Abstract
KRAS signaling is associated with cancer progression in several cancers. Upregulation of KRAS signaling is often seen in cancers that harbor high KRAS mutation rate, such as pancreatic cancer and non-small cell lung cancer (NSCLC). Less than 2% of breast cancers have KRAS mutation, however, the alteration of the effector signaling such as PI3K/AKT and MAPK pathways are well known. Mutated KRAS is known to function as immune suppressor in other cancers, but the role of KRAS signaling on tumor immune microenvironment (TIME) in breast cancer is not known. We hypothesize that the enrichment of KRAS signaling is associated with reduced patient survival as well as TIME in triple negative breast cancer (TNBC). Patient cohorts from Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; n = 1903) and The Cancer Genome Atlas (TCGA; n = 982) were used. Higher expression of KRAS in breast cancer cell-lines (MCF7, BT474, and MDA-MB231) compared to MCF10A, which is a model of benign mammary cells was found. Both MEK and PI3K inhibitors suppressed MB231 cell proliferation in dose dependent manner. Gene Set Variant Analysis (GSVA) of the patient cohorts demonstrated two peaks by KRAS_SIGNALING_UP gene sets which were divided into KRAS-high and -low groups using median cutoff. There was no difference in KRAS mutation between KRAS-high and low. Despite its cell proliferation promoting role, KRAS-high patients demonstrated significantly better Disease-Free Survival and Overall Survival in triple negative breast cancer (TNBC). KRAS-high TNBC was associated with favorable tumor immune microenvironment with elevated B cells and CD8 T cells, monocytes, or M1 macrophage. It was associated with decreased CD4 central memory T-cells, but not Regulatory T-cells, or M2 macrophage detected by xCell. To elucidate the mechanism of this association, Gene Set Enrichment Analysis was performed. Inflammatory response, IL6/JAK-STAT3 signaling, and Interferon gamma response gene sets were enriched in KRAS-high TNBC patients in both METABRIC and TCGA cohorts. In agreement, cytolytic activity score, interferon gamma response score, and lymphocyte infiltrating signature score, were all significantly elevated in KRAS-high TNBC. In conclusion, we found that patients with enrichment of KRAS signaling gene sets were associated with inflammation and favorable tumor immune microenvironment as well as improved survival in TNBC.
Keywords: KRAS, triple negative, breast cancer, tumor immune microenvironment, KRAS signaling, TCGA, METABRIC, adaptive immune cells, innate immune cells
Introduction
KRAS signaling is associated with cancer progression in several cancers [1,2]. Upregulation of KRAS signaling is often seen in cancers that harbor high KRAS mutation rate, such as pancreatic cancer and non-small cell lung cancer (NSCLC) [3-5]. In breast cancer, only less than 2% were reported to have KRAS mutation [6]; however, alteration of the effector signaling such as PI3K/AKT and MAPK pathways are well known [7]. PI3K/AKT signaling pathway is the most aberrant pathway in breast cancer. Although the alteration of this pathway is mainly observed in hormone receptor positive cancer, recent studies reveal that the alteration is also found in approximately 30% of triple negative breast cancer (TNBC) patients [8-10]. In general, TNBC is the most aggressive and lethal breast cancer subtype. The role of tumor immune microenvironment (TIME) in TNBC is well recognized [11]. The efficacy of adjuvant and neoadjuvant chemotherapy and prognosis is associated with infiltration of immune cells in TNBC patients [11,12]. KRAS mutant cells have been reported to create an immunosuppressive tumor microenvironment in colorectal cancer, making them less responsive to immune check point inhibition in mice model [13]. Also, other cancers which harbor high KRAS mutation rate such as pancreatic cancer and NSCLC are reported to have immunosuppressive tumor microenvironment [14]. Unlike those cancers the mutation rate of KRAS in breast cancer is low. Therefore, the clinical relevance of upregulation of KRAS signaling pathways remains elusive.
Gene set enrichment analysis (GSEA) is a computational algorithm that enables one to analyze the molecular profiles of the data set [15]. This analysis assesses the effect of certain gene to the biological activity of gene set of interest [16,17]. Gene Set Variant Analysis (GSVA) is the analysis to further explore the biological activity of a signaling pathway [18]. Utilizing GSVA, we can line up the patients in the order of how much KRAS signaling is upregulated.
In the current study, we hypothesize that the upregulation of KRAS signaling is associated with TNBC poor survival and its tumor immune microenvironment.
