Abstract
The human microbiome, often termed as “the forgotten organ”, is an aggregation of microorganisms and their genomes that forms a mutualistic complex with the host. Recent research has shown the symbiotic merits of a microbiome ecosystem and its crucial role in the hosts’ physiological functions. Disruption of this symbiotic relationship is prone to cause a broad spectrum of ailments, including cancer. The compositional and environmental factors that tip the scales from beneficial co-existence to the development of malignancy is actively investigated. Herein we review the latest research in knowledge regarding the association between the vaginal microbiomes and oncogenesis, with a particular focus on ovarian carcinoma.
Keywords: Vaginal microbiome, ovarian cancer, cervical cancer, HPV, immune response
Introduction
The human microbiota is a consortium of bacteria that reside within different body sites and ecological niches, whereas the human microbiome is the collective genomes of all microbial species and their environment [1]. The plurality of these microbiotas subsists in a mutualistic association with their human host. Human microbiota has demonstrated a crucial role in our body’s immunity, metabolism, and endocrine [2,3]. Given the emphasis on microbiomes in gastrointestinal disease development [4], recent studies are beginning to support the interactive role of vaginal microbiomes in gynecological diseases. For example, Prevotella species are often associated with bacterial vaginosis and cervicitis [5,6], while vaginal bacterial communities dominated by Lactobacillus gasseri are correlated with increased clearance of HPV infection [7]. Lactobacillus iners are linked to an increased risk for Chlamydia trachomatis infection and also prevails in the occurrence of HPV infection and cervical intraepithelial neoplasia (CIN) [8-10]. Despite progress in understanding the role of the microbiomes in cervical cancer, investigations regarding the role of microbiome in ovarian cancers are limited. Ovarian cancer is the fifth most commonly diagnosed cancer among women in the United States [11]. Among them, ovarian cancer accounts for more death than any other reproductive cancer, with an estimated 22,530 new cases and nearly 13,980 death in 2019. Despite the high prevalence and public health significance, the etiology of this disease remains largely elusive.
Microbiomes are essential in the prevention of pathogen invasion; therefore disruption of the dynamics between the microbiome and the host vaginal ecosystem is prone to cause vaginal tract infection and cancer [12]. In this review, we discuss the possible roles of vaginal microbiota in carcinogenesis, highlighting the relationship of micro-organisms and viral infection in ovarian cancer.
Vaginal microbiota: an overview
Although the physiological role of the gut microbiota has been explored for decades [13], investigations of microbial compositions have recently extended to the female reproductive system (Table 1). Through molecular amplification techniques such as qPCR and DNA sequencing, studies have identified lactic acid-producing Lactobacillus crispatus, L. gasseri, L. iners and Lactobacillus jensenii, which dominate the vaginal communities of most reproductive-age healthy women [14]. Besides the Lactobacillus genus, a heterogeneous group of strictly anaerobic bacteria was also reported, including Prevotella, Atopobium, Gardnerella, Dialister, Sneathia, Megasphaera, Peptoniphilus, Finegoldia, Eggerthella, and Aerococcus. [15]. These microbial florae were classified as five main community state types (CSTs) by Ravel et al. [15].
Table 1.
Summary of major microbiome studies involving gynecological cancers
| Author | Disease | Microbiome specimen | Year | Microbiome evaluation | Microbial change |
|---|---|---|---|---|---|
| Shannon | HPV infection | Endocervical cytobrushes | 2017 | 16S rRNA sequencing | ↑Anaerobes |
| ↓L. gasseri, Fusobacterium nucleatum | |||||
| Brotman | HPV infection | Midvaginal swabs | 2014 | 16S rRNA sequencing | ↓Lactobacillus spp. |
| Chao | HPV infection | Posterior vaginal fornix | 2019 | 16S rRNA sequencing | ↑Leptotrichia and Prevetella |
| ↓ Lactobacillus spp. | |||||
| Lee | HPV infection | Endocervical brush | 2013 | 16S rRNA sequencing | ↑Sneathia spp. |
| Paola | HPV persistence | Cervico-vaginal samples | 2017 | 16S rRNA sequencing | ↑Atopobium spp., G. vaginalis |
| Wu | HPV persistence | Cyto-brush | 2018 | 16S rRNA sequencing | ↑Prevotella, Dialister |
| Adebamowo | HPV persistence | Mid-vaginal swabs | 2017 | 16S rRNA sequencing | ↑Mycoplasma hominis |
| Piyathilake | HSIL | Merocel ophthalmic sponges placed in cervical os | 2016 | 16S rRNA sequencing | ↑L. iners |
| Kwasniewski | LSIL | Cervical swabs | 2018 | 16S rRNA sequencing | ↑L. acidophilus, L. iners |
| ↓L. crispatus | |||||
| Kwasniewski | HSIL | Cervical swabs | 2018 | 16S rRNA sequencing | ↑G. vaginalis , L. acidophilus |
| ↓L. crispatus, L. taiwanensis, L. iners | |||||
| Oh | CIN | Cervical Sampler Brush | 2015 | 16S rRNA sequencing | ↑A. vaginae, L. iners, G. vaginalis |
| ↓L. crispatus | |||||
| Godoy-Vitorino | CIN | Cervical samples (posterior fornix) | 2018 | 16S rRNA sequencing | ↑A. vaginae, G. vaginalis |
| Bhatla | Cervical cancer | Cytobrush | 2013 | 16S rRNA sequencing | ↑C. trachomatis |
| Zhao | Ovarian cancer | Fresh ovarian cancer tissues | 2019 | 16S rRNA sequencing and qPCR | ↑Proteobacteria, Firmicute |
| Banerjee | Ovarian cancer | Ovarian cancer tissue | 2017 | PathoChip Array | ↑Proteobacteria, Firmicutes, Brucella, Chlamydia, Mycoplasma |
| Emara | Ovarian cancer | Ovarian cystic fluid and ovarian cancer tissue | 2007 | ovarian cystic fluid culture | ↑Brucella |
| Shanmughapriya | Ovarian cancer | Fresh ovarian tissues | 2012 | nested PCR-based assay | ↑Chlamydia |
| Chan | Ovarian cancer | Human ovarian cancer tissue | 2012 | PCR-ELISA | ↑Mycoplasma |
| Trabert | Ovarian cancer | Serum samples | 2018 | multiplex, fluorescent bead-based assay | ↑C. trachomatis |
| Di Giovanni | Ovarian cancer | Ovarian cancer tissue | 2016 | Bacterialogical Culture | ↑Mycobacterium spp. |
| Nene | Ovarian cancer | Cervical smear samples | 2019 | 16S rRNA sequencing and qPCR | ↓Lactobacillus spp. |
| Ness | Ovarian cancer | Blood samples | 2003 | Serologic testing for IgG antibodies | ↑Chlamydia |
| Walther | Endometrial cancer | Vaginal and cervical swabs | 2017 | 16S rRNA sequencing and qPCR | ↑A. vaginae, Porphyromonas spp. |
| Hokenstad | Endometrial cancer | Vaginal and cervical swabs | 2017 | RT-PCR, FISH | ↑Porphyromonas somerae |
The vaginal microbiota is diverse in population and ethnicity. A healthy vagina microbiome was previously thought to be dominated by Lactobacillus species [16]. However, advanced technologies and additional studies using an ethnically diverse cohort of women have revealed a more complex landscape [15,17]. Some studies indicated that the vaginal microbiomes in some healthy women are composed of Gardnerella, Atopobium, Prevotella, Pseudomonas, or Streptococcus species rather than Lactobacillus [18]. A study evaluated the microbiota composition of the six largest ethnic groups (African Surinamese, Dutch, Ghanaian, Moroccan, South-Asian Surinamese, and Turkish) in Amsterdam, the results showed African Surinamese ethnicity, and Ghanaian ethnicity were correlated with vaginal microbiotas containing Gardnerella vaginalis, and African Surinamese ethnicity with vaginal microbiotas dominated by L. iners [19]. Ravel et al. [20] studied the vaginal microbiome of 396 North American women from four ethnic backgrounds (Asian, Black, Hispanic, and White). Their study showed a significant discrepancy in vaginal microbiome composition. Vaginal bacterial communities dominated by species of Lactobacillus were found in 89.7% and 80.2% of White and Asian women, but in only 59.6% and 61.9% of Hispanic and black women. In contrast, a lower prevalence of communities dominated by Lactobacillus spp. was seen in Hispanic and black women. Another research showed that L. iners, L. jensenii, and G. vaginalis were prevalent in Canadian women [21]. Similarly, the study on Belgian women observed a prevalence of L. crispatus, L. iners, and Prevotella [22]. These observational reports further illuminate the broad spectrum of vaginal microbiota composition across different demographic backgrounds.
