Abstract
Retinoblastoma binding protein 4 (RBBP4) plays an important role in transcription, cell cycle, and proliferation. Immunohistochemistry was performed to assess HDAC1 and RBBP4 expression in 240 BC patients. The expression of HDAC1 and RBBP4 in 12 pairs of BC tissues and their normal tissues was determined by western blotting. Kaplan-Meier analysis and Cox’s proportional hazards regression were applied to evaluate the prognostic significance of HDAC1 and RBBP4. HDAC1 and RBBP4 expression in BC was significantly higher than that in normal tissues. HDAC1 was positively correlated with RBBP4 in breast cancer. HDAC1 and RBBP4 were negatively correlated with ER and PR in BC, respectively. The patients with high expression of RBBP4 had a worse overall survival time. The expression of RBBP4 was found to be significantly correlated with lymph node metastasis. RBBP4 may play a major role though HDAC1 in the development, metastasis, and prognosis of BC.
Keywords: RBBP4, HDAC1, breast cancer, prognosis
Introduction
Breast cancer (BC) is the most common cancer in women nowadays. According to the latest statistics from the National Cancer Center, breast cancer accounts for 17.10% of the total number of new tumors in women, and it has become the leading cause of death in women under 45 years due to malignant tumors [1]. Biomarkers for BC can predict the prognosis of BC patients. In recent years, with the development of molecular studies, breast cancer molecular typing has gradually become hot, which provides an important basis for solving the heterogeneity of tumors, the accuracy of prognosis judgment, and the individualization of clinical treatment. At present, breast cancer is often classified into Luminal A, Luminal B, HER2+, and Triple Negative Breast Cancer (TNBC). Luminal A has the best prognosis and TNBC has a poor prognosis, but its pathogenesis still needs further study. Therefore, it is of great significance to study the mechanism of occurrence, development, and prognostic factors of different breast cancers to find ideas for individualized treatment.
There are many studies on the role of epigenetics in the development of breast cancer. Histone Deacetylases (HDACs) can remove acetyl, leading to chromatin structure compression and subsequent suppression of gene transcription [2,3]. HDAC1 (Histone deacetylase 1) is a histone deacetylase found in mammals. It can mediate structural changes of nucleosomes, regulate gene expression, and inhibit gene transcription, thus affecting tumor proliferation, metastasis, differentiation and invasion [4,5]. Research has shown that inhibiting HDACs can induce re-expression of ER in ER-deficient cells [6]. However, the mechanism of HDAC1 to regulate ER, PR and HER-2 transcription is not clear in breast cancer. The expression of HDAC1 in different types of breast cancer is rarely reported. Therefore, it is important to study how its upstream factors regulate its deacetylation and further affect ER, PR, and HER-2.
Retinoblastoma binding protein 4 (RBBP4), a 48 kD new tumor-specific protein, was found in the lysate of Hela cells [7,8]. RBBP4 is a nuclear protein belonging to a highly conserved subfamily with four WD-repeat sequences. RBBP4 was named due to its ability to bind to retinoblastoma protein in vivo and in vitro. Protein sequence domain analysis showed that RBBP4 is a conserved protein, especially in the WD40 domain [9].This retinoblastoma-binding protein plays an important role in nucleosome assembly and histone modification, which influences gene transcription and regulates cell cycle and proliferation [10]. RBBP4 plays an important role in chromatin metabolism, nucleosome assembly, and histone modification, which can regulate gene transcription, cell cycle, and proliferation [11]. Moreover, RBBP4 has gained attention for its potential involvement in the mechanism of carcinomas, such as liver cancer [12], lung cancer [13] and glioma [14,15]. However, its role and mechanism in breast cancer have not been reported.
The RBBP4 was the first identified in the peak of proteins related to different collections of chromatin containing assembly and nucleosome modifying compounds, including histone deacetylase (HDAC) compounds [16,17]. Thus, it plays an important role in chromatin metabolism, nucleosome assembly, and histone modification and participates in HDAC-mediated transcription [18]. In the presence of Rb and HDAC1, RBBP4 was shown to be able to associate with E2F1 and participate in cell cycle regulation [19]. The Rb protein can inhibit gene transcription, while RBBP4 can cause blocking the inhibition of Rb protein, through HDAC1 which directly binds Rb protein. However, whether RBBP4 in breast cancer can affect the mechanism of the occurrence and development of different subtypes of breast cancer by HDAC1, and then affect the prognosis and therapeutic efficacy of patients is unknown. In this study, the expression of HDAC1 and RBBP4 was detected by IHC and western blot, and the correlation between HDAC1, RBBP4 and ER, PR, HER-2 was explored. We assess the relationship between them and clinicopathological factors and prognosis.