Materials and methods
Cell culture
MCF10A, MCF7, SKBR3, BT474, and MB231 cells were obtained from the JCRB (Japanese Collection of Research Bioresources) Cell Bank. All cells were cultured in accordance to the manufacturer’s guidelines.
Inhibitor experiment and cell viability
On the day before the administration of inhibitors, MB231 cells were seeded into 6-well plates to a concentration of 0.5×105/well. PI3K inhibitor LY294002 (CST, USA) and MEK inhibitor PD98059 (Calbiochem, USA) were used for the transfection of the cells as previously reported [19].
The trypan-blue dye exclusion test was performed to examine the percentage of viable cells.
Clinical data acquisition
The gene expression levels of Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) as well as The Cancer Genome Atlas (TCGA) were retrieved through cBioPortal as described previously [20-22]. Total of 1903 patients from METABRIC cohort and 982 patients from TCGA cohorts were included in this study. The Institutional Review Board at Roswell Park Comprehensive Cancer Center was waived for this study because we used the de-identified and publicly available data bases such as METABRIC and TCGA.
Gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA)
Gene Set Variation Analysis (GSVA) is a method to estimate variation of gene set enrichment through the samples of expression data set [18]. The patients were divided by the median cutoff of GSVA scoring of KRAS_SIGNALING_UP gene set variation analysis as either KRAS-high or -low group.
Gene Set Enrichment Analysis (GSEA) was conducted in order to examine the relationship between KRAS_SIGNALING_UP and other Hallmark gene sets. This analysis was performed using the publicly available software provided by Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) as previously reported [15-17,22-26].
xCell, cytolytic activity score (CYT) and other immunological factors
As previously described, we used a computational algorithm, xCell to estimate the cell composition of immune cells within tumor [27]. CYT score was calculated using the expression values of granzyme A and perforin as previously reported [20,21,24,28-31]. IFN-gamma response scoring and Lymphococyte Signature scoring were utilized as described previously [32-34].
Statistical analyses
All of the statistical analyses were conducted by using R software (http://www.r-project.org/), Bioconductor (https://www.r-project.org/). For the survival analysis, Kaplan-Meier method with the log-rank test was performed to compare the survival curves between KRAS_SIGNALING_UP high group and low groups as previously described [35]. In all analyses of this study, P < 0.05 was considered statistically significant.
Results
KRAS was expressed higher in breast cancer cell-lines compared from MCF10A, and both MEK and PI3K inhibitors suppressed MB231 cell proliferation in dose dependent manner
To assess whether KRAS signaling is functioning in breast cancer cell lines, we compared the KRAS expression levels of breast cancer cell lines- MCF7, SKBR3, BT474, MB231 with MCF10A, which models a normal mammary cell. We found that the expression levels of KRAS were upregulated in MCF7, BT474, and MB231 cells when compared with MCF10A (Figure 1A). To further explore the function of KRAS signaling in breast cancer cells, we inhibited the downstream of KRAS pathway by administering PI3K inhibitor (LY294002) and MEK inhibitor (PD98059) and the number of viable cells were counted. As a result, both PI3K inhibitor and MEK inhibitor significantly suppressed the number of MB231 cells in dose dependent manner. These results suggest that KRAS is expressed higher in breast cancer cell-lines compared from MCF10A, and that KRAS signaling promote MB231 cell proliferation (Figure 1B).
Figure 1.
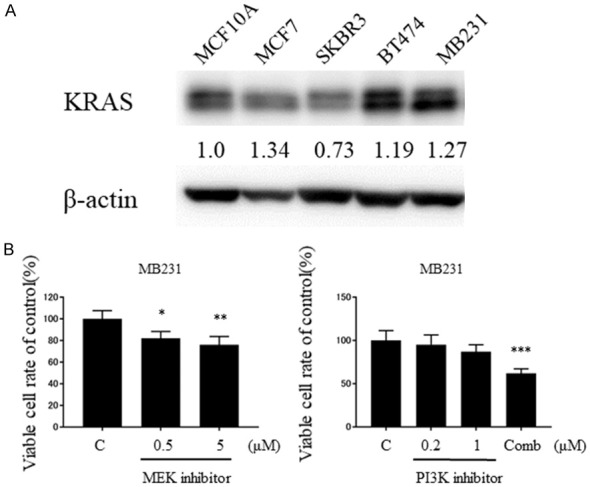
KRAS signaling was functioning in the breast cancer cell lines. A. Up-regulation of KRAS expression in breast cancer cell lines when compared with MCF10A. Densitometric values were calculated for KRAS. B. Cell growth suppression after transfection of MB231 cells with MEK inhibitor or PI3K inhibitor at 72 H. Comb: PI3Ki 1 + MEKi 5 (μM). *P < 0.05; **P < 0.01; ***P < 0.001.