A healthy vaginal microbiome is considered the first line of defense. Lactobacillus up-regulates tight junction proteins that inhibit pathogen migration and improves epithelial integrity. The possibility of controlling pathogenesis by co-aggregating Lactobacillus with pathogens and thereby tying up the latter’s ability to spread across surfaces, or by interfering with virulence expression such as the production of toxins, has also been considered [23]. In addition, L. crispatus and L. jensenii could minimize the effect of inflammation through inhibiting the release of pro-inflammatory mediators from vaginal epithelial cells [24]. The metabolites of the Lactobacillus species, besides lactic acid and other acidic compounds, hydrogen peroxide, and bacteriocin-like compounds, may be pivotal to cervicovaginal homeostasis [25].
Changes in vaginal microbiomes are associated with host reproductive fitness. A recent study on sub-Saharan African women found significant differences in vaginal community composition in those developing bacterial vaginosis. Of these women, the composition of the Lactobacillus genus and Lactobacillus vaginalis were significantly lower, and the composition of G. vaginalis, A. vaginae, and P. bivia were higher after developing bacterial vaginosis [26]. Recent research also identified Mycoplasma genitalium as a possible cause for pelvic inflammatory disease (PID), a contributor to epithelial ovarian cancer [27]. Alterations in the vaginal composition are associated with bacterial dysbiosis, which could give rise to cancer-promoting virulence factors.
Vaginal microbiomes and gynecological cancer
Ovarian cancer
Ovarian cancer is a major threat to female health, ranking seventh in the most commonly diagnosed cancer among women worldwide [28]. The early stage of the disease is often asymptomatic, and most patients remain undiagnosed until advanced stages of cancer [29], thus finding specific biomarkers for early diagnosis is of utmost importance. Despite well-characterized risk factors (e.g., family history, age, inflammation, reproductive factors, benign gynecologic conditions, and gynecologic surgery) and genetic susceptibility (e.g., mutations in BRCA1 and BRCA2 genes) [28,30], its etiology is not fully understood. Recent studies have indicated that many microorganisms are involved in the development of ovarian cancer (Figure 1A) [31,32]. Zhao [33] and colleagues compared ovarian cancer tissue sample (n = 25) with tissues from normal distal fallopian tubes (n = 25) using 16s RNA sequencing. Comparatively, the microbiome diversity and richness was significantly decreased in the ovarian cancer samples. At the phylum level, a significant increase in Proteobacteria and Firmicutes abundance was seen in ovarian cancer tissue samples, suggesting an association between microbiome compositional change and ovarian cancer development (Figure 2). Similarly, in a study by Banerjee et al. [34], a microarray-based approach was applied to identify microbial signatures unique to ovarian cancer. Specifically, two predominant bacterial phyla were detected, consisting of Proteobacteria (52%) and Firmicutes (22%). Additionally, they detected Brucella, Chlamydia, and Mycoplasma in over 60% of the ovarian cancer sample screened. Their finding is consistent with previous investigations, also highlighting an enrichment of Brucella [35], Chamydia [36], and Mycoplasma [37] in ovarian cancer tissues. Researches investigating the role of M. genitalium, C. trachomatis, or Neisseria gonorrhoeae in ovarian cancer suggest a relationship between these microbes and the development of ovarian cancer. A multicenter study evaluated more than 1,100 samples collected from the association of the serologic markers of C. trachomatis and ovarian cancer in two separate populations. One is a case-control study in Poland containing 800 subjects, and another is a prospective case-control study in the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial consisting of 319 subjects. They found an association between doubled ovarian cancer risk with Pgp3, the antibodies against Chlamydia plasmid-encoded protein, also the gold standard in determining Chlamydia infection [38]. There is also a report of endometrial tuberculosis stimulating ovarian cancer [39]. Nevertheless, contradictory results were reported, which found no positive indications for the presence of C trachomatis, N. Gonorrhoeae, M genitalium, and HPV from a cohort of 186 women with ovarian cancer, borderline tumors, or benign conditions [40]. The variance of the sample probably leads to inconsistent results, since ovarian cancer exhibits various histologic types and differs from pathogenesis and tumor microenvironment [41].
Figure 1.
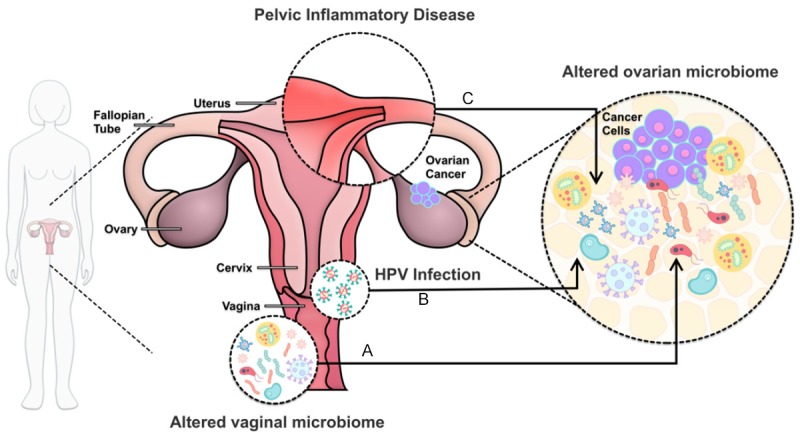
Interactions between female reproductive tract microbiota and the development of ovarian cancer. A. Microbiome compositional alterations in both ovarian and cervicovaginal mircroenvironment have been shown to correlate with the occurence of ovarian cancer. B. HPV genotypes may contribute to ovarian cancer progression. C. PID is related to the etiology of ovarian carcinoma, specifically pathogens like Chlamydia are associated with higher risk of developing ovarian cancer.