Materials and methods
Patients and tissue samples
Formalin-fixed and paraffin-embedded (FFPE) breast cancer specimens and normal tissues were retrieved from the archive of Binzhou Medical University Hospital, China. A total of 240 patients with BC from January 2011 to December 2013 are collected. These patients had median age of 54 years from 34 to 78. There are 60 patients in each group, and there are four groups: Luminal A, Luminal B, HER-2+ and Triple Negative Breast Cancer (TNBC). None of the patients underwent any types of treatments before surgery. Clinicopathologic features of all the patients were collected retrospectively by reviewing medical records.
Fresh-frozen tumor tissues and the corresponding normal tissues were used for western blotting extraction. These tissues were collected from 12 patients with BC who underwent curative surgery between August 2018 and June 2019. Tumor samples were obtained at surgery and stored at -80°C. The corresponding normal tissues were considered as the control. There were 3 patients in each group.
This research was approved by the Ethics Committee of Binzhou Medical University Hospital.
Follow up
All patients were followed up by telephone or outpatient clinics.
Immunohistochemistry (IHC)
The expression of HDAC1 and RBBP4 in BC and normal tissues was determined through IHC. FFPE archived tissues were cut into 3-μm sections, dewaxed, rehydrated, and blocked with 3% hydrogen peroxide. The sections were subjected to heat for antigen retrieval. The sections were then incubated with rabbit polyclonal antibody against human HDAC1 and RBBP4, respectively, overnight at 4°C. The sections were washed with PBS and incubated with horseradish peroxidase-labeled secondary antibody for 30 min. Subsequently, all sections were visualized with DAB kit (Zhong Shan Golden Bridge Biotechnology, Beijing, China) and the nucleus was counterstained with hematoxylin. In addition, all the primary antibodies were are purchased from Abcam, USA. A positive control was supplied by Abcam, and negative controls were prepared by replacing the primary antibody with PBS.
The results of final IHC were evaluated according to the intensity and percentage of positively stained cells. The staining intensity was graded as the followings: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The percentage of positive cells was assigned as the followings: 1 (≤25%), 2 (26%-50%), 3 (51%-75%), 4 (>76%). The final IHC score is calculated by multiplying the score of staining intensity and the score of percentage of positive cells. The score lower than 4 was regarded as low expression and the score higher than 4 was regarded as high expression. The results of IHC were blindly evaluated by two pathologists.
Western blotting
The fresh tissue was extracted on the ice. Protein concentrations were measured by BCA assay. The protein extracts were separated through SDS-PAGE and then transferred onto PVDF membranes (Millipore, USA). The membranes were blocked with 5% nonfat milk at room temperature for 90 min. The membranes were then incubated with HDAC1, RBBP4 and GAPDH, respectively, overnight at 4°C. Next, the membranes were incubated at the room temperature for 1.5 hours with secondary antibodies. The results were observed with enhanced chemiluminescence (ECL; Thermo Fisher, USA) by chemiluminescence detection system.
Statistical analysis
SPSS 23.0 was used to conduct all statistical analyses. The correlation between the proteins of different groups was performed using Spearman’s correlation analyses. The correlation of the protein expression with clinical parameters was analyzed by Pearson chi-squared test. Overall survival was plotted using the Kaplan-Meier method. Log-Rank method was used to compare the statistical difference and the independent prognostic factor for BC was assessed using a multivariate multivariate survival analysis. P<0.05 was considered significant.
Results
Expression of HDAC1 and RBBP4 and their correlation in breast cancer
The expression of HDAC1 and RBBP4 are assessed in breast cancer by both IHC and western blotting.
By IHC (Figure 1A), the positive rates of HDAC1 and RBBP4 in BC (73.33% and 70.42%, respectively) were higher than those in normal tissues (17.92% and 15.42%, respectively, P<0.05, Table 1). By western blotting, the expressions of HDAC1 and RBBP4 in BC are significantly higher than those in normal (P<0.05, Figure 1B).
Figure 1.
