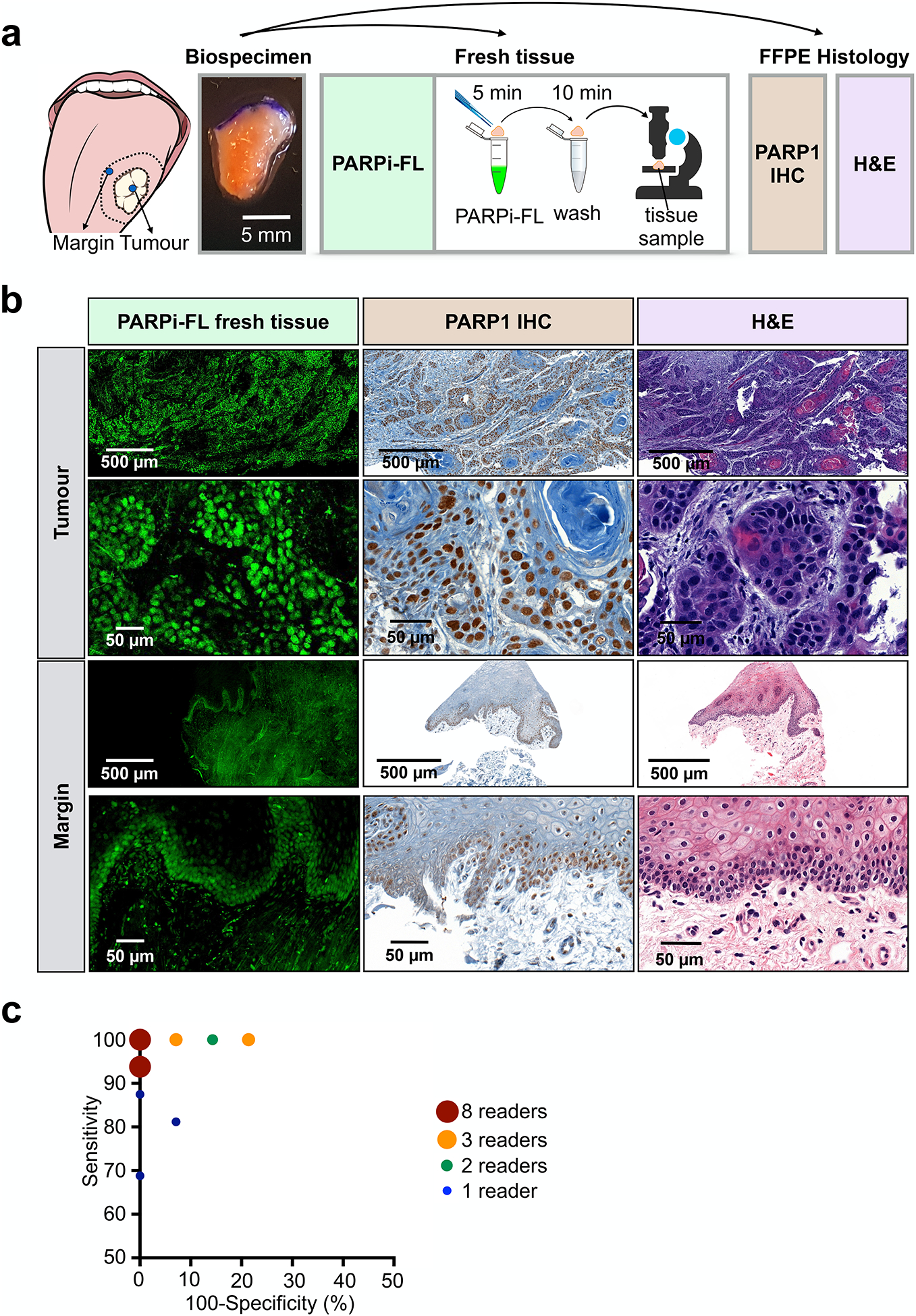
a, Workflow of rapid PARPi-FL staining of fresh human biospecimens. Biospecimens were split and separately processed for fresh tissue staining as well as histopathology. b, Examples of PARPi-FL staining of a tumour and margin sample and corresponding histopathology of the same sample. We aimed to scan the entire fresh tissue in a high-resolution tile scan. Lower and higher magnification images showcase PARPi-FL staining and corresponding PARP1 expression in the specimens. A total of 22 tissues (n=12 tumours and n=10 benign tissues adjacent to tumour from 13 individual patients) were stained and analysed. The full data set is provided in supplementary Materials PDF files 1 and 2. c, In a blinded study, readers (n=27) scored 30 cases (n=12 tumours, n=10 margins, n=8 duplicates (4 tumours, 4 margins)) as tumour or margins (see fig. S9 for study design). Pairs of sensitivity and specificity for each reader are represented in the graph.
Confocal microscopy images of tumour samples showed abundant nuclear PARPi-FL in PARP1-expressing tumour cells (Fig. 5b). In benign samples, nuclear PARPi-FL staining was confined to the thin PARP1-expressing basal layer of the epithelium (Fig. 5b). Endogenous collagen-related autofluorescence, which was observed in the submucosa of the clinical samples, was discriminated from the exogenous PARPi-FL signal by two-channel imaging at 488 nm and 543 nm (fig. S10a). Confocal images were acquired from fresh biopsies of 12 patients and compiled into a study set of 30 cases (n=12 tumours, n=10 benign tissues adjacent to tumour, n=8 duplicates (4 tumours, 4 benign tissues adjacent to tumour)).
The images were read by volunteers (n=27) in a blinded study; each specimen was scored as tumour or benign tissue adjacent to tumour (see fig. S10b for study design and the Supplementary files for the blinded study training dataset and test dataset). Of a total of 810 ratings, 361/378 negative margins and 414/432 tumours were assigned correctly (fig. S10, c and d). Pairs of sensitivity and specificity for each reader are reported in Fig. 5c. We found an overall sensitivity of 95.8%, an average specificity of 95.5%, a positive predictive value of 96.1% and a negative predictive value of 95.3% (Table 1). Overall intrareader agreement was 97.2% (Table 1 and fig. S10e). We confirmed the non-destructive nature of the staining to biopsy tissue by carrying out H&E histopathology following PARPi-FL staining and imaging and found no evidence of perturbed tissue quality (fig. S11). Thus, PARPi-FL allowed for rapid identification of tumour cells in fresh biopsies with high sensitivity and specificity, and without impeding the use of the sample for regular processing afterwards.
Table 1 |.
Rapid PARPi-FL staining on fresh biospecimens-blinded study analysis.
| Overall (%) | 95% Confidence Interval (CI) (%) | |
|---|---|---|
| Sensitivity | 95.8 | 93.5 – 97.4 |
| Specificity | 95.5 | 92.9 – 97.2 |
| Positive predictive value (PPV) | 96.1 | 93.7 – 97.5 |
| Negative predictive value (NPV) | 95.3 | 92.6 – 97.0 |
| Correctly assigned cases | 95.7 | 94.0 – 96.9 |
| Overall intrareader agreement | 97.2 | 91.4 – 99.1 |
| Tumour agreement | 94.5 | 83.4 – 98.3 |
| Margin agreement | 94.7 | 84.7 – 98.3 |
The purpose of the presented dataset was to validate our PARPi-FL staining method and identify if samples containing tumour (in the clinical setting: positive margins) can be reliably distinguished from samples not containing tumour (negative margins). Most importantly, considering an intraoperative setting, our data also suggests that focal tumour invasion into a suspected negative margin can be clearly identified and demarcated from surrounding normal tissue (fig. S12).