Patient distribution with the gene sets KRAS_SIGNALING_UP scoring demonstrated bimodality and higher scores in TNBC
To investigate the clinical relevance of the KRAS signaling in breast cancer patients, gene sets variation analysis (GSVA) was performed to assess the patient distribution of KRAS_SIGNALING_UP gene sets. Interestingly, the histogram of METABRIC cohort demonstrated bimodality (Figure 2A). Since the histogram demonstrated two peaks, we chose median as cutoff for dividing the group into KRAS-high and KRAS-low groups.
Figure 2.
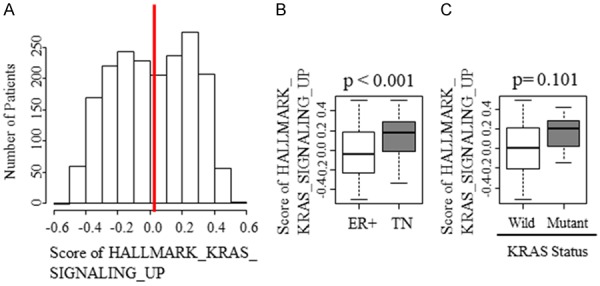
The patient distribution demonstrated bimodality with the scoring of gene set KRAS_SIGNALING_UP and higher scores in TNBC. A. Histogram of KRAS_SIGNALING_UP patients. Red line demonstrates median cutoff. B. The difference in scoring between estrogen receptor positive (ER+) and triple negative (TN) subgroups. C. The difference in scoring between KRAS wildtype and mutant.
We also found that the score of KRAS_SIGNALING_UP was higher in TNBC compared with estrogen receptor (ER) positive subgroup (Figure 2B). This result implied that KRAS signaling is more clinically relevant in TNBC.
Since KRAS mutation is highly relevant in other cancers, the relationship between KRAS mutation and upregulation of KRAS signaling was assessed, but there was no difference in KRAS_SCORING_UP score between KRAS wildtype and mutant (Figure 2C).
KRAS-high score was associated with improved disease-free survival (DFS) and overall survival (OS) in TNBC
Since KRAS signaling promotes cell proliferation in breast cancer, we expected that KRAS-high patients to have worse survival. To our surprise, KRAS-high group demonstrated significantly better disease-free survival when compared with low group in TNBC (Figure 3; P < 0.045). This was consistent with overall survival (OS) in TNBC. However, the whole cohort demonstrated significantly better survival with OS (Figure 3; P = 0.002) alone and not with DFS (Figure 3; P = 0.904).
Figure 3.
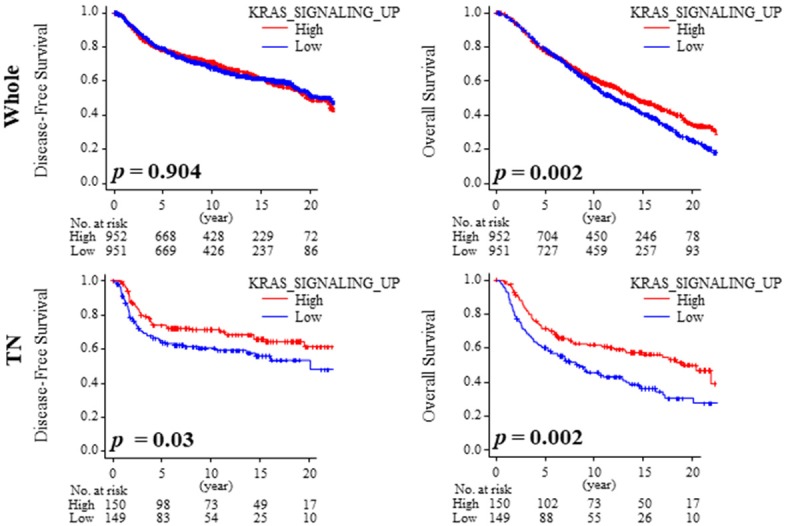
KRAS-high TNBC demonstrated favorable prognostic outcome. Kaplan-Meier analysis of Disease-Free Survival (DFS) and Overall Survival (OS) in whole and TNBC.