Figure 2.
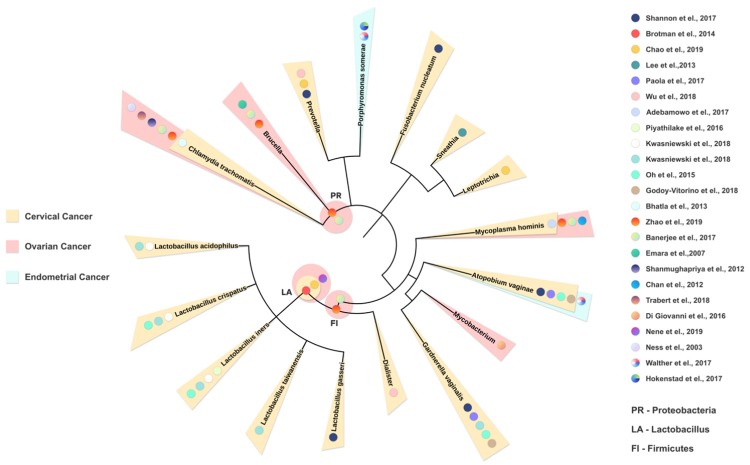
Phylogenetic tree summarizing established links between female reproductive tract microbiome and gynecological cancers. We constructed a phylogenetic tree using evolutionary distance with phyloT software [95] to describe the phylogenetic similarity of all microbiome reported to be associated with gynecological cancer in human studies. From the inside out, representing from the wider (kingdom) to the more specific (species) taxonomy. Based on the phenotype of the various studies included, the bacterial taxonomy is labeled according to the source of publication (colored dots) and shaded (OC = ovarian cancer, light red; CC = cervical cancer, light yellow; EC = endometrial cancer, light blue) based on the cancer types.
More recently, Martin and colleagues, utilizing 16s RNA, compared the cervicovaginal microbial profile of patients with ovarian cancer or patients with BRCA1 mutations with healthy matched controls. Identified microorganism compositions were then classified based on Lactobacilli species proportion. Those in which Lactobacilli species accounted for at least 50% were labeled community type L, and vice versa community type O. This study demonstrated that ovarian cancer or its risk factors (i.e., age and BRCA mutations) is significantly correlated with a community type O cervicovaginal microbiota [32]. These finds suggest cervicovaginal microbiota dysbiosis may play a role in ovarian cancer tumorigenesis. However, whether such dysbiosis could be altered through the re-installment of community type L microbiome [32], or whether changes in microbiome composition could translate into ovarian cancer protection, remains to be investigated.
The association between ovarian cancer and hrHPV were extensively studied, despite inconsistent results (Figure 1B). HPV was found associated with malignant transformation of mature cystic teratoma (MCT) into primary squamous cell carcinoma (SCC) [42]. A recent study was conducted in Hunan province to investigate the prevalence of HPV infection in epithelial ovarian cancer (EOC). They found HPV18 was positive in 7.76% malignant EOC patients, 9.09% in benign ovarian cancers, while only 1.01% in healthy persons. HPV33 was positive in 12.11% of malignant EOC samples, and 6.06% benign samples, whereas merely 1.51% in healthy people. These results showed a high correlation of HPV 18 and 33 with the ovarian cancer development [43]. Another study achieved similar results that HPV infection was associated with advanced stages of ovarian cancer [44]. In this study, HPV was detected in 10% EOC cases, with the most prevalent genotypes being HPV16 and 18 followed by HPV33. For patients infected with HR-HPV genotypes, they also suggested the CADM1, MAL, PAX1, and ADCYAP1 genes promoter hypermethylation as one of the possible mechanisms contributing to ovarian carcinogenesis. Sangria banerjee [34] detected molecular signatures of both high-risk HPV16 and 18, along low-risk HPVs in the ovarian cancer samples. They found only low-risk HPVs were related to tumor-negative controls, implicating that hrHPV might be the origin of cancer. Interestingly, they observed widespread integration of viral sequences into various intronic regions, or at intergenic regions within 56 kb upstream of numerous cancer-related human genes. HPV integrations of E1 or E2 regions were reported to inactivate the transcription. However, a system-level analysis is further needed to understand the functional interaction between specific phenotypic traits of the microbiome in different ovarian cancer subtypes and HPV infection.
Pelvic inflammatory disease (PID) is widely established as a risk factor for epithelial ovarian cancer (Figure 1C) [45]. Among the organisms responsible for PID onset, Chlamydia is the most common, followed by Nesseria gonorrhea [46,47]. Previous studies have found that antibodies against Chlamydia infection are associated with ovarian cancer [48]. More recently, Trabert [38] and colleagues evaluated the associations between serologic markers and ovarian cancer risk in two independent populations. Specifically, in a Polish case-control study (244 ovarian cancer/566 control subject), antibodies against Chlamydia (Pgp3 protein) were associated with an elevation in ovarian cancer risk (OR = 1.63, 95% [CI] : 1.20 to 2.22). Similarly, in another case-control study in the PLCO Cancer Screening Trial (160 ovarian cancers/159 control subjects), Pgp3 antibodies were also found to be associated with increased ovarian cancer risk (OR = 1.43, 95% [CI] : 0.78 to 2.63). However, in both studies, no associations were found between antibodies against other infectious agents and ovarian cancer. Meanwhile, accumulated studies have ascribed PID to mixed infections by organisms in the vaginal, including pathogenic microorganisms responsible for bacterial vaginosis [46,49-51]. These microbiomes are linked to the etiology of PID and could thus contribute to the development of ovarian cancer.
Cervical cancer
Cervical cancer ranks second in the cause of cancer death in women between 20 to 39 years old [11]. Human Papillomavirus (HPV) persistent infection is pivotal for cervical carcinogenesis. Despite being the most pervasive sexually transmitted infection (STI) worldwide, the majority of 100 subtypes of HPV are non-carcinogenic. However, there are at least 13 high-risk subtypes involved in the pathogenesis of malignancies [52,53]. Of these subtypes, HPV16 and HPV18 are the most prevalent and account for 70% of the cases [54]. Interestingly, a recent study showed that specific cervicovaginal microbiota composition correlated with the acquisition of high-risk HPV (hrHPV) types [55].