Admittedly, the rate-limiting step of this technology would be image acquisition with a state-of-the-art confocal microscope. Reasoning that a quicker, more user-friendly readout would be preferable, we tested a confocal scanner, which is designed to rapidly analyse fresh tissue specimens by creating a high-resolution strip-mosaic (Vivascope 2500, Caliber ID, Andover, MA). We stained fresh FaDu xenograft tissue with PARPi-FL, placed the tissue on the scanner, and scanned the field of view of 15×15 mm in about 150 s. The high-resolution images allowed for clear identification of PARPi-FL-stained nuclei (fig. S13a). This was confirmed using human biospecimens (fig. S13b, compare with Fig. 5b for regular confocal microscopy). The Vivascope 2500 could be used as a back-table instrument in a surgical suite, allowing for ad hoc PARPi-FL images of freshly excised tissues, before tissue is passed on for other in-depth analyses such as cryosections, IHC, or sequencing.
We also explored the feasibility of macroscopic and microscopic in vivo imaging. Here, we validated an in vivo imaging device, the ViewnVivo (Optiscan, Mulgrave, Australia), a miniaturized handheld confocal endomicroscope with an adjustable focal plane (Fig. 6a). The device is equipped with a 488 nm laser—the optimal wavelength for PARPi-FL detection. The choice of different filters allows for discrimination between PARPi-FL fluorescence and autofluorescence. The z-mechanism was able to detect PARPi-FL at up to 50 μm tissue depths (fig. S14, a and b). Images of the same human biospecimens as presented in fig. S13B revealed comparable image quality in tumour and margin samples (Fig. 6b). Intuitively, the ViewnVivo has the potential to be used for intraoperative, PARPi-FL-based in vivo imaging without tissue excision. For this specific application – imaging in the surgical cavity – intravenous instead of topical delivery would be required, since distribution of PARPi-FL in the entire tumour mass would enable surgical guidance across the whole surgical bed. PARPi-FL has been used intravenously in mouse models before26, but not in large animals. For potential translation to the clinic, key questions in this regard are therefore the necessary injected mass and surgical window for imaging. Intravenous injection of 0.05 mg/kg PARPi-FL in a healthy pig (fig. S15a) resulted in a blood half-life of 1.6 ± 0.48 min and simultaneously revealed that it was possible to detect PARPi-FL specifically in the epithelial basal layer between 5 min and 120 min post injection (fig. S15, b and c). This suggests feasibility of PARPi-FL injection during surgery to provide visual guidance, to identify tumour margins and to detect focal lesions in the surgical cavity.
Fig. 6 |. Feasibility of microscopic and macroscopic in-human imaging of PARPi-FL.
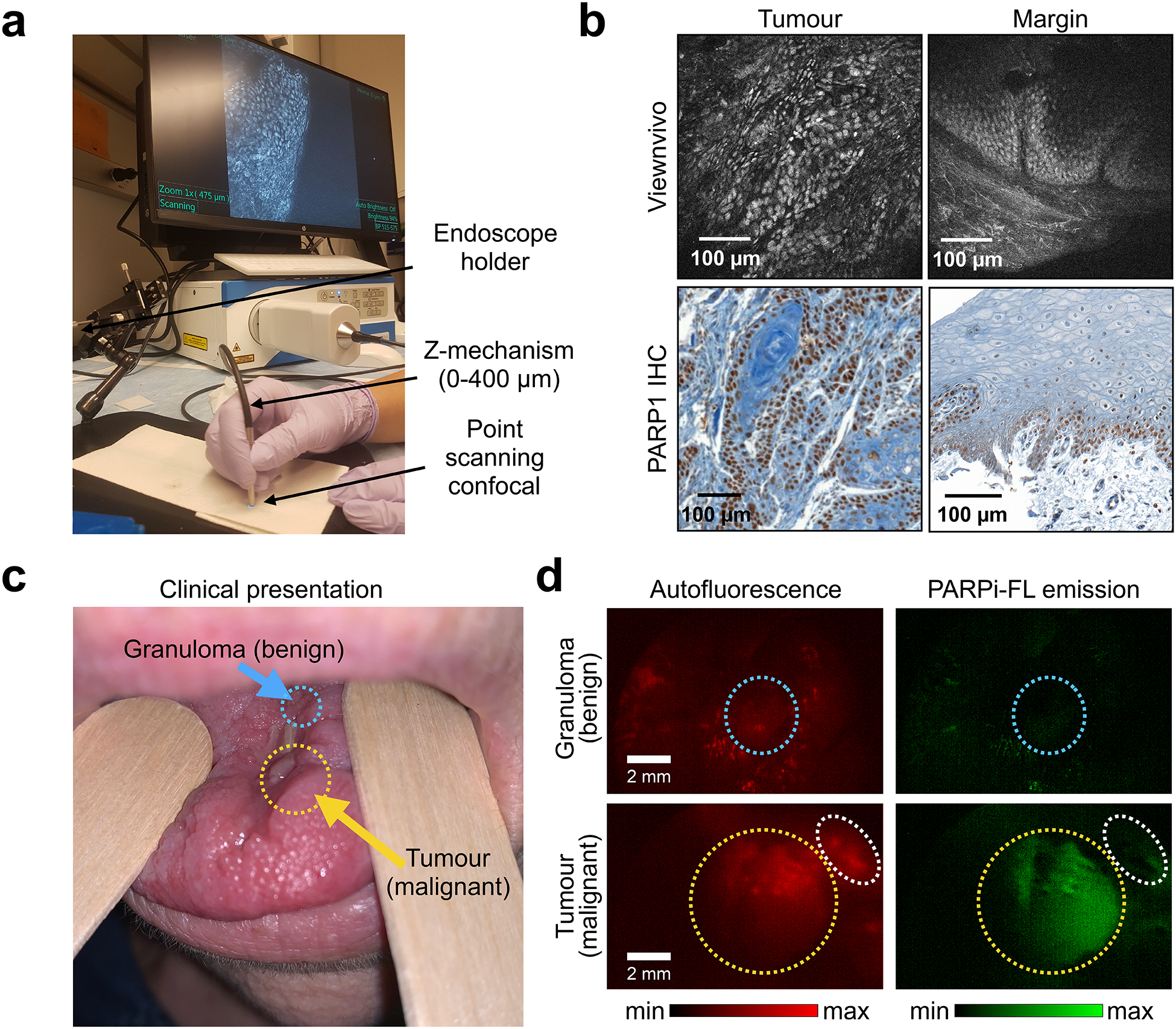
a, Imaging setup for the ViewnVivo point scanning confocal endomicroscope suitable for in vivo imaging. b, Fluorescence images from a patient sample stained with 100 nM PARPi-FL and corresponding PARP1 IHC. c, PARPi-FL first-in-human imaging (NCT03085147), showing a patient who presented with a malignant recurrent squamous cell carcinoma (yellow arrow) and an adjacent benign granuloma (blue arrow). d, Using a Quest Spectrum imaging device with a laparoscopic camera and PARPi-FL optimized laser/filter system, the tumour area of the patient was imaged after gargling a 500 nM PARPi-FL solution. Granuloma (blue circles) and tumour (yellow circles) areas showed distinct patterns of PARPi-FL accumulation, and specific PARPi-FL uptake was only detectable in the tumour. The white circles highlight a non-malignant region which showed autofluorescence, but no PARPi-FL uptake.
PARPi-FL imaging in a human patient.
Lastly, while the first-in-human clinical trial using PARPi-FL (NCT03085147) is ongoing, feasibility of in-human imaging has been shown using a topically applied mouthwash. Fig. 6c shows a patient with a benign granuloma and a recurrent malignant oral squamous cell carcinoma who gargled a 500 nM solution of PARPi-FL for one minute. Using a custom-made laparoscopic imaging system, we did not observe any retention of the imaging agent in the granuloma, whereas the tumour showed specific uptake of PARPi-FL. Autofluorescence was removed by subtracting the system’s red from its green emission channel (Fig. S16). This suggests that PARPi-FL can be used clinically to distinguish benign and malignant lesions, both ex vivo as well as in vivo (Fig. 6d).