KRAS-high TNBC enrich immune related gene sets in both METABRIC and TCGA cohorts
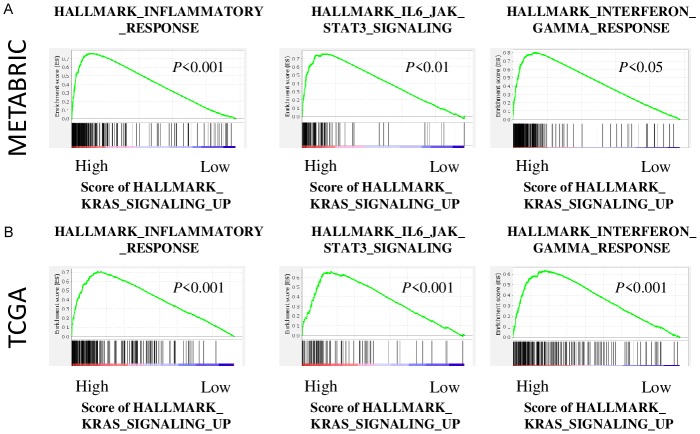
Given the unexpected better survival of KRAS-high patients, we investigated its mechanism by performing GSEA on TNBC, which has significantly high enrichment of KRAS signaling. Interestingly, KRAS-high group significantly enriched the immune related gene sets, INFLAMMATORY_RESPONSE (P < 0.001, NES = 1.78, FDR < 0.01), IL6_JAK_STAT3_SIGNALING (P < 0.01, NES = 1.71, FDR < 0.01), and INTERFERON_GAMMA_RESPONSE (P < 0.05, NES = 1.61, FDR < 0.05) compared with KRAS-low group in METABRIC cohort (Figure 4A). These results were validated with TCGA cohort, where INFLAMMATORY_RESPONSE (P < 0.001, NES = 2.57, FDR < 0.001), IL6_JAK_STAT3_SIGNALING (P < 0.001, NES = 2.33, FDR < 0.001), and INTERFERON_GAMMA_RESPONS (P < 0.001, NES = 2.15, FDR < 0.001) were significantly enriched in KRAS-high group (Figure 4B).
Figure 4.
KRAS-high group enriched immune related gene sets in both METABRIC and TCGA cohorts. A. The enriched gene sets of METABRC cohort. B. The enriched gene sets of TCGA cohort.
KRAS-high group demonstrated higher cytolytic activity score (CYT), interferon gamma response score and lymphocyte infiltrating signature score
To further elucidate the effect of KRAS signaling on cancer immunity, multiple scores that reflect immune activity were measured. Interferon (IFN) gamma response score, which also reflects immune activity, was significantly higher in KRAS-high group in both whole cohort and TNBC of TCGA cohort (Figure 5A; P < 0.001 and P < 0.05, respectively). The similar finding was observed in lymphocyte infiltrating signature score that reflect the amount of tumor infiltrating lymphocytes of METABRIC cohort (Figure 5B; P < 0.001 and P < 0.001, respectively). Furthermore, CYT score, which demonstrates immune cytolytic activity, strikingly echoed the results of GSEA as CYT scores of KRAS-high group were significantly higher in both whole cohort and TNBC compared with low group (Figure 5C; P < 0.001 and P < 0.001, respectively). This finding was validated with TCGA cohort (Figure 5D; P < 0.001 and P < 0.001, respectively). Taken together, cancer immunity is enhanced in KRAS-high TNBC.
Figure 5.
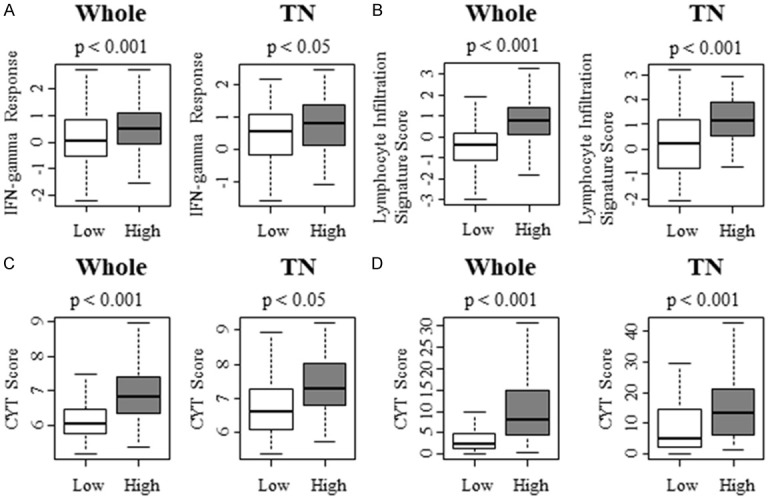
KRAS-high group demonstrated higher immune related scoring and cytolytic activity score (CYT) in whole and TN subgroup. A. IFN gamma Response Score of whole and TN subgroup with TCGA cohort. B. Lymphocyte Infiltrating Signature Score of whole and TN subgroup with TCGA cohort. C. Cytolytic activity score (CYT) of whole and TN subgroup with METABRIC cohort. D. CYT scoring of whole and TN subgroup with TCGA cohort.