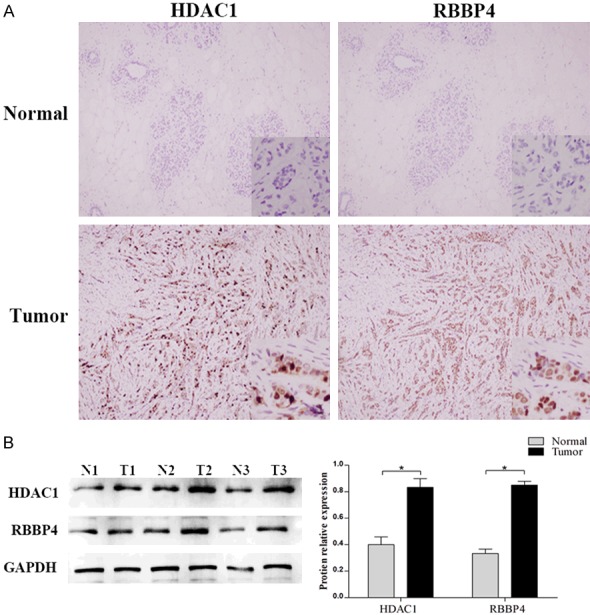
Expression of HDAC1 and RBBP4 in BC and normal tissues. A. Expression of HDAC1 and RBBP4 in BC and normal tissues by IHC; B. Expression of HDAC1 and RBBP4 in BC and normal tissues by western blot.
Table 1.
Expression of HDAC1 and RBBP4 in breast cancer and normal tissues [n (%)]
| n | HDAC1 | P | RBBP4 | P | |
|---|---|---|---|---|---|
| Normal | 240 | 43 (17.92) | <0.001* | 37 (15.42) | <0.001* |
| Cancer | 240 | 176 (73.33) | 169 (70.42) |
P<0.05.
Spearman analysis shows that HDAC1 is positively correlated with RBBP4 in breast cancer (P<0.05, Table 2). In other words, RBBP4 was also highly expressed in breast cancer with high expression of HDAC1.
Table 2.
Expression of HDAC1 and RBBP4 in breast cancer
| n | RBBP4 | P | |||
|---|---|---|---|---|---|
|
| |||||
| - | + | ||||
| HDAC1 | - | 64 | 27 | 37 | 0.010* |
| + | 176 | 44 | 132 | ||
P<0.05.
Expressions of HDAC1 and RBBP4 in different types of breast cancer
According to the expression of ER, PR, and HER-2 by IHC, the BC were further divided into four groups: Luminal A (ER+, PR+, HER-2-), Luminal B (ER+, PR+, HER-2+), HER-2+ (ER-, PR-, HER-2-) and Triple Negative Breast Cancer (TNBC, ER-, PR-, HER-2-).
According to the result of IHC, the expression of HDAC1 in Luminal A and Luminal B was significantly lower than that in HER-2+ and TNBC (P<0.05, Table 3; Figures 2, 3A). In Luminal A, RBBP4 was lower than HER-2+. RBBP4 was higher than Luminal B in HER-2+ and TNBC. However, there was no differences in the expression of HDAC1 and RBBP4 between Luminal A and Luminal B. At the same time, there were no differences between HER-2+ and TNBC.
Table 3.
Expression of HDAC1 and RBBP4 in different types of breast cancer
| n | HDAC1 | P | RBBP4 | P | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| - | + | - | + | ||||
| Luminal A | 60 | 24 | 36 | 0.001* | 22 | 48 | 0.020* |
| Luminal B | 60 | 27 | 33 | 24 | 36 | ||
| HER-2+ | 60 | 4 | 56 | 11 | 49 | ||
| TNBC | 60 | 9 | 51 | 14 | 46 | ||
P<0.05.
Figure 2.