Discussion
In this study, supported by both clinical and preclinical specimens, we designed and validated a fluorescence-driven imaging approach for oral, oropharyngeal, and oesophageal cancer. In human biospecimens, and across all three investigated cancer types, we confirm that malignant tissues express significantly higher levels of PARP1 than normal epithelium and submucosal tissue, supporting our central hypothesis that PARP1 will yield high-contrast imaging as a diagnostic and intraoperative biomarker. We experimentally demonstrate how PARPi-FL staining and high-resolution imaging has the potential to improve early diagnosis and surgical removal of tumours. Of particular translational importance, PARPi-FL-based differentiation of tumour and normal tissue in fresh human oral cancer biopsies could be achieved with >95% sensitivity and specificity. PARPi-FL imaging uses fresh tissue for staining and is non-destructive and contemporaneous, all features that promote effortless clinical integration. Intuitively, the absence of terminal processing preserves the tissue for further applications following PARPi-FL imaging, such as histopathological, proteomic, or genomic analyses.
We investigated PARP1 expression in oesophageal cancer, consisting of adeno- and squamous cell carcinomas (stage T1–T3). We found that all tumours had higher levels of PARP1 expression than normal epithelium and submucosal (deep margin) tissue, representing a clear quantitative separation between normal and tumour tissue. Similarly, we found a range of different PARP1 expression levels in our preclinical models, where increasing PARP1 expression resulted in higher PARPi-FL uptake and tumour-to-oesophagus ratios of up to 20. The mouse model with the lowest PARP1 expression had a six-fold higher PARPi-FL signal in the tumour than in the normal oesophagus. These ratios suggest it might be possible to delineate EAC in the oesophagus with high contrast using a non-invasive endoscopic imaging approach.
Considering current clinical practice in EAC, including screening and surveillance protocols in patient populations with an increased risk to develop the disease—specifically in patients with Barrett’s oesophagus (BE)—a non-invasive imaging approach could have a significant impact on screening and early detection. In addition to white light endoscopy, current surveillance protocols require invasive four-quadrant biopsies every 2–5 years. Sampling errors complicate this approach, since progression to cancer usually occurs in only a small fraction of the area presenting Barrett’s mucosa, while excisional quadrant biopsies cover only about 4% of the area affected by BE28. Conceivably, complementing white light endoscopy with PARPi-FL imaging could guide biopsy site selection and enhance non-invasive identification of suspicious areas, including flat, small, and developing superficial lesions.
Existing diagnostic aids, including acetic acid, methylene blue29,30, or Lugol’s iodine31 do not provide a biomarker-specific signal. As their benefit for biopsy site selection is limited, their clinical adoption rate is low. Clinical translation and integration of PARPi-FL imaging into regular white light endoscopy workflows can be optimized by using a topical application approach, as has been suggested for oral cancer imaging26. To reach the oesophagus, we tested an alternative route of administration using a balloon applicator. Recently, balloon-based devices have been introduced to retrieve cells32 or DNA33 from the oesophagus in order to detect BE and EAC. In our case, we deployed a balloon-based applicator with highly absorbent Lubrizol stripes, which are collapsed inside of folds when the balloon is deflated but exposed and pressed onto the oesophageal surface upon inflation. In a healthy pig, we were able to show that, within three minutes, PARPi-FL penetrated up to 800 μm deep into the tissue and bound to mucus glands that showed low levels of physiological PARP1 expression. These results corroborate the small molecule’s ability to penetrate well beyond the epithelial layers where EAC originates.
Novel methods for early detection, surveillance, and surgical guidance are also urgently needed in oral and oropharyngeal cancer. For both malignancies the incidence is increasing, while survival has shown only modest improvement over the last three decades34. Although the oral cavity is easily accessible for regular inspection, about two-thirds of all patients in the US present with advanced-stage disease. Another complicating factor for improving the standard of care is that socioeconomic groups with high oral cancer incidence are also the least likely to have dental or health insurance, further delaying diagnosis of their often asymptomatic disease35,36. The global situation is now more dire than ever, with respect to both incidence and number of associated deaths (300,000 and 145,000, respectively37). In India, where oral cancer is the third most common malignancy in men, the disease often remains untreated for extended amounts of time. Here, it is less HPV or smoking/drinking that drives the disease, but rather the consumption of paan/gutka (chewing tobacco), both common mild stimulants that contain betel leaf and areca nuts. Very few patients in India survive if their disease is discovered with regional or distant metastases (18% and 1.1%, respectively).
Currently, no diagnostic adjuncts have shown a proven clinical benefit for detection or surveillance of oral cancer. However, considering the various emerging optical imaging methods11, we believe that molecularly specific fluorescence contrast, as provided by PARPi-FL, combined with novel analytical devices and methodologies, has the clear potential to improve delineation and detection of oral and oropharyngeal cancer. One such novel combination is PARPi-FL brush cytology. Brush cytology, also known as exfoliative cytology or brush biopsy, is a minimally invasive technique in which cells are collected between the surface and basement membrane of the epithelium, rather than using invasive incisional biopsy, followed by cytopathological evaluation7,38. Despite extensive clinical evaluation, adoption into clinical practice is still limited, partially due to the high cost of cytopathology as well as mixed reports about its sensitivity and specificity39.
We evaluated whether a combination of brush biopsy and PARPi-FL staining is feasible in oropharyngeal cancer to identify and quantify tumour cells in a tissue sample. Analysis of PARP1 expression via IHC quantification in oropharyngeal cancer biospecimens revealed consistently high PARP1 expression, while normal epithelial expression resembled other physiological backgrounds. Located behind the oral cavity, oro- and hypopharyngeal cancers are less accessible to visual inspection than other oral cancer types, such as tongue cancer. Brush cytology, however, could be easily performed in this location. To establish a PARPi-FL staining method for brush cytology-like samples, we prepared a single-cell solution from solid xenograft tumour, as it would be derived during the brush cytology process,26; (Fig. 2). We then stained this complex tissue sample with PARPi-FL and analysed the percentage of PARPi-FL-positive cells using flow cytometry. We found a significantly higher number of PARPi-FL-stained cells in xenograft samples than in non-neoplastic tongue tissue samples. Importantly, due to the homology of human and murine PARP1, PARPi-FL targets both human and murine tissues. A baseline level of PARPi-FL-positive cells in normal tongue samples was not surprising, since PARP1 expression in the epithelium’s basal layer is well-established.
These proof-of-principle results warrant further studies on combining PARPi-FL with brush cytology to establish baselines for PARPi-FL-positive cells in preclinical and human samples. A flow-based technology has the advantage of investigating suspicious lesions in a minimally invasive, quantitative, and automatable approach. In addition to pharyngeal cancer, application of PARPi-FL brush cytology is promising in all locations where brush cytology is currently explored, including other sites inside the oral cavity40 and oesophagus41,42.
While PARPi-FL brush cytology is a technology tailored for the screening and surveillance setting, PARPi-FL tissue staining is a promising approach for both pre- and intraoperative diagnostics. In several patient datasets from the US (Memorial Sloan Kettering Cancer Center; MSK) and India (Mazumdar Shaw Medical Center, Narayana Health; MSMC), we were able to establish that PARP1 expression at the epithelium undergoes statistically significant increases during disease progression. Considering the potential clinical impact, our most essential finding was that PARP1-based diagnosis could differentiate benign and potentially malignant lesions from normal epithelium and cancerous lesions that require surgical treatment, allowing for surveillance and chemoprevention to reduce the risk of cancer development43.