KRAS-high TNBC is associated with anti-tumor immune microenvironment
Previous studies have demonstrated that tumor infiltrating lymphocyte are a positive predictive biomarker for breast cancer [36]. To this end, we hypothesize that KRAS-high TNBC attract adaptive and innate immune cells. We analyzed the intra-tumoral immune cell composition using a computational algorithm, xCell, on transcriptomic profiles of METABRIC and TCGA cohort. Interestingly, anti-tumor infiltrating lymphocytes, such as B cells and CD8 T cells were higher in KRAS-high TNBC with METABRIC cohort (Figure 6A; P < 0.001 and P < 0.001, respectively). These results were echoed with TCGA cohort (Figure 6B; P < 0.001 and P < 0.001, respectively). On the contrary, pro-cancer immune cells, CD4 central memory T cells (Tcm) were significantly lower in KRAS-high TNBC in both cohorts (Figure 6A; P < 0.001 and Figure 6B; P < 0.001, respectively), whereas regulatory T cells (Treg) were not consistent with two cohorts. With METBARIC, Treg was significantly higher in KRAS-high TNBC (Figure 6A; P < 0.001), however, that did not reflect in TCGA (Figure 6B; n.s.).
Figure 6.
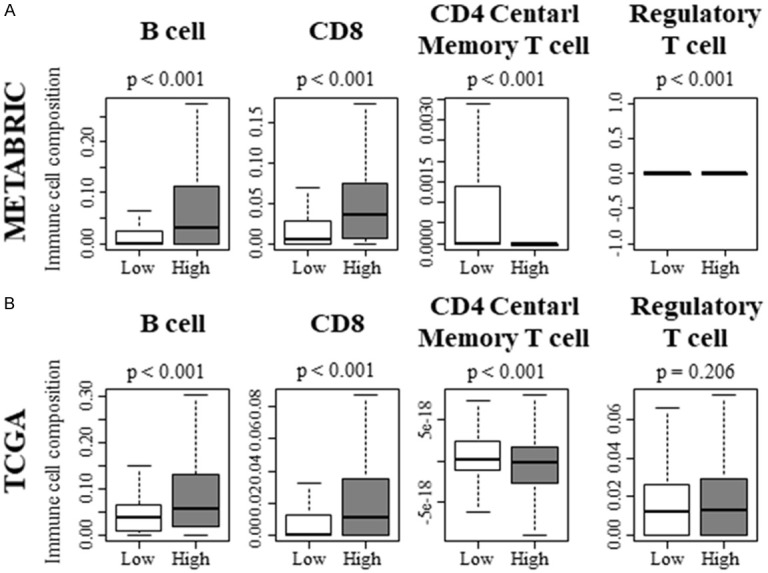
Adaptive immune cells contribute to the shift of tumor immune microenvironment toward anti-cancer environment in KRAS-high TNBC. A. Adaptive immune cell composition within a tumor of METABRIC cohort. B. Adaptive immune cell composition within a tumor of TCGA cohort.
Regarding the innate immune cells, monocytes and anti-tumor macrophage M1, pro-tumor macrophage M2 were significantly higher in KRAS-high TNBC with METABRIC cohort (Figure 7A; P < 0.001, P < 0.001 and P < 0.001, respectively). However, only monocytes and M1 were validated with TCGA cohort (Figure 7B; P < 0.001 and P < 0.001, respectively). These results implicate that KRAS-high TNBC possess higher overall anti-cancer immunity.
Figure 7.
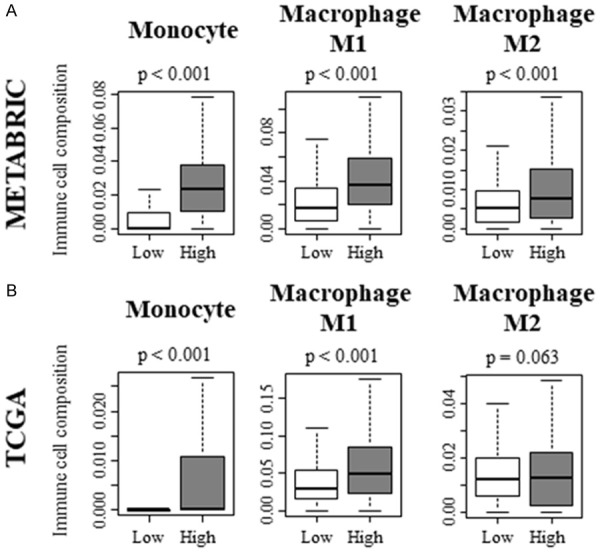
Innate immune cells contribute to the shift of tumor immune microenvironment toward anti-cancer environment in KRAS-high TNBC. A. Innate immune cell composition within a tumor of METABRIC cohort. B. Innate immune cell composition within a tumor of TCGA cohort.
