Abstract
Dgcr8 is involved in the biogenesis of canonical miRNAs to process pri-miRNA into pre-miRNA. Previous studies have provided evidence that Dgcr8 plays an essential role in different biological processes. However, the function of maternal and zygotic Dgcr8 in early embryonic development remains largely unknown. Recently, we have reported a novel approach for generating germline-specific deletions in zebrafish. This germline knockout model offers an opportunity to investigate into the differential roles of maternal or zygotic Dgcr8. Although germline specific dgcr8 deletion has no influence on gonad development, maternal or zygotic dgcr8 is essential for embryonic development in the offspring. Both maternal dgcr8 (Mdgcr8) and maternal zygotic dgcr8 (MZdgcr8) mutants display multiple developmental defects and die within 1 week. Moreover, MZdcgr8 mutant displays more severe morphogenesis defects. However, when a miR-430 duplex (the most abundantly expressed miRNA in early embryonic stage) is used to rescue the maternal mutant phenotype, the Mdgcr8 embryos could be rescued successfully and grow into adulthood and achieve sexual maturation, whereas the MZdgcr8 embryos are only partially rescued and they all die within 1 week. The differential phenotypes between the Mdgcr8 and MZdgcr8 embryos provide us with an opportunity to study the roles of individual miRNAs during early development.
Keywords: maternal dgcr8, zygotic dgcr8, microRNAs, zebrafish, embryonic development
Introduction
The model organism zebrafish (Danio rerio) is an excellent system for studying the developmental process of organogenesis. However, its application is limited due to the lack of a conditional knockout (cKO) platform. Recently, we have developed a novel approach for achieving cKO in zebrafish (Liu et al., 2017), making it possible to study the maternal genes in specific tissues or cells. Using this platform, maternal gene knockout zebrafish can be constructed by specifically deleting maternal genes in the oocytes. Furthermore, maternal mutant models cannot be constructed using the usual knockout methods because of the lethality of maternal homozygous gene knockouts. Therefore, our approach makes it possible to study the function of maternal factors during embryonic development in zebrafish.
Generally, maternal factors are important components in early embryonic development during the cleavage stage to maintain normal meiosis prior to the activation of the embryonic genome (Dosch et al., 2004; Wagner et al., 2004; Harvey et al., 2013). In zebrafish, the first maternal-effect mutant called janus was discovered, and this mutant appears to have a phenotype with a partially penetrant axis duplication, but the mechanism of this phenotypic change was not known at that time (Abdelilah et al., 1994). Subsequently, numerous maternal effect genes were discovered in the 1990s (Haffter et al., 1996; Kane et al., 1996; Gritsman et al., 1999; Miller-Bertaglio et al., 1999; Sirotkin et al., 2000). In mammals, thanks to the application of gene knock-out technology in mice, many studies have demonstrated that mammalian embryogenesis also needs maternal regulation, and numerous maternal factors have been identified in mice (Christians et al., 2000; Tong et al., 2000; Payer et al., 2003; Wu et al., 2003; Wagner et al., 2004).
Usually, maternal mutants have no influence on the mother, but normal zygotic development is seriously disrupted even if the mutated allele fails to pass to the offspring (Marlow, 2010). In zebrafish, several maternal genes have been identified by mutagenesis screening (Solnica-Krezel et al., 1996; Dosch et al., 2004; Wagner et al., 2004). Studies of the maternal mutants have revealed that maternal factors not only promote early embryonic cell division but also direct cell fate and organize the embryonic body plan (Tadros and Lipshitz, 2009; Marlow, 2010).
In the present study, we have used a number of approaches to examine the difference in Mdgcr8 and MZdgcr8 after rescuing. The absence of maternal Dgcr8 resulted in severe defects, including the disruption of gastrulation or epiboly movement, brain malformation, hematopoiesis defects, heart defects, and body curvature changes. Our findings also demonstrate that most of these defects could be rescued except hematopoiesis and heart defects in MZdgcr8. Further analysis demonstrated that miR-430 partially rescued hematopoiesis and heart function, suggesting that a multitude of miRNAs including miR-430 are the key factors in maintaining development of the heart and hematopoiesis system in early embryonic development.
Materials and Methods
Generation of Mdgcr8 and MZdgcr8 Zebrafish
We obtained Mdgcr8 and MZdgcr8 mutant zebrafish using kop: Cre-UTR-nanos3, dgcr8 cKO females crossed with wild-type male and kop: Cre-UTR-nanos3, dgcr8 cKO male, respectively. Mdgcr8 and MZdgcr8 embryos were collected from natural spawning in the zebrafish facility system with a 14 h/10 h light/dark cycle, and embryos were collected and kept in 28.5°C incubators with a light/dark cycle. The developmental staging of the embryos was classified according to the universal principle (Kimmel et al., 1995).
RNA Isolation and Real-Time PCR
Total RNA samples were isolated from zebrafish embryos at several developmental stages (shield, 75%-epiboly, prim-6, prim-16) with an RNeasy Mini Kit (Qiagen). The amount and purity of the RNA samples were determined by NanoDrop 2000 spectrophotometry (Thermo Fisher Scientific). The cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara). Real-time PCR was performed on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems) using the SYBR Green I Kit (Applied Biosystem). The primers used in this study are listed in Supplementary Table S1. The mRNA transcript levels were normalized against the ef14α transcript level.
Whole-Mount in situ Hybridization
Whole mount in situ hybridization (ISH) was performed as described (Li et al., 2014). cDNA fragments were amplified by RT-PCR with specific primers (Supplementary Table S1), followed by in vitro transcription with either T7 RNA polymerase to generate the antisense probe using the DIG RNA Labeling Kit (Roche, United States). Images were captured using a SZX16 stereomicroscope with fluorescence imaging (Olympus, Japan).
Morphological and Histological Analysis
After anesthetization and dissection of adult zebrafish, gonads including testes and ovaries were carefully obtained and then transferred into a culture dish containing 60% L-15 medium for investigation. After fixed in Bouin’s fixative buffer (Sigma, United States) or 4% PFA (Sigma, United States) overnight at 4°C, the gonad samples were dehydrated and embedded in paraffin, and then sectioned at 5 μm thickness on a Leica microtome. After drying overnight, the slides were stained with hematoxylin and eosin (H&E) according to the standard protocol (Sabaliauskas et al., 2006). Folliculogenesis (Wang and Ge, 2004) and spermatogenesis (Leal et al., 2009) were staged accordingly.
Mitotic Centrosome Detection
Embryos were fixed with 4% PFA overnight at 4°C and dehydrated in absolute methanol for at least 20 min at −20°C. After washing thrice with 1 × PBST, the embryos were then permeabilized in −20°C acetone for 8 min. After washing, the embryos were blocked with blocking buffer (2% lamb serum, 0.1% dimethyl sulfoxide, 0.1% bovine serum albumin, and 0.2% Triton-X100 in PBS) for 1 h at room temperature. The embryos were treated with anti-γ-tubulin antibody (Sigma) at a dilution of 1:1000 in blocking buffer overnight at 4°C. After washing with PBST for 30 min three to five times at room temperature, the embryos were incubated in fluorescein-conjugated secondary goat antirabbit antibody (Alexa-fluor 555, 1:500, Life Technology, United States) at 1:1000 dilution for 2 h at room temperature. The embryos were then washed eight times in PBST for 5 min each. The whole embryos were mounted in a confocal dish with DAPI Fluoromount-G medium (Southern Biotech) and incubated for 5 min prior to imaging.
