Abstract
The chemokine (C-X-C motif) ligand (CXCL) family plays an important role in inflammation. In order to understand the role of CXC chemokine family in carcinogenesis, this study explored a group of early gastric cancer (GC) patients, and assessed the level of CXC chemokine ligand (CXCL) in blood samples of patients representing systemic circulation and tumor microenvironment, detected the expression of CXC chemokine receptor (CXCR) in tumor tissues, and measured tumor infiltrating immune cell subsets. 69 patients with GC were included in a single center prospective study and were followed up for 6 years. The level of CXCL1-14 was determined by ELISA and the concentration gradient of chemokine was calculated. Western blot was used to detect the expression of CXCR1, CXCR2, CXCR3, and CXCR4 in tumor tissue. CXCL1-14 expression was inhibited by siRNA in HGC27 cells and then the migration ability of HGC27 cells was detected by cell scratch test. The results of this study showed that the chemokine concentrations of CXCL1, CXCL2, CXCL5, CXCL8, CXCL11, and CXCL13 in peripheral blood and tumor drainage blood of patients without recurrence after treatment were significantly lower than those before treatment. The concentrations of CXCL1, CXCL2, CXCL4, CXCL5, CXCL7, CXCL8, CXCL9, CXCL10, CXCL12, CXCL13, and CXCL14 in peripheral blood and tumor drainage blood were significantly higher than those in patients without recurrence. Patients with low expression of CXCR1 and CXCR3 had lower AFP (alpha fetoprotein), smaller tumor volume, and lower TNM tumor stage. Patients with lower expression of CXCR2 and CXCR4 had higher AFP (alpha fetoprotein) level, larger tumor volume, and higher TNM tumor stage. After down-regulation of CXCLs expression, the migration ability of most cell lines was significantly inhibited. This study suggests that CXCL chemokine family plays an important role in the pathogenesis of GC and can be used as a marker for the development of GC.
Keywords: CXCL, CXCR, gastric cancer, chemokine, prognosis
Introduction
Gastric cancer (GC) is the fourth most common malignant tumor in the world and ranks the third in the mortality rate of male and female cancer. The 5-year survival rate of GC is usually lower than 30% [1]. Although the global incidence of GC has declined dramatically in the past decades, most patients with GC are still in the middle and late stages of the diagnosis. In recent years, cancer treatment strategies tend to the research of tumor molecular targeted therapy [2,3].
The growth, proliferation, and metastasis of tumor cells is a continuous process, which needs a continuous energy supply. The formation of blood vessels provides a material basis for this process. Understanding the occurrence of cancer requires a holistic approach, including the analysis of epidemiological and clinical data, molecular detection of blood biomarkers, and thorough analysis of tumor cells and immune cell infiltration [4,5]. The data obtained in the past decades confirmed that cancer, as a disease, has heterogeneity; which means that there is no single perfect biomarker for GC, but a group of multi parameters may be a more objective diagnostic and prognostic tool [6]. Solid tumors like GC contain not only tumor cells, but also various types of stromal cells, such as fibroblasts and endothelial cells [7]. In addition, the tumor is infiltrated by inflammatory cells, including neutrophils, macrophages, and lymphocytes. Tumor cells, stromal cells, and tumor related leukocytes contribute to the local production of chemokines in tumor and affect the level of systemic circulating chemokines [8]. Chemokines are a kind of cytokines that control cell migration. The abnormal expression and function of chemokines and their receptors will lead to immune cells unable to perform the right function in the right position [9,10].
In our study, we focused on a group of CXC chemokines, which are members of the cytokine family. Previous studies have shown that CXC chemokines family is involved in tumor growth regulation and metastasis [11,12]. Our team’s previous studies confirmed the diagnostic and prognostic value of CXC chemokines as biomarkers [13]. The in-depth exploration of CXC chemokine family can provide valuable scientific basis for clinical and transformation research of GC patients undergoing surgical treatment and provide opportunities for tumor microenvironment research. In order to better understand the role of CXC chemokines in tumorigenesis, this study detected the CXC chemokine ligand (CXCL) levels in blood samples of patients who represent the systemic and tumor microenvironment to determine whether chemokines are involved in the carcinogenesis process. To ensure the overall treatment, our additional goal is to determine the expression intensity of CXC chemokine receptor (CXCR) in tumor tissue and to evaluate the components of tumor immune cell (TIC) infiltration.
Materials and methods
Experimental object
From January 2012 to December 2013, 69 patients with GC underwent radical resection and a single center prospective study with clinical, radiological, and pathological stages of adenocarcinoma (n=48) or squamous cell carcinoma (n=21). There was no new adjuvant and/or adjuvant therapy in all patients. The patients were followed up for 6 years. During the follow-up period, the general condition of the patient and information about the outcome and recurrence of the disease were obtained. All patients signed the informed consent for the experimental content.