Discussion
We demonstrated that KRAS is expressed higher in breast cancer cell-lines compared to MCF10A, and both MEK and PI3K inhibitors suppressed MB231 cell proliferation in dose dependent manner. GSVA demonstrated two peaks by KRAS_SIGNALING_UP gene sets and was divided into KRAS-high and -low groups using median cutoff. TNBC had significantly higher KRAS signaling compared with ER positive BC. There was no difference in KRAS mutation between those groups. Unexpectedly, we found that KRAS-high TNBC was associated with better DFS and OS. Utilizing xCell, KRAS signaling associate with anti-tumor immune microenvironment in TNBC. KRAS-high TNBC enriched immune related gene sets, inflammatory response, IL6/JAK-STAT3 signaling, interferon gamma response in both METABRIC and TCGA cohorts. To further clarify the contribution of KRAS signaling to the tumor immune microenvironment, we assessed CYT score, IFN gamma response score and lymphocyte infiltrating signature score which were significantly higher in KRAS-high group versus KRAS-low group in the cohorts analyzed.
To date, we have published multiple reports utilizing GSEA [22-25,30,37,38]. This analysis allows us to demonstrate how many biological pathways associate with the different gene expression levels of interest [15]. GSVA possesses the ability to detect the underlying mechanism over a sample population [18]. To best of our knowledge, this is the first study to utilize GSVA for elucidating the clinical relevance of KRAS signaling pathway and its association with TIME.
It was quite unexpected to find that TNBC with high KRAS signaling to be associated with better survival. Since it is well recognized that infiltration of certain immune cells is associated with better outcome for breast cancer patients, we hypothesized that KRAS-high TNBC is associated with favorable TIME. It is well established that infiltration of CD8 cells to the tumor microenvironment is associated with favorable prognostic outcome. Also, according to previous study, high infiltration of B cells is associated with better survival [39]. Our results demonstrated that KRAS-high TNBC exhibits high number of infiltrating B cells and CD8 T cells in both METABRIC and TCGA. Furthermore, the infiltration of M1, innate immune cells which associate with anti-cancer TIME, was higher with KRAS-high TNBC.
On the contrary, some immune cells act as pro-cancer TIME. The higher percentage of CD4 central memory T cell (Tcm) and regulatory T cell contribute to form the immunosuppressive TIME and hence worse outcome for breast cancer patients [40,41]. Also, high fraction of M2 macrophage has been reported to be associated with worse survival in breast cancer patients [42]. Of those pro-cancer immune cells, only Tcm was significantly upregulated in both METABRIC and TCGA cohorts. In agreement with previous reports, we found that lower percentage of Tcm in KRAS-high TNBC, which had better OS and DFS. KRAS-high TNBC demonstrated higher number of infiltrating Treg and M2 in METABRIC. However, these results were not supported in TCGA, where there was no significance between KRAS-high and KRAS-low TNBC. Overall, TIME shifted towards anti-tumor TIME and we speculate that this was reason behind those survival benefits in TNBC.
We found that KRAS signaling enriched cancer immunity related HALLMARK gene sets, such as, INFLAMMATORY_RESPONSE, IL6_JAK_STAT3_SIGNALING, and INTERFERON_GAMMA_RESPONSE. These results prompted us to further explore the association between KRAS signaling pathway and TIME by performing analysis of immune related scoring system. KRAS mutation is reported to be associated with the immunosuppressive TIME in other cancers [14]. However, in this current study upregulation of KRAS signaling was associated with better TIME in all and TNBC patients. The favorable TIME was demonstrated by the high scores of CYT scoring within KRAS-high group in both METABRIC and TCGA cohorts. This demonstrates that the ability to attack the cancer cells is increased in KRAS-high group. Also, this was consistent with IFN gamma response score and Lymphocyte Infiltrating Signature Score which suggest that the TIME shifts to anti-tumor immune microenvironment.