Some studies suggest that dysbiosis occurs in HPV infection, and a broad microbial alteration pattern was revealed a reduction of Lactobacillus spp. The diminution of these commensal microbes is concomitant with a loss of their protective capabilities and could yield a substantial impact on the onset and progression of the disease. The ability of commensal microbes to produce lactic acid is a crucial benefit to the women’s genital tract. Lactic acid acidifies the vaginal environment, benefiting the proliferation of Lactobacillus while inhibiting the growth of infection-associated organisms. In a recent study, Shannon and co-worker [7] studied a cohort of 65 African/Caribbean women to assess microbiome composition and structure through 16S rRNA sequencing. They concluded that participants with HPV infection are associated with a vaginal microbiome consistent with CST-IV, characterized by a paucity of Lactobacillus spp. and a wide array of anaerobes (58.8% vs. 29.4%; P = 0.043). They also observed a step-wise lowering in the relative abundance of L. gasseri, Fusobacterium nucleatum, Cornybacterium accolens, Anaerococcus tetradius, Finegoldia magna, Peptoniphilus harei, and Raoultella planticola. This result is in accordance with previous observations that women with hrHPV infections have decreased the abundance of Lactobacillus spp [56]. Meanwhile, another study carried out on Nigerian women also suggested a correlation between hrHPV infection and a decreased concentration of Lactobacillus spp [57]. This association is also linked with an increased correlation of anaerobes, particularly of the genera Leptotrichia and Prevotella [58]. The relative abundance of Sneathia spp. in persons with HPV infection is increased. A Korean twin study by Lee et al. [59] found an abundance of Sneathia spp. in HPV positive groups, emphasizing its potential as a biomarker for the prediction of HPV infection. Another study also identified Sneathia spp. as the most copious species in the cervix of women with squamous intraepithelial lesion and associated the presence of Sneathia spp. with HPV-positive squamous intraepithelial lesion [60]. Alterations of these bacterial populations and concomitant variability of lactic acid production may have profound results on host regulation of inflammation.
Persistent HPV infection could result in precancerous lesions in the female genital tract [61]. A recent study conducted by Paola and colleagues [62] used next-generation sequencing to examine cervicovaginal microbiota in 55 HPV positive women. They found the abundance of CST IV subgroup, including bacterial genera such as Prevotella, Gardnerella, Atopobium, Megasphoera, strongly correlated with HPV persistence. This study also identified Atopobium spp. and sialidase gene from G. vaginalis as feasible microbial markers for HPV persistence. Wu [63] also found 22 taxa to be associated with HPV persistence, and of these, 5 taxa belong to Prevotella and 1 taxon belongs to Dialister. Likewise, another study examined 194 Nigerian women and identified a strong correlation between persistent Mycoplasma hominis infection and persistent hrHPV (OR 8.78, 95%, P 0.01) [64]. Only small amounts of women would have persistent HPV infection and progress to cervical lesions [65] and the vaginal microbiome might play a role during this process. HPV infection has adversary impact on the host’s immune defenses and mucosal metabolism. This leads to the dysbiosis of the vaginal microbiota, and thus promoting viral persistence and disease progression [66]. Further longitudinal studies are needed to investigate whether and how the microbiome helps maintain the persistent infection and develop to CIN or cervical cancer.
Microbiomes also directly linked to cervical cancer (Figure 2). Studies to date have documented an overall increase in diversity. Mitra et al. [67] suggest that cervical intraepithelial neoplasia (CIN) progression is correlated with increasing vaginal microbiota diversity. This increased diversity was possibly because of the epithelial barrier rupture and the host’s immune dysregulation. Lactobacillus is the most abundant genus in vaginal microbiotas and can affect the host dichotomously. While the abundance of some Lactobacillus spp. is reduced in cervical cancer or precancerous diseases. A cervical microbiome characterized by a prevalence of L. iners is associated with high-grade cervical intraepithelial neoplasia in women infected with hrHPVs [8]. A more recent study observed that women with low-grade squamous intraepithelial lesion (LSIL) were characterized by high prevalence of Lactobacillus acidophilus and L. iners, and no presence of L. crispatus. In contrast, women with high-grade squamous intraepithelial lesion (HSIL) were marked with high proportion of G. vaginalis and L. acidophilus, and no L. crispatus, Lactobacillus taiwanensis, or L. iners detected [68]. Pathogenic bacteria, including A. vaginae, G. vaginalis are increasingly observed in CIN or cervical cancer. Oh et al. [69] studied the cervical microbiota of a Korean cohort of 120 women, 70 with CIN and 50 as the control. The investigators observed a predominance of A. vaginae, L. iners, G. vaginalis and an accompanied dearth of L. crispatus in women with high CIN risk. This result is in accordance with another study that enrichment of A. vaginae and G. vaginalis was found in patients with CIN3 [70]. Other pathogens, such as C. trachomatis, has also been identified as a cofactor of carcinogenesis, with a higher rate of infection in patients with cervical cancer [71,72]. Though the results are inconsistent among published studies [73], partly owing to specimen variance or analysis methods. Associations were established both for single and multiple hrHPV genotype infections, supporting the hypothesis that a C. trachomatis infection contributes to cervical cancer, together with inflammation and HPV [74]. These non-commensal microbiotas may induce inflammatory cytokines, especially in coinfection with HPV. However, how inflammatory cytokines induced by these non-commensal microbiotas are associated with cervical cancer progression needs further elucidation.
Endometrial cancer
Researches have also identified potential microbiotas contributing to the genesis of endometrial cancer (Figure 2). A recently identified A. vaginae and Porphyromonas spp. in the reproductive tract in combination with a high vaginal pH to be statistically related to the occurrence of endometrial cancer [75]. It was further demonstrated that Porphyromonas spp. combined with high pH in the vagina could be a promising biomarker for endometrial cancer [76]. These findings are significant, as they put forth a promising biomarker for early detection and pave the way for possible primary preventive interventions. Molecular mechanisms underlying the interaction between microbiome and pathogenesis of endometrial cancer still need elucidation.