Program
During the operation, the peripheral blood samples were taken from the elbow vein and put into a 5 ml vacuum tube. The tubes were centrifuged at 1500 g for 10 minutes at room temperature (RT). After centrifugation, the plasma was immediately stored at -80°C for use. Before the ELISA test, the samples were thawed at RT and treated in accordance with the instructions provided by the ELISA kit manufacturer. The level of CXCL1-14 (Abnova, USA) was determined by ELISA Kit with sensitivity of 1 pg/ml and detection range of 1-15000 pg/ml. This was repeated 3 times for each sample. The concentration of CXCLs in plasma was calculated according to the standard curve. The mean value of CXCLs level was stated with 95% confidence interval.
The difference of CXCLs concentration between systemic vascular bed and tumor vascular bed circulation can be described by CXCLs concentration gradient. The gradient of CXCLs was calculated as: [(TCV-PCV)/PCV] × 100, in which TCV was the representative value of tumor circulation and PCV was the representative value of peripheral circulation.
Quantitative real-time PCR
Total RNA was extracted and isolated from tissue samples using Trizol reagent (Invitrogen, USA) following the user manual. SYBR PremixExTaqTM (Takara, Japan) was used in Quantitative real-time PCR (qRT-PCR) on BIO-RAD MY IQ (USA). All primer sequences are shown in Table 1. The relative expression of CXCLs was shown as fold difference relative to GAPDH.
Table 1.
Primer sequences of products expression
| Gene name | Primer name | Primer sequence |
|---|---|---|
| CXCL1 | forward primer | 5’ CGCTACAGCGACGTGAAGAA 3’ |
| reverse primer | 5’ GTTCCAGGCGTTGTACCAC 3’ | |
| CXCL2 | forward primer | 5’ GCTTGTC TCAACCCCGCATC 3’ |
| reverse primer | 5’ TGGATTTGCCATTTTTCAGCATCTT 3’ | |
| CXCL3 | forward primer | 5’ GCAGGGAATTCACCTCAAGA 3’ |
| reverse primer | 5’ TTTTCGATGATTTTCTGAACCA 3’ | |
| CXCL4 | forward primer | 5’ TGAAGAATGGAAGGAAAATTTGC 3’ |
| reverse primer | 5’ CAAATGCACACACGTAGGCAGCT 3’ | |
| CXCL5 | forward primer | 5’ AGCTGCGTTGCGTTTGTTTAC 3’ |
| reverse primer | 5’ TGGCGAACACTTGCAGATTAC 3’ | |
| CXCL6 | forward primer | 5’ AGAGCTGCGTTGCACTTGTT 3’ |
| reverse primer | 5’ GCAGTTTACCAATCGTTTTGGGG 3’ | |
| CXCL7 | forward primer | 5’ CTGGCTTCCTCCACCAAAGG 3’ |
| reverse primer | 5’ GACTTGGTTGCAATGGGTTCC 3’ | |
| CXCL8 | forward primer | 5’ CTTTGTCCATTCCCACTTCTGA 3’ |
| reverse primer | 5’ TCCCTAACGGTTGCCTTTGTAT 3’ | |
| CXCL9 | forward primer | 5’ CCAGTAGTGAGAAAGGGTCGC 3’ |
| reverse primer | 5’ AGGGCTTGGGGCAAATTGTT 3’ | |
| CXCL10 | forward primer | 5’ GTGGCATTCAAGGAGTACCTC 3’ |
| reverse primer | 5’ TGATGGCCTTCGATTCTGGATT 3’ | |
| CXCL11 | forward primer | 5’ GACGCTGTCTTTGCATAGGC 3’ |
| reverse primer | 5’ GGATTTAGGCATCGTTGTCCTTT 3’ | |
| CXCL12 | forward primer | 5’ ATTCTCAACACTCCAAACTGTGC 3’ |
| reverse primer | 5’ ACTTTAGCTTCGGGTCAATGC 3’ | |
| CXCL13 | forward primer | 5’ GCTTGAGGTGTAGATGTGTCC 3’ |
| reverse primer | 5’ CCCACGGGGCAAGATTTGAA 3’ | |
| CXCL14 | forward primer | 5’ CGCTACAGCGACGTGAAGAA 3’ |
| reverse primer | 5’ GTTCCAGGCGTTGTACCAC 3’ | |
| GAPDH | forward primer | 5’ ATGTCGTGGAGTCTACTGGC 3’ |
| reverse primer | 5’ TGACCTTGCCCACAGCCTTG 3’ |
Western blot assays
In order to study the relationship between CXC chemokine receptor (CXCR), immune cells, and GC, western blot was used to detect the expression of CXCRs. The total protein was extracted by RIPA method and the samples were electrophoretic by 10% SDS-PAGE. The amount of sample on each pore was 20 μL. After the membrane was transferred by semi dry electric transfer instrument, 5% skimmed milk powder was sealed for 1 hour, then the diluted Rabbit anti CXCR1 (1:1000), CXCR2 (1:1000), CXCR3 (1:5000), and CXCR4 (1:1000) antibody was added. After incubation at night, 4°C, 0.1% TBST washed the membrane three times, 5 min/time, added 1:10000 diluted Goat anti rabbit IgG, incubated in the greenhouse for 30 min, washed the membrane, added the chemiluminescent solution, developed, and analyzed the strip light density with ImageJ software.