The current study has a few limitations. First, this study was retrospective study conducted using the public accessible databases, METABRIC and TCGA. Even though these databases offer significant amount of clinical information, these databases also lack some clinical data for certain number of patients. TNBC is less prevalent when compared to ER positive subtype, which may result in a decrease of reliability of the results.
In conclusion, we found that enrichment of genes related with KRAS signaling is associated with improved DFS and OS in TNBC patients. KRAS_SIGNALING_UP high TNBC was found to be associated with anti-tumor immune microenvironment, which was demonstrated by immune cell composition analysis, GSEA, CYT, interferon gamma response score as well as lymphocyte infiltration signature score.
Acknowledgements
This work was supported by NIH grant R01CA160688 to K.T., and National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Resources.
Disclosure of conflict of interest
None.
Abbreviations
- CYT
Cytolitic activity score
- METABRIC
Molecular Taxonomy of Breast Cancer International Consortium
- GSEA
Gene Set Enrichment Analysis
- GSVA
Gene Set Variant Analysis
- TCGA
The Cancer Genome Atlas
- Tcm
CD4 central memory T cell
- TN
Triple negative
- TNBC
Triple negative breast cancer
- Treg
Regulatory T cell
References
- 1.Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8 doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akao Y, Kumazaki M, Shinohara H, Sugito N, Kuranaga Y, Tsujino T, Yoshikawa Y, Kitade Y. Impairment of K-Ras signaling networks and increased efficacy of epidermal growth factor receptor inhibitors by a novel synthetic miR-143. Cancer Sci. 2018;109:1455–1467. doi: 10.1111/cas.13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy A, Pongor LS, Szabo A, Santarpia M, Gyorffy B. KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer. Int J Cancer. 2017;140:930–937. doi: 10.1002/ijc.30509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan P, He XH, Rong YF, Cao J, Li Y, Hu YP, Liu Y, Li D, Lou W, Liu MF. KRAS/NF-κB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res. 2017;77:100–111. doi: 10.1158/0008-5472.CAN-16-1898. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giltnane JM, Balko JM. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med. 2014;17:275–283. [PubMed] [Google Scholar]
- 8.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, Yamashita T, Lu YS, Inoue K, Takahashi M, Papai Z, Longin AS, Mills D, Wilke C, Hirawat S, Juric D. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 9.Cossu-Rocca P, Orru S, Muroni MR, Sanges F, Sotgiu G, Ena S, Pira G, Murgia L, Manca A, Uras MG, Sarobba MG, Urru S, De Miglio MR. Analysis of PIK3CA mutations and activation pathways in triple negative breast cancer. PLoS One. 2015;10:e0141763. doi: 10.1371/journal.pone.0141763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriegsmann M, Endris V, Wolf T, Pfarr N, Stenzinger A, Loibl S, Denkert C, Schneeweiss A, Budczies J, Sinn P, Weichert W. Mutational profiles in triple-negative breast cancer defined by ultradeep multigene sequencing show high rates of PI3K pathway alterations and clinically relevant entity subgroup specific differences. Oncotarget. 2014;5:9952–9965. doi: 10.18632/oncotarget.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgado R, Loi S. Tumour infiltrating lymphocytes in breast cancer: increasing clinical relevance. Lancet Oncol. 2018;19:3–5. doi: 10.1016/S1470-2045(17)30905-1. [DOI] [PubMed] [Google Scholar]
- 12.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, Loi S. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 13.Hanggi K, Ruffell B. Oncogenic KRAS drives immune suppression in colorectal cancer. Cancer Cell. 2019;35:535–537. doi: 10.1016/j.ccell.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Cullis J, Das S, Bar-Sagi D. Kras and tumor immunity: friend or foe? Cold Spring Harb Perspect Med. 2018;8 doi: 10.1101/cshperspect.a031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi H, Katsuta E, Yan L, Dasgupta S, Takabe K. High expression of Annexin A2 is associated with DNA repair, metabolic alteration, and worse survival in pancreatic ductal adenocarcinoma. Surgery. 2019;166:150–156. doi: 10.1016/j.surg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan S, Kawaguchi T, Peng X, Qi Q, Liu S, Yan L, Takabe K. Tumor infiltrating lymphocytes and macrophages improve survival in microsatellite unstable colorectal cancer. Sci Rep. 2019;9:13455. doi: 10.1038/s41598-019-49878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshikawa Y, Taniguchi K, Tsujino T, Heishima K, Inamoto T, Takai T, Minami K, Azuma H, Miyata K, Hayashi K, Kataoka K, Akao Y. Anti-cancer effects of a chemically modified miR-143 on bladder cancer by either systemic or intravesical treatment. Mol Ther Methods Clin Dev. 2019;13:290–302. doi: 10.1016/j.omtm.