Phosphorylation of Histone H3 (PH3) Antibody Staining
Zebrafish embryos were first fixed at 4°C in 4% PFA overnight. After washing three times with PBST (PBS + 0.2% Triton-X100) for 5 min each, the embryos were permeabilized for 8 min in −20°C acetone, washed in PBST three times, blocked for 30 min at room temperature with blocking buffer (2% lamb serum, 0.1% dimethyl sulfoxide, 0.1% bovine serum albumin, and 0.2% Triton-X100 in PBS), and then incubated overnight at 4°C in polyclonal rabbit anti-phospho-histone H3 antibody (Cell Signaling Technology, United States) at a concentration of 1:1000. After washing five times with PBST for 5 min each, the embryos were incubated for 2 h at room temperature in secondary goat antirabbit antibody (Alexa-fluor 555, 1:500, Life Technology, United States). Then, the embryos were washed eight times with PBST for 5 min each and processed for imaging. Changes in the numbers of mitotic cell were quantified by counting the number of phospho-histone 3 (PH3) positive cells in the whole body of the embryo taking the mean of three embryos.
Rescue of Mdgcr8 and MZdgcr8 Using miR-430 Duplex
To perform the rescue of the Mdgcr8 and MZdgcr8 phenotypes, the miR-430 duplex mimics were synthesized by Shanghai GenePharma Co., Ltd. Working solutions were prepared in RNase free water at 20 μM and stored at −20°C. For the rescue, 1–2 nl of miR-430 duplexes (a mixture of miR-430a, miR-430b, and miR-430c at a ratio of 1:1:1) were injected into one-cell stage Mdgcr8 and MZdgcr8 embryos. The phenotypic changes in embryo development were recorded on a stereomicroscope.
Quantifying Heart Rate
From the digital video of 52 h post fertilization (hpf) or 4 dpf embryos, the number of heartbeats were counted for 20 s. Heart rate (beats/min) was calculated by multiplying the number of beats counted by three. Four embryos were counted per group.
Touch Sensitivity Assay
At approximately 48 hpf of development, wild-type, Mdgcr8 and MZdgcr8 mutant embryos were touched at the trunk region adjacent to the yolk extension with a glass needle (Giraldez et al., 2005). The response was recorded for three consecutive stimuli and assessed by a touch response behavioral assay (Granato et al., 1996). No response was determined based on the absence of tail movement after touching the embryo three times. A weak response was determined as a wiggle of the tail after stimulation. A strong response was defined as when the fish tail gave a c-bend opposite to the site of the touch. The whole touch procedure was captured on digital video. Different groups of embryos were assessed twice.
O-Dianisidine Staining
O-dianisidine solution was prepared at 0.7 mg/ml in 100% ethanol and protected from exposure to light. The working stain solution was prepared by mixing 1 ml of 0.7 mg/ml O-dianisidine, 1 ml of ddH2O, 250 μl of 100 mM sodium acetate, and 50 μl of 30% hydrogen peroxide (Giraldez et al., 2005). After anesthetization, the embryos were transferred into a 24-well plate and 0.5 ml of the working solution was added. Embryos were incubated in the dark at room temperature for 20 min, washed three times with PBS, and fixed with 4% PFA overnight. Embryos were placed in 100% glycerol for imaging. The images were acquired on a SZX16 stereomicroscope microscope (Olympus, Japan).
mRNA Sequencing and Small RNAs Sequencing
For mRNA and small RNA sequencing, total RNA was extracted from MZdcgr8 and control embryos at 30% epiboly stage of early development representing 4.5 hpf using miRNeasy Mini Kit (Qiagen). RNA integrity was assessed on an Agilent Bioanalyzer 2100 (Agilent). Each group was consisted of 50 embryos. Qualified total RNA was further purified by RNA Clean XP Kit (Beckman) and RNase-Free DNase Set (Qiagen). Library construction and sequencing was performed by Shanghai Biotechnology Corporation. The 30% epiboly stage was chosen because it is a key period in MZT during early vertebrate development. At this stage, the embryo starts to shift from utilization of maternal mRNAs and factors, and initiates zygotic transcription.
Statistical Analysis
In this study, all raw data were analyzed by the GraphPad Instat software (GraphPad Software, United States). Mean values ± SEM. P < 0.05 were considered statistically significant using one-way ANOVA. Tukey test was used for multiple comparisons to determine statistical differences. All experiments were performed at least three times to confirm reproducibility.
Results
Germline Specific Dgcr8 Deletion Has No Functional Influence on Gonad Development
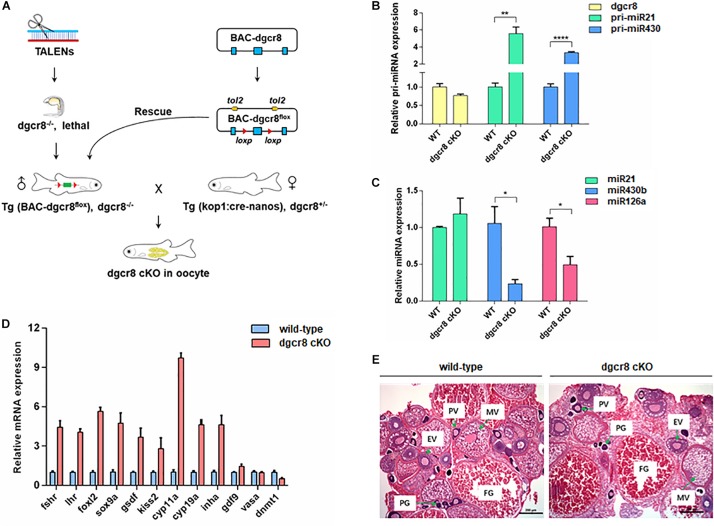
As shown in the schematic diagram in Figure 1A, we obtained germline specific dgcr8 cKO zebrafish using Tg(BAC-dgcr8flox), dgcr8–/– male crossed with Tg (kop: cre-UTR-nanos3, CMV: EGFP), dgcr8± female. To determine the expression levels of dgcr8 and individual miRNAs in adult ovary of the germline specific dgcr8 cKO, qRT-PCR was employed, and the results suggested that dgcr8 was not significantly downregulated, but primary miRNAs were upregulated significantly due to the absence of dgcr8 processing in the adult ovary (Figure 1B), and mature miRNAs were downregulated sharply because of disruption of the miRNA biogenesis pathway (Figure 1C). The majority of the germline markers were upregulated in the ovary of dgcr8 cKO except vasa and dnmt1 (Figure 1D). The dgcr8 cKO ovary morphology was also validated by histology, with all the ovarian follicle stages found in the dgcr8 cKO ovary with no obvious difference compared with the wild-type ovarian follicles (Figure 1E).
FIGURE 1.
Germline specific dgcr8 deletion has no functional influence on gonad development. (A) A basic schematic diagram of generating dgcr8 germline conditional knockout zebrafish. (B) Real-time analysis of the expression of dgcr8 mRNA and primary miRNAs in ovary of dgcr8 kop-cKO fish. (C) Real-time PCR analysis of the expression of mature miRNAs in ovary of dgcr8 kop-cKO fish. (D) Expression of germline markers in the ovary of dgcr8 kop-cKO. (E) Histology of ovaries in the wild-type and dgcr8 kop-cKO female zebrafish. *p < 0.05, **p < 0.01, and ****p < 0.0001.