Manipulation of the microbiome in gynecological cancer therapeutics
Chemotherapy resistance has long been a problem for patients with ovarian cancer. We previously found Helicase POLQ-like (HELQ) as a promising indicator of cisplatin chemo-resistance for epithelial ovarian cancer [77]. Recently, growing evidence implicates that human microbiomes influence cancer therapy mainly through two aspects: modulating cancer therapeutic response and mediating treatment-related toxity [78]. In preclinical models, the response to oxaliplatin depends on the expression of proinflammatory genes of the microbial flora and the generation of reactive oxygen species by myeloid cells in the tumor microenvironment [79]. Contrarily, the response to gemcitabine can be compromised by Mycoplasma, through its pyrimidine nucleoside phosphorylase and cytidine deaminase enzymes, which influences cytostatic activity [80]. For some patients with recurrent or persistent, metastatic gynecological cancer, programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) inhibitors are a possible choice to enhance the clinical outcomes [81]. Recent progress has emphasized the role of the microbiome in regulating tumor responses to chemotherapeutic agents as well as immunotherapies targeting PD-L1 or cytotoxic T lymphocyte-associated protein 4 (CTLA-4) [82]. Furthermore, Routy et al. [83] showed that antibiotic usage is associated with abnormal responses to immunotherapeutic PD-1 blockade. Through profiling samples from lung and kidney cancer patients, they found that patients nonresponding to PD-1 inhibitors had lower levels of the bacterium Akkermansia muciniphila. After oral supplementation of bacteria in antibiotic-treated mice, response to immunotherapy was restored. Matson et al. [84] and Gopalakrishnan et al. [85] studied PD-1 blockade in melanoma patients and found a higher concentration of favorable bacteria in the guts of responding patients. They also found an imbalance of gut flora composition in nonresponders, which is associated with impaired immune cell activity. These observations are in conjunction with the hypothesis that the microbiome may play an important role in immunotherapy. However, whether or not cervicovaginal microbiotas could influence the efficacy of chemotherapy in gynecological cancers still needs to be investigated.
Manipulation of the vaginal microbiota is essential for women’s health. Probiotic L. rhamnosus GR-1 was shown to mediate adhesion to the vaginal epithelium and mediates the attachment of urogenital pathogens [86]. Studies indicate that strains of Lactobacillus can inhibit the growth of G. vaginalis [87]. L. crispatus has also demonstrated potentials as a hopeful probiotic in preventing N. gonorrhoeae infections through counteracting N. gonorrhoeae viability [88]. Recently, a research implementing L. rhamnosus BMX 54 in 57 women for at least 6 months has shown positive outcomes in controlling HPV infection [89]. Additional research shows that supernatants of L. gasseri, L. jensenii, and L. crispatus could inhibit the activity of cervical cancer cells via regulation of HPV oncogenes [90]. L. gasseri and L. crispatus has also been reported to exert cytotoxic effects on cervical tumor cells selectively, but not normal cells [91]. These results are tantalizing, but further studies are necessary to explore the underlying mechanisms for successful applications in humans. Another possible application of probiotics is to restore a healthy genital microbial community after some gynecological procedures.
Conclusion and future direction
Studies to date are encouraging, despite further research on the human microbiome and gynecological cancer are needed. The current studies have given us insights into this field, yet with drawbacks and contradictions. For instance, the studies on the HPV infection, ovarian cancer, and endometrial cancer showed conflicting results. One possible reason is the difference in detection methods. In a meta-analysis mentioned previously [27], the meta-regression suggested that the HPV prevalence was closely related to HPV DNA detection method; racial and local differentiations are another reason behind these discrepancies [18]; individual variances might also account for the different results [92]. Co-factors related to an individual’s lifestyle, such as tobacco usage and hormonal contraceptives, as well as multiple sex partners and early sexual activities, are associated with SCC [93]. The difference in location of the analyzed samples also accounts for the inconsistent results, Chen et al. [94] has reported the different compositions of microbiota in the cervical canal, uterus, and vagina.
Present researches have focused on the relationship between microbiota and gynecologic malignancy, with little regard for cause and effect. Some microbiome presence may facilitate the HPV infection, but the reverse scenario is also possible that HPV infection harbors a salubrious environment that satisfies microbial needs. Another quandary is although highly pervasive, only a small percentage of women with persistent HPV infection subsequently acquire clinically significant diseases. It remains to be solved what roles these microbiotas could play in HPV persistent infection. Investigation of unique microbiome signature in different cancers paves the way for diagnostic biomarkers as well as provide insights for prognosis, prevention, and the development of treatment.
Current research is promising, owing to technological advances in sequencing technology and improved in vitro models. New knowledge in this field shed light on the diagnosis for early detection and therapeutic potential of cancer. As demonstrated in this review, significant gaps in knowledge remain, especially regarding the microbiome and gynecologic cancers. To have a translational impact, additionally, it is crucial to develop a system-level understanding of health and disease by measuring biological components of a system using a statistical and metagenomics framework. Also, further investigations are needed to improve treatment and develop new interventions for women’s health.
Acknowledgements
The authors received funding from the Hunan Provincial Gynecological Cancer Diagnosis and Treatment Engineering Research Center, China.
Disclosure of conflict of interest
None.
References
- 1.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, Padavattan N, Ismail N, Moodley A, Sabatini ME, Ghebremichael MS, Nusbaum C, Huttenhower C, Virgin HW, Ndung’u T, Dong KL, Walker BD, Fichorova RN, Kwon DS. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng W, Shen J, Bo T, Peng L, Xu H, Nasser MI, Zhuang Q, Zhao M. Cutting edge: probiotics and fecal microbiota transplantation in immunomodulation. J Immunol Res. 2019;2019:1603758. doi: 10.1155/2019/1603758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng J, Xiao X, Hu M, Zhang X. Interaction between gut microbiome and cardiovascular disease. Life Sci. 2018;214:153–157. doi: 10.1016/j.lfs.2018.10.063. [DOI] [PubMed] [Google Scholar]