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeshita T, Yan L, Asaoka M, Rashid O, Takabe K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci Rep. 2019;9:16942. doi: 10.1038/s41598-019-53482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeshita T, Asaoka M, Katsuta E, Photiadis SJ, Narayanan S, Yan L, Takabe K. High expression of polo-like kinase 1 is associated with TP53 inactivation, DNA repair deficiency, and worse prognosis in ER positive Her2 negative breast cancer. Am J Transl Res. 2019;11:6507–6521. [PMC free article] [PubMed] [Google Scholar]
- 23.Hoki T, Katsuta E, Yan L, Takabe K, Ito F. Low DMT1 expression associates with increased oxidative phosphorylation and early recurrence in hepatocellular carcinoma. J Surg Res. 2019;234:343–352. doi: 10.1016/j.jss.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, Opyrchal M, Yan L, Patnaik S, Otsuji E, Takabe K. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019;26:2191–2199. doi: 10.1245/s10434-019-07338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsuta E, Maawy AA, Yan L, Takabe K. High expression of bone morphogenetic protein (BMP) 6 and BMP7 are associated with higher immune cell infiltration and better survival in estrogen receptor-positive breast cancer. Oncol Rep. 2019;42:1413–1421. doi: 10.3892/or.2019.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katsuta E, Yan L, Takeshita T, McDonald KA, Dasgupta S, Opyrchal M, Takabe K. High MYC mRNA expression is more clinically relevant than MYC DNA amplification in triple-negative breast cancer. Int J Mol Sci. 2019;21 doi: 10.3390/ijms21010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asaoka M, Ishikawa T, Takabe K, Patnaik SK. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20225621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf DM, Lenburg ME, Yau C, Boudreau A, van ’t Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9:e88309. doi: 10.1371/journal.pone.0088309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calabrò A, Beissbarth T, Kuner R, Stojanov M, Benner A, Asslaber M, Ploner F, Zatloukal K, Samonigg H, Poustka A, Sültmann H. Effects of infiltrating lymphocytes and estrogen receptor on gene expression and prognosis in breast cancer. Breast Cancer Res Treat. 2009;116:69–77. doi: 10.1007/s10549-008-0105-3. [DOI] [PubMed] [Google Scholar]
- 35.Terakawa T, Katsuta E, Yan L, Turaga N, McDonald KA, Fujisawa M, Guru KA, Takabe K. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget. 2018;9:14207–14218. doi: 10.18632/oncotarget.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, de Azambuja E, Quinaux E, Di Leo A, Michiels S, Piccart MJ, Sotiriou C. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi T, Yan L, Qi Q, Peng X, Edge SB, Young J, Yao S, Liu S, Otsuji E, Takabe K. Novel microRNA-based risk score identified by integrated analyses to predict metastasis and poor prognosis in breast cancer. Ann Surg Oncol. 2018;25:4037–4046. doi: 10.1245/s10434-018-6859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young J, Kawaguchi T, Yan L, Qi Q, Liu S, Takabe K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget. 2017;8:99978–99989. doi: 10.18632/oncotarget.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoud SM, Lee AH, Paish EC, Macmillan RD, Ellis IO, Green AR. The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Res Treat. 2012;132:545–553. doi: 10.1007/s10549-011-1620-1. [DOI] [PubMed] [Google Scholar]
- 40.Deng L, Lu D, Bai Y, Wang Y, Bu H, Zheng H. Immune profiles of tumor microenvironment and clinical prognosis among women with triple-negative breast cancer. Cancer Epidemiol Biomarkers Prev. 2019;28:1977–1985. doi: 10.1158/1055-9965.EPI-19-0469. [DOI] [PubMed] [Google Scholar]
- 41.Bates JP, Derakhshandeh R, Jones L, Webb TJ. Mechanisms of immune evasion in breast cancer. BMC Cancer. 2018;18:556. doi: 10.1186/s12885-018-4441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiainen S, Tumelius R, Rilla K, Hämäläinen K, Tammi M, Tammi R, Kosma VM, Oikari S, Auvinen P. High numbers of macrophages, especially M2-like (CD163-positive), correlate with hyaluronan accumulation and poor outcome in breast cancer. Histopathology. 2015;66:873–883. doi: 10.1111/his.12607. [DOI] [PubMed] [Google Scholar]