MZdcgr8 Embryos Display More Severe Morphogenesis Defects Than Mdcgr8
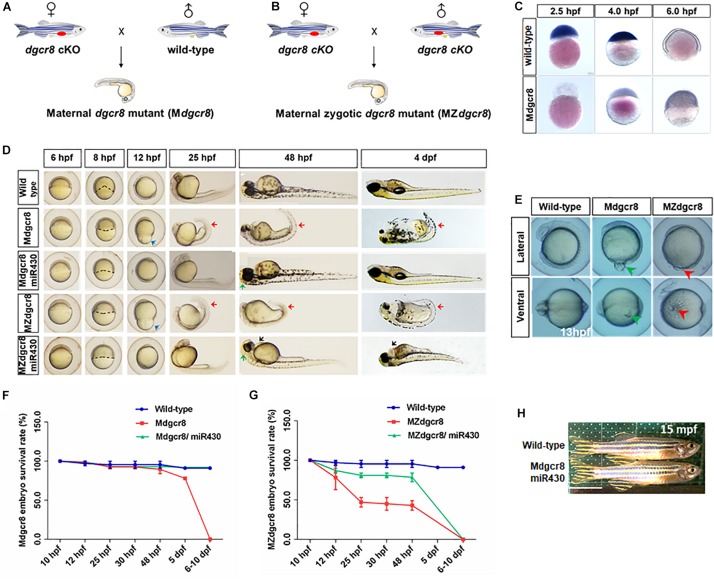
To further determine the necessity of maternal Dgcr8 in the early developmental process, we obtained a fish line with maternal Dgcr8 deletion (Mdgcr8 and MZdgcr8) by crossing dgcr8 cKO female fish with wild-type male fish (Figure 2A) and dgcr8 cKO male fish (Figure 2B). Mdgcr8 could provide direct evidence for the role of maternal factors because of the presence of zygotic Dgcr8 of paternal origin in the early developmental stage. The ISH signal of dgcr8 transcripts decreased from 2.5 to 6 hpf in the wild-type embryos (Figure 2C), but the signal in the Mdgcr8 embryos increased due to the presence of zygotic dgcr8 (Figure 2C). Real time-PCR analysis showed that dgcr8 transcripts were decreased during the developmental process from the 256-cell stage to the 75% epiboly stage in the MZdgcr8 embryos (Supplementary Figure S1).
FIGURE 2.
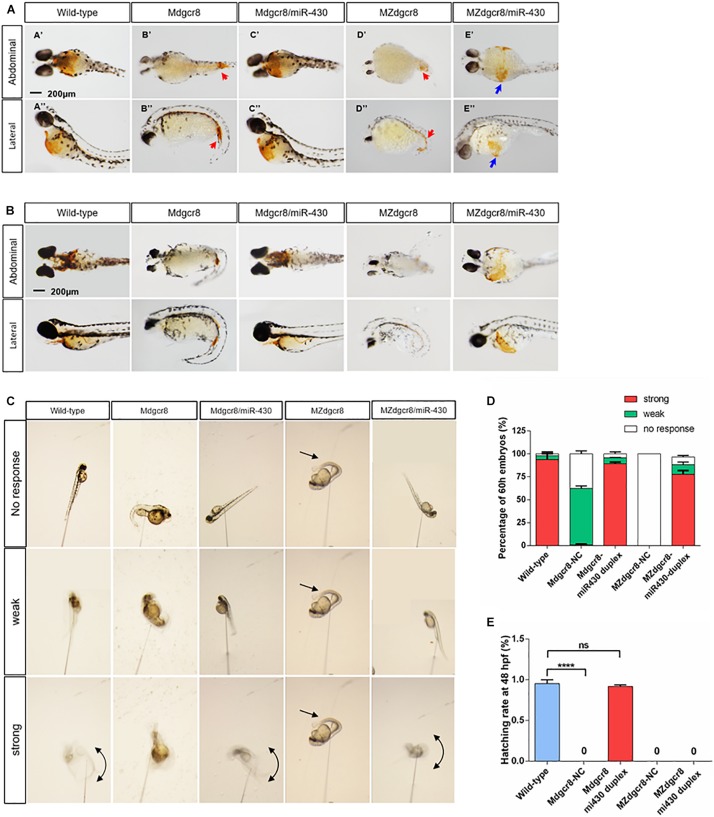
Rescue of Mdgcr8 and MZdgcr8 using miR-430 duplex. (A) A schematic diagram of generating Mdgcr8 mutant zebrafish by out-crossing with wild-type male. (B) A schematic diagram of generating MZdgcr8 mutant zebrafish by in-crossing. (C) The expression of dgcr8 was analyzed by in situ hybridization in early embryonic stage. (D) Mdgcr8 and MZdgcr8 exhibit gastrulation defect, brain malformations, body curvature, and heart developmental defects compared with wild-type. miR-430 duplex was used to rescue the defects. (E) Epiboly defects in Mdgcr8 and MZdgcr8. At 14 hpf, the phenotype of yolk excision could be observed in MZdgcr8 mutant (5 X), but not in Mdgcr8. (F) Mdgcr8 embryonic survival rate become normal after rescue. (G) MZdgcr8 mutant embryos only survived up to 6–10 dpf after rescue. (H) Mdgcr8 mutant embryos after rescue could develop to adults and exhibit proper sex ratio. Scale bar 1 cm.
From the shield stage onward, Mdgcr8 and MZdgcr8 mutant embryos exhibited development delay and developed more slowly than the wild-type embryos by approximately 3–4 h at 25 hpf (Figure 2D). Their epiboly movements were disrupted with a longer animal-vegetal axis but a shorter dorsal-ventral axis, and the epiboly level was decreased significantly compared to the wild-type embryos at 12 hpf (Figure 2D). Intriguingly, the part of the yolk was excised to induce the next developmental stage in MZdgcr8 mutants. The accumulation of cells in the region of the anterior axial mesendoderm showed a reduced extent at 13 hpf, but yolk excision did not happen in the Mdgcr8 mutant (Figure 2E), suggesting that zygotic dgcr8 might partially make up for the epiboly defects. From 25 hpf, MZdgcr8 and Mdgcr8 mutant embryos exhibited tail curvature until death on 6–10 dpf (Figure 2D).
miR-430 Duplex Successfully Rescues Mdgcr8 but Not MZdgcr8
In zebrafish, the miR-430 family is the most abundant miRNA family during early embryogenesis (Chen et al., 2005). During the maternal to zygotic transition (MZT), miR-430 is responsible for targeting and clearing more than 200 mRNAs in zebrafish embryos (Giraldez et al., 2006). In this study, we found that the Mdcgr8 mutant could be saved and they could even survive to adulthood after rescue, but MZdgcr8 could only be partially rescued and they died on 6–10 dpf (Figure 2D). At 8 hpf, the epiboly level of Mdgcr8 and MZdgcr8 decreased compared with the wild-type, but significantly improved after rescue (Figure 2D). In particular, the epiboly defects showing a longer animal-vegetal axis and a shorter dorsal-ventral axis (Figure 2D, blue arrows) were rescued completely by miR-430 duplex at 12 hpf. Furthermore, the body curvature (Figure 2D, red arrows) was rescued successfully in Mdgcr8 and MZdgcr8 at 25 and 48 hpf. However, miR-430 could rescue Mdgcr8 heart defects but failed to do so in the MZdgcr8 embryos and hydropericardium (hp) was observed clearly in the MZdgcr8/miR-430 group at 48 hpf (Figure 2D, black arrows). Moreover, the brain malformation was rescued successfully in Mdgcr8 and MZdgcr8 embryos at 48 hpf (Figure 2D).