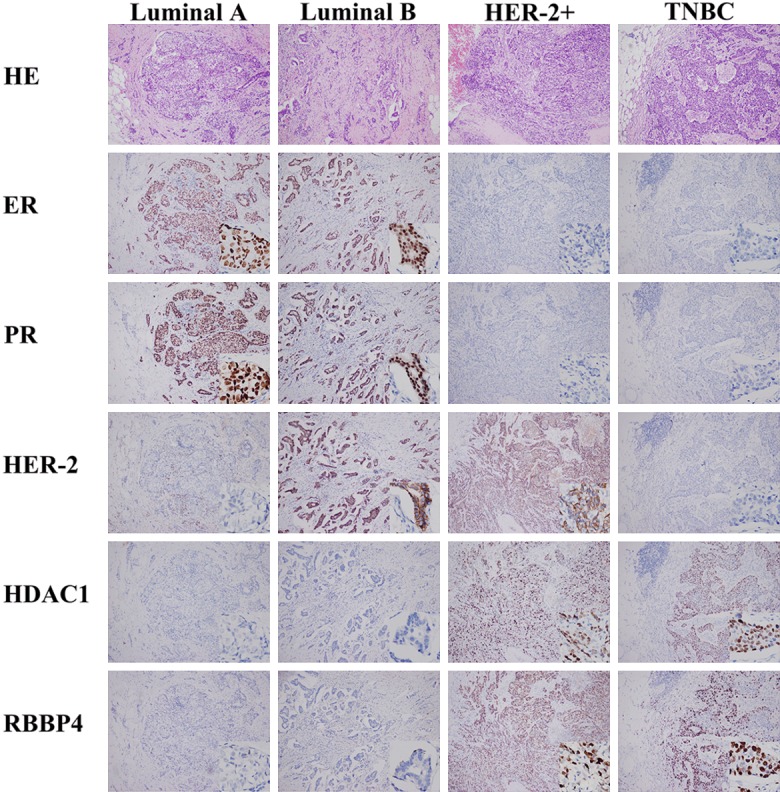
Expression of ER, PR, HER-2, HDAC1 and RBBP4 in different types of breast cancer by IHC.
Figure 3.
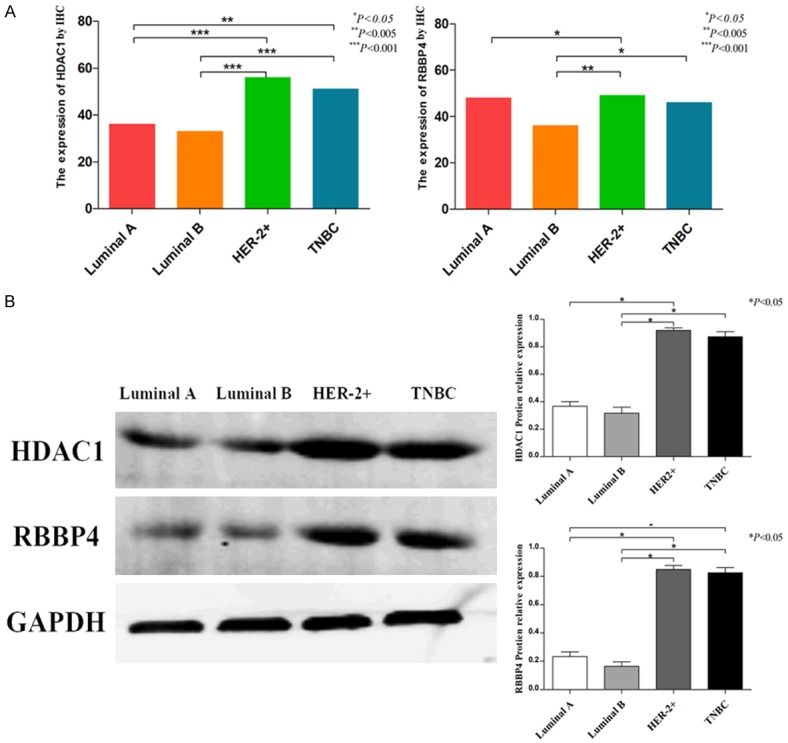
Expression of HDAC1 and RBBP4 in different types of breast cancer.
By western blot, the expression of HDAC1 and RBBP4 in Luminal A and Luminal B was significantly lower than that in HER-2+ and TNBC (P<0.05, Figure 3B). There was no difference in expression between Luminal A and Luminal B. There were no differences between HER-2+ and TNBC.
Correlation between HDAC1, RBBP4 and ER, PR, HER-2 in breast cancer
Spearman analysis showed that HDAC1 and RBBP4 is negatively correlated with ER and PR in BC, respectively (P<0.05, Table 4); however, there was no correlation between HDAC1 and HER-2 in BC, nor RBBP4. (P>0.05, Table 4).
Table 4.