- 5.Si J, You HJ, Yu J, Sung J, Ko G. Prevotella as a Hub for vaginal microbiota under the influence of host genetics and their association with obesity. Cell Host Microbe. 2017;21:97–105. doi: 10.1016/j.chom.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Gottschick C, Deng ZL, Vital M, Masur C, Abels C, Pieper DH, Rohde M, Mendling W, Wagner-Dobler I. Treatment of biofilms in bacterial vaginosis by an amphoteric tenside pessary-clinical study and microbiota analysis. Microbiome. 2017;5:119. doi: 10.1186/s40168-017-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shannon B, Yi TJ, Perusini S, Gajer P, Ma B, Humphrys MS, Thomas-Pavanel J, Chieza L, Janakiram P, Saunders M, Tharao W, Huibner S, Shahabi K, Ravel J, Rebbapragada A, Kaul R. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol. 2017;10:1310–1319. doi: 10.1038/mi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila) 2016;9:357–366. doi: 10.1158/1940-6207.CAPR-15-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Houdt R, Ma B, Bruisten SM, Speksnijder A, Ravel J, de Vries HJC. Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to Chlamydia trachomatis infection in Dutch women: a case-control study. Sex Transm Infect. 2018;94:117–123. doi: 10.1136/sextrans-2017-053133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Veer C, Bruisten SM, van der Helm JJ, de Vries HJC, van Houdt R. A33 The cervico-vaginale microbiota in chlamydia trachomtais notified women: a case-control study at the sexually transmitted infection outpatient clinic in Amsterdam. Virus Evol. 2017;3 doi: 10.1093/ve/vew036.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 12.Unemo M, Bradshaw CS, Hocking JS, de Vries HJC, Francis SC, Mabey D, Marrazzo JM, Sonder GJB, Schwebke JR, Hoornenborg E, Peeling RW, Philip SS, Low N, Fairley CK. Sexually transmitted infections: challenges ahead. Lancet Infect Dis. 2017;17:e235–e279. doi: 10.1016/S1473-3099(17)30310-9. [DOI] [PubMed] [Google Scholar]
- 13.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Wijgert JH, Borgdorff H, Verhelst R, Crucitti T, Francis S, Verstraelen H, Jespers V. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One. 2014;9:e105998. doi: 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PS, Chen IM, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, DeSantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Dunne WM, Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Haake SK, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, O’Laughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang Z, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu Y, Wylie KM, Wylie T, Yandava C, Ye L, Ye Y, Yooseph S, Youmans BP, Zhang L, Zhou Y, Zhu Y, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methé BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. 2012;66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF 3rd, Vaginal Microbiome C, Jefferson KK, Buck GA. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borgdorff H, van der Veer C, van Houdt R, Alberts CJ, de Vries HJ, Bruisten SM, Snijder MB, Prins M, Geerlings SE, Schim van der Loeff MF, van de Wijgert J. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One. 2017;12:e0181135. doi: 10.1371/journal.pone.0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert AY, Chaban B, Wagner EC, Schellenberg JJ, Links MG, van Schalkwyk J, Reid G, Hemmingsen SM, Hill JE, Money D, Group VR. A study of the vaginal microbiome in healthy canadian women utilizing cpn60-based molecular profiling reveals distinct gardnerella subgroup community state types. PLoS One. 2015;10:e0135620. doi: 10.1371/journal.pone.0135620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verstraelen H, Vilchez-Vargas R, Desimpel F, Jauregui R, Vankeirsbilck N, Weyers S, Verhelst R, De Sutter P, Pieper DH, Van De Wiele T. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. PeerJ. 2016;4:e1602. doi: 10.7717/peerj.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid G. Therapeutic opportunities in the vaginal microbiome. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.BAD-0001-2016. [DOI] [PubMed] [Google Scholar]
- 24.Aldunate M, Srbinovski D, Hearps AC, Latham CF, Ramsland PA, Gugasyan R, Cone RA, Tachedjian G. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front Physiol. 2015;6:164. doi: 10.3389/fphys.2015.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid G. Cervicovaginal microbiomes-threats and possibilities. Trends Endocrinol Metab. 2016;27:446–454. doi: 10.1016/j.tem.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Jespers V, Kyongo J, Joseph S, Hardy L, Cools P, Crucitti T, Mwaura M, Ndayisaba G, Delany-Moretlwe S, Buyze J, Vanham G, van de Wijgert J. A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa. Sci Rep. 2017;7:11974. doi: 10.1038/s41598-017-12198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lis R, Rowhani-Rahbar A, Manhart LE. Mycoplasma genitalium infection and female reproductive tract disease: a meta-analysis. Clin Infect Dis. 2015;61:418–426. doi: 10.1093/cid/civ312. [DOI] [PubMed] [Google Scholar]
- 28.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med. 2017;14:9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terry KL, Schock H, Fortner RT, Husing A, Fichorova RN, Yamamoto HS, Vitonis AF, Johnson T, Overvad K, Tjonneland A, Boutron-Ruault MC, Mesrine S, Severi G, Dossus L, Rinaldi S, Boeing H, Benetou V, Lagiou P, Trichopoulou A, Krogh V, Kuhn E, Panico S, Bueno-de-Mesquita HB, Onland-Moret NC, Peeters PH, Gram IT, Weiderpass E, Duell EJ, Sanchez MJ, Ardanaz E, Etxezarreta N, Navarro C, Idahl A, Lundin E, Jirstrom K, Manjer J, Wareham NJ, Khaw KT, Byrne KS, Travis RC, Gunter MJ, Merritt MA, Riboli E, Cramer DW, Kaaks R. A prospective evaluation of early detection biomarkers for ovarian cancer in the European EPIC cohort. Clin Cancer Res. 2016;22:4664–4675. doi: 10.1158/1078-0432.CCR-16-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YB, Mei Y, Long J, Zhang Y, Hu DL, Zhou HH. RIF1 promotes human epithelial ovarian cancer growth and progression via activating human telomerase reverse transcriptase expression. J Exp Clin Cancer Res. 2018;37:182. doi: 10.1186/s13046-018-0854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X, Yang M, Ding Y, Chen J. Microbial infection, inflammation and epithelial ovarian cancer. Oncol Lett. 2017;14:1911–1919. doi: 10.3892/ol.2017.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nene NR, Reisel D, Leimbach A, Franchi D, Jones A, Evans I, Knapp S, Ryan A, Ghazali S, Timms JF, Paprotka T, Bjorge L, Zikan M, Cibula D, Colombo N, Widschwendter M. Association between the cervicovaginal microbiome, BRCA1 mutation status, and risk of ovarian cancer: a case-control study. Lancet Oncol. 2019;20:1171–1182. doi: 10.1016/S1470-2045(19)30340-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Sun C, Huang J, Xia M, Guo E, Li N, Lu H, Shan W, Wu Y, Li Y, Xu X, Weng D, Meng L, Hu J, Gao Q, Ma D, Chen G. The biodiversity composition of microbiome in ovarian carcinoma patients. Sci Rep. 2019;9:1691. doi: 10.1038/s41598-018-38031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, Coukos G, Robertson ES. The ovarian cancer oncobiome. Oncotarget. 2017;8:36225–36245. doi: 10.18632/oncotarget.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emara MM, Vyas V, al Awadi S, Jaroslav N, Khodry A, Essam T, Rouf Y, Amanguno H, Purohit P. Synchronous occurrence of brucellosis and ovarian cancer - a case report. Austral-Asian J Cancer. 2007;6:257–259. [Google Scholar]