After rescue, the Mdgcr8 embryonic survival rate became normal compared to the wild-type (Figure 2F), but no MZdgcr8/miR-430 embryo survived beyond 10 dpf (Figure 2G), indicating that dgcr8 or miRNAs other than miR-430 were necessary for the later developmental processes. At 15 months post fertilization (mpf), the morphology of the Mdgcr8/miR-430 mutants were normal and exhibited a proper sex ratio (male: female = 12:6) (Figure 2H), and they could also produce offspring normally by in-cross. Taken together, the miR-430 duplex could rescue Mdgcr8 embryos completely but not MZdgcr8.
Maternal Dgcr8 Regulates Mitosis During Early Embryonic Development
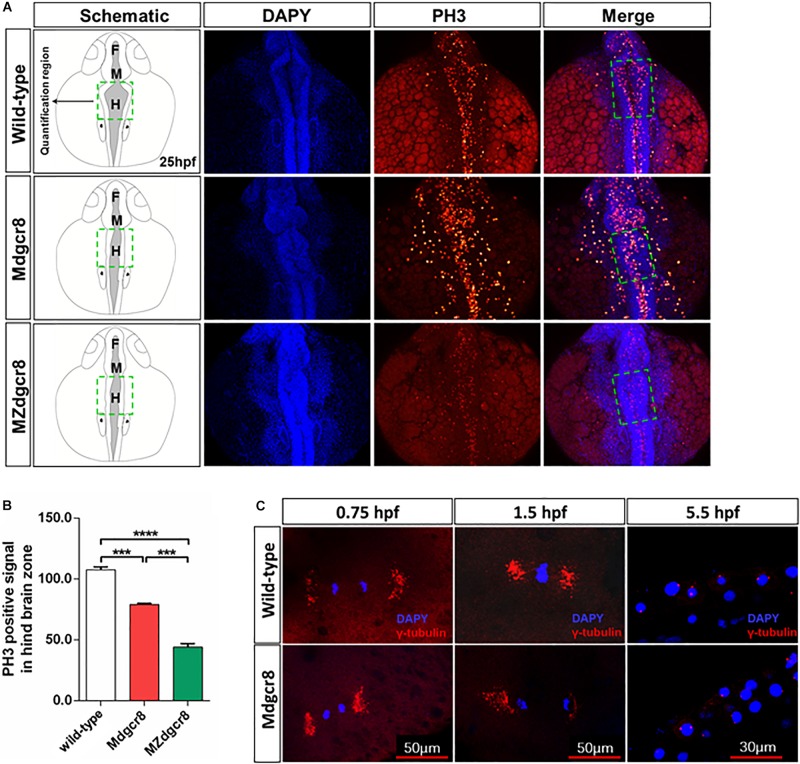
To investigate the developmental delay in Mdgcr8 mutant embryos, proliferation was assayed by a PH3 immunolabeling G2/M phase mitosis marker. At 25 hpf, the hindbrain zone of the Mdgcr8 embryos showed significant decreases in the PH3 immunolabeling of the positive cells compared to wild-type embryos, and MZdgcr8 exhibited a significant decrease in comparison with Mdgcr8 (Figures 3A,B). Moreover, the body of Mdgcr8 and MZdgcr8 were smaller than the wild-type embryos (Figure 3A). To further examine the organization of maternal mutant mitotic centrosomes, γ-tubulin immunostaining was used. Compared with wild-type, Mdgcr8 centrosomes appeared normal at 0.75, 1.5, and 5.5 hpf during early developmental stage (Figure 3C). These results suggested that maternal Dgcr8 might regulate cell proliferation through spindle organization other than centrosome organization.
FIGURE 3.
Maternal Dgcr8 regulates mitosis during early embryonic development in Mdgcr8 and MZdgcr8. (A) Representative images of 25 hpf WT, Mdgcr8 and MZdgcr8 embryos showing the distribution of PH3 immunolabeling cells (B) and quantification of positive cells in the hindbrain before the otic vesicle in the dashed box region (N = 3). (C) Mitotic centrosomes were detected using γ-tubulin immunostaining. ***p < 0.001 and ****p < 0.0001.
Loss of Maternal or Zygotic Dgcr8 Causes Cardiac Defects
In zebrafish, the cardiac progenitor cells are localized in the anterior lateral plate mesoderm as two parts on both sides of the embryo at the 5-somite stage (∼12 hpf). Then the two parts of the cardiac progenitor cells migrate toward the middle-line and combine together at the 18-somite stage (∼18 hpf). Next they reorganize to constitute a primitive heart tube that starts peristaltic contraction at the 26-somite stage (∼22 hpf), and finally develop into two cardiac chambers, namely, the atrium and ventricle, which become more organized at the long-pec stage (∼48 hpf) (Huang et al., 2009).
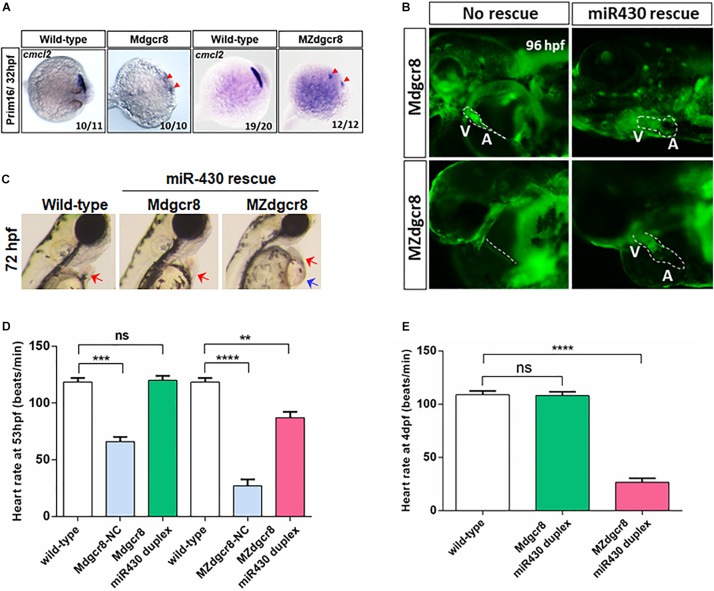
To investigate the process of cardiac development, we used ISH to detect the cardiac progenitor cells and found that these cells (marked by cmcl2) were specified but failed to migrate toward the middle-line at the Prim-16 stage (32 hpf) in the Mdgcr8 and MZ dgcr8 mutant, but wild-type embryos formed a complete heart tube (Figure 4A). In terms of morphology, the rescued Mdgcr8 exhibited a normal cardiac structure compared to the wild-type (Figures 4B,C), but the rescued MZdgcr8 only had a thinner cardiac tube and developed a hp syndrome (blue arrow) at 72 hpf (Figures 4B,C).
FIGURE 4.