Correlation between the expressions of HDAC1 and RBBP4 with ER, PR, and HER-2 in breast cancer
| n | HDAC1 | P | RBBP4 | P | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| - | + | - | + | |||||
| ER | - | 120 | 13 | 107 | 0.001* | 25 | 95 | 0.003* |
| + | 120 | 51 | 69 | 46 | 74 | |||
| PR | - | 120 | 13 | 107 | 0.001* | 25 | 95 | 0.003* |
| + | 120 | 51 | 69 | 46 | 74 | |||
| HER-2 | - | 120 | 33 | 87 | 0.770 | 36 | 84 | 0.888 |
| + | 120 | 31 | 89 | 35 | 85 | |||
P<0.05.
High expression of RBBP4 is an independent biomarker of poor prognosis in BC patients
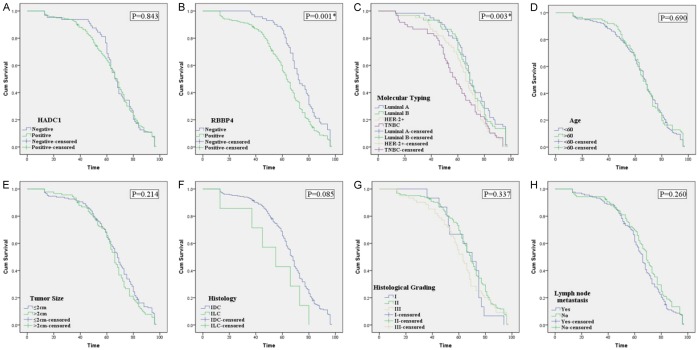
All 240 patients with BC had complete follow-up data: the median overall survival (OS) time is 66.5 months, and the 5-year survival rate is 67.5%. Kaplan-Meier analysis showed that patients with high expression of RBBP4 had a worse OS time than that of patients with low expression of RBBP4; and a worse OS is associated with TNBC. Luminal A has the best prognosis (P<0.05, Figure 4). Interestingly, HDAC1 is unrelated to prognosis (P>0.05, Figure 4). Multivariate survival analysis showed that RBBP4, molecular typing, and lymph node metastasis were independent prognostic factors for BC patients (Table 5).
Figure 4.
Kaplan-Meier survival analysis in BC. The log-rank test was used to calculate P value.
Table 5.
Multivariate survival analysis of clinicopathologic measures with OS by Cox proportional hazards regression
| B | SE | Wald | df | Exp (B) | 95% CI | P | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lower | Upper | |||||||
| RBBP4 | 0.493 | 0.152 | 10.572 | 1 | 1.637 | 1.216 | 2.204 | 0.001* |
| HDAC1 | -0.287 | 0.165 | 3.017 | 1 | 0.751 | 0.543 | 1.037 | 0.082 |
| Molecular typing | 0.250 | 0.068 | 13.428 | 1 | 1.284 | 1.123 | 1.467 | 0.001* |
| Age | -0.018 | 0.137 | 0.017 | 1 | 0.982 | 0.750 | 1.286 | 0.897 |
| Histology | 0.453 | 0.389 | 1.354 | 1 | 1.572 | 0.734 | 3.369 | 0.245 |
| Tumor size | 0.204 | 0.138 | 2.182 | 1 | 1.226 | 0.935 | 1.608 | 0.140 |
| Histologic grade | 0.131 | 0.132 | 0.984 | 1 | 1.140 | 0.880 | 1.476 | 0.321 |
| Lymph node metastasis | 0.310 | 0.140 | 4.937 | 1 | 0.733 | 0.558 | 0.964 | 0.026* |
P<0.05.
Correlation of HDAC1 and RBBP4 with clinicopathologic features
We analyzed the correlation between the HDAC1 and RBBP4 in BC and a set of clinicopathologic measures, including age, histology, tumor site, histological grade, and lymph node metastasis (Table 6). The expression of RBBP4 was found to be significantly correlated with lymph node metastasis (P=0.008). Other characteristics, such as age, gender, tumor size, and histology, were not associated with RBBP4 expression. The expression of HDAC1 was not correlated with clinicopathologic measures (P>0.05, Table 6).
Table 6.