Construction of CXCLs low expression cell line
CXCL1 (Gen bank gene ID: 2919), CXCL2 (Gen bank gene ID: 2920), CXCL3 (Gen bank gene ID: 2921), CXCL4 (Gen bank gene ID: 5196), CXCL5 (Gen bank gene ID: 6374), CXCL6 (Gen bank gene ID: 6372), CXCL7 (Gen bank gene ID: 5473), CXCL8 (Gen bank gene ID: 3576), CXCL9 (Gen bank gene ID: 4283), CXCL10 (Gen- Bank gene ID: 3627), CXCL11 (Gen bank gene ID: 6373), CXCL12 (Gen bank gene ID: 6387), CXCL13 (Gen bank gene ID: 10563), CXCL14 (Gen bank gene ID: 9547) gene sequences were designed by Invitrogen online software. The designed siRNA was synthesized by Ribobio. The siRNA was transfected into HGC27 cells by Lipofectamine 2000 (Invitrogen). After 6 hours of cell culture, the transfection effect was verified by western blot.
Cell scratch test
The cells were verified to obtain CXCLs low expression cell lines. After the cells were cultured for 12 hours, a 2 mm wide scratch was scratched along the middle line of the culture plate with a 1 ml pipette head. The culture plate was washed with PBS buffer solution and no cells were found in the scratches through observation under the microscope. After adding the culture medium, the culture continued for 24 hours. Suck the culture supernatant, add 1 ml of 4% paraformaldehyde into each hole, and take photos under the optical microscope 30 minutes later.
Statistical analysis
Data are presented as mean ± SEM. A Pearson chi-squared test was applied to determine clinicopathological correlations. The Kaplan-Meier method was used to calculate the survival curves. Significance was determined by the log-rank test. The difference among groups was determined by ANOVA analysis and comparison between two groups was analyzed by the Student’s t-test using GraphPad software version 5.0 (GraphPad Software, CA). Differences were considered significant when P<0.05.
Results
The relationship between CXCLs concentration and prognosis of GC patients
The final data collection in December 2018 showed that out of 69 GC patients who received surgical treatment, a total of 31 patients recurred. For further analysis, we divided the patients into two groups according to the follow-up data, taking the recurrence of cancer as a reference. We compared CXCL concentrations in paired blood samples representing systemic circulation and tumor bed circulation, as shown in Table 2. Compared with before treatment, the concentrations of CXCL1, CXCL2, CXCL5, CXCL8, CXCL11, and CXCL13 in peripheral blood and tumor drainage blood of patients without recurrence after treatment were significantly lower. The concentrations of CXCL1, CXCL2, CXCL4, CXCL5, CXCL7, CXCL8, CXCL9, CXCL10, CXCL12, CXCL13, and CXCL14 in peripheral blood and tumor drainage blood were significantly higher than those in patients without recurrence.
Table 2.