Heart defects was successfully rescued in Mdgcr8, but not in MZdgcr8. (A) The cardiac progenitor cells failed to migrate to the middle-line at prim-16 stage (32 hpf) in Mdgcr8 and MZdgcr8 (red arrows). (B) After miR430 rescue, cardiogenesis appeared normal including structure of atrium and ventricle. (C) MZdgcr8 failed to be rescued by mir430, exhibiting hydropericardium phenotype. (D,E) Heart-rate results indicated that the heart defects were completely rescued in Mdgcr8, but only partially in MZdgcr8, and MZdgcr8 heart rate gradually decreased at 4 days. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To determine whether maternal Dgcr8 participates in heart development or cardiac function, we investigated the heart rate of the embryos. Mdgcr8 and MZdgcr8 had significantly decreased heart rates compared to the wild-type, but no significant changes were observed in the rescued Mdgcr8, suggesting that miR-430 duplex successfully rescued the heart defects (Figure 4D). The mean heart rate in MZdgcr8 embryos was only 26 beats/min and this could reach up to 87 beats/min after rescue, which was still much lower than the wild-type (118 beats/min), indicating that the heart defects could be rescued partially by miR-430 in MZdgcr8 (Figure 4D). At 4 dpf, the heart rate of the rescued Mdgcr8 was normal, but the heart rate of the rescued MZdgcr8 decreased gradually because of body malformations (Figure 4E). It could thus be concluded that both maternal and zygotic dgcr8 are essential for heart development and cardiac function.
Loss of Maternal or Zygotic Dgcr8 Leads to Hematopoietic Defects
In zebrafish, hematopoietic development is divided into two waves. The first wave, named the primitive wave, mainly encompasses induction of erythrocytes and myeloid cells. Primitive blood cells begin to circulate throughout the body of the embryo at 24 hpf (Orkin and Zon, 2008; Paik and Zon, 2010). The second wave is the definitive wave, which occurs in later development and mainly produces the hematopoietic stem and progenitor cells (HSPCs) and multipotent progenitors. Subsequently, HSPCs differentiate into all of the mature blood cells to maintain the embryos throughout life (Ellett and Lieschke, 2010).
To analyze functional hemoglobin in mature primitive erythrocytes in rescued embryos, we employed O-dianisidine staining and found that no O-dianisidine-positive erythroid cells were observed in the yolk sac, but staining accumulation was observed in the tail caudal vein of the maternal mutants at 50 hpf (Figures 5A–B’,B”D’,D”, red arrows). These phenotypes were consistent with qualitative visual observation of a slower circulation and decreased heart rate in Dgcr8 maternal mutant embryos. Dgcr8 maternal mutants displayed a developmental delay that became more severe as the embryos grew older, and O-dianisidine staining was observed at 74 hpf but with no improvement in hemoglobin function (Figure 5B). Moreover, the level of O-dianisidine staining in the 74 hpf mutant embryos was still lower than that of the wild-type at 48 hpf, indicating that the decrease in O-dianisidine staining was unlikely due to the developmental delay alone. After rescuing by the miR-430 duplex, Mdgcr8 could be rescued completely but was only partially rescued in MZdgcr8 (Figures 5A–C’,C”,E’,E”), indicating that the hematopoiesis defects were successfully rescued in Mdgcr8 but not in MZdgcr8.
FIGURE 5.
Maternal mutants exhibit reduced erythropoiesis and could be rescued in Mdgcr8, but not in MZdgcr8. Whole embryo O-dianisidine staining assay was used for assessing functional hemoglobin in mature primitive erythrocytes in wild-type, Mdgcr8, MZdgcr8, and miR-430 rescued groups at 50 (A) and 74 hpf (B). Hemoglobinized cells accumulate in the yolk sac in wild-type but mainly accumulate in the caudal vein (red arrows) in MZdgcr8 and MZdgcr8 mutants. 10–15 embryos per groups. Scale bar: 200 μm. (C) The touch response was categorized as strong, weak, and no response based on the analysis of the video frames. (D) At 60 hpf, the rescue rate was determined in wild-type and maternal mutant embryos. (E) At 48 hpf, the hatching rate was detected in wild-type and maternal mutant embryos. ****p < 0.0001.
Touch-Induced Response Decreases in Mdgcr8 and MZdgcr8
At approximately 2 dpf, the wild-type larvae hatch and start swimming under external stimulation. Swimming occurs by increasing the fin beat amplitude and rhythmic left-right axial cycle response of the tail (Granato et al., 1996). To investigate the role of Dgcr8 in neuronal development, a touch sensitivity assay was employed to determine the level of the escape response. At approximately 60 hpf, wild-type embryos responded to mechanosensory stimulation with a contralateral movement of the tail against where the stimulus was applied, and the tail then exhibited a characteristic c-bend (Granato et al., 1996). The touch response was then categorized as strong, weak, or no response. Analysis of the video frames showed that embryos were almost motionless and failed to respond to touch in Mdgcr8 (no response, 37.65%, n = 15) and MZdgcr8 (no response, 100%, n = 23); or generated a little twitch of the tail in Mdgcr8 (weak, 61%, n = 25) and MZdgcr8 (weak, 0%) (Figures 5C,D). Only 0.9% of the Mdgcr8 embryos displayed a strong response of the tail upon stimulation, and no MZdgcr8 embryos showed a strong movement (Figures 5C,D). After rescuing by the miR-430 duplex, the level of touch response improved significantly in both Mdgcr8 (strong, 89%, n = 14) and MZdgcr8 (strong, 77%, n = 15) embryos (Figures 5C,D). Moreover, the hatching rate of embryos is related to the level of spontaneous coiling movement inside the chorion in addition to the secreted proteolytic enzymes required for chorion-softening. Mdgcr8 and MZdgcr8 failed to hatch from the chorion naturally and no coiling movements were observed. Interestingly, the hatch rate of Mdgcr8 could be rescued completely but not the hatch rate of MZdgcr8 (Figure 5E). These results revealed essential roles of Dgcr8 during zebrafish neurogenesis.
Expression Changes of Downstream Genes in miRNA Biogenesis Pathway
In Mdgcr8 mutant embryos, maternal dgcr8 mRNA could not be detected in early developmental stage (Supplementary Figure S1), the expression of other genes downstream of the canonical miRNA biogenesis pathway (drosha, dicer1, xpo5, and ago2) was significantly upregulated at the 256-cell and sphere stage (Supplementary Figure S1). This might be the cumulative outcome of ablation of pre-miRNAs due to the lack of maternal Dgcr8. Surprisingly, such a cumulative phenotype was not found in the MZdgcr8 mutant (Supplementary Figure S1). The expression level of pri-miR-430 was nearly sixfold that of the wild-type at the sphere stage (Supplementary Figure S1). Pri-miR-430 in Mdgcr8 embryos was also accumulated abundantly due to the lack of Dgcr8 processing.
Similarly, dgcr8 mRNA was also not detected in the MZdgcr8 mutant embryos (Supplementary Figure S2), but the expression level of other genes downstream of the canonical miRNA biogenesis pathway was normal (Supplementary Figure S2). These results indicated that all of the miRNA biogenesis transcripts were of maternal origin and were decreased during the developmental process. However, the primary miRNAs were not of maternal origin, and they might begin to be transcribed during the MZT period. Pri-miR-430 was only detected in the sphere stage (Supplementary Figure S2), while expression of pri-miR-21 peaked in the shield stage (Supplementary Figure S2). Both pri-miR-430 and pri-miR-21 in MZdgcr8 embryos accumulated abundantly due to the lack of Dgcr8 processing compared to the wild-type embryos (Supplementary Figure S2). In conclusion, these results showed that the canonical miRNA biogenesis pathway was disrupted because of Dgcr8 ablation in MZdgcr8 mutant embryos.