Despite a strong statistical separation of advanced disease from normal tissue, a certain overlap can be observed between groups, which we attribute to i) inter-patient heterogeneity of PARP1 expression and ii) imperfect histopathologic diagnostic separation of different stages of dysplasia. This is supported by our data where only paired data on PARP1 expression are analysed – here, no intra-patient overlap between tumour and normal epithelium was observed. PARP1 IHC quantification also showed that surgical tumour margins were clearly delineated by PARP1 expression. This is significant, since almost all oral cancers are surgically treated. Complete removal, defined by clear margins, is a major determining factor for local recurrence3,44,45. The current gold standard for negative tumour margins is clearance greater than 5 mm on permanent histopathology46, which is not available until days after surgery. The only method currently available to surgeons for intraoperative feedback before surgical closure and reconstruction is frozen section analysis of surgical margins. However, there is no standardized practice for frozen sections in head and neck cancer47. Moreover, many view frozen sections critically, since accuracy is reduced as compared to paraffin sections, positive effects on outcome have not been reported, and the cost:benefit ratio is 20:148. The PARPi-FL fresh tissue staining method reported here offers a promising alternative or addition to frozen section analysis, and could provide much faster feedback. We identified several cases of focal invasion of tumour cells into an otherwise normal area based on the PARPi-FL signal. PARPi-FL staining patterns strongly represent PARP1 staining on IHC but were achieved within minutes of biopsy (as quickly as one minute). If positive margins are found, the surgeon could perform re-excision and margin evaluation without delay. We determined an excellent sensitivity and specificity of >95% for the identification of tumour and margin samples using blinded readers who received minimal training (10 min), supporting our claim that this method could be implemented in the clinic without the need for highly specialized personnel. A unique feature of our PARPi-FL staining method is its tissue-preserving nature, which would allow for integration into existing workflows, since PARPi-FL stained tissue would remain available for either frozen or permanent pathology or other techniques, including sequencing.
In addition to ex vivo imaging, we have shown feasibility of PARPi-FL in vivo diagnostics, based on our findings that intravenous injection provides a broad surgical window of at least 5–120 min post injection and can be detected with a handheld confocal endomicroscope. In a clinical scenario, this could enable a targeted search for a positive margin within the tumour bed. Currently, correlating the exact location of a positive margin, identified histologically ex vivo, to its counterpart in the surgical cavity is challenging due to significant shrinking and distortion that the tissue undergoes upon resection. In addition, we show feasibility of macroscopic first-in-human PARPi-FL staining and imaging with a topically applied PARPi-FL solution, which is currently under clinical evaluation (NCT03085147).
It is frequently suggested that a near infrared version of PARPi-FL would be advantageous over the green fluorescent variant. However, the fact that PARP1 is an intranuclear target puts several design constraints on a functional PARP1 imaging agent, which have been discussed previously49. Longer wavelength PARP imaging agents were introduced in vitro, but the available data suggests pronounced effects on nuclear penetration and clearance affecting their in vivo performance50,51. In our opinion, overall pharmacokinetics of PARPi-FL and its ability to serve as a sensor for PARP1 outweighs the wavelength trade-offs. This is supported by numerous clinical studies involving green fluorescent dyes for surgical guidance, including the first-in-human study of a molecularly targeted optical imaging agent52.
Together, the versatility of PARPi-FL with respect to application, imaging settings and technologies enables a combination of micro- and macroscopic evaluation techniques and thus a large range of applications, including screening, surveillance, biopsy guidance, fresh biopsy staining, in vivo diagnostic capability, and intraoperative margin delineation. Thus, PARPi-FL offers a whole host of diagnostic and intraoperative applications that can also be combined with each other, which could not be achieved with previously introduced contrast agents or label free diagnostic methods.
Outlook
In future, extensive studies will be necessary to further confirm the clinical value of PARPi-FL, following guidelines specifically developed for fluorescent contrast agents to characterize their procedural benefit over existing practices27,53. Our next logical step towards standard-of-care clinical use of PARPi-FL will be blinded sampling of fresh tissue specimens at multiple clinical sites, with a focus on recruiting patients from different backgrounds and of varying disease stages. Although PARPi-FL could conceivably be used in a surgical setting, its ultimate clinical value as a field diagnostic cannot be confirmed until a larger, multi-institutional study has been completed.
Methods
PARP1 expression analysis.
Sample collection of formalin-fixed, paraffin-embedded biospecimens. The analysis of PARP1 expression was conducted on banked or newly collected paraffin embedded biospecimens at MSK and MSMC. Specimen selection and collection was approved by the Institutional Review Boards (IRB) at MSK and MSMC and the Independent Ethics Committee (MSMC). All diagnoses were conducted by the pathology department on H&E stained sections from the same samples. We analysed specimens of oral cancer (n=84 patients, 3 separate studies), oropharyngeal cancer (n=9) and oesophageal cancer (n=7) patients (a more detailed description can be found in Table S2).
PARP1 staining.
PARP1 IHC staining was carried out using a primary polyclonal rabbit anti-PARP1 antibody (sc-7150, 0.4 μg/mL, Santa Cruz Biotechnology). At MSK, the staining was carried out using the automated Discovery XT processor (Ventana Medical Systems) at the Molecular Cytology Core Facility as described previously26. At MSMC, the same anti-PARP1 antibody was used (0.4 μg/mL, 1 h) in combination with the DAKO Real Envision kit (K5007, DAKO). Since the PARP1 antibody was discontinued by Santa Cruz, we tested and validated PA5–16452 (Invitrogen) as potential alternative.
Quantification of PARP1 in automated protocol.
For PARP1 protein quantification, PARP1 IHC slides were digitalized using a MIRAX Slide Scanner (3DHISTECH, Budapest, Hungary). In surgical specimens, we separately analysed tumour, normal epithelium and deep margin if present. On biopsy specimen (tumour or margin) we analysed tumour on the tumour biopsy and normal epithelium and deep margin on the margin biopsy, if present. On at least 3 fields of view per area of interest (tumour, epithelium, deep margin) (20x magnification), PARP1 presence was quantified using ImageJ/FIJI. Diaminobenzidine (DAB) and Hematoxylin stainings were separated using the Color Deconvolution algorithm and appropriate threshold levels were set to measure the area of specific DAB staining (=PARP1 area) as well as hematoxylin staining (=tissue area) to calculate the relative PARP1 positive area on each image. The thresholds were kept constant within each dataset. We report means and standard deviation for each sample. For grouped analysis, we pooled the means of all samples. ROC curves were generated using GraphPad Prism 7.0.
Quantification in manual protocol.
Additional manual scoring was conducted on PARP1 IHC cases from MSMC. Therefore, the staining intensity was evaluated on a scale of 1–4 (Fig. S6a) and multiplied with the % PARP1-positive cells (1–100%, visual evaluation), resulting in a final score of 1–400. All slides were scored by three people. We report the mean score for each slide and the ROC curve comparing benign/mild dysplasia to severe dysplasia/malignant cases.
Synthesis of PARP1-targeted imaging agents.
The synthesis of PARPi-FL and [18F]PARPi have been carried out as described previously54. PARPi-FL was prepared as 1.5 mM stock solution in PEG300 and was diluted to its final concentration in 30%PEG300/PBS. [18F]PARPi was formulated in 0.9% sterile saline with 10% EtOH.
Small animals and xenografting.