Concentration of CXCLs in peripheral and tumor draining blood samples (pg/mL) ± SD
| CXCLs | Before treatment (n=69) | After treatment (n=69) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Peripheral | Tumor draining blood | No recurrence (n=38) | Relapse (n=31) | |||
|
|
|
|||||
| Peripheral | Tumor draining blood | Peripheral | Tumor draining blood | |||
| CXCL1 | 156.23±47.36 | 137.46±32.57 | 36.78±9.87* | 26.22±11.45* | 239.13±103.27# | 198.47±34.21# |
| CXCL2 | 246.89±35.12 | 309.82±49.44 | 50.17±12.89* | 66.17±25.13* | 451.213±89.24# | 486.53±157.16# |
| CXCL3 | 486.79±108.55 | 389.21±67.15 | 419.21±75.11 | 513.16±65.24 | 637.99±79.46 | 789.31±109.44 |
| CXCL4 | 2672.28±389.77 | 2107.26±416.34 | 2145.35±479.39 | 1954.17±581.1 | 3657.71±897.33# | 4016.64±1023.38# |
| CXCL5 | 568.79±108.11 | 541.26±200.37 | 357.65±79.15* | 311.37±127.68* | 986.32±177.64# | 1089.21±208.42# |
| CXCL6 | 301.35±63.17 | 289.47±78.21 | 219.68±111.29 | 256.17±101.35 | 416.37±109.57 | 468.18±148.39 |
| CXCL7 | 1255.27±227.34 | 1536.15±242.45 | 1419.88±239.65 | 1549.58±309.17 | 1002.41±218.29# | 889.53±108.57# |
| CXCL8 | 58.24±9.78 | 76.29±7.88 | 21.89±11.13* | 29.15±7.43* | 164.45±17.66# | 186.64±29.69# |
| CXCL9 | 754.14±109.85 | 656.67±98.33 | 538.22±125.75 | 678.26±218.31 | 1176.11±210.45# | 1258.17±117.46# |
| CXCL10 | 86.46±19.89 | 112.29±23.73 | 56.54±11.24 | 64.76±11.68 | 189.43±35.51# | 214.61±45.28# |
| CXCL11 | 35.26±5.79 | 46.28±11.37 | 86.18±27.78* | 96.27±33.18* | 67.56±8.91 | 72.79±11.61 |
| CXCL12 | 26.21±6.76 | 31.27±10.07 | 56.57±27.63 | 76.62±18.97 | 15.11±4.38# | 19.41±8.19# |
| CXCL13 | 489.11±87.43 | 556.17±109.46 | 201.77±49.78* | 196.39±57.66* | 866.15±78.93# | 796.84±174.51# |
| CXCL14 | 87.27±20.31 | 96.64±19.39 | 138.23±27.76 | 186.78±50.33 | 36.16±7.95# | 29.69±9.61# |
Compared with before treatment;
P < 0.05.
Compared with no recurrence;
P < 0.05.
The relationship between the expression of CXCRs and the prognosis of GC
Using the method of retrospective cohort study, qRT-PCR was used to detect the expression level of CXCRs in 69 pairs of GC tissues and matched adjacent tissues. Compared with the adjacent tissues, the expression level of CXCRs was significantly different in GC tissues. According to the middle value of CXCRs expression level, the studied population was divided into two parts: CXCRs high expression and CXCRs low expression. As shown in Table 3, there is no significant correlation between CXCRs expression and age and gender in GC patients. Patients with low expression of CXCR1 and CXCR3 had lower AFP (alpha fetoprotein), smaller tumor volume, and lower TNM tumor stage. Patients with lower expression of CXCR2 and CXCR4 had higher AFP (alpha fetoprotein) level, larger tumor volume, and higher TNM tumor stage.
Table 3.
Correlation of the expression of CXCRs with clinicopathologic features
| Clinicopathologic features | n (69) | CXCR1 expression | P | CXCR2 expression | P | CXCR3 expression | P | CXCR4 expression | P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| Low | High | Low | High | Low | High | Low | High | ||||||
| Age (year) | |||||||||||||
| <50 | 28 | 15 | 13 | 0.669 | 11 | 18 | 0.835 | 12 | 16 | 0.581 | 13 | 16 | 0.617 |
| ≥50 | 41 | 19 | 22 | 23 | 18 | 21 | 20 | 18 | 23 | ||||
| Sex | |||||||||||||
| Male | 37 | 20 | 17 | 0.812 | 16 | 21 | 0.396 | 15 | 22 | 0.327 | 22 | 15 | 0.723 |
| Female | 32 | 14 | 18 | 21 | 11 | 19 | 13 | 16 | 16 | ||||
| Serum AFP level (ng/mL) | |||||||||||||
| <20 | 21 | 15 | 6 | 0.015* | 3 | 18 | 0.026* | 17 | 10 | 0.037* | 13 | 7 | 0.029* |
| ≥20 | 48 | 9 | 41 | 30 | 18 | 11 | 37 | 26 | 22 | ||||
| Tumor size (cm) | |||||||||||||
| <5 | 31 | 23 | 8 | 0.023* | 12 | 19 | 0.052 | 15 | 16 | 0.039* | 13 | 18 | 0.041* |
| ≥5 | 38 | 11 | 27 | 21 | 17 | 9 | 28 | 29 | 9 | ||||
| TNM tumor stage | |||||||||||||
| I+II | 45 | 31 | 14 | 0.008* | 9 | 36 | 0.026* | 24 | 21 | 0.043* | 13 | 32 | 0.035* |
| III+IV | 24 | 3 | 21 | 19 | 5 | 6 | 18 | 20 | 4 | ||||
The asterisk indicate P<0.05.