Small RNA-Seq Analysis
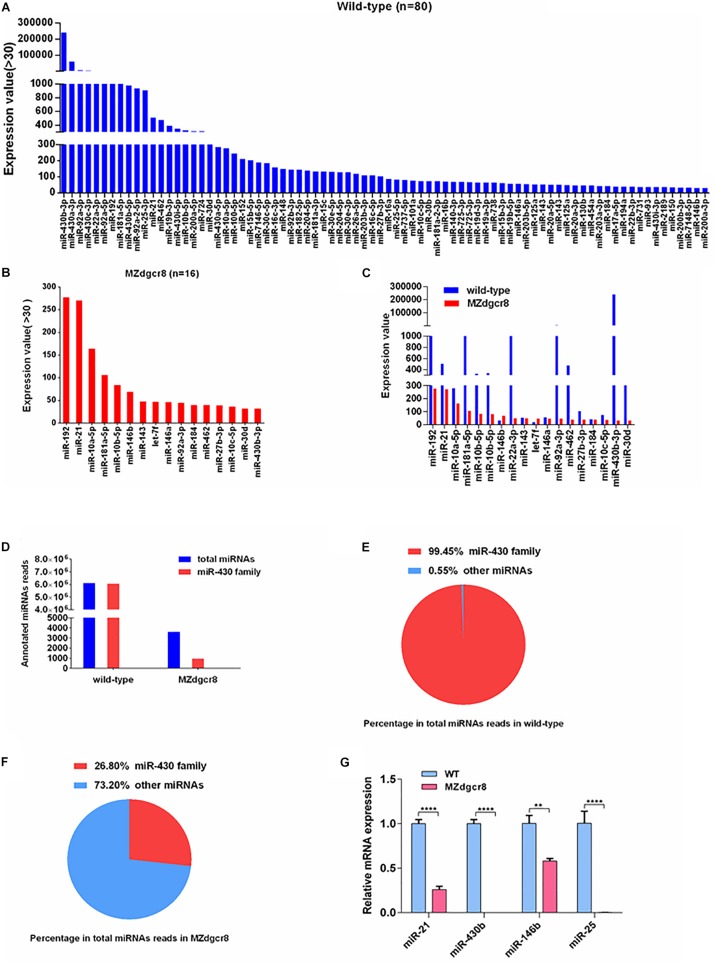
We have also analyzed the early embryonic transcriptomes and small RNA at the 30% epiboly stage (4.5 hpf) using small RNA-seq MZdgcr8 embryos. A total of 13,846,020 raw reads and 12,087,801 raw reads were obtained for the wild-type and MZdgcr8 embryos, respectively. We finally obtained 13,759,185 clean effective reads in wild-type and 12,011,626 in MZdgcr8 after removal of the reads of the adaptor sequence and low-quality sequences, including those smaller than 18 nt in length and smaller than 10 D in base molecular weight. By blasting with the Danio miRbase library, 5830 annotated counts were obtained in the wild-type embryos, but only 591 counts were annotated in MZdgcr8 (Supplementary Figures S3A,B). In the wild-type embryos, small RNAs with lengths of 23 nt were detected in the largest ratio, followed by 21 and 22 nt (Supplementary Figure S3C), and this is consistent with the 21–23 nt range of mature miRNAs. However, small RNAs with 21–23 nt in length failed to be detected in MZdgcr8 embryos (Supplementary Figure S3C). From the sum value reads, we also know that small nucleolar RNA (snoRNA) was downregulated significantly (Supplementary Figure S3D), and this is consistent with a previous report that DGCR8 regulated snoRNA biogenesis (Macias et al., 2012). These data revealed that almost all the miRNAs failed to express due to the ablation of maternal and zygotic Dgcr8 in early embryos.
To investigate the expression level or variation of specific miRNAs in early embryonic development, miRNAs with expression values higher than 30 (reads count) were chosen for further analysis. We obtained 80 miRNAs in the wild-type embryos and only 16 miRNAs in MZdgcr8 for values greater than 30 (Figures 6A,B). In wild-type embryos, the miR-430b is the most abundant miRNA (reads = 240,430) (Figure 6A) and in MZdgcr8 embryos, the most abundant miRNA is miR-192 (reads = 277) (Figure 6B). Although most of the abundant miRNAs were depleted, several miRNAs still displayed a high residual expression level in MZdgcr8 embryos, such as miR-192 (reads = 277), miR-21 (reads = 270), and miR-10a-5p (reads = 164) (Figure 6C). The residual expression of miR-21 was also validated using real-time PCR in this study (Figure 1C) although pri-miR21 was up-regulated about fivefold (Figure 1B). The residual expression of miR-21 was reported in the previous study in dgcr8 knockout mice (Yi et al., 2009). These results also revealed that there might be an alternative processing pathway independent of Dgcr8 for these miRNAs.
FIGURE 6.
Expression value of miRNAs in wild-type and MZdgcr8 embryos and qPCR validations. (A) 80 miRNAs for values greater than 30 were obtained in wild-type embryos. (B) 16 miRNAs for values greater than 30 were obtained in MZdgcr8 embryos. (C) miRNAs with higher values in wild-type and MZdgcr8. (D) Total miRNAs read count and miR-430 family read count in wild-type and MZdgcr8. (E) Percentage of miR-430 family in total miRNAs read count in wild-type. (F) Percentage of miR-430 family in total miRNAs read count in MZdgcr8. (G) Validation of some mature miRNAs by real-time PCR. **p < 0.01 and ****p < 0.0001.
The miR-430 family, the most abundant miRNAs family during early stages, accounted for 99.45% of all the annotated miRNA reads in wild-type embryos (Figures 6D,E). However, the miR-430 family was severely depleted in MZdgcr8 embryos (Figures 6D,F). These RNA-seq data suggested that the miR-430 family is the most important member in embryonic development. We have also validated the results of some individual mature miRNAs using real-time PCR (Figure 6G).