All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of MSK and followed the National Institutes of Health guidelines for animal welfare. For in vivo and ex vivo experiments, we grew subcutaneous xenografts in 8–10 week old female athymic nude mice (NCr-Foxn1nu, Taconic) using human pharynx squamous cell carcinoma (FaDu) and oesophageal adenocarcinoma (EAC) cell lines (ESO51, OE33, SKGT4, OE19) cancer cells by injecting 2×106 cells in a mixture of culture medium (50 μl) and Matrigel™ (BD Biosciences) (50 μl). Experiment were conducted 12–16 days after xenografting. We used 12 nude mice for imaging of EAC models, 9 nude mice for brush biopsy experiments, 10 nude mice for PARPi-FL fresh tissue staining optimization and 3 nude mice for rapid fresh tissue imaging on a strip-mosaic confocal scanner (Vivascope 2500). FaDu cells were ordered from ATCC, EAC cell lines were ordered from DSMZ. To prepare cells for xenografting, they were cultivated in a monolayer culture at 37°C in a 5% CO2 humidified atmosphere, following standard procedures. They were maintained in their respective growth medium (Roswell Park Memorial Institute medium for ESO51, OE33, SKGT4 and OE19) and minimum essential medium for FaDu), containing 10% (v/v) fetal bovine serum and 1% penicillin/streptavidin.
Imaging of PARP1 expression in EAC models via PARPi-FL.
To determine PARPi-FL uptake in EAC xenograft models, we intravenously injected PARPi-FL (75 nmol in 167 μl, 30%PEG300/PBS) in mice bearing ESO51, OE33, SKGT4 or OE19 xenografts (n=3/group). For injection, lines were not flushed. 90 min post-injection, animals were sacrificed and tumours, the oesophagus and thigh muscle were excised and were imaged immediately in the epifluorescence system IVIS (PerkinElmer, Waltham, MA) using the standard filter set for GFP imaging (excitation: 465/30 nm, emission: 520–580 nm). Autofluorescence removal and quantification were carried out as described previously26,55. Semiquantitative analysis of the PARPi-FL signal was conducted by measuring the average radiant efficiency in regions of interest (ROIs) that were placed on the excised tumours under white light guidance. Average radiant efficiency [p/s/cm2/sr]/[μW/cm2] is defined as the number of photons per second leaving a square centimetre of tissue and radiating into a solid angle of one steradian (sr). Resulting numbers are normalized for the integration time, binning, f/stop, field of view, illumination intensity, and the ROI area, making measurements comparable among each other. After epifluorescence imaging, the freshly excised whole tumours were imaged using a confocal microscope. Tissues were placed on a cover slip with a freshly cut surface facing the cover slip and all images were taken using identical settings and 488 nm laser excitation (LSM880, Zeiss, Jena, Germany). Subsequently, tumours were fixed in 4% Paraformaldehyde for 24 h and then paraffin embedded. H&E staining and PARP1 IHC staining and quantification were carried out as described above.
Western Blot (EAC).
PARP1 protein expression was measured in ESO51, OE19, SKGT4 and OE33 cell lysates using Western blot analysis as described before56. Briefly, proteins were isolated from cells and 30 μg of protein per sample were separated with SDS/PAGE gel electrophoresis and transferred to a Nitrocellulose membrane. Proteins were detected using antibodies specific for PARP1 (1:1000; sc-7150, Santa Cruz) and b-actin (1:2000; A3854, Sigma-Aldrich) with a corresponding horseradish peroxidase (HRP) conjugated secondary antibody (1:10,000, sc-2004, Santa-Cruz). Detection was performed using a chemiluminescent substrate (Thermo Scientific #34077, SuperSignal West Pico). Since distribution of the primary anti-PARP1 antibody was discontinued, we tested and validated PA5–16452 (Invitrogen) as potential alternative.
Topical application and tissue penetration of PARPi-FL in the oesophagus.
A topical application strategy of PARPi-FL for the human oesophagus was developed using a Yucatan mini swine (Sus scrofa domesticus), which reflects the human anatomy more closely than small animals. Pig experiments were carried out at CBSet, Inc. (Lexington, MA) and were approved by their IACUC. The animal was pre-medicated and anaesthetized throughout the procedure. The pig received Telazol (4–6 mg/kg) intramuscular once, isoflurane as inhalant (1–5% in O2) throughout the procedure and Buprenorphrine as intramuscular injection (0,01–0,03 mg/kg). Topical application was realized using a balloon applicator (developed by Aero-Di-Namics, New York, NY; produced by Medical Murray, North Barrington, IL). The balloons are made of silicone with 12 Lubrizol stripes (electrospun Lubrizol tecophilic TN TG 500) and have a nominal diameter of 7 mm, which extends to 20–25 mm upon inflation (Fig. 2G). The Lubrizol strips are protected inside of pleats during device delivery and removal and are only exposed when inflated. The Lubrizol stripes were loaded with 2 mL of a 1 μM PARPi-FL solution. The balloon was inserted into the anesthetized pigs oesophagus in a deflated state under white light endoscopic guidance. Inflation led to exposure of the Lubrizol stripes and a pressure induced release of PARPi-FL onto the oesophagus wall of the anesthetized pig for a duration of 3 min. 10 min after the application, the pig was sacrificed followed by necropsy and cryopreservation of the oesophagus in 10×10 mm tiles. Cryosections were cut perpendicular to the oesophagus surface and imaged using a confocal microscope to detect penetration depths and binding of PARPi-FL from the topical application. Adjacent slides were H&E stained to identify morphological structures. We used one pig and applied PARPi-FL to three different sites by re-loading and re-insertion of the balloon.
PARPi-FL staining and flow cytometry of tissue derived cell suspensions.
Brush biopsy samples, obtained from tissue derived cell suspensions were used for PARPi-FL staining. Specifically, FaDu (n=9) and OE19 (n=12) xenografts were grown in female athymic nude mice. Tumours and tongues were harvested, and single cell suspensions were obtained via tissue dissociation using the gentleMACS Octo Dissociator with Heaters (Miltenyi Biotech) in combination with a tissue dissociation kit (#130-095-929, Miltenyi Biotech), following the manufacturer’s instructions. Cells were then resuspended and stained with either 100 nM PARPi-FL, 100 μM olaparib mixed with 100 nM PARPi-FL or PBS for 1 hour at room temperature with occasional shaking, followed by a 1 hour wash in PBS at room temperature with occasional shaking. After resuspension in fresh PBS, cells were analysed on a Fortessa flow cytometer (BD Biosciences). DAPI was added to exclude dead cells from the analysis. Side scatter and forward scatter were used to select single cells, excluding doublets, aggregates and debris. Fluorescence was measured in the DAPI channel (DAPI live/dead stain), FITC channel (PARPi-FL) and PE channel (to control for near FITC laser bleeding). The appropriate gates were selected on unstained and single stained controls (Fig. S5a). The cell populations were analysed for the percentage of PARPi-FL positive cells.
Fresh biospecimen staining.