Western blot results showed that before treatment, the expression of CXCR1 and CXCR3 was significantly up-regulated, while that of CXCR2 and CXCR4 was significantly down regulated in GC tissue samples compared with the adjacent tissues (Figure 1). After treatment, the patient was divided into two parts. In the relapse free group, no tumor sample was taken, so the expression of CXCRs in tumor tissue could not be detected. However, in the tumor tissues of the recurrent group, the expression of CXCR1 and CXCR3 was significantly up-regulated, while that of CXCR2 and CXCR4 was significantly down regulated. This trend is more obvious than before treatment. This suggests that cancer cells may develop immune tolerance.
Figure 1.
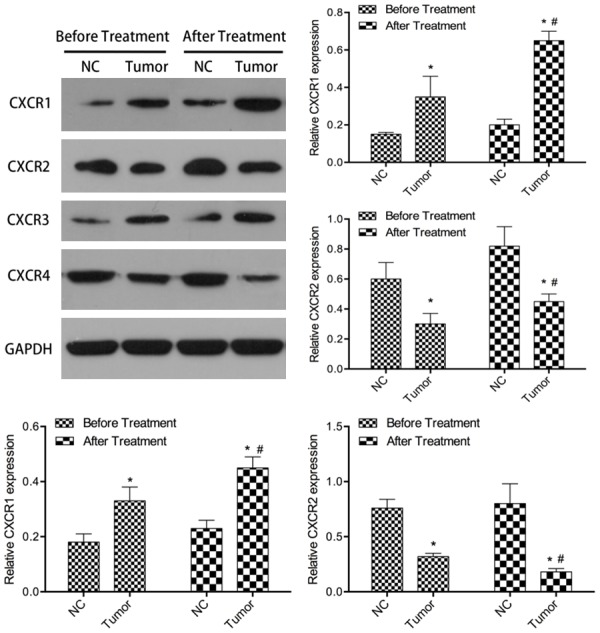
Relative CXCRs expression levels in GC tissues. CXCRs were detected in 69 pairs of GC tissues with western blot. NC, normal control tissues, Tumor, GC tissues. Compared with NC *P<0.05. Compared with before treatment #P<0.05.
The relationship between CXCRs expression and tumor invasion
The expression intensity of CXCR1 was related to the level of CXCL6 and CXCL13, the number of macrophage infiltrated tumor and tumor infiltrated immune cells. The expression intensity of CXCR2 was related to the level of CXCL1, CXCL2, CXCL5, and CXCL8 in the whole body and the infiltration of tumor plasma cells. The expression intensity of CXCR3 was correlated with the tumor infiltration level of CXCL4, CXCL9, CXCL10, CXCL11, B cells, T helper cells, and T cytotoxic cells. The expression intensity of CXCR4 was related to the level of CXCL14 in the whole body and the infiltration of tumor plasma cells.
CXCL1 gradient is related to the absolute number of B cells; CXCL2 gradient is related to the absolute number of B cells; CXCL3 gradient is related to the absolute number of macrophages and the total number of tumor infiltrating immune cells; CXCL4 gradient is related to the absolute number of macrophages and the total number of tumor infiltrating immune cells; CXCL5 gradient is related to the absolute number of Thelper cells, the percentage of T-helper cells, and the infiltration of t-toxic cells. CXCL6 gradients were correlated with the absolute number of B cells, CXCL7 gradients with the absolute number of T helper cells, CXCL8 gradients with the absolute number of T cytotoxic cells and the total number of tumor infiltrating cells. There was no significant correlation between CXCL9 gradient and immune cell subsets. There was no significant correlation between CXCL10 gradient and immune cell subsets. CXCL11 gradients were associated with the absolute number of T-cell toxic cells and the total number of tumor infiltrating cells. There was no significant correlation between CXCL12 gradient and T helper cell subsets. CXCL14 gradients were correlated with the absolute number of macrophages and the total number of tumor infiltrating immune cells.
Detection of cell migration in vitro by scratch test
After 6 h of transfection, western blot was used to verify the transfection effect. The experimental results showed that the CXCLs low expression cell line was successfully obtained (Figure 2). After 24 hours of cell culture, cell scratch test was used to detect the migration ability of HGC27 inhibited by CXCLs siRNA. The experimental results showed that after the down-regulation of CXCLs expression, the migration ability of most cell lines was significantly inhibited, but the migration ability of some cell lines did not change in detail (Figure 3).
Figure 2.