mRNA-Seq Analysis
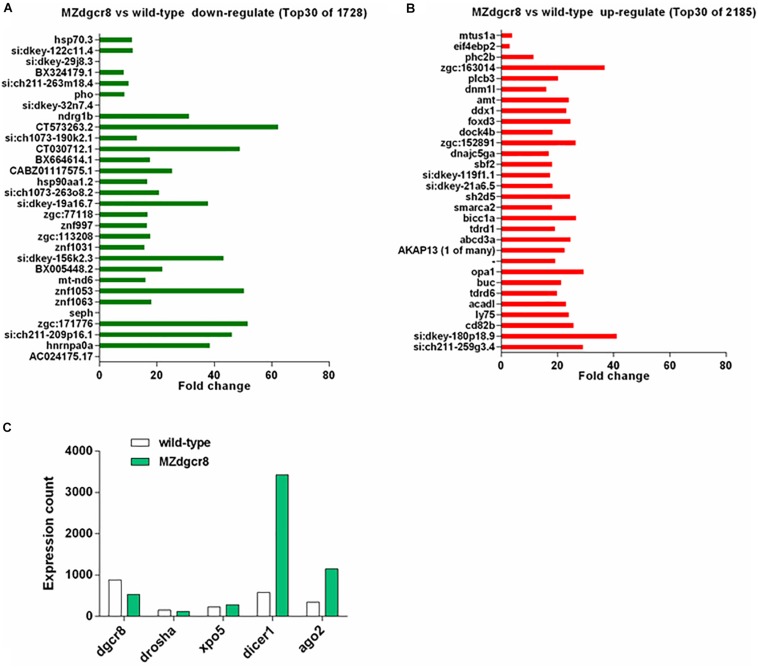
In addition to miRNAs, we also analyzed the mRNAs using RNA sequencing. We found that 2185 genes were upregulated, and 1728 genes were downregulated during the 30% epiboly stage (Figures 7A,B). According to the fold change, the top 30 of the upregulated or downregulated genes were further analyzed. The nucleoid binding protein hnrnpa0a decreased more than 30-fold (Figure 7A), while seph, an essential gene regulating organ development in zebrafish, was absent in the MZdgcr8 embryo (Figure 7A). For the upregulated genes, the targets of miR-430 (cd82b and gstm) were significantly upregulated in MZdgcr8, and this was verified using real-time PCR as described in our previous study (Liu et al., 2017). The opa1 required for proper mitochondrial metabolism in early development was also upregulated. During the 30% epiboly stage, the Dgcr8 mRNA could be detected in MZdgcr8 embryos after deleting Exon 3 and Exon 4 of the dgcr8 gene. The expression of other miRNA biogenesis genes including xpo5, dicer1, and ago2 was also increased (Figure 7C).
FIGURE 7.
mRNA expression variation in wild-type and MZdgcr8 embryos during 30% epiboly stage. (A) Top 30 downregulated genes from RNA-seq data. (B) Top 30 upregulated genes from RNA-seq data. (C) Expression count of miRNAs biogenesis related genes.
To assess whether maternal Dgcr8 was directly related to different biological processes or some cellular components, Kyoto Encyclopedia of Genes and Genomes (KEGG) gene enrichment analysis was performed based on annotations of the zebrafish genes (GRCz10, ENSEMBL). Through KEGG pathway analysis, the scatter plot results displayed highly significant enrichment of genes belonging to “the protein processing in endoplasmic reticulum” (Supplementary Figure S4), with a P-value of 0.00075. The protein processing pathway was activated due to the upregulation of related genes, and this might be induced by the accumulation of mRNAs owing to the deficiency of Dgcr8 or miRNAs.
Discussion
Mdgcr8 and MZdgcr8 mutants were generated using a BACK approach by germline specific deletion of Dgcr8 in zebrafish. Both Mdgcr8 and MZdgcr8 were malformed and finally causing embryo lethality. These observations indicate the essentiality of maternal Dgcr8 in early embryonic development. Previous studies observed that MZdicer zebrafish exhibit developmental defects in gastrulation, brain development, and heart development (Giraldez et al., 2005). Compared with other miRNA biogenesis enzymes, Dgcr8 is the only member that specifically processes miRNAs because Dicer is also responsible for processing other small endogenous RNAs. Our data suggest that the canonical miRNAs and other small RNAs processed by Dicer play important roles in early development.
We found that most gene expression patterns were similar in Mdgcr8 and MZdgcr8 embryos. Compared with MZdcgr8, the erythroid progenitors (marked by gata1) in Mdgcr8 were not severely affected at the 6-somite stage (12 hpf). Consistently, the erythroid cells were accumulated in the tail caudal vein of the Mdgcr8 embryos, but few erythroid cells were observed in MZdcgr8. These results suggest that the specification of erythroid progenitors is regulated by zygotic Dgcr8 in addition to maternal Dgcr8.
The miR-430 family is the most abundant miRNA expressed during early embryonic development, accounting for 99.45% of all the annotated miRNA reads at the 30% epiboly stage using miRNA sequencing. Therefore, we have employed a miR-430 mimic to rescue the Mdgcr8 and MZdgcr8 mutants and found that Mdgcr8 mutants could be completely rescued by miR-430, but MZdgcr8 could only be partially rescued. A previous study observed that the developmental defects in MZdicer mutants could be partially rescued by miR-430 (Giraldez et al., 2005), suggesting that miR-430 plays an important role in early development. However, rescue of the MZdgcr8 mutants by miR-430 is not sufficient for the MZdgcr8 mutant to survive to adulthood, indicating that other miRNAs processed by zygotic Dgcr8 are required for the later biological processes.
We further investigated the processes of neurogenesis, cardiogenesis, and hematopoiesis in the Mdgcr8 and MZdgcr8 mutants after miR-430 rescue. These results suggest that neurogenesis was completely rescued in both Mdgcr8 and MZdgcr8 mutants. Remarkably, brain morphogenesis was normal, and the touch response rate was not significantly changed in Mdgcr8 and MZdgcr8 mutants after rescue. It has been demonstrated that miR-430 could rescue brain morphogenesis successfully in MZdicer mutants (Giraldez et al., 2005). These observations indicate that miR-430 is both necessary and sufficient for neurogenesis. In contrast, the defects in the development of the heart and hematopoiesis were not rescued by miR-430 in MZdgcr8, consistent with previous study (Giraldez et al., 2005). It has been reported that miR-23 is essential for excessive endocardial cushion cell differentiation in zebrafish embryonic hearts (Lagendijk et al., 2011), and miR-218 mediates the formation of the linear heart tube in zebrafish during heart field migration (Fish et al., 2011). Studies have also demonstrated that miR-451 plays a crucial role in promoting erythroid maturation via its target transcript gata2 in hematopoiesis (Pase et al., 2009; Cifuentes et al., 2010). These observations show that other individual miRNAs besides miR-430 participate in the process of cardiogenesis and hematopoiesis in early development.
To investigate the role of Dgcr8 in miRNA processing, we sequenced the small RNAs at the 30% epiboly stage in MZdgcr8 mutant embryos. Comparing mature miRNA reads between wild-type and MZdgcr8 mutants revealed a large decrease (by 90%) of miRNA reads in the MZdgcr8 mutants. Interestingly, although most of the microRNAs were depleted in the early embryonic stage, three miRNAs still displayed a high residual expression level in MZdgcr8 embryos, including miR-192, miR-21, and miR-10a-5p. These results revealed that these miRNAs might have alternative miRNA processing pathways independent of Dgcr8. A similar conclusion was reached in another study, suggesting that residual expression of miR-21 was detected in Dgcr8 knockout mice (Yi et al., 2009).
Conclusion
In summary, we have demonstrated that Dgcr8 or global miRNAs are essential for embryonic development. And miR-430 was sufficient to rescue the Mdgcr8 mutant but only partially rescued the MZdgcr8. The MZdgcr8 rescue experiments provide us a platform for identifying the novel miRNA candidates in organogenesis, especially cardiogenesis and hematopoiesis.
Data Availability Statement
GEO reference number: GSE146606 is the reference Series for our publication: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE146606.
Ethics Statement
All animal procedures were approved by the Animal Experimentation Ethics Committee of the Chinese University of Hong Kong and were performed according to the animal license issued and endorsed by Department of Health, the Government of the Hong Kong Special Administrative Region.