- 36.Shanmughapriya S, Senthilkumar G, Vinodhini K, Das BC, Vasanthi N, Natarajaseenivasan K. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur J Clin Microbiol Infect Dis. 2012;31:2311–2317. doi: 10.1007/s10096-012-1570-5. [DOI] [PubMed] [Google Scholar]
- 37.Chan PJ, Seraj IM, Kalugdan TH, King A. Prevalence of mycoplasma conserved DNA in malignant ovarian cancer detected using sensitive PCR-ELISA. Gynecol Oncol. 1996;63:258–260. doi: 10.1006/gyno.1996.0316. [DOI] [PubMed] [Google Scholar]
- 38.Trabert B, Waterboer T, Idahl A, Brenner N, Brinton LA, Butt J, Coburn SB, Hartge P, Hufnagel K, Inturrisi F, Lissowska J, Mentzer A, Peplonska B, Sherman ME, Wills GS, Woodhall SC, Pawlita M, Wentzensen N. Antibodies against chlamydia trachomatis and ovarian cancer risk in two independent populations. J Natl Cancer Inst. 2019;111:129–136. doi: 10.1093/jnci/djy084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Giovanni SE, Cunha TM, Duarte AL, Alves I. Endometrial tuberculosis simulating an ovarian cancer: a case report. Acta Med Port. 2016;29:412–415. doi: 10.20344/amp.7706. [DOI] [PubMed] [Google Scholar]
- 40.Idahl A, Lundin E, Elgh F, Jurstrand M, Moller JK, Marklund I, Lindgren P, Ottander U. Chlamydia trachomatis, Mycoplasma genitalium, Neisseria gonorrhoeae, human papillomavirus, and polyomavirus are not detectable in human tissue with epithelial ovarian cancer, borderline tumor, or benign conditions. Am J Obstet Gynecol. 2010;202:71, e71–76. doi: 10.1016/j.ajog.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 41.Kurman RJ, Shih IeM. The origin and pathogenesis of epithelial ovarian cancer-a proposed unifying theory. Am J Surg Pathol. 2010;34:433. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang AJ, Chen DR, Cheng JT, Chang TH. Detection of human papillomavirus in squamous cell carcinoma arising from dermoid cysts. Taiwan J Obstet Gynecol. 2015;54:559–566. doi: 10.1016/j.tjog.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Zhang PP, Zhou L, Cao JS, Li YP, Zeng Z, Sun N, Shen L, Zhu HY, Ruan Y, Zha WT, Wang XY, Zhang KQ, Zhang R. Possible epithelial ovarian cancer association with HPV18 or HPV33 infection. Asian Pac J Cancer Prev. 2016;17:2959–2964. [PubMed] [Google Scholar]
- 44.Hassan ZK, Hafez MM, Kamel MM, Zekri AR. Human papillomavirus genotypes and methylation of CADM1, PAX1, MAL and ADCYAP1 genes in epithelial ovarian cancer patients. Asian Pac J Cancer Prev. 2017;18:169–176. doi: 10.22034/APJCP.2017.18.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Z, Zeng F, Yuan J, Tang J, Colditz GA, Tworoger SS, Trabert B, Su X. Pelvic inflammatory disease and the risk of ovarian cancer: a meta-analysis. Cancer Causes Control. 2017;28:415–428. doi: 10.1007/s10552-017-0873-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma H, Tal R, Clark NA, Segars JH. Microbiota and pelvic inflammatory disease. Semin Reprod Med. 2014;32:43–49. doi: 10.1055/s-0033-1361822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassenaar TM, Panigrahi P. Is a foetus developing in a sterile environment? Lett Appl Microbiol. 2014;59:572–579. doi: 10.1111/lam.12334. [DOI] [PubMed] [Google Scholar]
- 48.Ness RB, Goodman MT, Shen C, Brunham RC. Serologic evidence of past infection with Chlamydia trachomatis, in relation to ovarian cancer. J Infect Dis. 2003;187:1147–1152. doi: 10.1086/368380. [DOI] [PubMed] [Google Scholar]
- 49.Power ML, Quaglieri C, Schulkin J. Reproductive microbiomes: a new thread in the microbial network. Reprod Sci. 2017;24:1482–1492. doi: 10.1177/1933719117698577. [DOI] [PubMed] [Google Scholar]
- 50.Risser WL, Risser JM, Risser AL. Current perspectives in the USA on the diagnosis and treatment of pelvic inflammatory disease in adolescents. Adolesc Health Med Ther. 2017;8:87–94. doi: 10.2147/AHMT.S115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Wijgert JH, Jespers V. The global health impact of vaginal dysbiosis. Res Microbiol. 2017;168:859–864. doi: 10.1016/j.resmic.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 53.Serrano B, Brotons M, Bosch FX, Bruni L. Epidemiology and burden of HPV-related disease. Best Pract Res Clin Obstet Gynaecol. 2018;47:14–26. doi: 10.1016/j.bpobgyn.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Munoz N, Bosch FX, Castellsague X, Diaz M, de Sanjose S, Hammouda D, Shah KV, Meijer CJ. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 55.Huang X, Li C, Li F, Zhao J, Wan X, Wang K. Cervicovaginal microbiota composition correlates with the acquisition of high-risk human papillomavirus types. Int J Cancer. 2018;143:621–634. doi: 10.1002/ijc.31342. [DOI] [PubMed] [Google Scholar]
- 56.Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, Gravitt PE. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dareng EO, Ma B, Famooto AO, Adebamowo SN, Offiong RA, Olaniyan O, Dakum PS, Wheeler CM, Fadrosh D, Yang H, Gajer P, Brotman RM, Ravel J, Adebamowo CA. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol Infect. 2016;144:123–137. doi: 10.1017/S0950268815000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chao XP, Sun TT, Wang S, Fan QB, Shi HH, Zhu L, Lang JH. Correlation between the diversity of vaginal microbiota and the risk of high-risk human papillomavirus infection. Int J Gynecol Cancer. 2019;29:28–34. doi: 10.1136/ijgc-2018-000032. [DOI] [PubMed] [Google Scholar]
- 59.Lee JE, Lee S, Lee H, Song YM, Lee K, Han MJ, Sung J, Ko G. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One. 2013;8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, López-Estrada G, Delgado-Romero K, Burguete-García AI, Cantú D, García-Carrancá A, Madrid-Marina V. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One. 2016;11:e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sand FL, Munk C, Frederiksen K, Junge J, Iftner T, Dehlendorff C, Kjaer SK. Risk of CIN3 or worse with persistence of 13 individual oncogenic HPV types. Int J Cancer. 2019;144:1975–1982. doi: 10.1002/ijc.31883. [DOI] [PubMed] [Google Scholar]
- 62.Di Paola M, Sani C, Clemente AM, Iossa A, Perissi E, Castronovo G, Tanturli M, Rivero D, Cozzolino F, Cavalieri D, Carozzi F, De Filippo C, Torcia MG. Characterization of cervico-vaginal microbiota in women developing persistent high-risk human papillomavirus infection. Sci Rep. 2017;7:10200. doi: 10.1038/s41598-017-09842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritu W, Enqi W, Zheng S, Wang J, Ling Y, Wang Y. Evaluation of the associations between cervical microbiota and HPV infection, clearance, and persistence in cytologically normal women. Cancer Prev Res (Phila) 2019;12:43–56. doi: 10.1158/1940-6207.CAPR-18-0233. [DOI] [PubMed] [Google Scholar]