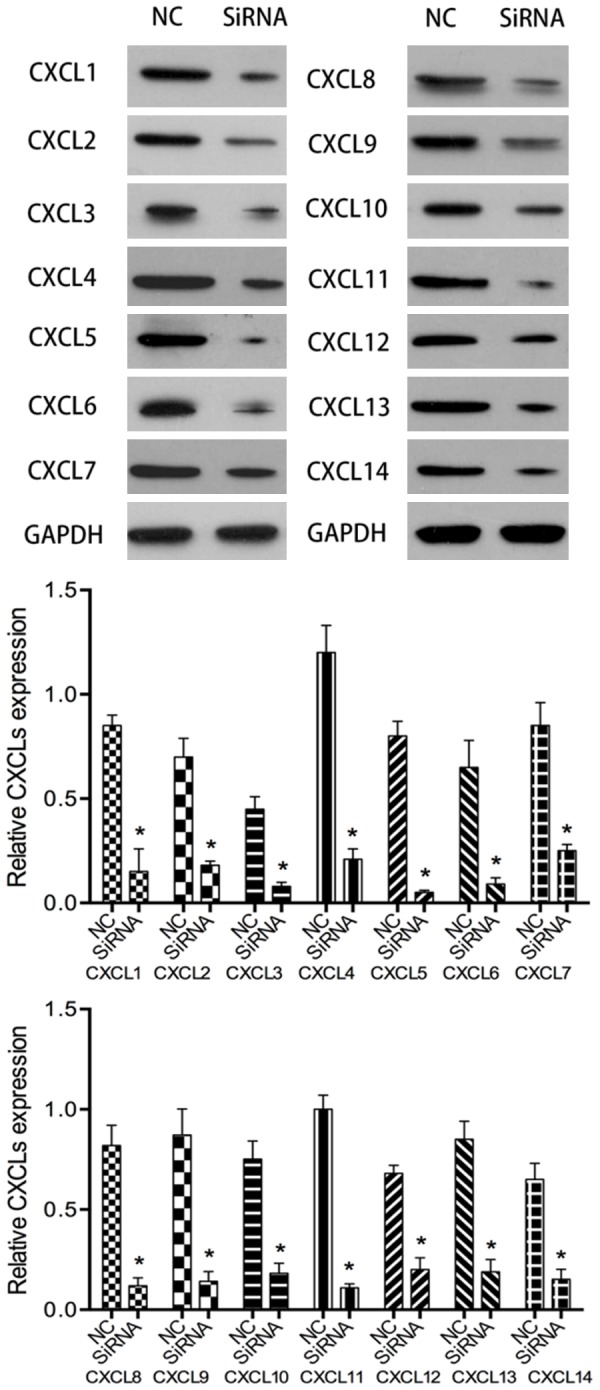
Western blot was used to verify the transfection effect of CXCLs siRNA. NC, normal control HGC27 cells. siRNA, HGC27 cells transfected with CXCLs siRNA. Compared with NC *P<0.05.
Figure 3.
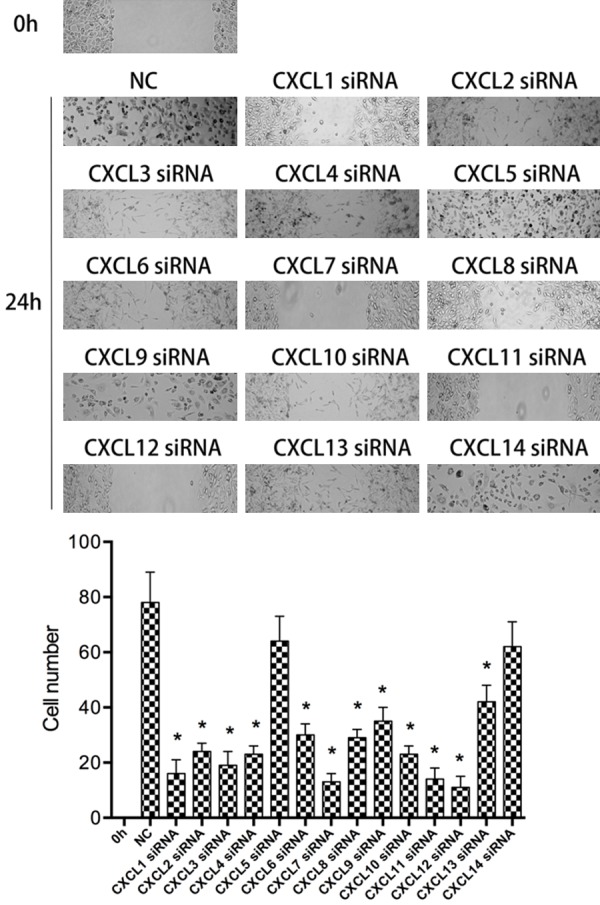
Scratch test was used to detect the migration ability of CXCLs siRNA to HGC27 cells. NC, normal control HGC27 cells. CXCL1-14 siRNA, HGC27 cells transfected with CXCLs siRNA. Compared with NC *P<0.05.