Author Contributions
CC, ZZ, and YL conceived and designed the research. ZZ, YL, WX, TL, and YX developed the methods and performed the experiments. KS prepared the reagents. ZZ analyzed the data and wrote the manuscript. CC and OS edited the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Sun Yonghua for providing Tg(kop: Cre-UTR-nanos3, CMV:EGFP) fishline in the China Zebrafish Resource Center at the Chinese Academy of Sciences. We also thank the Core Laboratories in the School of Biomedical Sciences in the Chinese University of Hong Kong for the provision of equipment and technical support.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grant No: 81773939) and Shenzhen Knowledge Innovation Program-Basic Research Project (Grant No: JCYJ20170818141120342). This work was funded by the General Research Fund (CUHK 14113119) of the Hong Kong Research Grants Council.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00299/full#supplementary-material
References
- Abdelilah S., Solnica-Krezel L., Stainier D. Y., Driever W. (1994). Implications for dorsoventral axis determination from the zebrafish mutation janus. Nature 370 468–471. [DOI] [PubMed] [Google Scholar]
- Chen P. Y., Manninga H., Slanchev K., Chien M., Russo J. J., Ju J., et al. (2005). The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 19 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E., Davis A. A., Thomas S. D., Benjamin I. J. (2000). Embryonic development - Maternal effect of Hsf1 on reproductive success. Nature 407 693–694. [DOI] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D. W., Patnode H., Mishima Y., Cheloufi S., et al. (2010). A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328 1694–1698. 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch R., Wagner D. S., Mintzer K. A., Runke G., Wiemelt A. P., Mullins M. C. (2004). Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev. Cell 6 771–780. [DOI] [PubMed] [Google Scholar]
- Ellett F., Lieschke G. J. (2010). Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin.Pharmacol. 10 563–570. 10.1016/j.coph.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Fish J. E., Wythe J. D., Xiao T., Bruneau B. G., Stainier D. Y. R., Srivastava D., et al. (2011). A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 138 1409–1419. 10.1242/dev.060046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308 833–838. [DOI] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312 75–79. [DOI] [PubMed] [Google Scholar]
- Granato M., Van Eeden F. J., Schach U., Trowe T., Brand M., Furutani-Seiki M., et al. (1996). Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123 399–413. [DOI] [PubMed] [Google Scholar]
- Gritsman K., Zhang J. J., Cheng S., Heckscher E., Talbot W. S., Schier A. F. (1999). The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97 121–132. [DOI] [PubMed] [Google Scholar]
- Haffter P., Granato M., Brand M., Mullins M. C., Hammerschmidt M., Kane D. A., et al. (1996). The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123 1–36. [DOI] [PubMed] [Google Scholar]
- Harvey S. A., Sealy I., Kettleborough R., Fenyes F., White R., Stemple D., et al. (2013). Identification of the zebrafish maternal and paternal transcriptomes. Development 140 2703–2710. 10.1242/dev.095091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhang R., Xu X. (2009). Myofibrillogenesis in the developing zebrafish heart: a functional study of tnnt2. Dev. Biol. 331 237–249. 10.1016/j.ydbio.2009.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane D. A., Hammerschmidt M., Mullins M. C., Maischein H. M., Brand M., Vaneeden F. J. M., et al. (1996). The zebrafish epiboly mutants. Development 123 47–55. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203 253–310. [DOI] [PubMed] [Google Scholar]
- Lagendijk A. K., Goumans M. J., Burkhard S. B., Bakkers J. (2011). MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 109 649–657. 10.1161/CIRCRESAHA.111.247635 [DOI] [PubMed] [Google Scholar]
- Leal M. C., Cardoso E. R., Nobrega R. H., Batlouni S. R., Bogerd J., Franca L. R., et al. (2009). Histological and stereological evaluation of zebrafish (Danio rerio) spermatogenesis with an emphasis on spermatogonial generations. Biol. Reprod. 81 177–187. 10.1095/biolreprod.109.076299 [DOI] [PubMed] [Google Scholar]
- Li J., Wu P., Liu Y., Wang D., Cheng C. H. (2014). Temporal and spatial expression of the four Igf ligands and two Igf type 1 receptors in zebrafish during early embryonic development. Gene Expr. Patterns 15 104–111. 10.1016/j.gep.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhu Z., Ho I. H. T., Shi Y., Xie Y., Li J., et al. (2017). Germline-specific dgcr8 knockout in zebrafish using a BACK approach. Cell Mol. Life Sci. 74 2503–2511. 10.1007/s00018-017-2471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias S., Plass M., Stajuda A., Michlewski G., Eyras E., Caceres J. F. (2012). DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 19 760–766. 10.1038/nsmb.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow F. L. (2010). Maternal Control of Development in Vertebrates: My Mother Made Me Do It!. Fort Wayne, IN: Biota Publishing. [PubMed] [Google Scholar]
- Miller-Bertaglio V., Carmany-Rampey A., Furthauer M., Gonzalez E. M., Thisse C., Thisse B., et al. (1999). Maternal and zygotic activity of the zebrafish ogon locus antagonizes BMP signaling. Dev. Biol. 214 72–86. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132 631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik E. J., Zon L. I. (2010). Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 54 1127–1137. [DOI] [PubMed] [Google Scholar]
- Pase L., Layton J. E., Kloosterman W. P., Carradice D., Waterhouse P. M., Lieschke G. J. (2009). miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood 113 1794–1804. 10.1182/blood-2008-05-155812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Saitou M., Barton S. C., Thresher R., Dixon J. P. C., Zahn D., et al. (2003). stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13 2110–2117. [DOI] [PubMed] [Google Scholar]
- Sabaliauskas N. A., Foutz C. A., Mest J. R., Budgeon L. R., Sidor A. T., Gershenson J. A., et al. (2006). High-throughput zebrafish histology. Methods 39 246–254. [DOI] [PubMed] [Google Scholar]
- Sirotkin H. I., Gates M. A., Kelly P. D., Schier A. F., Talbot W. S. (2000). fast1 is required for the development of dorsal axial structures in zebrafish. Curr. Biol. 10 1051–1054. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L., Stemple D. L., Mountcastle-Shah E., Rangini Z., Neuhauss S. C., Malicki J., et al. (1996). Mutations affecting cell fates and cellular rearrangements during gastrulation in zebrafish. Development 123 67–80. [DOI] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tong Z. B., Gold L., Pfeifer K. E., Dorward H., Lee E., Bondy C. A., et al. (2000). Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 26 267–268. [DOI] [PubMed] [Google Scholar]
- Wagner D. S., Dosch R., Mintzer K. A., Wiemelt A. P., Mullins M. C. (2004). Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev. Cell 6 781–790. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ge W. (2004). Developmental profiles of activin betaA, betaB, and follistatin expression in the zebrafish ovary: evidence for their differential roles during sexual maturation and ovulatory cycle. Biol. Reprod. 71 2056–2064. [DOI] [PubMed] [Google Scholar]
- Wu X. M., Viveiros M. M., Eppig J. J., Bai Y. C., Fitzpatrick S. L., Matzuk M. M. (2003). Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 33 187–191. [DOI] [PubMed] [Google Scholar]
- Yi R., Pasolli H. A., Landthaler M., Hafner M., Ojo T., Sheridan R., et al. (2009). DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl. Acad. Sci. U.S.A. 106 498–502. 10.1073/pnas.0810766105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEO reference number: GSE146606 is the reference Series for our publication: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE146606.