Optimization of the PARPi-FL fresh tissue staining protocol was carried out using FaDu xenografts (n=5) (Fig. S8). The goal of the optimization was to achieve a clearly visible staining only in the cell nucleus (no cytoplasmic signal), at the shortest possible staining time. The final protocol was used to stain fresh biopsies from oral cancer patients. 2 biopsies per patient (one biopsy was taken from the visible tumour and one biopsy at the 5 mm margin - see fig. 5A) were obtained pre-surgically under an IRB approved protocol. 13 patients were included in the study (see Table S3 for histopathological diagnoses; all margins were tumour negative). One tumour sample was retrospectively excluded due to a non-malignant, non-dysplastic histopathological diagnosis (verrucous hyperplasia). For 3 patients, margin samples were not available, resulting in a total of n=12 tumours and n=10 margins. Upon receipt, tissues were photographed and divided into three equal parts, each containing mucosa and submucosa. One part was used for immediate fresh tissue imaging, one part was cryoconserved and one part was fixed in 4% PFA for H&E and PARP1 IHC. The fresh tissue staining protocol consisted of staining for 5 min in 100 nM PARPi-FL (in 30%PEG300/PBS, room temperature) and a 10 min wash in 30%PEG300/PBS. Samples were then transferred to ice-cold PBS containing 10 μg/mL Hoechst 33342 until confocal imaging (LSM880, Zeiss, Germany). Tissues were placed between two cover glasses (48×60 mm, Brain Research Laboratories) and a three-channel tile scan of the entire tissue (at least 2×2 mm) was captured (405 nm to detect Hoechst 33342, 488 nm to detect PARPi-FL and 543 nm to detect autofluorescence) using a 20x objective at 0.6x zoom in a scan time under 10 min. The diagnostic value of PARPi-FL fresh tissue staining was determined in a blinded reader study (n=27 readers) (study design in Fig. S10B). Readers received a ~10 min training on characteristic features of PARPi-FL staining in tumour and margin samples (Supplementary Material X). They were then shown the study set, consisting of 30 cases (n=12 tumours, n=10 margins, n=8 duplicates (4 tumours, 4 margins) to determine sensitivity, specificity and intrareader variability. For each case, readers were presented 1–5 images for 5 sec/image, followed by a 10 sec decision window to classify a tissue as tumour or margin (full study set in Supplementary Material Y; each case was assigned a random ID (http://www.random.org) and cases were presented in ascending numbers to the readers). Images were presented at 488 nm and 488/543 nm overlay (to facilitate autofluorescence identification), where available (Fig. S10a). Statistical values were calculated in R. Biopsy samples from oesophageal cancer patients (n = 5), acquired under an IRB approved protocol, were stained with 100 nM PARPi-FL for 5 min, subjected to confocal microscopy scanning and were then fixed in 4% PFA, followed by paraffin embedding and H&E staining. H&E sections were evaluated by a pathologist.
Fresh biospecimen imaging with a strip-mosaic scanning confocal fresh tissue scanner.
To test the feasibility of tissue scanning at a faster speed than standard confocal microscopy, we tested a confocal scanner that was specifically designed to create high-resolution strip-mosaics of whole tissues and features a 488 nm laser excitation (Vivascope 2500, Caliber ID, Andover, MA). Following PARPi-FL staining of FaDu xenografts (0, 100, 250 nM PARPi-FL for 10 min) and human biospecimens (100 nM PARPi-FL for 10 min), tissues were placed on the specimen holder and images of a 15×15 mm field-of-view were acquired in about 150 sec. Images were compared to regular confocal microscopy and PARP1 IHC of the same samples.
Fresh biospecimen imaging with a point scanning miniaturized confocal endomicroscope.
In addition to scanning of excised biospecimens, imaging with a confocal endomicroscope would enable imaging within the surgical cavity/wound bed in real time during surgery. Therefore, we tested if a miniaturized point-scanning confocal endomicroscope (Optiscan, Mulgrave, Australia) can distinguish PARPi-FL stained tumour and normal biospecimens. The system uses a 488 nm laser with several bandpass and emission filters, which allow for identification of autofluorescence (Fig. S14a). The focal plane is adjustable between 0–400 μm depths (Fig. S14b). The same human biospecimens that were used for regular confocal imaging were also imaged using the ViewnVivo to determine if PARPi-FL stained nuclei can be clearly identified. The images were compared to PARP1 IHC images from the same patient.
PARPi-FL blood half-life and imaging after intravenous injection in a pig.
In a Yorkshire pig (50–55 kg), anaesthesia was induced using isoflurane inhalation anaesthesia under supervision (1–5% in O2) and the pig was placed on a table in the supine position. One intravenous catheter was placed in the auricular vein, where PARPi-FL (0.05 mg/kg, 10 mL, 30% PEG300 in PBS) was administered intravenously, followed by a 5 mL saline flush. Another catheter was placed in the cephalic vein for blood draws. Blood samples (1.0 mL) and tongue punch biopsies (3 mm) were taken at predetermined time points (5, 10, 30, 60, 90, and 120 minutes). The pig was euthanized with an intravenous overdose of pentobarbital sodium and phenytoin sodium (440 mg/ml). Tongue punch biopsies were stored in ice-cold PBS containing 10 μg/mL Hoechst 33342 until confocal imaging. The whole, fresh punch biopsies were imaged using a LSM880 confocal microscope (Zeiss, Germany) to identify PARPi-FL staining in the epithelial basal layer. Ice-cold acetonitrile (1.5 mL) was added to blood samples (1.0 mL) and vortexed for 30 s. After centrifugation for 10 min (4000 rpm), the supernatant (1.8 mL) was lyophilized overnight. Acetonitrile (200 uL) was added, vortexed for 15 s, and spun down for 5 min (4000 rpm). Then, supernatant (100 uL) of each time was analysed in a black bottom-less 96-well plate using a UV/VIS plate reader (Spectramax M5, Molecular Devices).
PARPi-FL first-in-human imaging.
To conduct PARPi-FL in-human imaging, we obtained IRB approval. Informed consent was obtained from each patient. The presented data are part of a clinical phase I/II trial (NCT03085147). The patient had histopathologically confirmed oral squamous cell carcinoma. Videos were acquired before and post contrast application using the Quest Spectrum platform (Quest Medical Imaging, Middenmeer, Netherlands) in combination with a laparoscope and a customized PARPi-FL optimized laser/filter system. The same instrument settings were used throughout the imaging procedure (30 ms exposure time, 100% laser power, gain: 25.5 dB). For contrast application, the patient gargled 15 mL of a 500 nM PARPi-FL solution (in 15% PEG300/PBS) for 60 sec, followed by a washing solution (15%PEG300/PBS) for 60 sec. No adverse events or discomfort were observed. For image processing, we subtracted the red channel (autofluorescence) from the green channel (autofluorescence and PARPi-FL), using a correction factor of 1.2.
PET Imaging with [18F]PARPi.
Animals bearing EAC xenografts were injected intravenously with 160–230 μCi of [18F]PARPi 2 hours before PET/CT imaging. To acquire PET/CT images (Inveon PET/CT, Siemens) animals were anesthetized with 2% isoflurane and positioned on the scanner bed. PET data were collected for 5–10 minutes, followed by CT. After acquisition, list mode emission data were sorted into two-dimensional sinograms via Fourier rebinning. Data were normalized to correct for non-uniform detector response, dead time count losses, and positron branching ratio, but no attenuation, scatter, or partial-volume averaging corrections were applied. Sinogram data were subsequently reconstructed into 128 × 128 × 159 matrix (0.78 × 0.78 × 0.80 mm3 voxel dimensions) using 2D ordered subset expectation maximization (OSEM2D; 4 iterations, 16 subsets). Image counts per voxel per second were converted to activity concentrations (Bq/cc or %IA/cc) using a system-specific calibration factor derived from imaging a mouse-sized water-equivalent phantom containing fluorine-18. CT scans were reconstructed using a modified Feldkamp cone beam reconstruction algorithm to generate 512 × 512 × 768 voxel image volumes (0.197 × 0.197 × 0.197 mm3 voxel dimensions). PET/CT images were processed using Inveon Research Workplace software, with a spherical VOI placed on the tumour or the thigh muscle using the CT.
H&E Histopathology after PARPi-FL staining.
We conducted formalin fixed paraffin embedded (FFPE) histopathology evaluation following PARPi-FL staining. In this case, PARPi-FL staining and imaging was conducted in approximately 60–90 min from receiving the fresh human biopsy. Then, the biopsy was fixed in 4% PFA for 24 hours at 4°C and processed for paraffin embedding, sectioning and H&E staining.
Preclinical staining optimization protocols.