Discussion
It is very important for the clinical study of cancer to explore the therapeutic target of cancer. GC is a kind of malignant tumor with high mortality and poor prognosis. Although the clinical prevention and treatment of GC has achieved important results, the early diagnosis of GC is still a big challenge [14-16]. The occurrence of GC is a very complex process. Irregular work and rest, eating habits, and unhealthy food are important external factors to induce GC. The disorder of human internal environment and the imbalance of cell self-monitoring mechanism are important internal factors to induce GC [17,18]. Therefore, it is very important to develop a new biomarker for early diagnosis of GC. Chemokine family plays an important role in the occurrence and development of a variety of tumors. The basic function of chemokines is the directional chemotaxis of cells expressing corresponding chemokine receptors [19-21].
Chemokines and their receptor families are important mediators for leukocyte migration to inflammatory sites. Long term inflammation may provide an ideal microenvironment for tumor cell development and growth [22]. Chemokine family can stimulate or inhibit the development of tumor by attracting tumor promoting and anti-tumor cells. Chemokines affect tumor transformation, survival, growth, invasion, and metastasis by regulating angiogenesis and tumor leukocyte interaction [23,24]. The function of chemokines is very complex. Some chemokines may be beneficial to the growth and development of tumor, while others may enhance the anti-tumor immunity [25,26].
In this study, we identified the level of differentially expressed proteins in a complex proteomic context. The concentration of CXCL in tumor tissue is a dynamic parameter. On the one hand, the circulating CXCL produced by other tissues may combine with the CXCR expressed by tumor or tumor infiltrating immune cells, so as to reduce the concentration of CXCL in tumor tissue blood drainage. On the other hand, CXCL can be produced by tumor or tumor infiltrating immune cells and then released to tumor tissue blood drainage in addition, the concentration of CXCL in tumor tissue was increased [27]. This phenomenon can be explained by the classical hypothesis of immune editing, that is, by recognizing tumor specific antigen, the immune system can not only clear tumor cells and protect the host, but also edit tumor genome to shape tumor development and generate tumor variants with reduced immunogenicity. In this process, immune cells and CXCL are in a state of dynamic balance [28]. In our study, we found that most chemokines (CXCL1, CXCL2, CXCL4, CXCL5, CXCL7, CXCL9, CXCL10, CXCL12, CXCL13, and CXCL14) concentrations changed significantly in patients with GC without postoperative recurrence. In these patients with GC recurrence, most of the chemokines in the blood concentration did not change significantly. After the next stage of tumor development, escape stage, patients will experience immunosuppression, so that tumor spread. We also found that the levels and gradients of chemokines were related to the expression of CXCR and the number of tumor infiltrating immune cell subsets, thus promoting the development of GC immune core.
Our study shows that CXCL family plays an important role in the pathogenesis of GC. It can be used as a marker for the development of GC. This method has a wide range of applications and can be included in the study of GC transformation.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81570495).
Disclosure of conflict of interest
None.
References
- 1.Niccolai E, Taddei A, Prisco D, Amedei A. Gastric cancer and the epoch of immunotherapy approaches. World J Gastroenterol. 2015;21:5778–93. doi: 10.3748/wjg.v21.i19.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otani Y, Okabayashi T, Shima Y, Shibuya Y, Ozaki K, Iwata J, Morita S, Iiyama T. Safety and efficacy of the surgical management of hemodialysis patients with gastric cancer. Acta Med Okayama. 2017;71:333–339. doi: 10.18926/AMO/55310. [DOI] [PubMed] [Google Scholar]
- 3.Fang M, Tao Y, Liu Z, Huang H, Lao M, Huang L, Zhu B. Meta-analysis of the relationship between NM23 expression to gastric cancer risk and clinical features. Biomed Res Int. 2017;2017:8047183. doi: 10.1155/2017/8047183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang TT, Zhao YL, Peng LS, Chen N, Chen W, Lv YP, Mao FY, Zhang JY, Cheng P, Teng YS, Fu XL, Yu PW, Guo G, Luo P, Zhuang Y, Zou QM. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut. 2017;66:1900–1911. doi: 10.1136/gutjnl-2016-313075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C, He H, Liu H, Li R, Chen Y, Qi Y, Jiang Q, Chen L, Zhang P, Zhang H, Li H, Zhang W, Sun Y, Xu J. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019;68:1764–1773. doi: 10.1136/gutjnl-2018-316324. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Zhang Q, Hu Y, Li T, Yu J, Zhao L, Ye G, Deng H, Mou T, Cai S, Zhou Z, Liu H, Chen G, Li G, Qi X. immunoscore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267:504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Gao X, Han L, Yu J, Li H. Identification of hub genes with prognostic values in gastric cancer by bioinformatics analysis. World J Surg Oncol. 2018;16:114. doi: 10.1186/s12957-018-1409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li TJ, Jiang YM, Hu YF, Huang L, Yu J, Zhao LY, Deng HJ, Mou TY, Liu H, Yang Y, Zhang Q, Li GX. Interleukin-17-producing neutrophils link inflammatory stimuli to disease progression by promoting angiogenesis in gastric cancer. Clin Cancer Res. 2017;23:1575–1585. doi: 10.1158/1078-0432.CCR-16-0617. [DOI] [PubMed] [Google Scholar]
- 9.Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2:1125–31. doi: 10.1158/2326-6066.CIR-14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng ZH, Shi YX, Yuan M, Xiong D, Zheng JH, Zhang ZY. Chemokines and their receptors in lung cancer progression and metastasis. J Zhejiang Univ Sci B. 2016;17:342–51. doi: 10.1631/jzus.B1500258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amedei A, Prisco D, D’Elios MM. The use of cytokines and chemokines in the cancer immunotherapy. Recent Pat Anticancer Drug Discov. 2013;8:126–42. [PubMed] [Google Scholar]
- 12.Roy I, Getschman AE, Volkman BF, Dwinell MB. Exploiting agonist biased signaling of chemokines to target cancer. Mol Carcinog. 2017;56:804–813. doi: 10.1002/mc.22571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen XY, Chen RP, Wu W, Huang ZM. MicroRNA-873 inhibits proliferation and induces apoptosis by targeting CXCL1 in gastric cancer. Int J Clin Exp Pathol. 2016;9:10011–10019. [Google Scholar]
- 14.Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842–62. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon H, Kim N. Diagnosis and management of high risk group for gastric cancer. Gut Liver. 2015;9:5–17. doi: 10.5009/gnl14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Ma X, Bi F, Liu M. Clinical significance of circulating tumor cells in gastric cancer patients. Oncotarget. 2017;8:25713–25720. doi: 10.18632/oncotarget.14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu DG, Choi CW, Kang DH, Kim HW, Park SB, Kim SJ, Nam HS. Predictive factors to diagnosis undifferentiated early gastric cancer after endoscopic submucosal dissection. Medicine (Baltimore) 2017;96:e8044. doi: 10.1097/MD.0000000000008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beeharry MK, Liu WT, Yan M, Zhu ZG. New blood markers detection technology: a leap in the diagnosis of gastric cancer. World J Gastroenterol. 2016;22:1202–12. doi: 10.3748/wjg.v22.i3.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcuzzi E, Angioni R, Molon B, Calì B. Chemokines and chemokine receptors: orchestrating tumor metastasization. Int J Mol Sci. 2018;20 doi: 10.3390/ijms20010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HJ, Song IC, Yun HJ, Jo DY, Kim S. CXC chemokines and chemokine receptors in gastric cancer: from basic findings towards therapeutic targeting. World J Gastroenterol. 2014;20:1681–93. doi: 10.3748/wjg.v20.i7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy I, Boyle KA, Vonderhaar EP, Zimmerman NP, Gorse E, Mackinnon AC, Hwang RF, Franco-Barraza J, Cukierman E, Tsai S, Evans DB, Dwinell MB. Cancer cell chemokines direct chemotaxis of activated stellate cells in pancreatic ductal adenocarcinoma. Lab Invest. 2017;97:302–317. doi: 10.1038/labinvest.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Ehling J, Tacke F. Role of chemokine pathways in hepatobiliary cancer. Cancer Lett. 2016;379:173–83. doi: 10.1016/j.canlet.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Allavena P, Germano G, Marchesi F, Mantovani A. Chemokines in cancer related inflammation. Exp Cell Res. 2011;317:664–73. doi: 10.1016/j.yexcr.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 25.O’Hayre M, Salanga CL, Handel TM, Allen SJ. Chemokines and cancer: migration, intracellular signalling and intercellular communication in the microenvironment. Biochem J. 2008;409:635–49. doi: 10.1042/BJ20071493. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X, Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 27.Singh R, Lillard JW Jr, Singh S. Chemokines: key players in cancer progression and metastasis. Front Biosci (Schol Ed) 2011;3:1569–82. doi: 10.2741/246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezaeeyan H, Shirzad R, McKee TD, Saki N. Role of chemokines in metastatic niche: new insights along with a diagnostic and prognostic approach. APMIS. 2018;126:359–370. doi: 10.1111/apm.12818. [DOI] [PubMed] [Google Scholar]