- 64.Adebamowo SN, Ma B, Zella D, Famooto A, Ravel J, Adebamowo C ACCME Research Group. Mycoplasma hominis and mycoplasma genitalium in the vaginal microbiota and persistent high-risk human papillomavirus infection. Front Public Health. 2017;5:140. doi: 10.3389/fpubh.2017.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sudenga SL, Shrestha S. Key considerations and current perspectives of epidemiological studies on human papillomavirus persistence, the intermediate phenotype to cervical cancer. Int J Infect Dis. 2013;17:e216–220. doi: 10.1016/j.ijid.2012.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kyrgiou M, Mitra A, Moscicki AB. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res. 2017;179:168–182. doi: 10.1016/j.trsl.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li JV, Holmes E, Nicholson JK, Bennett PR, Kyrgiou M. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwasniewski W, Wolun-Cholewa M, Kotarski J, Warchol W, Kuzma D, Kwasniewska A, Gozdzicka-Jozefiak A. Microbiota dysbiosis is associated with HPV-induced cervical carcinogenesis. Oncol Lett. 2018;16:7035–7047. doi: 10.3892/ol.2018.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh HY, Kim BS, Seo SS, Kong JS, Lee JK, Park SY, Hong KM, Kim HK, Kim MK. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect. 2015;21:674, e671–679. doi: 10.1016/j.cmi.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 70.Godoy-Vitorino F, Romaguera J, Zhao C, Vargas-Robles D, Ortiz-Morales G, Vazquez-Sanchez F, Sanchez-Vazquez M, de la Garza-Casillas M, Martinez-Ferrer M, White JR, Bittinger K, Dominguez-Bello MG, Blaser MJ. Cervicovaginal fungi and bacteria associated with cervical intraepithelial neoplasia and high-risk human papillomavirus infections in a hispanic population. Front Microbiol. 2018;9:2533. doi: 10.3389/fmicb.2018.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhatla N, Puri K, Joseph E, Kriplani A, Iyer VK, Sreenivas V. Association of Chlamydia trachomatis infection with human papillomavirus (HPV) & cervical intraepithelial neoplasia - a pilot study. Indian J Med Res. 2013;137:533–539. [PMC free article] [PubMed] [Google Scholar]
- 72.Silva J, Cerqueira F, Medeiros R. Chlamydia trachomatis infection: implications for HPV status and cervical cancer. Arch Gynecol Obstet. 2014;289:715–723. doi: 10.1007/s00404-013-3122-3. [DOI] [PubMed] [Google Scholar]
- 73.Smelov V, Gheit T, Sundstrom K, Ploner A, McKay-Chopin S, Eklund C, Tommasino M, Dillner J. Lack of significant effects of chlamydia trachomatis infection on cervical adenocarcinoma risk: nested case-control study. PLoS One. 2016;11:e0156215. doi: 10.1371/journal.pone.0156215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wohlmeister D, Vianna DR, Helfer VE, Gimenes F, Consolaro ME, Barcellos RB, Rossetti ML, Calil LN, Buffon A, Pilger DA. Association of human papillomavirus and Chlamydia trachomatis with intraepithelial alterations in cervix samples. Mem Inst Oswaldo Cruz. 2016;111:106–113. doi: 10.1590/0074-02760150330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walther-Antonio MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, Keeney GL, Creedon DJ, Nelson H, Mariani A, Chia N. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8:122. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hokenstad A, Mariani A, Walther-Antonio M. Vaginal detection of Porphyromonas somerae is indicative of endometrial cancer diagnosis. Gynecologic Oncology. 2017;145:76. [Google Scholar]
- 77.Long J, Zhu JY, Liu YB, Fu K, Tian Y, Li PY, Yang WQ, Yang SY, Yin JY, Yin G, Zhang Y. Helicase POLQ-like (HELQ) as a novel indicator of platinum-based chemoresistance for epithelial ovarian cancer. Gynecol Oncol. 2018;149:341–349. doi: 10.1016/j.ygyno.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 78.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 79.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vande Voorde J, Sabuncuoglu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, Balzarini J, Liekens S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem. 2014;289:13054–13065. doi: 10.1074/jbc.M114.558924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ventriglia J, Paciolla I, Pisano C, Cecere SC, Di Napoli M, Tambaro R, Califano D, Losito S, Scognamiglio G, Setola SV, Arenare L, Pignata S, Della Pepa C. Immunotherapy in ovarian, endometrial and cervical cancer: state of the art and future perspectives. Cancer Treat Rev. 2017;59:109–116. doi: 10.1016/j.ctrv.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Bérard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Doré J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 84.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petrova MI, Lievens E, Verhoeven TL, Macklaim JM, Gloor G, Schols D, Vanderleyden J, Reid G, Lebeer S. The lectin-like protein 1 in Lactobacillus rhamnosus GR-1 mediates tissue-specific adherence to vaginal epithelium and inhibits urogenital pathogens. Sci Rep. 2016;6:37437. doi: 10.1038/srep37437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim Y, Kang CH, Shin Y, Paek NS, So JS. Characterization and antimicrobial activity against Gardnerella vaginalis of vaginal Lactobacillus spp. isolated from Korean women. 2015;30:239–244. [Google Scholar]
- 88.Foschi C, Salvo M, Cevenini R, Parolin C, Vitali B, Marangoni A. Vaginal lactobacilli reduce Neisseria gonorrhoeae viability through multiple strategies: an in vitro study. Front Cell Infect Microbiol. 2017;7:502. doi: 10.3389/fcimb.2017.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palma E, Recine N, Domenici L, Giorgini M, Pierangeli A, Panici PB. Long-term Lactobacillus rhamnosus BMX 54 application to restore a balanced vaginal ecosystem: a promising solution against HPV-infection. BMC Infect Dis. 2018;18:13. doi: 10.1186/s12879-017-2938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang KD, Xu DJ, Wang BY, Yan DH, Lv Z, Su JR. Inhibitory effect of vaginal Lactobacillus supernatants on cervical cancer cells. Probiotics Antimicrob Proteins. 2018;10:236–242. doi: 10.1007/s12602-017-9339-x. [DOI] [PubMed] [Google Scholar]
- 91.Motevaseli E, Shirzad M, Akrami SM, Mousavi AS, Mirsalehian A, Modarressi MH. Normal and tumour cervical cells respond differently to vaginal lactobacilli, independent of pH and lactate. J Med Microbiol. 2013;62:1065–1072. doi: 10.1099/jmm.0.057521-0. [DOI] [PubMed] [Google Scholar]
- 92.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2:4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richard A, Rohrmann S, Schmid SM, Tirri BF, Huang DJ, Guth U, Eichholzer M. Lifestyle and health-related predictors of cervical cancer screening attendance in a Swiss population-based study. Cancer Epidemiol. 2015;39:870–876. doi: 10.1016/j.canep.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 94.Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, Xie H, Chen X, Zeng C, Wen B, Zeng L, Du H, Tang H, Xu C, Xia Y, Xia H, Yang H, Wang J, Wang J, Madsen L, Brix S, Kristiansen K, Xu X, Li J, Wu R, Jia H. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8:875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]