Correlation between clinicopathologic characteristics and the expression of HDAC1 and RBBP4 in breast cancer
| Measures | n | HDAC1 | r | P | RBBP4 | r | P |
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| + | + | ||||||
| Age (year) | |||||||
| <60 | 153 | 112 | 0.004 | 0.952 | 108 | -0.005 | 0.939 |
| ≥60 | 87 | 64 | 61 | ||||
| Histology | |||||||
| IDC | 233 | 171 | -0.007 | 0.908 | 163 | 0.058 | 0.369 |
| ILC | 7 | 5 | 6 | ||||
| Tumor size (cm) | |||||||
| <2 | 149 | 111 | -0.034 | 0.603 | 106 | -0.020 | 0.754 |
| ≥2 | 91 | 65 | 63 | ||||
| Histologic grading | |||||||
| I | 15 | 12 | 0.045 | 0.473 | 12 | 0.052 | 0.414 |
| II | 164 | 116 | 110 | ||||
| III | 61 | 48 | 47 | ||||
| Lymph node metastasis | |||||||
| No | 134 | 92 | 0.119 | 0.066 | 85 | 0.172 | 0.008* |
| Yes | 106 | 84 | 84 |
P<0.05.
Discussion
We herein demonstrated that RBBP4 is positively correlated with BC metastasis. RBBP4 is closely related to the prognosis, and it is an independent prognostic factor for BC. The survival time of patients with RBBP4 (+) was significantly shorter than for RBBP4 (-). Our results suggest that activation of RBBP4 may lead to tumorigenesis, and the high of expression significantly affects the prognosis of the BC. More importantly, RBBP4 may serve as avaluable independent prognostic biological marker.
BC is a heterogeneous disease given that its invasive process is associated with a variety of molecular alterations. At present, breast cancer is often classified into Luminal A, Luminal B, HER2+, and Triple Negative Breast Cancer (TNBC). There are many individualized therapies for different breast cancer, but there are no drugs related to epigenetics of breast cancer. Histone hypoacetylation and hypermethylation are the main characteristics of cancer cells. Among histone acetylation or deacetylation play a key role in the regulation of extensive gene expression. HDAC1 (Histone deacetylase 1) is a histone deacetylase found in mammals. It can affect tumor proliferation, metastasis, differentiation and invasion. It was found that HDAC1 was overexpressed in gastric cancer [20,21], breast cancer [22,23], colorectal cancer [24,25], pancreatic cancer [26], cervical cancer and other malignant tumors, which was closely related to cell apoptosis. Tharkar [27] found that HDAC1 plays an important role in the survival and proliferation of leukemia cells. Targeted inhibition of HDAC1 expression can play an anti-leukemia role. In this study, we find that HDAC1 has high expression in breast cancer. Spearman analysis shows that HDAC1 is negatively correlated with ER and PR in BC, bur not HER-2. We think that HDAC1 is involved in the transcription or translation of ER and PR, but does not participate in the transcription of HER-2. However, HDAC1 is unrelated to any clinicopathological factors and does not affect the prognosis of patients. We suppose that HDAC1 can only affect the expression of downstream genes, but not the prognosis of patients.
Qian [8] first discovered RBBP4 in HeLa cells, which is a new tumor-specific protein. RBBP4 is named for its ability to bind to retinoblastoma proteins in vivo and in vitro, also known as RbAp48. RBBP4 is a nuclear protein with four WD repeats at the C-terminal, belonging to a highly conserved protein family with such domains. Recent studies have shown that RBBP4 may be involved in many tumorigenesis mechanisms, such as hepatocellular carcinoma [28], gastric cancer [29] and acute myeloid leukemia [30]. According to the results of IHC, RBBP4 has high expression in breast cancer. At the same time, the expression of RBBP4 was found to be significantly correlated with lymph node metastasis. We suppose that RBBP4 is involved in the development and metastasis of breast cancer.
It has been found that RBBP4 binds to histone modification-related complexes, including histone deacetylases (HDACs). Schultz [19] found that RBBP4 could affect HDAC-mediated transcription. In the presence of Rb and HDAC1, RBBP4 can bind to e2F1 and participate in cell cycle regulation. We find that HDAC1 is positively correlated with RBBP4 in breast cancer. RBBP4 is negatively correlated with ER and PR in BC, bur not HER-2. This result is the same as for HDAC1. We think that RBBP4 is concerned with the expression of ER and PR through HDAC1.
Results of both IHC and western blot showed, that expression of HDAC1 in Luminal A and Luminal B was significantly lower than that in HER-2+ and TNBC. In Luminal A, RBBP4 was lower than in HER-2+. RBBP4 was higher than Luminal B in HER-2+ and TNBC. However, there was no difference in the expression of HDAC1 and RBBP4 between Luminal A and Luminal B. At the same time, there were no differences between HER-2+ and TNBC. We consider that this result is related to RBBP4 and HDAC1 only regulating ER and PR. Thus, the expression of HDAC1 and RBBP4 in HER-2+ and TNBC, which are ER- and PR-, was significantly higher than that in ER+/PR+.