PARPi-FL staining was optimized on fresh FaDu xenograft tissue. Freshly excised tumour tissue was cut into small pieces (2–3 mm diameter) before staining. We tested different staining protocols, varying PARPi-FL concentration (50, 100, 250 nM PARPi-FL in 30%PEG300/PBS), staining times (1, 5, 10 min) and washing steps (0, 2, 10 min in 30%PEG300/PBS). Samples were then transferred to ice-cold PBS containing 10 μg/mL Hoechst 33342 until confocal imaging (LSM880, Zeiss, Germany). To collect images, tissues were placed on top of a cover glass (48×60 mm, Brain Research Laboratories) and instrument settings were identical for all images.
Statistical analysis.
Statistical analysis was conducted using Graphpad Prism 7 and R. R was used to analyse the blinded study readings (packages used: base, stats and gee). In this regard, the confidence intervals for the diagnostic values were computed using a robust variance estimate accounting for a reader effect using a Generalized Estimating Equation (GEE) approach. With regard to statistical analysis with Graphpad Prism, we used the Wilcoxon test for analysis of paired samples, e.g., PARP1-positive area of tumour, epithelium, and deep margin. We used the Mann-Whitney test for analysis of unpaired samples, e.g., PARP1-positive area of different disease stages. We used an unpaired t-test for analyzing flow cytometry data and corrected for multiple comparison using the Holm-Sidak method, without assuming a consistent standard deviation (SD). Statistical significance was determined with alpha = 0.05. We specify which test was used and which level of significance was found for each result (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). ROC curves were generated with Graphpad Prism 7. All in vivo and ex vivo experiments subjected to statistical analysis consisted of group sizes of at least three. If not stated otherwise, data are presented as mean ± standard deviation.
Data availability.
The authors declare that the main data supporting the findings of this study are available within the paper and its supplementary information files. Associated raw data and step-by-step protocols can be made available from the corresponding author on reasonable request.
Code availability.
The ImageJ macro for the automated analysis of PARP1 expression on IHC slides is available on reasonable request.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgments:
We thank Aditi Sahu and Rachel Giese for supporting the work at MSMC. We thank Jay Budrewicz for his support of the experiments at CBSet, and Nora Katabi for providing their expertise in histopathology. We also thank Violeta Dokic for assistance during clinical work with PARPi-FL. We thank the Molecular Cytology Core Facility, Radiochemistry & Molecular Imaging Probes Core Facility, and Flow Cytometry Core Facility at Memorial Sloan Kettering Cancer Center. We also thank Garon Scott and Leah Bassity for editing the manuscript. Finally, we thank the participants of the blinded study, including (in alphabetical order): Adam Schulman, Aditi Sahu, Alexander Bolaender, Christian Mason, Edwin Pratt, Chrysafis Andreou, Fay Nicolson, Jack Berry, Jeroen Goos, Junior Gonzales, Kelly Henry, Luke Carter, Manu Jain, Marlena McGill, Navjot Guru, Nick Sobol, Patricia Ribeiro Pereira, Rustin Mirsafavi, Sheryl Roberts, Sophie Poty, Stephen Jannetti, Troy Crawford, Veronica Nagle, Xiancheng (Lewis) Wu, and Ahmad Sadique.
This work was supported by National Institutes of Health grants R01 CA204441, P30 CA008748, R43 CA228815, and K99 CA218875 (SK). The authors thank the Tow Foundation and MSK’s Center for Molecular Imaging & Nanotechnology, Imaging and Radiation Sciences Program, and Molecularly Targeted Intraoperative Imaging Fund.
Footnotes
Competing interests: S.K., S.P., C.B., and T.R. are shareholders of Summit Biomedical Imaging, LLC. S.K., S.P., and T.R. are co-inventors on filed U.S. patent (WO2016164771) held by MSK that covers methods of use for PARPi-FL. T.R. is a co-inventor on U.S. patent (WO2012074840) held by the General Hospital Corporation that covers the composition of PARPi-FL. B.C. and T.R. are co-inventors on the filed U.S. patent (WO2016033293) held by MSK that covers methods for the synthesis of [18F]PARPi. T.R. is a paid consultant for Theragnostics, Inc. M.S. is a co-founder of Aero-Di-Namics.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.SEER Cancer Stat Facts: Oral Cavity and Pharynx, based on November 2017 SEER data submission, posted to SEER website, April 2018. National Cancer Institute; Bethesda, MD, https://seer.cancer.gov/statfacts/html/oralcav.html. [Google Scholar]
- 2.SEER Cancer Stat Facts: Esophageal Cancer, based on November 2017 SEER data submission, posted to SEER website, April 2018. National Cancer Institute; Bethesda, MD: https://seer.cancer.gov/statfacts/html/esoph.html. [Google Scholar]
- 3.Sutton DN, Brown JS, Rogers SN, Vaughan ED & Woolgar JA The prognostic implications of the surgical margin in oral squamous cell carcinoma. Int J Oral Maxillofac Surg 32, 30–34, doi: 10.1054/ijom.2002.0313 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Ganly I, Patel S & Shah J Early stage squamous cell cancer of the oral tongue--clinicopathologic features affecting outcome. Cancer 118, 101–111, doi: 10.1002/cncr.26229 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Law S, Arcilla C, Chu KM & Wong J The significance of histologically infiltrated resection margin after esophagectomy for esophageal cancer. American journal of surgery 176, 286–290 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Fedele S Diagnostic aids in the screening of oral cancer. Head & neck oncology 1, 5, doi: 10.1186/1758-3284-1-5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lingen MW, Kalmar JR, Karrison T & Speight PM Critical evaluation of diagnostic aids for the detection of oral cancer. Oral oncology 44, 10–22, doi: 10.1016/j.oraloncology.2007.06.011 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller C et al. Adjunctive diagnostic techniques for oral lesions of unknown malignant potential: Systematic review with meta-analysis. Head & neck 37, 755–762, doi: 10.1002/hed.23667 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Olivo M, Bhuvaneswari R & Keogh I Advances in bio-optical imaging for the diagnosis of early oral cancer. Pharmaceutics 3, 354–378, doi: 10.3390/pharmaceutics3030354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strome A et al. Current Practice and Emerging Molecular Imaging Technologies in Oral Cancer Screening. Molecular imaging 17, 1536012118808644, doi: 10.1177/1536012118808644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang EC et al. Noninvasive diagnostic adjuncts for the evaluation of potentially premalignant oral epithelial lesions: current limitations and future directions. Oral Surg Oral Med Oral Pathol Oral Radiol 125, 670–681, doi: 10.1016/j.oooo.2018.02.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin D, Vigneswaran N, Gillenwater A & Richards-Kortum R Advances in fluorescence imaging techniques to detect oral cancer and its precursors. Future Oncol 6, 1143–1154, doi: 10.2217/fon.10.79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patton LL, Epstein JB & Kerr AR Adjunctive techniques for oral cancer examination and lesion diagnosis: a systematic review of the literature. J Am Dent Assoc 139, 896–905; quiz 993–894 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Nosho K et al. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. European journal of cancer 42, 2374–2381, doi: 10.1016/j.ejca.2006.01.061 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Staibano S et al. Poly(adenosine diphosphate-ribose) polymerase 1 expression in malignant melanomas from photoexposed areas of the head and neck region. Human pathology 36, 724–731, doi: 10.1016/j.humpath.2005.04.017 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Chow JP et al. PARP1 is overexpressed in nasopharyngeal carcinoma and its inhibition enhances radiotherapy. Molecular cancer therapeutics 12, 2517–2528, doi: 10.1158/1535-7163.MCT-13-0010 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Salemi M et al. Poly (ADP-ribose) polymerase 1 protein expression in normal and neoplastic prostatic tissue. European journal of histochemistry : EJH 57, e13, doi: 10.4081/ejh.2013.e13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green AR et al. Biological and clinical significance of PARP1 protein expression in breast cancer. Breast cancer research and treatment 149, 353–362, doi: 10.1007/s10549-014-3230-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dziaman T et al. PARP-1 expression is increased in colon adenoma and carcinoma and correlates with OGG1. PloS one 9, e115558, doi: 10.1371/journal.pone.0115558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossovskaya V, Koo IC, Kaldjian EP, Alvares C & Sherman BM Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumour Types. Genes & cancer 1, 812–821, doi: 10.1177/1947601910383418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michels J et al. Negative prognostic value of high levels of intracellular poly(ADP-ribose) in non-small cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 26, 2470–2477, doi: 10.1093/annonc/mdv393 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Rojo F et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 23, 1156–1164, doi: 10.1093/annonc/mdr361 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Pashaiefar H et al. PARP-1 Overexpression as an Independent Prognostic Factor in Adult Non-M3 Acute Myeloid Leukemia. Genet Test Mol Biomarkers 22, 343–349, doi: 10.1089/gtmb.2018.0085 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Li ZH et al. PARP1 is a novel independent prognostic factor for the poor prognosis of chordoma. Cancer Biomark 16, 633–639, doi: 10.3233/Cbm-160605 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Irwin CP et al. PARPi-FL--a fluorescent PARP1 inhibitor for glioblastoma imaging. Neoplasia 16, 432–440, doi: 10.1016/j.neo.2014.05.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kossatz S et al. Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent. Scientific reports 6, 21371, doi: 10.1038/srep21371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal EL et al. Successful Translation of Fluorescence Navigation During Oncologic Surgery: A Consensus Report. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 57, 144–150, doi: 10.2967/jnumed.115.158915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kariv R et al. The Seattle protocol does not more reliably predict the detection of cancer at the time of esophagectomy than a less intensive surveillance protocol. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 7, 653–658; quiz 606, doi: 10.1016/j.cgh.2008.11.024 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Canto MI et al. Methylene blue-directed biopsies improve detection of intestinal metaplasia and dysplasia in Barrett’s esophagus. Gastrointestinal endoscopy 51, 560–568 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Ngamruengphong S, Sharma VK & Das A Diagnostic yield of methylene blue chromoendoscopy for detecting specialized intestinal metaplasia and dysplasia in Barrett’s esophagus: a meta-analysis. Gastrointestinal endoscopy 69, 1021–1028, doi: 10.1016/j.gie.2008.06.056 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Shiozaki H et al. Endoscopic screening of early esophageal cancer with the Lugol dye method in patients with head and neck cancers. Cancer 66, 2068–2071 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Patel AA, Strome M & Blitzer A Directed balloon cytology of the esophagus: A novel device for obtaining circumferential cytologic sampling. Laryngoscope 127, 1032–1035, doi: 10.1002/lary.26472 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Moinova HR et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Science translational medicine 10, doi: 10.1126/scitranslmed.aao5848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warnakulasuriya S Global epidemiology of oral and oropharyngeal cancer. Oral oncology 45, 309–316, doi: 10.1016/j.oraloncology.2008.06.002 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Johnson S, Corsten MJ, McDonald JT & Chun J Socio-economic factors and stage at presentation of head and neck cancer patients in Ottawa, Canada: a logistic regression analysis. Oral oncology 46, 366–368, doi: 10.1016/j.oraloncology.2010.02.010 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Conway DI et al. Socioeconomic inequalities and oral cancer risk: a systematic review and meta-analysis of case-control studies. International journal of cancer. Journal international du cancer 122, 2811–2819, doi: 10.1002/ijc.23430 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Torre LA et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians 65, 87–108, doi: 10.3322/caac.21262 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Mehrotra R & Gupta DK Exciting new advances in oral cancer diagnosis: avenues to early detection. Head & neck oncology 3, 33, doi: 10.1186/1758-3284-3-33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenberg MS The “brush” controversy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 93, 217–218 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Mehrotra R The role of cytology in oral lesions: a review of recent improvements. Diagn Cytopathol 40, 73–83, doi: 10.1002/dc.21581 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Fusaroli P et al. Histology vs brush cytology (BC) in the diagnosis and follow up of Barrett’s esophagus (BE). Gastrointestinal endoscopy 61, Ab131–Ab131, doi:Doi 10.1016/S0016-5107(05)00776-5 (2005). [DOI] [Google Scholar]
- 42.Hardwick RH, Morgan RJ, Warren BF, Lott M & Alderson D Brush cytology in the diagnosis of neoplasia in Barrett’s esophagus. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus / I.S.D.E 10, 233–237 (1997). [DOI] [PubMed] [Google Scholar]
- 43.D’Souza S & Addepalli V Preventive measures in oral cancer: An overview. Biomed Pharmacother 107, 72–80, doi: 10.1016/j.biopha.2018.07.114 (2018). [DOI] [PubMed] [Google Scholar]
- 44.McMahon J et al. Influence of condition of surgical margins on local recurrence and disease-specific survival in oral and oropharyngeal cancer. Br J Oral Maxillofac Surg 41, 224–231 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Ravasz LA, Slootweg PJ, Hordijk GJ, Smit F & van der Tweel I The status of the resection margin as a prognostic factor in the treatment of head and neck carcinoma. J Craniomaxillofac Surg 19, 314–318 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Alicandri-Ciufelli M et al. Surgical margins in head and neck squamous cell carcinoma: what is ‘close’? Eur Arch Otorhinolaryngol 270, 2603–2609, doi: 10.1007/s00405-012-2317-8 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Black C, Marotti J, Zarovnaya E & Paydarfar J Critical evaluation of frozen section margins in head and neck cancer resections. Cancer 107, 2792–2800, doi: 10.1002/cncr.22347 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Pathak KA et al. Impact of use of frozen section assessment of operative margins on survival in oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 107, 235–239, doi: 10.1016/j.tripleo.2008.09.028 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Kossatz S, Weber W & Reiner T Detection and Delineation of Oral Cancer With a PARP1-Targeted Optical Imaging Agent. Molecular imaging 16, doi: 10.1177/1536012117723786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carney B, Kossatz S & Reiner T Molecular Imaging of PARP. Journal of Nuclear Medicine 58, 1025–1030, doi: 10.2967/jnumed.117.189936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thurber GM, Reiner T, Yang KS, Kohler RH & Weissleder R Effect of small-molecule modification on single-cell pharmacokinetics of PARP inhibitors. Molecular cancer therapeutics 13, 986–995, doi: 10.1158/1535-7163.MCT-13-0801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Dam GM et al. Intraoperative tumour-specific fluorescence imaging in ovarian cancer by folate receptor-alpha targeting: first in-human results. Nature medicine 17, 1315–1319, doi: 10.1038/nm.2472 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Tummers WS et al. Recommendations for reporting on emerging optical imaging agents to promote clinical approval. Theranostics 8, 5336–5347, doi: 10.7150/thno.27384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carney B et al. Target engagement imaging of PARP inhibitors in small-cell lung cancer. Nature communications 9, 176, doi: 10.1038/s41467-017-02096-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kossatz S, Weber WA & Reiner T Optical Imaging of PARP1 in Response to Radiation in Oral Squamous Cell Carcinoma. PloS one 11, e0147752, doi: 10.1371/journal.pone.0147752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kossatz S et al. Direct Imaging of Drug Distribution and Target Engagement of the PARP Inhibitor Rucaparib. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 59, 1316–1320, doi: 10.2967/jnumed.117.205765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the paper and its supplementary information files. Associated raw data and step-by-step protocols can be made available from the corresponding author on reasonable request.