In conclusion, RBBP4 has high expression in breast cancer, and may be involved in the occurrence and development of lymph node metastasis in breast cancer, and affect the prognosis of breast cancer patients. Its mechanism may be regulated transcription or translation of ER and PR by HDAC1. Therefore, it is important to further study the molecular mechanism of RBBP4. It is of great significance to judge the prognosis of breast cancer and to explore potential ways to inhibit tumor development. However, the study discussed only a correlation of protein expression and clinical significance, and we need to increase the sample quantity and cell experiments carried out to reveal its role and mechanism.
Acknowledgements
We would like to thank Shuhua Wu and Xiaoyang Xu for the pathological assistance. National Natural Science Foundation of China (NO. 81902702 and NO. 81173601), Natural Science Foundation of Shandong Province (NO. ZR2017LH072 and NO. ZR2017MH033), Projects of Binzhou technology development program (NO. 2015ZC0301), National Key Research and Development Project (NO. 2018YFC0114705), Scientific Research Staring Foundation of Binzhou Medical University (NO. BY2014KYQD36, NO. BY2014KJ36 and NO. BY2017KJ01). And the funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Nyante SJ, Lee SS, Benefield TS, Hoots TN, Henderson LM. The association between mammographic calcifications and breast cancer prognostic factors in a population-based registry cohort. Cancer. 2017;123:219–227. doi: 10.1002/cncr.30281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 3.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- 5.Jaworska J, Ziemka-Nalecz M, Zalewska T. Histone deacetylases 1 and 2 are required for brain development. Int J Dev Biol. 2015;59:171–177. doi: 10.1387/ijdb.150071tz. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Zhou C, Jiang H, Liang L, Shi W, Zhang Q, Sun P, Xiang R, Wang Y, Yang S. ZEB1 induces ER-α promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8:e2732. doi: 10.1038/cddis.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 8.Qian YW, Wang YC, Hollingsworth RE Jr, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 9.Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Müller CW. Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J Biol Chem. 2011;286:23388–23396. doi: 10.1074/jbc.M110.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naser R, Vandenbosch R, Omais S, Hayek D, Jaafar C, Al Lafi S, Saliba A, Baghdadi M, Skaf L, Ghanem N. Role of the retinoblastoma protein, Rb, during adult neurogenesis in the olfactory bulb. Sci Rep. 2016;6:20230. doi: 10.1038/srep20230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang J, Li L, Huang W, Sui C, Yang Y, Lin X, Hou G, Chen X, Fu J, Yuan S, Li S, Wen W, Tang S, Cao D, Wu M, Chen L, Wang H. MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial-mesenchymal transition. Cancer Lett. 2015;364:33–43. doi: 10.1016/j.canlet.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Lu Y, Stemmer PM, Zhang X, Bi Y, Yi Z, Chen F. The proteomic investigation reveals interaction of mdig protein with the machinery of DNA double-strand break repair. Oncotarget. 2015;6:28269–28281. doi: 10.18632/oncotarget.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong BC, Slack RS. RB: an essential player in adult neurogenesis. Neurogenesis (Austin) 2017;4:e1270382. doi: 10.1080/23262133.2016.1270382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Aubert A, Gomez de Segura JM, Karuppasamy M, Basu S, Murthy AS, Diamante A, Drury TA, Balmer J, Cramard J, Watson AA, Lando D, Lee SF, Palayret M, Kloet SL, Smits AH, Deery MJ, Vermeulen M, Hendrich B, Klenerman D, Schaffitzel C, Berger I, Laue ED. The nucleosome remodeling and deacetylase complex NuRD is built from preformed catalytically active sub-modules. J Mol Biol. 2016;428:2931–2942. doi: 10.1016/j.jmb.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alqarni SS, Murthy A, Zhang W, Przewloka MR, Silva AP, Watson AA, Lejon S, Pei XY, Smits AH, Kloet SL, Wang H, Shepherd NE, Stokes PH, Blobel GA, Vermeulen M, Glover DM, Mackay JP, Laue ED. Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J Biol Chem. 2014;289:21844–21855. doi: 10.1074/jbc.M114.558940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidberger JW, Sharifi Tabar M, Torrado M, Silva AP, Landsberg MJ, Brillault L, AlQarni S, Zeng YC, Parker BL, Low JK, Mackay JP. The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/7. Protein Sci. 2016;25:1472–82. doi: 10.1002/pro.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz LE, Haltom JA, Almeida MP, Wierson WA, Solin SL, Weiss TJ, Helmer JA, Sandquist EJ, Shive HR, McGrail M. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Dis Model Mech. 2018;11 doi: 10.1242/dmm.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao LL, Yue Z, Liu L, Pei L, Yin Y, Qin L, Zhao J, Liu H, Wang H, Jia M. The expression of histone deacetylase HDAC1 correlates with the progression and prognosis of gastrointestinal malignancy. Oncotarget. 2017;8:39241–39253. doi: 10.18632/oncotarget.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Z, Zeng J, Liu H, Wang T, Yu Z, Chen J. Role of HDAC1 in the progression of gastric cancer and the correlation with lncRNAs. Oncol Lett. 2019;17:3296–3304. doi: 10.3892/ol.2019.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byun JS, Park S, Yi DI, Shin JH, Hernandez SG, Hewitt SM, Nicklaus MC, Peach ML, Guasch L, Tang B, Wakefield LM, Yan T, Caban A, Jones A, Kabbout M, Vohra N, Nápoles AM, Singhal S, Yancey R, De Siervi A, Gardner K. Epigenetic re-wiring of breast cancer by pharmacological targeting of C-terminal binding protein. Cell Death Dis. 2019;10:689. doi: 10.1038/s41419-019-1892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montalto FI, Giordano F, Chiodo C, Marsico S, Mauro L, Sisci D, Aquila S, Lanzino M, Panno ML, Andò S, De Amicis F. Progesterone receptor B signaling reduces breast cancer cell aggressiveness: role of cyclin-D1/Cdk4 mediating paxillin phosphorylation. Cancers (Basel) 2019;11 doi: 10.3390/cancers11081201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B, Liu B, Chen D, Setroikromo R, Haisma HJ, Quax WJ. Histone deacetylase inhibitors sensitize TRAIL-induced apoptosis in colon cancer cells. Cancers (Basel) 2019;11 doi: 10.3390/cancers11050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundararajan V, Tan M, Tan TZ, Ye J, Thiery JP, Huang RY. SNAI1 recruits HDAC1 to suppress SNAI2 transcription during epithelial to mesenchymal transition. Sci Rep. 2019;9:8295. doi: 10.1038/s41598-019-44826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laschanzky RS, Humphrey LE, Ma J, Smith LM, Enke TJ, Shukla SK, Dasgupta A, Singh PK, Howell GM, Brattain MG, Ly QP, Black AR, Black JD. Selective inhibition of histone deacetylases 1/2/6 in combination with gemcitabine: a promising combination for pancreatic cancer therapy. Cancers (Basel) 2019;11 doi: 10.3390/cancers11091327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tharkar-Promod S, Johnson DP, Bennett SE, Dennis EM, Banowsky BG, Jones SS, Shearstone JR, Quayle SN, Min C, Jarpe M, Mosbruger T, Pomicter AD, Miles RR, Chen WY, Bhalla KN, Zweidler-McKay PA, Shrieve DC, Deininger MW, Chandrasekharan MB, Bhaskara S. HDAC1,2 inhibition and doxorubicin impair Mre11-dependent DNA repair and DISC to override BCR-ABL1-driven DSB repair in Philadelphia chromosome-positive B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32:49–60. doi: 10.1038/leu.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R, Tan Y, Fan J, Chang Y, Fu J, Jiang F, Chen C, Yang Y, Gu J, Wu D, Guo L, Cao D, Li H, Cao G, Wu M, Zhang MQ, Chen L, Wang H. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut. 2015;64:156–167. doi: 10.1136/gutjnl-2013-305715. [DOI] [PubMed] [Google Scholar]
- 29.Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, Li Z, Wei J, Liu M, Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Li F, Zhang B, Li S, Wu J, Shi Y. Structural basis of plant homeodomain finger 6 (PHF6) recognition by the retinoblastoma binding protein 4 (RBBP4) component of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2015;290:6630–6638. doi: 10.1074/jbc.M114.610196. [DOI] [PMC free article] [PubMed] [Google Scholar]